Abstract
Gene transfer agents (GTAs) are phage-like particles that can package and transfer a random piece of the producing cell’s genome, but are unable to transfer all the genes required for their own production. As such, GTAs represent an evolutionary conundrum: are they selfish genetic elements propagating through an unknown mechanism, defective viruses, or viral structures “repurposed” by cells for gene exchange, as their name implies? In Rhodobacter capsulatus, production of the R. capsulatus GTA (RcGTA) particles is associated with a cluster of genes resembling a small prophage. Utilizing transcriptomic, genetic and biochemical approaches, we report that the RcGTA “genome” consists of at least 24 genes distributed across five distinct loci. We demonstrate that, of these additional loci, two are involved in cell recognition and binding and one in the production and maturation of RcGTA particles. The five RcGTA “genome” loci are widespread within Rhodobacterales, but not all loci have the same evolutionary histories. Specifically, two of the loci have been subject to frequent, probably virus-mediated, gene transfer events. We argue that it is unlikely that RcGTA is a selfish genetic element. Instead, our findings are compatible with the scenario that RcGTA is a virus-derived element maintained by the producing organism due to a selective advantage of within-population gene exchange. The modularity of the RcGTA “genome” is presumably a result of selection on the host organism to retain GTA functionality.
Keywords: RcGTA, gene exchange, exaptation, prophage, virus, Rhodobacter.
Introduction
Viruses are traditionally viewed as the ultimate selfish genetic elements (Koonin and Dolja 2014). Their origin and antiquity is actively debated (e.g., Brüssow 2009; López-García 2012), but it is unquestionable that bacterial viruses (phages) and their hosts have long and intertwined evolutionary histories (Forterre and Prangishvili 2013; Koonin and Dolja 2013). Most sequenced bacterial genomes contain at least one integrated phage (prophage) (Casjens 2003; Canchaya et al. 2004). Some of these represent phages in their lysogenic cycle waiting for suitable environmental conditions to switch to a lytic cycle, whereas others are defective phages whose coding sequences are often decaying (Casjens 2003). A third category represents viral structures that were co-opted by bacteria to serve new functions, and these are now maintained in the bacterial genomes by natural selection (Bobay et al. 2014; Touchon et al. 2014). Examples of such fascinating exaptations include phage-derived structures for warfare (i.e., for killing prokaryotic competitors), horizontal gene transfer, and microbe–animal interactions (Okamoto et al. 1968; Nakayama et al. 2000; Sarris et al. 2014; Shikuma et al. 2014; Borgeaud et al. 2015), or instances where phage integrations into specific regions of genomes have come to serve as regulatory switches (Paul 2008; Feiner et al. 2015). Other known benefits of maintaining prophages include superinfection immunity against (or exclusion of) future infections by other phages (Wollman 1938; Rao 1968), acquisition of useful genes carried by phages (e.g., Davis et al. 2000; Brüssow et al. 2004), and faster growth under certain conditions (Edlin et al. 1975).
One class of phage-like elements, called gene transfer agents (GTAs) (Lang et al. 2012), promises to serve as another exemplar of a viral exaptation by bacterial and archaeal lineages. Phenotypically, GTAs are small phage particles that deliver their DNA content into a recipient bacterial or archaeal cell (Marrs 1974; Yen et al. 1979; Rapp and Wall 1987; Humphrey et al. 1997; Bertani 1999; Eiserling et al. 1999; Guy et al. 2013). However, there are several features that clearly distinguish GTAs from both typical and transducing phages (Lang and Beatty 2007; Stanton 2007). Here we focus on the GTA in Rhodobacter capsulatus, an alphaproteobacterium from the order Rhodobacterales where GTAs were first discovered (Marrs 1974). The R. capsulatus GTA (RcGTA) not only packages seemingly random pieces of host DNA (Solioz and Marrs 1977; Yen et al. 1979; Hynes et al. 2012), but is also biased against packaging genes responsible for its own production (Hynes et al. 2012). Production of RcGTA involves multiple host regulatory systems (Solioz et al. 1975; Lang and Beatty 2000; Schaefer et al. 2002; Leung et al. 2012; Mercer et al. 2012; Westbye et al. 2013; Mercer and Lang 2014; Kuchinski et al. 2016) and is intertwined with regulation of other cellular processes (Mercer et al. 2012), such as motility (Lang and Beatty 2002). As a result, RcGTA production is population-density dependent (Solioz 1975; Schaefer et al. 2002; Brimacombe et al. 2013) and is tightly controlled so that only approximately 1–3% of cells in a population end up producing and releasing RcGTA particles (Fogg et al. 2012; Hynes et al. 2012), whereas the remainder of the population becomes capable of receiving DNA from these particles (Brimacombe et al. 2013, 2014). This level of RcGTA production coordination by the host greatly exceeds the typical phage dependence upon host factors.
Genotypically, RcGTA production requires expression of an approximately 15-kb cluster of genes with indisputable sequence similarity to known phage structural genes and with the overall organization of a tailed-phage genome (Lang and Beatty 2000; Chen et al. 2008). Curiously, due to their small head size, RcGTA particles can only package approximately 4 kb of DNA (Solioz and Marrs 1977; Yen et al. 1979). Even more so than the aforementioned random DNA packaging, this restriction ensures that a typical RcGTA particle cannot transfer its genome to another cell, a defining characteristic that distinguishes GTAs from phages. RcGTA is unlikely to be simply a defective prophage. If this were the case, its genome should show signs of elevated rates of nucleotide substitutions and relatively limited taxonomic distribution. However, RcGTA-like gene clusters are widely distributed within the class Alphaproteobacteria (Lang and Beatty 2007), and are well conserved in the Rhodobacterales (Lang et al. 2002; Lang and Beatty 2007; Biers et al. 2008; Paul 2008; Lang et al. 2012). Furthermore, GTA genes in the Rhodobacterales are under purifying selection (Lang et al. 2012), suggesting the existence of selective pressure for maintenance of these genes.
Could RcGTA still be a selfish genetic element with a yet unknown mechanism for propagation of its 15-kb “genome”? Subsequent to the initial discovery of the abovementioned gene cluster (Lang and Beatty 2000), two genes needed for RcGTA release from cells were identified elsewhere in the R. capsulatus genome (Hynes et al. 2012; Westbye et al. 2013). Moreover, proteins encoded by additional genes from outside of the RcGTA cluster were detected in a proteomic study of RcGTA particles (Chen et al. 2008). Distribution of GTA genes across multiple loci is not unprecedented, as an unrelated GTA, VSH-1, produced by the spirochete Brachyspira hyodysenteriae (Humphrey et al. 1997), is encoded at two separate loci in the chromosome (Stanton et al. 2009). In this study, we present evidence that the RcGTA “genome” consists of at least five loci separated from each other by hundreds of kilobases on the R. capsulatus chromosome. Three of the five loci encode structural components of the particle. Moreover, we show that not all of these RcGTA genome loci share the same evolutionary history. We argue that our findings are not consistent with RcGTA being a selfish genetic element, and instead favor a scenario that RcGTA is a phage-derived “tool” widely maintained in the Rhodobacterales.
Results and Discussion
Identification of Candidate RcGTA Genes through Comparative Transcriptomics
Under laboratory conditions, RcGTA is produced primarily in the early stationary (ES) phase of growth (Solioz et al. 1975). By pairwise comparisons of the transcriptome of an R. capsulatus SB1003 culture in ES to the transcriptomes of mutant strains and growth phases either overproducing or with reduced production of RcGTA particles, we generated lists of open reading frames (ORFs) that are differentially expressed (fig. 1). Twenty-six ORFs from these lists were differentially expressed across all four pairwise transcriptome comparisons (fig. 1 and table 1; fold-change values in supplementary table S1, Supplementary Material online), suggesting a possible causal connection between the expression of these genes and RcGTA production. Two of the possible three-way comparisons share no ORFs in common, and the other two have only 6 and 13 ORFs each (fig. 1 and supplementary table S2, Supplementary Material online). The list of 26 ORFs includes the 17 ORFs that constitute the annotated RcGTA structural gene cluster (rcc01682–rcc01698) (Lang et al. 2012), establishing that our comparative transcriptomic approach successfully recovers genes associated with RcGTA production. The nine remaining ORFs are located in six separate loci outside the structural cluster (Table 1). Notably, two putatively RcGTA-associated ORFs are absent from the 26-gene list: A small ORF at the end of the RcGTA structural gene cluster (rcc01699) (Lang and Beatty 2000), and a plasmid ORF rcp00136 that encodes a protein found associated with RcGTA particles (Chen et al. 2008). This latter ORF was not identified in any of the pairwise transcriptome comparisons and is also absent from the RcGTA overproducer strain DE442, which lacks the plasmid carrying this gene (Hynes et al. 2012), and it was not investigated further.
Fig. 1.
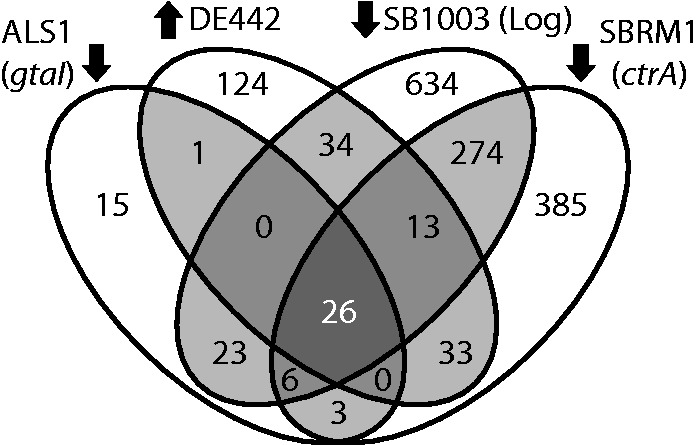
Identification of ORFs whose transcript levels vary with RcGTA production. The four ovals contain the number of ORFs with a >2-fold change in transcript level [relative to the SB1003 strain in the early stationary phase (ES) of growth] in the quorum-sensing mutant ALS1 (gtaI), the regulatory mutant SBRM1 (ctrA), SB1003 in logarithmic phase (Log), or the RcGTA-overproducer strain DE442 in ES. Arrows indicate the comparisons where RcGTA production is known to be higher (up) or lower (down) than in the reference. The central (darkest) region of overlap contains the ORFs whose transcript levels vary according to RcGTA production in all four comparisons.
Table 1.
Rhodobacter capsulatus ORFs Coregulated in Transcriptomic Comparisons.
| ORF | Genome Annotationb | Involvement in RcGTA Activity | Role |
|---|---|---|---|
| rcc00171a | CHP | Yes (this study) | Attachment |
| rcc00555 | HP | Yes (Hynes et al. 2012) | Release |
| rcc00556 | HP | Yes (Westbye et al. 2013) | Release |
| rcc01079a | CHP | Yes (this study; Westbye et al. 2016) | Adsorption |
| rcc01080a | CHP | Yes (this study; Westbye et al. 2016) | Adsorption |
| rcc01682 | CHP | ||
| rcc01683 | Terminase-like | Yes (Lang and Beatty 2000) | DNA packaging |
| rcc01684a | Portal, HK97 family | ||
| rcc01685 | CHP | No (Hynes et al. 2012) | Unknown |
| rcc01686 | Prohead protease, HK97 family | Yes (Lang and Beatty 2000) | Virion maturation |
| rcc01687a | Major capsid protein, HK97 family | Yes (Lang and Beatty 2000; Florizone 2006) | Structural protein |
| rcc01688a | Phage CHP | ||
| rcc01689a | Phage CHP | ||
| rcc01690a | CHP | ||
| rcc01691a | Major tail protein, TP901-1 family | ||
| rcc01692 | CHP | ||
| rcc01693 | Phage CHP | ||
| rcc01694a | Phage CHP | ||
| rcc01695 | Phage CHP | Yes (Lang and Beatty 2000) | Unknown |
| rcc01696a | Phage CHP | ||
| rcc01697 | Cell wall peptidase, NlpC/P60 family | Yes (Fogg et al. 2012) | DNA entry |
| rcc01698a | Phage CHP | Yes (Lang and Beatty 2000) | |
| rcc01865 | HP | Yes (this study) | Gene regulation |
| rcc01866 | HP | Yes (this study) | Virion maturation |
| rcc02623 | HP | No (this study) | |
| rcc02730 | HP | No (this study) |
aThe encoded protein was found associated with purified RcGTA particles for these ORFs (Chen et al. 2008).
bAs provided in the current GenBank records; CHP, conserved hypothetical protein; HP, hypothetical protein.
Experimental Evidence That the Coexpressed Genes Are Involved in RcGTA Function
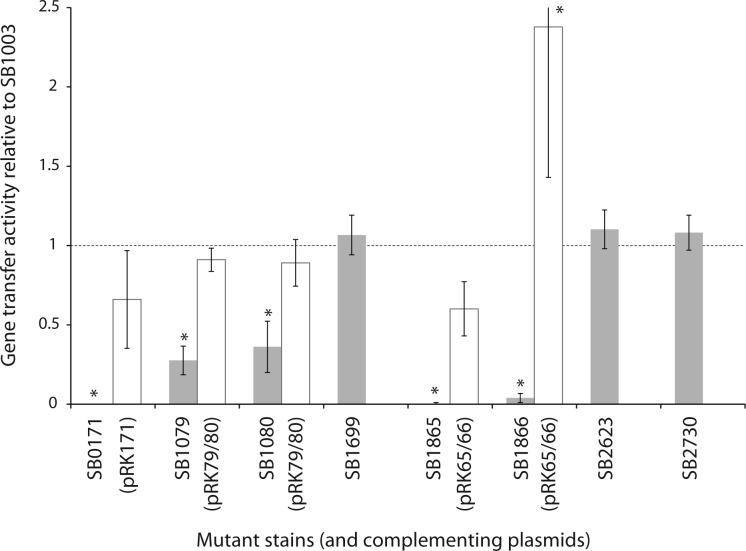
Among the nine identified ORFs that lie outside the RcGTA structural gene cluster, a locus of two ORFs (rcc00555 and rcc00556) was previously shown to be required for cell lysis and RcGTA release from cells (Fogg et al. 2012; Hynes et al. 2012; Westbye et al. 2013). We created mutant strains with disruptions of each of the seven remaining ORFs (Table 1), as well as of rcc01699 (orfg16). Of these, the strains with disruptions of rcc00171, rcc01079, rcc01080, rcc01865, and rcc01866 exhibited impaired RcGTA gene transfer activity (fig. 2). Specifically, disruption of rcc00171 or rcc01865 completely abolished gene transfer activity, whereas loss of rcc01866 reduced it >20-fold, and disruption of rcc01079 or rcc01080 reduced activity by approximately two-thirds. In trans complementation restored RcGTA gene transfer activity in the affected mutant strains (fig. 2). In complementation experiments with rcc01079 or rcc01080 mutant strains, it was necessary to introduce both genes to rescue gene transfer activity of either mutant. The rcc01079 nucleotide sequence is predicted to contain a -1 frameshift signal “GGGGAAAT” (identified by FSfinder; Moon et al. 2004) and we hypothesize that rcc01079 and rcc01080 produce a single product, which would explain the requirement for both genes to complement either mutation.
Fig. 2.
Effects of ORF disruptions and in trans complementation on RcGTA gene transfer activity. The activity is an average, across at least three replicate bioassays, of the amount of gene transfer activity for the mutant (gray) or its complement (white; where applicable) relative to that for the parental strain, SB1003. The activity of the parental strain is depicted by the dotted line, and the error bars represent the standard deviation. An asterisk (*) indicates that the strain’s gene transfer activity levels differed significantly from the parental strain (P < 0.05), as determined by ANOVA and Tukey HSD. Strains SB1699, SB2623, and SB2730 were not complemented as these mutant strains did not exhibit a significant difference in RcGTA gene transfer activity.
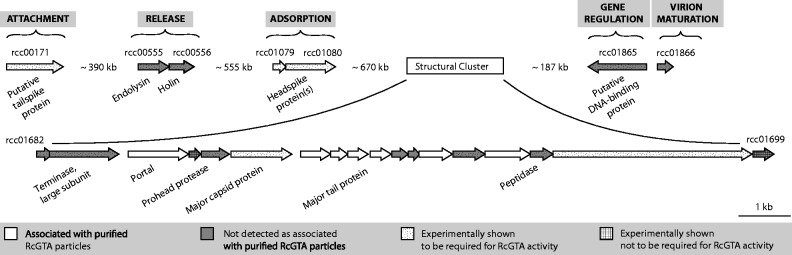
These findings indicate that RcGTA function depends on 24 genes distributed across five coregulated loci in the R. capsulatus genome, separated by hundreds of kilobases (fig. 3). Is this RcGTA “genome” a result of fragmentation of an ancestral single-locus RcGTA genome, co-option of independent phage genes, or recruitment of genes of cellular origin? We sought to address these questions by investigating the evolutionary history of each gene. Additionally, we determined the functions of the three previously uncharacterized loci.
Fig. 3.
The multilocus RcGTA “genome” and putative roles of its genes in particle production and gene transfer activity. Individual ORFs are drawn to scale, whereas distances between loci are shown in kilobases. The ORFs are shaded in gray if they were not found associated with RcGTA particles in a previous proteomic study (Chen et al. 2008). ORFs that were experimentally tested for their requirement in RcGTA activity are marked by one of the two hatch patterns (references for specific studies are in table 1). The indicated putative functions for genes outside of the structural cluster are discussed in detail in the text.
Three New RcGTA Genes Are Involved in Host Recognition
Based on previous proteomic identification (Chen et al. 2008) and our own bioinformatic analysis (fig. 3 and table 1, and detailed below), rcc00171, rcc01079, and rcc01080 were suspected to encode structural components of the RcGTA particle.
The rcc00171 gene is located directly downstream of three predicted extracellular polysaccharide (EPS) synthesis genes. Homologs of this gene are widely represented in Rhodobacterales genomes. The Rcc00171 amino acid sequence also has detectable similarity to several phage proteins, one of which is from RDJLφ1 (Huang et al. 2011) that infects the Rhodobacterales member Roseobacter denitrificans. In RDJLφ1, the gene is immediately adjacent to and downstream of homologs of the RcGTA genes orfg12-g15 (rcc01696–1698). The rcc00171 gene is annotated as “ribonuclease III,” as often are its alphaproteobacterial and phage homologs. However, we did not identify any significant similarity to bona fide ribonuclease proteins and cannot identify the basis of these annotations. Rcc00171 contains a conserved domain of unknown function, DUF2793 (Pfam PF10983), and DUF2793 domain-containing phage sequences are often annotated as either tail fiber or structural proteins. Additionally, homologs identified in Pseudomonas phages showed sequence similarity to the portion of the Rcc00171 sequence outside of the conserved DUF2793 domain, and these genes are also annotated as tail fiber or structural genes. The DUF2793 domain is occasionally found alongside a peptidase G2 domain, which is typically present in phage tailspike proteins (Schulz and Ficner 2011). Although no primary amino acid sequence similarity with known phage tailspike proteins is detectable, its predicted secondary structure is consistent with the experimentally determined structures of phage tailspike proteins (Fokine and Rossmann 2014), which contain the triple-stranded β-helix at a C-terminal region. Aside from an α-helical region at the N-terminus, Rcc00171 is predicted to consist exclusively of short β-strands, which is a hallmark of β-helices. Compositionally, Rcc00171 contains a large proportion of glycines, as well as of other small and aliphatic amino acids, which is also consistent with the composition of β-strand regions in known β-helical proteins (Iengar et al. 2006). Moreover, β-helices are common in lipopolysaccharide-binding proteins (Leiman et al. 2010) and one RcGTA receptor is known to be a capsular polysaccharide (Brimacombe et al. 2013). Taken together, our observations suggest that Rcc00171 is most likely a tailspike protein.
The rcc01079 and rcc01080 genes are located immediately upstream of genes required for production of the capsular polysaccharide RcGTA receptor (Brimacombe et al. 2013). Homologs of rcc01079 are patchily distributed within the Rhodobacterales, and one is present in the R. capsulatus phage RcapNL (accession number JQ066768), where it is annotated as “tail fiber protein” based on similarity to other phage sequences. Homologs of rcc01080 are patchily distributed in Proteobacteria and also found in some Cyanobacteria, Firmicutes, and Bacteroidetes. However, within the Rhodobacterales this gene was only identified in R. capsulatus. Notably, homologs of rcc01080 were also detected in RcapMu, which infects R. capsulatus (Fogg et al. 2011), and in phages infecting marine cyanobacterial and bacteroidetes hosts. The annotations of phage homologs vary and include “phage tail tip,” “minor phage tail,” and “putative carbohydrate-binding” proteins. The amino acid sequences of rcc01079 and rcc01080 lack any detectable conserved domains, but predicted secondary structures of both proteins are dominated by β strands, which is consistent with known structures of tail fiber proteins (Riede et al. 1987). The observed distribution of rcc01079 and rcc01080 homologs in bacteria and phages implies that these genes are of phage origin and have frequently been horizontally exchanged.
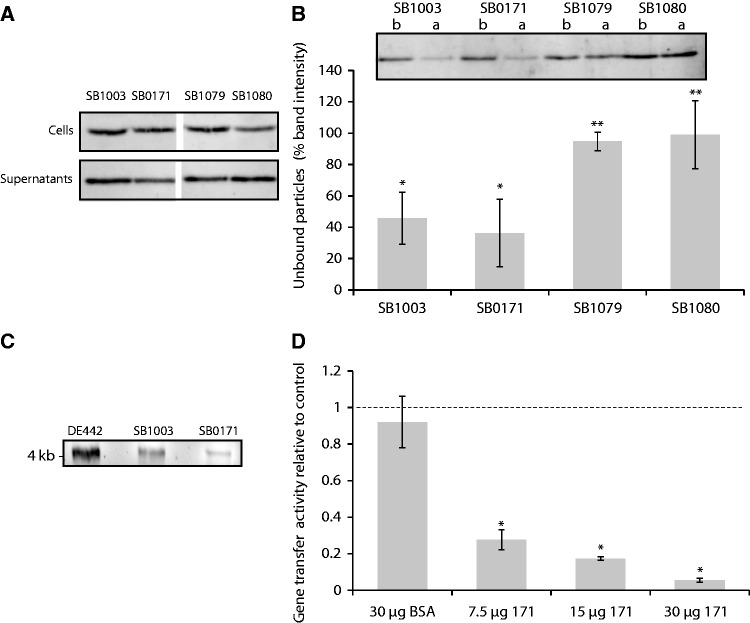
These in silico analyses suggest that all three genes might be involved in host recognition by GTA. Consistent with this hypothesis, disruption of each of the three genes did not affect the amount of intra- and extracellular RcGTA capsid protein compared with the parental strain (fig. 4A). As neither production nor release of the RcGTA particles was quantitatively affected, the particles were hypothesized to be defective in some other way, such as in their abilities to bind to recipient cells.
Fig. 4.
Roles of rcc0171, rcc01079 and rcc01080 in RcGTA gene transfer activity. (A) The relative amounts, as visualized by a representative western blot, of RcGTA capsid protein in the cells (top) and supernatants (bottom) of the indicated strains. This serves as a measure of RcGTA gene cluster expression and particle release from cells. All four strains show comparable RcGTA capsid protein levels inside and outside the cells. (B) Binding efficiency of RcGTA particles produced by SB1003, SB0171, SB1079, and SB1080. The binding activity is demonstrated by a representative western blot (top) comparing free RcGTA capsid protein before (b) and after (a) exposure to recipient cells followed by removal of the cells by centrifugation, and shown as quantified band intensity ratios of before/after over three replicate blots (bottom). RcGTA particles produced by the strains SB1079 and SB1080 bound poorly to recipient cells, and therefore were not removed when the cells were pelleted by centrifugation. Asterisks (* and **) represent statistically distinct groups in which every member is significantly different from those of the other group (ANOVA and Tukey HSD, P < 0.05), but not from those within its group. (C) DNA contents of RcGTA particles from DE442 (RcGTA overproducer), SB1003 and SB0171 strains, visualized by agarose gel electrophoresis. This qualitative assay confirmed the presence of the expected 4-kb DNA band within the particles. (D) Inhibition of gene transfer activity by purified Rcc00171. Gene transfer activity was determined as an average relative to a no-treatment control (dotted line) in three replicate bioassays, and bovine serum albumin (BSA) was used as a protein-addition control. Increased Rcc00171 concentration inhibited RcGTA activity. The error bars represent the standard deviation. An asterisk (*) denotes RcGTA gene transfer levels that differed significantly from the control (ANOVA and Tukey HSD, P < 0.05).
In binding assays, adsorption of particles from the rcc01079 and rcc01080 mutants to cells was significantly impaired (Tukey Honest Significant Difference [HSD] P < 0.05) relative to that of RcGTA from the parental strain SB1003 (fig. 4B). For these two mutants, the decreases in free capsid protein associated with binding were statistically insignificant (fig. 4B; Tukey HSD P > 0.1), consistent with impaired adsorption resulting from an absence of tail fibers. A concurrent study has determined that these fibers are, in fact, head-associated (Westbye et al. 2016).
The finding that neither production nor binding of the particles was affected in the rcc00171 mutant (fig. 4B) prompted us to investigate the possibility that the mutant particles lacked gene transfer activity because they do not contain DNA. This investigation was also partially motivated by the “ribonuclease III” annotation, which suggested a possible role for Rcc00171 in nucleic acid interactions. DNA extractions confirmed that RcGTA particles produced by the rcc00171 mutant contained DNA of the expected size (fig. 4C). The previous demonstration that capsule polysaccharide serves as a receptor for RcGTA attachment to cells (Brimacombe et al. 2013) and the location of the rcc00171 gene adjacent to genes predicted to be involved in polysaccharide export suggested the Rcc00171 protein might be involved in interaction with the R. capsulatus cell surface. We used the purified Rcc00171 protein to test its activity as a capsular or EPS lyase. Incubation of cells with the purified protein produced no detectable decrease in cell-associated sugars as measured by phenol–sulfuric acid carbohydrate quantification and no visible effects upon the capsular structure as observed by microscopy following capsule staining. Incubation of the purified protein with purified R. capsulatus EPS preparations also produced no evidence of breakdown of the sugars into smaller units when visualized on an acrylamide gel (data not shown).
Addition of the purified Rcc00171 protein to culture filtrates from the rcc00171 mutant did not rescue the gene transfer activity of these Rcc00171-deficient RcGTA particles. Surprisingly, however, addition of the protein inhibited the gene transfer activity of particles produced by the wild-type strain in a concentration-dependent manner, with addition of 7.5, 15 and 30 μg of protein resulting in approximately 27%, 17% and 5% activity relative to the untreated control, respectively (fig. 4D). The addition of extraneous Rcc00171 is presumably competing for and blocking attachment sites on the cell surface and therefore reducing the gene transfer rates in proportion to the concentration added. Therefore, we hypothesize that this protein is involved in attachment to a specific receptor following Rcc01079/Rcc01080-mediated adsorption. It is noteworthy that the rcc01079–rcc01080 genes are immediately upstream of, and coregulated with, genes responsible for producing the capsule structure to which RcGTA binds, whereas the rcc00171 gene is directly downstream of three genes predicted to encode proteins involved in EPS synthesis. A possible involvement of these proteins in RcGTA attachment remains to be investigated.
Two New RcGTA Genes Are Nonstructural
Neither Rcc01865 nor Rcc01866 had been identified as associated with mature RcGTA particles (Chen et al. 2008) and we therefore hypothesized that they are nonstructural contributors to RcGTA function.
Similar to rcc00171, a homolog of rcc01865 is widely represented in Rhodobacterales genomes, and is also present in the R. denitrificans phage RDJLφ1 (NC_015466). The latter homolog is annotated as encoding a DNA replication initiation ATPase protein, and the predicted protein sequence possesses a bacterial DnaA domain with a detectable helix–turn–helix (HTH) motif. The DnaA domain is not detectable within the Rcc01865 sequence, but an HTH motif is predicted in the corresponding region. Rcc01865 is highly charged (118 of 382 amino acids) and its predicted secondary structure is predominantly alpha-helical. These properties suggest the protein interacts with nucleic acids. Disruption of the rcc01865 gene resulted in loss of detectable gene transfer activity in the mutant strain (fig. 2), and RcGTA capsid protein was not detected inside or outside these cells (fig. 5A). Unsurprisingly, there was no residual intracellular gene transfer activity detected when the cells were manually lysed (fig. 5B). These data indicate that Rcc01865 is required for expression of the RcGTA structural gene cluster.
Fig. 5.
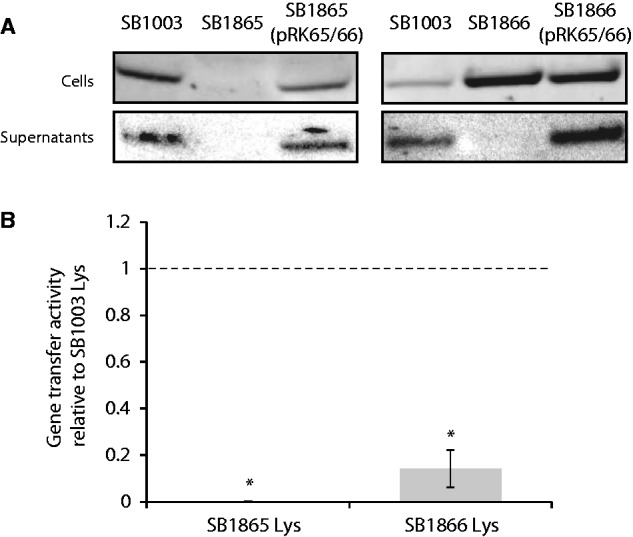
Roles of rcc01865 and rcc01866 in RcGTA production. (A) The relative amounts, as visualized by a representative western blot, of RcGTA capsid protein in cells and culture supernatants of the indicated strains. No capsid protein was detectable within SB1865 cells. In contrast, SB1866 cells contained a considerable amount of the capsid protein, but appeared incapable of releasing RcGTA particles from the cells. (B) RcGTA gene transfer activity released from cells by manual lysis. The gene transfer activity was determined as an average relative to the gene transfer activity from a lysed culture of SB1003 (the dotted line), in three replicate bioassays. No gene transfer was detected in the SB1865 cells. The RcGTA capsid protein accumulated within SB1866 cells does not correspond to functional RcGTA particles because very little gene transfer activity was detected when these cells were manually lysed. The error bars represent the standard deviation. An asterisk (*) indicates that the RcGTA gene transfer activities differed significantly from the control (ANOVA and Tukey HSD; P < 0.001).
Although it is conserved across the Rhodobacterales, the Rcc01866 sequence itself did not provide clues for us to speculate on its function. The genome of the aforementioned R. denitrificans phage RDJLφ1 also contains an rcc01866 homolog, but it is not immediately adjacent to the rcc01865 homolog. Intriguingly, the two homologs are separated by an ORF with amino acid sequence similarity to a portion of the CtrA response regulator, which is required for transcription of the RcGTA structural gene cluster (Lang and Beatty 2000; Mercer et al. 2010). How the RcGTA genes became part of the CtrA regulon remains a mystery, but the presence of this CtrA-related sequence in a phage that also has several RcGTA gene homologs (Huang et al. 2011) suggests the association might have roots in its ancestral phage state.
In the rcc01866 mutant, RcGTA capsid protein was detected within the cells but was undetectable outside (fig. 5A). Manual lysis of the SB1866 cells revealed the presence of some functional RcGTA particles within these cells (fig. 5B), but the amount was very low compared with what would be expected based on the amount of capsid protein observed. Therefore, Rcc01866 is likely important for either the maturation or assembly of functional RcGTA particles. Moreover, the release of the particles is dependent upon either Rcc01866 or the Rcc01866-mediated process. Disruption of the RcGTA-regulating sensor kinase CckA (Lang and Beatty 2000) resulted in an almost identical RcGTA phenotype (Westbye et al. 2013), which indicates CckA and Rcc01866 might act in a common regulatory pathway.
Distribution of the RcGTA “Genome” Loci in the Rhodobacterales
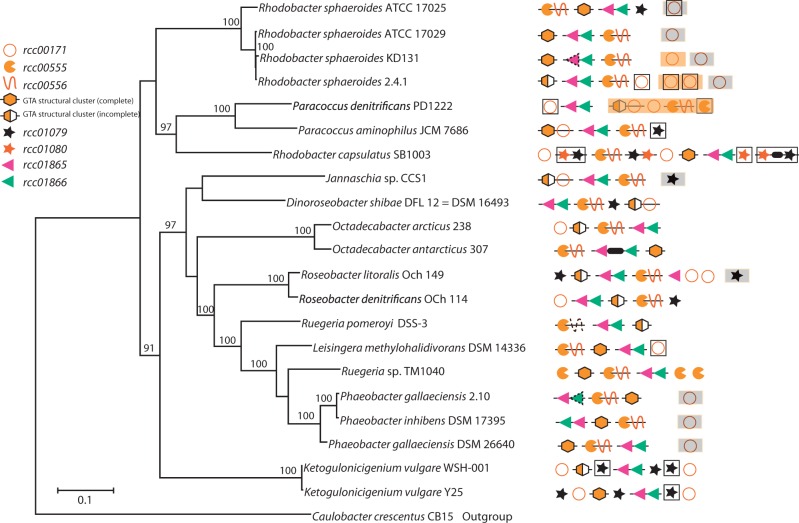
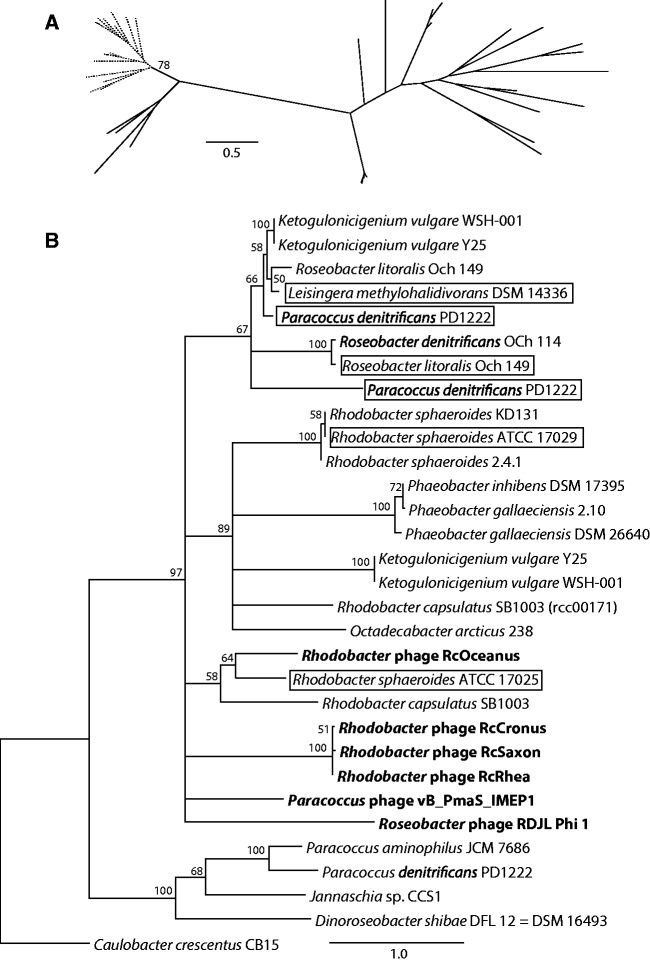
A search for homologs of the five RcGTA genome loci in 21 complete Rhodobacterales genomes (supplementary table S3, Supplementary Material online) revealed that these loci are widespread in these bacteria. The RcGTA-like structural cluster is present in all 21 examined genomes (fig. 6), and homologs of all 17 ORFs were detected in 12 of these. Three of the remaining four RcGTA loci are also well conserved: Homologs of rcc00555–rcc00556, rcc01865–rcc01866 and rcc00171 were detected in 19, 21 and 18 of the genomes, respectively. However, the rcc01079–rcc01080 locus is considerably less well conserved: Homologs of rcc01079 were detectable in only nine genomes, whereas rcc01080 had no identifiable homologs outside the R. capsulatus genome. As in the R. capsulatus SB1003 genome, the loci are scattered throughout each analyzed genome with no apparent conservation of relative order, even when comparing genomes within the same genus (fig. 6). Moreover, in several cases the loci are situated on different replicons. Specifically, the rcc00171 homologs reside on plasmids in seven genomes, the rcc01079 homologs are located on plasmids in both Roseobacter litoralis Och 149 and Jannaschia sp. CCS1, and in Paracoccus denitrificans PD1222 an rcc01865–rcc01866-like locus and the RcGTA-like structural gene cluster are found on separate chromosomes.
Fig. 6.
Conservation of the GTA loci across Rhodobacterales genomes. The presence and relative locations of RcGTA gene homologs are mapped onto the reference phylogenetic tree of the 21 analyzed Rhodobacterales genomes. The GTA “genome” loci are arranged as they appear in a genome relative to the annotated origin of replication. Adjacent genes are connected by lines. Genes that are found on a different chromosome or a plasmid are shown with orange and gray backgrounds, respectively. Regions outlined by black lines are located within or close to (<5 ORFs) predicted prophages. Symbols with a dotted contour indicate regions where the complete ORF is not present, but significant sequence similarity was nevertheless detected. A thick black line between genes represents the presence of an intervening ORF. An “incomplete” GTA structural cluster lacks at least one of the rcc01682–rcc01698 homologs. The reference phylogenetic tree was reconstructed by maximum likelihood from a concatenated alignment of 32 core genes. Caulobacter crescentus CB15 was used as an outgroup. Only bootstrap support values >70% are shown.
The R. capsulatus SB1003 genome contains multiple homologs of rcc01079 (rcc00962, rcc02905) and rcc01080 (rcc00961, rcc02899, rcc02903), and one homolog of rcc00171 (rcc00919). All of the rcc001079 and rcc01080 homologs are associated with prophage regions, one of which represents RcapMu (Fogg et al. 2011). Multiple homologs of the rcc00171 and rcc001079–rcc01080 loci were also found in several additional Rhodobacterales genomes, where they are also typically located within predicted prophage regions (fig. 6). The presence of multiple homologs for some RcGTA genes and their occasional occurrence on plasmids, sequence similarity to phage genes, close association with predicted and known prophage regions, as well as disjointed distribution across the examined genomes lead us to hypothesize that the five GTA loci may not share the same evolutionary history. To address this possibility we reconstructed phylogenetic histories of GTA genes, calculated their substitution rates relative to those of phage and cellular core genes, and examined the genomic contexts of the GTA loci.
Genomic Contexts of RcGTA Loci
Prokaryotic genomes experience frequent rearrangements, and therefore gene synteny is preserved only over short evolutionary distances (Rocha 2006; Novichkov et al. 2009). Therefore, it may not be surprising that the relative order of “GTA genome” loci is not conserved across Rhodobacterales genomes. However, local synteny (i.e., spanning only a small number of genes) is usually preserved for longer time scales (Tamames 2001; Rocha 2006). Hence, if the GTA loci have resided in the Rhodobacterales since their last common ancestor, rather than being horizontally transferred, we would expect flanking “cellular” genes to be conserved. We observe local synteny for three of the five GTA loci: The structural cluster, rcc00555–rcc00556, and rcc01865–rcc01866 (supplementary fig. S1, Supplementary Material online). Specifically, the structural gene cluster is universally flanked by the core Rhodobacterales genes, pabC and cysE, although in several cases additional ORFs are found between the structural cluster and these conserved flanking genes. The rcc00555–rcc00556 locus is flanked upstream by cycJ and downstream by a heme lyase-encoding gene in 19 and 12 genomes, respectively. Interestingly, the two Ketogulonicigenium vulgare genomes that lack the rcc00555–rcc00556 locus have these flanking genes adjacent to each other. Local synteny is also partially conserved near the rcc01865–rcc01866 locus. Fourteen of the 21 genomes have gua1 located downstream of this locus and lipA is found upstream in 16 genomes. In contrast, local synteny for the rcc00171 and rcc01079–rcc01080 loci is only conserved in closely related taxa, usually members of same genus.
Comparison of the Evolutionary Rates of GTA Genes and Their Phage Homologs
Nineteen of the 24 “GTA genome” genes have recognizable homologs in bona fide phages (supplementary fig. S2, Supplementary Material online). This raises the possibility that some of the RcGTA homologs across the Rhodobacterales are either prophage genes or genes of phage origin recently inserted into the genomes. If so, we would expect the RcGTA homologs to show substitution rates comparable to those of phage homologs and higher than those of “cellular” Rhodobacterales genes, because phage genes in general show much higher substitution rates, even when functions of the gene products are preserved (Sullivan et al. 2006). Furthermore, we would then also predict that the phage and RcGTA genes would not be clearly separated by a bipartition on a phylogenetic tree, although the accuracy of this analysis is limited by the availability of only a few genomes of phages infecting Rhodobacterales osts.
To test these hypotheses, we examined phylogenies of all RcGTA homologs in the analyzed Rhodobacterales genomes that had at least two phage homologs. The Rhodobacterales homologs for genes from the structural cluster, as well as for rcc01080 and rcc00555, form a separate group from their phage counterparts, even when homologs from phages known to infect members of the Rhodobacterales or other alphaproteobacterial hosts are present (fig. 7A and supplementary fig. S2, Supplementary Material online). In contrast, phage homologs of genes in two other loci, rcc00171 and rcc01079, are interspersed with the Rhodobacterales genomic sequences on the phylogenetic trees (fig. 7B and supplementary fig. S2, Supplementary Material online).
Fig. 7.
Examples of evolutionary relationships of phage and Rhodobacterales GTA gene homologs. (A) The phylogeny of a gene from within the structural cluster, rcc01696 (orfg13). All Rhodobacterales (branches in dotted lines) group separately from the bona fide phages (branches in solid lines). Bootstrap support is only shown for the bipartition separating the Rhodobacterales and phage homologs. (B) The phylogeny of a gene outside of the structural cluster, rcc00171. Phage homologs (taxa names in bold font) group within the Rhodobacterales, and the relationships among the Rhodobacterales homologs are incongruent with the reference tree (fig. 6). Outlined taxa likely represent homologs of phage origin, as they were found within or close to (<5 ORFs) a predicted prophage region. The tree was rooted using the Caulobacter crescentus CB15 sequence as an outgroup. Only bootstrap support values ≥50% are shown, and branches with <50% support were collapsed. Scale bars indicate the number of substitutions per site.
Phylogenetic trees of the structural cluster genes clearly indicate that the Rhodobacterales sequences have shorter branches than their phage homologs (supplementary fig. S2, Supplementary Material online), suggesting a slower evolutionary rate for the RcGTA genes. Using pairwise phylogenetic distances (PPD) as a proxy for substitution rates within a group of taxa, we compared the evolutionary rates of RcGTA genes, their phage homologs, and 839 conserved Rhodobacterales gene families (supplementary fig. S3, Supplementary Material online). Although the majority of the RcGTA genes evolve faster than the majority of the core genes, the median substitution rates of all RcGTA genes with detectable phage homologs are lower than those of the phage genes (supplementary fig. S3, Supplementary Material online; the differences are significant in all pairwise comparisons but one, supplementary table S4, Supplementary Material online). No phage-RcGTA comparisons could be made for rcc01080 (due to absence of homologs outside R. capsulatus), or for rcc00556, rcc01865, rcc01866 and five genes within the structural cluster (due to paucity of phage homologs). Based on the observed PPD values, the lack of detectable phage homologs for the latter eight genes is likely due to their rapid evolution. Notably, among the six fastest evolving RcGTA genes (rcc01682, rcc00171, rcc01866, rcc01079, rcc01865, rcc00556; listed in the decreasing order of PPD values), five lie outside the structural cluster.
Phylogenetic History of the rcc00171 Gene Reveals Phage-Mediated Horizontal Gene Transfer
If GTA genes were exchanged among the Rhodobacterales, their phylogenetic histories would exhibit topological incongruences with the phylogeny based on core genes (fig. 6). Unfortunately, phylogenetic trees reconstructed from four of the five faster-evolving GTA genes (rcc01079, rcc01080, rcc01865 and rcc01866) did not have sufficient bootstrap support values to allow a strong inference of either horizontal gene transfer or vertical inheritance (data not shown). The phylogenetic history of rcc00171, on the other hand, exhibits nine topological conflicts with the Rhodobacterales reference phylogeny (with ≥50% bootstrap support; fig. 7B), supporting the hypothesis that this gene has undergone horizontal transfer since the Rhodobacterales diverged from a common ancestor. Moreover, the phage homologs group within the Rhodobacterales sequences, indicating that phages are the likely mediators of the gene transfer.
Different Evolutionary Histories of RcGTA Loci Suggest an Ongoing Evolution of RcGTA
Taken together, the gene neighborhood and phylogenetic analyses indicate that the GTA structural cluster and endolysin–holin locus (rcc00555–rcc00556) have resided in the Rhodobacterales since their last common ancestor. The remaining three GTA loci (rcc01079–rcc01080, rcc01865–rcc01866, and rcc00171) are evolving much faster, and some rcc00171 homologs have likely been introduced by phages. Although rapid evolution of the rcc01079–rcc01080 and rcc01865–rcc01866 loci hints at a similar phage origin, this remains a conjecture until more genome sequences of phages infecting Rhodobacterales are available. It is noteworthy that, whereas this GTA has generally been conserved over a long time, two of these faster evolving loci are involved in cell recognition. One explanation could be the evolutionary pressures exerted on R. capsulatus populations by “true” phage predation. As in any evolutionary arms race between viruses and their hosts, R. capsulatus cells are under selection to modify structures used in viral entry in order to escape phage infection. Cell surface modification is a common means of spontaneous phage resistance (Labrie et al. 2010). Rhodobacter capsulatus cells are surrounded by a capsule and RcGTA likely shares its capsular polysaccharide receptor (Brimacombe et al. 2013) with contemporary phages. Modification of this structure might render cells incapable of receiving DNA from GTAs. If GTAs are indeed maintained by R. capsulatus due to a fitness advantage, GTA functionality will only be retained if its receptor interaction genes evolve equally quickly. Acquisition of an already functional host recognition module from a phage could be evolutionarily favored relative to de novo mutation of existing genes.
Concluding Remarks
The original discovery of the RcGTA structural gene cluster provided an undisputable evolutionary link between RcGTA and a phage ancestor. However, it is now clear that production and release of functional RcGTA particles requires loci scattered throughout the R. capsulatus genome, in addition to the multitude of cellular regulatory systems also encoded across the genome. Combined with the packaging constraints of the small particles, RcGTA appears completely incapable of self-propagation. Its continued maintenance is therefore presumably reliant on provision of benefit(s) to the host, the nature of which remains to be determined. These might include facilitation of homologous recombination, which is thought to benefit prokaryotic populations (Vos 2009; Takeuchi et al. 2014). Given that GTA particles package only a small DNA fragment, GTA-mediated DNA exchange might serve as a mechanism for “chromosomal curing” of genomes from parasitic genetic elements, as has been recently proposed to explain the costly maintenance of a natural competence system (Croucher et al. 2016). Regardless of its utility, different evolutionary histories of the distinct RcGTA genome loci suggest that while RcGTA has likely been associated with R. capsulatus since at least the time of divergence of the Rhodobacterales, RcGTA-encoding components are under different selective pressures, presumably in response to evolutionary forces experienced by R. capsulatus. The structural cluster and lysis cassette have been transmitted vertically within the Rhodobacterales, whereas genes involved in recipient cell recognition have been horizontally exchanged and were probably recruited more recently from phages. Therefore, RcGTA is a product of long-term vertical evolution from a (pro)phage ancestor combined with shorter-term evolutionary refinements resulting from recent horizontal gene transfer events.
Materials and Methods
Strains, Plasmids, and Growth Conditions
The strains and plasmids used in this study are listed in supplementary table S5, Supplementary Material online. Insertional disruptions of R. capsulatus genes were constructed by amplification of the gene of interest by polymerase chain reaction (PCR) using the primers indicated in supplementary table S6, Supplementary Material online, ligation of the resulting product into the pGEM-T Easy vector system (Promega, Madison WI), and subsequent insertion of the 1368-bp SmaI fragment of the KIXX cartridge (Barany 1985) encoding kanamycin resistance at the restriction site listed in supplementary table S6, Supplementary Material online. The pGEM construct with the disrupted gene was then transferred by conjugation from Escherichia coli C600 (pDT51) (Taylor et al. 1983) into the GTA-overproducing strain R. capsulatus DE442 and the marked gene disruption transferred into R. capsulatus SB1003 by RcGTA transfer (Scolnik and Haselkorn 1984). The disrupted gene replacements were confirmed by PCR with the original amplification primers and visualization of the size differences resulting from KIXX insertion.
Constructs to complement mutants in trans were created by amplification of the wild-type genes along with sufficient upstream regions to include the predicted promoter regions. The primers included 5′ KpnI sites to facilitate cloning into the plasmid pRK767, with resulting plasmids transferred to R. capsulatus by conjugation from E. coli S17-1 (Simon et al. 1983).
The pET28-171 expression plasmid was constructed by amplification of rcc00171 using the primers indicated in supplementary table S6, Supplementary Material online, which included 5′ restriction enzyme sites to directionally clone the insert into pET28a(+). The resulting construct was sequenced at The Centre for Applied Genomics (TCAG) (Toronto, Canada) using their T7 and T7 Term primers to confirm the in-frame fusion to the C-terminal His-tag and conservation of the original rcc00171 sequence.
Rhodobacter capsulatus cells were grown under aerobic conditions at 30 °C in RCV medium (Beatty and Gest 1981) for general culturing or at 35 °C under anaerobic photoheterotrophic conditions in YPS medium (Weaver et al. 1975) for higher RcGTA production in transductions and bioassays.
Identification of Candidate RcGTA Genes
Expression data from four microarrays were compared with those from the model strain, SB1003 (Strnad et al. 2010) in the ES growth phase, where RcGTA production is typically highest in laboratory conditions (Solioz et al. 1975; Hynes and Lang 2013). The strains and conditions for these array datasets were: SB1003 in logarithmic phase of growth, the quorum-sensing (gtaI) mutant ALS1 in ES, the RcGTA-overproducer DE442 in ES, and the ctrA mutant SBRM1 in ES. These were chosen for comparison because these are strains or growth conditions known to show differences in RcGTA production with respect to strain SB1003 in ES (Solioz et al. 1975; Yen et al. 1979; Schaefer et al. 2002; Mercer et al. 2010), and would therefore identify ORFs that have changes in transcript levels matching changes in RcGTA production. The microarray data were retrieved from the NCBI Gene Expression Omnibus database (accession numbers GSE18149, GSE33176, and GSE41014). Raw microarray data were first robust multiarray normalized (Irizarry et al. 2003), then normalized to the 50th percentile using GeneSpring 7.2 (Agilent Technologies, Santa Clara, CA). The signal intensities of the SB1003 ES sample were used as a reference to which data from other samples were compared pairwise. For each comparison, the genes with >2-fold change in signal intensity were selected. If replicate arrays were available for the strains or growth conditions, we chose the array whose comparison to the reference resulted in the inclusion of the RcGTA structural cluster genes and the greatest number of additional genes. This was performed in order to generate the largest possible gene list. Overlap of the resulting gene sets across pairwise comparisons was assessed, and genes present in all four comparisons were selected for further analyses.
RcGTA Gene Transfer Activity Bioassays
RcGTA gene transfer activity bioassays involve collecting a cell-free culture filtrate from an R. capsulatus strain, and quantifying the number of gene transfer events when the filtrate is incubated with a recipient strain that lacks a genetic marker present in the donor strain. Normalized inoculants were used to start anaerobic photoheterotrophic cultures of the strains to be analyzed for RcGTA gene transfer activity. These cultures were then grown for 48 h and the RcGTA gene transfer activity assayed by puhA transfer to DW5 cells as previously described (Hynes and Lang 2013). For lysis assays, cultures were centrifuged, resuspended in a lysis buffer, subjected to freeze-thaw cycles and then treated with DNase I prior to filtration, as described previously (Hynes et al. 2012). For protein complementation/inhibition assays, 3–30 µg of protein were added to recipient cells prior to the addition of RcGTA-containing filtrate. Controls received an equal volume of dialysate with 10% v/v glycerol, or a bovine serum albumin (BSA) solution in dialysate. All data were analyzed as the gene transfer activity relative to an untreated SB1003 control. The transfer rates were compared by one-way analysis of variance (ANOVA) and Tukey HSD tests.
Detection of RcGTA Major Capsid Protein by Western Blotting
Cells and culture filtrates (0.45 µm) from the cultures used in RcGTA gene transfer activity bioassays were assayed for the presence of RcGTA capsid protein by western blotting. Cell samples were prepared by centrifugation of a portion of the culture at 17,000 × g, removal of the supernatant and resuspension of the cells in an equal volume of TE buffer, pH 8.0. Samples of cell resuspensions (5 µl) or culture filtrates (10 µl) were subjected to SDS-PAGE, with blotting and detection of the capsid protein performed as previously described (Mercer et al. 2012). Images were captured on an ImageQuant LAS 4000 (GE Life Sciences, Baie D’Urfe, Canada) and band intensities quantified, where relevant, with ImageQuantTL v8.1. The acquired blot images were each uniformly adjusted for brightness and contrast, using Adobe Photoshop CS 6.0 (Adobe Systems Inc., San Jose, CA).
Assays of RcGTA Binding to Cells
To monitor the binding efficiency of RcGTA particles to R. capsulatus cells, RcGTA particles were added either to G buffer (Solioz and Marrs 1977), or to G buffer containing recipient cells as done for RcGTA activity bioassays. After 1 h of incubation the cells were pelleted by centrifugation at 17,000 × g for 1 min. The supernatants were collected and analyzed for the remaining amount of capsid protein by western blotting as described above.
DNA Extraction from RcGTA Particles
Cultures were grown for 48 h at 35 °C under anaerobic photoheterotrophic conditions in YPS medium. The DNA extractions were performed as previously described (Hynes et al. 2012). Briefly, centrifuged culture supernatants were filtered, the filtrates ultracentrifuged and the resulting resuspended in G buffer (Solioz and Marrs 1977), treated with nucleases and proteinase K, and the DNA purified by phenol:chloroform:isoamyl alcohol (25:24:1) extraction and ethanol precipitation. The DNA was then visualized by agarose gel electrophoresis.
Protein Purification
Overnight cultures of E. coli BL1(DE3) containing pET28-171 were used to inoculate 200 ml of LB broth containing 25 µg ml−1 kanamycin. The cultures were grown at 37 °C for 1 h, induced by addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 1 mM and allowed to grow for another 4 h. Cell pellets of induced cultures were resuspended in 4 ml lysis buffer [50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 0.1% (v/v) Benzonase nuclease (Qiagen), 1 mg ml−1 lysozyme; pH 8] and incubated on ice for 30 min. The lysates were centrifuged at 14,000 × g for 30 min at 4 °C and supernatants were mixed 4:1 (v/v) with Ni-NTA agarose (Qiagen) and incubated at 4 °C with slow shaking for 1 h. The samples were loaded into polypropylene columns, washed twice with wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole; pH 8), and the fusion protein eluted in elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole; pH 8). The purified protein was dialyzed into a coupling buffer (20 mM sodium phosphate, 500 mM NaCl; pH 7.5) and quantified relative to known BSA standards using an ND-1000 Nanodrop spectrophotometer. The success of the purification was evaluated by SDS-PAGE and Coomassie Blue staining with samples from the preinduced culture, induced culture, lysate, wash, and eluate. The dialyzed purified protein was split into aliquots and stored at 4 °C or, with the addition of glycerol to 10% (v/v), at −80 °C.
Polysaccharide Lyase Activity Assays
To test for potential polysaccharide lyase activity of the purified Rcc00171 protein, we exposed R. capsulatus SB1003 cells resuspended in G buffer, as normally used in gene transfer bioassays, to either 25 or 55 µg of BSA or purified Rcc00171. After 1 h, the samples were treated for phenol–sulfuric acid carbohydrate quantification as well as for a variation of Anthony’s capsule stain as described previously (Brimacombe et al. 2013). We also purified R. capsulatus SB1003 polysaccharide using cetyltrimethylammonium bromide as described previously (Clarke et al. 2000). Capsular polysaccharides (12.5 µg) were then treated with 0.018–0.9 µg of Rcc00171 protein for 30 min in G buffer, and the resulting polysaccharides were run on a 15% polyacrylamide gel and visualized by a combined Alcian blue-silver staining method (Fau and Cowman 1986).
Detection of Homologs of the RcGTA Genes in Rhodobacterales and Phages, and Reconstruction of Their Evolutionary Histories
The 26 candidate RcGTA genes were used as queries in BLASTP searches (v2.2.30+; E-value < 0.001; query and subject overlap by at least 40% of their length) (Altschul et al. 1997) against the database of 21 complete Rhodobacterales genomes (supplementary table S3, Supplementary Material online; downloaded from the NCBI FTP site at ftp://ftp.ncbi.nlm.nih.gov/genomes/Bacteria/). Additionally, for the 17 structural cluster ORFs, only homologs that had other structural cluster homologs within approximately 10 kb were retained, in order to minimize the inclusion of prophage homologs. To find more distant homologs, for each of the 26 genes, the retrieved homologs were used to build a Position Specific Scoring Matrix that was subsequently used as a query in PSI-BLAST (Altschul et al. 1997) searches (v2.2.30+; E-value < 0.001; ran until convergence, which varied from two to six iterations depending on the gene) against the same database.
To identify viral homologs, the detected Rhodobacterales homologs of the 26 candidate genes were used as queries in a BLASTP search (E-value < 0.001; query and subject overlap by at least 60% of their length) of a database consisting of the Viral RefSeq database (release 71) and the genomes of six additional Rhodobacter-infecting phages: Rhodobacter phage RcTitan (KR935213), Rhodobacter phage RcCronus (KR935217), Rhodobacter phage RcSpartan (KR935215), Rhodobacter phage RcRhea (KR935216), Rhodobacter phage RcSaxon (KT253150), and Rhodobacter phage RcOceanus (kindly provided by Dr David W. Bollivar). To identify more divergent viral homologs, these detected viral homologs were used as queries in the subsequent BLASTP search (E-value < 0.001; query and subject overlap by at least 40% of their length) of the same viral database.
Detected viral and Rhodobacterales homologs were combined into 26 gene sets and were aligned using MUSCLE v3.8.31 (Edgar 2004). Maximum-likelihood phylogenetic trees were reconstructed under the JTT + Γ substitution model and with 100 bootstrap replicates in RAxML v8.1.3 (Stamatakis 2014). PPD were also calculated under the JTT + Γ substitution model in RAxML v8.1.3 (Stamatakis 2014).
Rhodobacterales Gene Families and Reconstruction of a Reference Phylogeny
Gene families in the 21 Rhodobacterales genomes were detected using OrthoMCL v2.0.9 (Li et al. 2003) as follows: All coding sequences were subjected to all-against-all BLASTP searches (v2.2.30+; E-value < 0.0001), with reciprocal top-scoring BLASTP matches retained and their bitscores converted to a pairwise distance matrix, which was used to generate clusters through Markov clustering with the inflation parameter set to 1.2 (Enright et al. 2002). This procedure resulted in 8,297 gene families, 839 of which are single copy core gene families. A reference Rhodobacterales phylogeny was reconstructed using a subset of 32 single copy core gene families that were present 1) in the Caulobacter crescentus CB15 genome (outgroup) and 2) among 70 single copy genes previously used in phylogenetic analyses of Roseobacter lineages (Newton et al. 2010). Sequences in individual gene families were first aligned separately using MUSCLE v3.8.31 (Edgar 2004), and then concatenated into one alignment. A maximum-likelihood tree was reconstructed under the JTT + Γ substitution model with 100 bootstrap replicates using RAxML v8.1.3 (Stamatakis 2014).
Genomic Context Analysis
The genomic contexts of each locus were analyzed by comparing the sequence similarities of their surrounding regions (10–20 kb at both 5′- and 3′-ends) in all Rhodobacterales genomes (supplementary table S3, Supplementary Material online). The similarities were detected by TBLASTX searches (E-value < 0.001) (Altschul et al. 1997) of the nucleotide sequences of the regions against each other. Furthermore, five ORFs upstream and downstream of each locus were compared across genomes. The ORFs were designated as homologous if they belonged to same gene family (see previous section). The location of each locus in reference to prophage regions detected using PHAge Search Tool (Zhou et al. 2011) was recorded.
Refinement of Functional Annotations for Genes Involved in RcGTA Production by In Silico Analyses
The 26 candidate genes were used as queries to retrieve homologs from GenBank using BLINK, BLASTP, and/or PSI-BLAST, as implemented in the Entrez portal (accessed September 2015) (NCBI Coordinators 2016). The presence of conserved domains was assessed by searching profiles in the Conserved Domains Database (v3.14) using CD-Search (Marchler-Bauer and Bryant 2004). Domain architectures in other proteins with homologous domains were checked using CDART (Geer et al. 2002). Protein secondary structures were predicted using JPRED 4 server (Drozdetskiy et al. 2015). Isoelectric point, HTH domains, and transmembrane helices were predicted using the EMBOSS suite (Rice et al. 2000). Signal peptides were predicted using SignalP 4.1 server (Petersen et al. 2011) using a model trained on Gram-negative bacteria.
Supplementary Material
Supplementary figs. S1–S3 and tables S1–S5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the Natural Sciences and Engineering Research Council (NSERC) of Canada (341561 to A.S.L.); the Simons Foundation (Simons Investigator award to O.Z.); the National Science Foundation (NSF-DEB 1551674 to O.Z.); NSERC and Memorial University of Newfoundland School of Graduate Studies (fellowships to A.P.H. and R.G.M.); the Memorial University Biology Department Honours program (L.B., H.M., and M.E.P.); and the Memorial University of Newfoundland Faculty of Science (Science Undergraduate Research Award to F.D.). The authors thank Dr Michael Ragusa (Chemistry Department, Dartmouth College) for discussions of the Rcc00171 structural predictions and two anonymous reviewers for their helpful comments.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barany F. 1985. Single-stranded hexameric linkers: a system for in-phase insertion mutagenesis and protein engineering. Gene 37:111–123. [DOI] [PubMed] [Google Scholar]
- Beatty JT, Gest H. 1981. Generation of succinyl-coenzyme A in photosynthetic bacteria. Arch Microbiol. 129:335–340. [Google Scholar]
- Bertani G. 1999. Transduction-like gene transfer in the methanogen Methanococcus voltae. J Bacteriol. 181:2992–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biers EJ, Wang K, Pennington C, Belas R, Chen F, Moran MA. 2008. Occurrence and expression of gene transfer agent genes in marine bacterioplankton. Appl Environ Microbiol. 74:2933–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobay L-M, Touchon M, Rocha EPC. 2014. Pervasive domestication of defective prophages by bacteria. Proc Natl Acad Sci U S A. 111:12127–12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgeaud S, Metzger LC, Scrignari T, Blokesch M. 2015. The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science 347:63–67. [DOI] [PubMed] [Google Scholar]
- Brimacombe CA, Ding H, Beatty JT. 2014. Rhodobacter capsulatus DprA is essential for RecA-mediated gene transfer agent (RcGTA) recipient capability regulated by quorum-sensing and the CtrA response regulator. Mol Microbiol. 92:1260–1278. [DOI] [PubMed] [Google Scholar]
- Brimacombe CA, Stevens A, Jun D, Mercer R, Lang AS, Beatty JT. 2013. Quorum-sensing regulation of a capsular polysaccharide receptor for the Rhodobacter capsulatus gene transfer agent (RcGTA). Mol Microbiol. 87:802–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüssow H. 2009. The not so universal tree of life or the place of viruses in the living world. Philos T Roy Soc B. 364:2263–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüssow H, Canchaya C, Hardt W-D. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol R. 68:560–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canchaya C, Fournous G, Brüssow H. 2004. The impact of prophages on bacterial chromosomes. Mol Microbiol. 53:9–18. [DOI] [PubMed] [Google Scholar]
- Casjens S. 2003. Prophages and bacterial genomics: what have we learned so far? Mol Microbiol. 49:277–300. [DOI] [PubMed] [Google Scholar]
- Chen F, Spano A, Goodman BE, Blasier KR, Sabat A, Jeffery E, Norris A, Shabanowitz J, Hunt DF, Lebedev N. 2008. Proteomic analysis and identification of the structural and regulatory proteins of the Rhodobacter capsulatus gene transfer agent. J Proteome Res. 8:967–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke BR, Esumeh F, Roberts IS. 2000. Cloning, expression, and purification of the K5 capsular polysaccharide lyase (KflA) from coliphage K5A: evidence for two distinct K5 lyase enzymes. J Bacteriol. 182:3761–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher NJ, Mostowy R, Wymant C, Turner P, Bentley SD, Fraser C. 2016. Horizontal DNA transfer mechanisms of bacteria as weapons of intragenomic conflict. PLoS Biol. 14:e1002394.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BM, Moyer KE, Boyd EF, Waldor MK. 2000. CTX prophages in classical biotype Vibrio cholerae: functional phage genes but dysfunctional phage genomes. J Bacteriol. 182:6992–6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdetskiy A, Cole C, Procter J, Barton GJ. 2015. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 43:W389–W394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlin G, Lin LEO, Kudrna R. 1975. λ lysogens of E. coli reproduce more rapidly than non-lysogens. Nature 255:735–737. [DOI] [PubMed] [Google Scholar]
- Eiserling F, Pushkin A, Gingery M, Bertani G. 1999. Bacteriophage-like particles associated with the gene transfer agent of Methanococcus voltae PS. J Gen Virol. 80:3305–3308. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30:1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fau MH, Cowman MK. 1986. Combined alcian blue and silver staining of glycosaminoglycans in polyacrylamide gels: application to electrophoretic analysis of molecular weight distribution. Anal Biochem. 155:275–285. [DOI] [PubMed] [Google Scholar]
- Feiner R, Argov T, Rabinovich L, Sigal N, Borovok I, Herskovits AA. 2015. A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat Rev Microbiol. 13:641–650. [DOI] [PubMed] [Google Scholar]
- Florizone SM. 2006. Studies in the regulation of the gene transfer agent (GTA) of Rhodobacter capsulatus. MSc. Thesis. University of British Columbia. Vancouver, Canada.
- Fogg PCM, Hynes AP, Digby E, Lang AS, Beatty JT. 2011. Characterization of a newly discovered Mu-like bacteriophage, RcapMu, in Rhodobacter capsulatus strain SB1003. Virology 421:211–221. [DOI] [PubMed] [Google Scholar]
- Fogg PCM, Westbye AB, Beatty JT. 2012. One for all or all for one: heterogeneous expression and host cell lysis are key to gene transfer agent activity in Rhodobacter capsulatus. PLoS One 7:e43772.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokine A, Rossmann MG. 2014. Molecular architecture of tailed double-stranded DNA phages. Bacteriophage 4:e28281.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P, Prangishvili D. 2013. The major role of viruses in cellular evolution: facts and hypotheses. Curr Opin Virol. 3:558–565. [DOI] [PubMed] [Google Scholar]
- Geer LY, Domrachev M, Lipman DJ, Bryant SH. 2002. CDART: protein homology by domain architecture. Genome Res. 12:1619–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy L, Nystedt B, Toft C, Zaremba-Niedzwiedzka K, Berglund EC, Granberg F, Näslund K, Eriksson A-S, Andersson SGE. 2013. A gene transfer agent and a dynamic repertoire of secretion systems hold the keys to the explosive radiation of the emerging pathogen Bartonella. PLoS Genet. 9:e1003393.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Zhang Y, Chen F, Jiao N. 2011. Complete genome sequence of a marine roseophage provides evidence into the evolution of gene transfer agents in alphaproteobacteria. Virol J. 8:124.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey SB, Stanton TB, Jensen NS, Zuerner RL. 1997. Purification and characterization of VSH-1, a generalized transducing bacteriophage of Serpulina hyodysenteriae. J Bacteriol. 179:323–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes AP, Lang AS. 2013. Rhodobacter capsulatus gene transfer agent (RcGTA) activity bioassays. Bio-Protocols 3:e317. [Google Scholar]
- Hynes AP, Mercer RG, Watton DE, Buckley CB, Lang AS. 2012. DNA packaging bias and differential expression of gene transfer agent genes within a population during production and release of the Rhodobacter capsulatus gene transfer agent, RcGTA. Mol Microbiol. 85:314–325. [DOI] [PubMed] [Google Scholar]
- Iengar P, Joshi NV, Balaram P. 2006. Conformational and sequence signatures in beta helix proteins. Structure 14:529–542. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31:e15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Dolja VV. 2013. A virocentric perspective on the evolution of life. Curr Opin Virol. 3:546–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Dolja VV. 2014. Virus world as an evolutionary network of viruses and capsidless selfish elements. Microbiol Mol Biol R. 78:278–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchinski KS, Brimacombe CA, Westbye AB, Ding H, Beatty JT. 2016. The SOS response master regulator LexA regulates the gene transfer agent of Rhodobacter capsulatus and represses transcription of the signal transduction protein CckA. J Bacteriol. 198:1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat Rev Micro. 8:317–327. [DOI] [PubMed] [Google Scholar]
- Lang AS, Beatty JT. 2000. Genetic analysis of a bacterial genetic exchange element: the gene transfer agent of Rhodobacter capsulatus. Proc Natl Acad Sci U S A. 97:859–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang AS, Beatty JT. 2002. A bacterial signal transduction system controls genetic exchange and motility. J Bacteriol. 184:913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang AS, Beatty JT. 2007. Importance of widespread gene transfer agent genes in α-proteobacteria. Trends Microbiol. 15:54–62. [DOI] [PubMed] [Google Scholar]
- Lang AS, Taylor TA, Beatty JT. 2002. Evolutionary implications of phylogenetic analyses of the gene transfer agent (GTA) of Rhodobacter capsulatus. J Mol Evol. 55:534–543. [DOI] [PubMed] [Google Scholar]
- Lang AS, Zhaxybayeva O, Beatty JT. 2012. Gene transfer agents: phage-like elements of genetic exchange. Nat Rev Micro. 10:472–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiman PG, Arisaka F, van Raaij MJ, Kostyuchenko VA, Aksyuk AA, Kanamaru S, Rossmann MG. 2010. Morphogenesis of the T4 tail and tail fibers. Virol J. 7:355.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung MM, Brimacombe CA, Spiegelman GB, Beatty JT. 2012. The GtaR protein negatively regulates transcription of the gtaRI operon and modulates gene transfer agent (RcGTA) expression in Rhodobacter capsulatus. Mol Microbiol. 83:759–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-García P. 2012. The place of viruses in biology in light of the metabolism- versus-replication-first debate. Hist Philos Life Sci. 34:391–406. [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327–W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs BL. 1974. Genetic recombination in Rhodopseudomonas capsulata. Proc Natl Acad Sci U S A. 71:971–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer R, Lang A. 2014. Identification of a predicted partner-switching system that affects production of the gene transfer agent RcGTA and stationary phase viability in Rhodobacter capsulatus. BMC Microbiol. 14:71.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer RG, Callister SJ, Lipton MS, Pasa-Tolic L, Strnad H, Paces V, Beatty JT, Lang AS. 2010. Loss of the response regulator CtrA causes pleiotropic effects on gene expression but does not affect growth phase regulation in Rhodobacter capsulatus. J Bacteriol. 192:2701–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer RG, Quinlan M, Rose AR, Noll S, Beatty JT, Lang AS. 2012. Regulatory systems controlling motility and gene transfer agent production and release in Rhodobacter capsulatus. FEMS Microbiol Lett. 331:53–62. [DOI] [PubMed] [Google Scholar]
- Moon S, Byun Y, Kim H-J, Jeong S, Han K. 2004. Predicting genes expressed via -1 and +1 frameshifts. Nucleic Acids Res. 32:4884–4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Takashima K, Ishihara H, Shinomiya T, Kageyama M, Kanaya S, Ohnishi M, Murata T, Mori H, Hayashi T. 2000. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol Microbiol. 38:213–231. [DOI] [PubMed] [Google Scholar]
- NCBI Coordinators. 2016. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 44:D7–D19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton RJ, Griffin LE, Bowles KM, Meile C, Gifford S, Givens CE, Howard EC, King E, Oakley CA, Reisch CR, et al. 2010. Genome characteristics of a generalist marine bacterial lineage. ISME J. 4:784–798. [DOI] [PubMed] [Google Scholar]
- Novichkov PS, Wolf YI, Dubchak I, Koonin EV. 2009. Trends in prokaryotic evolution revealed by comparison of closely related bacterial and archaeal genomes. J Bacteriol. 91:65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Mudd JA, Mangan J, Huang WM, Subbaiah TV, Marmur J. 1968. Properties of the defective phage of Bacillus subtilis. J Mol Biol. 34:413–428. [DOI] [PubMed] [Google Scholar]
- Paul JH. 2008. Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas? ISME J. 2:579–589. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 8:785–786. [DOI] [PubMed] [Google Scholar]
- Rao RN. 1968. Bacteriophage P22 controlled exclusion in Salmonella typhimurium. J Mol Biol. 35:607–622. [DOI] [PubMed] [Google Scholar]
- Rapp BJ, Wall JD. 1987. Genetic transfer in Desulfovibrio desulfuricans. Proc Natl Acad Sci U S A. 84:9128–9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276–277. [DOI] [PubMed] [Google Scholar]
- Riede I, Schwarz H, Jahnig F. 1987. Predicted structure of tail-fiber proteins of T-even type phages. FEBS Lett. 215:145–150. [DOI] [PubMed] [Google Scholar]
- Rocha EPC. 2006. Inference and analysis of the relative stability of bacterial chromosomes. Mol Biol Evol. 23:513–522. [DOI] [PubMed] [Google Scholar]
- Sarris PF, Ladoukakis ED, Panopoulos NJ, Scoulica EV. 2014. A phage tail-derived element with wide distribution among both prokaryotic domains: a comparative genomic and phylogenetic study. Genome Biol Evol. 6:1739–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer AL, Taylor TA, Beatty JT, Greenberg EP. 2002. Long-chain acyl-homoserine lactone quorum-sensing regulation of Rhodobacter capsulatus gene transfer agent production. J Bacteriol. 184:6515–6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz EC, Ficner R. 2011. Knitting and snipping: chaperones in β-helix folding. Curr Opin Struc Biol. 21:232–239. [DOI] [PubMed] [Google Scholar]
- Scolnik PA, Haselkorn R. 1984. Activation of extra copies of genes coding for nitrogenase in Rhodopseudomonas capsulata. Nature 307:289–292. [DOI] [PubMed] [Google Scholar]
- Shikuma NJ, Pilhofer M, Weiss GL, Hadfield MG, Jensen GJ, Newman DK. 2014. Marine tubeworm metamorphosis induced by arrays of bacterial phage tail-like structures. Science 343:529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Nat Biotech. 1:37–45. [Google Scholar]
- Solioz M. 1975. The gene transfer agent of Rhodopseudomonas capsulata. Ph.D. Thesis. St. Louis: Saint Louis University.
- Solioz M, Marrs B. 1977. The gene transfer agent of Rhodopseudomonas capsulata: purification and characterization of its nucleic acid. Arch Biochem Biophys. 181:300–307. [DOI] [PubMed] [Google Scholar]
- Solioz M, Yen H-C, Marrs B. 1975. Release and uptake of gene transfer agent by Rhodopseudomonas capsulata. J Bacteriol. 123:651–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton TB. 2007. Prophage-like gene transfer agents–Novel mechanisms of gene exchange for Methanococcus, Desulfovibrio, Brachyspira, and Rhodobacter species. Anaerobe 13:43–49. [DOI] [PubMed] [Google Scholar]
- Stanton TB, Humphrey SB, Bayles DO, Zuerner RL. 2009. Identification of a divided genome for VSH-1, the prophage-like gene transfer agent of Brachyspira hyodysenteriae. J Bacteriol. 191:1719–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad H, Lapidus A, Paces J, Ulbrich P, Vlcek C, Paces V, Haselkorn R. 2010. Complete genome sequence of the photosynthetic purple nonsulfur bacterium Rhodobacter capsulatus SB 1003. J Bacteriol. 192:3545–3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MB, Lindell D, Lee JA, Thompson LR, Bielawski JP, Chisholm SW. 2006. Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. PLoS Biol. 4:1344–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi N, Kaneko K, Koonin EV. 2014. Horizontal gene transfer can rescue prokaryotes from Muller’s ratchet: benefit of DNA from dead cells and population subdivision. G3 (Bethesda) 4:325–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamames J. 2001. Evolution of gene order conservation in prokaryotes. Genome Biol. 2:research0020.1–0020.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DP, Cohen SN, Clark WG, Marrs BL. 1983. Alignment of the genetic and restriction maps of the photosynthetic region of the Rhodopseudomonas capsulata chromosome by a conjugation-mediated marker rescue technique. J Bacteriol. 154:580–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Bobay L-M, Rocha EP. 2014. The chromosomal accommodation and domestication of mobile genetic elements. Curr Opin Microbiol. 22:22–29. [DOI] [PubMed] [Google Scholar]
- Vos M. 2009. Why do bacteria engage in homologous recombination? Trends Microbiol. 17:226–232. [DOI] [PubMed] [Google Scholar]
- Weaver PF, Wall JD, Gest H. 1975. Characterization of Rhodopseudomonas capsulata. Arch Microbiol. 105:207–216. [DOI] [PubMed] [Google Scholar]
- Westbye AB, Kuchinski K, Yip CK, Beatty JT. 2016. The gene transfer agent RcGTA contains head spikes needed for binding to the Rhodobacter capsulatus polysaccharide cell capsule. J Mol Biol. 428:477–491. [DOI] [PubMed] [Google Scholar]
- Westbye AB, Leung MM, Florizone SM, Taylor TA, Johnson JA, Fogg PC, Beatty JT. 2013. Phosphate concentration and the putative sensor kinase protein CckA modulate cell lysis and release of the Rhodobacter capsulatus gene transfer agent. J Bacteriol. 195:5025–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman E. 1938. Recherches sur le phénoméne de Twort-d’Hérelle (Bactériophagie ou autolyse hérédo-Contagieuse). Ann Inst Pasteur Paris. 60:13–57. [Google Scholar]
- Yen H-C, Hu NT, Marrs BL. 1979. Characterization of the gene transfer agent made by an overproducer mutant of Rhodopseudomonas capsulata. J Mol Biol. 131:157–168. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res. 39:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.