Abstract
All living creatures change their gene expression program in response to nutrient availability and metabolic demands. Nutrients and metabolites can directly control transcription and activate second-messenger systems. More recent studies reveal that metabolites also affect post-transcriptional regulatory mechanisms. Here, we review the increasing number of connections between metabolism and post-transcriptional regulation in eukaryotic organisms. First, we present evidence that riboswitches, a common mechanism of metabolite sensing in bacteria, also function in eukaryotes. Next, we review an example of a double stranded RNA modifying enzyme that directly interacts with a metabolite, suggesting a link between RNA editing and metabolic state. Finally, we discuss work that shows some metabolic enzymes bind directly to RNA to affect mRNA stability or translation efficiency. These examples were discovered through gene-specific genetic, biochemical, and structural studies. A directed systems level approach will be necessary to determine whether they are anomalies of evolution or pioneer discoveries in what may be a broadly connected network of metabolism and post-transcriptional regulation.
Metabolite homeostasis is required for normal cellular and systemic function, and the loss of homeostasis often leads to disease. A classic example is type I diabetes, where a precipitous change in blood glucose levels, caused by a failure in insulin signaling, leads to blindness, sores, infections on extremities, and nerve damage. Homeostasis depends upon the ability of cells to change the expression of their genes in response to changes in metabolite concentration. The mechanisms by which metabolites regulate gene expression are incompletely understood.
There are numerous examples where metabolite concentration regulates the activity of factors involved in transcription 1,2. For example, the peroxisome proliferator-activated receptor (PPAR) family of nuclear hormone receptors bind directly to fatty acids and eicosanoids. Upon metabolite association, PPARs bind to specific DNA elements in promoters and act as transcription factors to regulate expression of a variety genes, including several involved in lipid metabolism 1-3. A number of potent synthetic PPAR ligands are used to treat diseases such as dyslipidemia and diabetes. In fact, metabolite responsive transcription factors are targeted in several therapeutic strategies, probably because of their importance in regulating of gene expression and their ability to bind to small molecule metabolite ligands 4.
Second messenger signaling also couples metabolic state to gene expression. Metabolites, hormones, gasses, and other small molecules bind and activate receptors on the cell surface to enact an intracellular signaling cascade that leads to the activation or repression of specific genes. An example of second-messenger signaling is the phosphoinositide 3-kinase (PI3K) pathway. PI3K catalyzes the formation of inositol trisphosphate (IP3) in response to a number of hormones and metabolites involved in cell growth and survival 5,6. IP3 regulates genes involved in cell cycling and apoptosis 5 via the mammalian target of rapamycin (mTOR) pathway. These complex signaling cascades have received extensive attention because activating or inhibiting metabolite and hormone surface receptors using small molecules can be a viable therapeutic strategy.
By contrast, there are relatively few examples of post-transcriptional metabolite sensing in eukaryotes. Here, we review those examples, and present competing hypotheses concerning the apparent lack of metabolite sensing in eukaryotes.
Riboswitches
Riboswitches are structured RNA elements often found in the 5′ untranslated region (UTR) of bacterial transcripts. They sense the concentration of specific metabolites through direct interactions. Metabolite binding typically induces a change in riboswitch conformation that in turn alters transcription termination or translation initiation efficiency. As such, riboswitches provide a negative feedback loop that shuts down production of metabolic enzymes and related factors required to produce the associated metabolite.
The first riboswitch was described in 2002 by Breaker and coworkers, who reported that in Escherichia coli, transcripts of the cobalamin (vitamin B12) biosynthesis operon btuB directly bind cobalamin to induce a structural change that prevents ribosome binding 7. Since then, many riboswitches have been identified and categorized into classes based upon the type of ligand and the secondary structure formed upon association with that ligand. To date more than 20 classes of riboswitches sensitive to a variety of metabolites have been identified in bacteria. In contrast, only one class of riboswitch has been discovered in eukaryotes. A number of excellent reviews on bacterial riboswitches have been published recently 8-10. Here, we will focus on the eukaryotic riboswitches.
TPP Riboswitches in Eukaryotes
The thiamine-responsive TPP riboswitch is the most common bacterial riboswitch, and it has now been identified in plants, fungi, and archaea (Figure 1) 11-14. Hanamoto et al. discovered the first eukaryotic TPP riboswitch in the 5′ UTR of the thiA gene of the filamentous fungus Aspergillus orzyae, used in the production of sake 11. An intron is present in the thiA 5′ UTR that contains two motifs highly conserved in fungal thiamine biosynthesis genes. Reporter studies and northern analyses revealed that thiamine concentration controls the extent of 5′ intron splicing in a manner that depended on both conserved elements. The increase in unspliced 5′ UTR mRNA in the presence of thiamine correlates with a decrease in the expression of thiA. Subsequent studies by Breaker et al. identified sequences that match a TPP riboswitch consensus descriptor in thiamine biosynthesis genes from Arabidopsis thaliana, Oriza sativa (rice), Poa secunda (bluegrass), and the fungi Neurospora crassa and Fusarium oxysporum 15 . Structural probing revealed that the element from Arabidopsis, found in the thiA 3′ UTR, adopts a TPP riboswitch-like structure in the presence of thiamine, suggesting that the element comprises a bona fide riboswitch similar to those observed in bacteria.
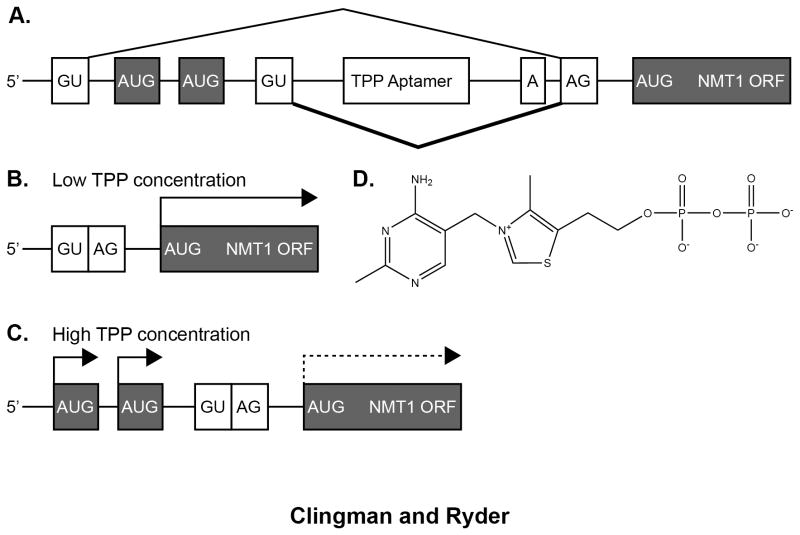
Figure 1.
TPP-regulated alternative splicing of the NMT1 gene. A) Schematic of the 5′ UTR intron structure. The thin line above the schematic denotes the predominant isoform in low TPP conditions. The thick line below the schematic denotes the pattern in high TPP conditions. B) The low TPP isoform contains a short 5′ UTR with a single initiation codon, leading to efficient translation of NMT1. C) The high TPP isoform contains a longer 5′ UTR with two uORFs, leading to reduced translation initiation of the cognate NMT1 ORF. D) Chemical structure of TPP, sensed by the riboswitch motif in the 5′ UTR intron.
In the case of the TPP riboswitch from the Neurospora crassa NMT1 5′-UTR (Figure 1), the riboswitch induces an alternative splicing event 12. In the presence of elevated TPP, an upstream 5′-splice site is used leading to production of an mRNA with a longer 5′-UTR. The TPP riboswitch motif is present within the intron sequence that is normally spliced out. The longer mRNA includes an upstream open reading frame (uORF) that decreases translation of the downstream gene product by competing with the authentic translation start site. The data reveal a mechanism whereby a structural change in the TPP riboswitch represses translation of the NMT1 gene via a change in the splice isoform ratio.
Additional studies of eukaryotic TPP riboswitches reveal a common role in modulating alternative mRNA splicing as a means of regulating gene expression 13,16,17. It is intriguing to note that the eukaryotic riboswitches work by modifying a eukaryote-specific process that occurs in the nucleus, rather than affecting transcription or translation initiation directly, as in bacteria. Although several examples of the TTP riboswitch have been characterized in eukaryotes, other riboswitch classes have not yet been identified. It is possible that they do not exist; however, it is likely that through refinement of bioinformatics and experimental techniques, additional examples will be found.
Identifying eukaryotic riboswitches
A number of groups have used riboswitch sequence and structure conservation to search bacterial genomes for additional riboswitches 18-22. Merino and colleagues published an algorithm that sorted and paired operons with Cluster of Orthologous Groups (COG) from the protein database 20,23. Next they used successive rounds of Multiple EM for Motif Elicitation (MEME) 24 followed by Motif Alignment and Search Tool (MAST) to identify over-represented motifs in each COG 20. A subsequent publication described the development of RibEx (riboswitch explorer), a tool that cross-references the genomes of non-redundant bacterial organisms to identify sequence motifs of putative riboswitches and other structural regulatory elements 20,23. RibEx eliminates the use of COGs in favor of using 145 complete genomes in the initial alignment. Unfortunately, a similar tool does not exist for eukaryotes.
Riboswitch identification in eukaryotes has proven challenging due to the increased complexity of the regulatory mechanisms governing gene expression. In bacteria, riboswitch aptamer domains have high sequence conservation, while expression platforms can have disparate sequence, and the regulatory mechanisms are often inferred by their location. These expression platforms are usually positioned to block ribosome binding sites or prevent formation of transcription terminating stems. Identifying eukaryotic riboswitch expression platforms is complicated by a number of factors. First, eukaryotic genes are not grouped into operons. Second, riboswitches could theoretically work at the level of mRNA processing, nuclear export, stability, or translation. Eukaryotic riboswitches are therefore likely to have a heterogeneous nature at the structural and sequence levels. Third, most prokaryotic genomes are roughly 88% protein-coding, while higher organisms such as humans are estimated to have as little as 2% protein-coding sequence 25. The increased size and complexity of eukaryotic genomes presents both challenges and possibilities for bioinformaticians. The large number of mRNA processing events identified in eukaryotes, combined with the possibility that riboswitches may exist in non-coding RNAs to regulate gene expression in trans leads to almost limitless possibilities for eukaryotic riboswitch mechanisms. It is therefore not surprising that bioinformatics approaches have been difficult to develop for eukaryotic riboswitch identification.
Several groups have begun to search for non-coding RNAs within intergenic sequences of bacteria, archea, and eukaryotes. Although these searches are not specifically targeting riboswitch identification, it is possible that the methodology and the precedent set by this type of bioinformatic search may influence future riboswitch discovery in organisms other than bacteria. Breaker and colleagues have been optimizing clustering techniques to discover novel non-coding RNAs in bacteria 26. Similarly, a recent bioinformatic survey of the archaean Pyrococcus abyssi used clustering of sequence, primary structure, and secondary structure to identify conserved non-coding RNAs located in intergenic regions. This study revealed several elements that share features with the SAM-I and lysine bacterial riboswitches, although this remains to be experimentally tested 27. Although most recent work has focused on ribozyme discovery, the recent success in identifying large non-coding RNAs may enable future application of similar methods to the discovery of non-coding RNAs, including riboswitches, in eukaryotes.
Because only one riboswitch class has been identified in eukaryotes, it is not clear whether eukaryotic riboswitches are rare, or have not been discovered due to the increased complexity of higher organisms. If higher organisms truly lack of riboswitches, perhaps proteins have taken over the metabolite-sensing functionality. Numerous proteins regulate gene expression at the post-transcriptional level, and a handful of canonical RNA-binding proteins have demonstrated metabolite sensitivity. Additionally, over the past two decades several metabolic enzymes have demonstrated RNA-binding activity.
Metabolite-Sensitive RNA-Binding Proteins
RNA-binding proteins alter gene expression by regulating pre-mRNA processing steps including alternative splicing, 5′ and 3′ end formation, and by controlling how the RNA sequence is edited. RNA binding proteins also regulate the subcellular location, stability, and translation efficiency of mature mRNAs. Recently, the activities of several RNA binding proteins have been shown to be effected by the concentration of cellular metabolites, allowing cells to quickly respond to changes in the environment.
ADAR
The increased complexity of higher organisms cannot be explained by the relatively small increase in the number of protein-coding genes relative to bacteria. Instead, genetic diversity can be attributed to an expansion in the number of gene products derived from each gene. Alternative pre-mRNA splicing and mRNA editing enable recoding of the information stored in the genome leading to production of multiple protein variants from a single gene.
One example of mRNA editing is the programmed conversion of select adenosine bases to inosine, catalyzed by enzymes termed adenosine deaminases that act on RNA (ADAR). ADARs were initially discovered in Xenopus laevis 28. Homologs have since been identified in most metazoa, but not in plants, fungi, or yeast 28. ADARs from different organisms are structurally similar; all contain several double-stranded RNA binding domains (dsRBDs) and a highly conserved c-terminal deaminase domain. ADARs deaminate specific adenosines to produce inosine at precise locations within an RNA sequence. Inosine pairs with cytidine and is therefore interpreted as a guanine base in biological settings. These editing events cause single-codon alterations, changes in alternative splicing, and modulation of mRNA stability 29.
RNA editing has been observed in pre-mRNA coding sequences, repetitive elements, and pri-miRNAs 29. Neurons use editing extensively to regulate specialized neurotransmitter receptors, ion channels, and surface protein isoforms 30. Mice stably expressing an shRNA to silence ADAR2 expression displayed increased neuronal sensitivity to restricted blood and nutrient supply, and subsequent neuronal degeneration 31. ADAR activity is also reported to affect subcellular compartmentalization of certain mRNAs 32,33, and self-editing of ADAR mRNA 34,35 is proposed to affect ADAR homo- and hetero-dimerization 36 which in turn regulates mRNA editing efficiency. ADAR editing is implicated in a number of disease states including schizophrenia, neuromuscular disorders, and certain cancers 30,31,37.
ADAR associates with IP6, a biologically relevant metabolite
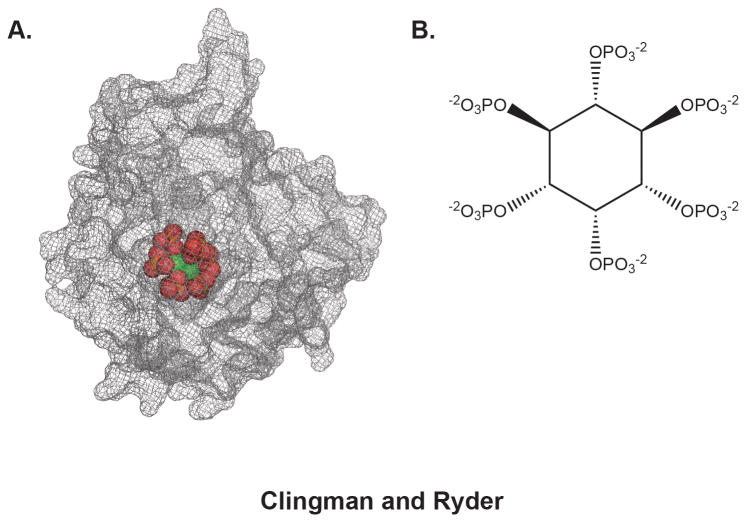
The Bass group determined the high resolution crystal structure of the catalytic domain of ADAR2 and discovered an inositol hexakisphosphate (IP6) molecule required for proper folding of the enzyme 38 (Figure 2). Subsequent experiments showed that an ADAR substrate is edited in wild type yeast expressing hADAR2, but not in yeast that lack the IPK1 gene and thus do not produce IP6 38. The crystal structure revealed that IP6 is intimately associated with protein, buried in an internal cavity lined with basic residues. IP6 was not added during purification or crystallization, indicating that endogenous IP6 co-purified with the protein from yeast 38. This suggests that the association between ADAR and IP6 is likely very tight, although to our knowledge the dissociation constant has not been measured.
Figure 2.
The structure of human ADAR2 bound to IP6. A) The surface of hADAR2 is rendered in mesh, revealing the deep internal cavity that coordinates IP6 (rendered in spheres). The structure was rendered from coordinate file 1ZY738. B) Chemical structure of IP6, sensed by ADAR.
It is probably that ADAR biogenesis is governed by intracellular IP6 concentration 6. IP6 is abundant in healthy cells, where it is involved in signaling pathways including proliferation, differentiation, DNA repair, energy transduction, and RNA export 39. IP6 serves as a phosphate donor in many signaling pathways. It is possible that IP6 levels influence the amount and stability of ADAR available to modify specific genes. This intriguing hypothesis requires additional biochemical and in vivo studies, especially in light of data that suggests that loss of ADAR editing activity can lead to cancer, while abundant IP6 can help prevent cancers 30,39. Measurement of the extent of mRNA editing in the presence and absence of IP6, and determining the effect of editing on cellular physiology, will be necessary first steps towards demonstrating that ADAR is a biologically relevant sensor of IP6 concentration.
Metabolic enzymes with RNA-binding activity
Metabolic enzymes are integral to maintaining homeostasis because they catalyze the chemical reactions necessary to produce or use nutrients. Metabolic enzymes must be tightly regulated through feedback loops to ensure appropriate metabolic flux. Some metabolic enzymes have been reported to ‘moonlight’ as RNA-binding proteins, regulating gene expression at the post-transcriptional level in addition to catalyzing chemical reactions 40. Hentze and Preiss termed this interplay between RNA, Enzymes and Metabolites the ‘REM’ phase of RNA regulation 40. In this section, we will introduce several examples of metabolic enzymes that act as RNA-binding proteins, and discuss the possibility that the REM network may be more extensive than is currently realized.
Cytosolic aconitase
Iron is an essential metabolite involved in processes including oxygen transport, cellular respiration and heme synthesis. Iron is also a cofactor for numerous metalloenzymes involved in a wide range of biological processes. Failure to regulate intracellular iron levels results in iron deficiency or toxicity. Free iron catalyzes free radical generation, which can damage DNA, lipids, and proteins. Therefore, most intracellular iron exists in complex with enzymes and carrier proteins. Cytosolic aconitase is an iron-sensitive enzyme that conditionally doubles as an RNA-binding protein to regulate iron homeostasis 41-47. Interestingly, the functionality of cytosolic aconitase in metabolic regulation was not understood until the discovery that iron-responsive protein-1 (IRP1) and cytosolic aconitase are the same protein with two distinct functions.
Aconitase is a tricarboxylic acid (TCA) cycle isomerase that catalyzes the conversion of citrate to isocitrate. Cytosolic aconitase (IRP1) has similar activity to that of mitochondrial aconitase. It was initially unclear why cells would contain two distinct copies of a functionally similar enzyme 48. When intracellular iron is low, IRP1 binds iron-responsive RNA elements (IREs) in iron regulatory genes to modulate their expression. The iron regulatory genes include the iron storage factor ferritin, iron uptake factors like TfR and DMT-1, and the iron export factor ferroportin 44. When IRP1 was purified, sequenced, and cloned, it was found to have approximately 30% homology to Saccharomyces cerevisiae and porcine mitochondrial aconitase 49-54. Subsequent work showed that human recombinant IRP1 has aconitase activity in the presence of iron, and bovine cytosolic aconitase binds to IREs in low iron conditions 55-57. Further experiments from the Hentze and Klausner groups confirmed that cytosolic aconitase and IRP1 are the same protein, with distinct functional activities depending upon intracellular iron concentrations.
In the presence of high intracellular iron the [4Fe-4S] catalytic cluster assembles to enable aconitase functionality 58. When intracellular iron decreases, the protein undergoes and allosteric change to reveal an iron responsive element (IRE)-binding site, enabling functionality as the RNA-binding protein IRP1 59. The IRE is a highly-conserved hairpin present in the 3′ and 5′ UTRs of several genes related to iron homeostasis 60. Interaction between IREs and IRP1 induces repression of ferritin mRNA translation and transferritin receptor mRNA stabilization 61.
How important is iron concentration sensing by IRP1? Interestingly, IRP1-/- mice develop normally without an apparent phenotype 62. In contrast, IRP2-/- mice display microcytic anemia, increased red cell protoporphyrin IX levels, and neurodegeneration, but are otherwise normal 63,64. While mice that lack IRP1 or IRP2 are viable, mice that lack both fail in embryogenesis prior to implantation 65. The mouse mutants reveal IRP activity is required for animal viability, highlighting the importance of regulating iron homeostasis at the post-transcriptional level. They also suggest that IRP2 can compensate for loss of IRP1, while IRP1 is not sufficient to compensate for loss of IRP2 65. Additional work is needed to understand the basis for this difference.
GAPDH
GAPDH has traditionally been labeled a housekeeping protein because it is involved in glycolysis. However, several studies indicate that its biological activity is more diverse 66,67. GAPDH has been shown to bind a range of RNA species, including mRNA, tRNA, rRNA and viral RNA, and may participate in such activities as RNA export and regulation of RNA stability 68-70. GAPDH directly associates with AU-rich elements (AREs) present in the 3′ UTRs of number of RNA species 71. In most cases, however, the physiological relevance of RNA-binding activity by GAPDH has not been validated.
One proposed role for GAPDH RNA-binding activity lies in the regulation of cytokine and endothelin expression. Cytokines and endothelins are small bioactive proteins that modulate the immune response and effect blood vessel constriction, respectively. GAPDH stabilizes mRNA encoding colony-stimulating factor 1 (CSF1), which is implicated in tumorigenesis 72. Conversely, GAPDH association promotes turnover of transcripts encoding the endothelial vasoconstrictor endothelin (ET) 73. The mechanism of GAPDH-mediated regulation of mRNA stability is not known. There is growing evidence that GAPDH regulates lymphokine translation by binding lymphokine transcripts in polysomes 71. Again, the mechanism of regulation is not known.
In vivo, GAPDH is predicted to exist in two conformations: an RNA-binding form that is not active in glycolysis, and an NAD+-binding form that is active in glycolysis 74. The ratio of the two conformations may be regulated by the local concentration of NAD+, NADH, and ATP 74,75. Oxidation state is also predicted to differentially affect the RNA- or NAD+-binding activity of GAPDH 74,75. As such, GAPDH may sense the concentration of oxidized NAD+ in order to control expression of mRNA targets. More work is needed to understand the mechanism by which GAPDH converts from an active metabolic enzyme to an RNA-binding factor, and exactly how changes in metabolic state affect the various transcripts with which it associates 76.
Identifying metabolite-sensing proteins with RNA-binding activity
Recent advances in proteome-wide assays are enabling rapid identification of both metabolite-sensitive proteins with putative RNA-binding activity and RNA-binding proteins with putative metabolite-sensing activity. Several techniques that survey the RNA interactome and/or proteome can be used to assess changes upon alteration of metabolic state. These high-throughput approaches, combined with experimental validation, may lead to a the identification of additonal metabolite-sensing RNA-binding proteins
For proteins with known RNA-binding activity, several methods are available to identify associated transcripts in cells. These include HITS-CLIP, PAR-CLIP, and iCLIP, each of which rely on crosslinking of the protein to associated RNAs in cells, followed by immunopreciptation and deep sequencing to identify the associated RNA 77-79. To identify novel mRNA-binding proteins, Hentze, Krijgsveld, and colleagues adapated the HITS-CLIP and PAR-CLIP methods to monitor the entire mRNA interactome 80. In this technique, termed “interactome capture,” proteins are first cross-linked to associated RNAs in cells. Next, cells are lysed and oligo d(T) beads are used to capture polyadenylated RNA species. The protein interactome is then identified through mass spectrometry. In HeLa cells, over 300 novel RNA-binding proteins were identified by this approach, 46 of which are enzymes involved in intermediary metabolism. This indicates that metabolic enzymes that also bind to RNA are more widespread than previously thought. Because these enzymes function in intermediary metabolic pathways, they are logical candidates for coupling metabolite sensing to RNA regulation.
It will be more complicated to identify metabolite responsive RNA-binding proteins that are not established metabolic enzymes. It is possible that by employing the methods outlined above while manipulating metabolite concentrations, some proteins will display altered RNA-binding activity and/or target recognition. The most obvious candidates to use in this type of experiment are proteins that have already been shown to regulate genes involved in metabolite homeostasis. Proteins demonstrating changes in RNA-binding activity would then require in vitro and in vivo experimentation to determine the mechanisms employed to sense and responds to metabolic state.
Conclusion
Changes in metabolic state induce compensatory changes in gene expression. In Eukaryotes, transcriptional control of gene expression in response to metabolic changes has been widely studied. Metabolites including fatty acids, vitamins, alcohols, amino acids, and nucleotides and derivatives are known to regulate transcription. It is surprising that more is not known about the effect that intermediary metabolites have on post-transcriptional regulation. We have highlighted several examples of post-transcriptional mechanisms that respond to intermediary metabolites. These examples may represent unique, non-pervasive mechanisms of post-transcriptional regulation. However, it is likely that new systems level approaches will identify additional examples. If metabolic control of post-transcriptional regulation is indeed widespread in eukaryotes as it is in bacteria, there will be broad implications for development of novel therapeutics that target metabolite sensitive RNA-binding proteins involved in disease processes.
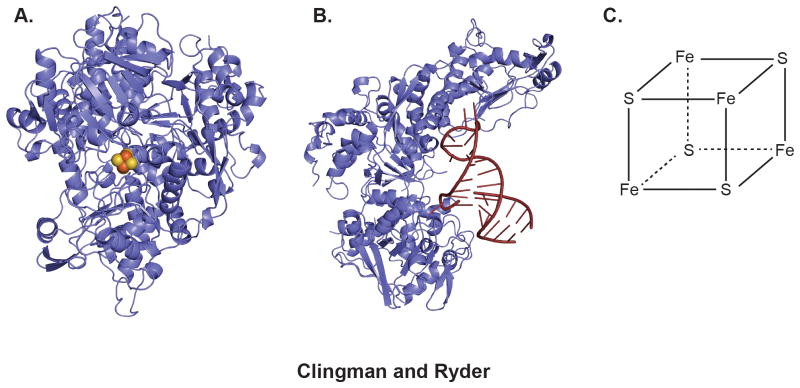
Figure 3.
The two forms of IRP1. A) structure of IRP1 in the iron replete state adopts the canonical aconite fold. The protein structure is rendered as a cartoon, the 4Fe-FS cluster is rendered as spheres. The structure was rendered form coordinate file 2B3X58. B) In iron deficient state, IRP1 adopts an alternative conformation that binds to a specific stem loop RNA structure (red). The structure was rendered from coordinate file 3SNP59. C) Chemical structure of the 4Fe-4S cluster sensed by IRP1.
Acknowledgments
Notes: The authors thank Ruth Zearfoss for critical comments concerning the manuscript. Work in the Ryder lab is supported by NIH Grant GM098643 to S.P.R.
Contributor Information
Carina C Clingman, Department of Biochemistry and Molecular Pharmacology, University of Massachusetts Medical School, Worcester, MA, 01605, USA.
Sean P Ryder, Email: Sean.Ryder@umassmed.edu, Department of Biochemistry and Molecular Pharmacology, University of Massachusetts Medical School, Worcester, MA, 01605, USA, Phone: 508-856-1372, Fax: 508-856-6464.
References
- 1.Kliewer SA, Sundseth SS, Jones SA, Brown PJ, Wisely GB, Koble CS, Devchand P, Wahli W, Willson TM, Lenhard JM, Lehmann JM. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors alpha and gamma. P Natl Acad Sci Usa. 1997;94:4318–23. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X, Sato R, Brown MS, Hua X, Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 3.Berger J, Moller DE. The mechanisms of action of PPARs. Annu Rev Med. 2002;53:409–35. doi: 10.1146/annurev.med.53.082901.104018. [DOI] [PubMed] [Google Scholar]
- 4.Zeng LR, Xie JP. Molecular basis underlying LuxR family transcription factors and function diversity and implications for novel antibiotic drug targets. J Cell Biochem. 2011;112:3079–84. doi: 10.1002/jcb.23262. [DOI] [PubMed] [Google Scholar]
- 5.Kim S, Kim SF, Maag D, Maxwell MJ, Resnick AC, Juluri KR, Chakraborty A, Koldobskiy MA, Cha SH, Barrow R, Snowman AM, Snyder SH. Amino acid signaling to mTOR mediated by inositol polyphosphate multikinase. Cell Metab. 2011;13:215–21. doi: 10.1016/j.cmet.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alcázar-Román AR, Wente SR. Inositol polyphosphates: a new frontier for regulating gene expression. Chromosoma. 2007;117:1–13. doi: 10.1007/s00412-007-0126-4. [DOI] [PubMed] [Google Scholar]
- 7.Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chem Biol. 2002;9:1043. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 8.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 9.Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem. 2009;78:305–34. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breaker RR. Riboswitches and the RNA World. Cold Spring Harbor Perspectives in Biology. 2012;4:a003566–6. doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kubodera T, Watanabe M, Yoshiuchi K, Yamashita N, Nishimura A, Nakai S, Gomi K, Hanamoto H. Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5′-UTR. FEBS letters. 2003;555:516–20. doi: 10.1016/s0014-5793(03)01335-8. [DOI] [PubMed] [Google Scholar]
- 12.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 13.Croft MT, Moulin M, Webb ME, Smith AG. Thiamine biosynthesis in algae is regulated by riboswitches. Proceedings of the National Academy of Sciences. 2007;104:20770–5. doi: 10.1073/pnas.0705786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bocobza SE, Aharoni A. Switching the light on plant riboswitches. Trends Plant Sci. 2008;13:526–33. doi: 10.1016/j.tplants.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Sudarsan N, Barrick JE, Breaker RR. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003;9:644–7. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thore S, Frick C, Ban N. Structural basis of thiamine pyrophosphate analogues binding to the eukaryotic riboswitch. J Am Chem Soc. 2008;130:8116–7. doi: 10.1021/ja801708e. [DOI] [PubMed] [Google Scholar]
- 17.Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–11. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- 18.Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. Regulation of the vitamin B12 metabolism and transport in bacteria by a conserved RNA structural element. RNA. 2003;9:1084–97. doi: 10.1261/rna.5710303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, Wickiser JK, Breaker RR. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. P Natl Acad Sci Usa. 2004;101:6421–6. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abreu-Goodger C, Ontiveros-Palacios N, Ciria R, Merino E. Conserved regulatory motifs in bacteria: riboswitches and beyond. Trends Genet. 2004;20:475–9. doi: 10.1016/j.tig.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N, Neph S, Tompa M, Ruzzo WL, Breaker RR. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007;35:4809–19. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer MM, Roth A, Chervin SM, Garcia GA, Breaker RR. Confirmation of a second natural preQ1 aptamer class in Streptococcaceae bacteria. RNA. 2008;14:685–95. doi: 10.1261/rna.937308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abreu-Goodger C, Merino E. RibEx: a web server for locating riboswitches and other conserved bacterial regulatory elements. Nucleic Acids Res. 2005;33:W690–2. doi: 10.1093/nar/gki445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dinger ME, Gascoigne DK, Mattick JS. The evolution of RNAs with multiple functions. Biochimie. 2011;93:2013–8. doi: 10.1016/j.biochi.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 26.Weinberg Z, Perreault J, Meyer MM, Breaker RR. Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis. Nature. 2009;462:656–9. doi: 10.1038/nature08586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phok K, Moisan A, Rinaldi D, Brucato N, Carpousis AJ, Gaspin C, Clouet-d'Orval B. Identification of CRISPR and riboswitch related RNAs among novel noncoding RNAs of the euryarchaeon Pyrococcus abyssi. BMC Genomics. 2011;12:312. doi: 10.1186/1471-2164-12-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barraud P, T FH. Allain. ADAR proteins: double-stranded RNA and Z-DNA binding domains. Curr Top Microbiol Immunol. 2012;353:35–60. doi: 10.1007/82_2011_145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farajollahi S, Maas S. Molecular diversity through RNA editing: a balancing act. Trends in Genetics. 2010 doi: 10.1016/j.tig.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maas S, Kawahara Y, Tamburro KM, Nishikura K. A-to-I RNA editing and human disease. RNA Biol. 2006;3:1–9. doi: 10.4161/rna.3.1.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng PL, Zhong X, Tu W, Soundarapandian MM, Molner P, Zhu D, Lau L, Liu S, Liu F, Lu Y. ADAR2-dependent RNA editing of AMPA receptor subunit GluR2 determines vulnerability of neurons in forebrain ischemia. Neuron. 2006;49:719–33. doi: 10.1016/j.neuron.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 32.Sansam CL, Wells KS, Emeson RB. Modulation of RNA editing by functional nucleolar sequestration of ADAR2. P Natl Acad Sci Usa. 2003;100:14018–23. doi: 10.1073/pnas.2336131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maas S, Gommans WM. Identification of a selective nuclear import signal in adenosine deaminases acting on RNA. 2009 doi: 10.1093/nar/gkp599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rueter SM, Dawson TR, Emeson RB. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- 35.Palladino MJ, Keegan LP, O'Connell MA, Reenan RA. dADAR, a Drosophila double-stranded RNA-specific adenosine deaminase is highly developmentally regulated and is itself a target for RNA editing. RNA. 2000;6:1004–18. doi: 10.1017/s1355838200000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valente L, Nishikura K. RNA binding-independent dimerization of adenosine deaminases acting on RNA and dominant negative effects of nonfunctional subunits on dimer functions. J Biol Chem. 2007;282:16054–61. doi: 10.1074/jbc.M611392200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmauss C. Regulation of Serotonin 2C Receptor Pre-mRNA Editing by Serotonin A2 . International Review of Neurobiology. Academic Press. 2005:83–100T2. doi: 10.1016/S0074-7742(05)63004-8. [DOI] [PubMed] [Google Scholar]
- 38.Macbeth MR, Schubert HL, Vandemark AP, Lingam AT, Hill CP, Bass BL. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science. 2005;309:1534–9. doi: 10.1126/science.1113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalam Shamsuddin A, Bose S. IP6 (Inositol Hexaphosphate) as a Signaling Molecule. Current Signal Transduction Therapy. 2012;7:289–304. [Google Scholar]
- 40.Hentze MW, Preiss T. The REM phase of gene regulation. Trends in Biochemical Sciences. 2010;35:423–6. doi: 10.1016/j.tibs.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 41.Hentze MW, Kühn LC. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. P Natl Acad Sci Usa. 1996;93:8175–82. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eisenstein RS. Iron regulatory proteins and the molecular control of mammalian iron metabolism. Annu Rev Nutr. 2000;20:627–62. doi: 10.1146/annurev.nutr.20.1.627. [DOI] [PubMed] [Google Scholar]
- 43.Schneider BD, Leibold EA. Regulation of mammalian iron homeostasis. Current Opinion in Clinical Nutrition & Metabolic Care. 2000;3:267–73. doi: 10.1097/00075197-200007000-00005. [DOI] [PubMed] [Google Scholar]
- 44.Theil EC, Eisenstein RS. Combinatorial mRNA regulation: iron regulatory proteins and iso-iron-responsive elements (Iso-IREs) J Biol Chem. 2000;275:40659–62. doi: 10.1074/jbc.R000019200. [DOI] [PubMed] [Google Scholar]
- 45.Klausner RD, Rouault TA, Harford JB. Regulating the fate of mRNA: the control of cellular iron metabolism. Cell. 1993;72:19–28. doi: 10.1016/0092-8674(93)90046-s. [DOI] [PubMed] [Google Scholar]
- 46.Rouault TA. Post-transcriptional regulation of human iron metabolism by iron regulatory proteins. Blood Cells Mol Dis. 2002;29:309–14. doi: 10.1006/bcmd.2002.0571. [DOI] [PubMed] [Google Scholar]
- 47.Cairo G, Recalcati S, Pietrangelo A, Minotti G. The iron regulatory proteins: targets and modulators of free radical reactions and oxidative damage. Free Radic Biol Med. 2002;32:1237–43. doi: 10.1016/s0891-5849(02)00825-0. [DOI] [PubMed] [Google Scholar]
- 48.Klausner RD, Rouault TA. A double life: cytosolic aconitase as a regulatory RNA binding protein. Mol Biol Cell. 1993;4:1–5. doi: 10.1091/mbc.4.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rouault TA, Hentze MW, Haile DJ, Harford JB, Klausner RD. The Iron-Responsive Element Binding Protein: A Method for the Affinity Purification of a Regulatory RNA-Binding Protein. P Natl Acad Sci Usa National Academy of Sciences. 1989;86:5768–72. doi: 10.1073/pnas.86.15.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neupert B, Thompson NA, Meyer C, Kühn LC. A high yield affinity purification method for specific RNA-binding proteins: isolation of the iron regulatory factor from human placenta. Nucleic Acids Res. 1990;18:51–5. doi: 10.1093/nar/18.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rouault TA, Tang CK, Kaptain S, Burgess WH, Haile DJ, Samaniego F, McBride OW, Harford JB, Klausner RD. Cloning of the cDNA encoding an RNA regulatory protein--the human iron-responsive element-binding protein. P Natl Acad Sci Usa. 1990;87:7958–62. doi: 10.1073/pnas.87.20.7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hirling H, Emery-Goodman A, Thompson N, Neupert B, Seiser C, Kühn LC. Expression of active iron regulatory factor from a full-length human cDNA by in vitro transcription/translation. Nucleic Acids Res. 1992;20:33–9. doi: 10.1093/nar/20.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hentze MW, Argos P. Homology between IRE-BP, a regulatory RNA-binding protein, aconitase, and isopropylmalate isomerase. Nucleic Acids Res [Internet] 1991;19:1739–40. doi: 10.1093/nar/19.8.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rouault TA, Stout CD, Kaptain S, Harford JB, Klausner RD. Structural relationship between an iron-regulated RNA-binding protein (IRE-BP) and aconitase: functional implications. Cell. 1991;64:881–3. doi: 10.1016/0092-8674(91)90312-m. [DOI] [PubMed] [Google Scholar]
- 55.Kaptain S, Downey WE, Tang C, Philpott C, Haile D, Orloff DG, Harford JB, Rouault TA, Klausner RD. A regulated RNA binding protein also possesses aconitase activity. P Natl Acad Sci Usa. 1991;88:10109–13. doi: 10.1073/pnas.88.22.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haile DJ, Rouault TA, Tang CK, Chin J, Harford JB, Klausner RD. Reciprocal control of RNA-binding and aconitase activity in the regulation of the iron-responsive element binding protein: role of the iron-sulfur cluster. P Natl Acad Sci Usa. 1992;89:7536–40. doi: 10.1073/pnas.89.16.7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kennedy MC, Mende-Mueller L, Blondin GA, Beinert H. Purification and characterization of cytosolic aconitase from beef liver and its relationship to the iron-responsive element binding protein. P Natl Acad Sci Usa. 1992;89:11730–4. doi: 10.1073/pnas.89.24.11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dupuy J, Volbeda A, Carpentier P, Darnault C, Moulis JM, Fontecilla-Camps JC. Crystal structure of human iron regulatory protein 1 as cytosolic aconitase. Structure. 2006;14:129–39. doi: 10.1016/j.str.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 59.Walden WE, Selezneva AI, Dupuy J, Volbeda A, Fontecilla-Camps JC, Theil EC, Volz K. Structure of dual function iron regulatory protein 1 complexed with ferritin IRE-RNA. Science. 2006;314:1903–8. doi: 10.1126/science.1133116. [DOI] [PubMed] [Google Scholar]
- 60.Henderson BR, Menotti E, Bonnard C, Kühn LC. Optimal sequence and structure of iron-responsive elements. Selection of RNA stem-loops with high affinity for iron regulatory factor. J Biol Chem. 1994;269:17481–9. [PubMed] [Google Scholar]
- 61.Butt J, Kim HY, Basilion JP, Cohen S, Iwai K, Philpott CC, Altschul S, Klausner RD, Rouault TA. Differences in the RNA binding sites of iron regulatory proteins and potential target diversity. P Natl Acad Sci Usa National Academy of Sciences. 1996;93:4345–9. doi: 10.1073/pnas.93.9.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meyron-Holtz EG, Ghosh MC, Iwai K, LaVaute T, Brazzolotto X, Berger UV, Land W, Ollivierre-Wilson H, Grinberg A, Love P, Rouault TA. Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. EMBO J. 2004;23:386–95. doi: 10.1038/sj.emboj.7600041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.LaVaute T, Smith S, Cooperman S, Iwai K, Land W, Meyron-Holtz E, Drake SK, Miller G, Abu-Asab M, Tsokos M, Switzer R, 3, Grinberg A, Love P, Tresser N, Rouault TA. Targeted deletion of the gene encoding iron regulatory protein-2 causes misregulation of iron metabolism and neurodegenerative disease in mice. Nature genetics. 2001;27:209–14. doi: 10.1038/84859. [DOI] [PubMed] [Google Scholar]
- 64.Cooperman SS, Meyron-Holtz EG, Olivierre-Wilson H, Ghosh MC, McConnell JP, Rouault TA. Microcytic anemia, erythropoietic protoporphyria, and neurodegeneration in mice with targeted deletion of iron-regulatory protein 2. Blood. 2005;106:1084–91. doi: 10.1182/blood-2004-12-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith SR, Ghosh MC, Ollivierre-Wilson H, Hang Tong W, Rouault TA. Complete loss of iron regulatory proteins 1 and 2 prevents viability of murine zygotes beyond the blastocyst stage of embryonic development. Blood Cells Mol Dis. 2006;36:283–7. doi: 10.1016/j.bcmd.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 66.Sirover MA. New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim Biophys Acta. 1999;1432:159–84. doi: 10.1016/s0167-4838(99)00119-3. [DOI] [PubMed] [Google Scholar]
- 67.Nicholls C, Li H, Liu JP. GAPDH: a common enzyme with uncommon functions. Clin Exp Pharmacol Physiol. 2012;39:674–9. doi: 10.1111/j.1440-1681.2011.05599.x. [DOI] [PubMed] [Google Scholar]
- 68.Ryazanov AG. Glyceraldehyde-3-phosphate dehydrogenase is one of the three major RNA-binding proteins of rabbit reticulocytes. FEBS letters. 1985;192:131–4. doi: 10.1016/0014-5793(85)80058-2. [DOI] [PubMed] [Google Scholar]
- 69.Singh R, Green MR. Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science. 1993;259:365–8. doi: 10.1126/science.8420004. [DOI] [PubMed] [Google Scholar]
- 70.Dollenmaier G. Interaction of glyceraldehyde-3-phosphate dehydrogenase with secondary and tertiary RNA structural elements of the hepatitis A virus 3′ translated and non-translated regions. Journal of General Virology. 2003;84:403–14. doi: 10.1099/vir.0.18501-0. [DOI] [PubMed] [Google Scholar]
- 71.Nagy E, Rigby WF. Glyceraldehyde-3-phosphate dehydrogenase selectively binds AU-rich RNA in the NAD(+)-binding region (Rossmann fold) J Biol Chem. 1995;270:2755–63. doi: 10.1074/jbc.270.6.2755. [DOI] [PubMed] [Google Scholar]
- 72.Lin EY, Nguyen AV, Russell RG, Pollard JW. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J Exp Med. 2001;193:727–40. doi: 10.1084/jem.193.6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodríguez-Pascual F, Redondo-Horcajo M, Magán-Marchal N, Lagares D, Martínez-Ruiz A, Kleinert H, Lamas S. Glyceraldehyde-3-phosphate dehydrogenase regulates endothelin-1 expression by a novel, redox-sensitive mechanism involving mRNA stability. Molecular and Cellular Biology. 2008;28:7139–55. doi: 10.1128/MCB.01145-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nagy E, Henics T, Eckert M, Miseta A, Lightowlers RN, Kellermayer M. Identification of the NAD(+)-binding fold of glyceraldehyde-3-phosphate dehydrogenase as a novel RNA-binding domain. Biochem Biophys Res Commun. 2000;275:253–60. doi: 10.1006/bbrc.2000.3246. [DOI] [PubMed] [Google Scholar]
- 75.Arutyunova EI, Danshina PV, Domnina LV, Pleten AP, Muronetz VI. Oxidation of glyceraldehyde-3-phosphate dehydrogenase enhances its binding to nucleic acids. Biochem Biophys Res Commun. 2003;307:547–52. doi: 10.1016/s0006-291x(03)01222-1. [DOI] [PubMed] [Google Scholar]
- 76.Cieśla J. Metabolic enzymes that bind RNA: yet another level of cellular regulatory network? Acta Biochim Pol. 2006;53:11–32. [PubMed] [Google Scholar]
- 77.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–9. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. PAR-CliP - A Method to Identify Transcriptome-wide the Binding Sites of RNA Binding Proteins. JoVE MyJoVE Corporation. 2010 doi: 10.3791/2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP--transcriptome-wide mapping of protein-RNA interactions with individual nucleotide resolution. JoVE. 2011 doi: 10.3791/2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, Krijgsveld J, Hentze MW. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]