Abstract
Mythimna separata walker (Lepidoptera: Noctuidae) is a polyphagous pest of nearly 100 families of more than 300 kinds of food and industrial crops. So far, both nucleotide and protein sequence information has been rarely available in database for M. separata, strictly limiting molecular biology research in this insect species. In this study, we carried out a transcriptome sequencing for M. separata. The sequencing and subsequent bioinformatics analysis yielded 69,238 unigenes, among which 45,227 unigenes were annotated to corresponding functions by blasting with high homologous genes in database, giving annotation rate of 65.32%. Several lepidopteran insects gave best matches with the transcriptome data. To gain insight into the mechanism of insecticide resistance in M. separata, 15 families of genes encoding insecticide resistance-related proteins were investigated. Substantial numbers of unigenes in these families were identified in the transcriptome data, and 17 out of 21 selected unigenes were successfully amplified. Expressions of most of these genes were detected at larval stages and in gut tissue, as was consistent with their putative involvement in insecticide resistance. Our study provides most comprehensive transcriptome data for M. separata to date, and also provides reference sequence information for other Noctuidae family insects.
Keywords: insecticide resistance, Mythimna separata, Noctuidae, transcriptome, unigene
The genomic information for insect species has been available for only some well-studied model species to date. This seriously restricted the study for the elucidation of gene functions in non-model insect species, and also hindered the development of pest control strategy based on characterization and exploitation of genes related to several important factors including insecticide action mechanism and resistance development. With the advent of next generation sequencing (NGS) technology and cost decrease of it with the technical advancement, it has become a valuable tool for researchers to obtain vast amounts of genetic information from non-model organisms without any sequence knowledge. In fact, the genomic data are expanding at an unprecedented rate for more and more insect species of different orders and families relying on this technique (http://www.entsoc.org/press-releases/entomologists-launch-5000-insect-genome-project-i5). Transcriptome sequencing data offer a whole set of expression sequence tags for sample of interest and so directly represent the transcribed sequence information. Transcriptome sequencing is a basis for unigene identification, and it is also a powerful research tool for gene expression analysis and functional characterization (Oppenheim et al. 2015).
Noctuidae species comprise the largest family in the lepidopteran order with more than 35,000 known species in more than 4,200 genera. Many of the Noctuidae family insects are economically important crop pests. Despite its large family members, widespread distribution, and economic importance, Noctuidae family insects are under-represented in the accessible transcriptomic and genomic sequence information of insect species. Transcriptome and even genome-sequencing projects have been reported for the representative species in other families in lepidopteran insects such as Plutella xylostella (Plutellidae family) (You et al. 2013), Bombyx mori (Bombycidae family) (Xia et al. 2008), and Danaus plexippus (Nymphalidae family) (Zhan et al. 2011). But very less of such efforts are reported for Noctuidae family, which offers us a new research orientation to pursue this issue.
Mythimna separata walker (Lepidoptera: Noctuidae) is becoming a devastating threat for the production of corn especially in northern China (Feng et al. 2008). Besides corn, M. separata is also a documented pest of nearly 100 families of more than 300 kinds of food and industrial crops. Until now, the control strategy toward this insect pest heavily relies on the usage of chemically synthesized pesticides (Jiang et al. 2011, Rashid et al. 2013). But the long-term and dominant use of chemical insecticides caused serious environmental problem and cropland deterioration, and also led to the development of insect resistance toward commonly applied chemicals (Yun et al. 2004). Therefore, more targeted control method based on the exploitation of M. separata cognate genes related to its growth, development and especially those involved in insecticide resistance is urgently needed. So far, only mitochondria genome of M. separata was sequenced, and only 43 ESTs and hundreds of gene sequences were available for this insect, far from sufficient to look for and manipulate the genes useful for the above regard (Li et al. 2015). De novo sequencing for M. separata transcriptome was performed here. The transcriptome sequence reads of M. separata were assembled to 69,238 unigenes after assembly. In total, 45,227 unigenes were annotated and were highly homologous with the reported genes, reaching at the annotation frequency of 65.32%. Specific attention was focused on some gene families encoding insecticide resistance related proteins. Many family members of these genes were retrieved from transcriptome data, and some of them were verified by PCR amplification. Expression patterns of the selected unigenes showed universal transcript accumulation at larval stages and in gut tissue.
Materials and Methods
Insect Culture and Sample Collections
Eggs of laboratory-adapted M. separata were obtained from The Institute of Plant Protection (IPP), Chinese Academy of Agricultural Sciences (CAAS, Beijing, China). Eggs were stored at 4°C on wet gauze in a petri dish and were transferred to room temperature for hatching. The fertilized nymphs were reared on fresh corn leaves in a plastic cage aerated via aperture at 25°C, 70% relative humidity, and a photoperiod of 14 h:10 h (light:dark). Development of the insects was monitored daily, insects of different developmental stages were collected, intestinal content was cleared, and the remaining parts were stored in liquid nitrogen for following experiment.
Total RNA Isolation and Quality Checking
Mixture samples of M. separata at different developmental stages including egg, second, third, fourth, fifth instar larvae, pupa, and adult were ground to powder in liquid nitrogen. RNA extraction was carried out using TRIzon Reagent (CWBio, China) following the manufacturer’s instructions. Possibly contaminated genomic DNA was removed by DNase (Invitrogen, USA) treatment. The OD260:OD280 ratio of total RNA was detected with Nanodrop 2000 (Thermo Scientific Inc., USA). One microgram of total RNA was resolved on 1% agarose gel. The gel was stained with Goldview I nucleic acid dye (SolarBio, Life Sciences, USA), and the nucleic acid band was visualized under GelDoc-It2 Imager (UVP, USA). DM2000 DNA ladder (CWBio) was run on the same gel to estimate the size of rRNA bands.
Construction of cDNA Library and Illumina Sequencing
RNA sample was shipped to BGI (Shenzhen, China) for sequencing. Briefly, 10 µg of the total RNA was used for cDNA library construction. Poly (A) mRNA was enriched with Oligo (dT) attached to magnetic beads (Promega, USA). RNA was fragmented in Thermomixer with the addition of fragmentation reagent. cDNA was synthesized with reverse transcriptase (Invitrogen) by the priming of random hexamer. cDNA was purified, sticky end was repaired, and the base “A” and adaptor sequence were ligated to the 3’ end of cDNA. Size selection and PCR amplification were performed, and the quality was checked with Agilent 2100 Bioanalyzer and ABI StepOnePlus Real-Time PCR system (ABI, USA). The cDNA library was subjected to sequencing on Illumina HiSeq 2000 platform (Illumina, Inc., USA).
De Novo Assembly and Bioinformatics Analysis
The raw reads were filtered to remove the unqualified reads. The clean reads were assembled into contigs with the SOAPdenovo software and were further connected into scaffolds. After paired-end joining and gap filling, the scaffolds were assembled into unigenes. Finally, the unigenes were screened against the databases such as non-redundant (NR), nucleotide (NT), Swiss-Prot, GO, KEGG, and COG for functional characterization with the cut-off value of e−5.
Screening of the Genes of Interest and Expression Pattern Analysis
The unigenes coding for proteins related to insecticide resistance were retrieved from the assembled sequences. Local BLAST was also performed by searching these putative genes using the sequences of homologous genes from other lepidopteran insects. The longest unigenes representing each family of genes were chosen for further analysis. PCR primers were designed using primer 3 software and were synthesized by Sangon Tech company in China. Total RNAs were isolated from M. separata at different developmental stages including egg, second, third, fourth, fifth instar larvae, pupa, and adult. Total RNAs were also isolated from different tissues of fourth instar larvae including leg, gut, salivary gland, reproductive organ, abdomen, and head, respectively. cDNAs were synthesized from above RNAs, respectively. The expression profiles of each selected unigenes in above samples were performed by PCR amplification. Abundance of β-actin (Accession number GQ856238) transcript amplified by the primers 5′-ccacgagaccacctacaact-3′ and 5′-ggaatcgacaatgttccgca-3′ was used as the internal control. Equal amounts of PCR products were run on agarose gel. The gel was stained with Goldview I nucleic acid dye, and was visualized under GelDoc-It2 Imager. DM2000 DNA ladder was run on the same gel to estimate the size of PCR fragment. The PCR fragment was purified from the gel using gel purification kit and ligated into pGEM-T vector by TA cloning method. The recombinant vectors were sequenced to verify the insert sequences.
Results
Quality Assessment of Total RNA for Illumina Sequencing
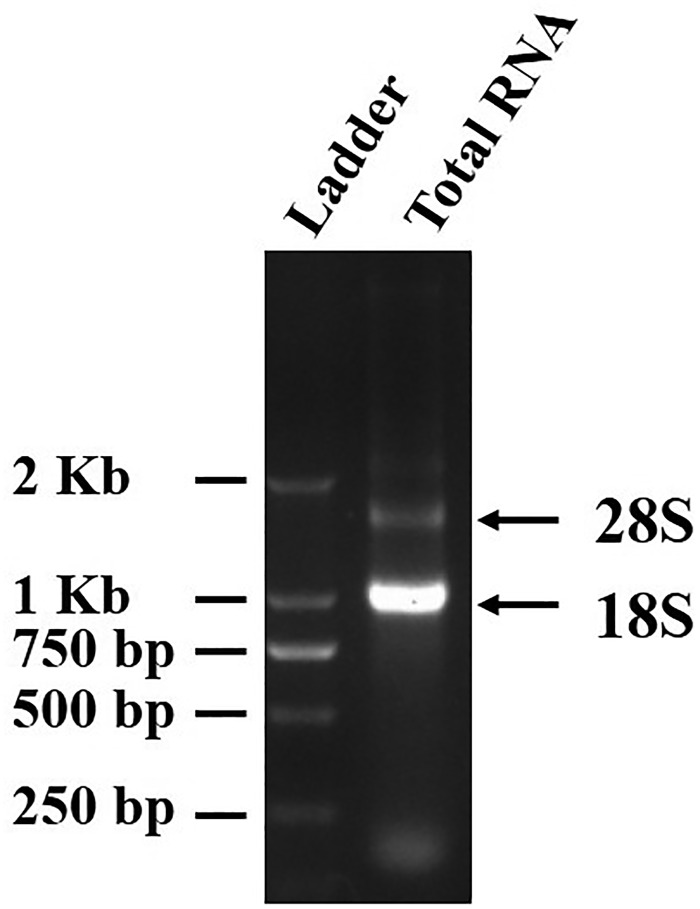
Total RNA was isolated from mixture samples of M. separata at different developmental stages. To assess the quality of total RNA sample, 1 µg of RNA was resolved on 1% agarose gel. We observed two main bands of 28S and 18S rRNA wherein 18S rRNA band was much brighter than 28S rRNA band (Fig. 1). We did not observe degraded RNA with low molecular weight, demonstrating the integrity of the RNA sample.
Fig. 1.
Electrophoresis detection of Mythimna separata total RNA. Ladder: size of the DNA band in DM2000 marker was indicated; Total RNA: 28S and 18S rRNAs were indicated by arrows.
Illumina Sequencing and Sequence Assembly
Owing to the fact that available information of the M. separata sequence data is very limited, its transcriptome was sequenced by Illumina NGS platform of BGI in Shenzhen, China. A library (SRP073346) from mixture RNA of M. separata was generated using Illumina sequencing platform in a single run. After sequencing we got 57,399,954 raw reads, among which the 51,618,686 clean reads were assembled into contigs with the Trinity software (SOAPdenovo). The raw sequencing results were shown in Table 1. The short reads were assembled into 173,691 contigs with the mean length of 265 bp. The raw reads were then mapped back to the contigs to confirm the assembly confidence of the contigs originated from same transcript. Finally, we got 69,238 unigenes with 15,657 clusters and 53,581 singletons, by filling the gap of scaffolds with paired-end reads, sequence assembly and elimination of the redundant sequences using the clustering software TGICL. The mean length of unigenes is 551 bp. Length distribution of contigs and unigenes was shown in Table 2; 14,247 (20.58%) of the unigenes have the length of more than 800 bp. Summary statistics of the contigs and unigenes was shown in Table 3.
Table 1.
Output statistics of sequencing results
| Names of sequences | Numbers |
|---|---|
| Total raw reads | 57,399,954 |
| Total clean reads | 51,618,686 |
| Total clean nucleotides (NT) | 4,645,681,740 |
| Q20 percentage | 97.35% |
| N percentage | 0.01% |
| GC percentage | 45.25% |
Total clean reads and total clean nucleotides represented the number of filtered reads and bases; Q20 percentage shows the percentage of base error rate below 0.01; N percentage represented the uncertain base pairs ratio; GC percentage shows filtered base number of G and C accounts for the proportion of the total number of bases.
Table 2.
The length distribution of contigs and unigenes
| Length (bp) | Contigs | Unigenes |
|---|---|---|
| 200–499 | 148,988 | 38,514 |
| 500–799 | 14,158 | 16,477 |
| 800–1,099 | 47,136 | 056 |
| 1,100–1,399 | 23,553 | 143 |
| 1,400–1,699 | 12,801 | 860 |
| 1,700–1,999 | 8,131 | 123 |
| 2,000–2,299 | 482 | 726 |
| 2,300–2,599 | 295 | 465 |
| 2,600–2,899 | 192 | 301 |
| 2,900–3,000 | 110 | 156 |
| ≥3,000 | 305 | 417 |
Table 3.
Summary statistics of assembly quality
| Contig | Unigene | |
|---|---|---|
| Sample | Mse | Mse |
| Total number | 173,691 | 69,238 |
| Total length (nt) | 46,005,554 | 38,164,980 |
| Mean length (nt) | 265 | 551 |
| Minimum contig length (bp) | 200 | 300 |
| Maximum contig length (bp) | 7,755 | 10,412 |
| Total consensus sequences | – | 69,238 |
| Distinct clusters | – | 15,657 |
| Distinct singletons | – | 53,581 |
Total consensus sequences show the assembled unigenes; distinct clusters show the clustered unigene, and in one cluster there are several high-identify (over 70%) unigenes that probably come from the same gene or homologous gene; distinct singletons show the genes belong to single gene.
Annotation Summary
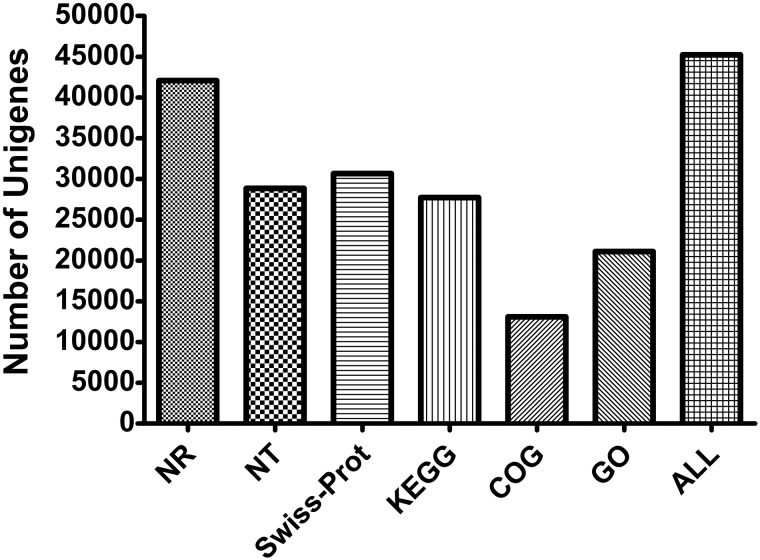
The unigene sequences were annotated by searching the NR NCBI protein database BLASTX with the cut-off e-value of e−5. Then the sequences were subjected to further analysis to verify their functions. The unigenes were also BLASTNed to NT database (e-value < e−5). Finally, we got the highest similarity proteins with our unigenes and obtained the functional annotation information. Among 69,238 unigenes, 45,227 unigenes had higher homology with known genes and gained the corresponding functional annotation information with the annotation rate of 65.32%. The statistical analysis of annotation result was shown in Figure 2. Briefly, 42,064 (60.75%) unigenes were annotated into NR database, 30,666 (44.29%) unigenes were annotated into Swiss-Prot database, 28,864 (41.69%) unigenes were annotated into NT database, 27,719 (40.03%) unigenes were annotated into KEGG database, 21,110 (30.49%) unigenes were annotated into GO database, and 13,083 (18.90%) unigenes were annotated into COG database. Sequence information of the unigenes and annotation results were provided in Supp dataset 1 (online only).
Fig. 2.
Histogram presentation of the annotation results of Mythimna separata unigenes against different databases. The number of unigenes with annotation in different databases was shown.
NR Annotation
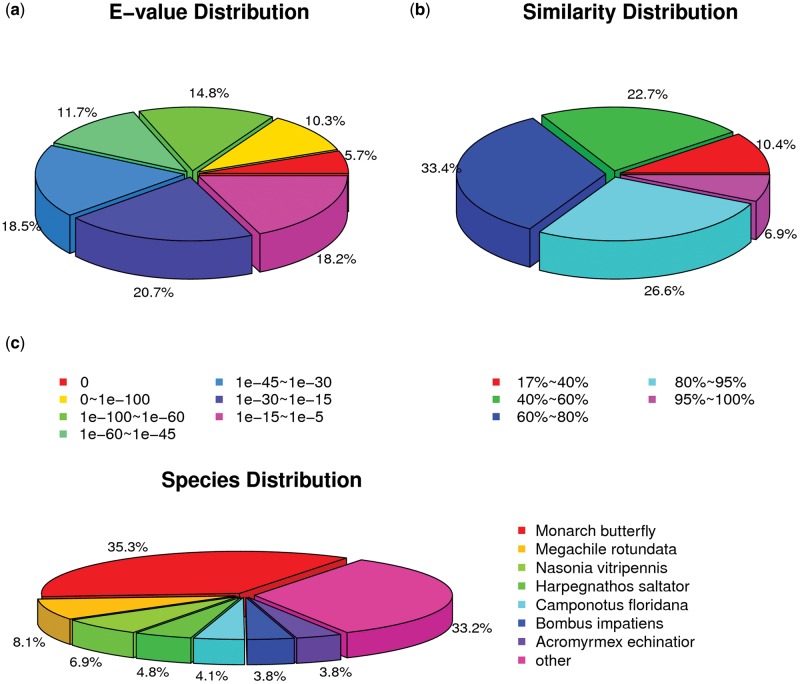
From the total, 42,064 (60.75%) unigenes matched to known proteins in the NR protein database of NCBI. Figure 3A showed the statistical analysis for e-value of blast result. e-Value of 30.80% NR annotated unigenes are lower than e−60, demonstrating high degree of confidence. Then we did the quantitative analysis which showed the similarity of NR annotation. The similarity distribution was shown in Figure 3B. Among the genes matched to NR database, 6.9% of genes have similarity over 95%, 60% have similarity between 60 and 95%, 22.7% have similarity between 40 and 60%, and only 10.4% have similarity less than 40%. Next, homology sequence BLASTing was carried out to evaluate the integrity and quality of the transcriptome. As shown in Figure 3C, seven species returned most homologous sequences with our data. Monarch butterfly (Danaus plexippus) was the species that gave the most BLAST hit of 35.3%, which was as expected in that full genome of monarch butterfly was recently sequenced and it represented the most comprehensive sequence information available in lepidopteran insects (Zhan et al. 2011). The other species that gave high match followed by monarch butterfly were the hymenopteran insects such as Megachile rotundata, Nasonia vitripennis, Harpegnathos saltator, Camponotus floridanus, Bombus impatiens, and Acromyrmex echinatior, which belonged to the same order with M. separata and possessed rich sequence information accessible in database.
Fig. 3.
Characteristics of homology search of the Mythimna separata unigenes against the non redundant (NR) protein database of NCBI. (A) e-Value distribution of the BLAST hits for the unigenes with a cut-off e-value of e−5. (B) Similarity distribution of the BLAST hits for the unigenes. (C) Species distribution of the unigene BLASTX matches against the NR protein database with a cut-off e-value of e−5. First hit of each unigenes was used for analysis and the proportions of homologous sequences in each species were shown.
Clusters of Orthologous Groups Annotation
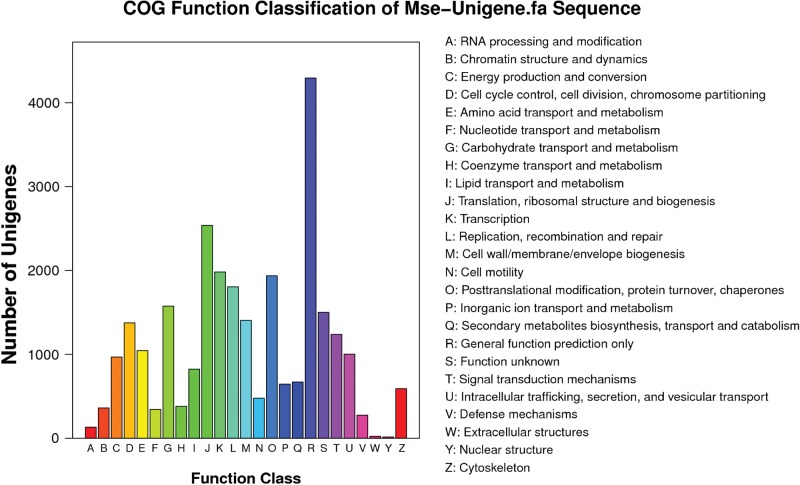
Clusters of orthologous groups (COG) database could classify orthology for the gene products. Assignments of COG were used to predict and categorize the functions of M. separata unigenes. Totally, 13,083 (18.90%) of the total unigenes were annotated and grouped into 25 specific categories based on the sequence homology. Among them, “General Function Prediction Only” represented the largest group containing 4,294 unigenes (15.67%), followed by “Translation, ribosomal structure and biogenesis”, “Transcription”, “Posttranslational modification, protein turnover, chaperones”, and “Replication, recombination, and repair” clusters containing unigenes of 2,538 (9.26%), 1,982 (7.23%), 1,937 (7.07%), and 1,805 (6.59%), respectively. The categories “Extracellular structures” and “Nuclear structure” possessed the smallest numbers of unigenes of 23 (0.08%) and 15 (0.05%), respectively (Fig. 4).
Fig. 4.
Classification of the clusters of orthologous groups for the transcriptome of Mythimna separata. 13,083 (18.90%) of the total unigenes were annotated and divided into 25 specific categories based on the sequence homology.
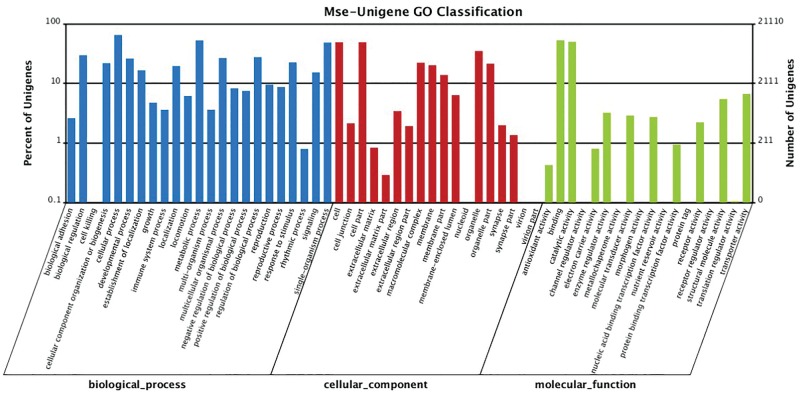
Gene Ontology Assignment
Gene Ontology (GO) is a normalized category system of gene function. All the NR annotated unigenes were subjected to GO analysis by Blast2GO software, and the retrieved annotation was processed by WEGO software to get functional classification. By sequence homology, 21,110 (30.49%) genes were categorized into 59 function groups (Fig. 5). In general, the unigenes are at the level of molecular function, cellular component, and biological process, respectively. In all of the three main categories of GO terms, “Cellular process”, “Binding”, “Metabolic process”, “Catalytic activity”, “Cell”, and “Cell part” terms have the highest gene numbers of 13,705 (64.9%), 11,146 (52.8%), 11,112 (52.64%), 10,529 (49.87%), 10,347 (49.01%), and 10,344 (49.00%), respectively. The least represented GO terms include “Nutrient reservoir activity” (2 unigenes), “Receptor regulator activity” (3 unigenes), and “Protein tag” (4 unigenes).
Fig. 5.
Classification of the gene ontology for the transcriptome of Mythimna separata. 21,110 (30.49%) of the total unigenes were categorized into 59 function groups.
Unigene Metabolic Pathway Analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG) can predict the potential metabolic pathway in which a specific unigene is involved and so provides a source for understanding of the complexity of gene function. Metabolic pathway analysis for M. separata unigenes was conducted using the KEGG annotation system. Totally, 27,719 (40.03%) unigenes were assigned to 258 KEGG pathways, among which “Signal transduction and metabolic pathways” was most highly represented with unigene number of 4,361 (15.73%), followed by “RNA transport” with unigene number of 998 (3.6%; Supp Table 1 [online only]).
Screening of Insecticide Resistance-Related Genes and Expression Profile Analysis
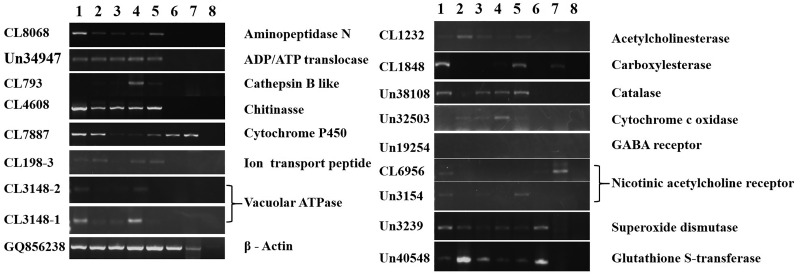
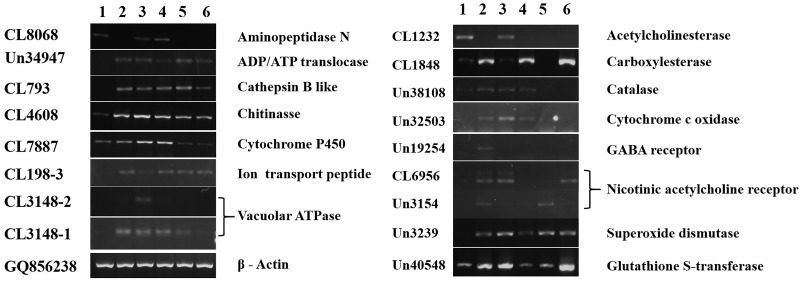
Besides enlarging M. separata sequence information, another aim of this study is to find some genes of interest, especially those related to insecticide resistance and detoxification. Cytochrome P450s are a family of enzymes implicated in detoxification and metabolism (Liu et al. 2015). Aminopeptidase N is involved in Bt resistance in insects (Chen et al. 2013). Cathepsin B is a cysteine protease involved in insect development such as moulting, and is also implicated in insecticide resistance (Silva et al.2012). V-ATPase, ADP/ATP translocase, ion transport peptide proteins are responsible for energy transfer and ion transport, and they are important for insect development and insecticide resistance (Wieczorek et al. 2003, Heinrich 2009, Upadhyay et al. 2011). Chitinase is involved in chitin synthesis and has long been studied as the biocontrol molecule agent toward insect pest (Chitvan et al. 2010). Acetylcholinesterase, carboxylesterase, catalase, cytochrome c oxidase, GABA receptor, nicotinic acetylcholine receptor, superoxide dismutase, and gluthione S-transferase are all reported to be involved in insecticide resistance (He et al. 2012). Substantial numbers of unigenes encoding these 15 family proteins were identified in the transcriptome data (Supp Table 2 [online only]), among which 21 genes were selected for further analysis. The number, annotation and primers of these unigenes were listed in Supp Table 3 (online only). We designed primers in the conserved regions and used the primers to amplify the unigene fragments. Seventeen out of 21 selected unigenes were successfully amplified and were verified by direct sequencing. Four of the unigenes were not successfully amplified, maybe because of their low expression, or the incorrect assembly of these unigenes. The successfully amplified unigenes were then subjected to phylogenetic analysis by aligning with the homologous sequences from other insect species. This phylogenetic analysis confirmed the annotation results as these unigenes showed substantial sequence identity with the homologous genes from other insect species (Supp Fig. 1 [online only]). Then the expression patterns of them were analyzed at different life stages. As shown in Figure 6, these genes displayed diverged expression patterns at different life stages. Overall, expressions of most of these unigenes were detected in egg and second to fifth instar larvae, while were not detected in pupa and adult. Further, the expression pattern of these unigenes was analyzed in variety of tissues of fourth instar larvae, which also showed their specificity of expression in different tissues. Generally, expressions of these unigenes were easily detectable in the tissues-like gut and salivary gland, while were not detected in the tissues like head and especially leg (Fig. 7).
Fig. 6.
Expression patterns of the unigenes at different life stages. 1, egg; 2, second instar larvae; 3, third instar larvae; 4, fourth instar larvae; 5, fifth instar larvae; 6, pupa; 7, adult; 8, water as a template for negative control. Steady state level of Mythimna separata β-actin gene served as the internal control.
Fig. 7.
Expression patterns of the selected unigenes in different tissues. 1, leg; 2, gut; 3, salivary gland; 4, reproductive organ; 5, abdomen; 6, head. Steady state level of Mythimna separata β-actin gene served as the internal control.
Discussion
The oriental armyworm M. separata is one of the most important pests of cereal crops including corn, wheat, and rice. To date, many research works on M. separata have focused on the elucidation of the biological and ecological aspects, while very less study aimed at understanding the mechanistic aspects, especially from the view of molecular biology. Few genetic resources are available for M. separata and other Noctuidae family insects, limiting the use of molecular biology approach to efficiently survey and manage the spread of these insects. To get a more comprehensive knowledge of the molecular mechanism underlying M. separata biology, transcriptome sequencing and de novo assembly of the transcripts were performed for pool of samples at different life stages. Quality, completeness, and depth of the sequencing data were assessed by different methods. As some insects including M. separata have the phenomenon of hidden-break, 18S rRNA is more abundant than 28S rRNA in Figure 1 (Fujiwara and Ishikawa 1986). Hybrid assembly of the 51,618,686 clean read sequences allowed us to assemble 69,238 unigenes of high quality. Annotation rate of the unigenes is 65.32%, which is comparable to or higher than those of other insects in lepidopteran order and especially in Noctuidae family such as Helicoverpa armigera (50.8%), Helicoverpa assulta (54.0%), Spodoptera frugiperda (51.1%), and Athetis lepigone (41.5%) (Li et al. 2013, Nascimento et al. 2015, Zhang et al. 2015). Most of these unigenes showed homology to seven closely related insect species, including monarch butterfly and six hymenopteran species Megachile rotundata, Nasonia vitripennis, Harpegnathos saltator, Camponotus floridanus, Bombus impatiens, and Acromyrmex echinatior. At the time we began the transcriptome analysis, we did not have enough Noctuidae family insect sequence data available in the database. When we compared our unigene data with the present database of Noctuidae family insects by local basting, we found higher hit match with them even most of the sequence information come from EST database, which could not cover the whole sequence information of these species. For example, Athetis lepigone (with 159,903 nucleotide sequences), Spodoptera frugiperda (with 95,191 nucleotide sequences), and Helicoverpa armigera (with 30,656 EST sequences) gave the blast hit of 59.62, 52.06, and 20.14%, respectively. These values are higher than the blast hit (3.8–8.1%) of the above six hymenopteran species. As these three species belong to the same family of Noctuidae with M. separata, it supported the genuity of the origin of our unigene sequences from Noctuidae species, i.e., M. separata. Substantial overlap with sequences information from these insect species indicates the high quality and reliability of our data. Unigene length is another factor determining the quality of transcriptome sequencing data. In this study, the number of unigenes with the sizes of 200–1,099 bp is 61,047, among which 34,564 (56.62%) unigenes matched described sequence in public database; the number of unigenes with the sizes of 1,100–1,999 bp is 6,126, among which 5,752 (93.89%) unigenes matched described sequence in public database; the number of unigenes with the sizes of 2,000–3,000 bp is 1,648, among which 1,332 (80.82%) unigenes matched described sequence in public database; the number of unigenes with the sizes of more than 3,000 bp is 417, among which 416 (99.76%) unigenes matched described sequence in public database. We observed a substantial percentage (34.68%) of unigenes that displayed unexpected functions or no similarity to known proteins, among them 97.46% had the length of less than 1,000 bp. We speculates that small proportion of the unigenes with no known match can be assigned as the novel genes in this species and they are particularly be of interest in the future studies.
With the transcriptome data and high-quality unigene sequences reported here, the number of genetic resources substantially increased from hundreds to hundred thousands in M. separata. These unigenes were classified into diversity of molecular functions and metabolic pathways covering many basic aspects of insect biology. The large number of genes involved in diversity of biological process would provide molecular resources and foundational knowledge for better understanding of its biology. As M. separata is an economically important pest, special attention was directed to find some transcripts coding for insecticide resistance related proteins. Twenty-one unigenes were selected for verification and expression pattern analysis, among which 17 (81%) unigenes were successfully amplified, indicating the reliability of the sequencing and assembly process. Interestingly, most of these unigenes were expressed at larval stages of M. separata and in the tissues like gut and salivary gland. It is consistent with the role of these insecticide resistance related genes, as the insects are most voracious at larval stages and therefore need to dispose of the consumed insecticides by the activity of these genes, and also because gut is the first barrier that encountered the ingested insecticides. Comparative analysis of these genes with their homologs from other crop pests, especially those of lepidopteran and Noctuidae family insects would provide the prospect for understanding of molecular mechanisms underlying the adaptation and resistance response shared between and common to related species. Expression pattern analysis of those putative insecticide resistance related genes under insecticide stress condition to verify possible involvement in insect-host communication and further investigation of their functions by reverse genetic manipulation tool like RNAi approach will be helpful for deeper understanding of these genes. Although difficulty of RNAi experiment in lepidopteran larva was reported (Terenius et al. 2011, Shukla et al. 2016), successful larval RNAi was reported in M. separata by dsRNA injection (Zhou et al. 2016). In fact, we newly developed a feeding based RNAi protocol for M. separata and will utilized it for the characterization of functional genes (unpublished data). It should be noted that most of the unigenes are partial fragments of specific genes. Further study like RACE technology is required to get a full length ORF sequence of a targeted gene of interest. In our experience, alignment of the unigene with the homologous genes from several closely related organisms to find most consensus sequence region and design the primers in this region is still the best way to successfully amplify the unigene fragments for some instances.
In summary, molecular mechanisms responsible for the biology of M. separata remain unclear to date. Biological and physiological mechanisms underlying its interaction with the host plant are therefore poorly understood. Supplementing the genetic resources of M. separata by our transcriptome data are essential to motivate further investigations to explore the functions of important genes and to better understand its molecular biology. Future development of management strategies toward this insect will greatly benefit from the present study and on-going research such as RNAi control of this insect.
Supplementary data
Supplementary data are available at Journal of Insect Science online.
Acknowledgments
This work was supported by funding from National Natural Science Foundation of China (31460036) and Program of Higher-level Talents of Inner Mongolia University (30105-125127).
References Cited
- Chen J., Supaporn L., Aimanova K. G., Gill S. S. 2013. A 104 kDa Aedes aegypti aminopeptidase N is a putative receptor for the Cry11Aa toxin from Bacillus thuringiensis subsp. israelensis. Insect Biochem. Mol. Biol. 43: 1201–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitvan K., Buschman L. L., Chen M., Subbaratnam M., Zhu K. 2010. A gut-specific chitinase gene essential for regulation of chitin content of peritrophic matrix and growth of Ostrinia nubilalis larvae. Insect Bioch. Mol. Biol.. 40: 621–629. [DOI] [PubMed] [Google Scholar]
- Feng H., Zhao X., Wu X., Wu B., Wu K., Cheng D., Guo Y. 2008. Autumn migration of Mythimna separata (Lepidoptera: Noctuidae) over the Bohai Sea in northern China. Environ. Entomol.. 37: 774–781. [DOI] [PubMed] [Google Scholar]
- Fujiwara H., Ishikawa H. 1986. Molecular mechanism of introduction of the hidden break into the 28S rRNA of insects: implication based on structural studies. Nucleic Acids Res. 14: 6393–6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich D. 2009. Insect ion transport peptides are derived from alternatively spliced genes and differentially expressed in the central and peripheral nervous system. J. Exp. Biol. 212: 401–412. [DOI] [PubMed] [Google Scholar]
- He W., You M., Vasseur L., Yang G., Xie M., Cui K., Bai J., Liu C., Li X., Xu X. 2012. Developmental and insecticide-resistant insights from the de novo assembled transcriptome of the diamondback moth, Plutella xylostella. Genomics 99: 169–177. [DOI] [PubMed] [Google Scholar]
- Jiang X., Luo L., Zhang L., Sappington T. W., Hu Y. 2011. Regulation of migration in Mythimna separata (Walker) in China: a review integrating environmental, physiological, hormonal, genetic, and molecular factors. Environ. Entomol. 40: 516–533. [DOI] [PubMed] [Google Scholar]
- Li F., Wang W., Zhang H., Shen W., Xu X., Chen J., Meng Z. 2015. Complete mitochondrial genome of the oriental armyworm Mythimna separata (Walker) (Lepidoptera: Noctuidae). Mitochondrial DNA. 26: 881–882. [DOI] [PubMed] [Google Scholar]
- Li L., Zhu Y., Ma J., Li Z., Dong Z. 2013. An analysis of the Athetis lepigone transcriptome from four developmental stages. PLoS One 8: e73911.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Ming L., Gong Y., Feng L., Li T. 2015. Cytochrome P450s–their expression, regulation, and role in insecticide resistance. Pesticide Biochem. Physiol. 120: 77–81. [DOI] [PubMed] [Google Scholar]
- Nascimento A. R. B. D., Fresia P., Cônsoli F. L., Omoto C. 2015. Comparative transcriptome analysis of lufenuron-resistant and susceptible strains of Spodoptera frugiperda (Lepidoptera: Noctuidae). BMC Genomics 16: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheim S. J., Baker R. H., Simon S., Desalle R. 2015. We can't all be supermodels: the value of comparative transcriptomics to the study of non-model insects. Insect Mol. Biol. 24: 139–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid M., Khan R. A., Zhang Y. 2013. Physiological and population responses of armyworm Mythimna separata (Lepidoptera: Noctuidae) to a sublethal dose of cantharidin-AC. J. Econ. Entomol. 106: 2177–2182. [DOI] [PubMed] [Google Scholar]
- Shukla J. N., Kalsi M., Sethi A., Narva K. E., Fishilevich E., Singh S., Mogilicherla K., Palli S. R. 2016. Reduced stability and intracellular transport of dsRNA contribute to poor RNAi response in lepidopteran insects. RNA Biol. 31: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A. X., Bacigalupe L. D., Luna-Rudloff M., Figueroa C. C. 2012. Insecticide resistance mechanisms in the Green Peach Aphid Myzus persicae (Hemiptera: Aphididae) II: costs and benefits. PLoS One 7: 880–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenius O., Papanicolaou A., Garbutt J. S., Eleftherianos I., Huvenne H., Kanginakudru S., Albrechtsen M., An C., Aymeric J. L., Barthel A. 2011. RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J. Insect Physiol. 57: 231–245. [DOI] [PubMed] [Google Scholar]
- Upadhyay S. K., Chandrashekar K., Thakur N., Verma P. C., Borgio J. F., Singh P. K., Tuli R. 2011. RNA interference for the control of whiteflies (Bemisia tabaci) by oral route. J. Biosci. 36: 153–161. [DOI] [PubMed] [Google Scholar]
- Wieczorek H., Huss M., Merzendorfer H., Reineke S., Vitavska O., Zeiske W. 2003. The insect plasma membrane H+ V-ATPase: intra-, inter-, and supramolecular aspects. J. Bioenerg. Biomembr. 35: 359–366. [DOI] [PubMed] [Google Scholar]
- Xia Q., Wang J., Zhou Z., Li R., Fan W., Cheng D., Cheng T., Qin J., Duana J., Xu H. 2008. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 38: 1036–1045. [DOI] [PubMed] [Google Scholar]
- You M., Yue Z., He W., Yang X., Yang G., Xie M., Zhan D., Baxter S. W., Liette V., Gurr G. M. 2013. A heterozygous moth genome provides insights into herbivory and detoxification. Nat. Genet. 45: 220–225. [DOI] [PubMed] [Google Scholar]
- Yun G., Deng S., Zhang Q., Huan-Li X. U., Cai Q. 2004. The resistance of Bt corn (MG95) to Pseudaletia separata. Entomol. Knowled. 41: 422–426. [Google Scholar]
- Zhang J., Wang B., Dong S., Cao D., Dong J., Walker W. B., Liu Y., Wang G. 2015. Antennal transcriptome analysis and comparison of chemosensory gene families in two closely related noctuidae moths, Helicoverpa armigera and H. assulta. PLoS One 10: A840.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan S., Merlin C., Boore J., Reppert S. 2011. The Monarch Butterfly genome yields insights into long-distance migration. Cell 147: 1171–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Fan D., Zhao K. 2016. Characterization of trypsin-like and chymotrypsin-like serine proteases from midgut of Mythimna Separata Walker. Arch. Insect Biochem. Physiol. 3: 173–191. [DOI] [PubMed] [Google Scholar]