Abstract
We have identified a fixed nonsynonymous sequence difference between humans (Val381; derived variant) and Neandertals (Ala381; ancestral variant) in the ligand-binding domain of the aryl hydrocarbon receptor (AHR) gene. In an exome sequence analysis of four Neandertal and Denisovan individuals compared with nine modern humans, there are only 90 total nucleotide sites genome-wide for which archaic hominins are fixed for the ancestral nonsynonymous variant and the modern humans are fixed for the derived variant. Of those sites, only 27, including Val381 in the AHR, also have no reported variability in the human dbSNP database, further suggesting that this highly conserved functional variant is a rare event. Functional analysis of the amino acid variant Ala381 within the AHR carried by Neandertals and nonhuman primates indicate enhanced polycyclic aromatic hydrocarbon (PAH) binding, DNA binding capacity, and AHR mediated transcriptional activity compared with the human AHR. Also relative to human AHR, the Neandertal AHR exhibited 150–1000 times greater sensitivity to induction of Cyp1a1 and Cyp1b1 expression by PAHs (e.g., benzo(a)pyrene). The resulting CYP1A1/CYP1B1 enzymes are responsible for PAH first pass metabolism, which can result in the generation of toxic intermediates and perhaps AHR-associated toxicities. In contrast, the human AHR retains the ancestral sensitivity observed in primates to nontoxic endogenous AHR ligands (e.g., indole, indoxyl sulfate). Our findings reveal that a functionally significant change in the AHR occurred uniquely in humans, relative to other primates, that would attenuate the response to many environmental pollutants, including chemicals present in smoke from fire use during cooking.
Keywords: Neandertal, Denisovan, Ah receptor, AHR, benzo(a)pyrene.
Introduction
The aryl hydrocarbon receptor (AHR) was initially identified as the receptor that bound with high affinity 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD or dioxin). The AHR is the only member of the bHLH-PAS transcription factor family that is activated through binding ligands (Beischlag, et al. 2008). The unliganded AHR resides in the cytoplasm bound to heat shock protein 90, X-associated protein 2 and p23. Upon ligand activation, the AHR complex translocates into the nucleus where a fraction of the receptor pool heterodimerizes with the Ah receptor nuclear translocator (ARNT). The AHR/ARNT heterodimer binds to dioxin responsive elements and induces the expression of genes involved in xenobiotic metabolism. The AHR is the predominant regulator, in conjunction with its dimerization partner ARNT, of CYP1A1/1A2/1B1 expression. Numerous hydrophobic planar chemicals in the environment can activate the AHR and induce phase one metabolism of these activators, as a means to enhance clearance. Polycyclic aromatic hydrocarbons (PAHs) are major products formed from partial combustion of organic matter (e.g., wood) and are characterized agonists for the AHR. PAH exposure through inhalation, dermal, or dietary routes can lead to AHR-mediated induction of CYP1A1 and CYP1B1, which results in the formation of hydroxylated and reactive epoxide containing metabolites (Gelboin 1980). These latter metabolites at sufficient concentrations can cause overt toxicity and death in a CYP1A1-dependent fashion (Uno, et al. 2001).
Although the AHR is often regarded as a xenobiotic receptor, it is becoming increasingly clear this receptor exhibits activity influencing numerous endogenous functions, including energy metabolism, cell cycle, lipid metabolism, and immune function (Esser and Rannug 2015; Tian, et al. 2015). In particular, the AHR plays an important role in both barrier function and immune surveillance in epithelial barrier tissues in skin and the gastrointestinal tract. The ability of commensal yeast and bacteria to produce AHR ligands suggests that the host AHR has evolved to respond to the presence of the microbiota resulting in modulation of immune function (Gaitanis, et al. 2008; Zelante, et al. 2013). In the gastrointestinal tract, activation of the AHR by dietary ligands during organogenesis enhances intestinal lymphoid follicle development (Kiss, et al. 2011). Studies in Ahr−/− mice point to a role for the AHR in the maintenance of normal homeostasis in a variety of physiological pathways (Gonzalez and Fernandez-Salguero 1998). These observations would suggest that activation of AHR by endogenous and dietary ligands would be beneficial and likely conserved through evolution.
The rodent AHR homolog (e.g., AHRb in mice) is known to bind exogenous ligands such as TCDD and PAHs with ∼10-fold higher affinity than the human AHR (Harper, et al. 1988; Ramadoss and Perdew 2004). This difference has been attributed to an amino acid difference in the ligand-binding pocket, which is a valine at position 381 in human AHR instead of an alanine residue as observed in rodents at this position (Ramadoss and Perdew 2004). These studies led to the concept that the human AHR possesses a global loss of ligand affinity relative to the rodent homolog. However, more recent reports have discovered that the human AHR has relatively higher affinity for a number of tryptophan-derived ligands, such as indirubin, indoxyl sufate, indole, and kynurenic acid (Flaveny, et al. 2009; DiNatale, Murray, et al. 2010; Schroeder, et al. 2010; Hubbard, et al. 2015). These latter studies would suggest that the ligand-binding pocket between rodents and humans exhibit differential ligand selectivity. The cause of differential AHR ligand selectivity between these species and the possible selective pressures that have led to these differences is not known.
Interestingly, studies examining fish populations within highly polluted habitats provide strong evidence for the notion that vertebrates can adapt to evolutionary pressure by persistent exposure to toxic environmental AHR ligands. The industrial release of highly concentrated planar polychlorinated biphenyls (PCBs), known to be potent AHR ligands, into the Hudson River has resulted in populations of Atlantic Tomcod that exhibit dramatically reduced ligand-mediated AHR transcriptional activity (Yuan, et al. 2006). Mechanistic studies revealed that 99% of the Hudson River Tomcod have an AHR-2 gene containing a 6 base pair deletion within the ligand-binding domain (LBD), that appears to explain the reduction in AHR activation (Wirgin, et al. 2011). Similarly, Atlantic killifish populations from PCB contaminated habitats also show significantly attenuated AHR-mediated transcriptional activity (Bello, et al. 2001). These studies illustrate that toxic AHR ligands can mediate rapid change within a relatively small number of generations. In the fish examples discussed above, the evolutionary processes were likely driven by acute embryo mortality under conditions of AHR ligand overexposure.
Here we examine the molecular evolution of the AHR in primates and, more specifically, compare the transcriptional activity of Neandertal and human AHR. Analyses of AHR nucleotide sequences in various primate species revealed that the encoded valine at residue 381 that mediates low affinity for PAHs is present only in modern humans. Yet, our functional experiments demonstrate that the human AHR retains the sensitivity to endogenous ligand(s) observed in other primate AHR. These observations indicate that the human AHR has a reduced affinity for exogenous AHR ligands, perhaps conferring tolerance to environmental AHR ligand exposures.
Results
The Human AHR Contains a V381 Residue that is Unique among Primates Examined
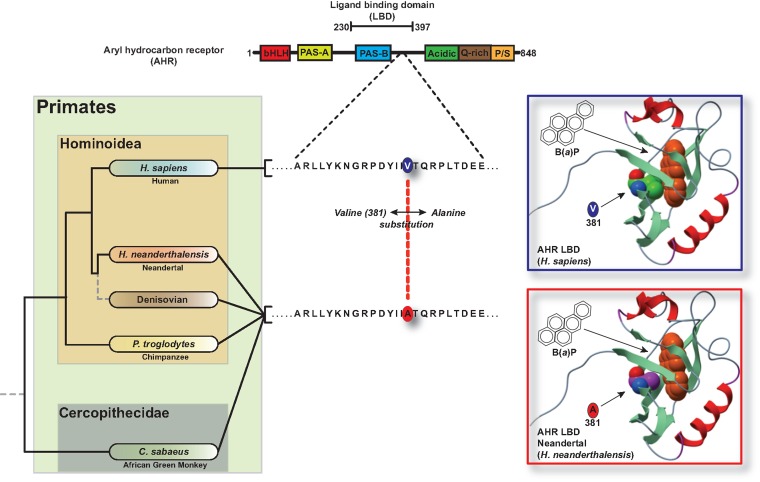
Previous studies from our laboratory have identified the location of a single amino acid residue at position 381 within the human AHR ligand-binding domain that is permissive for high or low exogenous ligand binding affinity (Ema, et al. 1994; Ramadoss and Perdew 2004). The human AHR gene encodes V381, resulting in a 10-fold reduction in the affinity of the AHR for a radiolabeled dioxin analog, compared with the A381 variant observed in mouse. These observations prompted an examination of AHR sequences from archaic hominins and nonhuman primates to examine the phylogenetic history of diminished PAH sensitivity. We first considered a database of genome-wide gene coding region (exome) variants among population samples of humans (n = 9 individuals or n = 18 diploid chromosomes), Neandertals (n = 3 individuals; n = 6 chromosomes), and one Denisovan individual (an archaic hominin population more closely related to Neandertals than modern humans; n = 2 chromosomes), based on paleogenomic sequence data (Meyer, et al. 2012; Castellano, et al. 2014; Prufer, et al. 2014). We found that, first, whereas the AHR is highly conserved overall (supplementary fig. S1, Supplementary Material online), Neandertals and the Denisovan individual are fixed for the ancestral variant at codon 554 (predicted amino acid K554), whereas humans are variable at this position (R554 or K554) (Wong, et al. 2001). Currently, no functional consequence has been ascribed to the R↔K554 polymorphism in humans. Second, and most interestingly, based on the presently available paleogenomic sequence data, Neandertals and Denisovans are fixed for the ancestral variant at codon 381 (A381) whereas humans are fixed for a derived variant at this position (V381) (fig. 1). In fact, this amino acid residue is invariant among any human whose genome has yet been sequenced (e.g., via the 1000 Genomes project, or reported in the dbSNP database) and the oldest (45, 000 y) anatomically modern human individual sequenced to date is also homozygous for the variant that encodes V381 (Fu, et al. 2014). This situation is unusual across the genome. In the Neandertal and Denisovan paleogenomic database (Castellano, et al. 2014), there are only 90 total sites across the exome for which the archaic hominin sample (n = 8 chromosomes) is fixed for an ancestral variant and the modern human sample (n = 18 chromosomes) is fixed for a derived nonsynonymous variant (supplementary table S1, Supplementary Material online). Of those sites, only 27, including the variant that encodes V381 in the AHR, also have no reported dbSNP variability (table 1 and supplementary table S2, Supplementary Material online). Furthermore, the AHR was only protein on this list that is involved in adaptation to environmental stimuli (e.g., xenobiotic metabolism).
Fig. 1.
Human AHR ligand-binding domain segregates from other primates through a single amino acid substitution. (A) Map of AHR functional domains and the single amino acid variation in the human AHR relative to other Hominoidea. (B) Homology modeling of the orientation of benzo(a)pyrene in the ligand binding pocket of the human and Neandertal AHR.
Table 1.
Exomes that are fixed for a derived nonsynonymous variant in humans compared with Neandertal/Denisovan that also exhibit no reported dbSNP variability.
| Gene | Peptide | Ref/ | Gene | Peptide | Ref/ |
|---|---|---|---|---|---|
| Alt Bases | Alt Bases | ||||
| SPAG 17 | Sperm associated antigen 17 | T/C | CCDC15 | Coiled-coil domain containing 15 | A/C |
| SPAG17 | Sperm associated antigen 17 | A/C | PRDM10 | PR domain containing 10 | T/G |
| SLC27A3 | Solute carrier family 27 member 3 | G/T | NOVA1 | Neuro-oncological ventral antigen 1 | C/T |
| ZBTB24 | Zinc finger and BTB domain containing 24 | C/G | GPR132 | G protein-coupled receptor 132 | C/G |
| AHR | Aryl hydrocarbon receptor | T/C | KIAA1199 | Cell migration inducing protein, hyaluronan binding | G/A |
| DNAH11 | Dynein, axonemal, heavy chain 11 | T/C | CDH16 | Cadherin 16, KSP-cadherin | T/C |
| ADAM18 | ADAM metallopeptidase domain 18 | G/A | SPAG5 | Sperm associated antigen 5 | A/G |
| RB1CC1 | RB1-inducible Coiled-Coil 1 | C/T | SSH2 | Slingshot protein phosphatase 2 | T/C |
| NEK6 | NIMA-related kinase 6 | C/G | RFNG | RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase | G/A |
| NR6A1 | Nuclear receptor subfamily 6, group A, member 1 | T/G | KIAA1772 | Growth regulation by estrogen in breast cancer-like | A/G |
| C9orf86 | RAB, member RAS oncogene family-like 6 | C/G | C19orf28 | Major facilitator superfamily domain containing 12 | C/G |
| ARRDC1 | Arrestin domain containing 1 | T/C | RSPH1 | Radial spoke head 1 homolog (chlamydomonas) | G/T |
| FAM178A | SMC5-SMC6 complex localization factor 2 | G/A | ADSL | Adenylosuccinate lyase | T/C |
| DCHS1 | Dachsous cadherin-related 1 | C/T |
The Human AHR Exhibits Reduced Activation by PAHs Compared with the Neandertal Receptor
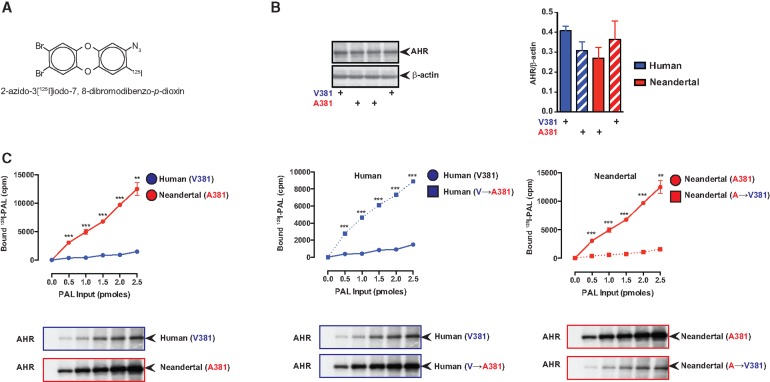
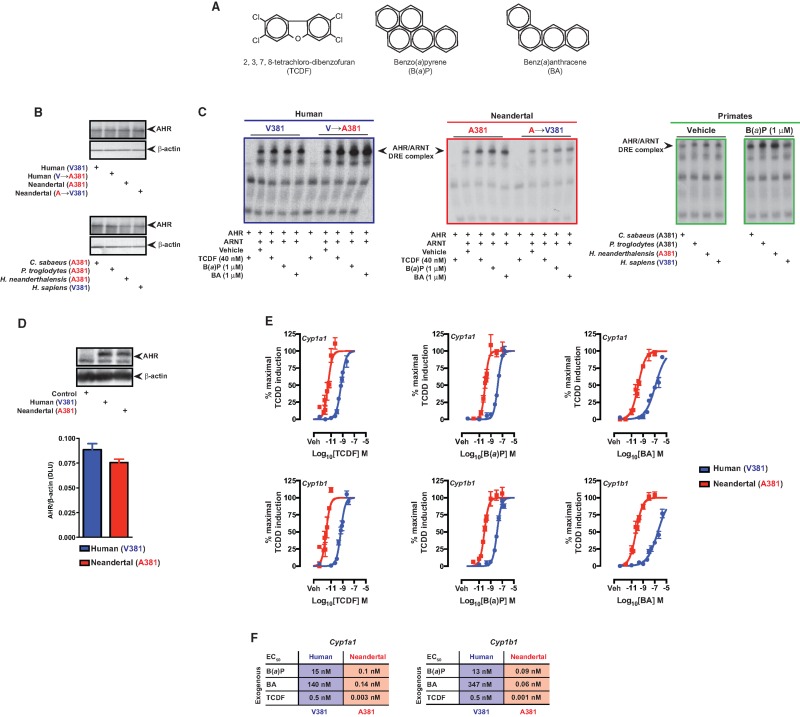
The archaic hominin A381 variant, located within the AHR LBD, is shared with nonhuman primates and other vertebrates, for example, rodents, which exhibit enhanced TCDD and PAH binding compared with humans (Harper, et al. 1988; Poland, et al. 1994; Ramadoss and Perdew 2004). It is important to point out that there are a number of nonconserved amino acid differences within the human compared with mouse AHR LBD (supplementary fig. S2, Supplementary Material online). In silico molecular modeling of the Neandertal AHR LBD in complex with B(a)P indicates that A381 is centrally localized in the ligand-binding pocket and may be more permissive for B(a)P binding than the human V381 counterpart (fig. 1B). To empirically examine ligand binding to the Neandertal and human AHR, cell-based ligand binding assays were performed (fig. 2). AHR-deficient COS-1 cells, upon transfection with AHR expression constructs expressed equal amounts of human or Neandertal AHR (fig. 2A). Results revealed enhanced covalent binding of photo-affinity ligand (PAL), 2-azido-3[125I]iodo-7,8-dibromodibenzo-p-dioxin (fig. 2B) to Neandertal AHR relative to human (fig. 2C). This radioligand is structurally similar to the widespread environmental contaminants chlorinated dibenzo-p-dioxins and dibenzofurans. Reciprocal mutation of V↔A381 within human/Neandertal AHR confirmed A381 as the sole determinant for enhanced ligand binding by Neandertal and human AHR (fig. 2C). Due to the high radiospecific activity of the photoaffinity ligand the amount of ligand used was significantly below the level that would be needed to saturate the LBD. Ligand binding to AHR initiates a process of receptor transformation, which facilitates nuclear translocation and dimerization with ARNT. Association of AHR/ARNT promotes binding to cognate DNA response elements within the regulatory regions of AHR target genes, including those responsible for PAH biotransformation, that is, CYP1A1/1B1. Polycyclic aromatic ligands ([2,3,7,8-tetrachlorodibenzofuran; TCDF], B(a)P and benz(a)anthracene) (fig. 3A) stimulated transformation of the AHR and subsequent DNA binding using equivalent amounts of in vitro translated human and Neandertal AHR (fig. 3B). In addition, their respective reciprocal V↔A381 mutants were also investigated by electrophoretic mobility shift assays (fig. 3C). Regardless of species, the presence of A381 (Neandertal or mutated human) resulted in enhanced DNA binding following exposure to each ligand. Conversely, V381 in each species (mutated Neandertal or human) resulted in diminished DNA binding. Further examination of B(a)P stimulated DNA binding by equivalent amounts of AHR expressed by various members of the order Primate identified a pattern of elevated DNA binding in those species encoding A381 (i.e., Cercopithecus sabeus, Pan troglodytes, and Neandertal) not observed in humans (fig. 3C and fig. 2B). Examination of the contribution of the V↔A381 fixed difference between human and Neandertal AHR to PAH-induced transcriptional activity was performed by dose-responsive quantitative polymerase chain reaction (PCR) analysis of Cyp1a1/1b1 expression in transiently transfected AHR-deficient BP8 cells that exhibited equal AHR expression levels (fig. 3D and E). AHR deficient BP8 cells were selected for these experiments because there is essentially no induction of AHR target genes in cells that are not transfected with AHR expression constructs (supplemental fig. S3, Supplementary Material online). Transcriptional activation of Cyp1a1/1b1 proved dose-responsive with each polycyclic aromatic ligand examined in the context of human and Neandertal AHR expression. However, Neandertal AHR exhibited at least a two orders of magnitude lower sensitivity to ligand-mediated induction of Cyp1a1/1b1 over human AHR, as evidenced by a decrease in half maximal effective concentration (EC50) values (fig. 3F). The relative levels of Cyp1a1 and Cyp1b1 induced by the Neandertal and human AHR were similar upon exposure to a 10 nM dose of TCDD, which yielded a maximal level of induction (supplementary fig. S4, Supplementary Material online).
Fig. 2.
Neandertal AHR exhibits enhanced dioxin analog binding relative to the human AHR. (A) AHR expression in COS1 cells was measured by western blot analysis. (B) Structure of the photoaffinity ligand. (C) COS1 cells transiently expressing either the Neandertal or human AHR were incubated with increasing concentration of 2-azido-3-[125I]iodo-7,8-dibromodibenzo-p-dioxin (PAL), exposed to UV light and cellular extracts analyzed by SDS-PAGE. Radioactive AHR bands were excised and the amount of radioactivity quantitated.
Fig. 3.
Comparative analysis of PAH mediated DNA binding potential and transcriptional activity of the Neandertal and human AHR. (A) Structures of AHR ligands tested. (B) AHR expression in in vitro translations was measured by western blot analysis. (C) In vitro translated AHR from the appropriate species and ARNT were utilized in electrophoretic mobility shift assays to assess AHR ligand-mediated AHR/ARNT/DNA complex formation. (D) AHR expression in transfected BP8 cells was measured by western blot analysis. (E) Dose-dependent induction of Cyp1a1 and Cyp1b1 expression mediated by the Neandertal or human AHR in transfected BP8 cells. (F) Summary of the EC50 values for the dose-response experiments in panel C.
The Human and Neandertal AHR Exhibit Similar levels of Activation by Endogenous Ligands
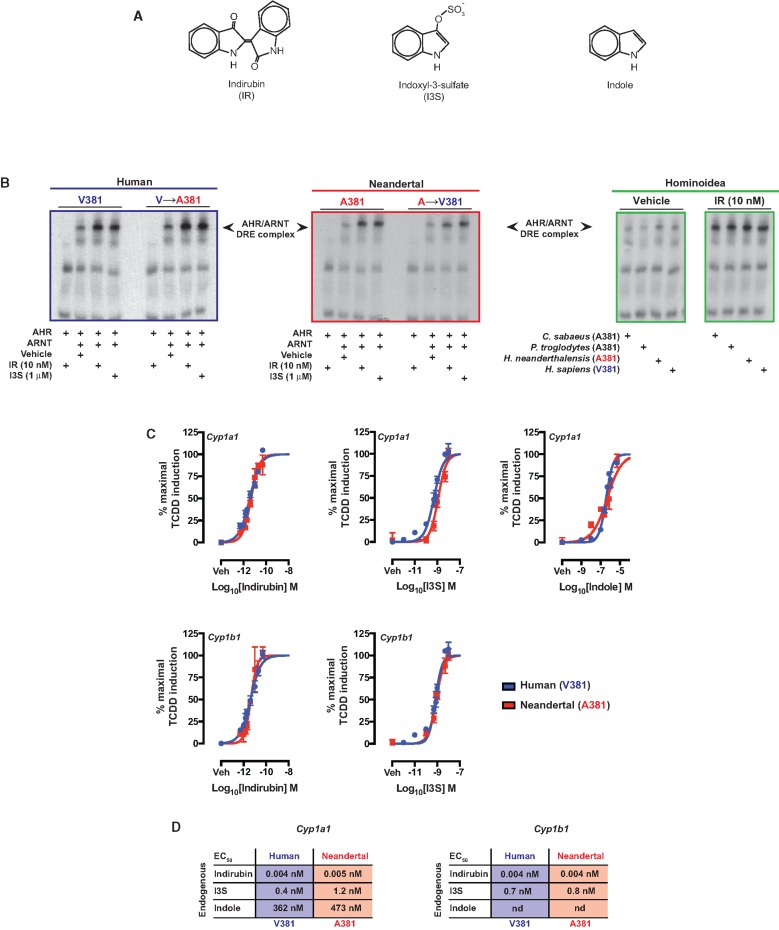
As a xenobiotic sensor, the AHR is activated by many polycyclic aromatic ligands and other xenobiotics, however, the increasing number of physiological functions attributed to AHR is activated by endogenous and diet-derived ligands, such as kynurenic acid, indirubin, indoxyl-3-sulfate, and indole. We therefore examined whether the adaptive A→V381 desensitization to polycyclic aromatic ligands observed in human AHR also impacted sensitivity to endogenous ligands, compared with the AHR in Neandertal and other nonhuman primates. Studies using indirubin, indoxyl-3-sulfate, and indole (fig. 4A), previously reported to exhibit enhanced binding to human AHR over mouse AHR (Flaveny, et al. 2009; Hubbard, et al. 2015), revealed no apparent difference between human and Neandertal AHR in their capacity to associate with ARNT and facilitate DNA binding in response to either indirubin, indole, or indoxyl-3-sulfate (fig 4B). Reciprocal mutations V↔A381 in each species proved refractory, failing to impact the previously observed DNA binding capacity (fig. 4B). Furthermore, the DNA binding capacity of other A381 encoding Hominoidea (C. sabeus and P. troglodytes) proved similar to that exhibited by human and Neandertal in response to indirubin, suggesting that the polycyclic aromatic ligands sensitivity determinant V/A381 is not critical to establishing endogenous ligand sensitivity. Examination of the contribution of the V↔A381 polymorphic nature of human and Neandertal AHR to endogenous ligand-induced transcriptional activity was performed by dose-responsive quantitative PCR analysis of Cyp1a1/1b1 expression in transiently transfected AHR-deficient cells (fig. 4C). Expression of Cyp1a1/1b1 proved dose-responsive with each endogenous ligand (e.g., indirubin, indole and indoxyl-3- sulfate) in the context of human and Neandertal AHR. Both human and Neandertal AHR exhibited near identical ligand mediated induction of Cyp1a1/1b1, as evidenced by similar EC50 (fig. 4D).
Fig. 4.
Comparative analysis of endogenous ligand mediated DNA binding potential and transcriptional activity of the Neandertal and human AHR. (A) Structures of AHR ligands tested. (B) In vitro translated AHR from the appropriate species and ARNT were utilized in electrophoretic mobility shift assays to assess AHR ligand-mediated AHR/ARNT/DNA complex formation. (C) Dose-dependent induction of Cyp1a1 and Cyp1b1 expression mediated by the Neandertal or human AHR in transfected BP8 cells. (D) Summary of the EC50 values for the dose-response experiments in panel C.
Discussion
Depending on the polycyclic aromatic ligand tested, Neandertal AHR induced Cyp1a1 activity between 150-fold and 1000-fold lower EC50 than that observed with the human AHR. Importantly, our cell transfection experiments showed that the marked difference in transcriptional activity was solely attributable to a switch from alanine to valine 381 in the LBD of human AHR. The AHR LBD in mice encodes A375, which is homologous to A381 in Neandertal and other nonhuman primates. However, quantitative ligand binding studies reveal only an order of magnitude difference in binding between A375 mouse AHR and V381 of the human AHR (Harper, et al. 1988). Such discrepancies suggest that ligand binding/affinity is not the sole determinant dictating AHR transcriptional activity; therefore we speculate that additional factors, such as receptor transformation potential, nuclear translocation, efficiency, and protein stability, may also have an effect in determining net transcriptional activity. Increased PAH binding and DNA binding, in conjunction with enhanced Cyp1a1/1b1 expression, provides evidence that Neandertals, expressing the A381 AHR variant, may have been more susceptible than modern humans to exogenous AHR ligand mediated toxicities. Thus, modern humans or their immediate ancestors acquired and retained a specific loss of sensitivity to exogenous ligand-mediated activation of the AHR acquired through mutation of A→V381 during the course of hominin evolution. Importantly, the V381 variant is fixed among all modern humans within the available genome sequence data, despite evidence of human-Neandertal admixture and some introgression of Neandertal alleles into the gene pools of non-African modern humans (Vattathil and Akey 2015), raising the possibility of a selective advantage associated with the V381 AHR variant.
Given the loss of sensitivity to exogenous ligands in humans, it is probable that the metabolic processes governed by the AHR would be altered. It is evident that humans are capable of effectively metabolizing PAHs leading to metabolic elimination, such as is the case for smokers. However, a greater degree of induction of CYP1A1 metabolism in nonhuman primates may lead to a relatively high level of toxic intermediates, without necessarily achieving a greater overall clearance rate, leading to toxic outcomes. Therefore, we postulate that exposure to sources of potentially toxic environmental AHR ligands may have been associated with strong selection pressure for mutations affecting the relative desensitization of AHR to exogenous ligands.
The precise evolutionary origin of the highly conserved A→V381 AHR mutation is difficult to assess due to the current absence of anatomically modern human lineage paleogenomic sequences prior to 45,000 ya. Nevertheless, the AHR underwent a change within its LBD, perhaps facilitating a degree of resistance to the consequences of environmental exposures. In contrast, the similar sensitivity to endogenous ligands within Hominoidea suggests that human AHR responsiveness to these ligands was acquired prior to divergence of the primate lineage and has persisted ever since; perhaps supporting the pivotal role for endogenous ligand-mediated AHR physiological functions in primates, including humans. This fact also supports the high level of specificity in the mutation that actually occurred. Furthermore, these data could represent one of the first examples of a functional evolutionary adaptation to environmental stress in humans. The potential fitness effects associated with fixation of the V381 variant in humans are not directly known and would be impossible to address with the tools currently available, especially considering that all humans have the V381 AHR variant. Nevertheless, one plausible explanation is that this mutation provides a degree of protection against the deleterious effects of toxic environmental AHR ligand exposure.
Many of the genetic, environmental, and sociocultural factors that shaped the evolution of the genus Homo, particularly Homo sapiens (humans), have not been firmly established. Potential sources of environmental AHR ligands include those associated with diet (e.g., plant metabolites, cooked foods), and those associated with combustion products from controlled or uncontrolled fire (e.g., cooking fires, wildfires, tar pits). Importantly, the ability to control fire is considered one of the seminal innovations of human prehistory. The use of fire results in exposure to smoke that is rich in AHR ligands, such as PAHs. The earliest occurrences of controlled fire are associated with Homo erectus (sensu lato) in Eurasia and Africa, and date to at least 250 ka and possibly as old as 760 ka or more (Hallegouet, et al. 1992; Goren-Inbar, et al. 2004; Gowlett and Wrangham 2013). The common ancestors of Homo neanderthalensis (Neandertals) and humans, living approximately 300–500 ka (Endicott, et al. 2010), were probably able to control fire (Gowlett and Wrangham 2013). The controlled uses of fire likely expanded the hominin niche, with uses for warmth, cooking, the heat treatment of tools and making pigments, and potentially landscape modification (Hovers, et al. 2003; Brown, et al. 2009; Parker, et al. 2016). Localization of fire in caves was common, indicating a partly troglodytic existence, and placement of central hearths in the depths of caves appears to have been associated with intensity of cooking, flint knapping, and animal processing activities (Hardy, et al. 2016). Evidence of microfragments of charcoal in Lower Paleolithic hominin dental calculus is consistent with smoke inhalation, and suggests that smoke remediation in caves and other structures may have been a problem (Hardy, et al. 2016; Monge, et al. 2015). It has been theorized that cooking improved nutrient availability that may have led to an increase in brain volume and improve fitness early in human evolution (Wrangham and Conklin-Brittain 2003; Fonseca-Azevedo and Herculano-Houzel 2012).
The routine use of controlled fire carried a cost. Smoke derived from the incomplete combustion of wood and other organic material generates particulates containing a multitude of irritants and toxic chemicals, including PAHs, which at high concentrations can result in acute toxic responses and subsequent chronic toxicities, leading to morbidity and exacerbation of comorbidities (Zelikoff, et al. 2002). Among morbidities associated with chronic smoke inhalation are acute respiratory infection and a high risk of low-birth weight and infant mortality due to maternal exposure (Smith, et al. 2000). Indeed, exposure to AHR ligands can mediate adaptive immune suppression in part through induction of Treg cells, which may increase susceptibility to viral or bacterial infections (Huang, et al. 2013; Bruhs, et al. 2015; Yang, et al. 2016). In addition, PAH exposure can mediate an increased time to pregnancy, reduced spermatogenesis, and increased apoptosis of oocytes (MacKenzie and Angevine 1981; Matikainen, et al. 2001; Revel, et al. 2001; Esakky and Moley 2016). Many PAHs elicit cellular toxicity through damaging electrophilic intermediates generated as a consequence of metabolism. The prototypical example of such a PAH is benzo(a)pyrene (BaP), which is capable of inducing its own metabolism to generate highly reactive B(a)P epoxides. Upon respiratory and dermal exposure to excessive smoke-derived PAHs, AHR/ARNT-mediated CYP1A1/1B1 expression is elevated to supraphysiological levels, resulting in relatively high concentrations of toxic PAH metabolic intermediates. Excessive rapid metabolism of PAHs to reactive intermediates can result in cellular necrosis and death in rodents (Uno, et al. 2001).
Thus, it is probable that Middle/Upper Paleolithic hominins, who used fire extensively in the absence of smoke remediation, may have been subject to persistent respiratory/dermal/dietary PAH exposure and consequently were at high risk of acquiring PAH-mediated reproductive/developmental/respiratory toxicities, possibly affecting individual fitness. It is very likely that both humans and Neandertals controlled and used fire. However, under these shared environmental conditions, the AHR V381 mutation that we describe and characterize in this study may have represented a gene-culture evolutionary advantage for humans. Whether the carriers of this derived mutation acquired tolerance to environmental AHR ligands to a degree that led to a selective advantage and the ultimate fixation of this allele among modern humans will be difficult to determine. Nonetheless, our observation that during hominin evolution the human AHR acquired a mutation linked to diminished PAHs binding and other exogenous AHR ligands (e.g., halogenated dioxins) that has persisted and become fixed in the modern human lineage is supportive of this concept.
Materials and Methods
Reagents
TCDF was obtained from AccuStandard. 1,2-benz(a)anthracene and benzo(a)pyrene were purchased from Sigma. Indoxyl-3-sulfate and indirubin were purchased from Alfa Aesar and Enzo Life Sciences, respectively. Indole was purchased from Sigma and purified as previously described (Hubbard, et al. 2015). The photoaffinity ligand 2-azido-3-[125I]iodo-7,8-dibromodibenzo-p-dioxin was synthesized as described (Poland, et al. 1986). Cultured supernantant from monoclonal antibody hybridoma clone RPT1 was produced as previously described (Perdew, et al. 1995). COS-1 and BP8 cells were obtained from American Type Culture collection (Manassas, VA) and Martin Göttlicher (Institute of Toxicology and Genetics, Eggenstein, Germany), respectively. BP8 cells are deficient in AHR expression and were generated as described (Weiss, et al. 1996).
Plasmids and Mutagenesis
The H. sapiens, P. trogloytes, and C. sabaeus AHR cDNA optimized for mammalian codon use and minimal secondary mRNA structure were synthesized by GenScript (Piscataway, NJ) and were inserted into pcDNA3. The nucleotide sequences for these cDNAs are shown in supplementary figure S5, Supplementary Material online. The plasmid pSV-Sport1-ARNT was kindly provided by Oliver Hankinson (University of California, Los Angeles).
Electrophoretic Mobility Shift Assays
Electrophoretic mobility shift assays were performed using in vitro translated AHR of a given specific species as indicated and human ARNT. All other aspects of these assays were performed as previously described (Chiaro, et al. 2008).
Cell Culture
COS-1 and BP8 cell lines were maintained in α-modified essential media (Sigma-Aldrich, St. Louis, MO) supplemented with 10% fetal bovine serum (Hyclone Laboratories, Logan UT). Cells were cultured at 37 °C in a humidified atmosphere composed of 95% air and 5% CO2 in the presence of 100 IU/ml penicillin and 100 μg/ml streptomycin (Sigma-Aldrich).
AHR Ligand Binding Assay
COS-1 cells were grown to 90% confluency in six well cell culture plates. Cells were transfected using the Lipofectamine® 2000 transfection reagent and PLUSTM reagent (Life Technologies, Carlsbad, CA) with 1.5 μg of AHR plasmid construct per well, according to the manufacturer’s protocol. All binding experiments were performed in the dark prior to photocrosslinking. These experiments were done essentially as described previously (Swanson and Perdew 1991; Ramadoss and Perdew 2004). Media from the transfected cells in six well plates was removed, and fresh media containing the appropriate amount of photoaffinity ligand was added to each well. Fresh media containing 0, 0.5, 1.0, 1.5, 2.0, or 2.5 pmoles of AHR photoaffinity ligand, 2-azido-3-[125I]iodo-7,8-dibromodibenzo-p-dioxin was applied to cells for 1 h (in duplicate) at 37 °C in 95% air/5% carbon dioxide. The bound PAL was cross linked to AHR via ultraviolet (UV) cross linking (302 nM) for 4 min at a distance of 8 cm. Cells were then lysed in 200 µL of lysis buffer (MENG, pH 7.28, 20 mM sodium molybdate, 500 mM sodium chloride, protease inhibitor cocktail, and 1%(v/v) NP-40). Samples incubated on ice and were spun at 13,000 × g for 10 min. A volume of 50 μL of the lysate supernatant per sample was resolved upon an 8% Tricine/sodium dodecyl sulfate (SDS) polyacrylamide gel. Protein and bound PAL were transferred to polyvinylidene fluoride membrane and visualized by autoradiography. Band intensities were quantified by filmless autoradiographic analysis using a Cyclone storage phosphor screen instrument (PerkinElmer Life and Analytical Sciences). Band intensities were determined by γ-counting or filmless autoradiographic analysis, and both methods yield similar results.
BP8 Cell Transfection/AHR Ligand Dose-Response Assays
Rat hepatoma BP8 cells that lack AHR expression were plated to 90% confluency in 12-well cell culture plates and transfected using the Lipofectamine® 3000 transfection reagent (Life Technologies, Carlsbad, CA) with a total of 1 μg of plasmid DNA per well. Transient transfections utilized pcDNA3.syn.hAHR or pcDNA3.syn.NeAHR were titered for equal protein expression between constructs. Upon completion of the transfection at 6 h, Lipofectamine reagents were removed and replaced with Opti-MEM® (Life Technologies, Carlsbad, CA) supplemented with 2% fetal bovine serum. After 17 h post transfection, cells were treated as indicated for 4 h. Total RNA was isolated from cells using TRI Reagent (Sigma-Aldrich), followed by reverse transcription using the High Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s protocols. PerfeCTa SYBR Green SuperMix for iQ (Quanta Biosciences, Gaithersburg, MD) was used to determine mRNA levels, and analysis was conducted using MyIQ software, in conjunction with a CFX ConnectTM Real-Time System (Bio-Rad Laboratories, Hercules, CA). To account for potential differences in AHR expression or transfection efficiency, quantification of ligand-induced Cyp1a1/1b1 mRNA level was transformed to represent percentage of maximal induction elicited by a saturating dose of TCDD (20 nM) for H. sapiens and H. Neandertalensis AHR transfected cells, respectively. Data were subjected to nonlinear regression using a four parameter variable slope log10[ligand] versus response curve-fitting model to yield final dose-response plots and EC50 values (GraphPad Prism).
Real-Time PCR Primers
Rat Cyp1a1-F 5′- CCC TAA CTC TTC CCT GGA TGC-3′
Rat Cyp1a1-R 5′- GGA TGT GGC CCT TCT CAA ATG-3′
Rat Cyp1b1-F 5′- GAC ATC TTTG GAG CCA GCC A -3′
Rat Cyp1b1-R 5′- TCC GGG TAT CTG GTA AAG AGG A -3′
Rat Rpl13a-F 5′- AAG CAG CTC TTG AGG CTA AGG -3′
Rat Rpl13a-R 5′- TGG GTT CAC ACC AAG AGT CC -3′
Plasmid Mutagenesis
The Neandertal AHR expression and mutant constructs were synthesized by QuikChange Site-Directed Mutagenesis (Agilent Technologies, Wilmington, DE) of the corresponding synthetic human AHR construct (pcDNA3.syn.hAHR) according to manufacturer’s protocols. The V381A and R554K codons were altered to generate the Neandertal AHR cDNA.
Mutagenesis Primers
hAHR-V381A-F 5′- GAT TAC ATC ATC GCT ACC CAG CGG CCC-3′
hAHR-V381A-R 5′-GGG CCG CTG GGT AGC GAT GAT GTA ATC -3′
hAHR-R554K-F 5′-CCT GGG CAT CGA TTT CGA AGA CAT CAA GCA CAT GCA GAA CG -3′
hAHR-R554K-R 5′-CGT TCT GCA TGT GCT TGA TGT CTT CGA AAT CGA TGC CCA GG -3′
Protein Electrophoresis and Blot Analysis
Cells were lysed in RIPA buffer as previously described (DiNatale, Schroeder, et al. 2010), protein resolved by Tricine SDS-polyacrylamide gel electrophoresis (PAGE), and transferred to polyvinylidene difluoride membrane. The relative level of AHR was determined using mAb RPT 1, a biotinylated goat antimouse IgG secondary antibody and [125I]streptavidin. B-actin detected with mAb B-actin (C4, Santa Cruz Biotechnology) was utilized as a loading control.
In Silico Molecular Modeling
The homology model of the human AhR-PASB-LBD is based on the nuclear magnetic resonance (NMR) apo structure of the Hypoxia inducible factor (HIF)-2a Per-ARNT-Sim-B (PASB) domain Protein database (PDB 1P97) was prepared and optimized as described (Perkins, et al. 2014). Molecular docking was run as previously described (Hubbard, et al. 2015).
Statistical Analysis
Data in the ligand binding assays were analyzed using one-way analysis of variance with Tukey’s multiple comparison posttest using GraphPad Prism (v.5.01) software to determine statistical significance between treatments. Data represent the mean change in a given endpoint ± s.e.m. (n = 2/treatment group) and were analyzed to determine significance (**P < 0.01; ***P < 0.001).
Genomics/Database Analysis
We first queried the database of archaic hominin and modern human “exome” genetic variation produced by Castellano et al. 2014. The nucleotide variants within this database were identified based on paleogenomic sequence data from three Neandertal individuals (Altai, El Sidron, and Vindija) and one individual from the Denisovan archaic hominin population, plus genome sequence data from nine modern human individuals (three individuals each with subSaharan African, European, and Asian ancestry). Genotypes were estimated for only those positions covered by a minimum of six independent sequencing reads in each individual, and ancestral and derived variant states were determined via comparisons to the gorilla and orangutan genome sequences. We used the program PolyPhen-2 (Adzhubei, et al. 2010) to infer whether each nucleotide variant in the database was nonsynonymous (amino acid changing; potentially functional) or synonymous (resulting in no change to the amino acid sequence; typically neutral with respect to fitness). Of the 15,383 nonsynonymous SNPs in the database for which ancestral and derived states could also be determined, only 90 were fixed for the derived allele in the modern human sample but the ancestral allele in the archaic hominin sample (Neandertals plus the Denisovan individual). We then queried the dbSNP database (Build 144) to discover that there is no record of variation in modern humans for 27 of these positions. We also queried the 1000 genomes database ([http://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/]; 2/19/15) for record of modern human AHR gene variation. We further examined the full AHR-binding region of one Neandertal individual (Prufer, et al. 2014) and one Denisovan individual (Meyer, et al. 2012), for which high-coverage genome sequence data are available (these data are also represented in Castellano et al. 2014 database, but with the omission of some lower coverage sites for the other two Neandertal individuals). For the Neandertal individual, we downloaded the read alignment file from the European Nucleotide Archive (accession ERP002097; L9105.bam) and used the variant detector program called freebayes to reconstruct the Neandertal reference AHR protein sequence (Garrison and Marth 2012). For the Denisovan sequence, we used an alignment track from the UCSC genome browser (https://genome.ucsc.edu/cgi-bin/hgGateway). We also downloaded the genome sequencing read alignment file for a 45 ky modern human individual (European Nucleotide Archive PRJEB6622) (Fu, et al. 2014) and used freebayes for variant calling. Multiple sequence alignments were performed using the program Clustal Omega (Sievers, et al. 2011).
Supplementary Material
Supplementary figures S1–S5 and tables S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Theresa Wilson for technical assistance and Marcia Perdew for editorial advice. We also thank Istvan Albert for his assistance with the genomics analyses. This work was supported by grants from the National Institutes of Health Grants ES004869 and ES019964. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. 2010. A method and server for predicting damaging missense mutations. Nat Methods. 7:248–249. Available from: http://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/, https://genome.ucsc.edu/cgi-bin/hgGateway, last accessed July 2, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beischlag TV, Luis Morales J, Hollingshead BD, Perdew GH. 2008. The aryl hydrocarbon receptor complex and the control of gene expression. Crit Rev Eukaryot Gene Expr. 18:207–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello SM, Franks DG, Stegeman JJ, Hahn ME. 2001. Acquired resistance to Ah receptor agonists in a population of Atlantic killifish (Fundulus heteroclitus) inhabiting a marine superfund site: in vivo and in vitro studies on the inducibility of xenobiotic metabolizing enzymes. Toxicol Sci. 60:77–91. [DOI] [PubMed] [Google Scholar]
- Brown KS, Marean CW, Herries AIR, Jacobs Z, Tribolo C, Braun D, Roberts DL, Meyer MC, Bernatchez J. 2009. Fire as an engineering tool of early modern humans. Science 325:859–862. [DOI] [PubMed] [Google Scholar]
- Bruhs A, Haarmann-Stemmann T, Frauenstein K, Krutmann J, Schwarz T, Schwarz A. 2015. Activation of the arylhydrocarbon receptor causes immunosuppression primarily by modulating dendritic cells. J Invest Dermatol. 135:435–444. [DOI] [PubMed] [Google Scholar]
- Castellano S, Parra G, Sanchez-Quinto FA, Racimo F, Kuhlwilm M, Kircher M, Sawyer S, Fu Q, Heinze A, Nickel B, et al. 2014. Patterns of coding variation in the complete exomes of three Neandertals. Proc Natl Acad Sci U S A. 111:6666–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaro CR, Morales JL, Prabhu KS, Perdew GH. 2008. Leukotriene A4 metabolites are endogenous ligands for the Ah receptor. Biochemistry 47:8445–8455. [DOI] [PubMed] [Google Scholar]
- DiNatale BC, Murray IA, Schroeder JC, Flaveny CA, Lahoti TS, Laurenzana EM, Omiecinski CJ, Perdew GH. 2010. Kynurenic acid is a potent endogenous aryl hydrocarbon receptor ligand that synergistically induces interleukin-6 in the presence of inflammatory signaling. Toxicol Sci. 115:89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNatale BC, Schroeder JC, Francey LJ, Kusnadi A, Perdew GH. 2010. Mechanistic insights into the events that lead to synergistic induction of interleukin 6 transcription upon activation of the aryl hydrocarbon receptor and inflammatory signaling. J Biol Chem. 285:24388–24397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema M, Ohe N, Suzuki M, Mimura J, Sogawa K, Ikawa S, Fujii-Kuriyama Y. 1994. Dioxin binding activities of polymorphic forms of mouse and human arylhydrocarbon receptors. J Biol Chem. 269:27337–27343. [PubMed] [Google Scholar]
- Endicott P, Ho SYW, Stringer C. 2010. Using genetic evidence to evaluate four palaeoanthropological hypotheses for the timing of Neanderthal and modern human origins. J Hum Evol. 59:87–95. [DOI] [PubMed] [Google Scholar]
- Esakky P, Moley KH. 2016. Preventing germ cell death by inactivating aryl hydrocarbon receptor (AHR). Cell Death Dis. 7:e2116.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser C, Rannug A. 2015. The aryl hydrocarbon receptor in barrier organ physiology, immunology, and toxicology. Pharmacol Rev 67:259–279. [DOI] [PubMed] [Google Scholar]
- Flaveny CA, Murray IA, Chiaro CR, Perdew GH. 2009. Ligand selectivity and gene regulation by the human aryl hydrocarbon receptor in transgenic mice. Mol Pharmacol 75:1412–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca-Azevedo K, Herculano-Houzel S. 2012. Metabolic constraint imposes tradeoff between body size and number of brain neurons in human evolution. Proc Natl Acad Sci U S A. 109:18571–18576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q, Li H, Moorjani P, Jay F, Slepchenko SM, Bondarev AA, Johnson PL, Aximu-Petri A, Prufer K, de Filippo C, et al. 2014. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature 514:445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitanis G, Magiatis P, Stathopoulou K, Bassukas ID, Alexopoulos EC, Velegraki A, Skaltsounis AL. 2008. AhR ligands, malassezin, and indolo[3,2-b]carbazole are selectively produced by Malassezia furfur strains isolated from seborrheic dermatitis. J Invest Dermatol. 128:1620–1625. [DOI] [PubMed] [Google Scholar]
- Garrison E, Marth G. 2012. Haplotype-based variant detection from short read sequencing. Preprint arXiv 1207.3907v2 [q-bio.GN].
- Gelboin HV. 1980. Benzo[alpha]pyrene metabolism, activation and carcinogenesis: role and regulation of mixed-function oxidases and related enzymes. Physiol Rev. 60:1107–1166. [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ, Fernandez-Salguero P. 1998. The aryl hydrocarbon receptor: studies using the AHR-null mice. Drug Metab Dispos. 26:1194–1198. [PubMed] [Google Scholar]
- Goren-Inbar N, Alperson N, Kislev ME, Simchoni O, Melamed Y, Ben-Nun A, Werker E. 2004. Evidence of hominin control of fire at Gesher Benot Ya’aqov, Israel. Science 304:725–727. [DOI] [PubMed] [Google Scholar]
- Gowlett JAJ, Wrangham RW. 2013. Earliest fire in Africa: towards the convergence of archaeological evidence and the cooking hypothesis. Azania: Archaeol Res Afr. 48:5–30. [Google Scholar]
- Hallegouet B, Hinguant S, Gebhardt A, Monnier J-L. 1992. Le gisement Paléolithique inférieur de Ménez-Drégan 1 (Plouhinec, Finistère) Premiers résultats des fouilles. Bull Soc Préhist Fr. 89:77–81. [Google Scholar]
- Hardy K, Radini A, Buckley S, Sarig R, Copeland L, Gopher A, Barkai R. 2016. Dental calculus reveals potential respiratory irritants and ingestion of essential plant-based nutrients at Lower Palaeolithic Qesem Cave Israel. Quaternary International. 398:129–135. [Google Scholar]
- Harper PA, Golas CL, Okey AB. 1988. Characterization of the Ah receptor and aryl hydrocarbon hydroxylase induction by 2,3,7,8-tetrachlorodibenzo-p-dioxin and benz(a)anthracene in the human A431 squamous cell carcinoma line. Cancer Res. 48:2388–2395. [PubMed] [Google Scholar]
- Hovers E, Ilani S, Bar-Yosef O, Vandermeersch B. 2003. An early case of color symbolism: ochre use by modern humans in Qafzeh Cave. Curr Anthropol. 44:491–522. [Google Scholar]
- Huang Z, Jiang Y, Yang Y, Shao J, Sun X, Chen J, Dong L, Zhang J. 2013. 3,3'-Diindolylmethane alleviates oxazolone-induced colitis through Th2/Th17 suppression and Treg induction. Mol Immunol. 53:335–344. [DOI] [PubMed] [Google Scholar]
- Hubbard TD, Murray IA, Bisson WH, Lahoti TS, Gowda K, Amin SG, Patterson AD, Perdew GH. 2015. Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci Rep. 5:12689.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss EA, Vonarbourg C, Kopfmann S, Hobeika E, Finke D, Esser C, Diefenbach A. 2011. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science 334:1561–1565. [DOI] [PubMed] [Google Scholar]
- MacKenzie KM, Angevine DM. 1981. Infertility in mice exposed in utero to benzo(a)pyrene. Biol Reprod. 24:183–191. [DOI] [PubMed] [Google Scholar]
- Matikainen T, Perez GI, Jurisicova A, Pru JK, Schlezinger JJ, Ryu HY, Laine J, Sakai T, Korsmeyer SJ, Casper RF, et al. 2001. Aromatic hydrocarbon receptor-driven bax gene expression is required for premature ovarian failure caused by biohazardous environmental chemicals. Nat Genet. 28:355–360. [DOI] [PubMed] [Google Scholar]
- Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prufer K, de Filippo C, et al. 2012. A high-coverage genome sequence from an archaic Denisovan individual. Science 338:222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monge G, Jimenez-Espejo FJ, García-Alix A, Martínez-Ruiz F, Mattielli N, Finlayson C, Ohkouchi N, Sánchez MC, de Castro JMB, Blasco R, et al. 2015. Earliest evidence of pollution by heavy metals in archaeological sites. Sci Rep. 5:14252.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker CH, Keefe ER, Herzog NM, O'Connell JF, Hawkes K. 2016. The pyrophilic primate hypothesis. Evol Anthropol. 25:54–63. [DOI] [PubMed] [Google Scholar]
- Perdew GH, Abbott B, Stanker LH. 1995. Production and characterization of monoclonal antibodies directed against the Ah receptor. Hybridoma 14:279–283. [DOI] [PubMed] [Google Scholar]
- Perkins A, Phillips JL, Kerkvliet NI, Tanguay RL, Perdew GH, Kolluri SK, Bisson WH. 2014. A structural switch between agonist and antagonist bound conformations for a ligand-optimized model of the human aryl hydrocarbon receptor ligand binding domain. Biology (Basel) 3:645–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland A, Glover E, Ebetino FH, Kende AS. 1986. Photoaffinity labeling of the Ah receptor. J Biol Chem. 261:6352–6365. [PubMed] [Google Scholar]
- Poland A, Palen D, Glover E. 1994. Analysis of the four alleles of the murine aryl hydrocarbon receptor. Mol Pharmacol. 46:915–921. [PubMed] [Google Scholar]
- Prufer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, et al. 2014. The complete genome sequence of a Neanderthal from the Altai mountains. Nature 505:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadoss P, Perdew GH. 2004. Use of 2-azido-3-[125I]iodo-7,8-dibromodibenzo-p-dioxin as a probe to determine the relative ligand affinity of human versus mouse aryl hydrocarbon receptor in cultured cells. Mol Pharmacol. 66:129–136. [DOI] [PubMed] [Google Scholar]
- Revel A, Raanani H, Younglai E, Xu J, Han R, Savouret JF, Casper RF. 2001. Resveratrol, a natural aryl hydrocarbon receptor antagonist, protects sperm from DNA damage and apoptosis caused by benzo(a)pyrene. Reprod Toxicol. 15:479–486. [DOI] [PubMed] [Google Scholar]
- Schroeder JC, Dinatale BC, Murray IA, Flaveny CA, Liu Q, Laurenzana EM, Lin JM, Strom SC, Omiecinski CJ, Amin S, et al. 2010. The uremic toxin 3-indoxyl sulfate is a potent endogenous agonist for the human aryl hydrocarbon receptor. Biochemistry 49:393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KR, Samet JM, Romieu I, Bruce N. 2000. Indoor air pollution in developing countries and acute lower respiratory infections in children. Thorax 55:518–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson HI, Perdew GH. 1991. Detection of the Ah receptor in rainbow trout: use of 2-azido-3-[125I]iodo-7,8-dibromodibenzo-p-dioxin in cell culture. Toxicol Lett. 58:85–95. [DOI] [PubMed] [Google Scholar]
- Tian J, Feng Y, Fu H, Xie HQ, Jiang JX, Zhao B. 2015. The aryl hydrocarbon receptor: a key bridging molecule of external and internal chemical signals. Environ Sci Technol. 49:9518–9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno S, Dalton TP, Shertzer HG, Genter MB, Warshawsky D, Talaska G, Nebert DW. 2001. Benzo[a]pyrene-induced toxicity: paradoxical protection in Cyp1a1(-/-) knockout mice having increased hepatic BaP-DNA adduct levels. Biochem Biophys Res Commun. 289:1049–1056. [DOI] [PubMed] [Google Scholar]
- Vattathil S, Akey JM. 2015. Small amounts of archaic admixture provide big insights into Human history. Cell 163:281–284. [DOI] [PubMed] [Google Scholar]
- Weiss C, Kolluri SK, Kiefer F, Gottlicher M. 1996. Complementation of Ah receptor deficiency in hepatoma cells: negative feedback regulation and cell cycle control by the Ah receptor. Exp Cell Res. 226:154–163. [DOI] [PubMed] [Google Scholar]
- Wirgin I, Roy NK, Loftus M, Chambers RC, Franks DG, Hahn ME. 2011. Mechanistic basis of resistance to PCBs in Atlantic tomcod from the Hudson River. Science 331:1322–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JM, Harper PA, Meyer UA, Bock KW, Morike K, Lagueux J, Ayotte P, Tyndale RF, Sellers EM, Manchester DK, et al. 2001. Ethnic variability in the allelic distribution of human aryl hydrocarbon receptor codon 554 and assessment of variant receptor function in vitro. Pharmacogenetics 11:85–94. [DOI] [PubMed] [Google Scholar]
- Wrangham R, Conklin-Brittain N. 2003. Cooking as a biological trait. Comp Biochem Physiol A Mol Integr Physiol. 136:35–46. [DOI] [PubMed] [Google Scholar]
- Yang EJ, Stokes JV, Kummari E, Eells J, Kaplan BL. 2016. Immunomodulation by subchronic low dose 2,3,7,8-tetrachlorodibenzo-p-dioxin in experimental autoimmune encephalomyelitis in the absence of pertussis toxin. Toxicol Sci. 151(1):35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Courtenay S, Wirgin I. 2006. Comparison of hepatic and extra hepatic induction of cytochrome P4501A by graded doses of aryl hydrocarbon receptor agonists in Atlantic tomcod from two populations. Aquat Toxicol. 76:306–320. [DOI] [PubMed] [Google Scholar]
- Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D’Angelo C, Massi-Benedetti C, Fallarino F, et al. 2013. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39:372–385. [DOI] [PubMed] [Google Scholar]
- Zelikoff JT, Chen LC, Cohen MD, Schlesinger RB. 2002. The toxicology of inhaled woodsmoke. J Toxicol Environ Health B Crit Rev. 5:269–282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.