Abstract
Transient receptor potential (TRP) channels are an ancient family of cation channels, working as metabotropic triggers, which respond to physical and chemical environmental cues. Perception of chemical signals mediate reproductive behaviors and is therefore an important target for sustainable management tactics against the codling moth Cydia pomonella L. (Lepidoptera: Tortricidae). However, olfactory behavior strongly depends on diel periodicity and correlation of chemical with physical cues, like temperature, and physical cues thus essentially contribute to the generation of behavioral response. From an antennal transcriptome generated by next generation sequencing, we characterized five candidate TRPs in the codling moth. The coding DNA sequence of one of these was extended to full length, and phylogenetic investigation revealed it to be orthologous of the TRPA5 genes, reported in several insect genomes as members of the insect TRPA group with unknown function but closely related to the thermal sensor pyrexia. Reverse transcription PCR revealed the existence of five alternate splice forms of CpTRPA5. Identification of a novel TRPA and its splice forms in codling moth antennae open for investigation of their possible sensory roles and implications in behavioral responses related to olfaction.
Keywords: transient receptor potential cation channel, TRPA subfamily, TRPA5, splice form, Cydia pomonella
Transmembrane cation channels from the transient receptor potential (TRP) family are key for multiple sensory modalities, including vision, hearing, chemosensation, thermosensation, and mechanosensation (Liedtke 2007; Fowler and Montell 2013), thus allowing the animals to achieve vital behaviors like avoidance of noxious temperatures (Tracey et al. 2003) or detection of heat emitted from hosts (Wang et al. 2009). TRPs in insects have been divided into seven subfamilies, of which four (TRPC, TRPV, TRPA and TRPN) play roles in insect sensory systems (Fowler and Montell 2013). Most insects appear to possess around a dozen TRP genes, approximately half the number of genes found in most mammals (Matsuura et al. 2009). In part, this may be compensated by a wider response spectrum, as there are reported cases where single insect TRP channels are responsible for detecting multiple sensory stimuli. For example, the Drosophila TRPV channel Nanchung (Nan) is essential for hearing (Kim et al. 2003; Gong et al. 2004) and hygrosensation (Liu et al. 2007). In Drosophila, some channels of the TRPC subfamily also function both in vision (Hardie and Minke 1992; Niemeyer et al. 1996) and in cold-avoidance (Rosenzweig et al. 2008). Furthermore, the Drosophila TRPN channel NompC is associated with touch sensation (Walker et al. 2000) as well as hearing (Eberl et al. 2000; Göpfert et al. 2006). This versatility, where single receptors detect multiple sensory stimuli, make TRPs interesting targets for insect control (Nesterov et al. 2015).
In contrast to mammals, in which only one TRPA channel has been identified (Clapham 2003; Wu et al. 2010), insects appear to have an expanded TRPA subfamily, with four or five genes per species (Matsuura et al. 2009; Peng et al. 2015). Like other TRP subfamilies in insects, the TRPAs appear to be versatile. For example, several D. melanogaster TRPA channels (dTRPA1, Pyrexia, Painless) detect different ranges of temperature and are involved in thermotaxis (Viswanath et al. 2003; Lee et al. 2005; Sokabe and Tominaga 2009), but Pyrexia and Painless are also involved in negative geotaxis, by contributing to gravity sensing (Sun et al. 2009). A fourth TRPA channel, Water witch (Wtrw), is involved in hygrosensation (Liu et al. 2007). Interestingly, insect TRPA channels are also involved in chemosensation (Kwon et al. 2010; Kang et al. 2010). Notably, the Drosophila TRPA Painless, initially identified as a nociceptive heat sensor (Tracey et al. 2003), was later found to be involved in the detection of allyl-isothiocyanates found in wasabi (Al-Anzi et al. 2006), and fructose (Xu et al. 2008). Furthermore, TRPA1 in the crop pest moth Helicoverpa armigera also detects repellent chemicals (Wei et al. 2015). In a more recent study we demonstrated that compounds emitted by the plant Perilla frutescens L. (Lamiales: Lamiaceae), which are reported to be active on rat TRPA1 (Bassoli et al. 2013), are detected by the olfactory system of the tortricid pest Lobesia botrana (Cattaneo et al. 2014). It could be speculated that these compounds might interact with antennal TRPA channels in L. botrana. Plants emitting compounds active on TRPAs are usually repellent to insects, via the activation of their olfactory systems (Leung and Foster 1996; Barnard 1999), which indicates that members of the insect TRPA subfamily represent potential targets for pest control strategies.
The codling moth, Cydia pomonella (L.) (Lepidoptera: Tortricidae), is a major pest of commercial crops such as apple, pear and walnuts in Palearctic and Nearctic regions (Witzgall et al. 2008). Whereas olfaction-based pest control methods have been developed (Ridgway et al. 1990; Witzgall et al. 2008), a better understanding of the molecular mechanisms of the olfactory process in this species may lead to the identification of new targets for olfactory disruption. In that search, we have previously sequenced the antennal transcriptome of this moth, and notably identified candidate olfactory and pheromone receptors (Bengtsson et al. 2012; Walker et al. 2016), functionally characterizing two of them (Bengtsson et al. 2014; Gonzalez et al. 2015). Using these transcriptomic data, we identified five candidate TRPs belonging to the TRPA and TRPC subfamilies. Among the TRPAs, we have notably characterized a C. pomonella orthologue of TRPA5 genes, which has been identified in several insect genomes (Peng et al. 2005). By performing RACE-PCR and searching a preliminary genome obtained by shotgun sequencing, we obtained the full-length coding sequence of CpTRPA5. We investigated its expression in male and female adult body parts by reverse transcription (RT)-PCR. This led to the identification of alternative splice forms with different expression patterns among genders and body parts, which were verified by intron/exon prediction using a genomic overview based on gDNA and RNA-sequencing.
Material and Methods
Dissection, Nucleic Acid Extraction
C. pomonella pupae were obtained from a laboratory rearing (Andermatt Biocontrol, Grossdietwil, Switzerland), and adults were allowed to emerge in cages kept at 23 °C, 70 ± 5% RH, 16 h: 8 h light: dark cycle and fed with 10% sugar solution. As previously reported (Bengtsson et al. 2014), dissection of 2–3 day old female and male insects was performed using sharp forceps: antennae were removed at the base of the pedicel of 100 insects per sample. Legs were removed at the coxa of 20 insects. For thorax samples, head, wings, legs and abdomen were removed from five insects. Wings were removed at their base from five insects, and the abdomen removed at the connection to the thorax of three insects. All body parts were immediately flash-frozen using liquid nitrogen, and thereafter kept at −80 °C. RNA was extracted using the RNeasy kit (Qiagen, Hilden, Germany), that included a DNase step to remove genomic DNA contamination. A gDNA sample was extracted from one male and one female adult insect using the DNeasy kit (Qiagen) following the recommended protocol. Body-part RNAs and gDNA were quantified using Nanodrop (Nanodrop 8000 UV-vis Spectrophotometer, Thermo Scientific, Wilmington, DE, USA). Aliquots of 1.0 µg total RNA were used for reverse transcription using Advantage RT-for-PCR kit (Clontech, Mountain View, CA) according with the manufacturer’s protocol.
cDNA Library Construction and Bioinformatics
Male and female contigs previously obtained (Bengtsson et al. 2012) were analyzed through bioinformatics, in search of candidate TRPs. Tblastn searches were performed using available amino acid sequences of Lepidoptera and other insect TRPs. Contigs presenting similarity to TRP genes were further assembled using Cap3 (http://pbil.univ-lyon1.fr/cap3.php). Open reading frames (ORFs) were searched and translated to amino acid sequences using ExPASy (http://www.expasy.org/translate/), and Blastx on the Genbank non-redundant database (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to verify their annotation. For CpTRPA5 and its splice forms, transmembrane domains were predicted from translated sequences using TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) and TMPred (http://www.ch.embnet.org/software/TMPRED_form.html) (Hofmann and Stoffel 1993). Topology configurations were predicted with TOPO 2.0 (http://www.sacs.ucsf.edu/cgi-bin/open-topo2.py). For the candidate CpTRPA5_F1117 splice form, which was predicted to lack transmembrane segments, we predicted the tertiary structure of the related polypeptide using the Proteus structure prediction server 2.0 (http://www.proteus2.ca/proteus2/). To perform 3D-representation, Rastop 2.2 was used (available for the public domain at http://www.geneinfinity.org/rastop/).
Phylogenetic Investigation of CpTRPs
TRP sequences of Rattus norvegicus Berkenhout, Caenorhabditis elegans Maupas and Drosophila melanogaster Meigen were downloaded from their proper genome browsers (http://rgd.mcw.edu/; http://www.wormbase.org/; http://flybase.org/). In addition to C. pomonella TRP sequences, TRP sequences were searched in two other lepidopteran species (the silk moth Bombyx mori L. and the monarch butterfly Danaus plexippus L.), as well as in the insects Apis mellifera L., Tribolium castaneum Herbst and Acyrthosiphon pisum Harris using NCBI-blast (http://blast.ncbi.nlm.nih.gov/Blast.cgi) with the amino acid sequences of D. melanogaster TRPs as a query. The partial amino acid sequence of a C. pomonella TRP-subunit, previously identified by BAC-FISH mapping on the Z-chromosome and reported to be the CpNan TRPV-candidate (Nguyen et al. 2013) was also included in the dataset. The 131 amino acid sequences were aligned using MAFFT version 7 (http://mafft.cbrc.jp/alignment/server/) (Katoh and Toh 2010) and a neighbor-joining tree was built using the BioNJ algorithm as implemented in SeaView v.4 (Gouy et al. 2010). Node support was assessed using a bootstrap procedure based on 1000 replicates. The figure was created using the iTOL web server (Letunic and Bork 2007).
Rapid Amplification of cDNA Ends (RACE) PCR of CpTRPA5
To extend CpTRPA5 by RACE-PCR in 5′ and in 3′ direction, 5′ and 3′ cDNAs were created from 1.0 µg antennal RNA using First Choice RLM-RACE kit (Ambion, Life technologies, Grand Island, NY USA) and SMARTer kit (Clontech). Amplifications were conducted according to the recommended protocols. Primers were designed by hand using existing contig data as reference. Thermodynamic features were checked by OligoEvaluator (Sygma Genosys, http://www.oligoevaluator.com), and putative oligodimerization was checked by OligoAnalyzer 3.1 (Integrated DNA Technologies, http://eu.idtdna.com/calc/analyzer). Primer melting temperatures were estimated using the salt-adjusted algorithm on the OligoCalc website (ww.basic.northwestern.edu/biotools/OligoCalc.html). For primers, the goal was a GC% 40-60, Tm < 70 °C, and to create a product with at least 150 bases of overlap with existing contig data. However, in some cases, it was necessary to compromise on one or several of these conditions (Table 1).
Table 1.
Sequences and estimated Tm for primers
| RACE primer (5′ or 3′) | Sequence | Tm (°C) |
|---|---|---|
| 5′_CpTRPA5_1 | AGCGGAACTGGATCATGAAG | 64.3 |
| 5′_CpTRPA5_2 | GAGATGGTGATGGCTGCAGGAAGGAGGG | 65.0 |
| 3′_CpTRPA5-1 | CAGGAAAACCAAGATGGAGGCACG | 66.9 |
| 3′_CpTRPA5-2 | GAGACGCCATTTTAGACAAAGCTCAAGCTC | 63.5 |
|
CDS extension (Fw or Rv) | ||
| Fw_CpTRPA5 | attB1-ATGGCAGCTTTATCAGGCGGCG | 65.8 |
| Rv_CpTRPA5 | attB2-TTATTTACTTAACTTACTTTCTAATCTTAACAA | 61.4 |
|
Sequencing primers (Fw or Rv) | ||
| M13 Fw | GTAAAACGACGGCCAGT | 52.4 |
| M13 Rv | CAGGAAACAGCTATGACC | 53.8 |
| Seq1-Fw | ATGATGGAGAGACTCCAATCCATTC | 64.1 |
| Seq2-Fw | ATGGGCTGGTTCCCTTTACATACAG | 65.8 |
| Seq3-Fw | TGCTGGCATGGTTAGAGATG | 58.4 |
Starting from RLM-RACE 5′-cDNA, 5′ CpTRPA5 was extended using the 5′_CpTRPA5_1 gene-specific primer together with the 5′ RACE Outer primer, supplied with the kit. Amplification was performed with the supplied thermostable DNA polymerase using a temperature program of 94 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, Tm of the gene-specific primer for 30 s, 72 °C for 3 min, and a final elongation of 72 °C for 7 min. An aliquot of 1.0 µl of the reaction mix was used as template to perform the nested amplification using the 5′_CpTRPA5_2 gene specific primer together with 5′ RACE Inner primer, supplied with the kit.
Starting from SMARTer-RACE 3′-cDNA, PCR amplification of 3′ CpTRPA5 was performed using the 3′_CpTRPA5-1 primer combined with the Universal primer A mix supplied with the kit. Amplification was performed with Advantage 2 polymerase (Clontech) using a temperature program of 95 °C for 5 min, followed by 35 cycles of 95 °C for 45 s, Tm of the gene specific primer for 1 min, 68 °C for 90 s, and a final elongation of 68 °C for 7 min. To perform the nested amplification, the 3′_CpTRPA5-2 gene-specific primer was combined with the Nested Universal Primer A, also supplied with the kit.
PCR products were analyzed by electrophoresis on a 1.5% agarose gel, stained with ethidium bromide, and visualized using a Gel Doc XR (Bio-Rad, Hercules, CA, USA). Relevant bands were excised and purified using the QIAquick Gel extraction kit (Qiagen). Quantification was conducted using a Nanodrop 3300 Fluorospectrometer (Thermo Scientific) with the PicoGreen® dsDNA reagent kit (Molecular Probes, Life Technologies). Samples were sequenced (Sanger sequencer, 3730xl Applied Biosystems, Life Technologies) using gene specific primers. Alignment of amplicon sequences from RACE-PCR amplifications was performed using Multalin (http://multalin.toulouse.inra.fr/multalin/) (Corpet 1988). The 5′ and 3′ sequenced regions were assembled with existing contig data to generate a partial CDS-template of 1677 bp. Despite being partial, this sequence was checked using the online tool ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/orfig.cgi).
Querying Genome and Transcriptome Assemblies
In order to identify the full-length CDS of the CpTRPA5 TRP, we used Blast (Altschul et al. 1990) on preliminary assemblies of a whole shotgun sequenced genome and an Illumina sequenced transcriptome (which are still under analysis and will be published elsewhere). We blasted the assemblies using the 1677 bp CpTRPA5 TRP from RACE-PCR as query in tblastn searches. Scaffolds that passed a threshold of e-30 were mapped against the query and manually assembled by hand using BioEdit v7.2.5 (Hall 1999) into a single scaffold that contained the putative full length CDS. The five genome scaffolds and the four transcripts that matched our RACE-PCR template are available for download and inspection at https://www.researchgate.net (DOI: 10.13140/RG.2.1.3056.8726). Sequencing and preliminary assembly of the CpTRPA5 genomic locus and comparison with antennal RNA-seq returned an overview of intron/exon boundaries within the CpTRPA5 locus. The final CDS provided by the locus was checked using ORF Finder.
Identification of Candidate CpTRPA5 Splice Forms
The full length CDS of CpTRPA5 was amplified from male and female antennal cDNA with Fw_CpTRPA5 and Rv_CpTRPA5 primers (Table 1), and attB regions were attached (attB1 forward region: 5’-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAACA-3’; attB2 reverse region: 5’-GGGGACCACTTTGTACAAGAAAGCTGGGT-3’, Gateway Technology, Invitrogen, Life technologies, Waltham, MA, USA), suitable for cloning into pDONR221. Amplification was performed with Phusion (New England Biolabs, Ipswich, MA, USA) using a temperature program of 94 °C for 2 min, followed by 35 cycles of 94 °C for 30 s, 58 °C for 15 s, 68 °C for 3 min and 10 s, and a final elongation step of 68 °C for 4 min. A 4.0 µl PCR volume was mixed with 1.0 µl BP-clonase (Gateway Technology, Invitrogen) and 150 ng pDONR221, to be incubated 4 h at 25 °C. A 2.0 µl volume of the reaction was used to transform TOP10 competent cells, 50 µl of which were plated on 50 µg/ml Kanamycin selective media and incubated overnight.
Colonies were screened by picking individual colonies from plates and dissolving it in 50 µl of 50 µg/ml Kanamycin LB selective media, which was incubated for 2 h at 37 °C and 225 rpm. PCRs were conducted using 1.0 µl of this culture using the Fw_CpTRPA5 and Rv_CpTRPA5 primers and amplifying with GoTaq Green Master Mix (Promega, Fitchburg, WI). Amplifications were conducted with a temperature program of 95 °C for 15 min, followed by 35 cycles of 95 °C for 30 s, 55 °C for 15 s, 72 °C for 3 min 10 s, and a final elongation step of 72 °C for 4 min. Samples were analyzed as described above. Cultures giving clear bands were grown at 37 °C and 225 rpm overnight in selective LB media with 50 µg/mL Kanamycin, after which plasmids were purified using the QIAprep Spin Miniprep kit (Qiagen). Quantification was conducted using Nanodrop 3300 Fluorospectrometer with PicoGreen® dsDNA reagent kit. Samples were sequenced using Sanger sequencer and universal M13-primers, as well as Fw_CpTRPA5, Rv_CpTRPA5 and other primers designed on the CDS (Table 1).
Alternative splice forms were verified by reverse-transcription (RT) PCR on cDNA samples from insect body parts, followed by sequencing of amplified bands. Positions of intron/exon boundaries within the CpTRPA5 locus were compared with splicing site positions of verified splice forms. Graphical intron/exon representation of splice forms was done using the online tool Exon-Intron Graphic Maker version 4 (http://wormweb.org/exonintron).
Reverse Transcription (RT)-PCR
To investigate transcripts of CpTRPA5 and its splice forms, RT-PCR was performed on cDNA samples prepared from antenna, thorax, abdomen, leg and wing total RNAs.
Amplifications were performed using the GoTaq Green Master Mix (Promega), splice form-specific forward primers designed based on Sanger sequencing data of pDONR221 clones (Table 2) and the reverse primer Rv_CpTRPA5, previously used to amplify the final assembled CDS (Table 1).
Table 2.
List of forward primers designed to verify the existence of candidate CpTRPA5 splice forms
| Splice form | Forward primer | Tm (°C) |
|---|---|---|
| CpTRPA5_M4* | TAGACTTGCAAAACAATTTG | 50.2 |
| CpTRPA5_M17* | AAGTTTGGCTCCATCGGCC | 59.5 |
| CpTRPA5_M41 | ACTGACGGCCCTAAGAAATTC | 59.5 |
| CpTRPA5_M43* | CTCGTATTGATTCAGGAAAAC | 54.4 |
| CpTRPA5_M415 | TGGAAGAAGTTTTAGACTTGC | 55.4 |
| CpTRPA5_M417 | TTTATCTACGTTTGTGGCGTT | 55.4 |
| CpTRPA5_M418* | TAGTTTTAGGTACCTATAAGC | 53.0 |
| CpTRPA5_F1111 | TTAGTTGAGAGTTTCCTAACT | 53.4 |
| CpTRPA5_F1115 | TTGTTGCTAAAAGATGGCGCC | 59.5 |
| CpTRPA5_F1117* | TTTTACACTATTATAGCCATT | 49.0 |
| CpTRPA5_F1124 | TGAATACTTGGAAGAAGTTTA | 51.7 |
*Splice forms verified by RT-PCR.
Except for CpTRPA5_M17 and CpTRPA5_M418 cases, all forward primers were designed to overlap splicing sites, the position of which were identified by sequencing pDONR221-clones and comparing these to the sequence of the final assembled CDS. For the final assembled CDS form, parallel amplifications were conducted using the Fw_CpTRPA5 and Rv_CpTRPA5 primers (Table 1). Positive control of cDNA synthesis consisted of amplification of the housekeeping gene rpl8 using degenerated primers (rpl8_Fw: 5′-GAGTCATCCGAGCTCARMGNAARGG-3’; rpl8_Rv: CCAGCAGTTTCGCTTNACYTTRTA; Tm = 54 °C). A temperature program with an initial 5-min step at 95 °C, and then 45 cycles of 95 °C for 1 min, primer melting temperature for 1 min, 72 °C for 3 min 10 s, and a final 7-min step at 72 °C was used. Each PCR reaction was repeated at least three times and controls consisted of no template PCRs. All PCRs were performed in parallel on a genomic DNA (gDNA) template. No amplification or amplifications of larger size products were observed in most cases, indicating that no significant gDNA contamination occurred in our cDNA preparations. Amplifications were analyzed as described above and product identity was confirmed by direct sequencing (Sanger) following gel extraction and quantification. RT-PCR from male and female body parts cDNAs was compared to verify the presence of candidate splice forms.
For the other TRP candidate RT-PCRs, primers were designed based on partial 454-contigs (Table 3). In addition to rpl8, the codling moth olfactory co-receptor (CpOrco, Bengtsson et al. 2012) was used as an antenna positive control. RT-PCRs were conducted as described above, except for minor adjustments of the temperature settings, using a 72 °C extension for 1 min 45 s. To verify the identity of the amplicons, bands were gel-purified and Sanger-sequenced using gene specific primers.
Table 3.
List of forward and reverse primers designed to validate the expression of candidate Cp-TRPs
| Gene | Forward primer | Reverse primer | Tm (°C) |
|---|---|---|---|
| CpTRPA5 | ATGGGCTGGTTCCCTTTACATACAG | TTATTTACTTAACTTACTTTCTAATCTTAACAA | 61.4 |
| CpPyx | TACCCAGCGTTCCAACTACC | CATGAGAGCAGCGAACTGAA | 63.2 |
| CpWtrw | TAGCCGGTTACTCCACCATC | AAAACAGGGGAGGGTCATTC | 63.2 |
| CpTRP | ATTCCCTCAGGCACTCACAA | CATGAAAGCTGGAAGGCTGT | 64.3 |
| CpTRPC | GGGAGACCAAGTCAACGGTATGC | GATGCGTTCAGTGTACGTGTGC | 65.4 |
| CpOrco | CCGGAGCCCACTGATATAGA | CCTCAGAACCGTCGTACCAT | 64.3 |
Results
Antennal TRP Repertoire in C. pomonella
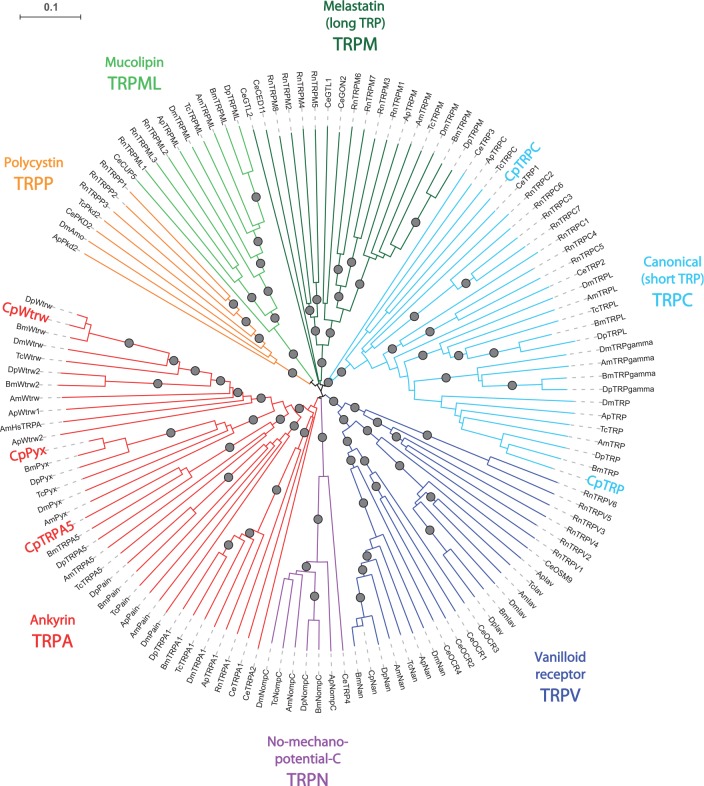
Using BLAST search on the C. pomonella antennal transcriptome (for details, see Bengtsson et al. 2012), we identified the partial sequences of five candidate TRPs that were judged to be incomplete at both 5′ and 3′ parts because of the lack of start and stop codons in the open reading frame. In order to assign each of these candidate TRPs to one of the seven TRP sub-families (Fowler and Montell 2013), we built a neighbor-joining phylogeny including TRP repertoires of two other lepidopteran species, four species from other insect orders and two model species from outside insects (C. elegans and R. norvegicus). This analysis revealed that two of these candidates—named CpTRP and CpTRPC—belong to the TRPC subfamily (Fig. 1): CpTRP is orthologous to the conserved TRP gene identified in multiple insects, which is involved in phototransduction in Drosophila (Hardie and Minke 1992; Niemeyer et al. 1996), while CpTRPC is orthologous to TRPC genes identified in genomes of T. castaneum and A. pisum. The three other candidate TRPs found in the C. pomonella antennal transcriptome belong to the TRPA sub-family, which has undergone expansion in insects (Fig. 1). CpPyx and CpWtrw are orthologous to the D. melanogaster TRPAs pyrexia (pyx) and water witch (wtrw), involved in thermosensation (Lee et al. 2005) and hygrosensation (Liu et al. 2007), respectively. CpTRPA5 appears to be an orthologue of TRPA5 genes found in several insect genomes (Peng et al. 2015), with the notable exception of D. melanogaster. The previously described CpNan (Nguyen et al. 2013) is part of the TRPV subfamily, and is an orthologue of the D. melanogaster Nanchung gene.
Fig. 1.
Neighbor-joining tree of metazoan candidate TRPs. C. pomonella candidate TRPs identified by transcriptome analysis in bold. Cp: Cydia pomonella L.; Rn: Rattus norvegicus Berkenhout; Ce: Caenorhabditis elegans Maupas; Dm: Drosophila melanogaster Meigen; Bm: Bombyx mori L.; Dp: Danaus plexippus L.; Am: Apis mellifera L.; Tc: Tribolium castaneum Herbst; Ap: Acyrthosiphon pisum Harris. Circles: bootstrap values > 80. Accession numbers are given in Supplementary Table S1.
RT-PCR on Adult Body Parts of C. pomonella TRPs
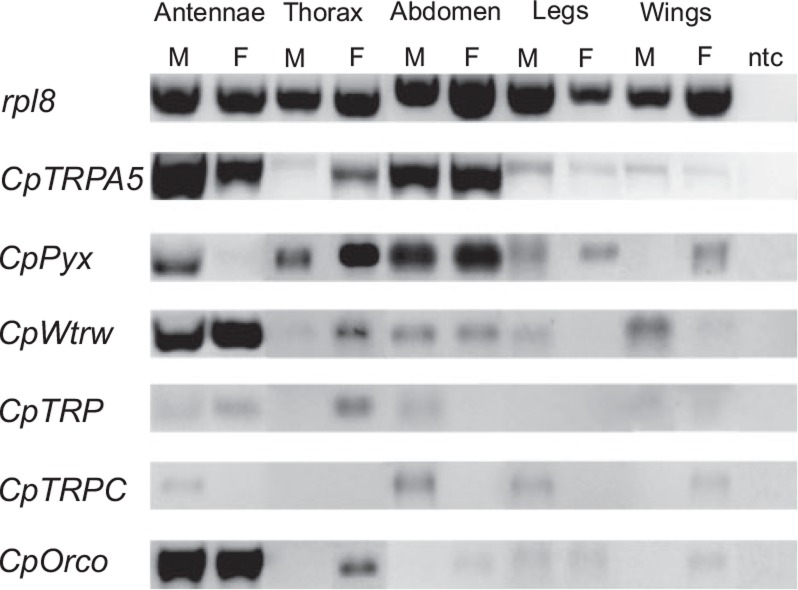
Performing RT-PCR amplifications of C. pomonella TRPs (TRPA5, pyrexia, water witch, TRP and TRPC), we found body-wide expression for TRPAs (TRPA5, pyrexia, water witch) and absent expressions in some body parts of both sexes for TRPCs (TRP and TRPC) (Fig. 2). TRPA5 is expressed in all body parts for both males and females. Similarly, RT-PCR revealed pyrexia and water witch expression in all body parts, except male wings and female antennae for pyrexia, and female legs for water witch. TRP is expressed in antennae of either sex, in female thorax and in male abdomen, and apparently no expression was found elsewhere. TRPC appeared to be expressed in male antennae, male abdomen, male legs and female wings.
Fig. 2.
RT-PCR of C. pomonella TRPs. Reverse transcription PCR of candidate Cp-TRP channels (CpTRPA5, CpPyr, CpWtrw, CpTRP, CpTRPC) in male and female C. pomonella body parts (Antennae, Thorax, Abdomen, Legs, Wings); ntc: no-template control; M: male; F: female; rpl8: positive control; CpOrco: antennal control. PCR volume loaded: 10 μl.
Assembly of the Open Reading Frame of the Codling Moth TRPA5
Since the reported mRNA length of its orthologues (e.g. B. mori XM_004926128.2; 3764 bp) was more than three times longer than our CpTRPA5 TRP contig (1012 bp), which also lacked of start and stop codons in frame, we judged the latter to be incomplete at both 5′ and 3′ ends. In an attempt to extend the sequence to full length, we performed 5′ and 3′- RACE-PCR. A partial coding sequence (CDS) of 1677 bp was generated by merging a 703 bp 5′-RACE-PCR product and a 432 bp 3′-RACE-PCR product with the 1012 bp contig, with an expected stop codon but without any candidate start codon. This was used as a template to query non-annotated C. pomonella sequences from genome and transcriptome data (unpublished), which led to the identification of a final CDS of 3126 bp including a start codon. To confirm the stop codon, we extended the sequence 4166 bp from the 3′ end by RACE-PCR, but no additional stop codon in frame with the partial CDS appeared. The full sequence of CpTRPA5 has been submitted to Genbank (accession number KU130118).
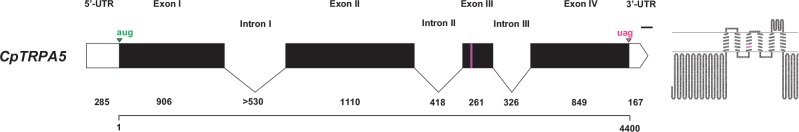
Comparison of the preliminary CpTRPA5 genomic locus with antennal RNA-seq data produced an overview of intron/exon boundaries within the CpTRPA5 locus (Fig. 3). The locus is constituted of four exons separated by three medium-sized introns. Interestingly, RNA-seq but not preliminary genomic assembly revealed 15 additional nucleotides (5′-CTCCATCGGCCTGGC-3′) within the third exon, indicating that they could originate from mRNA editing. For the 1041 length translated amino acid sequence of CpTRPA5, the TMHMM model predicted six transmembrane domains (between amino acids 619 and 641; 654 and 676; 691 and 713; 720 and 738; 753 and 775; 826 and 848), an N-terminal cytoplasmic region (from 1 to 618) and a C-terminal cytoplasmic region (from 849 to 1041), all of which would be expected for TRPs.
Fig. 3.
Graphical overview of the TRPA5 genomic locus. Left: Introns and exons representation. White rectangles: UTRs; black rectangles: exons; lines: introns; magenta rectangle: additional nucleotides from mRNA-editing (5′-CTCCATCGGCCTGGC-3′); green arrowhead and letters: start codon; magenta arrowhead and letters: stop codon; numbers: bp-lengths of UTRs, exons, introns; scale bar: 100 bp. Right: Topology representation of the translated polypeptide. Magenta amino acids: additional amino acids translated from edited mRNA (Nt-GSIGLA-Ct).
RT-PCR on Adult Body Parts and Confirmation of CpTRPA5 Splice Forms
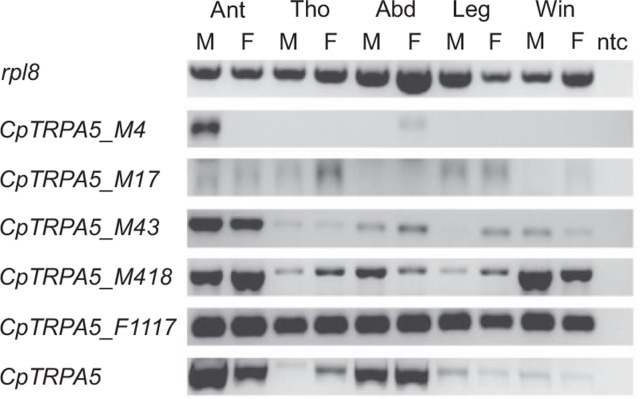
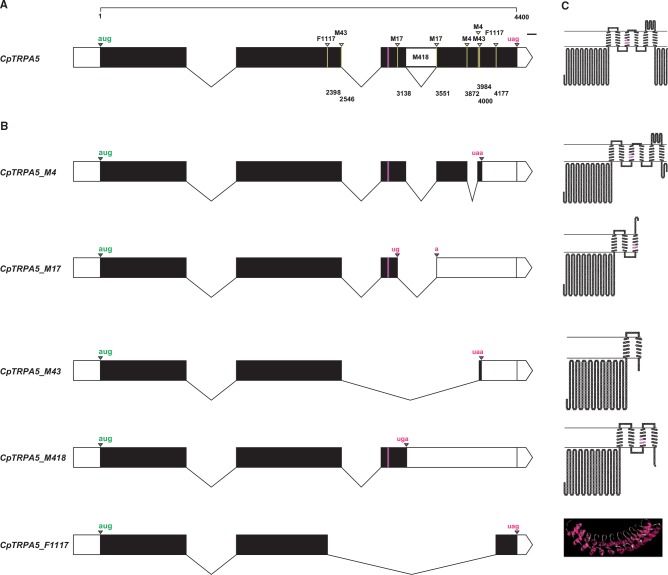
PCR of E.coli colonies transformed with pDONR221 containing the expected full-length CDS of CpTRPA5 revealed several positive clones containing what appeared to be different size transcripts of CpTRPA5 (data not shown). Sequencing of plasmids purified from positive clones revealed eleven rearrangements of the CDS, suggesting possible generation of splice forms from the original CpTRPA5. RT-PCR on adult male and female body parts confirmed the existence of five out of the eleven potential splice forms (CpTRPA5_M4, CpTRPA5_M17, CpTRPA5_M43, CpTRPA5_M418 and CpTRPA5_F1117) in at least two cDNA samples reverse-transcribed from independent body part RNAs. Sequences of these five splice forms have been submitted to Genbank (accession numbers KU130119, KU130120, KU130121, KU130122, and KU130123 respectively). Lack of amplification for six out of the eleven potential splice forms in all cDNA samples (CpTRPA5_M41, CpTRPA5_M415, CpTRPA5_M417, CpTRPA5_F1111, CpTRPA5_F1115, CpTRPA5_F1124) means that at this point, we cannot confirm their existence (Fig. 4).
Fig. 4.
Validation of CpTRPA5-splice forms in male and female body parts by RT-PCR. For comparison, the full-length CpTRPA5 gene was also included. The housekeeping gene rpl8 was used as a positive control in all body parts. Ant: antennae; Tho: thorax; Abd: abdomen; Leg: legs; Win: wings; ntc: no-template control; M: male; F: female. PCR volume loaded: 10 μl.
Genome and transcriptome data was used to verify positions of splicing sites expected for the generation of splice forms previously confirmed by RT-PCR (CpTRPA5_M4, CpTRPA5_M17, CpTRPA5_M43, CpTRPA5_M418 and CpTRPA5_F1117, Fig. 5A and B). The CpTRPA5_M4 splice form is generated by the excision of a short 112 bp fragment within the fourth exon, between positions 3872 and 3984 counted from ATG. This splicing is responsible for the termination at a premature UAA stop codon, shortening the C-terminal domain of the transmembrane protein. The downstream splicing site generating CpTRPA5_M17 form is expected to be in coincidence with the 3′ end of the third intron, at position 3,551. Interestingly, the premature excision of 88 bp upstream of the third intron at position 3,138 affects the open reading frame to generate a premature UGA-stop codon by combination of nucleotides U3136, G3137 and A3552 located on splicing sites boundaries. This prematurely terminates translation and it is potentially responsible for the generation of a truncated polypeptide having only three transmembrane domains, compared to six translated from the full CDS. The CpTRPA5_M43 splice form is generated by excision of a 1,454 bp fragment between position 2,546 at the end of the second exon, in coincidence with the second intron, and position 4,000 in the fourth exon. The excision shortens the CDS and modifies the open reading frame to an anticipated termination at a candidate UAA stop codon, at position 4,022. Sequencing of the pDONR221 clone of CpTRPA5_M418 splice form revealed an extra CDS region, which genomic overview revealed to correspond with the third un-spliced intron. The presence of the third intron in CpTRPA5_M418 likely makes it an incomplete splice form, the CDS of which is characterized by a premature termination due to an alternative candidate UGA stop codon located in the sequence of the un-spliced intron. Possible splicing sites in the second and fourth exons at positions 2,398 and 4,177 respectively, determines excision for a 1,779 bp fragment, which could generate the CpTRPA5_F1117 splice form. This candidate splice form lacks most of the coding sequence between exon II and exon IV, which codes for transmembrane domains, and it makes the potential translated protein soluble.
Fig. 5.
Graphic representation of introns, exons and topology of the translated polypeptides, of the fulllength CpTRPA5 variant and verified splice forms. (A) Introns and exons for the full-length CpTRPA5 variant. White rectangles: 5’-UTR, M418 additional region, 3’-UTR; black rectangles: exons; lines: introns; magenta rectangle: additional nucleotides from mRNA-editing; green arrowhead and letters: start codon; magenta arrowhead and letters: stop codon; yellow arrowheads and bars: splicing sites of RT-PCR verified splice forms (abbreviations indicated above arrowheads); numbers: splicing sites positions, counted from the start codon; scale bar: 100 bp. (B) Introns and exons for CpTRPA5 splice forms (CpTRPA5_M4, CpTRPA5_M17, CpTRPA5_M43, CpTRPA5_M418 and CpTRPA5_F1117). (C) Topology of the translated polypeptides of the full length CDS CpTRPA5 and of splice forms CpTRPA5_M4, CpTRPA5_M17, CpTRPA5_M43 and CpTRPA5_M418. Magenta: amino acids translated from edited-mRNA; 3D-prediction (Rastop): CpTRPA5_F1117.
For most splice forms, a topological transmembrane representation was predicted (Fig. 5C). Concerning the CpTRPA5_F1117 splice form, for which the transmembrane prediction returned a lack of transmembrane domains, 3D prediction of the tertiary structure was necessary. Among proposed 3D-models (Protein Database, http://www.rcsb.org/pdb/home/home.do) the model 1N11.pdb related with the D34-region of a human ankyrin was chosen. This model was reported by the server to be the best candidate to represent the 3D structure of the CpTRPA5_F1117 protein (e-value = 2.0E−31).
Discussion
Insect TRPA genes constitute a subfamily of several sensors responding to different types of stimuli (Tracey et al. 2003; Lee et al. 2005; Liu et al. 2007; Wang et al. 2009, Sun et al. 2009; Kang et al. 2010; Wang et al. 2011; Wei et al. 2015). In contrast, only one TRPA gene is expressed in mammals (Story et al. 2003; Bandell et al. 2004; Jordt et al. 2004; Macpherson et al. 2007; Nilius and Flockerzi 2014). As there is currently a dearth of functional data available, the sensory implications of these additional insect TRPA members are still unclear. With the wide range of functional specificity of insect TRPs, the deorphanization of these currently orphan TRPAs could give fresh insight to our understanding of insect sensing, and initial studies implicate roles for some TRPAs in nociception and thermal sensing (Braun 2012).
By searching the antennal transcriptome of the tortricid pest C. pomonella, we identified 5 candidate TRPs belonging to several orthology groups, i.e. Pyrexia, TRPA5, Water witch (within the TRPA subfamily), TRP and TRPC (within the TRPC subfamily) (Fig. 1). We also demonstrated the existence of different splice forms for CpTRPA5, which appears to have sex- and body part-specific expression patterns.
Evolutionary studies suggest that TRPAs underwent expansion in the common ancestor of crustaceans and insects, leading to four paralogous lineages (Peng et al. 2015). A fifth lineage, corresponding to the water witch genes, may have appeared in insects through a retrotransposition event from pyrexia (Matsuura et al. 2009). Compared to painless and TRPA1 that have been more conserved, pyrexia, water witch and TRPA5 genes experienced several gene gain and loss events in insects (Matsuura et al. 2009; Peng et al. 2015). As hypothesized for HsTRPA of Hymenoptera (Matsuura et al. 2009), several genes arising from TRPA duplications may be involved in gaining thermosensitive properties which can be speculated as a possible role for the TRPA5 orthologue we have identified in C. pomonella.
While we did not find in C. pomonella antennae some of the TRPs canonically involved in thermal sensing (TRPA1, Painless), we did find TRPs previously reported to be involved in sensing of heat (Pyrexia), cold (TRP), as well as hygroscopic sensing (Water witch). This suggests the existence of thermal and hygroscopic modalities in codling moth antennae, as recently demonstrated in Drosophila (Gallio et al. 2011; Liu et al. 2007), hymenoptera (Ruchty et al. 2010) and previously reported for other insects (Altner and Loftus 1985). Notably, TRPA activation in antennae could contribute, together with the olfactory system, to host finding, as demonstrated by recent studies on insect thermotaxis (Wang et al. 2009; Corfas and Voshall 2015). Furthermore, in orchards treated with pheromones for mating disruption of codling moth, the onset of male diel flight was strongly correlated with temperature: male were active at temperatures between 19 °C and 14 °C (Witzgall et al. 1999). Lack of correlation between flight onset and light intensity further suggested the contribution of temperature sensing to odor-guided behavioral responses in the codling moth.
In Drosophila, Pyrexia-expressing neurons appear to be widely distributed throughout the fly body, and are most likely involved in the detection of high temperatures (Lee et al. 2005). We observed a similar pattern of body-wide expression for the C. pomonella TRPA5, where RT-PCR indicated that it was expressed in all body parts. Similarly, C. pomonella pyrexia and water witch were expressed in all body parts, except male wings and female antennae for pyrexia and female legs for water witch, while a slight band for the latter in male thorax and female wings may suggest, although limited, a possible expression of water witch in these body parts (Fig. 2). Functional studies of Pyrexia in D. melanogaster also demonstrated different temperature sensitivities for different Pyrexia isoforms and for their combinations (Lee et al. 2005). When co-expressed, immunostaining showed both isoforms to be co-localized, suggesting a possible hetero-tetramerization of the quaternary structure of the channel, with a possible functional role in sensing specific temperature ranges. While speculative, a possible explanation for the existence of multiple TRPA5 splice forms in C. pomonella could be a combinatorial-based system for thermal sensing, similar to that observed for the Drosophila Pyrexia.
In addition, topology predictions indicated that certain CpTRPA5 splice forms lack most of the voltage-sensing domain (TM1–TM4), possibly leading to a shift in function (Fig. 5C). For CpTRPA5_M43, most of the voltage-sensing domain was lacking (TM2–TM6). For CpTRPA5_M418, TM5 and TM6 were missing, which form the central cation-conducting pore. Expression patterns of the full length CpTRPA5 and variants CpTRPA5_M43 and CpTRPA5_M418 were mostly similar, with the exception of a high expression in the wings for CpTRPA5_M418 (Fig. 4). Genomic overview further confirmed the existence of these splice forms. Indeed, the position of the upstream splicing site for CpTRPA5_M43 corresponds with the intron/exon boundary between exon II and intron II and CpTRPA5_M418 is the un-spliced variant generated by lack of intron III excision (Fig. 5B). Similar expression patterns observed with the full-length variant motivates further investigations to validate any possible functionality of these splice forms.
CpTRPA5_M17 and CpTRPA5_F1117 splice forms were observed in most body parts. The CpTRPA5_F1117 splice form lacks most of the CDS between exons II and IV coding for a soluble ankyrin-repeat module. While our knowledge is limited regarding the role of ankyrin domains of transmembrane proteins of eukaryote organisms, e.g. TRPs, they are known to be associated with the cytoskeleton (Zhang et al. 2015) by protein/protein interactions involving spectrin/actine complexes in the cytosolic side of the plasma membrane (Baines 2010). Although expression and purification of soluble ankyrins has been reported (Binz et al. 2003), the expression of free ankyrins derived from rearrangement of the CDS of transmembrane proteins is currently unknown.
In insects, it has been demonstrated that epigenetic regulation occurring by DNA methylation may regulate expression of specific genes by causing widespread and diverse changes in alternative splicing (Foret et al. 2012; Li-Byarlay et al. 2013). In this context, the identification of splice forms in all body parts without a specific pattern may have possible implications with silencing translation of the original CDS, by generating non-functional forms like CpTRPA5_F1117 and CpTRPA5_M17. Further investigations might focus on functional characterization of these splice forms such as their possible implications with silencing mechanisms.
Other splice forms may translate proteins functioning as TRPA sensors in the codling moth. For instance, among splice forms, CpTRPA5_M4 appears to be expressed only in male antennae and in the female abdomen. The existence of this splice form is confirmed by the identification of its downstream splicing site proximal with the one identified for CpTRPA5_M43, as validated by genomic overview (see above). Transmembrane predictions indicate that this splice form retains six transmembrane domains. Only a shorter re-arranged C-terminal distinguished it from the original full-length TRPA5. Although the N- and C-terminal regions are reported to be important to mediate control of channel gating (Hoffman et al. 2002) and activations of thermal-TRP-channels (Brauchi et al. 2006), this CpTRPA5_M4 form may be the best candidate functional channel, together with the full-length variant. CpTRPA5_M4 preserves a complete ankyrin-repeats N-terminal, required for TRPA-activation (Macpherson et al. 2007), and a six-TM structure, including TM5, TM6 and their connecting loop forming the central cation-conducting pore. As recently demonstrated, the existence of different isoforms seems to be the key of TRPA sensory discrimination of insects, depending on neuronal cell segregation (Kang et al. 2012). Whereas the full length CpTRPA5 is possibly involved in a general transduction pathway because of its wide expression in all insect body parts, the CpTRPA5_M4 isoform may be involved in thermal sensing in male antennae and the female abdomen, possibly the ovipositor.
Our data also suggest the occurrence of mRNA editing within the coding region of the third transmembrane domain of CpTRPA5 (Fig. 3). Current findings report mRNA editing occurring for K+ channels in multiple organisms, including insects (Holmgren and Rosenthal 2015). For instance, in Drosophila, editing generates multiple isoforms. Their frequency varies between different parts of the adult’s anatomy (Ingleby et al. 2009), having a variety of functional effects, including changes to activation, deactivation and inactivation kinetics, and some small shifts to the channel voltage sensitivity (Ryan et al. 2008). Pyrexia is known to be a thermal-gated K+ channel (Lee et al. 2005) and the relatedness of CpTRPA5 with pyrexia supports our mRNA-editing findings for CpTRPA5. Post-transcriptional modifications such as mRNA editing and generation of multiple splice forms observed in RT-PCR follow a general pattern of regulation of TRP function at multiple post-transcriptional levels (Voolstra and Huber 2014) and raise the question of potential functional effects.
Given the possible role of CpTRPA5 in temperature sensing in antennae, the present work opens perspectives for investigating the contribution of different sensory modalities in harmful behaviors of crop pest moths. Furthermore, better understanding the contribution of alternative splicing and post-transcriptional modifications to the function of CpTRPA5 would increase our knowledge on mechanisms that remain largely unexplored in the field of insect neuroethology.
Acknowledgments
We thank Andrea Campisano, Research and Innovation Centre - Fondazione Edmund Mach, for suggestions on RT-PCR. We thank Ed Genomics, Edinburg (Scotland, UK) for sequencing and for access to unpublished data. Partial financial support for this project was provided by the direction of the PhD course in Chemistry, Biochemistry and Ecology of Pesticides (CBEA) of DeFENS: Department of Food, Nutritional and Environmental Sciences, Università degli Studi di Milano. Additional funding was provided by the Italian Ministry of Education, Universities and Research (MIUR) [project “Analisi integrate dei processi molecolari e cellulari responsabili dell’elaborazione di segnali sensoriali in condizioni normali e patologiche” (no. 2010599KBR_005, prin 2011)]. We also want to acknowledge Università di Cagliari and the government of Regione Autonoma della Sardegna (Italy) for further financial support in phase of manuscript preparation [project “L’endemismo sardo-corso Papilio hospiton e sue piante-ospite: studio integrato per la conservazione” (L.R. 7/2007 - Annualità 2012, CUP F71J12000950002)].
Supplementary Data
Supplementary data are available at Journal of Insect Science online.
References Cited
- Al-Anzi B., Tracey W. D., Benzer S. 2006. Response of Drosophila to wasabi is mediated by painless, the fly homolog of mammalian TRPA1/ANKTM1. Curr. Biol. 16: 1034–1040. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Altner H., Loftus R. 1985. Ultrastructure and function of insect thermo- and hygroreceptors. Annu. Rev. Entomol. 30: 273–295. [Google Scholar]
- Baines A. J. 2010. The spectrin-ankyrin-4.1-adducin membrane skeleton: adapting eukaryotic cells to the demands of animal life. Protoplasma. 244: 99–131. [DOI] [PubMed] [Google Scholar]
- Bandell M., Story, Hwang G. M. S. W., Viswanath V., Eid S. R., Petrus M. J., Earley T. J., Patapoutian A. 2004. Noxious cold ion channel TRPA1 is activated by pungent compounds and bradykinin. Neuron. 41: 849–857. [DOI] [PubMed] [Google Scholar]
- Barnard D. R. 1999. Repellency of essential oils to mosquitoes (Diptera: Culicidae). J. Med. Entomol. 36: 625–629. [DOI] [PubMed] [Google Scholar]
- Bassoli A., Borgonovo G., Morini G., De Petrocellis L., Schiano Moriello A., Di Marzo V. 2013. Analogues of perillaketone has highly potent agonists of TRPA1 channel. Food. Chem. 141: 2044–2051. [DOI] [PubMed] [Google Scholar]
- Bengtsson J. M., Gonzalez F., Cattaneo A. M., Montagné N., Walker W. B., Bengtsson M., Anfora G., Ignell R., Jacquin-Joly E., Witzgall P. 2014. A predicted sex pheromone receptor of codling moth Cydia pomonella detects the plant volatile pear ester. Front. Ecol. Evol. 2: Article 33. [Google Scholar]
- Bengtsson J. M., Trona F., Montagné N., Anfora G., Ignell R., Witzgall P., Jacquin-Joly E. 2012. Putative chemosensory receptors of the Codling Moth, Cydia pomonella, identified by antennal transcriptome analysis. PLoS. One. 7: e31620.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binz H. K., Stumpp M. T., Forrer P., Amstutz P., Plüchthun A. 2003. Designing repeat proteins: well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. J. Mol. Biol. 332: 489–503. [DOI] [PubMed] [Google Scholar]
- Brauchi S., Orta G., Salazar M., Rosenmann E., Latorre R. 2006. A hot-sensing cold receptor: C-terminal domain determines thermosensation in Transient Receptor Potential channels. J. Neurosci. 26: 4835–4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun A. P. 2012. Structural remodeling of the N-terminus tunes TRPA1 channel activation and regulates behavioral responses in Drosophila. Channels. (Austin). 6: 50–51. [DOI] [PubMed] [Google Scholar]
- Cattaneo A. M., Bengtsson J. M., Borgonovo G., Bassoli A., Anfora G. 2014. Response of the European Grapevine Moth Lobesia botrana to somatosensory-active volatiles emitted by the non-host plant Perilla frutescens. Physiol. Entomol. 39: 229–236. [Google Scholar]
- Clapham D. E. 2003. TRP channels as cellular sensors. Nature. 426: 517–524. [DOI] [PubMed] [Google Scholar]
- Corfas R. A., Vosshall L. B. 2009. The cation channel TRPA1 tunes mosquito thermotaxis to host temperatures. eLife. 4: e11750. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic. Acids. Res. 16: 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl D. F., Hardy R. W., Kernan M. J. 2000. Genetically similar transduction mechanisms for touch and hearing in Drosophila. J. Neurosci. 20: 5981–5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler M. A., Montell C. C. 2013. Drosophila TRP channels and animal behavior. Life. Sci. 92: 394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foret S., Kucharski R., Pellegrini M., Feng S., Jacobsen S. E., Robinson G. E., Maleszka R. 2012. DNA methylation dynamics, metabolic fluxes, gene splicing, and alternative phenotypes in honey bees. PNAS. 109: 4968–4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallio M., Ofstad T. A., Macpherson L. J., Wang J. W., Zuker C. S. 2011. The coding of temperature in the Drosophila brain. Cell. 144: 614–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Son W., Chung Y. D., Kim J., Shin D. W., McClung C. A., Lee Y., Lee H. W., Chang D. J., Kaang B. K., Cho H., Oh U., Hirsh J., Kernan M. J., Kim C. 2004. Two interdependent TRPV channel subunits, Inactive and Nanchung, mediate hearing in Drosophila. J. Neurosci. 24: 9059–9066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F., Bengtsson J. M., Walker W. B., Sousa M. -F. R., Cattaneo A. M., Montagné N., Fouchier A., Anfora G., Jacquin-Joly E., Witzgall P., Ignell R., Bengtsson M. 2015. A conserved odorant receptor detects the same substituted indan compounds in a totricid and a noctuid moth. Front. Ecol. Evol. 3: article 131. [Google Scholar]
- Göpfert M. C., Albert J. T., Nadrowski B., Kamikouchi A. 2006. Specification of auditory sensitivity by Drosophila TRP channels. Nat. Neurosci. 9: 999–1000. [DOI] [PubMed] [Google Scholar]
- Gouy M., Guindon S., Gascuel O. 2010. ScanView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27: 221–224. [DOI] [PubMed] [Google Scholar]
- Hall T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows95/98/NT. Nucleic Acids Symp. Ser. 41: 95–98. [Google Scholar]
- Hardie R. C., Minke B. 1992. The TRP gene is essential for a light activated Ca2+ channel in Drosophila photoreceptors. Neuron. 8: 643–651. [DOI] [PubMed] [Google Scholar]
- Hoffman T., Schaefer M., Schultz G., Gudermann T. 2002. Subunit composition of mammalian transient receptor potential channels in living cells. PNAS. 99: 7461–7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K., Stoffel W. 1993. TMbase - A database of membrane spanning proteins segments Biol. Chem. Hoppe-Seyler 374, 166.
- Holmgren M., Rosenthal J. J. 2015. Regulation of ion channel and transporter function through RNA editing. Curr. Iss. Mol. Biol. 17: 23–36. [PMC free article] [PubMed] [Google Scholar]
- Ingleby L., Maloney R., Jepson J., Horn R., Reenan R. Regulated RNA editing and functional epistasis in Shaker potassium channels. J. Gen. Physiol. 133: 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordt S. E., Bautista D. M., Chuang H. H., McKemy D. D., Zygmunt P. M., Hogestatt E. D., Meng I. D., Julius D. 2004. Mustard oils and cannabinoids excite sensory nerve fibres through the TRP channel ANKTM1. Nature. 427: 260–265. [DOI] [PubMed] [Google Scholar]
- Katoh K., Toh H. 2010. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics. 26: 1899–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Chung Y. D., Park D. Y., Choi S., Shin D. W., Soh H., Lee H. W., Son W., Yim J., Park C. S., Kernan M. J., Kim C. 2003. A TRPV family ion channel required for hearing in Drosophila. Nature. 424: 81–84. [DOI] [PubMed] [Google Scholar]
- Kwon Y., Kim S. H., Ronderos D. S., Lee Y., Akitake B., Woodward O. M., Guggino W. B., Smith D. P., Montell C. 2010. Drosophila TRPA1 channel is required to avoid the naturally occurring insect repellent citronellal. Cb. 20: 1672–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K., Panzano C. V., Chang E. C., Ni L., Dainis A. M., Jenkins A. M., Regna K., Muskavitch M. A. T., Garrity P. A. 2012. Modulation of TRPA1 thermal sensitivity enables sensory discrimination in Drosophila. Nature. 481: 76–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K., Pulver S. R., Panzano V. C., Chang E. C., Griffith L. C., Theobald D. L., Garrity P. A. 2010. Analysis of Drosophila TRPA1 reveals an ancient origin for human chemical nociception. Nature. 464: 597–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Lee Y., Lee J., Bang S., Hyun S., Kang J., Hong S. T., Bae E., Kaang B. K., Kim J. 2005. Pyrexia is a new thermal transient receptor potential channel endowing tolerance to high temperatures in Drosophila melanogaster. Nat. Genet. 37: 305–310. [DOI] [PubMed] [Google Scholar]
- Letunic I., Bork P. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 23: 127–128. [DOI] [PubMed] [Google Scholar]
- Leung A. Y., Foster S. 1996. Encyclopedia of common natural ingredients used in food, drugs and cosmetics, 2nd edition John Wiley and Sons Inc, New York, NY. [Google Scholar]
- Li-Byarlay H., Li Y., Stroud H., Feng S., Newman T. C., Kaneda M., Hou K. K., Worley K. C., Elsik C. G., Wickline S. A., Jacobsen S. E., Ma J., Robinson G. E. 2013. RNA interference knockdown of DNA methyl-transferase 3 affects gene alternative splicing in the honeybee. PNAS. 110: 12750–12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedtke W. B. 2007. TRP ion channels function in sensory transduction and cellular signaling cascades. CRC Press, Boca Raton, FL. [PubMed] [Google Scholar]
- Liu L., Li Y., Wang R., Yin C., Dong Q., Hing H., Kim C., Welsh M. J. 2007. Drosophila hygrosensation requires the TRP channels water witch and nanchung. Nature. 450: 294–298. [DOI] [PubMed] [Google Scholar]
- Macpherson L. J., Dubin A. E., Evans M. J., Marr F., Schultz P. G., Cravatt B. F., Patapoutian A. 2007. Noxious compounds activate TRPA1 ion channels through covalent modification of cysteines. Nat. Lett. 445: 541–545. [DOI] [PubMed] [Google Scholar]
- Matsuura H., Sokabe T., Kohno K., Tominaga M., Kadowaki T. 2009. Evolutionary conservation and changes in insect TRP channels. BMC. Evol. Biol. 9: 228.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesterov A., Spalthoff C., Kandasamy R., Katana R., Rankl N. B., Andrés M., Jähde P., Dorsh J. A., Stam L. F., Braun F. -J., Warren B., Salgado V. L., Göpfert M. C. 2015. TRP channels in insect stretch receptors as insecticide targets. Neuron. 86: 665–671. [DOI] [PubMed] [Google Scholar]
- Nguyen P., Sýkorová M., Šíchová J., Kůta V., Dalíková M., Čapková Frydrychová R., Neven L. G., Sahara K., Marec F. 2013. Neo-sex chromosomes and adaptive potential in tortricid pests. PNAS. 110: 6931–6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemeyer B. A., Suzuki E., Scott K., Jalink K., Zuker C. S. 1996. The Drosophila light-activated conductance is composed of the two channels TRP and TRPL. Cell. 85: 651–659. [DOI] [PubMed] [Google Scholar]
- Nilius B., Flockerzi V. 2014. Mammalian Transient Receptor Potential (TRP) cation channels. Handbook of experimental pharmacology, Volume II, Springer Cham Heidelberg, New York, NY. [PubMed] [Google Scholar]
- Peng G., Xiao S., Kadowaki T. T. 2015. Evolution of TRP channels inferred by their classification in diverse animal species. Mol. Phylogenet. Evol. 84: 145–57. [DOI] [PubMed] [Google Scholar]
- Ridgway M., Silverstein R. L., Inscoe M. N. 1990. Behavior-modifying chemicals for insect management: applications of pheromones and other attractants. Marcel Dekker, New York, NY. [Google Scholar]
- Rosenzweig M., Kang K., Garrity P. A. 2008. Distinct TRP channels are required for warm and cool avoidance in Drosophila melanogaster. PNAS. 105: 14668–14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchty M., Helmchen F., Wehner R., Kleineidam C. J. 2010. Representation of thermal information in the antennal lobe of leaf-cutting ants. Front. Behav. Neurosci. 4: 174.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M. Y., Maloney R., Reenan R., Horn R. 2008. Characterization of five RNA editing sites in Shab potassium channels. Channels. (Austin). 2: 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokabe T., Tominaga M. 2009. A temperature-sensitive TRP ion channel, Painless, functions as a noxious heat sensor in fruit flies. Comm. Integr. Biol. 2: 170–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Story G. M., Peier A. M., Reeve A. J., Eid S. R., Mosbacher J., Hricik T. R., Earley T. J., Hergarden A. C., Andersson D. A., Hwang S. W., McIntyre P., Jegla T., Bevan S., Patapoutian A. 2003. ANKTM1, a TRP-like channel expressed in nociceptive neurons, is activated by cold temperatures. Cell. 112: 819–829. [DOI] [PubMed] [Google Scholar]
- Sun Y., Liu L., Ben-Shahar Y., Jacobs J. S., Eberl D. F., Welsh M. J. 2009. TRPA channels distinguish gravity sensing from hearing in Johnston's organ. PNAS. 106: 13606–13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracey W. D., Jr, Wilson R. I., Laurent G., Benzer S. 2003. painless, a Drosophila gene essential for nociception. Cell. 113: 261–273. [DOI] [PubMed] [Google Scholar]
- Viswanath V., Story G. M., Peier A. M., Petrus M. J., Lee V. M., Hwang S. W., Patapoutian A., Jegla T. 2003. Opposite thermosensor in fruitfly and mouse. Nature. 423: 822–823. [DOI] [PubMed] [Google Scholar]
- Voolstra O., Huber A. 2014. Post-translational modifications of TRP channels. Cells. 3: 258–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker R. G., Willingham A. T., Zuker C. S. 2000. A Drosophila mechanosensory transduction channel. Science. 287: 2229–2234. [DOI] [PubMed] [Google Scholar]
- Walker W. B. I. I. I., Gonzalez F., Garczynski S. F., Witzgall P. 2016. The chemosensory receptors of codling moth Cydia pomonella expression in larvae and adults. Scientific Reports. 6: 23518.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Qiu Y. T., Lu T., Kwon H. W., Pitts R. J., Van Loon J. J., Takken W., Zwiebel L. J. 2009. Anopheles gambiae TRPA1 is a heat-activated channel expressed in thermosensitive sensilla of female antennae. Eur. J. Neurosci. 30: 967–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Guo Y., Wang F., Wang Z. 2011. Drosophila TRPA channel Painless inhibits male-male courtship behavior through modulating olfactory sensation. PLoS. One. 6: e25890.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J. J., Fu T., Yang T., Liu Y., Wang G. R. 2015. A TRPA1 channel that senses thermal stimulus and irritating chemicals in Helicoverpa armigera. Insect. Mol. Biol. 24: 412–421. [DOI] [PubMed] [Google Scholar]
- Witzgall P., Stelinski L., Gut L., Thomson D. 2008. Codling moth management and chemical ecology. Annu. Rev. Entomol. 53: 503–522. [DOI] [PubMed] [Google Scholar]
- Witzgall P., Bäckman A. C., Svensson M., Koch U., Rama F., El-Sayed A., Brauchli J., Arn H., Bengtsson M., Löfqvist J. 1999. Behavioral observations of codling moth, Cydia pomonella, in orchards permeated with synthetic pheromone. Bio. Control. 44: 211–237. [Google Scholar]
- Wu L. J., Sweet T. B., Clapham D. E. 2010. International Union of Basic and Clinical Pharmacology. LXXVI. Current progress in the mammalian TRP ion channel family. Pharmacol. Rev. 62: 381–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Sornborger A. T., Lee J. K., Shen P. 2008. Drosophila TRPA channel modulates sugar-stimulated neural excitation, avoidance and social response. Nat. Neurosci. 11: 676–682. [DOI] [PubMed] [Google Scholar]
- Zhang W., Cheng L. E., Kittelmann M., Li J., Petkovic M., Cheng T., Jin P., Guo Z., Göpfert M. C., Jan L. Y. Y. N. 2015. Ankyrin repeats convey force to gate the NOMPC mechanotransduction channel. Cell. 162: 1391–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]