Abstract
Mitochondria are considered to be the “power plants” of the cell, but can also initiate and execute cell death, stimulated by oxidative stress (OS). OS induced mitochondrial dysfunction is characterized by a loss in oxygen consumption and reduced ATP production. Curcumin, as a potential therapeutic, has been explored as a candidate for mitochondrial OS suppression, but rapid metabolism and aqueous insolubility has prevented it from being effective. Further, efficient delivery of curcumin via the incorporation into nanocarriers has again been limited due to low drug loading capacities and/or significant burst release, resulting in acute cytotoxicity. Hence, to increase the therapeutic potential and reduce the toxic effects of curcumin, curcumin conjugated poly(β-amino ester) nanogels (CNGs) were synthesized using Michael addition chemistry. This approach provided easy control over the nanogel size, with CNGs showing a uniform release of active curcumin over 48 hours with no burst release. This controlled release system significantly increased the safety limit for curcumin, with a ten fold increase in the cytotoxic threshold, as compared to free curcumin. Further, real-time mitochondrial response analysis with the Seahorse XF96 showed effective and prolonged suppression of H2O2 induced mitochondrial oxidative stress upon pre-treating endothelial cells with CNGs and this potential of nanogels was studied at different pre-treatment times prior to H2O2 exposure.
Keywords: Curcumin release, poly(β-amino esters), nanogels, Seahorse Bioscience XF96
Graphical abstract
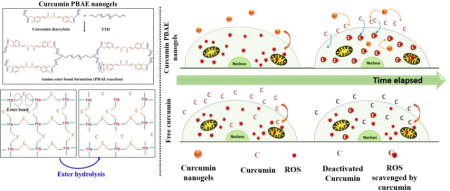
1. INTRODUCTION
Given its ubiquity in a number of disease states, oxidative stress (OS) and its consequent effects are extensively researched topics in biological and pharmaceutical disciplines. Its cascading effects are seen in diabetes, cancers, neurological disorders, cardiac disorders and many more (Maritim, Sanders et al. 2003, Reuter, Gupta et al. 2010, Hsieh and Yang 2013). OS occurs when the natural balance of oxidants and antioxidants is disturbed, leading to the excess production of reactive oxygen species (ROS) and reactive nitrogen species (RNS). Higher levels of ROS and RNS lead to cellular damage by lipid peroxidation, protein carbonylation, and DNA damage, etc. (Marc W. Fariss 2005, Orrenius, Gogvadze et al. 2007). The mitochondrion, the central system for energy production pathways and metabolism in most cells, is also a key organelle for ROS generation (Chen, Vazquez et al. 2003, Drose and Brandt 2012). Interestingly, mitochondria are not just a key generating source of free radicals but also a sensitive target to ROS and RNS damage (Marchi, Giorgi et al. 2012). Mitochondria are believed to be specifically affected by lipid peroxidation, leading to the generation of hydrogen peroxide and superoxide radicals, hence resulting in additional ROS production (Madrigal, Olivenza et al. 2001). This cycle cascades in a feed forward fashion leading to cell necrosis and apoptosis (Madrigal, Olivenza et al. 2001, Zhong and Yin 2015). Therefore, it is believed that when a cell is exposed to any free radical or ROS, the mitochondria are the first of the cellular organelles to be targeted by ROS. This leads to mitochondrial dysfunction, disturbing the normal respiration mechanism and loss in ATP production before finally losing a major energy pathway of the cells. Mitochondrial protein, lipids and DNA (mtDNA) are considered the primary target of ROS. As a result, mitochondrial oxidative stress (mitochondrial OS) becomes the indirect cause of many diseases, primarily neurogenerative disorders such as Alzheimer’s and Parkinson’s disease, numerous metabolic disturbances, cardiomyopathies and metabolic complexities in patients suffering from diabetes mellitus (Rolo and Palmeira 2006, Victor, Apostolova et al. 2009, Federico, Cardaioli et al. 2012, Huang, Partridge et al. 2012, Butterfield, Di Domenico et al. 2014, Ichikawa, Ghanefar et al. 2014).
Because mitochondria become the source of more ROS, it is important to control the damage to the mitochondrial structure and function when the cells encounter increased ROS/RNS. One potential solution to prevent mitochondrial OS could be through scavenging ROS via antioxidant supplementation. Most attempts to accomplish this have failed in vivo due to non-specific delivery of antioxidants to mitochondria below therapeutic limits as well as in vivo molecular instability issues with most of the antioxidants. Among the molecules/compounds studied, curcumin has shown protective effects, including cytoprotection against aluminum induced neurogenerative disorders (Sood, Nahar et al. 2011), maleate induced oxidative stress (Tapia, Sánchez-Lozada et al. 2014), and prevention of mitochondrial OS in high fat diet induced obese mice (Kuo, Chang et al. 2012). Despite these promising findings in animal models, most clinical studies performed with high and frequent oral doses up to 4 g/day for 48 weeks, specifically for Alzheimer’s, have failed to show clinically significant improvement in reducing symptoms for the disease (Ringman, Frautschy et al. 2012). Among limitations for curcumin, poor aqueous solubility and extremely low bioavailability are most significant because pure curcumin has shown to be below detectable serum concentrations within a few minutes of administration (Garcea, Jones et al. 2004). Attempts have been made to incorporate curcumin into many viable polymeric or similar nanocarriers with the aim for higher curcumin loading, sustained release characteristics and ability to perform its antioxidant function with no cytotoxic effect. Unfortunately, curcumin drug loading capacities of most nanocarriers have not exceeded more than 4 wt%, resulting in higher amount of carrier material but minimal doses of functional drug (Anand, Nair et al. 2010). Furthermore, little work has been pursued using curcumin as a treatment targeted towards stressed endothelial cells for cancer and tumor treatment.
In our approach to fight OS (specifically mitochondrial OS), we synthesized curcumin conjugated poly(β-amino ester) nanogels, where curcumin is chemically conjugated to the polymer backbone and upon ester hydrolysis results in uniform release pure curcumin. While covalent conjugation of antioxidants into PBAE crosslinked bulk gel systems has been previously demonstrated by Wattamwar et. al. (Wattamwar, Biswal et al. 2012), further translation of the chemistry into quercetin conjugated PBAE nanogels has been shown by Gupta et. al. (Gupta, Authimoolam et al. 2015). The route of covalent conjugation into the PBAE matrix gives curcumin the structural stability and prevents deactivation prior to hydrolysis. Therefore, uniform drug release over 36–40 hours resulted in protected functionality of curcumin for a longer period. Further, a higher IC50 value of more than 100 μg/ml of loaded curcumin in contrast to free curcumin (IC50 = 5.3 μg/ml) indirectly increases the safety window of this potential drug. Increased safety window gives higher flexibility to work in therapeutic levels and reach adequate curcumin levels to treat and or prevent OS. In contrast, free curcumin administered upon potential therapeutic treatment amounts will kill endothelial cells. A mitochondrial stress assay using Seahorse XF96 analyzer with the ability to monitor real time mitochondrial bioenergetics was employed to analyze mitochondrial bioenergetics post induced oxidative stress and assessed the protective effect of C-PBAE nanogels (CNGs) versus free curcumin. CNGs with higher, yet viable, treatment concentrations (5 μg/ml) were able to shield against dropping ATP production or basal respiration rates due to hydrogen peroxide induced mitochondrial OS. On the other hand, free curcumin proved to be fatal at these high treatment concentrations and showed no protection at safe concentrations of 1 μg/ml against mitochondrial OS.
2. METHODS AND MATERIALS
2.1 Reagents
Curcumin was purchased from Chem-impex Int’l ‘Inc, (Wood Dale, IL). 4,7,10-Trioxatridecane-1,13-diamine (TTD), acryloyl chloride, 2,2′-azinobis-(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), triethylamine (TEA) and potassium carbonate were bought from Sigma Aldrich (St. Louis, MO). Sodium chloride, potassium phosphate, sodium phosphate and sodium dodecyl sulfate (SDS) were purchased from Fisher Scientific (Fair Lawn, NJ). Human Umbilical Vein Endothelial Cell line (HUVECs), EBM-2 basal medium and its growth factors were purchased from Lonza, (Walkersville, MD). All solvents were purchased from Pharmco-aaper (Shelbyville, KY). All media kit and cell culture plates for XF96 Seahorse experiments were purchased from Seahorse Bioscience (Massachusetts, USA). Calcein AM red-orange tracer was bought from Invitrogen (Grand Island, NY).
2.2 Curcumin acrylate (CA) synthesis
Acrylation of the curcumin phenolic group was carried out via reaction of curcumin with acryloyl chloride in the molar ratio of 1:3 with anhydrous THF as the reaction medium. Triethyl amine (TEA) was added in same molar ratio as acryloyl chloride to capture byproduct hydrogen chloride by forming a chloride salt with the progression of reaction. Reaction was carried out for 15 hours at room temperature in dark (aluminum foiled). The acrylated product was filtered from the salts formed during the reaction. The product was further purified via a solvent extraction process to remove any unreacted monomers then vacuum dried to obtain dry curcumin acrylate product. Obtained curcumin acrylate was analyzed using HPLC to verify product quality.
2.3 Synthesis of curcumin PBAE nanogels
CA solubilized in acetonitrile at known concentrations was reacted with TTD keeping to ratio of acrylate to reactive amine hydrogen to 1:1. Curcumin has two phenolic groups and upon acrylate functionalization, forms curcumin diacrylate. TTD has four reactive amine hydrogens. Therefore, for each mole of TTD (molecular weight 220.31 g/mol), two moles of CA (molecular weight ~ 476 g/mol) are reacted to get the required acrylate to reactive hydrogen amine ratio of 1:1. To formulate 1 ml of curcumin conjugated PBAE nanogels with the starting CA feed concentration of 10 mg/ml, 2.3 mg of TTD was added and allowed to react for 24 hours. Feed reaction concentrations of CA were varied from 0.75 to 10 mg/ml, hence the amount of TTD added to the reaction system was changed accordingly to keep the stoichiometric ratio constant. After 24 hours and self-precipitation of CA and TTD into C-PBAE nanogels (CNGs), suspension was diluted in anhydrous THF without any inhibitors in a 1:5 ratio v/v (1 ml CNG suspension with 4 ml THF) as a purification step. An hour later, the system was centrifuged at 6.7 RCF for 20 minutes. Supernatant was removed and another wash with THF was given to remove any remaining unreacted monomers. After the second purification step, the centrifuged pellet was re-suspended in 1 ml acetonitrile via sonication and then freeze dried overnight to get dry C-PBAE nanogels (CNGs).
2.4 Yield of reaction, nanogel size and morphology characterization
Yield
Yield of the C-PBAE nanoparticle reaction synthesis was determined by calculating the unreacted CA concentration in supernatant after nanoparticle centrifugation. The unreacted CA concentration was determined by analyzing the supernatant using a Varian Cary 50 UV-Vis spectrophotometer and quantifying the absorbance at the curcumin characteristic wavelength of 420 nm. The yield was determined by subtracting the amount of unreacted CA from the starting amount added. An assumption that CA and TTD reacted in the same ratio was taken into account while calculating the yield.
Size and morphology
Hydrodynamic radii of the different CNGs formulations were measured using dynamic light scattering (DLS) (Malvern Zetasizer ZS90) after suspending dried nanoparticles in acetonitrile and setting the measurement parameters according to the solvent used. Additionally, size of the nanogels was also analyzed in aqueous medium by suspending nanogels in PBS buffer (0.1% SDS wt/vol) using bath and probe followed by hydrodynamic diameter measurement DLS. CNGs were also scanned with electron microscopy to determine their shape and morphology. To prepare the samples, synthesized nanoparticles were first suspended in acetonitrile at a concentration of 100 μg/ml using probe sonication. 10 μl of CNG suspension was put on ODT treated gold substrate and allowed to dry for 2 hours. The sample was then sputter coated using gold-palladium alloy and images were taken at various magnifications using S-4300 Hitachi Scanning Electron Microscope.
2.5 Hydrolytic degradation of C-PBAE nanogels and the subsequent curcumin release profile
In order to test the degradation of these nanogels with subsequent release of pure curcumin upon ester hydrolysis, nanoparticles synthesized with 5 mg/ml as the CA feed concentration (CNG (5)) were suspended in PBS buffer containing 0.1% SDS wt/vol (pH 7.4). SDS was added to the buffer to enhance the solubility of released curcumin or its oligomeric products for further quantification. The nanogels were suspended in the buffer at a concentration of 100 μg/ml. At time points 0, 2, 4, 6, 8, 12, 24, 30, 36, 52 and 71 hours, the suspension was centrifuged at 6.7 RCF for 20 minutes and supernatant was collected and stored at −20° C for further analysis. The centrifuged pellet was re-suspended in fresh buffer maintaining appropriate sink conditions. A degradation study was carried out over 72 hours under sink conditions at 37°C. To determine the release rate of curcumin during hydrolytic degradation of CNGs, UV-Vis spectrophotometry was used to analyze the curcumin concentration in the collected supernatants. Absorbance of the supernatants was measured at 420 nm using Varian Cary 50 Bio UV-Vis spectrophotometer followed by quantification of the amount of curcumin released using a standard calibration curve. Rest of the characterization studies were also carried out with CNGs synthesized with 5 mg/ml as feed CA concentration and is abbreviated as only CNGs in figures, captions and text.
2.6 Antioxidant activity of released curcumin
It was important to assess that the released curcumin from CNGs was still active with its antioxidant potential intact as its phenolic groups were chemically altered during the nanoparticle synthesis. Therefore, trolox equivalent antioxidant capacity (TEAC) assay was performed in order to verify the antioxidant potential or radical scavenging property of released degradation products. TEAC is a colorimetric assay based on scavenging of 2, 2′-azinobis-(3-ethylbenzothiazoline-6-sulfonate) radical anions (ABTS.-) in presence of any antioxidant. Briefly, 7 mM ABTS radical cation stock solution was prepared by mixing 1 ml of 8 mg/ml of ABTS solution with 1 ml of 1.32 mg/ml of potassium persulfate solution in DI water overnight. This concentrated ABTS radical stock solution was diluted in PBS to an absorbance of 0.4 AU at 734 nm to prepare working solution after baseline correction. Trolox, with known concentrations ranging from 0 to 0.27 mM, was used for calibration. The assay was carried out in a 96-well plate and 10 μl of the sample was added to each well, followed by 200 μl of ABTS radical working solution. Five minutes later, absorbance was read at 734 nm using Varian Cary 50 Bio UV-Vis spectrophotometer. A trolox calibration curve was used to determine equivalent trolox concentrations in the supernatant degradation products.
2.7 Dose dependent cytotoxicity of C-PBAE nanoparticles towards HUVECs
HUVECs were cultured in EBM basal medium (phenol red free) with EBM-2 growth factors to an 80% confluence in a 96-well plate overnight. Curcumin and CNGs (CA feed concentration = 5 mg/ml) suspension in media with equivalent curcumin concentrations ranging from 0 to 70 μg/ml were prepared. Wells containing HUVECs received and were treated with antioxidant/CNG solution for 24 hours (n=5). After 24 hours, media with antioxidant was removed from the wells, then incubated in 1 mM calcein AM red-orange live cell tracer. After 1 hour, cells were washed and fresh media was added to each well. The well-plate was subjected to fluorescence measurement with 540/590 nm as excitation and emission wavelength, respectively, using BioTek Synergy Mx, Gen5 2.0, Winooski, VT to measure dose dependent toxicity of C-PBAE nanoparticles.
2.8 Effect of CNGs exposure on H2O2 induced cellular OS: cell viability analysis
HUVECs cultured in EBM-2 basal medium were seeded in a 96-well plate and incubated overnight to achieve 80% confluency with respect to well area. CNGs (CA feed concentration = 5 mg/ml) at a concentration of 7.5 μg/ml (5 μg/ml equivalent curcumin loading) were suspended uniformly in EBM-2 basal medium with the aid of bath sonication. The media from the cells was removed and 200 μl of the CNG suspension was added to the well of interest. Similarly, curcumin solution in EBM medium was prepared at two concentrations, 1 and 5 μg/ml and added to the respective wells. 10 μl of 10 mM H2O2 stock solution prepared in EBM media was added simultaneously to the CNGs/curcumin treated groups to get final exposure concentration of 0.5mM. A control group of only CNGs, curcumin (1 and 5 μg/ml) and only H2O2 exposed cells was also prepared. To all the wells, 10 μl of DCF-DA prepared in EBM-2 media was added to get a final cell exposure concentration of 10 μM. 24 hours later, DCF fluorescence was read at 490/525 nm as excitation/emission wavelength using BioTek Synergy Mx, Gen5 2.0, Winooski, VT spectrophotometer. Post fluorescence measurement, CNGs ± H2O2 solutions were removed and wells were washed with fresh media, followed by addition of 1 mM Calcein AM red-orange live cell tracer. Cell viability was measured by following the similar protocol as given in section 2.7. Estimation of oxidative stress (DCF fluorescence) was reported as specific DCF values calculated as DCF fluorescence/cell viability of the respective groups.
2.9 Measurement of mitochondrial OS changes after treatment with C-PBAE nanogels using XF-96 extracellular flux analyzer
Seahorse XF-96 Flux Analyzer (Seahorse Bioscience, North Billerica, MA) was used to analyze the effect of C-PBAE nanogels on cellular bioenergetics. HUVECs were seeded in a Seahorse 96-well tissue culture plate at a seeding density of approximately 35,000 cells/well. Cells were then allowed to adhere and grow for 24 hours. Prior to the mitochondrial stress assay for all the experiments, the cell growth media was replaced with 175 uL of FX media (FX Assay Modified DMEM from Seahorse Bioscience with 5.5 mM glucose, 1 mM pyruvate and 2 mM glutamix). Wells and cells were washed once, media exchange was completed and then cells were equilibrated for 60 minutes at 37° C in non-CO2 incubator. The 96-well plate was then subjected to Mitochondrial stress assay to analyze mitochondrial energetics and cellular metabolic profile. The mitochondrial stress assay consisted of subsequent injections of three mitochondrial inhibitor drugs: oligomycin A (1 μM), which act as ATP synthase inhibitor; FCCP (1 μM), an uncoupling agent; and a mixture of 1 μM rotenone and 1 μM mitochondrial respiratory chain complex I and III to shut down the mitochondria oxygen consumption. Oxygen consumption rates (OCR) and extracellular acidification rates (ECAR) were measured using measurement periods of 3 minutes minimum and 3 minutes maximum with drug injection series. Once OCR/ECAR measurements were completed, the FX media was carefully removed from all individual wells and replaced with 25 uL of cell lysis buffer containing 0.32 Sucrose, 2 mM EDTA, 2 mM EGTA, 20 mM HEPES, pH 7.4, containing protease inhibitors 4 μg/mL leupeptin, 4 μg/mL pepstatin, 5 μg/mL aprotinin, and 0.2 mM PMSF. The plates then put in −20°C freezer overnight to disrupt cells membranes. The next day, plates were taken out to room temperature and put on orbital shaker for an hour. Unknown protein levels (samples) of the wells were measured using modified BCA protein assay (Thermo Scientific, Rockford IL). Known amounts of protein standards (2.5 μg, 5.0 μg, 10 μg etc.) were added in the background wells of the plate to construct an optical density standard curve. Protein levels were determined after the optical densities were measured on iMark plate reader (Bio-Rad, Hercules, CA). The OCR pmol/min were normalized to protein levels of pmol/min/μg or calculated as percent change (%OCR) from the third basal measurement for each group.
2.9.1 Mitochondrial bioenergetics response towards free curcumin or CNG exposure
To analyze the mitochondrial response towards free curcumin and CNGs, a 96-well plate seeded with 35,000 cells/well (HUVECs) was treated with free curcumin or CNG (synthesized with 5mg/ml as feed reactant concentration) at equivalent curcumin concentrations of 1 and 5μg/ml. Free curcumin or CNGs were suspended in HUVECs cell growth media before administering them to the cells. After 24 hours of treatment, cell growth media was removed and washed with XF media once. The cells were then incubated in XF media for 1 hour at 37°C. Subsequently, plate was then subjected to mitochondrial stress assay to analyze mitochondrial energetics and metabolic profile using Seahorse XF96 analyzer. Metabolic respiration rates were measured and quantified as OCR (pmol/min) at this step.
2.9.2 Protection from H2O2 induced mitochondrial OS due to CNGs pre-treatment
After analyzing different concentrations of curcumin and CNGs having same equivalent curcumin concentrations for mitochondrial respiration rates, viable concentrations of curcumin (1 μg/ml) and CNGs (5 μg/ml) were used for further analysis of protective effect against mitochondrial OS. HUVECs in 96-well plates were first pre-treated with free curcumin or CNGs at specified concentrations (1 and 5 μg/ml) for 0, 12 and 24 hours, then followed by introduction of 0.5 mM H2O2 for 2 hours. Next, cells were washed and replaced with un-buffered XF media and kept at 37° C for 1 hour, followed by mitochondrial stress assay.
2.9.3 Statistical Analysis
Data for figures 1 (A), (B), 2 (A) and (C), 3 (A) is presented as mean ± St. Dev, while data for figures 3 (A), (C), (D); 4 (B); 5 (A), (B), (C) and 6 (A), (B) and (C) is presented as mean ± SEM. The statistically significant differences between the mean values were calculated using one-way ANOVA test followed by Bon-ferroni post hoc test with the help of SigmaPlot software. A p value of <0.050 was considered to be representative of significant difference between the means under comparison.
Figure 1. Size and morphology of CNGs.
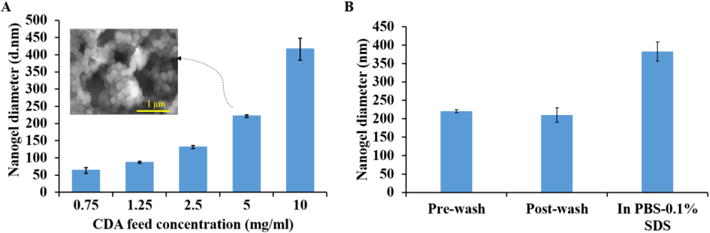
(A) Hydrodynamic diameter and SEM of precipitated curcumin PBAE nanogels after 24-hour reaction of CDA with TTD in acetonitrile at different CDA feed reactant concentrations. Acrylate to reactive hydrogen amine ratio kept constant at 1:1 for all the formulations Inset: SEM image of nanogels corresponding to 5mg/ml feed concentration. (B) Hydrodynamic diameter of nanogels pre- and post-wash with THF as a purification step showing no significant difference at both the stages. Size measurement also done in aqueous buffer (PBS-0.1% SDS. N=3, error bars: std. dev.
Figure 2. Degradation of CNGs in PBS buffer with 0.1% SDS (pH=7.4) at 37° C in a shaker bath.
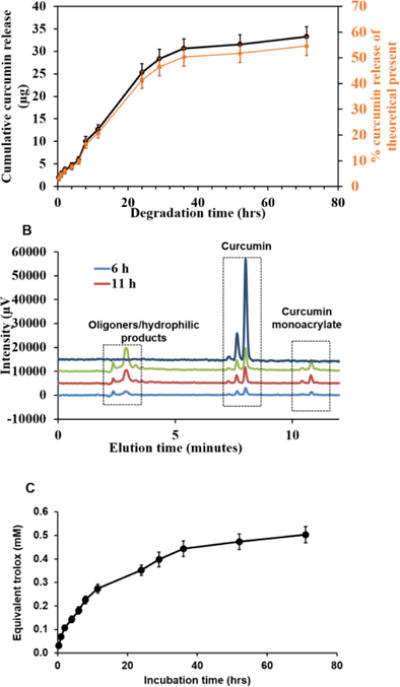
(A) Curcumin release profile during hydrolytic degradation. (B) HPLC chromatogram of nanogel released products recorded at 420 nm showing elution of pure curcumin with minor amount of monoacrylate. (C) Equivalent trolox amount of the released products representing the antioxidant activity analyzed using TEAC assay. N=3 replicates were used for each sample. Error bars: std. dev.
Figure 3. Dose dependent cytotoxicity of CNGs towards HUVECs after 24 hours of treatment and comparison with free curcumin.
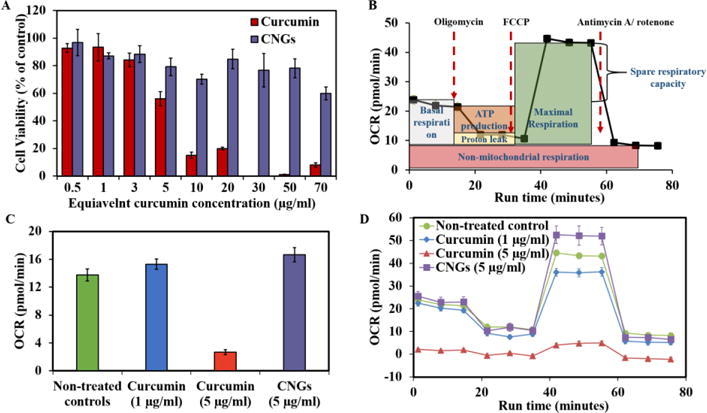
(A) Cell viability calculated after carrying out Calcein AM red-orange live cell tracer assay (B) Mitochondrial oxygen consumption rate profile of a typical mitochondrial stress assay conducted using Seahorse XF96 instrument. (C) Basal respiration rates of curcumin and CNGs represented at pmol/min after the calculations as depicted in Figure 3A. N=5 replicates were used for each group. (D) OCR (profile) of the controls and free curcumin or CNG at different curcumin concentrations in response to mitochondrial stress assay reagents added in a series at selected time points. Error bars: (A) – std. dev.; (C), (D) – std. err.
Figure 4. Figure 4 Effect of CNGs (5 mg/ml system) and curcumin on HUVECs with simultaneous treatment of 0.5 mM H2O2 for 24 hours.
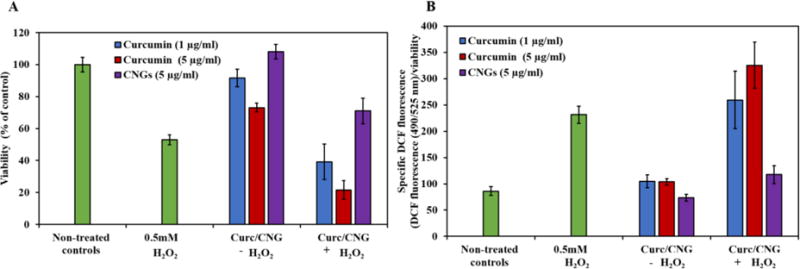
(A) Cell viability (B) Specific DCF fluorescence. CNGs with equivalent concentration of 5 μg/ml of loaded curcumin were administered to the cells. N=5, error bars: std. dev.
Figure 5. Mitochondrial function parameters after pre-treatment of free curcumin or CNG for 0, 12 and 24 hours followed by 2 hour exposure of 0.25 mM H2O2 which were subsequently subjected to mito stress assay with real time OCR measurements using Seahorse XF96 instrument.
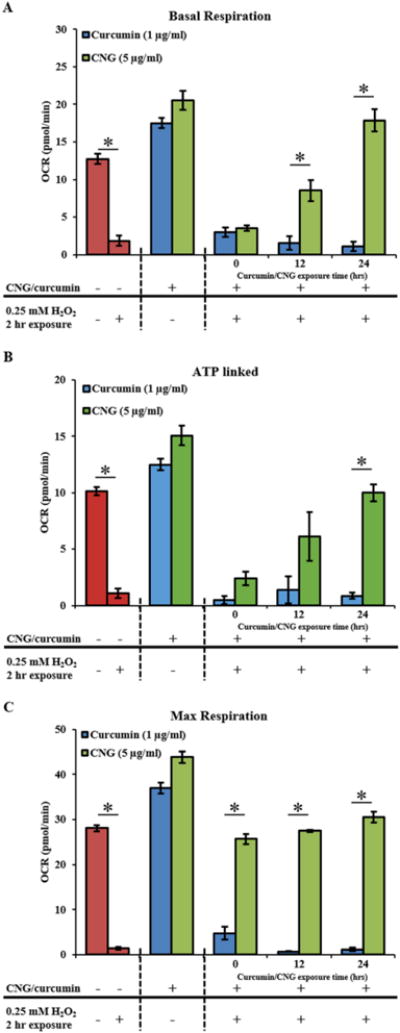
(A) Basal respiration. (B) ATP linked to mitochondria. (C) Maximum respiration. All OCR values are displayed as pmol/min. N=5 replicates were used for each group. Error bars: std. err.
Figure 6. Mitochondrial function parameters of the mitochondria presented as % OCR post total protein content normalization.
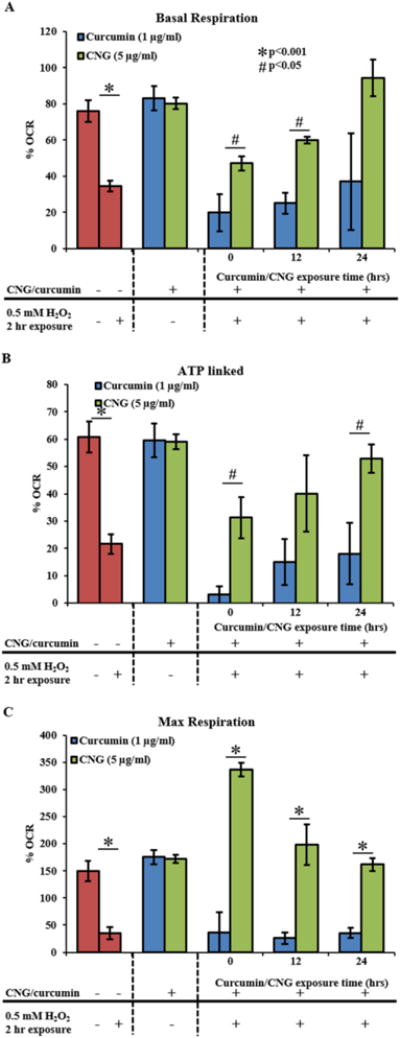
Pretreatment of free curcumin or CNG for 0, 12 and 24 hours followed by 2-hour exposure of 0.25mM H2O2, subsequently subjected to mitochondrial stress assay with real time OCR measurements using Seahorse CF96 instrument. (A) Basal respiration. (B) ATP linked to mitochondria. (C) Maximum respiration. All OCR values are calculated as pmol/min. N=5 replicates were used for each group. Error bars: std. err.
3. RESULTS
3.1 Curcumin nanogel size, yield and morphology
CA with 2 to 3 acrylate groups was set to react with TTD in acetonitrile under dilute conditions for 24 hours resulting in self-precipitated crosslinked nanogels. Varying the feed concentration of CA and maintaining the total acrylate to total reactive hydrogen amine ratio resulted in self-precipitated nanogels with variable hydrodynamic diameter. Upon UV-Vis analysis of the supernatants obtained after centrifugation, reaction yield was calculated to be 62.7%. Increasing the reactant concentration resulted in an increase in diameter. As shown in Figure 1A, hydrodynamic diameters of 64 ± 8, 87 ± 3, 131 ± 5, 221 ± 4, 416 ± 31 nm were obtained for CA feed reactant concentrations of 0.75, 1.25, 2.5, 5, 10 mg/ml respectively. These will be abbreviated as CNG (0.75), CNG (1.25), CNG (2.5), CNG (5) and CNG (10) throughout this text.
After the washing and purification step in anhydrous THF, CNG (5 mg/ml system) was again analyzed on DLS giving the hydrodynamic diameter of 210 ± 20, similar to the pre-wash diameter of the nanogels (Fig. 1B). This suggests little to no aggregation and no morphological change of nanogels post purification steps. The spherical morphology of the nanogel system was further confirmed after electron microscopy imaging (Fig. 1A inset) and an average diameter of 176 ± 26 nm was calculated using Image-J software. Post freeze-drying, nanogels showed the hydrodynamic diameter of 383 ± 23 (PDI = 0.24) nm in aqueous buffer (PBS with 0.1% SDS wt/vol) at t = 0 as shown in figure 1 B. This is a significant increase in size when compared to the diameter in acetonitrile or THF. Nanogel diameter measurement was also taken at other specified time intervals, showing the diameter of 331 ± 1, 354 ± 12, 354 ± 12, and 191 ± at 1, 5, 10 and 24 hours respectively within showing similar sizes upto 10 hours (Supplemental information Figure 1 S). The figure 1 S also includes polydispersity index (PDI) of the measurement at the corresponding stages above each bar graph, values ranging from 0.03 to 0.25 for first 10 hours. The increased diameter in aqueous buffer could be due to the swelling of the PBAE nanogels in aqueous medium or due to aggregation. However, very low polydispersity index (PDI) of the measurement at every stage indicates uniformity and low degree of aggregation indicating swelling as the contributing factor towards the increased size.
3.2 Degradation, curcumin release and its anti-oxidant activity
Curcumin nanogels (CNG (5)) were suspended in 0.1% SDS-PBS buffer at a concentration of 100 μg/ml. To dissolve released curcumin, 0.1% SDS wt/vol was added. Released curcumin in the supernatant was quantified using UV-Vis analytical method by recording the absorbance at 420 nm to determine sample concentration and cumulatively adding the obtained concentrations of each sample with increasing degradation time to obtain the degradation profile of CNGs. As shown in Figure 2A, 90% of the total curcumin release was uniformly distributed over 36 hours, and the remaining 10% release was spread throughout the next 30 hours. Meanwhile, taking into account 62.7% wt/wt as the theoretical loading of curcumin to nanogels, 55.58 ± 3% of the total curcumin loaded was recovered during degradation. This lag in recovered curcumin could be due to the release of oligomeric/monoacrylated products and may be underestimated in quantification during UV-Vis analysis. Also, the uniform release combined with similar size of nanogel for 1st 10 hours (Figure 1 B) implies bulk erosion profile instead of the surface erosion. HPLC analysis of the degradation products at 420 nm showed elution of pure curcumin (peak at 7.9 minutes) along with other products eluting at 11 mins (curcumin monoacrylates) and 2.9 minutes (β-amino acids-other half of PBAE degradation) absorbable at 420 nm (Fig. 2 B). When estimated based on peak area, the peak at 2.9 minutes contributes to about 50% of the total peak area of the three combined. Area of the peak at 7.9 minutes (i.e. curcumin) contributed to 33% of the total area combined, while the peak at 11 minutes contributed 17%. If calculating based on the total antioxidant released, 71% of the total peak area at 7.9 and 11 minutes eluted as curcumin and rest as curcumin acrylate. The peak areas cannot be directly correlated to the amounts of curcumin and its monoacrylated products as the extinction co-efficient for both would be different. Hence, only an estimation can be made on the relative quantities present. Carrying out TEAC assay on the released products supernatants gave an estimate of releasing curcumin’s antioxidant capacity over 70 hours. All the collected samples were assayed for antioxidant activity and quantified as mM of equivalent trolox because known concentrations of trolox were used as the standards. Finally, equivalent trolox amount were cumulatively added with respect to degradation time to obtain a uniform increase in the antioxidant activity profile with time from 0.031 mM at t=0 to 0.502 mM trolox after 70 hours of incubation. This implies that at each time point of hydrolytic ester cleavage, released curcumin was active to scavenge free radicals.
3.3 Dose dependent toxicity of CNGs on HUVECs and protection from oxidative stress
HUVECs were treated with variable concentrations of CNG (5) with equivalent curcumin concentrations ranging from 0.5 to 70 μg/ml. HUVECs were simultaneously, yet separately, treated with pure curcumin as a control. After 24 hours treatment, pure curcumin started showing toxicity towards HUVECs at 5 μg/ml (IC50 = 5.3 μg/ml) while CNGs were non-toxic up to 20 μg/ml of equivalent curcumin and cells were still 60% viable at concentration of 70 μg/ml (Fig. 3A). Upon external insult to HUVECs with the aid of 0.5mM H2O2, oxidative stress as well as its suppression potential due to CNG exposure was analyzed using viability and DCF fluorescence assay.
3.4 Protection from cellular oxidative stress
Cell viability for each treatment group was measured keeping non-treated control group as reference of 100% viability. Only H2O2 treated groups after 24-hour exposure resulted in 53 ± 3 % viability. CNG treated groups showed no significant change with respect to the control with percent viability of 108 ± 4 (Figure 4), while curcumin at 1 and 5 μg/ml showed 91.6 ± 5.4 and 73.1 ± 2.63 % viability respectively. Upon simultaneous exposure to 0.5mM H2O2 along with the antioxidant for 24 hours, CNG treated group resulted in 71 ± 4 % viability, which is significantly higher than only H2O2 treated groups. On the other hand, curcumin with 1 and 5 μg/ml with H2O2 did not show any improvement and rather lower viabilities at 39 ± 11 and 21.5 ± 5.9 % respectively. In order to get an estimate of oxidative stress, amount of free radical generation was measured with DCF fluorescence. Only H2O2 treated groups gave a measured specific DCF fluorescence of 231 ± 16.2, which is significantly higher than non-treated control group with fluorescence measurement of 86.2 ± 8.5. Only CNGs treated group did not result in a major difference w.r.t controls with specific DCF of 73.2 ± 6.54. An increase was seen with 1 and 5 μg/ml curcumin exposure with specific DCF measuring 104.3 ± 12 and 103 ± 6.3. Fluorescence measurement of CNGs + H2O2 group demonstrated suppression in oxidative stress due to H2O2 with lower values of specific DCF values of 117 ± 16.9 as compared to only H2O2 group. On the other hand, 1 and 5 μg/ml curcumin exposure along with H2O2 showed increased specific DCF as compared to non-treated controls giving the values of 259 ± 54 and 325 ± 43 respectively. High specific DCF fluorescence values could be a combination of lower viabilities and higher free radical production due to both H2O2 and curcumin exposure.
3.5 Protection from mitochondrial oxidative stress
To assess the potential of CNGs toward mitochondrial OS protection and its comparison with pure curcumin, HUVECs were incubated with both CNGs and curcumin at two different concentrations for 24 hours. This incubation was followed by basal respiration rate measurements and a mitochondrial stress test. Basal respiration rates, ATP linked respiration, proton leak, maximum respiration and Spare respiratory capacity were calculated from the mitochondrial stress test drug-response OCR values according to the calculation scheme shown in Figure 3B. The curcumin treatment at 1 μg/ml showed nominal OCR of 15.29 ± 0.73 pmol/min which is comparable to the non-treated controls: 13.7 ± 0.9but at 5 μg/ml of curcumin exposure, OCR values dropped down drastically to 2.68 ± 0.35 (Fig. 3C). This clearly implies the mitochondrial dysfunction at curcumin concentrations of 5 μg/ml.
On the other hand, HUVECs treatment with CNG at 7.97 μg/ml having equivalent curcumin concentration of 5μg/ml retained its OCR at 16.6 ± 1.0 pmol/min and showed no signs of mitochondrial dysfunction or damage to the cells. The overall response of the free curcumin or CNG treated cells towards mitochondrial stress assay can be seen in Figure 3D, where a healthy response similar to non-treated controls was seen for 1 μg/ml curcumin and CNG (5μg/ml equivalent curcumin loading). Curcumin with 5 μg/ml possessed no positive response towards the mitochondrial stress assay due to mitochondrial damage at that concentration.
Therefore, all additional studies of mitochondrial oxidative stress protection, viable concentration i.e. 1μg/ml for free curcumin and 5μg/ml of equivalent curcumin for CNGs were used. To assess and test the prolonged protection of curcumin PBAE nanogels against mitochondrial oxidative stress, two standards were set for the design of experiment: 1) antioxidant treatment concentrations as stated above; and 2) concentration and exposure time to cells of oxidative stress inducer, H2O2, which is 0.25mM and 2 hours. This parameter was set so only mitochondrial bioenergetics were disturbed due to exposure but not the onset of cell necrosis/apoptosis which could result in added artifacts during OCR recordings. Mitochondrial respiration rate was measured as OCR (pmol/min) and the bioenergetics parameters are represented as basic OCR (pmol/min) as well as %OCR (after base-line normalization) to eliminate the effect of any variation in starting cell density as shown in Figures 5 and 6. Two hour H2O2 exposure by itself led to a drop in basal respiration, ATP linked and maximal respiration respectively to 34.5 ± 3.0%, 21.6 ± 3.6% and 34.8 ± 11.3% OCR from 76.1 ± 6.0%, 60.7 ± 5.7% and 149.6 ± 20.0% of the non-treated controls as shown in Figure 5 A, B and C. Curcumin treatments at 0, 12 and 24 hours prior to H2O2 addition did not aid in recovery of the OCR, basal respiration or maximal respiration, which were observed to be 19.8 ± 10.2%, 25.0 ± 5.7% and 37.0 ± 26.7% respectively. In other words, viable concentrations of curcumin did not show any effective protection against mitochondrial OS. On the other hand, CNGs showed an enhanced OCR post H2O2 treatment at all exposure times giving basal OCR: 47.17 ± 3.93, 60.0 ± 2.0, 94.3 ± 10.0 as %OCR, ATP linked OCR: 31.2 ± 7.5, 40.0 ± 14, 52.8 ± 5.2 and maximum respiration rates of 336.5 ± 12.5, 198.6 ± 37.3 and 161.7 ± 11.7 respectively for 0, 12 and 24 hours of prior nanogel treatment.
4. DISCUSSION
There can be several ROS generating pathways such as NADP(H) oxidase, xanthine oxidase including mitochondrial electron chain transport, which leads to cellular oxidative stress condition (Basta, Lazzerini et al. 2005). But in the present study, mitochondrial response was analyzed because of its high sensitivity to disturbance in redox balance. Significant efforts have been made to improve the delivery of mitochondria oxidative stress targeting drugs (e.g., MitoQ) with the help of nanocarriers, for the purpose of suppressing mitochondrial oxidative stress and reducing mitochondrial dysfunction (Graham, Huynh et al. 2009, Zang, Sadek et al. 2012). MitoQ is an ubiquinol moiety linked to a lipophilic triphhenylphosphonium (TPP) by a ten-carbon alkyl chain. A part of the TPP chain helps in the rapid cellular uptake of the drug loaded carrier across the cell membrane and further accumulation in mitochondria (Robin J Smith 2011). In vivo, MitoQ and its analogs (MitoVitE, MitoPBN) have shown positive results against mitochondrial oxidative stress by inhibiting lipid peroxidation, ROS/RNS generation etc. (Sheu, Nauduri et al. 2006). TPP based targeted antioxidant delivery approaches are most effective in rapid accumulation of antioxidants within the mitochondria, where the toxicity of the hydrophobic moiety (TPP) can result in nonspecific mitochondrial dysfunction, resulting in disruption of membrane integrity, respiration, and ATP production rate. This non-specificity further depends on the amount of compound internalized. Therefore, the constraint is the amount of drug that can be delivered with a TTP based nanocarrier to ultimately reach therapeutic levels before reaching disruptive concentrations (Victor, Apostolova et al. 2009, Hirzel, Lindinger et al. 2013, Reily, Mitchell et al. 2013). Also, the ubiquinol based approaches may increase protection against peroxynitrite and superoxide radicals, but their efficacy has been reported to be small to negligible for one of the most significant oxidants (hydrogen peroxide H2O2) in cellular physiology (Smith, Adlam et al. 2008).
As an alternative, a number of studies have demonstrated the protective effect of curcumin towards mitochondrial dysfunction and mitochondrial oxidative stress (Zhu, Chen et al. 2004, Martinez-Morua, Soto-Urquieta et al. 2013). Most research has been concentrated on neurodegenerative diseases and finding new therapeutics for Alzheimer’s (Zhang, Fiala et al. 2006, Mishra and Palanivelu 2008). Curcumin has been shown to inhibit the depolarization of mitochondrial membrane potential as well as decreasing the levels of the pathological marker for the Alzheimer’s disease amyloid-β (Huang, Xu et al. 2012, Swomley, Förster et al. 2014). Similar results were observed in aluminum induced mitochondrial dysfunction studies and malate induced oxidative stress, commonly known as nephropathy etc. (Sood, Nahar et al. 2011) (Trujillo, Chirino et al. 2013). These positive results and outcomes from such approaches are reported only from in vitro models, but there is very little evidence of positive results in vivo. Faster in vivo metabolism and the inability of curcumin to reach therapeutic concentration in the targeted micro-locations may be the cause of failure in vivo rather than in vitro.
In theory, it is possible to overcome the delivery limitations to curcumin by developing a slow drug releasing polymeric systems. Further developing these in the nanocarrier form, nanogels would allow them to be administered through a wide range of delivery routes, either oral, inhalation, intravenous or percutaneous, depending upon the ultimate intended site of action. For instance, it is expected that for systemic use, approaches centered on oral or inhalation would be most beneficial. Using synthesized PBAE conjugated curcumin nanogels, our aim was to improve the delivery of curcumin in its active and stable form at a therapeutic, yet non-toxic concentration, with enough time to cure the site of action before metabolizing. Indeed, prior work with poly(trolox) demonstrated the ability to elicit new antioxidant benefits not observed in the parent compound, suggesting that these benefits may be seen for a wide variety of redox active materials (Wattamwar, Mo et al. 2010). Keeping this in mind, this work confirms that the benefits of forming an antioxidant polymer with its uniform release rate having an overall therapeutic effectiveness.
During the synthesis process, nanogel diameter could be varied from 50 to 250 nm with a mere change in monomer concentrations and keeping the molar ratio constant (Fig. 1 A). As no other competing monomer in front of curcumin acrylate was present to react with the tetra functional amine (TTD), we expect the theoretical starting monomer composition of nanogel reaction to be the final composition of synthesized nanogel, which when calculated gives 62.8% curcumin loading by mass. To our knowledge, this was the highest curcumin loading ever reported in the nanocarriers.
These spherical nanogels were synthesized by functionalizing the alcoholic group of curcumin into an acrylate followed by covalent conjugation with TTD forming cross-linked PBAEs. Further, the mechanism of ester hydrolysis adjacent to the antioxidant resulted in release of curcumin in its original structural form (Fig. 2B). The functionalization and conjugation step permits the structural stability of curcumin and prevents it from metabolizing until released, potentially solving one of the in vivo limitations. Adding to the stability advantage, slow and uniform degradation of CNGs avoids the occurrence of any burst release, even at initial time points as shown in Figure 2 A (15% of the total released in first 10 hours or 28% release in first 20 hours). This is an important point to consider because burst release from drug carriers or high bolus doses of free curcumin can initiate a pro-oxidant effect and, instead of acting as a treatment drug, become toxic in vitro to healthy tissues/endothelial cells. The uniform release profile of curcumin during degradation also helped increase the safety limit for curcumin treatment, giving IC50 values of more than 100 μg/ml as compared to free curcumin with IC50 values of 5.3 μg/ml, observed after 24 hours of treatment with HUVECs (Fig. 3A). This property would provide an option to treat an injury at higher and more effective dosage levels.
Antioxidant potential of these nanogels with respect to time during degradations was analyzed by carrying out TEAC antioxidant activity assay on all degradation samples. The cumulative increase in equivalent trolox content over time confirmed the protection of curcumin from losing its antioxidant activity in aqueous media until hydrolyzed from the polymer matrix (Fig. 2C). In other words, functionality of the drug was kept intact until released.
Mitochondrial functional levels confirmed lower dose dependent cytotoxicity compared to free curcumin and prolonged active antioxidant release of CNGs. Seahorse XF96 analyzer recorded real time mitochondrial OCR in response to the treatments and/or mitochondria stress assays specifically developed to determine basal respiration rates, ATP production, spare respiratory capacity, and maximal respiration. The dose dependent response post 24-hour treatment with 1 and 5 μg/ml of equivalent curcumin in CNGs showed no negative impact on OCR levels at either concentration. However, mitochondrial function/OCR was completely shut down with free curcumin at 5 μg/ml while 1 μg/ml was observed to be a safe concentration for mitochondria (Fig. 3C and Fig. 4D). These results comply with the cell viability assay (50% cell death at 5 μg/ml) but show a more acute response, thus providing an opportunity to analyze our material at more sensitive levels. With this understanding, non-fatal curcumin and curcumin nanogel concentrations, 1 and 5 μg/ml respectively (Fig. 3C and Fig. 4D), were used to specifically observe any change in mitochondrial function upon induced oxidative stress injury without causing cell death.
We used H2O2 for overall cellular and mitochondrial oxidative stress insult analysis, because H2O2 is believed to induce mitochondrial dysfunction via sudden increase of ROS inside the mitochondria. This activity results in oxidative stress initiating cell necrosis and apoptotic processes. Measuring the overall effect of H2O2 on HUVECs showed a decrease in cell viability and increase in specific DCF fluorescence at the same time, demonstrating the occurrence of oxidative stress due to H2O2. This effect was effectively suppressed by simultaneously exposing cells with CNGs, indicating the suppression of induced oxidative stress due to steady curcumin release from CNGs with no inherent toxicity. For the Seahorse XF96 studies, concentration and exposure time of H2O2 were selected to affect mitochondrial function without reaching the cells’ necrotic or apoptotic state. This allowed for mitochondrial respiration and energy production potential to be monitored with no cell death effects. H2O2 concentration of 0.5 mM with 2-hour exposure showed a decrease in ATP production (% OCR) from 60.8 ± 5.7 to 21.6 ± 3.6, and a reduction in mitochondrial respiration from 76.0 ± 6.0 to 34.5 ± 3.0 (Fig. 5A and B). Oligomycin, FCCP and antimycinA/rotenone were used as the titrant substrates (mitochondrial stress test assay) to analyze the functional state of the mitochondria post H2O2 ± CNG/curcumin treatment. Interestingly, upon pre-treating the cells with CNGs (5 μg/ml) or curcumin (1 μg/ml) for 0, 12, 24 hours before the 2-hour exposure to 0.25 mM H2O2, curcumin nanogels were able to retain and maintain the basal respiration rates in HUVECs, leveling to non-treated controls. Also, ATP linked and maximum respiration analyzed after running stress assays for all treatment time scales was also higher than the H2O2 injury control group. In contrast, groups with 0, 12 and 24 hours of curcumin pre-treatment did not help the mitochondrion maintain basal respiration, ATP linked production rates or maximum respiration rates, but were similarly low compared to H2O2 treated groups at those treatment times. As seen in Figures 5 and 6 A, B and C, mitochondrial bioenergetics worsened in curcumin groups simultaneously treated with H2O2 (t=0), relapsing to H2O2 levels pretreatment time increased. A synergistic effect of curcumin and H2O2, where the pro-oxidant effect might have increased in presence of another oxidant, may explain this, which was also seen with overall cell viability and DCF testing (Fig. 4 A and B). At t = 0, when curcumin is at 20% concentration of its IC50 concentration i.e., 1 μg/ml along with H2O2 starts showing pro-oxidant effect which decreases as curcumin active content starts reducing down with time due to curcumin molecular degradation and by 24 hour, cells would encounter the instant effect of H2O2 only. All this suggests that CNGs are benign to both the mitochondrion as a PBAE polymer matrix by itself, and at a higher equivalent curcumin loading. This is due to the slow, steady curcumin release property of CNGs. Prolonged protection from mitochondrial oxidative stress for at least 24 hours confirms the release of active curcumin against the no positive effects shown by free curcumin. Therefore, a continuous supply of active antioxidant at therapeutically effective yet non-fatal concentrations would be capable of scavenging excess free radicals and suppressing mitochondrial oxidative stress and cell apoptosis.
5. CONCLUSION
Curcumin conjugated PBAE nanogels were synthesized, resulting in a slow and steady release of active curcumin molecules and an increased limit for a safe concentration of the drug. This work demonstrates the importance of delivery and controlled release upon the functional impact of biologically unstable compounds such as polyphenolic antioxidants. These spherical nanogels effectively suppressed mitochondrial oxidative stress over a 24-hour period, which was confirmed by real time mitochondrial respiration state using Seahorse XF96 analyzer. The nanogels system can further be explored in the area of specific targeting towards different inflammatory marker for localized drug delivery.
Supplementary Material
Acknowledgments
We are thankful to Heather N Russell-Simmnons and Michael C. Alstott for help with manuscript preparation; the University of Kentucky Markey Cancer Center Redox Metabolism Shared Resource Facility for performing Seahorse experiments; and the University of Kentucky Markey Cancer Center Research Communications Office for text edits and figure formatting.
FUNDING SOURCE
This publication was supported in part by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant UL1TR000117, by the National Institute of Dental and Craniofacial Research, National Institutes of Health through Grant R43 DE02352301, and by the National Cancer Institute grant P30 CA177558 (for RM SRF resource facilities). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Abbreviations
- ABTS
2, 2′-azinobis-(3-ethylbenzothiazoline-6-sulfonate)
- ATP
Adenosine triphosphate
- CA
Curcumin acrylate
- CNG
Curcumin nanogels
- C-PBAE
Curcumin-poly(β amino ester)
- DCF
Dichlorofluorocein
- DCFDA
Dichlorofluorocein-diacetate
- DLS
Dynamic light scattering
- DMEM
Dubelco’s
- DNA
Deoxyribonucleic acid
- EBM
Endothelal basal medium
- ECAR
Extracellular acidification rate
- FCCP
Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone
- H2O2
Hydrogen peroxide
- HUVECs
Human umbilical vein endothelial cells
- Mitochondrial OS
Mitochondrial oxidative stress
- mtDNA
Mitochondrial deoxyribonucleic acid
- OCR
Oxygen consumption rate
- OS
Oxidative stress
- RCF
Relative centrifugal force
- RNS
Reactive nitrogen species
- ROS
Reactive oxygen species
- SDS
Sodium dodecyl sulfate
- SEM
Scanning electron microscopy
- TEA
Triethyl amine
- TEAC
Trolox equivalent antioxidant capacity
- THF
Tetrahydrofuran
- TPP
Triphhenylphosphonium
- TTD
4,7,10-Trioxatridecane-1,13-diamine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURES
The authors have no potential conflicts of interest.
References
- Anand P, Nair HB, Sung B, Kunnumakkara AB, Yadav VR, Tekmal RR, Aggarwal BB. Design of curcumin-loaded PLGA nanoparticles formulation with enhanced cellular uptake, and increased bioactivity in vitro and superior bioavailability in vivo. Biochemical Pharmacology. 2010;79(3):330–338. doi: 10.1016/j.bcp.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Basta G, Lazzerini G, Del Turco S, Ratto GM, Schmidt AM, De Caterina R. At least 2 distinct pathways generating reactive oxygen species mediate vascular cell adhesion molecule-1 induction by advanced glycation end products. Arterioscler Thromb Vasc Biol. 2005;25(7):1401–1407. doi: 10.1161/01.ATV.0000167522.48370.5e. [DOI] [PubMed] [Google Scholar]
- Butterfield DA, Di Domenico F, Barone E. Elevated risk of type 2 diabetes for development of Alzheimer disease: a key role for oxidative stress in brain. Biochim Biophys Acta. 2014;1842(9):1693–1706. doi: 10.1016/j.bbadis.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Vazquez EJ, Moghaddas S, Hoppel CL, Lesnefsky EJ. Production of reactive oxygen species by mitochondria: central role of complex III. J Biol Chem. 2003;278(38):36027–36031. doi: 10.1074/jbc.M304854200. [DOI] [PubMed] [Google Scholar]
- Drose S, Brandt U. Molecular mechanisms of superoxide production by the mitochondrial respiratory chain. Adv Exp Med Biol. 2012;748:145–169. doi: 10.1007/978-1-4614-3573-0_6. [DOI] [PubMed] [Google Scholar]
- Federico A, Cardaioli E, Da Pozzo P, Formichi P, Gallus GN, Radi E. Mitochondria, oxidative stress and neurodegeneration. J Neurol Sci. 2012;322(1–2):254–262. doi: 10.1016/j.jns.2012.05.030. [DOI] [PubMed] [Google Scholar]
- Garcea G, Jones DJ, Singh R, Dennison AR, Farmer PB, Sharma RA, Steward WP, Gescher AJ, Berry DP. Detection of curcumin and its metabolites in hepatic tissue and portal blood of patients following oral administration. Br J Cancer. 2004;90(5):1011–1015. doi: 10.1038/sj.bjc.6601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham D, Huynh NN, Hamilton CA, Beattie E, Smith RA, Cocheme HM, Murphy MP, Dominiczak AF. Mitochondria-targeted antioxidant MitoQ10 improves endothelial function and attenuates cardiac hypertrophy. Hypertension. 2009;54(2):322–328. doi: 10.1161/HYPERTENSIONAHA.109.130351. [DOI] [PubMed] [Google Scholar]
- Gupta P, Authimoolam SP, Hilt JZ, Dziubla TD. Quercetin conjugated poly(β-amino esters) nanogels for the treatment of cellular oxidative stress. Acta Biomaterialia. 2015;27:194–204. doi: 10.1016/j.actbio.2015.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirzel E, Lindinger PW, Maseneni S, Giese M, Rhein VV, Eckert A, Hoch M, Krahenbuhl S, Eberle AN. Differential modulation of ROS signals and other mitochondrial parameters by the antioxidants MitoQ, resveratrol and curcumin in human adipocytes. J Recept Signal Transduct Res. 2013;33(5):304–312. doi: 10.3109/10799893.2013.822887. [DOI] [PubMed] [Google Scholar]
- Hsieh H-L, Yang C-M. Role of Redox Signaling in Neuroinflammation and Neurodegenerative Diseases. BioMed Research International. 2013;2013:484613. doi: 10.1155/2013/484613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HC, Xu K, Jiang ZF. Curcumin-mediated neuroprotection against amyloid-beta-induced mitochondrial dysfunction involves the inhibition of GSK-3beta. J Alzheimers Dis. 2012;32(4):981–996. doi: 10.3233/JAD-2012-120688. [DOI] [PubMed] [Google Scholar]
- Huang SX, Partridge MA, Ghandhi SA, Davidson MM, Amundson SA, Hei TK. Mitochondria-derived reactive intermediate species mediate asbestos-induced genotoxicity and oxidative stress-responsive signaling pathways. Environ Health Perspect. 2012;120(6):840–847. doi: 10.1289/ehp.1104287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa Y, Ghanefar M, Bayeva M, Wu R, Khechaduri A, Prasad SVN, Mutharasan RK, Naik TJ, Ardehali H. Cardiotoxicity of doxorubicin is mediated through mitochondrial iron accumulation. The Journal of Clinical Investigation. 2014;124(2):617–630. doi: 10.1172/JCI72931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo JJ, Chang HH, Tsai TH, Lee TY. Positive effect of curcumin on inflammation and mitochondrial dysfunction in obese mice with liver steatosis. Int J Mol Med. 2012;30(3):673–679. doi: 10.3892/ijmm.2012.1049. [DOI] [PubMed] [Google Scholar]
- Madrigal JL, Olivenza R, Moro MA, Lizasoain I, Lorenzo P, Rodrigo J, Leza JC. Glutathione depletion, lipid peroxidation and mitochondrial dysfunction are induced by chronic stress in rat brain. Neuropsychopharmacology. 2001;24(4):420–429. doi: 10.1016/S0893-133X(00)00208-6. [DOI] [PubMed] [Google Scholar]
- Fariss Marc W, C CB, Patel Manisha, Van Houten Bennett, Orrenius Sten. Role of mitochondria in toxic oxidative stress. Molecular interventions. 2005;5(2):94–111. doi: 10.1124/mi.5.2.7. [DOI] [PubMed] [Google Scholar]
- Marchi S, Giorgi C, Suski JM, Agnoletto C, Bononi A, Bonora M, Marchi ED, Missiroli S, Patergnani S, Poletti F, Rimessi A, Duszynski J, Wieckowski MR, Pinton P. Mitochondria-Ros Crosstalk in the Control of Cell Death and Aging. Journal of Signal Transduction. 2012;2012 doi: 10.1155/2012/329635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maritim AC, Sanders RA, Watkins JB., 3rd Diabetes, oxidative stress, and antioxidants: a review. J Biochem Mol Toxicol. 2003;17(1):24–38. doi: 10.1002/jbt.10058. [DOI] [PubMed] [Google Scholar]
- Martinez-Morua A, Soto-Urquieta MG, Franco-Robles E, Zuniga-Trujillo I, Campos-Cervantes A, Perez-Vazquez V, Ramirez-Emiliano J. Curcumin decreases oxidative stress in mitochondria isolated from liver and kidneys of high-fat diet-induced obese mice. J Asian Nat Prod Res. 2013;15(8):905–915. doi: 10.1080/10286020.2013.802687. [DOI] [PubMed] [Google Scholar]
- Mishra S, Palanivelu K. The effect of curcumin (turmeric) on Alzheimer’s disease: An overview. Annals of Indian Academy of Neurology. 2008;11(1):13–19. doi: 10.4103/0972-2327.40220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrenius S, Gogvadze V, Zhivotovsky B. Mitochondrial Oxidative Stress: Implications for Cell Death. Annual Review of Pharmacology and Toxicology. 2007;47(1):143–183. doi: 10.1146/annurev.pharmtox.47.120505.105122. [DOI] [PubMed] [Google Scholar]
- Reily C, Mitchell T, Chacko BK, Benavides GA, Murphy MP, Darley-Usmar VM. Mitochondrially targeted compounds and their impact on cellular bioenergetics. Redox Biology. 2013;1(1):86–93. doi: 10.1016/j.redox.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative stress, inflammation, and cancer: How are they linked? Free radical biology & medicine. 2010;49(11):1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringman J, Frautschy S, Teng E, Begum A, Bardens J, Beigi M, Gylys K, Badmaev V, Heath D, Apostolova L, Porter V, Vanek Z, Marshall G, Hellemann G, Sugar C, Masterman D, Montine T, Cummings J, Cole G. Oral curcumin for Alzheimer’s disease: tolerability and efficacy in a 24-week randomized, double blind, placebo-controlled study. Alzheimer’s Research & Therapy. 2012;4(5):43. doi: 10.1186/alzrt146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Robin J, M MP. Mitochondria-targeted Antioxidants as Therapies. Discovery Medicine. 2011;11(57):106–114. [PubMed] [Google Scholar]
- Rolo AP, Palmeira CM. Diabetes and mitochondrial function: role of hyperglycemia and oxidative stress. Toxicol Appl Pharmacol. 2006;212(2):167–178. doi: 10.1016/j.taap.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Sheu S-S, Nauduri D, Anders MW. Targeting antioxidants to mitochondria: A new therapeutic direction. Biochimica et Biophysica Acta (BBA) – Molecular Basis of Disease. 2006;1762(2):256–265. doi: 10.1016/j.bbadis.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Smith RA, Adlam VJ, Blaikie FH, Manas AR, Porteous CM, James AM, Ross MF, Logan A, Cocheme HM, Trnka J, Prime TA, Abakumova I, Jones BA, Filipovska A, Murphy MP. Mitochondria-targeted antioxidants in the treatment of disease. Ann N Y Acad Sci. 2008;1147:105–111. doi: 10.1196/annals.1427.003. [DOI] [PubMed] [Google Scholar]
- Sood PK, Nahar U, Nehru B. Curcumin attenuates aluminum-induced oxidative stress and mitochondrial dysfunction in rat brain. Neurotox Res. 2011;20(4):351–361. doi: 10.1007/s12640-011-9249-8. [DOI] [PubMed] [Google Scholar]
- Swomley AM, Förster S, Keeney JT, Triplett J, Zhang Z, Sultana R, Butterfield DA. Abeta, oxidative stress in Alzheimer disease: Evidence based on proteomics studies. Biochimica et Biophysica Acta (BBA) – Molecular Basis of Disease. 2014;1842(8):1248–1257. doi: 10.1016/j.bbadis.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia E, Sánchez-Lozada LG, García-Niño WR, García E, Cerecedo A, García-Arroyo FE, Osorio H, Arellano A, Cristóbal-García M, Loredo ML, Molina-Jijón E, Hernández-Damián J, Negrette-Guzmán M, Zazueta C, Huerta-Yepez S, Reyes JL, Madero M, Pedraza-Chaverrí J. Curcumin prevents maleate-induced nephrotoxicity: Relation to hemodynamic alterations, oxidative stress, mitochondrial oxygen consumption and activity of respiratory complex I. Free Radical Research. 2014;48(11):1342–1354. doi: 10.3109/10715762.2014.954109. [DOI] [PubMed] [Google Scholar]
- Trujillo J, Chirino YI, Molina-Jijón E, Andérica-Romero AC, Tapia E, Pedraza-Chaverrí J. Renoprotective effect of the antioxidant curcumin: Recent findings. Redox Biology. 2013;1(1):448–456. doi: 10.1016/j.redox.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor VM, Apostolova N, Herance R, Hernandez-Mijares A, Rocha M. Oxidative stress and mitochondrial dysfunction in atherosclerosis: mitochondria-targeted antioxidants as potential therapy. Curr Med Chem. 2009;16(35):4654–4667. doi: 10.2174/092986709789878265. [DOI] [PubMed] [Google Scholar]
- Wattamwar PP, Biswal D, Cochran DB, Lyvers AC, Eitel RE, Anderson KW, Hilt JZ, Dziubla TD. Synthesis and characterization of poly(antioxidant beta-amino esters) for controlled release of polyphenolic antioxidants. Acta Biomater. 2012;8(7):2529–2537. doi: 10.1016/j.actbio.2012.03.022. [DOI] [PubMed] [Google Scholar]
- Wattamwar PP, Mo Y, Wan R, Palli R, Zhang Q, Dziubla TD. Antioxidant Activity of Degradable Polymer Poly(trolox ester) to Suppress Oxidative Stress Injury in the Cells. Advanced Functional Materials. 2010;20(1):147–154. [Google Scholar]
- Zang QS, Sadek H, Maass DL, Martinez B, Ma L, Kilgore JA, Williams NS, Frantz DE, Wigginton JG, Nwariaku FE, Wolf SE, Minei JP. Specific inhibition of mitochondrial oxidative stress suppresses inflammation and improves cardiac function in a rat pneumonia-related sepsis model. American Journal of Physiology – Heart and Circulatory Physiology. 2012;302(9):H1847–H1859. doi: 10.1152/ajpheart.00203.2011. [DOI] [PubMed] [Google Scholar]
- Zhang L, Fiala M, Cashman J, Sayre J, Espinosa A, Mahanian M, Zaghi J, Badmaev V, Graves MC, Bernard G, Rosenthal M. Curcuminoids enhance amyloid-beta uptake by macrophages of Alzheimer’s disease patients. J Alzheimers Dis. 2006;10(1):1–7. doi: 10.3233/jad-2006-10101. [DOI] [PubMed] [Google Scholar]
- Zhong H, Yin H. Role of lipid peroxidation derived 4-hydroxynonenal (4-HNE) in cancer: Focusing on mitochondria. Redox Biology. 2015;4(0):193–199. doi: 10.1016/j.redox.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YG, Chen XC, Chen ZZ, Zeng YQ, Shi GB, Su YH, Peng X. Curcumin protects mitochondria from oxidative damage and attenuates apoptosis in cortical neurons. Acta Pharmacol Sin. 2004;25(12):1606–1612. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


