Abstract
Harpellales, an early-diverging fungal lineage, is associated with the digestive tracts of aquatic arthropod hosts. Concurrent with the production and annotation of the first four Harpellales genomes, we discovered that Zancudomyces culisetae, one of the most widely distributed Harpellales species, encodes an insect-like polyubiquitin chain. Ubiquitin and ubiquitin-like proteins are universally involved in protein degradation and regulation of immune response in eukaryotic organisms. Phylogenetic analyses inferred that this polyubiquitin variant has a mosquito origin. In addition, its amino acid composition, animal-like secondary structure, as well as the fungal nature of flanking genes all further support this as a horizontal gene transfer event. The single-copy polyubiquitin gene from Z. culisetae has lower GC ratio compared with homologs of insect taxa, which implies homogenization of the gene since its putatively ancient transfer. The acquired polyubiquitin gene may have served to improve important functions within Z. culisetae, by perhaps exploiting the insect hosts’ ubiquitin-proteasome systems in the gut environment. Preliminary comparisons among the four Harpellales genomes highlight the reduced genome size of Z. culisetae, which corroborates its distinguishable symbiotic lifestyle. This is the first record of a horizontally transferred ubiquitin gene from disease-bearing insects to the gut-dwelling fungal endobiont and should invite further exploration in an evolutionary context.
Keywords: comparative genomics, evolution, Kickxellomycotina, pathogen, ubiquitination.
Introduction
Insects are hosts to various symbionts, including bacteria, fungi, and viruses (White et al. 2006; Moran 2007; Hedges et al. 2008) and these symbiotic interactions have spurred the interest of both ecologists and evolutionary biologists. As they evolve, reciprocal responses between hosts and symbionts may have reshaped both associates, possibly invoking morphological changes that accompany genetic signatures (Mandel et al. 2009; Moran and Jarvik 2010). With obligate symbiotic associations, there may be irreversible gene gain or loss events that correspond to functional changes as both insects and endobionts adapt over evolutionary timescales (Moran 2007; Mandel et al. 2009; Moran and Jarvik 2010; Selman et al. 2011).
Harpellales is an order of early-diverging fungi (James et al. 2006; White 2006; Hibbett et al. 2007), which commonly attach to the chitinous hindgut linings of immature stages of aquatic insects (lower Diptera, including black flies, midges, and mosquitoes, as well as mayflies and stoneflies), and are thus known as “gut fungi” (White and Lichtwardt 2004; Strongman et al. 2010; Valle and Cafaro 2010; Tretter et al. 2014; Wang et al. 2014). Members of the Harpellales are usually considered commensals, although at least one species has been reported to be fatal to its mosquito host (Sweeney 1981). Zancudomyces was recently established to accommodate Z. culisetae based on both molecular phylogenetic and morphological analyses (Wang et al. 2013). Formerly recognized as Smittium culisetae, the species has been shown to benefit the in vivo development of infested mosquito larvae under specific conditions (Horn and Lichtwardt 1981). In contrast, Z. culisetae can also lead to the death of mosquito larvae, in situations where the hosts hindgut becomes overgrown (Williams 2001).
Genome-wide data are providing the opportunity to critically assess symbiotic ontogenetic stages, from surface adhesion, host invasion, molecular interactions to genomic modifications (Moya et al. 2008). This also includes possibilities of investigating horizontal gene transfer (HGT) events. For example, whole-genome sequencing enabled the identification of several independent purine nucleotide phosphorylase HGT events between Encephalitozoon (Microsporidia) and arthropod donors (Selman et al. 2011). As an example of a fungi-donated gene, the carotenoid coding gene within the pea aphid genome has been shown to be laterally transferred from fungi (Moran and Jarvik 2010).
Ubiquitin is universally present in eukaryotes where it is widely known as posttranslational tag for the hydrolytic destruction of proteins (Goldstein et al. 1975; Welchman et al. 2005). Ubiquitin and ubiquitin-like proteins have also been found to play crucial roles in DNA transcription, autophagy, and inflammatory responses during pathogen defense by the host (Jiang and Chen 2012; Severo et al. 2013). For example, ubiquitination is involved in regulation of immune responses in mosquitoes, which are notorious vectors for spreading diseases like dengue, malaria, Zika fever, and West Nile encephalitis (Choy et al. 2013). Simultaneously, some pathogens seem to have countered with similar ubiquitin-dependent processes to facilitate entrance into the host (Collins and Brown 2010; Haldar et al. 2015). Ubiquitin may function in separate ways depending on how monoubiquitins are linked. Generally, K48-linked polyubiquitin chains target proteins destined for proteolysis, whereas K63-linked chains are involved in inflammatory response, protein trafficking, and ribosomal protein synthesis (Zhao and Ulrich 2010). Within gut linings of arthropods, the ubiquitin-proteasome system (UPS) is believed to function in food particle degradation and nutrient absorption, but may also simultaneously affect their immune responses (Severo et al. 2013). As gut-dwelling symbionts, Harpellales occupy an interface that presumably exposes them intimately and intensively to the hosts’ ubiquitination machinery.
In light of the aforementioned cases of HGT between insects and fungal symbionts, we investigated the existence of such HGT elements within Harpellales, which present an excellent study system due to their inflexible association with insect hosts. Four Harpellales genomes (Z. culisetae, S. mucronatum, and two strains of S. culicis) were sequenced and annotated for the present study. Through phylogenetic reconstruction and a series of comparative analyses of amino acid compositions, predicted secondary structures, and synteny across eukaryotic clades, we authenticate the first case of a polyubiquitin gene transfer event from mosquito hosts to the gut fungus, Z. culisetae.
Results
Harpellales Genome Features
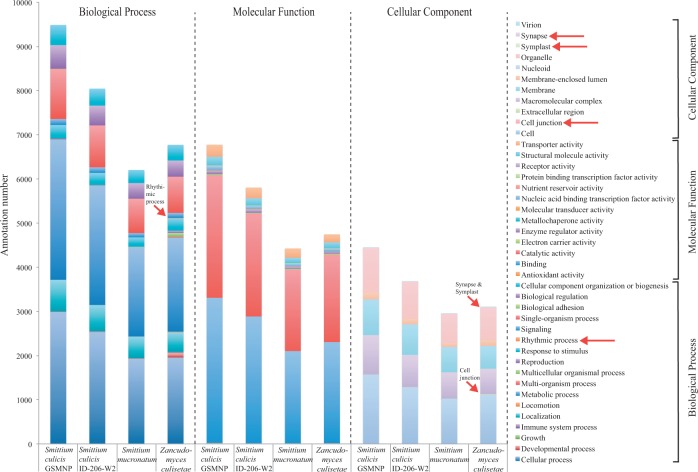
Genome assembly statistics and annotation features are presented in table 1. The Core Eukaryotic Genes Mapping Approach (CEGMA) recovered the presence of above 90% of core eukaryotic genes in all four assemblies. The genome size of Z. culisetae (28.7 Mb) is much smaller than those of Smittium (71.1–102.4 Mb) and genome-wide GC ratios of Smittium representatives were below 30%, whereas Z. culisetae was 35.5%. The ab initio gene predictions discovered approximately 12,000 genes in each strain of S. culicis, 8,410 genes in S. mucronatum, and 8,252 genes in Z. culisetae. On average, the Smittium genomes had more than two exons per gene, whereas Z. culisetae had less than two. Gene ontology analyses also indicated that Z. culisetae possesses several genes with unique (not found in Smittium) annotations for biological processes (rhythmic process) and cellular components (cell junction, symplast, and synapse) (fig. 1), suggesting that the genomic compositions may differ significantly between Z. culisetae and Smittium.
Table 1.
Broad Scale Comparison of Genome Features among the Four Gut Fungi (Harpellales).
| Taxa | Smittium culicis | Smittium culicis | Smittium mucronatum | Zancudomyces culisetae |
|---|---|---|---|---|
| Strain ID | GSMNP (ARSEF 9010) | ID-206-W2 | ALG-7-W6 (ARSEF 9090) | COL-18-3 (ARSEF 9012) |
| Coverage | 49× | 27× | 24× | 26× |
| Number of scaffolds (>1kb) | 6,137 | 7,749 | 7,797 | 1,954 |
| Genome size by scaffolds (Mb) | 77.12 | 71.05 | 102.35 | 28.70 |
| Repeats ratio | 3.34% | 3.64% | 2.94% | 4.29% |
| GC ratio | 28.61% | 29.46% | 26.05% | 35.52% |
| CEGMA complete (+incomplete) genes | 93.95% (97.98%) | 93.55% (97.58%) | 89.52% (93.55%) | 85.89% (92.74%) |
| Open reading frames | 16,101 | 15,575 | 11,486 | 9,667 |
| Protein-coding genes | 12,468 | 11,593 | 8,410 | 8,252 |
| Exons per gene | 2.26 | 2.20 | 2.28 | 1.78 |
| Gene density (genes per Mb) | 162 | 163 | 82 | 288 |
| Percentage of secreted proteins | 7.74% | 7.09% | 8.10% | 9.78% |
| Transmembrane helices | 5891 | 4912 | 4213 | 3214 |
| HGT (mapped to host genomes) | 59 (0) | 60 (0) | 27 (0) | 33 (5) |
Fig. 1.
Genome level comparisons of the Gene Ontology annotations (level 2) among the four Harpellales. Unique annotations for Zancudomyces culisetae are denoted with red arrows.
HGT Detection and Syntenic Analyses
A set of similarity searches (using BLASTp) (Altschul et al. 1990), as well as a customized Python script (“HGTfilter.py”, available from GitHub) was employed to recover putative HGT elements from the four Harpellales genomes. The analyses recovered 59, 60, 27, and 33 potential HGT events from S. culicis strain GSMNP, S. culicis strain ID-206-W2, S. mucronatum, and Z. culisetae, respectively (table 1). Among the total pool of 179 candidates, only five, all from Z. culisetae, could be adequately mapped back to the host genomes. One of these (supplementary table S1, Supplementary Material online), a triple-ubiquitin gene, demonstrated a conserved domain during BLASTp analyses, suggesting its appropriateness for further investigation into HGT-related events. Our results suggest that this gene occurs as a single copy in the Z. culisetae genome (although monoubiquitins occur on other scaffolds in the genome), and that the original fungal copy may have been lost at some point during evolution and interaction with the insect hosts.
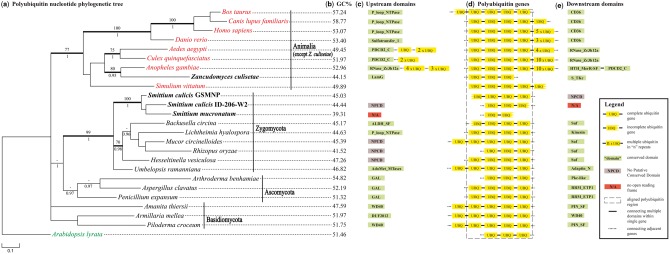
Multiple polyubiquitin candidates (with E-values < 1E −100) with varying repeat motifs were discovered in the examined eukaryotic genomes (supplementary table S2, Supplementary Material online). In order to infer homology, corresponding flanking genes were recovered for each included taxon by scanning the genomes and annotations manually. The rationale for this strategy is twofold. First, it will aid in revealing the HGT element within the Z. culisetae genome where the insert should be flanked by genes of fungal origin. Second, it would minimally allow for inference of homology on a clade-by-clade basis if the upstream and downstream genes were conserved throughout the clades. We found high levels of conservation of adjacent genes within clades (fig. 2c and e; supplementary table S2, Supplementary Material online), but rather high disparity among clades. The numbers of repeats in the polyubiquitin genes also varies across the diversity, with animals having more repeats than fungi in general (fig. 2d). The single-copy polyubiquitin gene of Z culisetae is flanked by two protein-coding genes of fungal origin, which contain conserved domains. The flanking upstream gene codes for “laminin globular (LamG)”; putative homologs of this gene were found to be conserved in the Smittium genomes but were located in different parts of the genome (i.e., gene order was not conserved). However, LamG was found to be lacking from all animal taxa (top BLASTp hit against a closely related zygomycotan fungus, Mortierella verticillata, and no hits for animal taxa). The flanking downstream gene contains “Serine/Threonine protein kinases, catalytic domain (S_TKc)” and is again conserved regarding amino acid structure but not regarding its genomic position in both Smittium and animal species (top BLASTp hit against Rhizophagus irregularis, a closely related glomeromycotan fungal species) (fig. 3); phylogenetic analysis of both fungal- and animal-derived S_TKc confirmed the fungal origin of the Z. culisetae-derived gene (supplementary fig. S1, Supplementary Material online). Interestingly, S_TKc is associated with apoptosis, focal adhesion, and metabolic pathways of ubiquitin-mediated proteolysis (Sanjo et al. 1998).
Fig. 2.
Phylogenetic and syntenic analyses of polyubiquitin nucleotide sequences. (a) Phylogenetic tree of polyubiquitin nucleotide sequences derived from the Bayesian analysis; MLBP ≥ 70% are shown above branches, whereas BPP ≥ 0.95 are shown below branches. Branches significantly supported by both are in bold. Animal taxa are noted in red, the outgroup in green, fungi in black, with Harpellales in bold. (b) GC ratios of the aligned polyubiquitin genes. (c–e) The aligned polyubiquitin gene region and adjacent domains. Regions inside the dotted box were included in the phylogenetic analyses.
Fig. 3.
Diagram showing the lengths, distances, and orientations of the horizontally transferred triple-ubiquitin gene (red) with flanking genes being of fungal origin (green), from the genome of Zancudomyces culisetae.
Phylogenetic Analyses of Polyubiquitin Sequences
The Bayesian inference analyses reached congruence after 1 million and 0.5 million generations, respectively, for amino acid and nucleotide sequences. Trees resulting from the maximum likelihood (ML) and Bayesian analyses fully agree on the topology, with the exception of the unresolved placement of Umbelopsis rammaniana (places as sister to Smittium in the ML tree but with negligible support). Both ML bootstrap proportions (MLBP 100%) and Bayesian posterior probabilities (BPP 1.00) significantly support the animal origin of the Z. culisetae polyubiquitin chain based on amino acid sequences (fig. 4a). The other three representatives of Harpellales (two S. culicis strains and S. mucronatum) clustered with other zygomycotan fungi with strong support (MLBP 94%, BPP 0.99). The multiple sequence alignment of polyubiquitin also revealed almost complete conservation of amino acids across the animal clade (including Z. culisetae) and their full conservation of a proline residue at position 19 versus a serine residue in both fungi and plant outgroup taxa (fig. 4b). The phylogenetic tree inferred from the nucleotide sequences demonstrated higher resolution and deeper structure (fig. 2a). Consistent with the amino acid tree, the animal origin of the Z. culisetae polyubiquitin gene is confirmed (MLBP 77%, BPP 1.00) and its position as the sister group to that of Anopheles gambiae is recovered with relatively strong support (MLBP 80%, BPP 0.95). The three Smittium species form a monophyletic group (MLBP 100%, BPP 1.00) together with representatives of Zygomycota (MLBP 99%, BPP 1.00). Consistent with the amino acid tree (fig. 2a), the phylogenetic analysis of the polyubiquitin nucleotide sequences failed to recover the Dikarya clade of higher fungi (Ascomycota + Basidiomycota).
Fig. 4.
Phylogeny and secondary structures inferred from (poly)ubiquitin amino acid sequences. (a) Phylogenetic tree of polyubiquitin amino acid sequences derived from the Bayesian analysis. Annotations as in figure 2a. (b) Partial view of the multiple amino acid sequence alignment, highlighting the critical residue difference (red box) at site 19 (of the full alignment including 76 amino acids in each unit). (c) Predicted monoubiquitin secondary structures.
Secondary Structures, Selection Analyses, and GC Ratio Variation
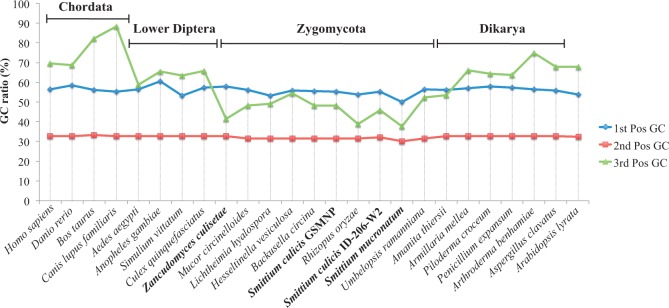
Secondary structure analyses predicted that ubiquitins show different structures between animals (including Z. culisetae) and fungi (fig. 4c). Specifically, the animal ubiquitins share a coil structure immediately adjacent to the first set of helices, in contrast to all other fungal members, which instead show an additional helix structure. The polyubiquitin genes were under strong purifying selection, with more than 94% of the codons showing negative selection, according to codon-specific analyses (table 2). Among the taxa examined, GC ratios of polyubiquitin genes (fig. 2b) were much elevated for animals (49.45–58.77%) compared with zygomycotan fungi (39.31%–47.26%). The GC ratio of Z. culisetae (44.15%) falls within the range of other Harpellales and zygomycotan representatives (fig. 2b), despite its animal origin (fig. 2a). Interestingly, the GC ratio range of the Zygomycota clade is also lower than that of “Dikarya” (47.59–54.82%). The GC ratios of the first and second codon positions are rather consistent across both animals and fungi, yet the ratios of the third codon position vary greatly (fig. 5). Two major categories emerged according to the third codon position GC ratios: one (Animalia and Dikarya except Amanita thiersii) shows higher third codon position GC ratios than either that of the first or second codon position; the other category (Zygomycota and Am. thiersii) shows lower third codon position GC ratios than the first codon position, but higher than the second codon position.
Table 2.
Codon-Based Tests of Selection Analyses for Polyubiquitin Genes.
| Selection Type | Overall Level |
Individual Sites (304 codons in total) |
|||
|---|---|---|---|---|---|
| Z-Test | PARRIS | REL | SLAC | FEL | |
| Purifying selection | Yes; P = 0 | N/A | 304 | 288 | 292 |
| Positive selection | No; P = 1 | No; P = 1 | 0 | 0 | 0 |
| Neutral | Not performed | N/A | 0 | 16 | 12 |
Note.—N/A, not applicable in the listed analyses.
Fig. 5.
GC ratios variation of the polyubiquitin genes across the 25 taxa, mapped by first, second, and third codon positions. The four Harpellales are in bold.
Discussion
With support provided by phylogenetic analyses, amino acid compositions, secondary structure prediction, and a variety of BLAST and BLAT analyses, our results all converge on the indication that the gut fungal symbiont, Z. culisetae, has acquired a single-copy polyubiquitin gene through horizontal transfer from an insect host, possibly from the ancestral lineage of A. gambiae. This represents the first report of HGT within Harpellales, notwithstanding that their symbiotic lifestyle presents an intimacy that is similar to other systems that have experienced such events. It is reasonable to expect that genetic modifications have occurred within Harpellales genomes as they adapted to the gut-dwelling lifestyle. All four Harpellales genomes present AT enrichment, and Z. culisetae shows a much reduced genome size when compared with the other representatives.
Homolog Detection and Phylogenetic Analyses
Representatives of Ascomycota and most Zygomycota (with the exception of Backusella, Hesseltinella, and Umbelopsis) possess a single-copy polyubiquitin gene (supplementary table S2, Supplementary Material online), whereas all Basidiomycota have at least two copies; for Basidiomycota, the retention by one of the copies of the WD40 domain in the adjacent upstream gene (downstream in Armillaria mellea, due to a putative reversal event) allowed for orthology inferences used in the phylogenetic reconstruction (fig. 2 and supplementary table S2, Supplementary Material online). Within animals, all genomes used here also possessed at least two copies of the gene, making orthology predictions particularly cumbersome. This was, in part, remedied by both BLAT host mapping results (supplementary table S1, Supplementary Material online) and the assessment of flanking genes on a clade-by-clade basis (supplementary table S2, Supplementary Material online). Phylogenetic analyses based on all copies (supplementary fig. S2, Supplementary Material online) suggest that the polyubiquitin gene is lacking the resolving power to recover the Dikarya clade of the higher fungi.
Selection Analyses and Potential Functions of the Horizontally Transferred Polyubiquitin Gene
The lower GC ratio of Z. culisetae, compared with other members of the animal clade (figs. 2b and 5), implies that the HGT event was followed by substantial homogenization of the gene region. Given the putative importance in function of the animal-like polyubiquitin gene found in Z. culisetae, it was not surprising to find high levels of negative selection acting across the gene, although other evolutionary forces keep working at the nucleotide level, leading to synonymous substitutions. This is reflected both in the GC ratio of the Z. culisetae polyubiquitin gene, which is consistent with other parts of the fungal genome (fig. 5), and in the higher resolution of the phylogenetic tree derived from the nucleotide alignment (fig. 2a).
Ubiquitin is critical in controlling the fate of eukaryotic proteins and previous studies have revealed other complex functions of polyubiquitin chains in eukaryotic systems (Collins and Brown 2010; Hagai and Levy 2010; Zhao and Ulrich 2010; Severo et al. 2013). A potential benefit of an insect-originated triple-ubiquitin gene might be to label and degrade certain insect proteins by hijacking their own UPS machinery. However, why only three out of the 14 A. gambiae ubiquitin repeats were found to be acquired by Z. culisetae remains to be answered. The significance and function of the repeat number is still unclear, although it has been found that doubling the polyubiquitin repeat units from four to eight did not accelerate the degradation process of proteins (Zhao and Ulrich 2010). This suggests either that the number of repeats bears no burden for the functionality of the protein (this characteristic should then be under neutral selection and may present itself in random constellations across clades, as seems to be the case) or that the 14-repeat ubiquitin genes in some animal taxa may serve other functions. The proteasome regulatory pathway is capable of degrading many kinds of proteins, though its efficiency varies greatly depending on the biophysical properties of the substrate (Baugh et al. 2009). The polyubiquitin chains (especially the K48 linked variant) alter the thermodynamic stability of the substrate, unwind its local structures, and help initiate its degradation (Hagai and Levy 2010). The proline residue found here for animals (including Z. culisetae) versus the serine residue of other lineages may be associated with specific substrate binding and unfolding, following signal transductions (fig. 4b and c).
The LamG and S_TKc domains on adjacent flanking genes (fig. 3) may amplify the potential of the triple-ubiquitin gene in serving its function. Laminin is a family of glycoproteins that are important parts of the basal lamina, involved in cell differentiation, migration, adhesion, and survival (Timpl et al. 1979), and are secreted and incorporated into cell-associated extracellular matrices. Laminins and laminin-binding domains are involved in adhesion of Aspergillus fumigatus conidia to host cell surfaces (Upadhyay et al. 2009). The exact function of the LamG domain remains unknown, although binding functions and disease progression have been ascribed to different LamG modules (Schéele et al. 2007). The LamG adjacent to the triple-ubiquitin of Z. culisetae is a transmembrane protein according to the TMHMM prediction (supplementary fig. S3, Supplementary Material online). The S_TKc domain serves in protein phosphorylation and ATP-binding processes (Hanks et al. 1988). The highly conserved catalytic domain is essential for catalyzing numerous related enzymes, several of which play important roles in ubiquitin-mediated proteolysis, apoptosis, and differentiation (Sanjo et al. 1998).
Based on our current knowledge, the mosquito-originated polyubiquitin gene in Z. culisetae may be useful during the invasive processes of the fungus, to induce the hosts UPS by labeling and degrading host cell membrane proteins. The upstream and downstream genes may also assist this process in differentiating, adhering, and catalyzing. An alternative use of the acquired mosquito-originated polyubiquitin gene could be that Z. culisetae uses it as a defense against bacteria, viruses, or other microbes that coexist in the insect guts, whether for its own competitive advantage or as an ally of the host. Recent research has shown that polyubiquitin has important roles in regulating the hosts’ immune and inflammatory responses (Jiang and Chen 2012; Severo et al. 2013) and is able to target nonself-entities (i.e., microbial pathogens) and assist selective autophagy (Collins and Brown 2010; Jiang and Chen 2012). In addition Haldar et al. (2015) reported that ubiquitin-centered mechanisms were involved in immune-response attacks on pathogen-containing vacuoles by the host.
Zancudomyces and Harpellales Genomic Studies
Zancudomyces culisetae was one of the first gut fungi to be isolated axenically and it is one of the most frequently encountered species of Harpellales from various regions globally (Lichtwardt et al. 1999; Valle and Santamaria 2004; White et al. 2006; Wang et al. 2013). Zancudomyces culisetae has been used in pioneering numerous research avenues (Williams 1983; Horn 1989; Grigg and Lichtwardt 1996; Gottlieb and Lichtwardt 2001; Tretter et al. 2013) as it demonstrates an intimate relationship with larval mosquitoes (Horn and Lichtwardt 1981; Williams 2001); the fungal spores present a delicate response mechanism, which is triggered by pH and ion changes along the digestive tract, and corresponding with germling release and development (Horn 1989). In this study, novel genome-level comparisons revealed that Z. culisetae has a considerably smaller genome size, greater gene density, and more unique gene ontology annotations compared with Smittium (table 1 and fig. 1). These genomic insights could indicate that Z. culisetae has evolved a tighter relationship with its hosts, either as a mutualistic symbiont or perhaps even with parasitic potential. The horizontally transferred triple-ubiquitin gene may also help secure the symbiotic relationship between Z. culisetae and mosquitoes, and to some extent, related aquatic Diptera, which may explain the exceptional success of Z. culisetae in light of its global distribution. Smittium mucronatum presents the largest genome size among the four, which may be related to its host specificity to Psectrocladius (Chironomidae). A similar result was recently recovered for an Aedes aegypti-specialized fungal pathogen, Edhazardia aedis, which shows a notably larger genome size (51 Mb) when compared with other Edhazardia species (2–9 Mb) (Desjardins et al. 2015). While such considerations are in their infancy, further studies relating to host specificity, secondary metabolites, genes specialization, and molecular interactions should shed light on these questions, as well as on how Harpellales have maintained their obligate gut-dwelling lifestyle in such an effective manner (Galagan et al. 2005; Staats et al. 2014).
Materials and Methods
Fungal Strains and DNA Extraction
Four Harpellales taxa were included in this study (table 1). Two Smittium culicis strains (GSMNP and ID-206-W2) were selected to represent divergent clades within the species complex (Wang et al. 2014). The type species S. mucronatum, S. culicis strain GSMNP, and Z. culisetae were obtained from USDA-ARS Collection of Entomopathogenic Fungal Cultures (ARSEF). Smittium culicis strain ID-206-W2 was recently isolated from the hindgut of a mosquito larva in Boise, ID, USA (MMW’s lab at Boise State University). Strains were cultured in broth tubes of Brain Heart Infusion Glucose Tryptone (BHIGTv) medium at room temperature (White et al. 2006), and the DNA extraction followed a standard CTAB protocol (Wang et al. 2014).
Genome Sequencing, Assembly, and Annotation
Paired-end libraries (with 500 bp insert size) were prepared and sequenced for all four strains at the Centre for Applied Genomics (Hospital for Sick Children, Toronto, Canada) using one lane of the Illumina HiSeq 2500 platform (2 × 150 bp read length). Raw sequence reads were quality trimmed and assembled with RAY v2.3.1 (Boisvert et al. 2010). Potential contamination was examined and characterized using the Blobology pipeline (Kumar et al. 2013). Satellites, simple repeats, and low-complexity sequences were annotated with RepeatMasker v4.0.5 (http://www.repeatmasker.org, last accessed September 18, 2015) and Tandem Repeat Finder v4.07b (Benson 1999), corresponding to the “Fungi” taxon. Gene prediction employed AUGUSTUS v3.1 (Keller et al. 2011) using the genome profiles of Conidiobolus coronatus (Entomophthoramycotina, Zygomycota) (Chang et al. 2015). As a corollary, TransDecoder (Haas et al. 2013) was used to predict open reading frames and enable a conservative comparison to estimate gene numbers. Gene functions of the AUGUSTUS prediction set were inferred from Blast2GO v3.0 (Conesa et al. 2005) and InterProScan v5.8-49.0 (Jones et al. 2014) against the nonredundant database in NCBI and protein signature databases in EBI. Secreted proteins were predicted by SignalP v4.1 without truncation (Petersen et al. 2011), and transmembrane helices were predicted through TMHMM Server v2.0 (Krogh et al. 2001). CEGMA v2.4.010312 (Parra et al. 2007) was used to identify the presence of core eukaryotic protein-coding genes for subsequent evaluation of genome coverage.
HGT Detection and Homolog Identification
The four Harpellales proteomes were BLASTed (using BLASTp) against a concatenated proteome database, consisting of 512 fungal representatives from Broad Institute and Joint Genome Institute (JGI), as well as five proteomes of insect (lower Diptera) hosts of Harpellales (Aedes aegypti, Anopheles gambiae, Culex quinquefasciatus, Chironomus tentans, and Simulium vittatum) from VectorBase, European Bioinformatics Institute (EBI), and the Human Genome Sequencing Center at Baylor College of Medicine (BCM-HGSC) (Holt et al. 2002; International Human Genome Sequencing Consortium 2004; Nene et al. 2007; Ma et al. 2009; Zimin et al. 2009; Arensburger et al. 2010; Burmester et al. 2011; Hu et al. 2011; Arnaud et al. 2012; Collins et al. 2013; Howe et al. 2013; Hoeppner et al. 2014; Kutsenko et al. 2014; Kohler et al. 2015). A customized Python script (“HGTfilter.py”, available from GitHub) was applied in order to identify promising HGT elements in the Harpellales genomes. The script works by comparing BLAST-based hits against both the 512 fungal genomes (in this case) and the five host genomes and lifting out hits that match insect-derived genes at a lower E-value than fungus-derived genes. Due to the employment of fungal genomes across the diversity of the kingdom, an insect-derived match necessarily had to “compete” with a broad swath of fungi in order to be deemed of insect origin. All corresponding nucleotide sequences of the filtered outputs were mapped back as queries to the host genomes using BLAT (Kent 2002), in order to robustly infer HGT events. Homologs among 12 fungi and 8 animals were selected (based on a 1E −50 cutoff) to represent Ascomycota, Basidiomycota, Zygomycota, and animal clades (table 3 and fig. 2a). To strengthen the inference of homology on a clade-by-clade basis, upstream and downstream genes for each homolog were recovered by manually scanning the genomes and annotations. The polyubiquitin gene for the outgroup taxon, Arabidopsis lyrata, was obtained from GenBank under accession number XM_002872723. The syntenic structure of triple-ubiquitin and adjacent genes within the Z. culisetae genome were plotted using the genoPlotR package in R (Guy et al. 2010).
Table 3.
Eukaryotic Organisms and Their Polyubiquitin Genes Included in the Phylogenetic Analyses.
| Clade | Species | Strain No. | Location | Source | Publication |
|---|---|---|---|---|---|
| Ascomycota | Aspergillus clavatus | NRRL 1 | Scaffold_1099423829800 | JGI | Arnaud et al. 2012 |
| Ascomycota | Arthroderma benhamiae | CBS 112371 | Supercontig_42 | JGI | Burmester et al. 2011 |
| Ascomycota | Penicillium expansum | ATCC 24692 | Scaffold_2 | JGI | N/A |
| Basidiomycota | Amanita thiersii | Skay4041 | Scaffold_1 | JGI | Kohler et al. 2015 |
| Basidiomycota | Armillaria mellea | DSM 3731 | Scaffold_NODE_108303 | JGI | Collins et al. 2013 |
| Basidiomycota | Piloderma croceum | F 1598 | Scaffold_00016 | JGI | Kohler et al. 2015 |
| Zygomycota | Backusella circina | FSU 941 | Scaffold_257 | JGI | N/A |
| Zygomycota | Hesseltinella vesiculosa | NRRL3301 | Scaffold_6 | JGI | N/A |
| Zygomycota | Mucor circinelloides | CBS277.49 | Scaffold_13 | JGI | N/A |
| Zygomycota | Lichtheimia hyalospora | FSU 10163 | Scaffold_4 | JGI | N/A |
| Zygomycota | Rhizopus oryzae | 99-880 | Scaffold_3.9 | Broad Institute | Ma et al. 2009 |
| Zygomycota | Umbelopsis ramanniana | AG | Scaffold_63 | JGI | N/A |
| Zygomycota | Zancudomyces culisetae | COL-18-3 | Scaffold_1672 | NEW | Produced in this study |
| Zygomycota | Smittium culicis | GSMNP | Scaffold_5123 | NEW | Produced in this study |
| Zygomycota | Smittium culicis | ID-206-W2 | Scaffold_922 | NEW | Produced in this study |
| Zygomycota | Smittium mucronatum | ALG-7-W6 | Scaffold_2577 | NEW | Produced in this study |
| Insect | Anopheles gambiae | PEST | Chr_2R | VectorBase | Holt et al. 2002 |
| Insect | Aedes aegypti | Liverpool | Supercontig_1.99 | VectorBase | Nene et al. 2007 |
| Insect | Culex quinquefasciatus | Johannesburg | Supercontig_3.50 | VectorBase | Arensburger et al. 2010 |
| Insect | Simulium vittatum | UGA | Scf7180000737758 | BCM-HGSC | N/A |
| Chordata | Homo sapiens | Assembly Version: GRCH38.p2 | Chr_NC_000012.12 | GenBank | International Human Genome Sequencing Consortium 2004 |
| Chordata | Bos Taurus | Assembly Version: UMD_3.1.1 | Chr_AC_000174.1 | GenBank | Zimin et al. 2009 |
| Chordata | Canis lupus familiaris | Assembly Version: CanFam3.1 | Chr_NC_006608.3 | GenBank | Hoeppner et al. 2014 |
| Chordata | Danio rerio | Assembly Version: GRCz10 | Chr_NC_007121.6 | GenBank | Howe et al. 2013 |
| Plant | Arabidopsis lyrata | MN47 | N/A | GenBank | Hu et al. 2011 |
Multiple Sequence Alignment, Model Test, and Phylogenetic Reconstruction
The polyubiquitin amino acid sequences were aligned using MUSCLE v3.8.31 (Edgar 2004), the result of which served as the guide for the nucleotide alignment. The RtREV + I+G and GTR + I+G models were suggested for the polyubiquitin amino acid and nucleotide sequence alignments by ProtTest v2.4 (Abascal et al. 2005) and JModelTest v2.1.3 (Posada 2008), respectively. ML analyses employed RAxML v8 (Stamatakis 2014) and Bayesian inferences were performed using MrBayes v3.2 (Ronquist et al. 2012) for both amino acid and codon-partitioned nucleotide sequences. The ML search used 1,000 initial addition sequences with 25 initial GAMMA rate categories and final optimization with four GAMMA shape categories. Support values for nodes were acquired through 1,000 pseudoreplicates with random seeds. For the Bayesian inference analysis, a total of eight chains (two runs, each with three hot and one cold chain) were performed for 50 million generations and Tracer v1.5 (http://beast.bio.ed.ac.uk/Tracer, last accessed December 2, 2015) was used to confirm convergence of the Bayesian chains and the sufficiency of the default burnin value (25%). Regarding the phylogenetic analysis of the upstream flanking S_TKc gene, only RAxML was used to infer the phylogeny, using the same settings as mentioned above.
Secondary Structure Prediction and Selection Analyses
Monoubiquitin secondary structures were predicted using the CFSSP server (Kumar 2013). Selection pressures on the polyubiquitin genes across both animal and fungal lineages were assessed on a molecule-wide basis. Both purifying and positive selection hypotheses were tested using the Z-test in MEGA v6.06 (Tamura et al. 2013) and positive selection was tested with the PARRIS method in HyPhy (Kosakovsky Pond et al. 2005) through the Datamonkey server (Delport et al. 2010). The Z-test was performed allowing both for 0% gaps and 30% gaps, using 1,000 replicates for the bootstraps and the Nei-Gojobori method (Nei and Gojobori 1986). Codon-specific selection was tested using codon-based likelihood ratio tests, including Single-Likelihood Ancestor Count (SLAC), Fixed Effects Likelihood (FEL), and Random Effects Likelihood (REL) on the Datamonkey server following the methods detailed in Kvist et al. (2013). Negative selection pressures (no signatures of positive selection were recovered) were inferred only for codons where SLAC, FEL, and REL agreed on this result.
Supplementary Material
Supplementary material is available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank R. Humber and K. Hansen for preparing and providing the fungal strains, D. Greenshields, J. Hess, T. James, K. O’Donnell A. Pringle, K. Seifert, J. Spatafora, J. Stajich, and ZyGoLife consortium as well as JGI and BCM-HGSC for permission to use genomes ahead of publication. The authors thank J. Anderson, D. Currie, and three anonymous reviewers for their critical comments on the research design. The authors also thank the SciNet staff at the University of Toronto for facilitating access to the supercomputing infrastructure. This work was supported by a University of Toronto Fellowship to Y.W. and Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant 453847 to J.-M.M. M.M.W. gratefully acknowledges support from the ZyGoLife project, a National Science Foundation (NSF) Award DEB1441715, for ongoing research. S.K. thanks Olle Engkvist Byggmästare Foundation for support. The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
References
- Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Arensburger P, Megy K, Waterhouse RM, Abrudan J, Amedeo P, Antelo B, Bartholomay L, Bidwell S, Caler E, Camara F, et al. 2010. Sequencing of Culex quinquefasciatus comparative genomics. Science 330:86–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud MB, Cerqueira GC, Inglis DO, Skrzypek MS, Binkley J, Chibucos MC, Crabtree J, Howarth C, Orvis J, Shah P, et al. 2012. The Aspergillus Genome Database (AspGD): recent developments in comprehensive multispecies curation, comparative genomics and community resources. Nucleic Acids Res. 40:653–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh JM, Viktorova EG, Pilipenko EV. 2009. Proteasomes can degrade a significant proportion of cellular proteins independent of ubiquitination. J Mol Biol. 386:814–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyse DNA sequences. Nucleic Acids Res. 27:573–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert S, Laviolette F, Corbeil J. 2010. Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol. 17:1519–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmester A, Shelest E, Glöckner G, Heddergott C, Schindler S, Staib P, Heidel A, Felder M, Petzold A, Szafranski K, et al. 2011. Comparative and functional genomics provide insights into the pathogenicity of dermatophytic fungi. Genome Biol. 12:R7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Wang S, Sekimoto S, Aerts A, Choi C, Clum A, LaButti K, Lindquist E, Ngan CY, Ohm RA, et al. 2015. Phylogenomic analyses indicate that early fungi evolved digesting cell walls of algal ancestors of land plants. Genome Biol Evol. 7:1590–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy A, Severo MS, Sun R, Girke T, Gillespie JJ, Pedra JHF. 2013. Decoding the ubiquitin-mediated pathway of arthropod disease vectors. PLoS One 8:e78077.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C, Keane TM, Turner DJ, O’Keeffe G, Fitzpatrick DA, Doyle S. 2013. Genomic and proteomic dissection of the ubiquitous plant pathogen, Armillaria mellea: toward a new infection model system. J Proteome Res. 12:2552–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CA, Brown EJ. 2010. Cytosol as battleground: ubiquitin as a weapon for both host and pathogen. Trends Cell Biol. 20:205–213. [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. [DOI] [PubMed] [Google Scholar]
- Delport W, Poon AFY, Frost SDW, Kosakovsky Pond SL. 2010. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26:2455–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins CA, Sanscrainte ND, Goldberg JM, Heiman D, Young S, Zeng Q, Madhani HD, Becnel JJ, Cuomo CA. 2015. Contrasting host-pathogen interactions and genome evolution in two generalist and specialist microsporidian pathogens of mosquitoes. Nat Commun. 6:7121.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan JE, Henn MR, Ma L-J, Cuomo CA, Birren B. 2005. Genomics of the fungal kingdom: insights into eukaryotic biology. Genome Res. 15:1620–1631. [DOI] [PubMed] [Google Scholar]
- Goldstein G, Scheid M, Hammerling U, Boyse EA, Schlesinger DH, Niall HD. 1975. Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc Natl Acad Sci U S A. 72:11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb AM, Lichtwardt RW. 2001. Molecular variation within and among species of Harpellales. Mycologia 93:66–81. [Google Scholar]
- Grigg R, Lichtwardt RW. 1996. Isozyme patterns in cultured Harpellales. Mycologia 88:219–229. [Google Scholar]
- Guy L, Roat Kultima J, Andersson SGE. 2010. genoPlotR: comparative gene and genome visualization in R. Bioinformatics 26:2334–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 8:1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagai T, Levy Y. 2010. Ubiquitin not only serves as a tag but also assists degradation by inducing protein unfolding. Proc Natl Acad Sci U S A. 107:2001–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar AK, Foltz C, Finethy R, Piro AS, Feeley EM, Pilla-Moffett DM, Komatsu M, Frickel E-M, Coers J. 2015. Ubiquitin systems mark pathogen-containing vacuoles as targets for host defense by guanylate binding proteins. Proc Natl Acad Sci U S A. 112:E5628–E5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T. 1988. The kinase family: conserved protein phylogeny features and deduced domains of the catalytic. Science 241:42–52. [DOI] [PubMed] [Google Scholar]
- Hedges LM, Brownlie JC, O’Neill SL, Johnson KN. 2008. Wolbachia and virus protection in insects. Science 322:702.. [DOI] [PubMed] [Google Scholar]
- Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, Huhndorf S, James T, Kirk PM, Lücking R, et al. 2007. A higher-level phylogenetic classification of the Fungi. Mycol Res. 111:509–547. [DOI] [PubMed] [Google Scholar]
- Hoeppner MP, Lundquist A, Pirun M, Meadows JRS, Zamani N, Johnson J, Sundström G, Cook A, FitzGerald MG, Swofford R, et al. 2014. An improved canine genome and a comprehensive catalogue of coding genes and non-coding transcripts. PLoS One 9:e91172.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt RA, Broder S, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JMC, et al. 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298:129–149. [DOI] [PubMed] [Google Scholar]
- Horn BW. 1989. Requirement for potassium and pH shift in host-mediated sporangiospore extrusion from trichospores of Smittium culisetae and other Smittium species. Mycol Res. 93:303–313. [Google Scholar]
- Horn BW, Lichtwardt RW. 1981. Studies on the nutritional relationship of larval Aedes aegypti (Diptera: Culicidae) with Smittium culisetae (Trichomycetes). Mycologia 73:724–740. [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, et al. 2013. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496:498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, Cao J, Cheng J-F, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, et al. 2011. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 43:476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2004. Finishing the euchromatic sequence of the human genome. Nature 431:931–945. [DOI] [PubMed] [Google Scholar]
- James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, et al. 2006. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443:818–822. [DOI] [PubMed] [Google Scholar]
- Jiang X, Chen ZJ. 2012. The role of ubiquitylation in immune defence and pathogen evasion. Nat Rev Immunol. 12:35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller O, Kollmar M, Stanke M, Waack S. 2011. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics 27:757–763. [DOI] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Kuo A, Nagy LG, Morin E, Barry KW, Buscot F, Canbäck B, Choi C, Cichocki N, Clum A, et al. 2015. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat Genet. 47:410–415. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305:567–580. [DOI] [PubMed] [Google Scholar]
- Kumar S, Jones M, Koutsovoulos G, Clarke M, Blaxter M. 2013. Blobology: exploring raw genome data for contaminants, symbionts and parasites using taxon-annotated GC-coverage plots. Front Genet. 4:237.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar TA. 2013. CFSSP: Chou and Fasman Secondary Structure Prediction server. Wide Spectr. 1:15–19. [Google Scholar]
- Kutsenko A, Svensson T, Nystedt B, Lundeberg J, Björk P, Sonnhammer E, Giacomello S, Visa N, Wieslander L. 2014. The Chironomus tentans genome sequence and the organization of the Balbiani ring genes. BMC Genomics 15:819.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvist S, Min GS, Siddall ME. 2013. Diversity and selective pressures of anticoagulants in three medicinal leeches (Hirudinida: Hirudinidae, Macrobdellidae). Ecol Evol. 3:918–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtwardt RW, Ferrington LC, Lastra CL. 1999. Trichomycetes in Argentinean aquatic insect larvae. Mycologia 91:1060. [Google Scholar]
- Ma L-J, Ibrahim AS, Skory C, Grabherr MG, Burger G, Butler M, Elias M, Idnurm A, Lang BF, Sone T, et al. 2009. Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole-genome duplication. PLoS Genet. 5:e1000549.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG. 2009. A single regulatory gene is sufficient to alter bacterial host range. Nature 458:215–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran N, Jarvik T. 2010. Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science 328:624–627. [DOI] [PubMed] [Google Scholar]
- Moran NA. 2007. Symbiosis as an adaptive process and source of phenotypic complexity. Proc Natl Acad Sci U S A. 104:8627–8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya A, Peretó J, Gil R, Latorre A. 2008. Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat Rev Genet. 9:218–229. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 3:418–426. [DOI] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi Z, Megy K, Grabherr M, et al. 2007. Genome sequence of Aedes aegypti, a major arbovirus vector. Science 316:1718–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23:1061–1067. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 8:785–786. [DOI] [PubMed] [Google Scholar]
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25:1253–1256. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjo H, Kawait T, Akira S. 1998. DRAKs, novel serine/threonine kinases related to death-associated protein kinase that trigger apoptosis. J Biol Chem. 273:29066–29071. [DOI] [PubMed] [Google Scholar]
- Schéele S, Nyström A, Durbeej M, Talts JF, Ekblom M, Ekblom P. 2007. Laminin isoforms in development and disease. J Mol Med. 85:825–836. [DOI] [PubMed] [Google Scholar]
- Selman M, Pombert J-F, Solter L, Farinelli L, Weiss LM, Keeling P, Corradi N. 2011. Acquisition of an animal gene by microsporidian intracellular parasites. Curr Biol. 21:576–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severo MS, Sakhon OS, Choy A, Stephens KD, Pedra JHF. 2013. The “ubiquitous” reality of vector immunology. Cell Microbiol. 15:1070–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staats CC, Junges A, Guedes RLM, Thompson CE, de Morais GL, Boldo JT, de Almeida LGP, Andreis FC, Gerber AL, Sbaraini N, et al. 2014. Comparative genome analysis of entomopathogenic fungi reveals a complex set of secreted proteins. BMC Genomics 15:822.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strongman DB, Wang J, Xu S. 2010. New trichomycetes from western China. Mycologia 102:174–184. [DOI] [PubMed] [Google Scholar]
- Sweeney AW. 1981. An undescribed species of Smittium (Trichomycetes) pathogenic to mosquito larvae in Australia. Trans Br Mycol Soc. 77:55–60. [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpl R, Rohde H, Robey PG, Rennard SI, Foidart J-M, Martin GR. 1979. Laminin-a glycoprotein from basement membranes. J Biol Chem. 254:9933–9937. [PubMed] [Google Scholar]
- Tretter ED, Johnson EM, Benny GL, Lichtwardt RW, Wang Y, Novak SJ, Smith JF. 2014. An eight-gene molecular phylogeny of the Kickxellomycotina, including the first phylogenetic placement of Asellariales. Mycologia 106:912–935. [DOI] [PubMed] [Google Scholar]
- Tretter ED, Johnson EM, Wang Y, Kandel P, White MM. 2013. Examining new phylogenetic markers to uncover the evolutionary history of early-diverging fungi: comparing MCM7, TSR1 and rRNA genes for single- and multi-gene analyses of the Kickxellomycotina. Persoonia 30:106–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay SK, Mahajan L, Ramjee S, Singh Y, Basir SF, Madan T. 2009. Identification and characterization of a laminin-binding protein of Aspergillus fumigatus: extracellular thaumatin domain protein (AfCalAp). J Med Microbiol. 58:714–722. [DOI] [PubMed] [Google Scholar]
- Valle LG, Cafaro MJ. 2010. First report of Harpellales from the Dominican Republic (Hispaniola) and the insular effect on gut fungi. Mycologia 102:363–373. [DOI] [PubMed] [Google Scholar]
- Valle LG, Santamaria S. 2004. The genus Smittium (Trichomycetes, Harpellales) in the Iberian Peninsula. Mycologia 96:682–701. [PubMed] [Google Scholar]
- Wang Y, Tretter ED, Johnson EM, Kandel P, Lichtwardt RW, Novak SJ, Smith JF, White MM. 2014. Using a five-gene phylogeny to test morphology-based hypotheses of Smittium and allies, endosymbiotic gut fungi (Harpellales) associated with arthropods. Mol Phylogenet Evol. 79:23–41. [DOI] [PubMed] [Google Scholar]
- Wang Y, Tretter ED, Lichtwardt RW, White MM. 2013. Overview of 75 years of Smittium research, establishing a new genus for Smittium culisetae, and prospects for future revisions of the “Smittium” Clade. Mycologia 105:90–111. [DOI] [PubMed] [Google Scholar]
- Welchman RL, Gordon C, Mayer RJ. 2005. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 6:599–609. [DOI] [PubMed] [Google Scholar]
- White MM. 2006. Evolutionary implications of a rRNA-based phylogeny of Harpellales. Mycol Res. 110:1011–1024. [DOI] [PubMed] [Google Scholar]
- White MM, Lichtwardt RW. 2004. Fungal symbionts (Harpellales) in Norwegian aquatic insect larvae. Mycologia 96:891–910. [DOI] [PubMed] [Google Scholar]
- White MM, Siri A, Lichtwardt RW. 2006. Trichomycete insect symbionts in Great Smoky Mountains National Park and vicinity. Mycologia 98:333–352. [DOI] [PubMed] [Google Scholar]
- Williams MC. 1983. Zygospores in Smittium culisetae (Trichomycetes) and observations on trichospore germination. Mycologia 75:251–256. [Google Scholar]
- Williams MC. 2001. Trichomycetes a brief review of research In: Misra JK, Horn B, editors. Trichomycetes and other fungal groups. Enfield, NH: Science Publishers Inc; p. 19. [Google Scholar]
- Zhao S, Ulrich HD. 2010. Distinct consequences of posttranslational modification by linear versus K63-linked polyubiquitin chains. Proc Natl Acad Sci U S A. 107:7704–7709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin AV, Delcher AL, Florea L, Kelley DR, Schatz MC, Puiu D, Hanrahan F, Pertea G, Van Tassell CP, Sonstegard TS, et al. 2009. A whole-genome assembly of the domestic cow, Bos taurus. Genome Biol. 10:R42.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.