Summary
The endolysosomal system is critical for the maintenance of cellular homeostasis. However, how endolysosomal compartment is regulated by mitochondrial function is largely unknown. We have generated a mouse model with defective mitochondrial function in CD4+ T lymphocytes by genetic deletion of the mitochondrial transcription factor A (Tfam). Mitochondrial respiration-deficiency impairs lysosome function, promotes p62 and sphingomyelin accumulation and disrupts endolysosomal trafficking pathways and autophagy, thus linking a primary mitochondrial dysfunction to a lysosomal storage disorder. The impaired lysosome function in Tfam-deficient cells subverts T cell differentiation toward pro-inflammatory subsets and exacerbates the in vivo inflammatory response. Restoration of NAD+ levels improves lysosome function and corrects the inflammatory defects in Tfam-deficient T cells. Our results uncover a mechanism by which mitochondria regulate lysosome function to preserve T cell differentiation and effector functions, and identify novel strategies for intervention in mitochondrial-related diseases.
Keywords: TFAM, mitochondria, metabolism, glycolysis, fatty acid oxidation, lysosome storage disorder, TFEB, sphingomyelin, T cell differentiation, inflammation, autophagy
Introduction
Mitochondria are multifunctional organelles that integrate cell metabolism by regulating important anabolic and catabolic pathways. Mitochondria integrate their diverse functions through multiple pathways that ultimately converge on proper function of the respiratory complexes of the electron transport chain (Friedman and Nunnari, 2014). The subunits of these mitochondrial complexes are encoded by both nuclear and mitochondrial DNA (mtDNA), and a correct balance among them is critical to maintaining the function of the electron transport chain, and is associated with organismal life-span (Gomes et al., 2013; Houtkooper et al., 2013). The mitochondrial transcription factor A (Tfam) is the most abundant mtDNA-associated protein in mammalian cells and its expression controls mtDNA copy number in cells, making it an essential player in the maintenance, transcription, and replication of mtDNA (Ekstrand et al., 2004; Larsson et al., 1998). Global deletion of Tfam is embryonically lethal, but tissue-specific ablation of this factor disrupts respiratory chain function in selected cell populations and generates a variety of alterations that recapitulate important phenotypes of human mitochondrial diseases (Vernochet et al., 2012; Viader et al., 2013).
Mitochondria are dynamic organelles that modify their distribution, structure and function in response to changing circumstances such as oxygen availability or stress (Acin-Perez et al., 2014; Detmer and Chan, 2007; Lapuente-Brun et al., 2013). Moreover, the crosstalk of mitochondria with other intracellular organelles is crucial during cell adaptation to stresses and for the preservation of organelle functionality (Rowland and Voeltz, 2012). Contacts between mitochondria and the endoplasmic reticulum form critical hubs implicated in the regulation of mitochondrial fission, calcium homeostasis, ATP production, apoptosis and lipid metabolism, and deregulation of these contacts can lead to disease (Arruda et al., 2014). In addition, mitochondrial contacts with the lysosome-related vacuole in yeast (Elbaz-Alon et al., 2014; Honscher et al., 2014) and with melanosomes in melanocytes (Daniele et al., 2014), have been shown to modulate lipid homeostasis and organelle function. Defining the intricate communication of mitochondria with other intracellular organelles is thus critical for understanding essential physiological processes and how their dysfunction might contribute to the development of human disease.
The endolysosomal system is a dynamic membrane compartment critical to the maintenance of cellular homeostasis. Endosomes, multivesicular bodies and lysosomes regulate signaling, secretion, and the recycling and degradation of protein and lipid cellular components. Lysosomes are critical organelles with degradative and recycling functions, but also mediate other fundamental processes such as plasma membrane repair, signaling and regulation of lipid metabolism (Settembre et al., 2013b). Lysosome biogenesis and function are subjected to transcriptional regulation by the transcription factor EB (TFEB), which allows adaptation of lysosomal function to different physiological conditions (Medina et al., 2015). Alterations to lysosome function and endolysosomal trafficking pathways are mostly found in lysosomal storage disorders (LSDs). LSDs are caused either by deficiency of lysosomal proteins or by changes in non-lysosomal proteins that result in the accumulation of undegraded substrates in lysosomes and abnormal storage of lipids such as sphingomyelins, triacylglycerides, glycosphingolipids or sphingosines (Futerman and van Meer, 2004). Remarkably, lysosomal dysfunction in Gaucher disease blocks mitochondrial turnover by mitophagy (Osellame et al., 2013), thus connecting the lysosome degradation capacity to the maintenance of a healthy mitochondrial repertoire. Whether mitochondria regulate the biogenesis and function of the endolysosomal compartment is still unknown.
During adaptive immune responses, naive CD4+ T cells that encounter a foreign antigen must reprogram their metabolism to meet the costly biosynthetic and bioenergetic demands of cellular activation, proliferation and effector function (Pearce et al., 2013). T cells reorient their metabolism toward aerobic glycolysis. Although less efficient at ATP synthesis, glycolysis generates intermediate metabolites that support biosynthetic pathways for effector functions, including cytokine production (Chang et al., 2013). Because of the glycolytic nature of T lymphocyte metabolism, the role of mitochondria in T cell function has been traditionally neglected. However, accumulating evidence indicates that T cell effector functions require mitochondrial oxygen consumption, ATP and ROS (Weinberg et al., 2015).
To investigate the mechanisms by which mitochondria influence T cell responses and ultimately affect immune disease progression, we have generated a mouse model in which mitochondrial function is specifically disrupted in the adaptive immune system by targeting Tfam in CD4+ T cells. Tfam-deficient cells have below-normal levels of mtDNA, diminished mitochondrial-respiration, and a metabolic signature characterized by increased anaerobic glycolysis and impaired fatty acid β-oxidation. Respiration-impaired cells show reduced lysosomal calcium mobilization and impaired lysosomal degradation capacity revealed by p62 and sphingomyelin accumulation, defects that cells try to compensate by inducing lysosome biogenesis through the transcription factor EB (TFEB). The impaired lysosome function in Tfam-deficient cells subverts T cell differentiation toward pro-inflammatory subsets and exacerbates the in vivo inflammatory response. Improvement of lysosome function by restoration of NAD+/NADH balance through NAD+ precursors corrected inflammatory defects in Tfam-deficient T cells. These data reveal a novel mechanism by which mitochondria regulate lysosome function to preserve T cell differentiation and effector functions.
Results
Tfam depletion impairs respiratory-chain function in T cells
To investigate the involvement of mitochondria in the regulation of endolysosomal function and T cell responses, we generated a mouse model of mitochondrial dysfunction by specifically deleting Tfam in CD4+ T cells. This was achieved by crossing mice with loxP-flanked Tfam alleles (Tfamfl/fl) with CD4-Cre mice. Consistent with gene deletion in the early stages of T cell development, Tfam deletion efficiently decreased the mRNA of Tfam in CD4+ and in CD8+ T naive lymphocytes (Figure S1A-B). CD4Cre+/wtTfamfl/fl mice (herein Tfam-/- mice) developed normally and showed similar frequency of CD4 and CD8 single positive, and double positive thymocytes to their control littermates (Figure 1A), indicating that Tfam is not required during early T cell development. Tfam-/- mice presented slightly lower percentages of CD4+ and CD8+ T cells in the spleen and peripheral lymph nodes (Figure 1B), but had similar numbers of splenocytes, B cells and dendritic cells to littermate CD4Crewt/wtTfamfl/fl controls (herein wt mice) (Figure S1C).
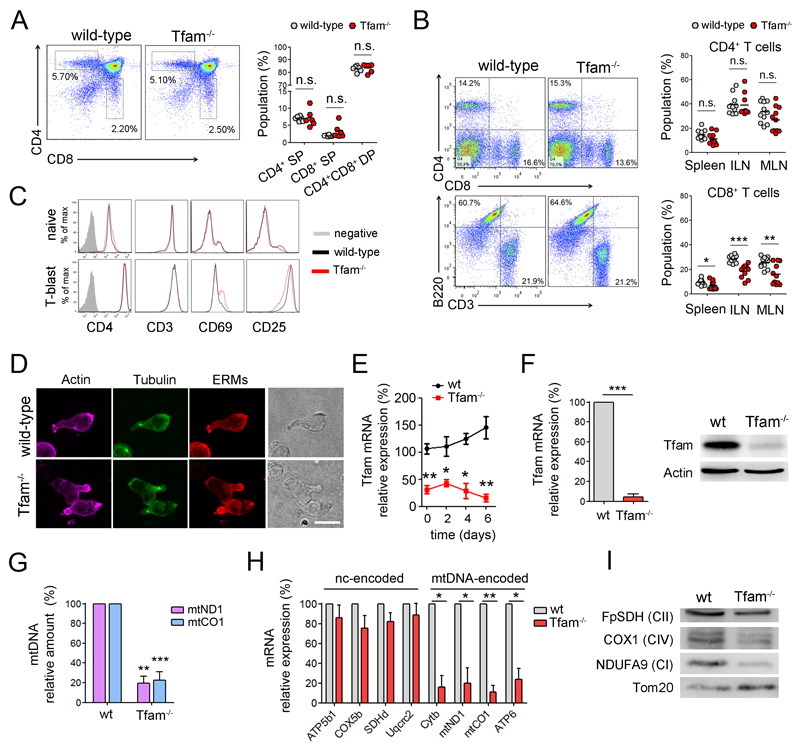
Figure 1. Tfam depletion induces severe mtDNA deficiency in T cells.
(A) Dot plots show CD4 and CD8 expression in thymocytes from wt and Tfam-/- mice. Right, percentage of CD4 and CD8 single positive (SP), and CD4/CD8 double positive (DP) cells. (B) Dot plots show CD4 and CD8 T cells, and CD3 and B220 cells from the spleens. Right, percentages of CD4 and CD8 cells in the spleen, inguinal (ILN) and mesenteric lymph nodes (MLN) (n=11). (C) Expression of cell surface markers in naive CD4 T cells and T-lymphoblasts differentiated with ConA (48 hr) and IL-2 (4 days). (D) Confocal images show the polarization of cytoskeletal components in T lymphoblasts by actin, tubulin and ERM (ezrin-radixin-moesin) staining. Scale bar represents 10μm. (E) Relative Tfam mRNA levels by RT-PCR in naive CD4 T cells (day 0) and during lymphoblast differentiation. (F) Tfam mRNA (left) and protein levels (right) in CD4 T lymphoblasts. (G) mtDNA levels (mtCO1 and mtND1) relative to nuclear DNA (SDH) in CD4 T lymphoblasts. (H) mRNA levels of mtDNA-encoded and genome-encoded mitochondrial subunits. (I) Immunoblot of T lymphoblast mitochondrial proteins. Complex I (CI) was detected with anti-NDUFA9, CII with anti-FpSDH, and CIV with anti-COX1. Tom20 was used as loading control. Data (B, E, F, G, H) are means ± SEM (n > 3); *p<0.05, **p<0.01 and ***p<0.001 (Student’s t-test).
Naive Tfam-/- CD4+ T cells expressed comparable levels of T cell surface markers to wt cells and responded normally to in vitro differentiation toward T lymphoblasts, adopting a polarized morphology (Figure 1C and 1D). The levels of Tfam were suppressed throughout lymphoblast differentiation, excluding the selection of Tfam-positive cells during in vitro expansion (Figure 1E and 1F). Consistent with the close relationship between levels of Tfam and mtDNA, lack of Tfam induced a severe decrease in mtDNA content, both in Tfam-/- CD4+ T lymphoblasts and in human Jurkat T cells stably transfected with Tfam short hairpin RNAs (shTfam) (Figure 1G and S1D). Tfam deficiency reduced the mRNA expression of mtDNA-encoded subunits of mitochondrial complexes I, III, IV and V, whereas nuclear-encoded subunits were not significantly affected (Figure 1H). Consistently, the protein levels of complexes I and IV, which include mtDNA-encoded subunits, were below normal, whereas the nuclear-encoded complex II was unaffected (Figure 1I).
In response to these respiratory-chain alterations, Tfam-deficient cells increased mitochondrial content (Figure 2A). However, electron microscopy analysis revealed severely aberrant mitochondrial morphology in Tfam-/- CD4+ T cells, accompanied by impaired cristae organization and loss of mitochondrial electron density (Figure 2B), consistent with a mitochondrial dysfunction caused by depletion of Tfam.
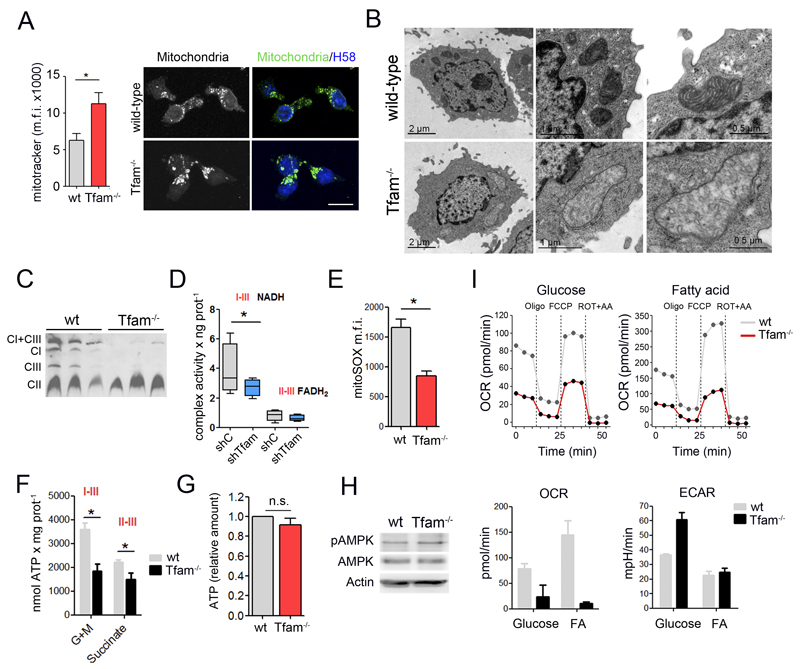
Figure 2. Tfam ablation induces OXPHOS deficiency in T cells.
(A) Left, flow cytometry analysis of Mitotracker staining. Right, confocal images of mitochondria (anti-MnSOD), and nuclei HOECHST58 (H58, blue) in T lymphoblasts. Scale bar represents 10μm. (B) Electron microscopy images in T lymphoblasts. (C) Blue-native gel electrophoresis analysis of electron-transport-chain complexes (detection of NDUFA9, FpSDH and Core1 for complexes I, II and III, respectively) from three wt and three Tfam-/- T cell lysates. (D) Combined mitochondrial complex activities in Jurkat T cells. Lines extending from the boxes indicate the variability outside the upper and lower quartiles. (E) Flow cytometry analysis of mROS in T lymphoblasts stained with MitoSOX. Data are presented as the mean fluorescence intensity. (F) Glutamate- and pyruvate-driven ATP-dependent production in Jurkat T cells. (G) Cellular ATP content in wt and Tfam-/- T cells. (H) Immunoblot analysis of AMPK phosphorylation at Thr172 in T lymphoblasts. Actin was used as a loading control. (I) CD3/CD28-activated CD4 T cells fed either with glucose or palmitate (FA) for 2 hr and oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) measured in real time (upper panels). Lower panels show the basal OCR and ECAR from a representative experiment. Data (D, E, F, G) are means ± SEM (n ≥ 3, *p<0.05, Student’s t-test).
Blue native gel electrophoresis (BNGE) of extracts from Tfam-silenced cells revealed severe loss of complexes I and III but not II, and associated alterations in mitochondrial supercomplex assembly (Figure 2C). Accordingly, Tfam-depleted cells showed stronger impairment of electron transport from NADH (complexes I+III) than from FADH2 (complexes II+III) (Figure 2D), demonstrating an imbalance in the NAD+/NADH ratio in respiratory-chain impaired cells. Connected with the reduced electron transport chain function, Tfam-deficient T cells also had below-normal content of mitochondrial reactive oxygen species (ROS) (Figure 2E), reduced mitochondrial ATP generation (Figure 2F), and impaired survival in galactose medium (Figure S1E). In addition, while oligomycin treatment caused a transient hyperpolarization in wt cells, it dissipated mitochondrial membrane potential in Tfam-/- suggesting reversal F1F0ATPase function as consequence of impaired electron transport chain function (Figure S1F). Despite the effect on mitochondrial ATP production, total cellular ATP content in primary CD4+ Tfam-/- T cells and Jurkat shTfam T cells was comparable with controls (Figure 2G and S1G). In addition, the phosphorylation level of the metabolic sensor AMP-activated protein kinase (AMPK) did not differ between Tfam-deficient and control cells (Figure 2H), indicating that global energy status is not compromised in Tfam-depleted cells.
We then analyzed the metabolic consequences of Tfam deletion on CD4+ T cells by flux analysis. In activated CD4+ T cells, we measured the extracellular acidification rate (ECAR), as an index of lactate production and glycolysis, and the oxygen consumption rate (OCR) as an indicator of mitochondrial oxidative phosphorylation (OXPHOS). Upon activation with anti-CD3 and anti-CD28, wild-type T cells used glycolysis and OXPHOS for glucose consumption, as described (Michalek et al., 2011; Pearce et al., 2013). In contrast, Tfam-/- T cells presented a low OCR and an ECAR above wild-type levels, demonstrating anaerobic glucose utilization (Figure 2I). Additionally, we examined mitochondrial fatty acid ß-oxidation (FAO) in respiration-deficient cells. Naive wt and Tfam-/- CD4+ T cells were activated over 48h and then incubated with fatty acids prior to flux analysis. In these conditions, activated wild-type CD4+ T cells showed increased OXPHOS and reduced glycolysis, thus relying on FAO and mitochondrial OXPHOS for energy production. In contrast, Tfam-/- T cells showed reduced OCR, supporting the conclusion that FAO is impaired in respiratory-chain deficient cells (Figure 2I).
Tfam deletion thus promotes loss of mtDNA, OXPHOS deficiency, and compromised mitochondrial function, but has no significant impact on cellular energy status or survival. Additionally, impaired mitochondrial respiration induces a metabolic reprogramming characterized by increased anaerobic glucose consumption and impaired FAO.
Respiration-impaired cells increase lysosomal compartment through TFEB
In addition to the profound morphological and functional mitochondrial alterations, electron microscopy studies revealed striking intracellular vesiculation in Tfam-deficient cells (Figure 3A). These abnormalities prompted us to explore the role of mitochondria in the abundance and organization of the endolysosomal compartment. By flow cytometry, the early endosome marker EEA1 showed no substantial differences, but expression of the late endosomal markers HRS and LBPA (lysobisphosphatidic acid) was elevated in respiratory-chain impaired cells (Figure 3B). Immunofluorescence analysis revealed enlargement of late endosomes in Tfam-deficient cells (Figures 3C). In addition, lysosomes, identified by staining for LAMP1 and Lysotracker, appeared to be more abundant in Tfam-deficient cells than in wild-types (Figure 3B-3C), indicating the expansion of the late endosome and lysosome compartment in cells that lack Tfam.
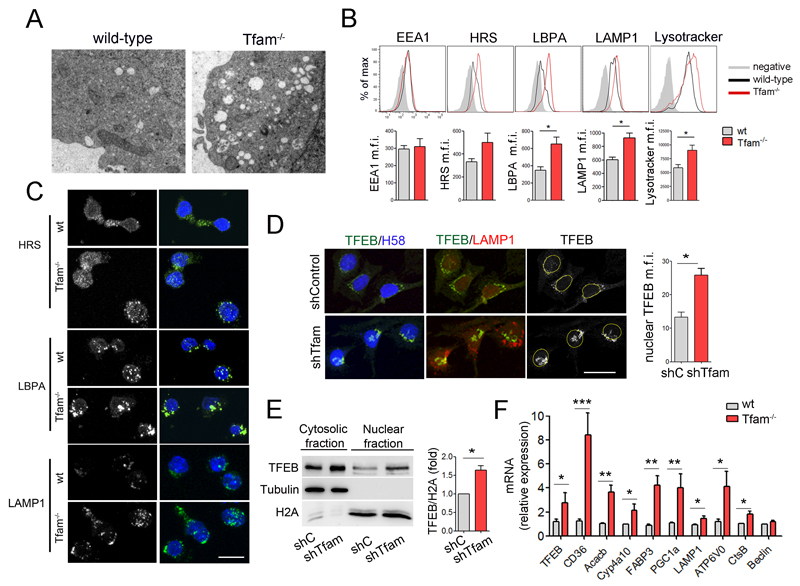
Figure 3. Tfam regulates lysosomal biogenesis through TFEB.
(A) Electron microscopy images show the abnormal intracellular accumulation of vesicles in Tfam-/- T lymphoblasts. (B) Flow cytometry analysis of EEA1, HRS, LBPA, LAMP1 and lysotracker content in CD4 T lymphoblasts. Results show mean fluorescence intensity (m.f.i.) (n=3). (C) Confocal images of HRS, LBPA, and LAMP1 in T lymphoblasts. Nuclei were stained with HOECHST58 (H58, blue). Scale bar represents 10μm. (D) Confocal images of TFEB (green), LAMP1 (red), H58 (blue) in Oli-Neu cells. Right, quantification of nuclear TFEB m.f.i. (E) Western blot analysis of TFEB subcellular location. Cytosolic and nuclear fractions of Jurkat cells were blotted for TFEB, tubulin, and histone 2A (H2A). Chart shows densitometry analysis of the ratio of nuclear TFEB to H2A (n=3). (F) Relative mRNA levels of TFEB and target genes in T lymphoblasts by RT-PCR (n=5). Data (B, D, E, F) are means ± SEM; *p<0.05, **p<0.01 and ***p<0.001 (Student’s t-test).
Lysosomal biogenesis is regulated by TFEB, a master regulator of the CLEAR (coordinated lysosomal expression and regulation) gene network (Sardiello et al., 2009). During lysosomal stress or starvation, cytosolic TFEB relocates to the nucleus and activates a transcriptional program aimed at increasing autophagy and lysosomal biogenesis and function (Settembre et al., 2013b). TFEB is also responsible for the transcriptional activation of PGC1α, which adjusts the content of mitochondrial and peroxisomal enzymes and the expression of components that increase lipid catabolic reactions (Settembre et al., 2013a). Confocal analysis revealed that TFEB translocates to the nucleus in Tfam-depleted cells (Figure 3D and S2A). Nuclear localization of TFEB was confirmed by cell fractionation assays (Figure 3E and S2B), and correlated with upregulated expression of the TFEB target genes LAMP1, ATP6V0, CtsB, FABP3, CD36, Acacb, Cyp4a10 and PGC1α (Figure 3F), which are involved in lipid uptake, lipid catabolism, lysosomal and mitochondrial function. These results suggest that the enlarged lysosomal compartment observed in respiration-impaired Tfam-/- cells is associated with TFEB activation.
Mitochondrial dysfunction promotes lysosomal disorder and sphingolipidosis
To further examine the impact of Tfam deficiency on lysosome function, we measured the levels of calcium in lysosomes after inducing its release with bafilomycin-A1 (Lloyd-Evans et al., 2008), a selective inhibitor of the vacuolar-type H+ ATPase. Lysosomal calcium mobilization was weaker in Tfam-deficient cells (Figure 4A), whereas the subsequent addition of ionomycin, which releases calcium from remaining stores, triggered comparable release in wt and Tfam-/- T cells. Measurement with a specific lysosomal pH-sensitive probe indicated reduced pH in lysosomes from respiration-impaired Tfam-/- cells (Figure S3A). The activity of the lysosomal endopeptidase cathepsin B was significantly reduced in Tfam-/- T cells (Figure 4B), whereas acid phosphatase activity showed no difference (Figure S3B).
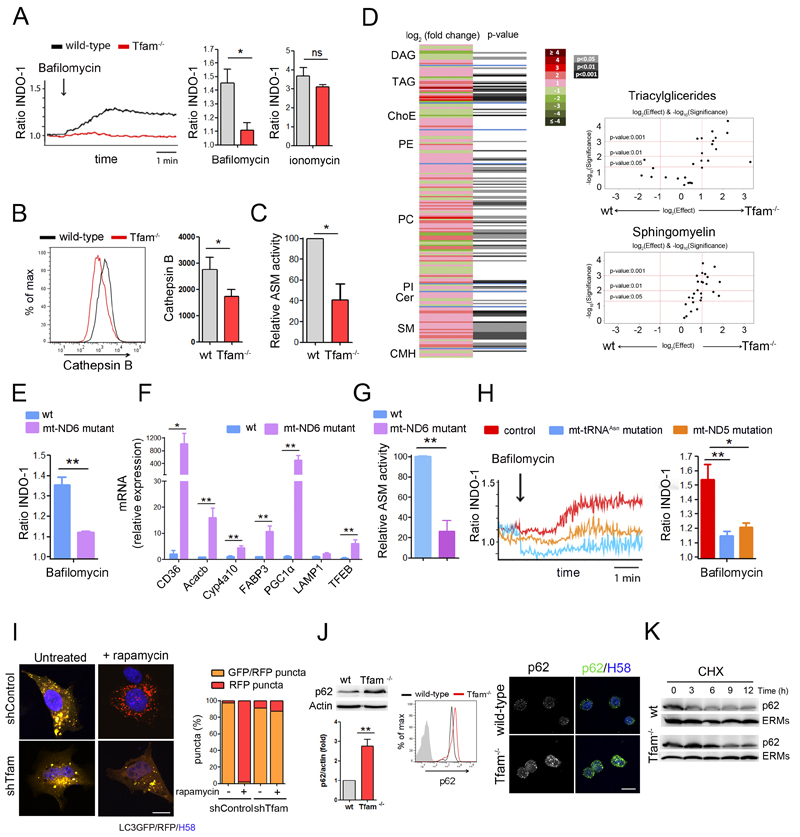
Figure 4. Tfam controls lysosomal function.
(A) Lysosomal calcium mobilization in T lymphoblasts loaded with Indo-1, treated with bafilomycin-A1 and detected by flow cytometry. The 340/380 nm ratio is shown. Right, quantification of Indo-1 signal in bafilomycin-A1- or ionomycin-treated cells relative to the basal level. (B) Histograms and quantification of Cathepsin B activity measured by flow cytometry (MagicRed). (C) Acid sphingomyelinase (ASM) activity in T lymphocytes. (D) Heatmap represents cell lipidomic signatures of T cells (n = 3). More intense colors indicate larger drops (green) or elevations (red) of the metabolite levels in Tfam-/- samples. DAG, diacylglycerides; TAG, triacylglycerides; ChoE, cholesteryl esters; PE, phosphatidylethanolamines; PC, phosphatidylcholines; PI; phosphatidylinositols; Cer, ceramides; SM, sphingomyelins; CMH, monohexosylceramides. Right, volcano plots [log10(p-value) vs. log2(fold change)] for the comparison of Tfam-/- and wt T cells for the indicated metabolites. (E) Lysosomal calcium in mt-ND6 Complex I (CI) mutant and wt fibroblasts treated as in A. Ratio of Indo-1 signal upon bafilomycin-A1 treatment relative to the basal level. (F) Relative mRNA levels of TFEB and selected target genes in wt and mt-ND6 mutant fibroblasts assessed by RT-PCR. (G) ASM activity in wt and mt-ND6 mutant fibroblasts. (H) Lysosomal calcium in fibroblasts from patients with point mutation in CI subunit mt-ND5 gene (m.13513G>A), point mutation in the mt-tRNAAsn gene (m.5658T>C) or control samples assessed as in A. (I) Left, confocal images show Oli-Neu transfected with LC3-GFP-RFP treated or not with rapamycin. Right, graph shows percentage of GFP/RFP puncta (yellow vesicles) and RFP puncta (red vesicles) from at least 100 vesicles in two independent experiments. (J) Analysis of p62 by western blot, flow cytometry and confocal immunofluorescence in T lymphoblasts. Chart shows densitometry analysis of p62 by western blot. (K) Western blot analysis of p62 protein turnover in cells treated with cycloheximide for the indicated times. ERMs are used as loading control. Data (A, B, C, E, F, G, H, J) are means ± SEM, n ≥3; *p<0.05 and **p<0.01 (Student’s t-test). Scala bars represent 10 μm.
Sphingomyelins (SMs) and other lipids such as cholesterol, glycosphingolipids and sphingosine accumulate abnormally in lysosomal storage disorders (LSDs) (Futerman and van Meer, 2004). Lipidomic analysis showed that CD4+ Tfam-/- T cells have an altered lipid profile compared with wild-types, affecting 90 out of 218 lipids analyzed and characterized by elevated levels of several lipid species (Figure 4D). Consistent with the reduced lysosomal degradation capacity, respiration-impaired Tfam-/- cells showed a significant accumulation of SMs and triacylglycerides (TAGs) (Figure 4D and S3C). Supporting lysosomal dysfunction and SM accumulation in respiration-impaired cells, Tfam-/-T cells showed significantly reduced acid sphingomyelinase (ASM) activity (Figure 4C).
To dissect how mitochondria regulate lysosomal function, we investigated whether cells containing point mutations in mtDNA genes for specific mitochondrial respiratory complexes undergo the same lysosomal calcium defects. Mouse fibroblasts that carry a high load (98%) of a null mutation in the mt-ND6 gene (complex I subunit) showed impaired lysosomal calcium mobilization, upregulation of TFEB target genes and reduced ASM activity (Figure 4E-4G). Notably, impaired lysosomal calcium mobilization was also detected in fibroblasts from two human patients, one carrying a high load (88%) of a point mutation in the mt-tRNAAsn gene (m.5658T>C), and another carrying a 50% load of a point mutation in the complex I subunit mt-ND5 gene (m.13513G>A) (Figure 4H). These results indicate that lysosomal calcium levels are decreased not only by the severe OXPHOS-deficiency associated with Tfam-depletion, but also by point mutations in mtDNA associated with complex I dysfunction, both in mice and in human patient samples.
Defects in lipid trafficking and autophagosome-lysosome fusion are commonly observed in sphingolipidoses. The reduced lysosomal calcium and abnormal SM accumulation detected in respiration-impaired cells prompted us to investigate the ability of lysosomes to fuse with autophagosomes. In cells transfected with the autophagosome marker LC3GFP-RFP, GFP fluorescence is decreased when autophagosomes fuse with lysosomes because it is more sensitive to pH than the RFP signal, which is maintained in acidic compartments. In these experimental settings, control cells showed increased RFP puncta upon induction of autophagy by rapamycin treatment (Figure 4I). In contrast, Tfam-deficient cells showed a significant reduction in the number of RFP-positive puncta, indicating impaired autophagosome-lysosome fusion in the absence of Tfam. Assessment of mitochondrial degradation by mitophagy upon acute membrane depolarization with CCCP revealed larger parkin-GFP aggregates in Tfam-deficient cells than in control cells (Figure S3D), indicating impaired parkin clearance and mitochondrial degradation.
In accordance with impaired autophagic flux, Tfam-depleted cells showed increased levels of the lipidated form of LC3 (LC3-II) (Figure S3E). Additionally, flow cytometry, western blot and confocal analysis showed a significant accumulation of the autophagic substrate p62 in Tfam-deficient cells (Figure 4J). Blocking protein synthesis with cycloheximide revealed slower p62 turnover in respiration-impaired cells than in wild-types, suggesting that the accumulation of p62 was caused by impaired lysosomal degradation and not by increased mRNA transcription (Figure 4K).
Cells that lack Tfam exhibit lysosomal defects similar to those seen in LSDs: abnormal lipid trafficking, altered calcium mobilization and defective autophagosome-lysosome fusion. Overall, these results reveal a novel link between a primary mitochondrial dysfunction and the acquisition of a lysosomal storage disorder.
Tfam deficiency exacerbates inflammatory responses
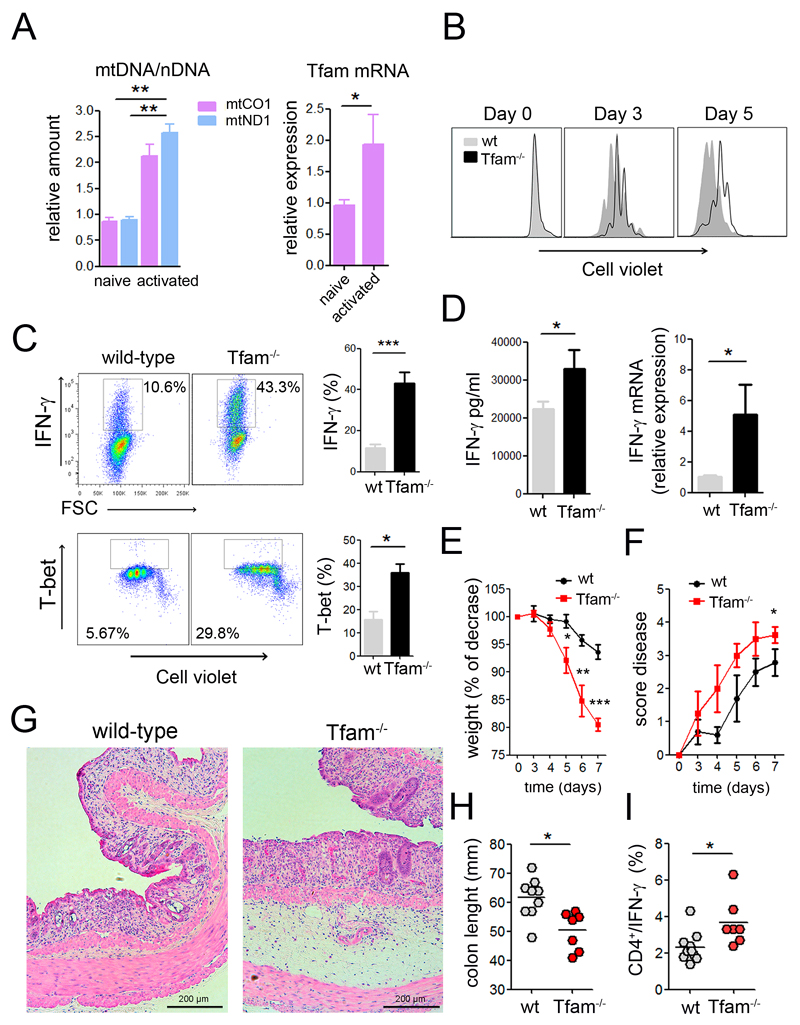
We next assessed how these mitochondrial and endolysosomal alterations caused by Tfam deficiency affect T cell effector function. Upon T cell activation, Tfam expression increased concomitantly with an increase in mtDNA levels (Figure 5A and S4A), suggesting that mitochondrial adaptation to T cell activation requires an increase in mtDNA levels. T cells from wild-type and Tfam-/- mice were labeled with the proliferation indicator Cell Violet, stimulated with anti-CD3 and anti-CD28 antibodies, and evaluated by flow cytometry. Tfam-/- T cells showed reduced proliferation (Figure 5B). Moreover, upon in vitro cell activation, there was no evidence of higher apoptosis in Tfam-/- T cells, as assessed by flow cytometry analysis of annexin V and HOECHST58 staining (Figure S4B). CD4+ Tfam-/- cells showed similar levels of the activation markers CD69 and CD25 (Figure S4C). Stimulated Tfam-/- CD4+ T lymphocytes showed increased numbers of IFN-γ- and T-bet-producing cells compared to wild-type cells (Figure 5C). RT-PCR analysis indicated strikingly elevated mRNA expression of the pro-inflammatory cytokines IFN-γ, IL-6, IL-1α and IL-1β and increased IFN-γ and IL-6 secretion were detected in the supernatant of Tfam-/- T cells (Figure 5D, S4D and S4E). In contrast, IL-4 mRNA levels and cytokine secretion of IL-4 and IL-10 were below-normal in Tfam-deficient cells (Figure S4D and S4E). These in vitro data highlight the requirement on mitochondrial function for the fine regulation of T cell responses.
Figure 5. Inhibition of OXPHOS exacerbates inflammatory responses in vitro and in vivo.
(A) Relative mtDNA and Tfam mRNA levels in naive and CD3/CD28 activated CD4 T cells. (B) Cell proliferation analysis by Cell Violet dilution of CD4 T cells activated with CD3/CD28. (C, D) CD4 T cells were activated with CD3/CD28 (6 days), and IFN-γ and T-bet intracellular levels were measured by flow cytometry (C), IFN-γ secretion by ELISA and relative IFN-γ mRNA levels by RT-PCR (D). (E) Weight and (F) disease score were monitored for the indicated times in mice fed with 3% dextran sulfate sodium (DSS) in the drinking water. H&E staining of colon sections (G) and colon length (H) on day 7 after DSS treatment. (I) IFN-γproducing CD4 T cells in the mesenteric lymph nodes. Data are means ± SEM, (A, C, D, n≥3), (E-I, at least seven mice in two independent experiments); *p<0.05 and **p<0.01, ***p<0.001 (Student’s t-test).
To assess the role of mitochondria in the regulation of the inflammatory function of T cells in vivo, we first examined a model of contact hypersensitivity (CHS), which is mainly mediated by IFN-γ. The specificity of the CHS response is generally defined as the difference between ear swelling responses to a hapten such as oxazolone in naive and sensitized animals. Upon sensitization to oxazolone, Tfam-/- mice showed larger increases in ear thickness, lymph node cellularity, and IFN-γ-producing CD4+ T lymphocytes (Figure S4F). In addition, we performed a model of DSS-induced colitis, which is driven by Th1 and Th17 cells and suppressed by Treg cells. Wild-type mice and Tfam-/- mice were fed with DSS to induce gut damage and trigger disease. Tfam-/- mice showed increased disease severity, as reflected by higher weight loss over time, increased disease score and shorter colon length than wt mice (Figure 5E-5H, S4G and S4H). Remarkably, Tfam-deficiency increased the frequency of IFN-γ- and T-bet-expressing CD4+ T cells in the mesenteric lymph nodes upon DSS-treatment (Figure 5I and S4I). These results indicate that respiration-impaired T cells develop a stronger inflammatory response in vitro and in vivo and support a critical role for mitochondria in the regulation of T cell effector functions during immune responses.
Mitochondrial dysfunction deviates T cell differentiation towards pro-inflammatory Th1 subsets
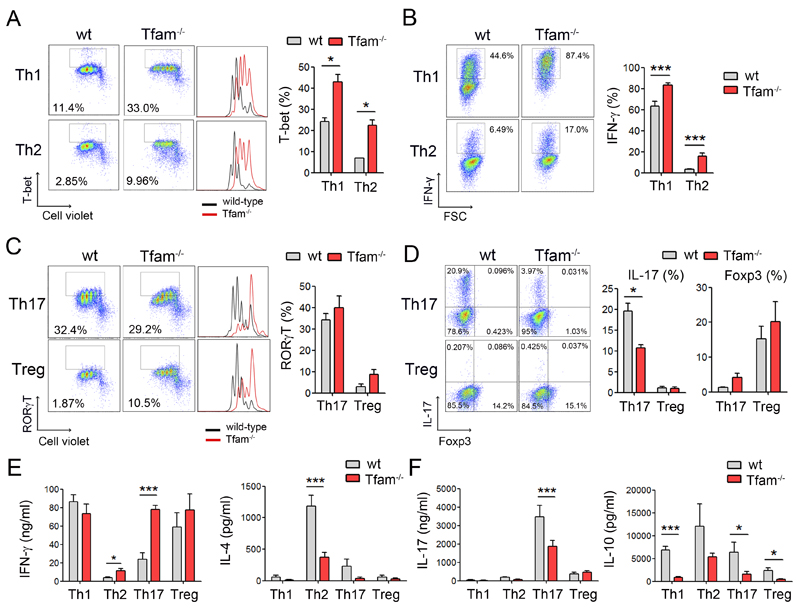
To provide further insights into how mitochondrial function controls T cell differentiation, we assessed the ability of Tfam-deficient T cells to differentiate into effector T cell subsets. The balance among different functional effector T cell subsets determines the outcome of the immune response and is essential to prevent undesired inflammation and autoimmune diseases. Naive CD4+ T cells were labeled with Cell Violet, differentiated in vitro with different cytokines and neutralizing antibodies to Th1, Th2, Th17 and regulatory (Treg) T cell subsets, and assessed for proliferation and expression of specific transcription factors and cytokines. Tfam deficiency greatly impaired proliferation in all the skewing conditions (Figure 6A and 6C). Tfam-depleted cells showed higher expression of the Th1-specific transcription T-bet, higher numbers of IFN-γ-producing cells and higher levels of IFN-γ secretion (Figure 6A, 6B and 6E). In all polarization conditions, Tfam-/- CD4+ T cells secreted less IL-4 than wt cells (Figure 6E), demonstrating a Th1/Th2 imbalance toward the Th1 response in the absence of Tfam.
Figure 6. Mitochondrial dysfunction subverts T cell differentiation toward Th1 cell subsets.
(A) Cell Violet dilution and T-bet expression in CD4 T cells cultured under Th1 and Th2 conditions over 3 days. Chart shows percentage of T-bet expressing cells (n=3). (B) Intracellular flow cytometry analysis of IFN-γ-producing cells under Th1 and Th2 conditions over 6 days. Graph, quantification of IFN-γ-producing cells (n=6). (C) Naive CD4 T cells were cultured under Th17 and Treg conditions over 3 days. Dot plots show RORγT-expressing cells and histograms show cell proliferation assessed by Cell Violet dilution. Chart shows percentage of RORγT-expressing cells (n=3). (D) Intracellular flow cytometry analysis of CD4 T cells producing IL-17 and Foxp3. Chart shows percentage of IL-17 and Foxp3-producing cells (n=6). (E, F) Polarized CD4 T cells at day 6 were cultured in equal numbers in fresh medium followed by activation with PMA/ionomycin for 16 hr. Cytokines were measured by ELISA in the supernatants. Data are means ± SEM (n ≥ 3); *p<0.05, **p<0.01 and ***p<0.001 (Student’s t-test).
When polarized toward Th17 T cell subsets, no substantial differences were found in the number of RORγt-expressing cells between Tfam-/- and wt cells (Figure 6C). However, impaired mitochondrial function in Tfam-/- CD4+ T cells significantly reduced the levels of IL-17-producing cells and impaired IL-17 and IL-10 secretion (Figure 6D and 6F), while increasing IFN-γ secretion (Figure 6E). In Treg conditions, no significant differences were found in the number of Foxp3-expressing cells, but Tfam-/- CD4+ T cells secreted reduced amounts of anti-inflammatory cytokine IL-10 (Figure 6D and 6F). Thus, Tfam-deficiency favoured the appearance Th17 cells which coexpress RORγt and T-bet, and produce high levels of IFN-γ but reduced amounts of IL-10, and which have been proposed to be pathogenic Th17 cells involved in autoimmune diseases in mice (Peters et al., 2011). Collectively, these results reveal a role for mitochondrial function in the regulation of T cell differentiation and indicate that mitochondrial dysfunction is associated with a deregulation of T cell differentiation toward pro-inflammatory Th1 cell subsets.
Increasing intracellular NAD+ content boosts lysosomal function and dampens Th1 responses in respiration-impaired cells
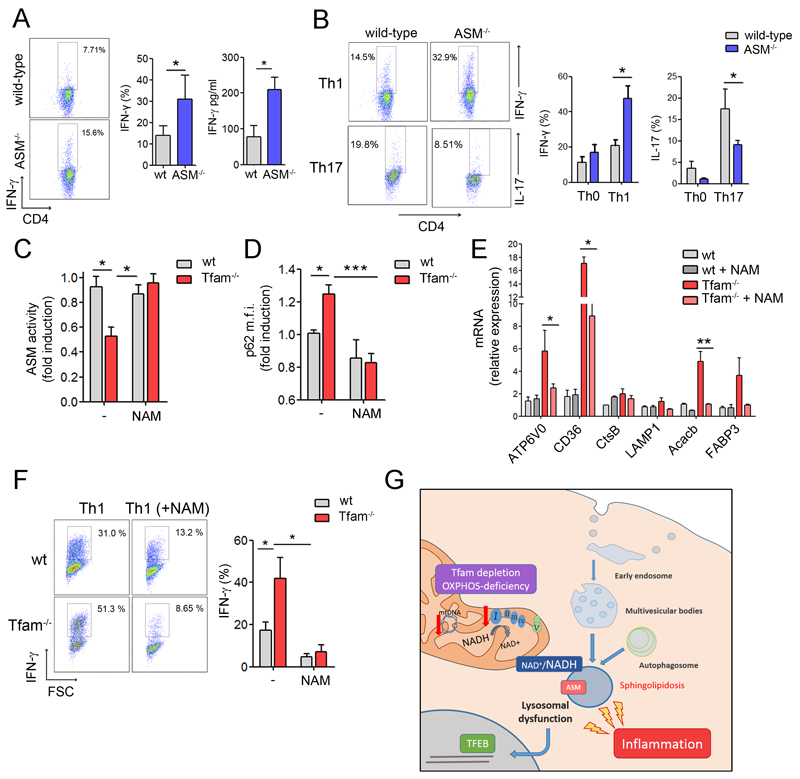
We then investigate whether lysosomal dysfunction caused by mitochondrial respiration deficiency is involved in the increased inflammatory responses observed in Tfam-deficient cells. Naive CD4+ T cells activated in the presence of the lysosomal inhibitor leupeptin or SM showed increased levels of IFN-γ mRNA (Figure S5A), suggesting that the lysosomal dysfunction might contribute to the increased pro-inflammatory profile of Tfam-/- CD4+ T cells. To ascertain the involvement of lysosome function in the regulation of T cell responses, we studied T cell activation and differentiation in mice lacking ASM, which mimics the LSD Niemann Pick disease Type A (NPA) and, similarly to Tfam-/-, shows SM accumulation, lysosomal disorder and autophagy impairment (Gabande-Rodriguez et al., 2014). Naive CD4+ T cells from ASM-/- mice activated in non-skewing conditions showed a higher frequency of IFN-γ-producing cells and higher levels of IFN-γ secretion than wild-types (Figure 7A). Similarly to respiration-impaired Tfam-/- cells, ASM-/- cells showed higher frequency of IFN-γ-producing cells in Th1 conditions, and reduced frequency of IL-17-producing cells in Th17 skewing conditions (Figure 7B). T cells from ASM-/- mice did not show appreciable mitochondrial alterations in terms of mitochondrial mass, mROS, ATP production and OCR (Figure S5B-S5D). Collectively, these results show that lysosome dysfunction is sufficient to deregulate T cell differentiation toward pro-inflammatory Th1 subsets.
Figure 7. Increasing NAD+ levels improves lysosome function and reduces inflammatory responses in Tfam-deficient cells.
(A) Dot plots show intracellular IFN-γ levels by flow cytometry in CD4 T cells activated with CD3/CD28 (6 days). Graphs show percentage of IFN-γ-producing cells and IFN-γ levels in the supernatant by ELISA. (B) CD4 T lymphocytes cultured under Th0, Th1 and Th17 condition for 6 days. Flow cytometry analysis of the frequency of CD4 T cells producing IFN-γ and IL-17. (C) ASM activity and (D) flow cytometry analysis of p62 levels in T-lymphoblasts treated with NAM (10mM) for 2 days. (E) Relative mRNA levels of TFEB target genes in wt and Tfam-/- T cells incubated or not with NAM. (F) CD4 T cells cultured towards Th1 in the presence or absence of NAM for 3 days. Flow cytometry analysis of the frequency of CD4 T cells producing IFN-γ. (G) Diagram representing the proposed mechanism. Data are means ± SEM (n≥3); *p<0.05, **p<0.01 and ***p<0.001 (Student’s t-test).
Increasing intracellular levels of NAD+ has been proposed to be a therapeutical strategy to restore mitochondrial function in respiratory-chain deficiencies (Cerutti et al., 2014; Gomes et al., 2013; Karamanlidis et al., 2013). To assess whether modulation of the cellular NAD+/NADH ratio may be involved in the regulation of lysosomal function, we treated Tfam-deficient T cells with the nicotinamide precursor NAM to reestablish NAD+/NADH balance, and assessed parameters of lysosome function. Addition of NAM increased ASM activity, reduced p62 accumulation, and downregulates TFEB target genes in Tfam-deficient cells (Figure 7C-7E), suggesting that modulation of mitochondrial NAD+/NADH levels impacts lysosome function. Finally, to determine whether restoring NAD+/NADH levels would limit the increased pro-inflammatory profile in respiration-impaired Tfam-/- T cells, naive CD4+ T cells were activated in the presence of NAM for 72h and their cytokine profile assessed. NAM treatment reduced the frequency of IFN-γ-producing Tfam-/- T cells (Figure S5E). Additionally, under Th1 polarization conditions, naive Tfam-/- CD4+ T cells cultured in the presence of NAM showed a reduced percentage of IFN-γ-producing cells (Figure 7F), indicating that improved lysosomal function reduces the exacerbated inflammatory response of mitochondrial respiration-impaired cells.
Discussion
Our findings reveal a novel mechanism through which mitochondrial respiration preserves cellular homeostasis by regulating the function of the endolysosomal compartment during essential biological processes, illustrated here in the regulation of T cell function. We show that lack of Tfam in T cells decreases cellular mtDNA content, alters mitochondrial metabolism and is associated with impaired endolysosomal function, abnormal accumulation of sphingomyelins and increased pro-inflammatory T cell responses (Figure 7G).
We provide here the first evidence for primary mitochondrial dysfunction leading to altered lysosomal function and sphingolipidosis. Cells lacking Tfam exhibit abnormal lipid trafficking, altered lysosomal Ca2+ mobilization, and autophagosome-lysosome fusion, defects similar to those seen in LSDs. In Niemann-Pick (NP) type A and B diseases (NPA and NPB), sphingomyelin (SM) accumulates in lysosomes due to insufficient activity of ASM. SM accumulation has been found to inhibit the main calcium channel in the lysosome, impairing endolysosomal trafficking, protein degradation and macroautophagy (Gabande-Rodriguez et al., 2014; Shen et al., 2012). Our findings demonstrate that mitochondrial dysfunction results in autophagosome-lysosome fusion defects and reduced mitochondrial clearance by mitophagy, which are commonly observed in almost all sphingolipidoses (Lieberman et al., 2012). A predominant nuclear localization of TFEB has been detected in cells from mouse models of LSD (Sardiello et al., 2009). Mitochondrial respiration-deficiency increases TFEB activation suggesting that the TFEB pathway is activated as a cellular response to lysosomal stress after intralysosomal storage of non-degraded molecules.
Lysosomal calcium level is impaired not only by Tfam deletion, which triggers a general lowering of the levels of mtDNA, but also in cells from patients with point mutations in mitochondrial respiratory complex I. Mitochondrial complex I is essential for maintenance of the NAD+/NADH ratio through NADH dehydrogenase activity, and an imbalanced NAD+/NADH ratio in cancer cells with mutations in mitochondrial complex I subunits is associated with impaired autophagy and p62 degradation (Santidrian et al., 2013). Inhibition of complex I activity with rotenone in a mouse model of Parkinson’s disease impairs autophagic flux and promotes p62 and α-synuclein accumulation (Wu et al., 2015). Our data demonstrate that NAD+ precursors restore ASM activity, prevent p62 accumulation and downregulate TFEB target genes in Tfam-deficient cells, thus supporting a role of mitochondrial respiratory complex I and NAD+/NADH ratio in the regulation of lysosomal function. These data support the notion that lysosomes adapt their biogenesis and function to the metabolic state of the cell by sensing intracellular NAD+/NADH levels. Further research is necessary to understand how NAD precursors reestablish ASM activity and control lysosomal function, and whether it relies on transcriptional regulation by sirtuins, NAD-dependent deacetylases.
Recent studies support the contribution of lysosomal homeostasis to the regulation of T cell function during adaptive immune responses (Valdor et al., 2014). The lysosomal dysfunction and elevated SM levels in Tfam-deficient cells might be responsible for the acquisition of a pro-inflammatory profile, since similar cytokine profile was found in ASM-/- T cells, which presented lysosomal dysfunction without altered mitochondrial respiration. Together, our results point that loss of lysosomal homeostasis by a primary mitochondrial dysfunction trigger pro-inflammatory T cell responses. Lysosomal function could regulate immune response by modulating cellular metabolism. By degrading key glycolytic enzymes, lysosome function regulates glycolysis and the production of effector cytokines, including IFN-γ and IL-1β (Lu et al., 2014).
Our results shed light on how metabolic reprogramming toward anaerobic glycolysis and the alterations to the endolysosomal compartment affect T cell differentiation to effector lineages. It has been reported that Th1, Th2 and Th17 lineages display a strong bias toward glycolysis over mitochondrial metabolism, whereas Treg cells have a mixed metabolism involving glycolysis, lipid oxidation, and OXPHOS (Gerriets et al., 2015; Michalek et al., 2011). Metabolic reprogramming towards glycolysis in Tfam-/- T cells could contribute to their pro-inflammatory phenotype, since glycolysis enhance IFN-γ expression by preventing GAPDH from its binding to the 3’UTR of IFN-γ mRNA (Chang et al., 2013). Glycolysis is important in driving Th17 differentiation through Hif-1α(Pearce et al., 2013). In addition, impaired mitochondrial respiration induces succinate accumulation, which impairs prolyl hydroxylase activity, stabilizing Hif-1α and enhancing IL-1β production (Tannahill et al., 2013). Tfam depletion favours the differentiation of Th1 subsets, dampens Th2 differentiation and inhibits Th17 T cell subsets. Interestingly, in Th17 skewing conditions, Tfam-deficient cells showed reduced IL-17 and IL-10 but increased IFN-γ secretion, which suggests that cues from mitochondria can modulate the differentiation toward pathogenic Th17 cells.
Defects in mitochondrial homeostasis are closely linked to the development of human pathologies such as cancer, inflammation, neurodegenerative diseases and metabolic disorders (Taylor and Turnbull, 2005), and are also a recurrent hallmark of aging (Lopez-Otin et al., 2013). In summary, our findings on the role of mitochondrial respiration as a regulator of endolysosomal function could therefore provide new potential insights into the involvement of mitochondria in human diseases and aging. These findings provide evidence for a novel functional link between mitochondrial respiration and the endolysosomal system, with implications for treating mitochondrial-related diseases.
Experimental Procedures
Mice, antibodies and reagents
All animal experiments were performed in the specific pathogen-free facilities at CNIC in accordance with European Union recommendations and institutional guidelines. Tfamfl/fl mice were kindly provided by N.G. Larsson (Larsson et al., 1998) and CD4Cre+/wt mice were purchased from the Jackson Laboratory. Double heterozygotes (Tfam+/fl, CD4Cre+/wt) were obtained and backcrossed to the Tfamfl/fl strain to generate tissue specific knockouts (CD4Cre+/wt Tfamfl/fl). ASM-/- mice were kindly donated by E.H. Schuchman (Mount Sinai School of Medicine, New York). Sources of chemical and biological reagents detailed in Supplemental Experimental Procedures.
Cells, plasmids and cell transfection
Naive CD4+ T cells were obtained by negative selection using the auto-MACS Pro Separator (Miltenyi Biotec). Where stated, cells were activated by stimulation on plates coated with anti-CD3 (5 μg ml−1) and soluble anti-CD28 (2 μg ml−1). To obtain differentiated T lymphoblasts, naive CD4+ cells were cultured for 36-48 hr with 2 μg ml−1 concanavalin A, and 50 U ml−1 IL-2 was added to the medium every 2 days. Balb/cJ and mt-ND6 (FG23-1) mutant cell line were described (Acin-Perez et al., 2014). Human fibroblasts from a control patient, a patient with a point mutation in the complex I subunit mt-ND5 gene (m.13513G>A), and a patient with a point mutation in the mt-tRNAAsn gene (m.5658T>C) were obtained from skin biopsies for research purposes after written consent in accordance with the Helsinki declaration. The human Jurkat T cell line and the oligodendroglia Oli-Neu cell line were silenced for Tfam by lentiviral infection as detailed in Supplemental Experimental Procedures.
T cell differentiation, proliferation, and intracellular staining
Naïve CD4+CD25- T cells were differentiated to the different effector subsets as detailed in Supplemental Experimental Procedures. For proliferation assays, cells were labeled Cell Violet (1 μM) and activated with anti-CD3/CD28 or differentiated as indicated. For intracellular staining of transcription factors, cells were differentiated during 3 days, collected with 5 mM EDTA in PBS, fixed, permeabilized and stained with anti-RORγT and anti-T-bet (BD biosciences). For intracellular cytokine staining, cells were restimulated for 4 hr with PMA and ionomycin in the presence of brefeldin, permeabilized with 0.5% saponin and stained with anti-CD4, anti-IFN-γ, anti-IL-17 and anti-Foxp3. Data were acquired on a FACSCanto flow cytometer (BD Bioscience) and analyzed using the FLOWJO software (Tree Star).
Statistical analyses
All values were expressed as the mean ± s.e.m. Samples were analyzed using Student’s t-test for two groups and ANOVA for multiple groups. For in vitro experiments, statistical analyses were calculated with Student’s t-test; for in vivo experiments, differences between groups were calculated using the Mann–Whitney U test for unpaired data (GraphPad Prism version 5.0). Differences were considered significant when * p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
Supplemental Information
Supplemental information includes five figures, Supplemental Experimental Procedures, and Supplemental References.
Ackowledgements
We thank N.G. Larsson (Max Planck Institute for Biology of Ageing) for providing Tfamfl/fl mice. F. Sanchez-Madrid for continuous support and advice. C. López-Otín for helpful comments. M. Navarro for the critical reading of the manuscript, the Electron Microscopy Service (Universidad Autónoma de Madrid) for electron microscopy studies and S. Bartlett for English editing. F.B is supported by ERC-2011-AdG 294340-GENTRIS. This work was supported by grants SAF2011-25834 from Ministerio de Economía y Competitividad-Spain, COST-Action BN1202, Cardiovascular Network RD12-0042-0056 (Instituto de Salud Carlos III). M. M. is supported by Contrato de Investigadores SNS Miguel Servet I (CP14/00219 and MS14/00219) from the Instituto de Salud Carlos III. The Centro Nacional de Investigaciones Cardiovasculares (CNIC, Spain) is supported by the Spanish Ministry of Science and Innovation and the Pro-CNIC Foundation.
Footnotes
Authors Contributions:
F.B., R. A-P, C.V-B, C.M., N. N-A, and M.M. performed experimental work. A.B., M.A.M., J.M.R., J.M.F., V.d.P., J.A.E. E.GR, and M.D.L provided critical materials. F.B, J.A.E., and M.M. designed research. F.B. and M.M. analyzed data and wrote the manuscript.
References
- Acin-Perez R, Carrascoso I, Baixauli F, Roche-Molina M, Latorre-Pellicer A, Fernandez-Silva P, Mittelbrunn M, Sanchez-Madrid F, Perez-Martos A, Lowell CA, et al. ROS-Triggered Phosphorylation of Complex II by Fgr Kinase Regulates Cellular Adaptation to Fuel Use. Cell Metab. 2014;19:1020–1033. doi: 10.1016/j.cmet.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arruda AP, Pers BM, Parlakgul G, Guney E, Inouye K, Hotamisligil GS. Chronic enrichment of hepatic endoplasmic reticulum-mitochondria contact leads to mitochondrial dysfunction in obesity. Nat Med. 2014;20:1427–1435. doi: 10.1038/nm.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti R, Pirinen E, Lamperti C, Marchet S, Sauve AA, Li W, Leoni V, Schon EA, Dantzer F, Auwerx J, et al. NAD(+)-dependent activation of Sirt1 corrects the phenotype in a mouse model of mitochondrial disease. Cell Metab. 2014;19:1042–1049. doi: 10.1016/j.cmet.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CH, Curtis JD, Maggi LB, Jr, Faubert B, Villarino AV, O'Sullivan D, Huang SC, van der Windt GJ, Blagih J, Qiu J, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniele T, Hurbain I, Vago R, Casari G, Raposo G, Tacchetti C, Schiaffino MV. Mitochondria and melanosomes establish physical contacts modulated by Mfn2 and involved in organelle biogenesis. Curr Biol. 2014;24:393–403. doi: 10.1016/j.cub.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Detmer SA, Chan DC. Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol. 2007;8:870–879. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, Rustin P, Gustafsson CM, Larsson NG. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- Elbaz-Alon Y, Rosenfeld-Gur E, Shinder V, Futerman AH, Geiger T, Schuldiner M. A Dynamic Interface between Vacuoles and Mitochondria in Yeast. Dev Cell. 2014;30:95–102. doi: 10.1016/j.devcel.2014.06.007. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Nunnari J. Mitochondrial form and function. Nature. 2014;505:335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futerman AH, van Meer G. The cell biology of lysosomal storage disorders. Nat Rev Mol Cell Biol. 2004;5:554–565. doi: 10.1038/nrm1423. [DOI] [PubMed] [Google Scholar]
- Gabande-Rodriguez E, Boya P, Labrador V, Dotti CG, Ledesma MD. High sphingomyelin levels induce lysosomal damage and autophagy dysfunction in Niemann Pick disease type A. Cell Death Differ. 2014;21:864–875. doi: 10.1038/cdd.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerriets VA, Kishton RJ, Nichols AG, Macintyre AN, Inoue M, Ilkayeva O, Winter PS, Liu X, Priyadharshini B, Slawinska ME, et al. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J Clin Invest. 2015;125:194–207. doi: 10.1172/JCI76012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honscher C, Mari M, Auffarth K, Bohnert M, Griffith J, Geerts W, van der Laan M, Cabrera M, Reggiori F, Ungermann C. Cellular Metabolism Regulates Contact Sites between Vacuoles and Mitochondria. Dev Cell. 2014;30:86–94. doi: 10.1016/j.devcel.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamanlidis G, Lee CF, Garcia-Menendez L, Kolwicz SC, Jr, Suthammarak W, Gong G, Sedensky MM, Morgan G, Wang W, Tian R. Mitochondrial complex I deficiency increases protein acetylation and accelerates heart failure. Cell Metab. 2013;18:239–250. doi: 10.1016/j.cmet.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapuente-Brun E, Moreno-Loshuertos R, Acin-Perez R, Latorre-Pellicer A, Colas C, Balsa E, Perales-Clemente E, Quiros PM, Calvo E, Rodriguez-Hernandez MA, et al. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science. 2013;340:1567–1570. doi: 10.1126/science.1230381. [DOI] [PubMed] [Google Scholar]
- Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- Lieberman AP, Puertollano R, Raben N, Slaugenhaupt S, Walkley SU, Ballabio A. Autophagy in lysosomal storage disorders. Autophagy. 2012;8:719–730. doi: 10.4161/auto.19469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Zhang Y, McDonald DO, Jing H, Carroll B, Robertson N, Zhang Q, Griffin H, Sanderson S, Lakey JH, et al. Dual proteolytic pathways govern glycolysis and immune competence. Cell. 2014;159:1578–1590. doi: 10.1016/j.cell.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Evans E, Morgan AJ, He X, Smith DA, Elliot-Smith E, Sillence DJ, Churchill GC, Schuchman EH, Galione A, Platt FM. Niemann-Pick disease type C1 is a sphingosine storage disease that causes deregulation of lysosomal calcium. Nat Med. 2008;14:1247–1255. doi: 10.1038/nm.1876. [DOI] [PubMed] [Google Scholar]
- Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto-Rosato A, Prezioso C, Forrester A, et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol. 2015;17:288–299. doi: 10.1038/ncb3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, Sullivan SA, Nichols AG, Rathmell JC. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osellame LD, Rahim AA, Hargreaves IP, Gegg ME, Richard-Londt A, Brandner S, Waddington SN, Schapira AH, Duchen MR. Mitochondria and quality control defects in a mouse model of Gaucher disease--links to Parkinson's disease. Cell Metab. 2013;17:941–953. doi: 10.1016/j.cmet.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: insights into metabolism and lymphocyte function. Science. 2013;342:1242454. doi: 10.1126/science.1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, Lee Y, Kuchroo VK. The many faces of Th17 cells. Curr Opin Immunol. 2011;23:702–706. doi: 10.1016/j.coi.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland AA, Voeltz GK. Endoplasmic reticulum-mitochondria contacts: function of the junction. Nat Rev Mol Cell Biol. 2012;13:607–625. doi: 10.1038/nrm3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santidrian AF, Matsuno-Yagi A, Ritland M, Seo BB, LeBoeuf SE, Gay LJ, Yagi T, Felding-Habermann B. Mitochondrial complex I activity and NAD+/NADH balance regulate breast cancer progression. J Clin Invest. 2013;123:1068–1081. doi: 10.1172/JCI64264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Klisch TJ, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013a;15:647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 2013b;14:283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D, Wang X, Li X, Zhang X, Yao Z, Dibble S, Dong XP, Yu T, Lieberman AP, Showalter HD, et al. Lipid storage disorders block lysosomal trafficking by inhibiting a TRP channel and lysosomal calcium release. Nat Commun. 2012;3:731. doi: 10.1038/ncomms1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdor R, Mocholi E, Botbol Y, Guerrero-Ros I, Chandra D, Koga H, Gravekamp C, Cuervo AM, Macian F. Chaperone-mediated autophagy regulates T cell responses through targeted degradation of negative regulators of T cell activation. Nat Immunol. 2014;15:1046–1054. doi: 10.1038/ni.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernochet C, Mourier A, Bezy O, Macotela Y, Boucher J, Rardin MJ, An D, Lee KY, Ilkayeva OR, Zingaretti CM, et al. Adipose-specific deletion of TFAM increases mitochondrial oxidation and protects mice against obesity and insulin resistance. Cell Metab. 2012;16:765–776. doi: 10.1016/j.cmet.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viader A, Sasaki Y, Kim S, Strickland A, Workman CS, Yang K, Gross RW, Milbrandt J. Aberrant Schwann cell lipid metabolism linked to mitochondrial deficits leads to axon degeneration and neuropathy. Neuron. 2013;77:886–898. doi: 10.1016/j.neuron.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg SE, Sena LA, Chandel NS. Mitochondria in the Regulation of Innate and Adaptive Immunity. Immunity. 2015;42:406–417. doi: 10.1016/j.immuni.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Xu HD, Guan JJ, Hou YS, Gu JH, Zhen XC, Qin ZH. Rotenone impairs autophagic flux and lysosomal functions in Parkinson's disease. Neuroscience. 2015;284:900–911. doi: 10.1016/j.neuroscience.2014.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.