Abstract
Cytochrome P450 2D6 (CYP2D6) is an enzyme of great importance for the metabolism of clinically used drugs. More than 100 variants of the CYP2D6 gene have been identified so far. The aim of this study was to investigate the allele distribution of CYP2D6 gene variants in 100 individuals of each of the Macedonian, Albanian and Romany population, by genotyping using long range polymerase chain reaction (PCR) and a multiplex single base extension method. The most frequent variants and almost equally distributed in the three groups were the fully functional alleles *1 and *2. The most common non functional allele in all groups was *4 that was found in 22.5% of the Albanians. The most common allele with decreased activity was *41 which was found in 23.0% of the Romany ethnic group, in 11.0% of the Macedonians and in 10.5% of the Albanians. Seven percent of the Albanians, 6.0% of the Romani and 4.0% of the Macedonians were poor metabolizers, while 5.0% of the Macedonians, 1.0% of Albanians and 1.0% of the Romanies were ultrarapid metabolizers. We concluded that the CYP2D6 gene locus is highly heterogeneous in these groups and that the prevalence of the CYP2D6 allele variants and genotypes in the Republic of Macedonia is in accordance with that of other European populations.
Keywords: Allele, CYP2D6 gene, CYP450, Ethnic groups, Extensive metabolizers (EMs), Intermediate metabolizers (IMs), Poor metabolizers (PMs), Ultrarapid metabolizers (UMs)
Introduction
Physiological responses to the same drug are known to vary substantially between different individuals. Although this may result from environmental and physiological factors or drug-drug interactions, in many cases the response is inherited and arises from a polymorphism in genes that encode drug transporters, drug receptors, and especially, drug-metabolizing enzymes [1]. Of the genes that encode drug-metabolizing enzymes, CYP2D6, a member of the cytochrome P450 superfamily (CYP450), is well characterized.
Approximately 57 CYP genes that encode cytochrome P450 proteins and 58 pseudogenes are present in the human genome and are classified into distinct families and subfamilies according to their sequence similarity [2]. The CYP2D subfamily comprises the CYP2D6 gene and two pseudogenes (CYP2D7 and CYP2D8), located in tandem on chromosome 22q13.1, at the 3’ end of the CYP2D cluster. The CYP2D6 gene contains nine exons comprised of 1461 codons. The evolution of this locus has involved elimination of three genes and inactivation of two neighboring genes (CYP2D7 and CYP2D8), all of which display 92.0-97.0% nucleotide similarity across their sequences [3]. The CYP2D6 gene mediates the metabolism of almost a quarter of drugs in common clinical use, including opiate analgesics, antiarrhythmics, antipsychotics, antidepressants, tamoxifen and β blockers [1].
The CYP2D6 gene is highly polymorphic, with more than 100 variations and numerous subvariants having been identified [4]. These variations include: single-base changes, short insertions and deletions, major deletions [5] and whole gene duplications [6].
There are four major phenotype classes: ultra-rapid metabolizers (UMs), extensive metabolizers (EMs), poor metabolizers (PMs) and intermediate metabolizers (IMs) and two subclasses: IMS to EMs and PMs to IMs. The sub-classification of the intermediate metabolizers is ascribed to the wide spectrum of metabolic activity that can range from marginally better than the PM phenotype to activity that is close to that of the EM phenotype (Table 1) [4,7].
Table 1.
| Predicted Drug Metabolizing Phenotypeb | Combination of Alleles |
|---|---|
| UM | A combination of one normal activity allele with one increased activity allele |
| EM | Two normal activity alleles; a combination of one increased activity allele with one decreased activity allele |
| IM to EM | A combination of one normal activity allele with one decreased activity allele; a combination of one increased activity allele with one null activity allele |
| IM | One normal activity allele with one null activity allele; two decreased activity alleles |
| PM to IM | A combination of one decreased activity allele with one null activity allele |
| PM | Only null activity alleles detected |
The phenotypes were derived from the Human Cytochrome P450 (CYP) Allele Nomenclature Committee website [4] and the PharmGKB website for the related Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2D6 [7].
UM: ultrarapid metabolizer; EM: extensive metabolizer; IM: intermediate metabolizer; PM: poor metabolizer.
The EM phenotype is expressed by the majority of the population and is therefore considered “the norm” [8]. Poor metabolizers inherit two null CY-P2D6 alleles that include at least 22 different alleles which do not encode a functional protein and show no detectable residual enzymatic activity. This leads to accumulation of high levels of unmetabolized drugs that are CYP2D6 substrates, greater potential for adverse effects and drug-drug interactions, and lower efficacy of drugs that require CYP2D6 activation [9]. The UM phenotype is caused by amplification of active CYP2D6 genes, primarily the CYP2D6*1 and CYP 2D6*2 alleles. Individuals with this phenotype metabolize drugs at an ultrarapid rate, which may lead to loss of therapeutic efficacy at standard doses [10]. Individuals who are heterozygous for a defective and a fully active CYP2D6 allele or are homozygous for an allele with decreased activity, for example alleles *10, *17, *36 and *41, often demonstrate an IM phenotype [11]. Previous genetic studies showed high levels of CYP2D6 polymorphism, both within and between populations [12], and a surprisingly high frequency of null and reduced function variants.
Poor metabolizers account for 5.0 to 10.0% of the Caucasian population and less than 1.0% of the Asian population [13]. In Caucasians, common deficient alleles include CYP2D6*3, *4, *5 and *6, accounting for about 98.0% of PMs [14]. On the other hand, the decreased activity allele *41 is predominantly present in the Middle East, with frequencies reaching up to 22.5% [15].
In contrast to PMs, UMs usually carry a duplicated, or even multiduplicated (up to 13 copies of CYP2D6) active CYP2D6 allele (CYP2D6*xN). The frequencies of CYP 2D6*xN vary greatly between races [16]. Both CYP2D6*5 and CYP2D6*xN result from CYP2D6 gene rearrangement [17] and comprise CYP2D6 gene copy number variations.
The CYP2D6 genotyping to predict metabolic status is considered a valid alternative to traditional phenotyping methods [18]. Assessing the CYP2D6 genotype also offers several distinct advantages over the experimental determination of a CYP2D6 phenotype [19]. Genotyping usually requires only a blood sample and can be done before a drug is given to a patient. It therefore may facilitate improved drug efficiency and diminished risk for adverse drug reactions [20]. The aim of this study was to investigate the allele distribution of CYP2D6 variants in Macedonian, Albanian and Romany populations as well in the Republic of Macedonia.
Materials and Methods
DNA Samples
DNA material for genotyping from Macedonians (n = 100), Albanians (n = 100) and Romanies (n = 100), was obtained from the DNA bank of the Research Centre for Genetic Engineering and Biotechnology “Georgi D. Efremov” at the Macedonian Academy of Science and Arts, Skopje, Republic of Macedonia. We decided to analyze 100 samples from each ethnicity, in order to be able to compare and statistically process the obtained results. The number of samples from each ethnic group does not reflect the actual representation of ethnicities in the Republic of Macedonia. The Ethics Committee of the Macedonian Academy of Science and Arts approved this study. The samples were anonymized after collection.
CYP2D6 Genotyping
The genotyping was performed following a recently described protocol by Sistonen et al. [21], based on a combination of long range polymerase chain reaction (PCR), to detect whole-gene deletion/ duplication and multiplex extension of unlabeled oligo-nucleotide primers with fluorescently labeled dideoxynucleotide triphosphates (ABI PRISM® SNaPshot Multiplex Kit; Life Technologies, Carlsbad, CA, USA) to characterize 11 relevant polymorphic positions in the coding region of CYP2D6. This made it possible to identify CYP 2D6 variants which are highly represented in different human populations (i.e.,*2, *4, *10, *17, *29, *39, *41), rare variants known to be responsible for low or null metabolic activity (i.e., *3, *6 and *9), and whole gene deletion (*5) and duplications (*1xN, *2xN). This permitted us to identify the mutations that are generally thought to comprise more than 90.0% of known variants in Europeans [22], and mutation 1023 (C>T) which is rare in Europe, but is common in African populations. The haplotypes which did not show any of these mutations were classified as *1.
Three parallel long range-PCRs were run for each sample using Expand Long Template PCR System (Roche Diagnostics, Basel, Switzerland).We obtained a 5.1 kb fragment containing all nine CYP2D6 exons using CYP 2D6-F [21] and CYP2D6-R primers [21]. This product was used as a template in order to be able to type 11 positions in one reaction, based on single-base primer extension with fluorescently labelled ddNTPs (ABI PRISM® SNaPshot Multiplex Kit; Life Technologies).
The 25 µL reaction mixture contained 2U enzyme mix, Expand Long Template Buffer 1, 2 × concentrated, with 17.5 mM MgCl2, 0.2 mM each dideoxynucleotide triphosphate, 0.4 µM of each primer, and 50 ng of genomic DNA. The PCR reaction was conducted as follows: denaturation at 94°C for 10 min., 10 cycles at 94°C for 30 seconds and 68°C for 30 seconds, 25 cycles at 94°C for 30 seconds and 68°C for 10 min. and 15 seconds, plus 15 seconds per cycle, and a final extension at 68°C for 30 min.
The long range-PCR products were analyzed on 0.8% agarose gels, and the 5.1 kb fragments were purified by use of 1 µL of ExoSAP-IT (USB, Affymetrix Inc., San Diego, CA, USA) overnight at 37°C (2 µL PCR product + 1 µL Exo SAP IT). The reaction ended with inactivation of the enzyme at 86°C for 20 min.
The purified 5.1 kb product was used as template in the SNaPshot (Life Technologies) reaction. In the following single-base extension reaction, the detection primers annealed adjacent to the single nucleotide polymorphism (SNP) position and was extended with fluorescently- labeled dideoxynucleotide triphosphates. The SNaPshot (Life Technologies) reaction contained 3.0 µL of purified PCR product, 1 µL of pooled detection primers [21], 1 µL water and 1 µL of SNaPshot (Life Technologies) ready reaction mixture in a final volume of 7 µL. The cycling profile was 25 cycles at 96°C for 10 seconds, 55°C for 10 seconds and 60°C for 30 seconds. After the reaction, 5-phosphoryl groups of unincorporated dideoxynucleotide triphosphates were removed by addition of SAP (shrimp alkaline phosphatase) (USB, Affymetrix Inc.) for 1 hour at 37°C, followed by enzyme inactivation at 86°C for 20 min. Capillary electrophoresis of samples was performed on the ABI PRISM® 3130 genetic analyzer with LIZ120 (Life Technologies) as a size standard. The obtained results were analyzed with GeneScan 4.0 software (Life Technologies).
Two additional long range-PCR reactions were used to analyze the major rearrangements, i.e., duplication or deletion of the entire CYP2D6 gene [21]. Both deletion and duplication PCR reactions were performed in a reaction volume of 12 µL containing 1U enzyme mix from the Expand Long Template PCR System (Roche Diagnostics), Long Template Buffer 1, 1 × concentrated, with 17.5 mM MgCl2, and 0.2 mM each dideoxynucleotide triphosphate. The primer concentrations were as follows: for the duplication-specific reaction, 0.3 µM CYP-207-F, 0.2 µM CYP-32-R, and 0.1 µM CYP-13-F; and for the deletion-specific reaction, 0.3 µM CYP-13-F, 0.2 µM CYP-24-R, and 0.1 µM CYP-207-F. Thecycling profile was as described above.
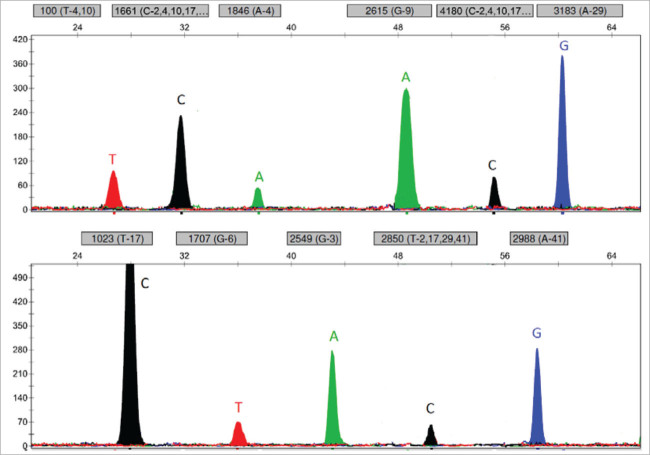
The primers CYP-13-F and CYP-24-R generated a deletion-specific fragment of 3.5 kb, while primers CYP-24-R and CYP-207-F yielded a control fragment of 3.0 kb. With the deletion-specific PCR we analyzed 94 homozygous samples. We analyzed only these samples because they had only one peak for each of the 11 analyzed polymorphic sites and we could not determine the difference between a homozygous set of polymorphic sites or a deleted allele (Figure 1). In comparison, the heterozygous patients had two peaks on at least one polymorphic site, indicating the presence of two alleles. We also analyzed 60 heterozygous samples in order to confirm our hypothesis.
Figure 1.
Representative electropherogram of homozygous patient with genotype *4/*4.
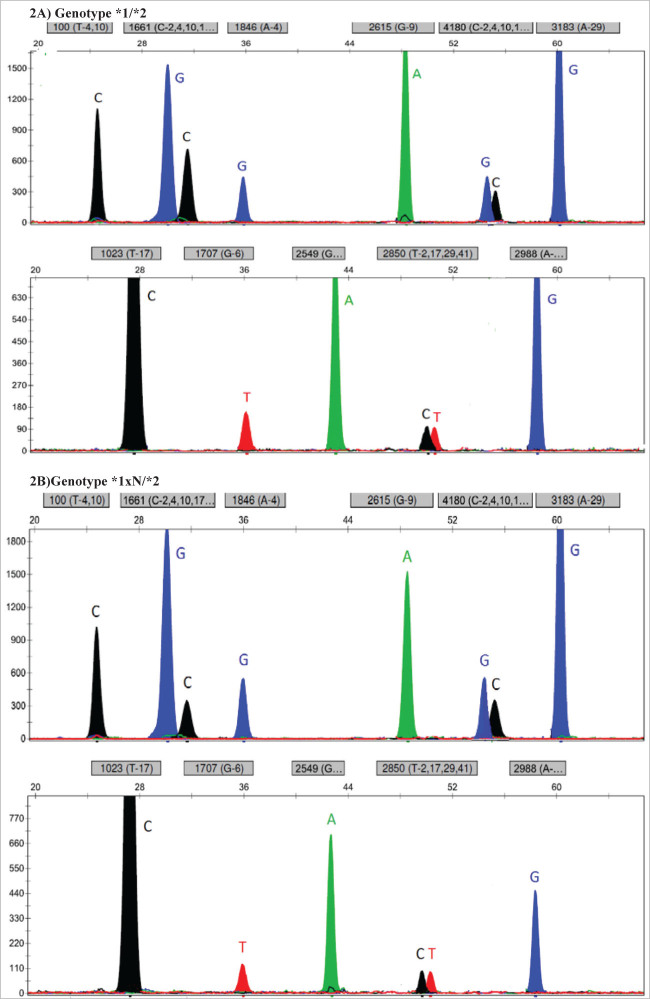
We looked for CYP2D6 gene duplications using the CYP-207-F and CYP-32-R primer pair. With these primers, we amplified a duplication-specific fragment of 3.2 kb. Simultaneously, a control fragment of 3.8 kb was amplified with the CYP-13-F forward primer. Apart from the duplication-specific PCR, we also confirmed the detected duplications with a comparison of the ratios of the two peaks that appeared at the same position for each separate polymorphic site. By comparing two electropherograms obtained from the SNaPshot (Life Technologies) analysis, we noticed that the duplicated allele could readily be identified as the one of interest, as it displayed higher signals in the polymorphic heterozygous sites (Figure 2A and 2B) [21].
Figure 2.
A) Representative electropherogram of a heterozygous patient without a duplication. B) Representative electropherogram of a heterozygous patient with a duplication.
The χ2 test and Fisher’s exact test were used to compare frequencies between populations. GraphPad InStat software (version 3.1; http://www.graphpad. com) was used for statistical analysis. A p value of 0.05 was considered significant.
Results
Table 2 summarizes the allele frequencies of the three groups. Of the fully functional alleles, the most frequent variant was the wild type allele *1 (in 36.5% of Macedonians, 33.0% of Romanies and 37.0% of Albanians), followed by allele *2 (in 25.5% of Macedonians, 21.5% of Romanies and 25.5% of Albanians).
Table 2.
CYP2D6 allele frequency in Macedonians, Albanians and Romanies.
| Allele and SNP | Location | Amino Acid Change | dbSNP ID | Functional Effect | Allele Frequency | ||
|---|---|---|---|---|---|---|---|
| Macedonians | Albanians | Romanies | |||||
| *1: wild type | – | – | – | normal | 0.365 | 0.370 | 0.330 |
| *2: 2850(C>T) 4180 (G>C) | exon 2 exon 6 | Arg296Cys Ser486Thr | rs16947 rs1135840 | normal | 0.255 | 0.215 | 0.255 |
| *3: 2549delA | exon 5 | frameshift | rs35742686 | none | 0.020 | 0.035 | 0.015 |
| *4: 1846 (G>A) | intron 3 - exon 4 junction | splicing defect | rs3892097 | none | 0.170 | 0.225 | 0.150 |
| *5 | ‒ | gene deletion | ‒ | none | 0.015 | 0.025 | 0.005 |
| *6: 1707delT | ‒ | frameshift | rs5030655 | none | 0.010 | 0.015 | 0.005 |
| *9: 2615-2617delAAG | ‒ | Lys281del | rs5030656 | decreased | 0.000 | 0.005 | 0.005 |
| *10: 100 (C>T) | exon 1 | Pro34Ser | rs1065852 | decreased | 0.015 | 0.005 | 0.000 |
| *17: 1023 (C>T) | exon 2 | Thr107Ile | rs28371706 | decreased | 0.000 | 0.000 | 0.000 |
| *29: 1661 (G>C) | ‒ | Arg36His | rs61736512 | decreased | 0.015 | 0.000 | 0.000 |
| *41: 2988 (G>A) | ‒ | Glu242Lys | rs28371725 | decreased | 0.110 | 0.105 | 0.230 |
| *1xN | ‒ | gene duplication | ‒ | increased | 0.020 | 0.000 | 0.005 |
| *2xN | ‒ | gene duplication | ‒ | increased | 0.005 | 0.005 | 0.000 |
SNP: single nucleotide polymorphism; dbSNP: database SNP.
The decreased activity alleles were observed in 23.5% of the Romany population, in 14.0% of the Macedonian and in 11.5% of the Albanian. Four decreased activity alleles were detected, i.e.,*9, *10, *29 and *41, from which allele *41 stood out as the most frequent variant. The frequency of the allele *41 in the Romanies (23.0%) is higher in comparison with the Macedonians (11.0%) and Albanians (10.5%), and this difference is statistically significant (p = 0.002 and p = 0.0012, respectively). The *9 allele was detected only in the Romany (0.5%) and the Albanian population (0.5%), whereas the *10 allele was present exclusively in the Macedonian and Albanian population with frequencies of 1.5 and 0.5%, respectively. The *29 allele was observed solely in the Macedonian ethnic group (1.5%).
Non functional alleles were found in 30.0% of the Albanians, 21.5% of the Macedonians and 17.5% of the Romanies. The non functional *4 allele was found in 22.5% of the Albanians, 17.0% of the Macedonians and 15.0% of the Romanies. Alleles *3, *5 and *6 were present in 3.5, 2.5 and 1.5%, respectively, of the Albanians, in 2.0, 1.5 and 1.0%, respectively, of the Macedonians, and in 1.5, 0.5 and 0.5%, respectively, of the Romanies.
Five duplications (four *1xN alleles and one *2xN allele) were identified in the Macedonian group. Only one duplication was found in each of the Albanian (*2xN) and Romany (*1xN) groups.
Table 3 summarizes the CYP2D6 genotypes and corresponding phenotypes found in the three groups. The *1/*2 genotype was present in 20.0% of the Macedonian and in 17.0% of the Romany group. The*1/*4 genotype was present in 17.0% of the Albanian group. We yielded statistically significant results for the frequency of the genotypes *1/*41 and *2/*41, which were found in 12.0 and 17.0%, respectively, of the Romany and in 5.0% for each genotype in the Macedonians (p = 0.013 and p = 0.0357, respectively) (Table 3).
Table 3.
Genotype frequency of determined CYP2D6 genotypes and corresponding phenotypes.
| Predicted | Genotype | Frequency in Macedonians (%) | Frequency in Albanians (%) | Frequency in Romanies (%) |
|---|---|---|---|---|
| Ultrarapid metabolizer | *1xN/*1 | 0.01 | 0.00 | 0.00 |
| *1xN/*2 | 0.03 | 0.00 | 0.01 | |
| *2xN/*2 | 0.01 | 0.01 | 0.00 | |
| Extensive metabolizer | *1/*1 | 0.14 | 0.15 | 0.13 |
| *1/*2 | 0.20 | 0.13 | 0.17 | |
| *1/*9 | 0.00 | 0.00 | 0.01 | |
| *1/*10 | 0.02 | 0.00 | 0.00 | |
| *1/*29 | 0.01 | 0.00 | 0.00 | |
| *2/*29 | 0.01 | 0.00 | 0.00 | |
| *1/*41 | 0.05 | 0.08 | 0.12 | |
| *2/*2 | 0.06 | 0.05 | 0.07 | |
| *2/*3 | 0.01 | 0.02 | 0.00 | |
| *2/*41 | 0.05 | 0.03 | 0.17 | |
| Intermediate metabolizer | *1/*3 | 0.01 | 0.03 | 0.01 |
| *1/*4 | 0.13 | 0.17 | 0.07 | |
| *1/*5 | 0.02 | 0.02 | 0.01 | |
| *1/*6 | 0.00 | 0.02 | 0.01 | |
| *2/*4 | 0.09 | 0.12 | 0.02 | |
| *2/*5 | 0.01 | 0.00 | 0.00 | |
| *2/*6 | 0.00 | 0.01 | 0.00 | |
| *41/*41 | 0.03 | 0.02 | 0.03 | |
| *9/*41 | 0.00 | 0.01 | 0.00 | |
| *4/*41 | 0.04 | 0.04 | 0.09 | |
| *4/*29 | 0.01 | 0.00 | 0.00 | |
| *6/*10 | 0.01 | 0.00 | 0.00 | |
| *6/*41 | 0.01 | 0.00 | 0.00 | |
| *3/*41 | 0.00 | 0.01 | 0.02 | |
| *4/*10 | 0.00 | 0.01 | 0.00 | |
| Poor metabolizer | *3/*3 | 0.01 | 0.00 | 0.00 |
| *3/*4 | 0.00 | 0.01 | 0.00 | |
| *4/*5 | 0.00 | 0.03 | 0.00 | |
| *4/*4 | 0.03 | 0.03 | 0.06 |
Discussion
The CYP2D6 gene is of great interest for clinical practice because it is responsible for the metabolism of many commonly used drugs and its genetic polymorphism can have a strong effect on the substrate and lead to wide inter-individual variation. Variation in CYP2D6 activity has important therapeutic consequences and can play a significant role in the development of adverse events or therapeutic failure in susceptible individuals.
The knowledge of how a drug is metabolized and which enzymes are involved helps to predict drug-drug interactions and how fast an individual patient may metabolize a specific drug. Because CYP2D6 metabolizes at least 25.0% of the marketed drugs, including selective serotonin reuptake inhibitors (SSRI), tricylic antidepressants (TCA), β blockers, opiates, neuroleptics, tamoxifen and antiar-rhythmics, there are possible issues of drug interaction in vivo which are of clinical significance.
The CYP2D6*3, CYP2D6*4 and CYP2D6*6 are the most important non functional alleles that are predominantly responsible for the poor metabolizing capacity. These alleles are found in 90.0-95.0% of the PMs in Europe [23]. The results obtained in our study showed that the prevalence of the CYP2D6*4 allele frequency is in accordance with that of other European populations, which vary between 12.0 and 21.0%. The frequency values for the allele *4 range between 15.0% in Romanies, 17.0% in Macedonians and 22.5% in Albanians (Table 4).
Table 4.
Comparison of CYP2D6 allele frequencies in different populations.
| Population | Refs | Functional | Non Functional | Reduced | Duplications | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| *1 | *2 | *3 | *4 | *5 | *6 | *9 | *10 | *17 | *29 | *41 | *1xN | *2xN | ||
| French | 15 | 28.00 | 32.00 | 0.00 | 16.00 | 4.00 | 0.00 | 4.00 | 2.00 | 0.00 | 0.00 | 10.00 | 2.00 | 0.00 |
| Sardinian | 15 | 28.60 | 35.70 | 1.80 | 21.40 | 1.80 | 0.00 | 0.00 | 5.40 | 0.00 | 0.00 | 3.60 | 0.00 | 1.80 |
| Russian | 15 | 32.00 | 34.00 | 0.00 | 20.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 8.00 | 0.00 | 4.00 |
| German | 32 | 36.40 | 32.40 | 2.00 | 20.70 | 2.00 | 0.90 | 1.80 | 1.50 | ‒ | ‒ | ‒ | 0.50 | 1.30 |
| Croatian | 29 | 76.50 | ‒ | 2.75 | 14.00 | 1.00 | 1.50 | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | 4.00 |
| Greek | 30 | ‒ | ‒ | 2.30 | 17.84 | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | 6.01 |
| Czech | 31 | ‒ | ‒ | 1.12 | 22.87 | 3.14 | 0.02 | ‒ | ‒ | ‒ | ‒ | ‒ | 3.14 | ‒ |
| Bedouin | 15 | 35.70 | 20.40 | 0.00 | 5.10 | 4.10 | 2.00 | 0.00 | 0.00 | 3.10 | 0.00 | 22.40 | 1.00 | 2.00 |
| Macedonian | 27 | 24.90 | 10.80 | 0.80 | 18.70 | 9.10 | 0.00 | 1.60 | 2.70 | ‒ | ‒ | 8.60 | 5.90a | |
| Macedonianb | ‒ | 36.50 | 25.50 | 2.00 | 17.00 | 1.50 | 1.00 | 0.00 | 1.50 | 0.00 | 1.50 | 11.00 | 2.00 | 0.50 |
| Albanianb | ‒ | 37.00 | 21.50 | 3.50 | 22.50 | 2.50 | 1.50 | 0.50 | 0.50 | 0.00 | 0.00 | 10.50 | 0.00 | 0.50 |
| Romanyb | ‒ | 33.00 | 25.40 | 1.50 | 15.00 | 0.50 | 0.50 | 0.50 | 0.00 | 0.00 | 0.00 | 23.00 | 0.50 | 0.00 |
The article does not clearly explain on which alleles the duplications occur.
This study.
From the alleles with severely reduced activity, the results we acquired show an increased frequency of allele *41 (23.0%) in Romanies. These frequencies are in correlation with the values obtained from several studies conducted in the Middle East [24-26]. These results are not surprising given the Indian origin of the Romany population and their migration through the Middle East, before they reached the Balkans.
Our results showed similar percentages of deletions and duplications in comparison with the other European populations. In the Albanian ethnic group we noticed the highest number of deletions, reaching frequency of 2.5% and in the Macedonian ethnic group the highest number of duplication (2.5%) (Table 4).
Our study is not the first one that has reviewed the frequency distribution of the CYP2D6 alleles in the Republic of Macedonia, still it is the first one that was conducted for each ethnicity separately. The prevalence values for the polymorphic alleles CYP2D6*3, *4, *6, *9 and *10 (0.008, 0.187, 0.0, 0.016 and 0.027, respectively) reported by Kapedanovska Nestorovska et al. [27] are in agreement with our results. However, the results for deletions and duplications in the two studies were not in concordance. Kapedanovska Nestorovska et al. [27] reported high frequencies of deletions and duplications (0.091 and 0.059, respectively) which are not in concordance with our study (Table 4). The method that Kapedanovska Nestorovska et al. [27] used for detection of the duplications and deletions was the quantitative real-time PCR method, using TaqMan Copy Number Assay (Thermo Fisher Scientific, Waltham, MA, USA). One of the main limitations of real-time PCR is the increased risk of false positive results, so this discordance is probably due to the lack of specificity and sensitivity of the method used.
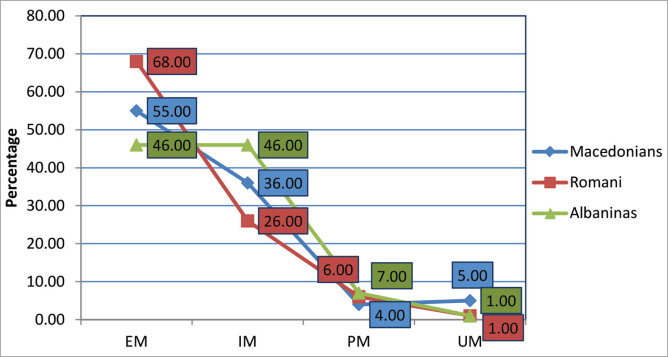
The metabolizing classes that are of pharmacological interest include the group of PMs and UMs. It is important to point out that the Albanians topped the list as the group with the highest number of PMs (7.0%), due to the accumulation of the *4 allele in their population, followed by the Romanies (6.0%) and the Macedonians (4.0%). The Macedonians stood out as the ethnic group with the highest number of UMs (5.0%), followed by the Albanians (1.0%) and the Romanies (1.0%) (Figure 3).
Figure 3.
The calculated percentage of metabolizing classes within the Macedonian, Albanian and Romany population (EM: extensive metabolizers; IM: intermediate metabolizers; PM: poor metabolizers; UM: ultrarapid metabolizers).
Concordant genotype-phenotype correlation provides a basis for predicting the phenotype based on genetic testing, which has the potential to achieve optimal pharmacotherapy. Predictive CYP2D6 genotyping is estimated to be beneficial for treatment of about 30.0-40.0% of CYP2D6 drug substrates, that is for about 7.0-10.0% of all drugs clinically used.
A study conducted in a psychiatric setting in the USA has shown that the CYP2D6 polymorphisms can have an effect on the cost of treating a patient; patients with ultrarapid and poor metabolizing capacity were found to cost between $4,000 and $6,000 per year more to treat than EM or IM individuals [28]. Thus, genotyping for individuals receiving single or multiple drugs that are metabolized by CYP2D6 may help clinicians to avoid adverse drug-drug interactions and to individualize better treatment with medications.
In addition, the Food and Drug Administration (FDA) approved testing is now available and an increasing number of medical centers provide this service to patients under their care. It will be increasingly important for doctors, physicians, pharmacists, and other care providers to be able to provide coherent therapeutic recommendations to patients with predetermined pharmacogenetic data.
Following the global trends in pharmacogenetics and the recommendations from the FDA, we decided to conduct this study in order to help with the introduction of pharmacogenetic testing for certain drugs in clinical practice, thus avoiding detrimental drug reactions, facilitating improved drug efficiency and moreover individualizing treatment for each patient. Our study is the first to assess the frequency distribution of the CYP2D6 alleles in the three major ethnic groups living in the Republic of Macedonia, with our findings displaying a genuine correspondence between the prevalence of CYP2D6 allele variants and genotypes in the Republic of Macedonia to other European populations.
Declaration of Interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
*Research Centre for Genetic Engineering and Biotechnology “Georgi D. Efremov,” Macedonian Academy of Sciences and Arts, Skopje, Republic of Macedonia
References
- 1.Gardiner SJ, Begg EJ. Pharmacogenetics, drug-metabolizing enzymes, and clinical practice. Pharmacol Rev. 2006;58(3):521–590. doi: 10.1124/pr.58.3.6. [DOI] [PubMed] [Google Scholar]
- 2.Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ, Wain HM, Nebert DW. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics. 2004;14(1):1–18. doi: 10.1097/00008571-200401000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Kimura S, Umeno M, Skoda RC, Meyer UA, Gonazalez FJ. The human debrisoquine 4-hydrolase(CYP2D)locus:Sequence and identification of the polymorphic CYP2D6 gene, a related gene, and a pseudogene. Am J Hum Genet. 1989;45(6):889–904. [PMC free article] [PubMed] [Google Scholar]
- 4.The Human Cytochrome P450 (CYP) Allele Nomenclature committee. ( http://www.cypalleles. ki.se/ cyp2d6.htm)
- 5.Steen VM, Andreassen OA, Daly AK, Tefre T, Borresen A-L, Idle JR. et al. Detection of the poor metab-olizer-associated CYP2D6(D) gene deletion allele by long-PCR technology. Pharmacogenetics. 1995;5(4):215–223. doi: 10.1097/00008571-199508000-00005. [DOI] [PubMed] [Google Scholar]
- 6.Johansson I, Lundqvist E, Bertilsson L, Dahl ML, Sjoqvist F, Ingelman-Sundberg M. Inherited amplification of an active gene in the cytochrome P450 CYP2D locus as a cause of ultrarapid metabolism of debrisoquine. Proc Natl Acad Sci USA. 1993;90(24):11825–11829. doi: 10.1073/pnas.90.24.11825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The Pharmacogenomics Knowledge Base (PharmGKB) ( https://www.pharmgkb.org/page/cpic)
- 8.Evans WE, Relling MV. Moving towards individualized medicine with pharmacogenomics. Nature. 2004;429(6990):464–468. doi: 10.1038/nature02626. [DOI] [PubMed] [Google Scholar]
- 9.Ma MK, Woo MH, McLeod HL. Genetic basis of drug metabolism. Am J Health Syst Pharm. 2002;59(21):2061–2069. doi: 10.1093/ajhp/59.21.2061. [DOI] [PubMed] [Google Scholar]
- 10.Ingelman-Sundberg M, Oscarson M, McLellan RA. Polymorphic human cytochrome P450 enzymes: An opportunity for individualized drug treatment. Trends Pharmacol Sci. 1999;20(8):342–349. doi: 10.1016/s0165-6147(99)01363-2. [DOI] [PubMed] [Google Scholar]
- 11.Meyer UA. Pharmacogenetics – five decades of therapeutic lessons from genetic diversity. Nat Rev Genet. 2004;5(9):669–676. doi: 10.1038/nrg1428. [DOI] [PubMed] [Google Scholar]
- 12.Bradford LD. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics. 2002;3(2):229–243. doi: 10.1517/14622416.3.2.229. [DOI] [PubMed] [Google Scholar]
- 13.Tateishi T, Chida M, Ariyoshi N, Mizorogi Y, Kamataki T, Kobayashi S. Analysis of the CYP2D6 gene in relation to dextromethorphan Odemethylation capacity in a Japanese population. Clin Pharmacol Ther. 1999;65(5):570–575. doi: 10.1016/S0009-9236(99)70077-9. [DOI] [PubMed] [Google Scholar]
- 14.Gaedigk A, Gotschall RR, Forbes NS, Simon SD, Kearns GL, Leeder JS. Optimization of cytochrome P4502D6 (CYP2D6) phenotype assignment using a genotyping algorithm based on allele frequency data. Phar-macogenetics. 1999;9(6):669–682. doi: 10.1097/01213011-199912000-00002. [DOI] [PubMed] [Google Scholar]
- 15.Sistonen J, Sajantila A, Lao O, Corander J, Barbujani G, Fuselli S. CYP2D6 worldwide genetic variation shows high frequency of altered activity variants and no continental structure. Pharmacogenet Genomics. 2007;17(2):93–101. doi: 10.1097/01.fpc.0000239974.69464.f2. [DOI] [PubMed] [Google Scholar]
- 16.Ingelman-Sundberg M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): Clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics J. 2004;5(1):6–13. doi: 10.1038/sj.tpj.6500285. [DOI] [PubMed] [Google Scholar]
- 17.Soderback E, Zackrisson AL, Lindblom B, Alderborn A. Determination of CYP2D6 gene copy number by pyrosequencing. Clin Chem. 2005;51(3):522–531. doi: 10.1373/clinchem.2004.043182. [DOI] [PubMed] [Google Scholar]
- 18.Lundqvist E, Johansson I, Ingelman-Sundberg M. Genetic mechanisms for duplication and multiduplication of the human CYP2D6 gene and methods for detection of duplicated CYP2D6 genes. Gene. 1999;226(2):27–338. doi: 10.1016/s0378-1119(98)00567-8. [DOI] [PubMed] [Google Scholar]
- 19.Lindpaintner K. Pharmacogenetics and the future of medical practice. Br J Clin Pharmacol. 2002;54(2):221–230. doi: 10.1046/j.1365-2125.2002.01630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirchheiner J, Nickchen K, Bauer M, Wong ML, Licinio J, Roots I. et al. Pharmacogenetics of antidepressants and antipsychotics: The contribution of allelic variations to the phenotype of drug response (Review) Mol Psychiatry. 2004;9(5):442–473. doi: 10.1038/sj.mp.4001494. [DOI] [PubMed] [Google Scholar]
- 21.Sistonen J, Fuselli S, Levo A, Sajantila A. CYP 2D6 genotyping by a multiplex primer extension reaction. Clin Chem. 2005;51(7):1291–1295. doi: 10.1373/clinchem.2004.046466. [DOI] [PubMed] [Google Scholar]
- 22.Goddard KA, Hopkins PJ, Hall JM, Witte JS. Linkage disequilibrium and allele-frequency distributions for 114 single-nucleotide polymerphisms in five populations. Am J Hum Genet. 2000;66(1):216–234. doi: 10.1086/302727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rebsamen MC, Desmeules J, Daali Y, Chiappe A, Diemand A, Rey C. et al. The AmpliChip CYP450 test: Cytochrome P450 2D6 genotype assessment and phenotype prediction. Pharmacogenomics J. 2009;9(1):34–41. doi: 10.1038/tpj.2008.7. [DOI] [PubMed] [Google Scholar]
- 24.Umamaheswaran G, Dhakchinamoorthi K, Chandrasekaran A. Distribution of genetic polymorphisms of genes encoding drug metabolizing enzymes & drug transporters – a review with Indian perspective. Indian J Med Res. 2014;139(1):27–65. [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Dosari M, Al-Jenoobi F, Alkharfy K, Alghamdi A, Bagulb K, Parvez M. et al. High prevalence of CYP 2D6*41 (G2988A) allele in Saudi Arabians. Environ Toxicol Pharmacol. 2013;36(3):1063–1067. doi: 10.1016/j.etap.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 26.Qumsieh RY, Ali BR, Abdulrazzaq YM, Osman O, Akawi NA, Bastaki SMA. Identification of new alleles and the determination of alleles and genotypes frequencies at the CYP2D6 gene in Emiratis. PLoS ONE. 2011;6(12):e28943. doi: 10.1371/journal.pone.0028943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapedanovska NestorovskaA, Jakovski K, Naumovska Z, Hiljadnikova Bajro M, Sterjev Z, Eftimov A. et al. Distribution of the most common genetic variants associated with a variable drug response in the population of the Republic of Macedonia. Balkan J Med Genet. 2014;17(2):5–14. doi: 10.2478/bjmg-2014-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chou WH1, Yan FX, de Leon J, Barnhill J, Rogers T, Cronin M. et al. Extension of a pilot study: Impact from the cytochrome P450 2D6 polymorphism on outcome and costs associated with severe mental illness. J Clin Psychopharmacol. 2000;20(2):246–251. doi: 10.1097/00004714-200004000-00019. [DOI] [PubMed] [Google Scholar]
- 29.Bozina N, Granić P, Lalić Z, Tramisak I, Lovrić M, Stavljenić-Rukavina A. Genetic polymorphisms of cytochromes P450: CYP2C9, CYP2C19, and CYP2D6 in Croatian population. Croat Med J. 2003;44(4):425–428. [PubMed] [Google Scholar]
- 30.Arvanitidis K, Ragia G, Iordanidou M, Kyriaki S, Xanthi A, Tavridou A. et al. Genetic polymorphisms of drug-metabolizing enzymes CYP2D6, CYP2C9, CYP2C19 and CYP3A5 in the Greek population. Fundam Clin Pharmacol. 2007;21(4):419–426. doi: 10.1111/j.1472-8206.2007.00510.x. [DOI] [PubMed] [Google Scholar]
- 31.Buzková H, Pechandová K, Slanar O, Perlik F. Frequency of single nucleotide polymorphisms of CYP-2D6 in the Czech population. Cell Biochem Funct. 2008;26(1):76–81. doi: 10.1002/cbf.1402. [DOI] [PubMed] [Google Scholar]
- 32.Sachse C, Brockmoller J, Bauer S, Roots I. Cytochrome P450 2D6 variants in a Caucasian population: Allele frequencies and phenotypic consequences. Am J Hum Genet. 1997;60(2):284–295. [PMC free article] [PubMed] [Google Scholar]