Abstract
Resistant KIT mutations have hindered the development of KIT kinase inhibitors for treatment of patients with systemic mastocytosis. The goal of this research was to characterize the synergistic effects of a novel combination therapy involving inhibition of KIT and calcineurin phosphatase, a nuclear factor of activated T cells (NFAT) regulator, using a panel of KIT-mutant mast cell lines. The effects of monotherapy or combination therapy on the cellular viability/survival of KIT-mutant mast cells were evaluated. In addition, NFAT-dependent transcriptional activity was monitored in a representative cell line to evaluate the mechanisms responsible for the efficacy of combination therapy. Finally, shRNA was used to stably knockdown calcineurin expression to confirm the role of calcineurin in the observed synergy. The combination of a KIT inhibitor and a calcineurin phosphatase inhibitor (CNPI) synergized to reduce cell viability and induce apoptosis in six distinct KIT-mutant mast cell lines. Both KIT inhibitors and CNPIs were found to decrease NFAT-dependent transcriptional activity. NFAT-specific inhibitors induced similar synergistic apoptosis induction as CNPIs when combined with a KIT inhibitor. Notably, NFAT was constitutively active in each KIT-mutant cell line tested. Knockdown of calcineurin subunit PPP3R1 sensitized cells to KIT inhibition and increased NFAT phosphorylation and cytoplasmic localization. Constitutive activation of NFAT appears to represent a novel and targetable characteristic of KIT-mutant mast cell disease. Our studies suggest that combining KIT inhibition with NFAT inhibition might represent a new treatment strategy for mast cell disease.
Introduction
Activating KIT mutations, most commonly KIT D816V, are a hallmark of human systemic mastocytosis (1, 2). Related mutations are also found in mast cell neoplasms of other vertebrates, including mice, rats, and dogs (3–7). Theoretically, treatment of advanced systemic mastocytosis with KIT kinase inhibitors should target the neoplastic cells and exert a therapeutic effect. However, to date, the use of KIT inhibitors against advanced systemic mastocytosis has had only minor clinical impact. There are several reasons for this. First, current tyrosine kinase inhibitors (TKI) have limited activity against KIT D816V (8). Second, patients who do respond to KIT TKI therapy can develop secondary resistance over time due to the emergence of secondary resistance mutations (9). Third, at least in the case of other KIT-mutant cancers, treatment with KIT inhibitors typically results in a pool of quiescent, persistent disease that can quickly progress following discontinuation of KIT TKI therapy (10). The immediate goal of this research was to investigate the effect of a new combination therapy, simultaneous inhibition of KIT and calcineurin, in KIT-mutant systemic mastocytosis. In addition, we sought to understand the underlying mechanism behind the efficacy of this combination such that improved therapies could be developed from targeting this novel biology. It is our belief that such novel combination therapies will lead to more effective treatment options that overcome both disease resistance and disease persistence in human systemic mastocytosis as well as other KIT-mutant cancers.
KIT is a type III receptor tyrosine kinase expressed on the surface of mast cells, hematopoietic stem and progenitor cells, melanocytes, interstitial cells of Cajal, and germ cells (11). KIT is activated by extracellular binding of its ligand, stem cell factor, and leads to KIT dimerization, tyrosine kinase activation, and subsequent activation of downstream signaling pathways: PI3K–AKT, MAPK, and JAK–STAT (12, 13). KIT signaling promotes cell growth, survival, and proliferation (14). Activating KIT mutations, such as D816V, result in ligand-independent kinase activity and constitutive activation of KIT-dependent downstream signaling pathways (12, 15). In addition to systemic mastocytosis, activating KIT mutations have been identified in gastrointestinal stromal tumors (16), acute myeloid leukemia (17), melanoma (18), and seminoma (19).
Although it is well recognized that constitutive KIT signaling is associated with systemic mastocytosis (3, 5, 20–23) and likely is the causative abnormality, it is also believed that KIT signaling alone is insufficient for disease progression (24). The molecular mechanisms leading to disease progression remain unknown. One potential mechanism of progression, which we explore in this article, is constitutive activation of the nuclear factor of activated T cells (NFAT) signaling pathway. Constitutive NFAT activity was recently identified in melanoma, colon cancer, and chronic myelogenous leukemia (CML; refs. 25–29). In the melanoma model, NFAT family members were not only found to be constitutively active, but also to regulate quiescence and proliferation of skin stem cells (30). Gregory and colleagues found that NFAT inhibition sensitized CML cells to imatinib treatment (26), and similarly, Spreafico and colleagues reported the benefits of combining NFAT and MEK inhibition in models of colorectal cancer (25).
NFAT is a family of transcription factors, related to the Rel-NF-κB family of transcription factors. Each NFAT protein has a Rel homology region that makes base-specific (GGAAA) contacts with DNA to regulate the transcription of a diverse number of genes involved in the regulation of cellular proliferation, differentiation, survival, and apoptosis. There are four calcium-responsive NFAT family members, NFAT1–4. On the basis of knockout studies in mice and siRNA knockdown studies in cell lines, NFAT1–4 appear to have somewhat overlapping functions; some of which are context specific (31–34).
In resting cells, NFAT is localized in the cytoplasm in a highly phosphorylated, inactive state. Increases in intra-cellular calcium lead to activation of calcineurin, which binds to the regulatory domain of NFAT and dephosphorylates NFAT. Dephosphorylation exposes the NFAT nuclear localization signal leading to rapid translocation into the nucleus, where it acts as a transcription factor. Within the nucleus, constitutively active kinases (i.e., GSK3, CK1, DYRK1) phosphorylate NFAT, resulting in translocation back into the cytoplasm (31, 33, 34).
Here, we provide evidence that NFAT is constitutively active in KIT-mutant mast cells and that dual inhibition of KIT kinase and calcineurin phosphatase activity leads to synergistic decreases in cell viability and increases in apoptosis. We propose that simultaneous inhibition of KIT and calcineurin signaling may improve treatment of KIT-mutant systemic mastocytosis.
Materials and Methods
Cell culture
P815 (35), HMC1.1, and HMC1.2 (36) cells were grown in DMEM (10% FBS, 1% L-glutamine, 1% penicillin-streptomycin). RBL2H3 cells (37) were grown in minimal essential media alpha (15% FBS, 1% L-glutamine, 1% penicillin-streptomycin). BR and C2 (38) cells were grown in DMEM (2% calf serum, 0.125 g histidine, 2.5% HEPES). All cells were grown at 37°C and 5% CO2. Morphology of each cell line was monitored and growth curve analysis and sequencing was performed biannually to ensure the identity of each line throughout the course of these studies. The P815 and RBL2H3 cells were purchased from ATCC. The BR and C2 cells were generously donated by Dr. Bill Raymond (University of California at San Francisco). The HMC1.1 and HMC1.2 cells were generously donated by Dr. J.H. Butterfield (Mayo Clinic, Rochester, MN) (Supplementary Table S1).
Immunoblotting studies
To prepare nuclear and cytoplasmic protein extracts, we used Thermo Scientific NE-PER Nuclear and Cytoplasmic Extraction Kit (#78833) and followed the manufacturer’s instructions.
Lysates were prepared using either the NE-PER Kit or routine detergent lysis (RIPA lysis buffer: 50 mmol/L Tris-HCl, 1% IGEPAL, 0.25% deoxycholate, 150 mmol/L NaCl, 5 mmol/L EDTA). The antibodies used for immunoblotting are listed in Supplementary Table S2.
Cell viability and caspase assays
Cells were plated at 10,000 cells per well at the same time they were treated in opaque, 96-well plates in a total volume of 50 μL/well. Cells were incubated for 48 hours then 50 μL of Cell Titer Glo substrate (Promega #G7571) was added to each well. Plates were rocked for 15 minutes and analyzed using the “CellTiterGlo” program ona Micro-plate luminometer GloMax-96 (Promega). Alternatively, after incubation, 50 μL of caspase-3/7 (Promega #G8091) or caspase-8 or -9 reagent (Promega #G8200, G8210) was added to each well. Caspases 8 and 9 were used in experiments with RBL2H3 cells. Plates were rocked for 60 minutes and read using the “Caspase-Glo” program on a Microplate luminometer GloMax-96 (Promega).
Long-term replating assay
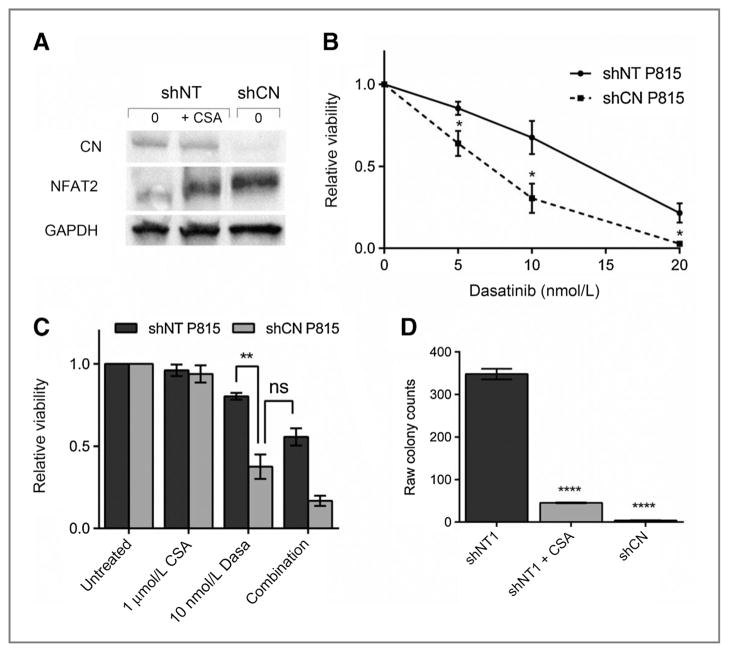
P815 cells were treated with 1 μmol/L cyclosporin A (CSA), 20 nmol/L dasatinib, or CSA plus dasatinib for 7 days. At day 7, the cells were washed, drug was removed, and cells were diluted1:500 and replated on to 6-well plates. Cells were allowed to recover. Cells treated with 1 μmol/L CSA reached confluency at 48 hours after plating and colonies were too numerous to count. Cells treated with dasatinib or combination were allowed to recover in drug-free media for 7 days at which point they were stained with 0.05% crystal violet and colonies were quantified.
NFAT-P815 and shCN-P815 cell lines
The NFAT-P815 line was created using virions containing the Cignal Lenti NFAT Reporter construct (SABioscience catalog # 336851, CLS-015L). P815 cells were transduced using 78 μL DMEM media (+15% FBS, 1% penicillin-streptomycin, 1% L-glutamine), 6 μg/mL polybrene (2 μL/well at 0.3 μg/μL), and 20 μL Cignal Lenti NFAT reporter. Stably transduced cells were selected for resistance using 1 μg/mL puromycin. Drug-resistant cells were tested with the Luciferase Assay System (Promega, #E1501) to quantify firefly luciferase expression. P815 cells with Ppp3r1 knockdown (shCN) or non-targeting knockdown (shNT) were created using pLKO.1 Mission lentiviral transduction particles from Sigma [Ppp3r1 shRNA (NM_024459.1-227s1c1), nontargeting shRNA (SHC002V)]. Briefly, P815 cells were transduced overnight with 85.5 μL media, 0.006 μg polybrene, and 12.5 μL of lentiviral particles. Following a 24-hour recovery, stably transduced clones were selected using 1 μg/mL puromycin.
NFAT-dependent transcription assays
NFAT-P815 cells were plated the day they were treated. After incubation, media were aspirated from wells and 20 μL of Passive Lysis Buffer (Promega) was added to each well. Plates were rocked for 15 minutes and analyzed with the “Luciferase Assay System with Injector” program, which injects 100 μL of firefly luciferase reagent into each well before quantifying luciferase protein. The luciferase readout was used as an indication of NFAT-dependent transcriptional activity and was normalized to cell viability (CTG Assay) for drug incubations lasting longer than 4 hours.
Real-time PCR
Total RNA was extracted from P815 cells using a Qiagen RNAeasy Plus Mini Kit combined with the Qiagen RNase-Free DNase Treatment. Single-stranded cDNA was prepared from 1 μg of total RNA in a 50 μL reaction using 60 μmol/L random hexamer primers, 0.5 mmol/L dNTPs, 100 U RNaseOUT, 5 mmol/L dithiothreitol, 1× First Strand buffer, and 500 U SuperScript III reverse transcriptase following manufacturer’s instructions (Invitrogen by Life Technologies).
Quantitative real-time polymerase chain reaction (qRT-PCR) was carried out in a 20 μL reaction using 1 μg single-stranded cDNA (corresponding to 40 ng initial total RNA) and 19 μL probes Master MIX (Roche), with a FAM-labeled hydrolysis probe specific to the reference, GAPDH. Cycling conditions on a Light Cycler 480 instrument (Roche) included 10 minutes at 95°C, 40 cycles of 95°C for 10 seconds, and 60°C for 20 seconds. NFAT1–4 and GAPDH (murine, canine, human, or rat) were detected using commercial TaqMan Gene Expression assays (Applied Biosystems by Life Technologies). Murine probes: Nfat2: Mm00479445_m1, Nfat3: Mm00477776_m1, Nfat4: Mm01249200_m1, Nfat3: Mm00452375_m1, Gapdh: Mm99999915_g1. Rat probes: Nfat2: Rn04280453_m1, Nfat1: Rn01750325_m1. Canine probes: Nfat2: Cf02648711_m1, Nfat4: Cf02703251_m1, Nfat3: Cf02703250_m1. Human probes: NFAT2: Hs00542678_m01, NFAT1: Hs00905451_m01, NFAT4: Hs00190046_m01, NFAT3: Hs00190037_m01. Expression results were analyzed using the comparative Ct method (also known as the 2−ΔΔCt method; ref. 39).
Statistical analysis
All error bars represent SE across at least three independent biologic replicates. Statistical analysis of results was performed using Prism 6 software (GraphPad). Unpaired two-tailed t tests were used to determine statistical significance between two groups.
Results
Combining a calcineurin inhibitor with a KIT inhibitor leads to synergistic decrease in cell viability and induction of apoptosis
On the basis of previous studies where calcineurin phosphatase inhibition synergized with kinase inhibition to decrease CML cell viability (26), we evaluated the effects of combining a KIT TKI with a CNPI in a number of KIT-mutant cell lines. For example, P815 cells (murine Kit D814Y) were treated with a KIT inhibitor (dasatinib 5, 10, or 20 nmol/L), calcineurin inhibitor (CSA, 1 μmol/L), or both for 48 hours before measuring cell viability (Fig. 1). In this dose range, dasatinib produced a dose-dependent decrease in cell viability. In contrast, single-agent CSA at 1 μmol/L had only a minimal effect on cellular viability (<15% decrease). There was a significant decrease in cell viability with combination treatment, as compared with either agent alone (Fig. 1A). The viability data was analyzed using Calcusyn software, which uses the method of Chou and Talalay (40) to quantify synergy through computation of combination index (CI) values. The average CI value in P815 cells for CSA plus dasatinib was 0.438, indicating a synergistic effect of combination therapy. This experiment was repeated in a total of six KIT-mutant mast cell lines (cell line characteristics summarized in Supplementary Table S1), with six different KIT TKIs and four different CNPIs and we observed similar degrees of synergy (Table 1).
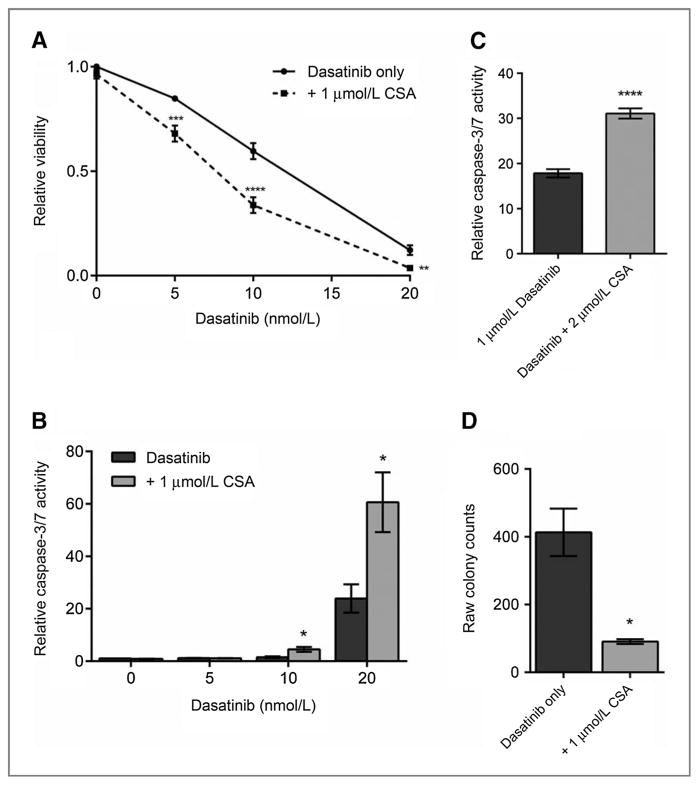
Figure 1.
Combining a calcineurin inhibitor with a KIT inhibitor leads to synergistic decrease in cell viability and induction of apoptosis. A, cell viability measured at 48 hours of single-agent or combination treatment with a calcineurin phosphatase inhibitor (CSA) and a KIT inhibitor (dasatinib) in mouse P815 cells. B, apoptosis induction measured by caspase-3/7 activity at 48 hours of single-agent or combination treatment of a calcineurin phosphatase inhibitor (CSA) and a KIT inhibitor (dasatinib) in mouse P815 cells. C, apoptosis induction measured by caspase-3/7 at 48 hours of single-agent or combination treatment of a calcineurin phosphatase inhibitor (CSA) and a KIT inhibitor (dasatinib) in human HMC1.2. D, colony growth of P815 cells treated with 20 nmol/L dasatinib alone or in combination with 1 μmol/L CSA. Cells were treated for a week and allowed to recover for a week before colonies were quantified. Error bars, SE; *, P < 0.05; **, P < 0.005; ***, P < 0.001; ****, P < 0.0001, compared with dasatinib treatment alone.
Table 1.
Summary of CI values for KIT-mutant mast cell lines from CNPI+KIT inhibition CI values calculated after 48-hour combination therapy. CI values calculated using the method of Chou and Talalay (40)
| Cell line | KIT inhibitor | Calcineurin inhibitor | CI value | Cell line | KIT inhibitor | Calcineurin inhibitor | CI value |
|---|---|---|---|---|---|---|---|
| p815 | Dasatinib | CSA | 0.438 | RBL2H3 | Dasatinib | FK506 | 0.61 |
| p815 | Dasatinib | Ascomycin | 0.829 | RBL2H3 | Dasatinib | CSA | 0.324 |
| p815 | Dasatinib | Pimecrolimus | 0.652 | RBL2H3 | Dasatinib | Ascomycin | 0.409 |
| p815 | Imatinib | FK506 | 0.659 | RBL2H3 | Dasatinib | Pimecrolimus | 0.909 |
| p815 | Sunitinib | CSA | 1.029 | RBL2H3 | Sunitinib | CSA | 0.643 |
| p815 | Nilotinib | CSA | 0.61 | RBL2H3 | Nilotinib | CSA | 0.227 |
| p815 | Sorafenib | CSA | 0.774 | RBL2H3 | Sorafenib | CSA | 0.973 |
| p815 | Crenolanib | CSA | 0.508 | RBL2H3 | Crenolanib | CSA | 0.602 |
| BR | Dasatinib | CSA | 0.261 | RBL2H3 | Imatinib | CSA | 0.657 |
| BR | Dasatinib | Ascomycin | 0.517 | HMC1.1 | Dasatinib | CSA | 0.504 |
| BR | Dasatinib | Pimecrolimus | 0.465 | HMC1.1 | Dasatinib | Ascomycin | 0.442 |
| BR | Imatinib | FK506 | 0.425 | HMC1.1 | Dasatinib | Pimecrolimus | 0.631 |
| BR | Sunitinib | CSA | 0.268 | HMC1.1 | Sunitinib | CSA | 0.854 |
| BR | Nilotinib | CSA | 0.175 | HMC1.1 | Nilotinib | CSA | 0.231 |
| BR | Sorafenib | CSA | 0.216 | HMC1.1 | Crenolanib | CSA | 0.719 |
| BR | Crenolanib | CSA | 0.593 | HMC1.2 | Dasatinib | CSA | 0.481 |
| C2 | Dasatinib | FK506 | 1.056 | HMC1.2 | Dasatinib | Ascomycin | 0.861 |
| C2 | Dasatinib | CSA | 0.546 | HMC1.2 | Dasatinib | Pimecrolimus | 0.153 |
| C2 | Dasatinib | Ascomycin | 0.682 | HMC1.2 | Sunitinib | CSA | 0.509 |
| C2 | Dasatinib | Pimecrolimus | 0.798 | HMC1.2 | Crenolanib | CSA | 0.771 |
| C2 | Sunitinib | CSA | 0.584 | ||||
| C2 | Nilotinib | CSA | 0.232 | ||||
| C2 | Sorafenib | CSA | 0.647 | ||||
| C2 | Crenolanib | CSA | 0.142 | ||||
| C2 | Imatinib | CSA | 0.466 |
In our cellular proliferation experiments, we noted changes in cell morphology consistent with cellular death. To measure the effects of combination therapy on induction of apoptosis (as assessed by activation of caspase activity), we performed single-agent versus combination therapy experiments. We repeated our previous combination experiments treating P815 cells with 5, 10, or 20nmol/L dasatinib alone or in combination with 1 μmol/L CSA. After 48 hours, caspase-3 and 7 activity was quantified as a measure of cellular apoptosis and normalized to cell viability (Fig. 1B).
Caspase-3/7 activity was significantly greater in cells treated with combination therapy compared with treatment with dasatinib or CSA alone. We saw similar degrees of caspase induction with each KIT-mutant cell line tested to date (data shown for HMC1.2; Fig. 1C). We expanded these studies to include additional CNPIs (FK506, ascomycin, fenvalerate, and pimecrolimus) and KIT inhibitors (imatinib, nilotinib, sunitinib, and ponatinib). Combining each of the KIT TKIs and CNPIs synergistically decreased cell viability and induced apoptosis (data not shown).
In addition to improving response to KIT inhibitor therapy in KIT-mutant systemic mastocytosis models, we also tested the ability of combination therapy to combat disease persistence. We examined colony-forming capacity of cells after extended drug exposure. As noted above, CSA alone had no effect on proliferation and replating of CSA-treated cells led to a confluent lawn of cells after only 2 days. Dasatinib treatment resulted in an average of 467 colonies, whereas combination therapy further reduced colony count to 82 for an 83% reduction in colony growth. This represents a significant decrease in colony growth over monotherapy with dasatinib (P = 0.006; Fig. 1D).
NFAT is constitutively active in KIT-mutant cell lines
On the basis of the above results, we hypothesized that CNPIs reduced the transcriptional activity of members of the NFAT family and that inhibition of NFAT-induced transcription was required for the observed synergy. It is well known that NFAT is a downstream target of calcineurin. However, treatment with CNPIs has a number of off-target effects that could be responsible for the observed synergy (41–43). To investigate whether NFAT was mediating the observed synergy between KIT inhibitors and CNPIs, we characterized NFAT expression and activity within KIT-mutant cells.
Given that there are four different calcium/calcineurin-responsive NFAT family members, we performed qRT-PCR in human, mouse, canine, and rat KIT-mutant cell lines to both quantify and qualify NFAT family member RNA transcripts. NFAT1, NFAT2, and NFAT4 were expressed in the murine P815 cell line, the human mast cell lines HMC1.1 and HMC1.2, and the rat RBL2H3 cell line (Supplementary Table S3). In canine BR and C2 cell lines, however, NFAT2 and NFAT4 were the only detectable NFAT species.
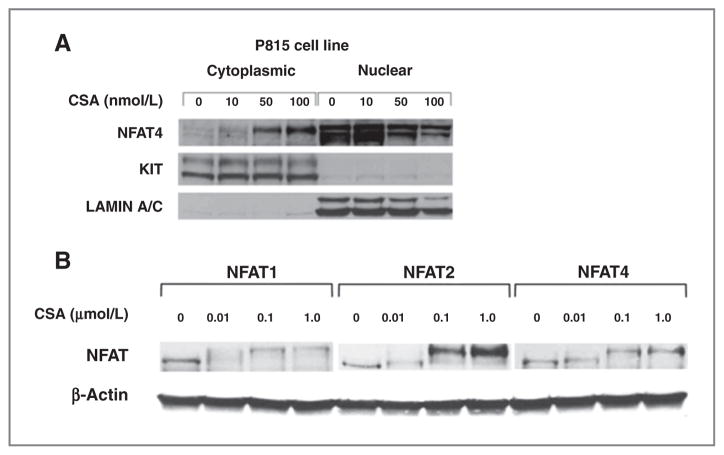
To evaluate NFAT phosphorylation and subcellular localization in KIT-mutant mast cell lines, we treated cells for 3 hours with increasing doses of CSA (Fig. 2A). Cells were fractionated and probed for NFAT family members. We observed nearly 100% of NFAT species localized in the nucleus of untreated P815 cells, indicating constitutive NFAT activation under resting conditions in these cells. After treatment with CSA, we saw a shift toward the higher molecular weight band indicative of the highly phosphorylated NFAT species. Concurrently, we observed NFAT translocation to the cytoplasm. The band shift and translocation were both observed with each of the three NFAT species expressed in P815 cells (Fig. 2B). These experiments not only confirmed our qRT-PCR expression results, but also suggested inactivation of NFAT by CSA as a possible mechanism underlying the synergy of KIT TKI and CNPI combination therapy.
Figure 2.
KIT-mutant mast cells express constitutively active NFAT species that are sensitive to inactivation by calcineurin phosphatase inhibitors. A, immunoblot of nuclear and cytoplasmic fractions from P815 cells treated for 3 hours with CSA. Total KIT is shown as a cytoplasmic loading control and Lamin A/C is shown as a nuclear loading control. B, immunoblot of cytoplasmic fractions from P815 cells treated for 3 hours with CSA. Membrane was probed for NFAT family members NFAT1, 2, and 4. The lower molecular weight band represents dephosphorylated, active NFAT and the higher molecular weight band represents phosphorylated, inactive NFAT. β-Actin is shown as a loading control.
We tested other KIT-mutant cell lines to determine whether constitutive NFAT activation was a common characteristic of mastocytosis cell lines. We also wanted to establish whether NFAT was maximally activated. In addition to P815 cells, we treated rat RBL2H3 and human HMC1.2 cells with 1 μmol/L CSA or 1 μmol/L ionomycin plus 100 nmol/L 12-O-tetradecanoylphorbol-13-acetate (TPA) for 2 hours. Ionomycin and TPA synergize to activate calcium flux that leads to downstream activation of calcineurin and NFAT. After incubation, the cells were fractionated and immunoblotting was used to visualize NFAT phosphorylation and localization. Each NFAT species expressed in HMC1.2 cells was found to be constitutively active based on band size and protein localization (Supplementary Fig. S1). The same pattern of constitutively active NFAT was observed in P815, RBL2H3, and HMC1.1 cells (data not shown). In each cell line we observed a minimal further translocation of NFAT species into the nucleus following induction of calcium flux, indicating maximal activation of NFAT species in KIT-mutant mast cells (data not shown). Because of a lack of antibodies suitable to detect canine NFAT species by immunoblotting, NFAT activation, phosphorylation, and subcellular localization could not be evaluated in the BR and C2 cell lines.
Constitutive activation of NFAT represents a novel finding in KIT-mutant mast cells that has not been previously reported. Having characterized NFAT gene and protein expression in these cell lines, we sought to assess the regulation of NFAT-dependent transcriptional activity by monotherapy and combination therapy.
Monotherapy and combination therapy decrease basal NFAT transcriptional activity in KIT-mutant cells
Because NFAT is a transcription factor, single-agent and combination treatment should also lead to changes in NFAT-dependent transcriptional activity. To investigate the modulation of NFAT transcriptional activity, we stably transduced an NFAT-dependent reporter construct into the P815 cell line. In this construct, transcription is driven from a tandem repeat of the NFAT consensus binding sequence (GGAAA) and a minimal CMV promoter to regulate the expression of firefly luciferase. There was a substantial basal level of reporter expression in this cell line, confirming constitutive basal NFAT activity.
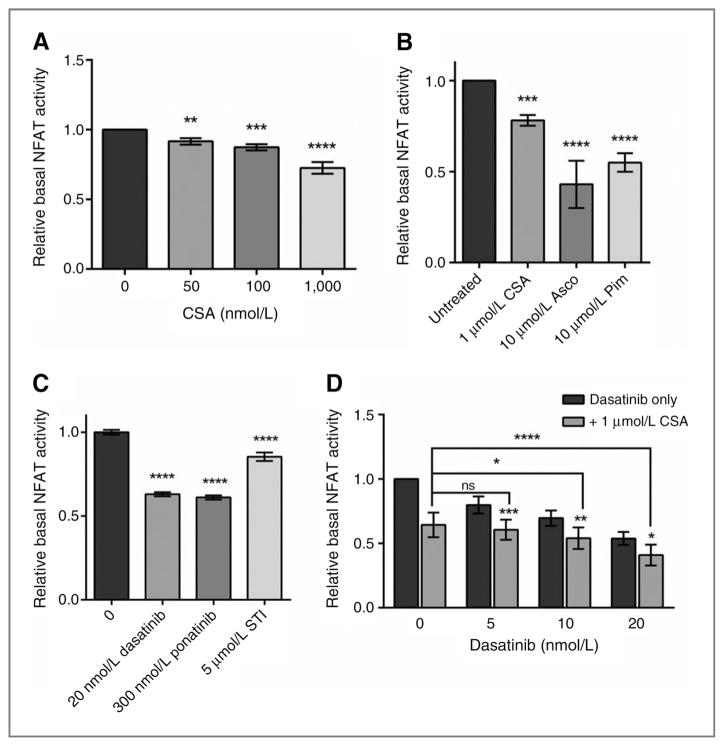
CSA treatment of the NFAT-dependent reporter-P815 cell line (NFAT-P815) induced a dose-dependent decrease in NFAT transcriptional activity as assessed by the level of firefly luciferase (Fig. 3A). We saw a similar decrease in reporter activity when the cells were treated with other CNPIs, including ascomycin, and pimecrolimus (Fig. 3B). Although we observed a dose-dependent decrease in NFAT transcriptional activity, we were unable to completely ablate reporter activity with doses of physiologically relevant CNPIs. We repeated the experiments with two other NFAT reporter systems and saw similar results (data not shown). To confirm that 1 μmol/L CSA was modulating transcriptional activity, we measured RNA transcript levels of downstream targets Ccnd1, Ccnd2, Pim2, Bcl6, Socs1, and Cish in P815 cells after 24 hours of CSA treatment. We saw a significant increase in Bcl6 transcript levels as expected, and decreases in the other target transcript levels (Supplementary Fig. S2). On the basis of these results, we believe the modest decrease in NFAT reporter activity does reflect meaningful changes in NFAT-dependent transcription of endogenous transcripts.
Figure 3.
Monotherapy and combination therapy decrease basal NFAT transcriptional activity in KIT-mutant cells. Basal NFAT transcriptional activity as measured in NFAT-dependent luciferase reporter stable P815 cells after 6-hour drug treatment. A, cells treated with increasing doses of CSA before lysis and assessment of luciferase activity. B, cells treated with 1 μmol/L CSA, ascomycin, or pimecrolimus before assessment of luciferase activity. C, cells treated with 10 nmol/L dasatinib, 0.3 μmol/L ponatinib, or 5 μmol/L imatinib before assessment of luciferase activity. D, cells treated with increasing doses of dasatinib as single agent or in combination with 1 μmol/L CSA before assessment of luciferase activity. Error bars, SE; *, P < 0.05;**, P < 0.005; ***, P < 0.001; ****, P < 0.0001, compared with either monotreatment.
Unexpectedly, treatment of NFAT-P815 cells with a KIT inhibitor (dasatinib, ponatinib, or imatinib) also resulted in decreased NFAT-dependent transcriptional activity (Fig. 3C). The KIT TKI doses used are those that produced synergy in the cellular viability assay when combined with 1 μmol/L CSA. Furthermore, combination treatment with CSA and dasatinib significantly decreased NFAT-dependent transcriptional activity compared with either drug alone (Fig. 3D), suggesting an intersection between the KIT and NFAT signaling pathways at the level of transcriptional control.
These data confirm constitutive calcineurin-dependent activation of NFAT-dependent transcriptional activity in KIT-mutant mast cell lines. Basal NFAT-dependent transcriptional activity was modulated by either calcineurin inhibitors or KIT inhibitors. Notably, combination therapy significantly decreased NFAT-dependent transcriptional activity compared with monotherapy, suggesting regulation of NFAT-dependent transcriptional activity as a common downstream target of this combination treatment.
KIT inhibition of NFAT-dependent transcriptional activity is independent of intracellular calcium levels
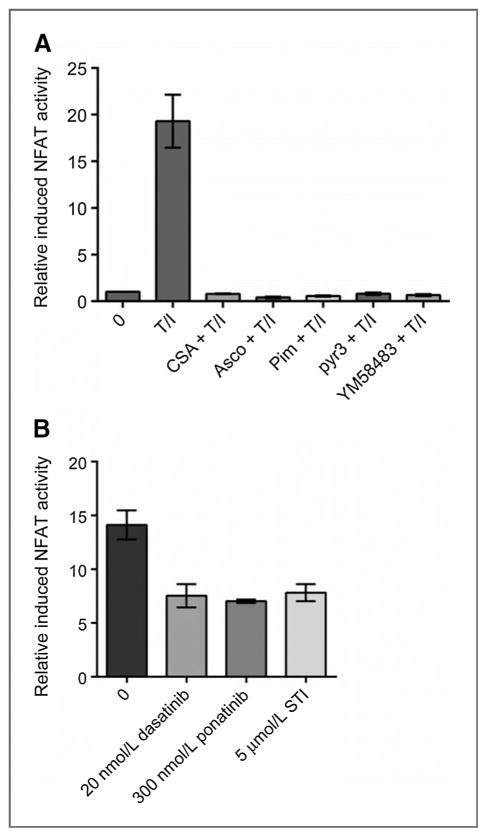
In previously described model systems, NFAT is regulated by intracellular calcium levels that activate cal-modulin/calcineurin. Although KIT signaling can activate calcium influx (44, 45), it is unknown whether this aspect of KIT activity is responsible for the inhibition of basal NFAT-dependent reporter activity we observe. To determine whether KIT inhibitors were acting through the same calcium-dependent pathway as calcineurin inhibitors, we evaluated induced NFAT transcriptional activity by treating NFAT-P815 cells with a combination of TPA and ionomycin (100 nmol/L and 1 μmol/L, respectively) for 2 hours to activate calcium flux–dependent signaling and increase NFAT-dependent reporter activity. There was an average 10- to 20-fold increase in NFAT-dependent reporter gene expression after TPA/ionomycin treatment. This increase was completely inhibited by pretreating the cells for 2 hours with a CNPI (CSA, pimecrolimus, or ascomycin; Fig. 4A). To confirm that the observed NFAT transcriptional activity is dependent upon calcium influx, NFAT-P815 cells were pretreated with calcium channel inhibitors YM58483 (a CRAC channel inhibitor; ref. 46) or pyr3 (a TRPC inhibitor; ref. 47) followed by a 2-hour treatment with TPA/ionomycin. Both calcium channel blockers completely inhibited the induction of NFAT-dependent reporter activity (Fig. 4A).
Figure 4.
KIT inhibition does not act through calcium signaling to decrease NFAT transcriptional activity. Induced NFAT transcriptional activity measured in stable NFAT-dependent luciferase reporter P815 cells. A, inhibition of induced NFAT activity with a CNPI (CSA, ascomycin, or pimecrolimus) or a calcium channel blocker (pyr3 or YM58483) is shown after a 2-hour pretreatment with a calcineurin inhibitor or a calcium channel blocker, followed by a 2-hour treatment with TPA/ionomycin (T+I). B, the effect of KIT inhibitors on induced NFAT-dependent reporter activity is shown after 2 hours of pretreatment with a KIT inhibitor (dasatinib, ponatinib, or imatinib), and 2 hours of TPA+ionomycin. Error bars, SE.
Finally, NFAT-P815 cells were treated with three different KIT inhibitors (dasatinib, ponatinib, or imatinib), followed by a 2-hour treatment with TPA/ionomycin (Fig. 4B). In contrast with the case with CNPIs, KIT inhibitors only partially reduced the TPA/ionomycin-induced increase in NFAT-dependent reporter activity, suggesting that KIT inhibitors do not effectively block induced calcium flux and subsequent calcineurin activation. However, the fact that dasatinib affects basal NFAT activity suggests an alternative mechanism by which KIT inhibitors regulate basal and induced NFAT activity in KIT-mutant mast cells. It appears that induced NFAT transcriptional activity is completely calcium-dependent, whereas basal NFAT transcriptional activity is modulated by additional KIT-dependent mechanisms, such as signaling through pathways downstream of KIT.
One explanation for the synergy observed in our combination treatment studies is that inhibition of calcineurin signaling modulates NFAT and this, in turn, modulates KIT expression or activity. Alternatively, KIT kinase inhibitors could modulate NFAT subcellular localization and/or phosphorylation through an unidentified mechanism. This latter model would explain why KIT inhibitors modulate NFAT-dependent reporter activity in KIT-mutant mast cells. To test this hypothesis, P815 cells were treated with CSA or FK506 (10, 100, 1,000 nmol/L). Protein lysates from treated cells were fractionated and probed for activation of tyrosine phosphorylated KIT, as well as the downstream KIT signaling targets ERK and AKT. CSA and FK506 had no effect on phospho-KIT, phospho-ERK, or phospho-AKT using doses that were sufficient to modulate NFAT activity and which had a synergistic effect in our cellular assays. This indicates that CNPIs do not directly affect KIT kinase activity or proximal signaling pathways (e.g., MAPK; Supplementary Fig. S3A). In additional experiments, P815 cells were treated with dasatinib or imatinib (1, 10, 50 nmol/L and 0.5, 1, 5 μmol/L, respectively) before cellular fractionation and immunoblotting for NFAT2 and NFAT4. Inhibition of KIT signaling by dasatinib or imatinib did not cause a shift in NFAT species to the higher molecular weight and highly phosphorylated state, nor did it cause any increase in cytoplasmic localization (Supplementary Fig. S3B).
Collectively, these data indicate that the effects of KIT inhibition on NFAT-dependent transcriptional activity and cell viability is exerted through an alternative mechanism, independent of calcium signaling or calcineurin activity. Notwithstanding the lack of direct crosstalk, these two pathways seem to converge at the transcriptional level.
Calcineurin knockdown sensitizes P815 cells to dasatinib treatment
To determine the mechanism mediating the synergy of combination therapy, we tested whether the effects of combination therapy were calcineurin-dependent. As discussed above, CSA can have additional off-target effects besides its well-characterized effect on inhibiting calcineurin phosphatase activity. To eliminate these off-target effects as mechanisms behind synergy, we used lentiviral transduction to deliver shRNA and create stable knockdown of the calcineurin subunit Ppp3r1. Calcineurin is a heterodimer, composed of a catalytic subunit and a regulatory subunit. Knockdown of either subunit leads to enzyme inactivation. In addition to three unique catalytic subunit isozymes (PPP3CA, PPP3CB, PPP3CC), there are two regulatory isozymes (PPP3R1, PPP3R2). Only PPP3R1 is expressed in P815 cells, so we were able to use a single knockdown of Ppp3r1 to inhibit calcineurin phosphatase activity.
We stably transduced P815 cells with a nontargeting mammalian shRNA or a Ppp3r1 shRNA. Puromycin-resistant cells were selected for further analysis. Immunoblotting of cells stably transduced with a Ppp3r1 shRNA demonstrated that knockdown of calcineurin led to a shift in NFAT localization, similar to that observed with 1 μmol/L CSA treatment (Fig. 5A). These effects were not seen with stable clones expressing a nontargeting shRNA (shNT). The stable shCN-P815 cells were tested with increasing doses of dasatinib. Compared with shNT-P815 cells, the shCN-P815 cells showed increased sensitivity to dasatinib treatment (Fig. 5B). Next, we treated shCN-P815 cells with 10 nmol/L dasatinib with and without 1 μmol/L CSA. The shCN knockdown cell line showed increased sensitivity to dasatinib treatment compared with shNT-P815 cells. The decrease in cell viability in the CN knockdown cells treated with dasatinib alone was comparable with the observed cell viability following combination therapy in shNT-P815 cells (Fig. 5C). Identical results were obtained using two independent clones transduced with the same Ppp3r1 shRNA.
Figure 5.
Ppp3r1 knockdown sensitizes P815 cells to dasatinib treatment to the extent equal to combination treatment in parental P815 cells. A, cytoplasmic extracts from P815 cells with stable knockdown of calcineurin subunit Ppp3r1 (shCN-P815) versus nontargeting shRNA (shNT-P815). B, 48-hour viability of stable shCN-P815 cells treated with increasing doses of dasatinib compared with shNT-P815 cells. C, the effect of single agent (dasatinib or CSA) or combination treatment on shNT-P815 cells or shCN-P815 cells at 48 hours. D, colony growth of shNT-P815 cells treated with 20 nmol/L dasatinib alone or in combination with 1 μmol/L CSA compared with shCN-P815 cells treated with 20 nmol/L dasatinib. Cells were treated for a week and allowed to recover for a week before colonies were quantified. Error bars, SE; *, P < 0.05; **, P < 0.005; ****, P < 0.0001, compared with NT-P815 cells; ns, not significant.
Finally, we examined the colony-forming capacity of shCN-P815 cells compared with shNT-P815 cells. As before, cells were treated for 1 week with 20 nmol/L dasatinib alone or in combination with 1 μmol/L CSA and allowed to recover in media for 1 week. The shCN-P815 cells showed decreased replating capacity compared with shNT-P815 cells when treated with dasatinib alone. The replating capacity of the shCN-P815 cells treated with dasatinib alone was similar to the capacity of the shNT-P815 cells following treatment with CSA plus dasatinib (Fig. 5E). On the basis of these results, we conclude that the observed synergy is calcineurin-dependent and not the result of inhibiting other CSA targets.
NFAT-specific inhibitors combine with KIT inhibitors to synergistically decrease cell viability and induce apoptosis
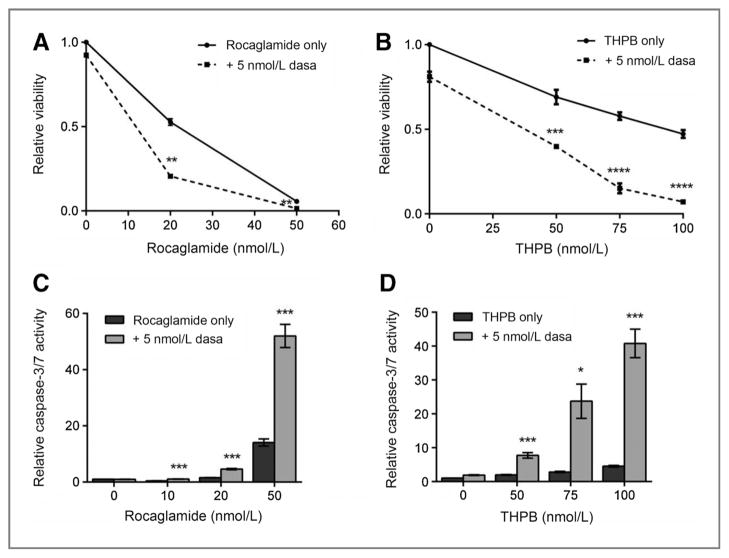
To further confirm that the observed synergy was both calcineurin- and NFAT-dependent, we tested the effects of two NFAT-specific inhibitors [rocaglamide (48) and tributylhexadecylphosphonium bromide (THPB; ref. 49], alone or in combination with dasatinib. Combination therapy with rocaglamide plus dasatinib or THPB plus dasatinib decreased cell viability more significantly than monotherapy (Fig. 6A and B). The average calculated CI value was 0.73 and 0.36 for rocaglamide plus dasatinib and THPB plus dasatinib, respectively, indicating synergy for both drug combinations.
Figure 6.
NFAT-specific inhibitors combine with KIT inhibitors to synergistically decrease cell viability and induce apoptosis. A and B, combination treatment of P815 cells with an NFAT-specific inhibitor, rocaglamide (A) or THBP (B) and a KIT inhibitor (dasatinib). Cell viability was measured at 48 hours. C and D, combination treatment with an NFAT-specific inhibitor, rocaglamide (C) or THBP (D) and a KIT inhibitor (dasatinib). Caspase-3/7 activity was measured at 48 hours. Error bars, SE; *, P < 0.05; **, P < 0.005; ***, P < 0.001; ****, P < 0.0001, compared with either monotreatment.
To determine whether NFAT-specific inhibitors induced apoptosis when combined with a KIT inhibitor, we evaluated apoptosis induction by caspase-3/7 assay after treating P815 cells for 48 hours with monotherapy versus combination therapy. For each NFAT inhibitor combination, there was a significant increase in caspase-3/7 activity when combined with a KIT TKI, particularly at doses above 20 nmol/L rocaglamide and 50 nmol/L THPB (Fig. 6C and D).
Our data with NFAT-specific inhibitors are in complete agreement with our results using calcineurin phosphatase inhibitors, or PPP3R1 knockdown, suggesting that the synergy between KIT inhibitors and calcineurin inhibitors is NFAT-dependent.
Discussion
Although targeted kinase therapy has transformed treatment of some KIT-mutant cancers, patients with KIT-mutant systemic mastocytosis have yet to fully benefit from these therapies. Unfortunately, most cases of human systemic mastocytosis harbor the KIT D816V mutation, which is minimally or at best partially inhibited by clinically available KIT TKIs. Therefore, novel treatment options are needed to sensitize these cells to KIT TKI therapy. In this article, we present data indicating that calcineurin/NFAT inhibition sensitizes KIT-mutant mast cells to KIT TKIs. Synergy was seen even when the KIT TKI was used at doses that had only a minimal antiproliferative effect as a single agent. This observation is of particular relevance to the treatment of systemic mastocytosis with an associated D816V mutation.
Notably, we report for the first time that NFAT family members are constitutively activated in all KIT-mutant mast cell lines tested to date, including the human HMC1.2 cell line harboring KIT V560G + D816V. This phenomenon has not previously been described and may contribute to the progression of mast cell disease. The cause of this constitutive NFAT activation is unknown, but NFAT activation and localization can be manipulated using CNPIs (CSA and FK506). Calcineurin phosphatase inhibitors alone were not sufficient to decrease cell viability or induce apoptosis, but combining KIT TKIs and calcineurin/NFAT inhibitors led to a synergistic antiproliferative and proapoptotic effect in KIT-mutant mast cells. This suggests that CNPIs cooperate with KIT inhibition to affect cell viability and apoptosis. In addition, we presented evidence that combination therapy may diminish disease persistence based on significantly reduced replating efficiency following simultaneous inhibition of KIT and calcineurin in KIT-mutant mast cell models.
It is known that CNPIs such as CSA have other targets in addition to calcineurin. One such reported target is the drug efflux pump PgP, which mediates multidrug resistance (41–43). If synergy was mediated via the inhibition of PgP by CSA, then we would expect to see a further decrease in KIT activation after treatment of KIT-mutant cells with CSA/dasatinib when compared with single agent dasatinib. However, combination therapy did not further modulate KIT autophosphorylation compared with dasatinib alone (Supplementary Fig. S4). In agreement with this observation, combination therapy and single-agent dasatinib had identical effects on KIT-dependent signaling through the PI3K and MAPK pathways (Supplementary Fig. S4). To confirm that the observed synergy was, in fact, NFAT-dependent, we knocked down the expression of the Ppp3r1 subunit of calcineurin. Ppp3r1 knockdown sensitized P815 cells to dasatinib treatment and eliminated the effect of adding 1 μmol/L CSA. The fact that calcineurin knockdown replaced the effects of adding CSA strongly supports our hypothesis that inhibition of calcineurin is required for the observed synergy. To further confirm these results, we tested NFAT-specific inhibitors in combination with KIT inhibitors. Our results replicated the results we saw with CSA combined with dasatinib, as determined by CI values; we therefore conclude that synergy between KIT inhibitors and CNPIs is NFAT-dependent.
We also found that KIT inhibitors did not affect NFAT phosphorylation or cellular localization. However, KIT inhibitors partially inhibited basal NFAT-dependent transcriptional activity in our NFAT-dependent reporter cell line, suggesting that these two pathways converge to regulate NFAT-dependent transcriptional activity. It has been reported that KIT signaling activates calcium signaling (45), so we hypothesized that inhibition of KIT signaling could affect NFAT through reduction of calcium flux. However, when we induced calcium influx with TPA and ionomycin, KIT inhibitors were unable to fully block the resultant increase in NFAT-dependent transcriptional activity. Furthermore, both calcium channel blockers and CNPIs were able to completely inhibit the increase in NFAT activity induced by increased calcium flux. Conversely, calcium channel blockers did not ablate basal NFAT-dependent transcriptional activity, as assessed with our NFAT-dependent reporter cell line model. Finally, KIT inhibitors partially blocked basal NFAT-dependent transcriptional activity. We conclude that constitutive NFAT activity in mast cells is only minimally dependent upon calcium flux.
In addition to KIT modulating NFAT-dependent transcriptional activity, we examined the possibility that NFAT activity could modulate KIT expression or kinase activity. However, we found no evidence that calcineurin inhibitors modulated levels of total or phospho-KIT (Supplementary Fig. S3A). To further investigate the mechanism behind modulation of NFAT-dependent transcription by inhibitors of these distinct pathways, we examined the effects of KIT inhibition on NFAT localization and phosphorylation. We found that neither dasatinib nor imatinib affected NFAT localization or phosphorylation at doses that were sufficient to inhibit KIT signaling. It is known that NFAT transcriptional activity synergizes with certain DNA-binding partners such as JUN (33, 34). We hypothesize that KIT inhibition modulates one or more NFAT-binding partners leading to the observed decrease in NFAT-dependent transcriptional activity.
Our work characterizing the effects of combining KIT TKI and CNPI in KIT-mutant mast cells leads us to hypothesize that such a combination treatment could be used to improve treatment options for patients with KIT-mutant systemic mastocytosis. Despite these promising preliminary in vitro results, there are concerns over the use of immunosuppressive agents (e.g., CSA, FK506) to treat patients with cancer. Theoretically, identification and targeting downstream effectors of the observed synergy could also be effective against KIT-mutant systemic mastocytosis, without the risk of immunosuppression. Further studies are indicated to identify other combination treatments that target the novel biology of KIT-mutant cancer cells that express constitutively activated NFAT species.
Supplementary Material
Acknowledgments
Grant Support
This work was supported in part by funding from a VA Merit Review grant (to M.C. Heinrich), the Life Raft Group (to M.C. Heinrich), and the GIST Cancer Research Fund (to M.C. Heinrich)
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
M.C. Heinrich has received speakers’ bureau honoraria from Novartis, Onyx, and Pfizer; has ownership interest (including patents) in MolecularMD and Novartis; and is a consultant/advisory board member for Novartis, Pfizer, and Ariad. No potential conflicts of interest were disclosed by the other authors.
Authors’ Contributions
Conception and design: A.C. Macleod, M.C. Heinrich
Development of methodology: A.C. Macleod, J. Patterson, M.C. Heinrich
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): A.C. Macleod, L.R. Klug, D.J. Griffith, A. Town, M.C. Heinrich
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): A.C. Macleod, L.R. Klug, J. Patterson, C. Beadling, M.C. Heinrich
Writing, review, and/or revision of the manuscript: A.C. Macleod, L.R. Klug, D.J. Griffith
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): J. Patterson, D.J. Griffith
References
- 1.Taylor ML, Dastych J, Sehgal D, Sundstrom M, Nilsson G, Akin C, et al. The Kit-activating mutation D816V enhances stem cell factor co-dependent factor. Blood. 2001;98:1195–9. doi: 10.1182/blood.v98.4.1195. [DOI] [PubMed] [Google Scholar]
- 2.Nagata H, Worobec AS, Semere T, Metcalfe DD. Elevated expression of the proto-oncogene c-kit in patients with mastocytosis. Leukemia. 1998;12:175–81. doi: 10.1038/sj.leu.2400906. [DOI] [PubMed] [Google Scholar]
- 3.Piao X, Bernstein A. A point mutation in the catalytic domain of c-kit induces growth factor independence, tumorigenicity, and differentiation of mast cells. Blood. 1996;87:3117–23. [PubMed] [Google Scholar]
- 4.Kitayama H, Kanakura Y, Furitsu T, Tsujimura T, Oritani K, Ikeda H, et al. Constitutively activating mutations of c-kit receptor tyrosine kinase confer factor-independent growth and tumorigenicity of factor-dependent hematopoietic cell lines. Blood. 1995;85:790–8. [PubMed] [Google Scholar]
- 5.Piao X, Paulson R, van der Geer P, Pawson T, Bernstein A. Oncogenic mutation in the Kit receptor tyrosine kinase alters substrate specificity and induces degradation of the protein tyrosine phosphatase SHP-1. Proc Natl Acad Sci. 1996;93:14665–9. doi: 10.1073/pnas.93.25.14665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peter B, Hadzijusufovic E, Blatt K, Gleixner KV, Pickl WF, Thaiwong T, et al. KIT polymorphisms and mutations determine responses of neoplastic mastacells to bafetinib (INNO-406) Exp Hematol. 2010;38:782–91. doi: 10.1016/j.exphem.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Tsujimura T, Furitsu T, Morimoto M, Kanayama Y, Nomura S, Matsuzawa Y, et al. Substitution of an aspartic acid results in constitutive activation of c-kit receptor tyrosine kinase in rat tumor mast cell line RBL-2H3. Int Arch Allergy Immunol. 1995;106:377–85. doi: 10.1159/000236870. [DOI] [PubMed] [Google Scholar]
- 8.Pardanani A, Elliott M, Reeder T, Li CY, Baxter EJ, Cross NCP, et al. Imatinib for systemic mast-cell disease. The Lancet. 2003;362:535–6. doi: 10.1016/s0140-6736(03)14115-3. [DOI] [PubMed] [Google Scholar]
- 9.Gotlib J, Berub C, Growney JD, Chen CC, George TI, Williams C, et al. Activity of the tyrosine kinase inhibitor PKC412 in a patient with mast cell leukemia with the D816V KIT mutation. Blood. 2005;106:2865–70. doi: 10.1182/blood-2005-04-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Cesne A, Ray-Coquard I, Bui BN, Adenis A, Rios M, Bertucci F, et al. Discontinuation of imatinib in patients with advanced gastrointestinal stromal tumours after 3 years of treatment: an open-label multicentre randomised phase 3 trial. Lancet Oncol. 2010;11:942–9. doi: 10.1016/S1470-2045(10)70222-9. [DOI] [PubMed] [Google Scholar]
- 11.Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 12.Huang E, Nocka K, Beier DR, Chu TY, Buck J, Lahm HW, et al. The hematopoietic growth factor KL is encoded by the SI locus and is the ligand of the c-kit receptor, the gene product of the W locus. Cell. 1990;63:225–33. doi: 10.1016/0092-8674(90)90303-v. [DOI] [PubMed] [Google Scholar]
- 13.Anette D, Fabiola M, Bryna M, Nora E, Joseph Dipak P, Samuel S, et al. Mechanisms of oncogenic KIT signal transduction in primary gastrointestinal stromal tumors (GISTs) Oncogene. 2004;23:3999–4006. doi: 10.1038/sj.onc.1207525. [DOI] [PubMed] [Google Scholar]
- 14.Matsui J, Wakabayashi T, Asada M, Yoshimatsu K, Okada M. Stem cell factor/c-kit signaling promotes the survival, migration, and capillary tube formation of human umbilical vein endothelial cells. J Biol Chem. 2004;279:18600–7. doi: 10.1074/jbc.M311643200. [DOI] [PubMed] [Google Scholar]
- 15.Furitsu T, Tsujimura T, Tono T, Ikeda H, Kitayama H, Koshimizu U, et al. Identification of mutations in the coding sequence of the proto-oncogene c-kit in a human mast cell leukemia cell line causing ligand-independent activation of c-kit product. J Clin Invest. 1993;92:1736–44. doi: 10.1172/JCI116761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lux ML, Rubin BP, Biase TL, Chen CJ, Maclure T, Demetri G, et al. KIT extracellular and kinase domain mutations in gastrointestinal stromal tumors. Am J Pathol. 2000;156:791–5. doi: 10.1016/S0002-9440(10)64946-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broudy VC, Smith FO, Lin N, Zsebo KM, Egrie J, Bernstein ID. Blasts from patients with acute myelogenous leukemia express functional receptors for stem cell factor. Blood. 1992;80:60–7. [PubMed] [Google Scholar]
- 18.Ohashi A, Funasaka Y, Ueda M, Ichihashi M. c-KIT receptor expression in cutaneous malignant melanoma and benign melanotic naevi. Melanoma Res. 1996;6:25–30. doi: 10.1097/00008390-199602000-00004. [DOI] [PubMed] [Google Scholar]
- 19.Meyts ER-D, Skakkebkk N. Expression of the c-kit protein product in carcinoma-in-situ and invasive testicular germ cell tumours. Int J Androl. 1994;17:85–92. doi: 10.1111/j.1365-2605.1994.tb01225.x. [DOI] [PubMed] [Google Scholar]
- 20.Longley BJ, Tyrrell L, Lu SZ, Ma YS, Langley K, Ding TG, et al. Somatic c-KIT activating mutation in urticaria pigmentosa and aggressive mastocytosis: establishment of clonality in a human mast cell neoplasm. Nat Genet. 1996;12:312–4. doi: 10.1038/ng0396-312. [DOI] [PubMed] [Google Scholar]
- 21.Nagata H, Worobec AS, Oh CK, Chowdhury BA, Tannenbaum S, Suzuki Y, et al. Identification of a point mutation in the catalytic domain of the protooncogene c-kit in peripheral blood mononuclear cells of patients who have mastocytosis with an associated hematologic disorder. Proc Natl Acad Sci U S A. 1995;92:10560–4. doi: 10.1073/pnas.92.23.10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerbaulet A, Wickenhauser C, Scholten J, Peschke K, Drube S, Horny HP, et al. Mast cell hyperplasia, B-cell malignancy, and intestinal inflammation in mice with conditional expression of a constitutively active kit. Blood. 2011;117:2012–21. doi: 10.1182/blood-2008-11-189605. [DOI] [PubMed] [Google Scholar]
- 23.Zappulla JP, Dubreuil P, Desbois S, Letard S, Hamouda NB, Daeron M, et al. Mastocytosis in mice expressing human Kit receptor with the activating Asp816Val mutation. J Exp Med. 2005;202:1635–41. doi: 10.1084/jem.20050807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valent P. Mastocytosis: a paradigmatic example of a rare disease with complex biology and pathology. Am J Cancer Res. 2013;3:159–72. [PMC free article] [PubMed] [Google Scholar]
- 25.Spreafico A, Tentler JJ, Pitts TM, Tan AC, Gregory MA, Arcaroli JJ, et al. Rational combination of a MEK inhibitor, selumetinib, and the Wnt/calcium pathway modulator, cyclosporin A, in preclinical models of colorectal cancer. Clin Cancer Res. 2013;19:4149–62. doi: 10.1158/1078-0432.CCR-12-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gregory MA, Phang TL, Neviani P, Alvarez-Calderon F, Eide CA, O’Hare T, et al. Wnt/Ca2+/NFAT signaling maintains survival of Ph+ leukemia cells upon inhibition of Bcr-Abl. Cancer Cell. 2010;18:74–87. doi: 10.1016/j.ccr.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dissanayake SK, Olkhanud PB, O’Connell MP, Carter A, French AD, Camilli TC, et al. Wnt5A regulates expression of tumor-associated antigens in melanoma via changes in signal transducers and activators of transcription 3 phosphorylation. Cancer Res. 2008;68:10205–14. doi: 10.1158/0008-5472.CAN-08-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flockhart RJ, Armstrong JL, Reynolds NJ, Lovat PE. NFAT signalling is a novel target of oncogenic BRAF in metastatic melanoma. Br J Cancer. 2009;101:1448–55. doi: 10.1038/sj.bjc.6605277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perotti V, Baldassari P, Bersani I, Molla A, Vegetti C, Tassi E, et al. NFATc2 is a potential therapeutic target in human melanoma. J Invest Dermatol. 2012;132:2652–60. doi: 10.1038/jid.2012.179. [DOI] [PubMed] [Google Scholar]
- 30.Robbs BK, Cruz AL, Werneck MB, Mognol GP, Viola JP. Dual roles for NFAT transcription factor genes as oncogenes and tumor suppressors. Mol Cell Biol. 2008;28:7168–81. doi: 10.1128/MCB.00256-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller MR, Rao A. NFAT, immunity and cancer: a transcription factor comes of age. Nat Rev Immunol. 2010;10:645–56. doi: 10.1038/nri2818. [DOI] [PubMed] [Google Scholar]
- 32.Viola JP, Carvalho LD, Fonseca BP, Teixeira LK. NFAT transcription factors: from cell cycle to tumor development. Braz J Med Biol Res. 2005;38:335–44. doi: 10.1590/s0100-879x2005000300003. [DOI] [PubMed] [Google Scholar]
- 33.Macian F, Lopez-Rodriguez C, Rao A. Partners in transcription: NFAT and AP-1. Oncogene. 2001;20:2476–89. doi: 10.1038/sj.onc.1204386. [DOI] [PubMed] [Google Scholar]
- 34.Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17:2205–32. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- 35.Ralph P, Moore MA, Nilsson K. Lysozyme synthesis by established human and murine histiocytic lymphoma cell lines. J Exp Med. 1976;143:1528–33. doi: 10.1084/jem.143.6.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butterfield JH, Weiler D, Dewald G, Gleich G. Establishment of an immature mast cell line from a patient with mast cell leukemia. Leuk Res. 1988;12:345–55. doi: 10.1016/0145-2126(88)90050-1. [DOI] [PubMed] [Google Scholar]
- 37.Seldin D, Adelman S, Austen K, Stevens R, Hein A, Caulfield J, et al. Homology of the rat basophilic leukemia cell and the rat mucosal mast cell. Proc Natl Acad Sci U S A. 1985;82:3871–5. doi: 10.1073/pnas.82.11.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeVinney R, Gold WM. Establishment of two dog mastocytoma cell lines in continuous culture. Am J Respir Cell Mol Biol. 1990;3:413–20. doi: 10.1165/ajrcmb/3.5.413. [DOI] [PubMed] [Google Scholar]
- 39.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protocols. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 40.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 41.Coley HM, Twentyman PR, Workman P. Identification of anthracyclines and related agents that retain preferential activity over adriamycin in multidrug-resistant cell lines, and further resistance modification by verapamil and cyclosporin A. Cancer Chemother Pharmacol. 1989;24:284–90. doi: 10.1007/BF00304759. [DOI] [PubMed] [Google Scholar]
- 42.Gaveriaux C, Boesch D, Boelsterli JJ, Bollinger P, Eberle MK, Hiestand P, et al. Overcoming multidrug resistance in Chinese hamster ovary cells in vitro by cyclosporin A (Sandimmune) and non-immunosuppressive derivatives. Br J Cancer. 1989;60:867–71. doi: 10.1038/bjc.1989.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nooter K, Sonneveld P, Janssen A, Oostrum R, Boersma T, Herweijer H, et al. Expression of the mdr3 gene in prolymphocytic leukemia: association with cyclosporin-A-induced increase in drug accumulation. Int J Cancer. 1990;45:626–31. doi: 10.1002/ijc.2910450409. [DOI] [PubMed] [Google Scholar]
- 44.Sivalenka RR, Jessberger R. SWAP-70 regulates c-kit-induced mast cell activation, cell-cell adhesion, and migration. Mol Cell Biol. 2004;24:10277–88. doi: 10.1128/MCB.24.23.10277-10288.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ueda S, Mizuki M, Ikeda H, Tsujimura T, Matsumura I, Nakano K, et al. Critical roles of c-Kit tyrosine residues 567 and 719 in stem cell factor-induced chemotaxis: contribution of src family kinase and PI3-kinase on calcium mobilization and cell migration. Blood. 2002;99:3342–9. doi: 10.1182/blood.v99.9.3342. [DOI] [PubMed] [Google Scholar]
- 46.Ishikawa J, Ohga K, Yoshino T, Takezawa R, Ichikawa A, Kubota H, et al. A pyrazole derivative, YM-58483, potently inhibits store-operated sustained Ca2 +Influx and IL-2 production in T lymphocytes. J Immunol. 2003;170:4441–9. doi: 10.4049/jimmunol.170.9.4441. [DOI] [PubMed] [Google Scholar]
- 47.Kiyonaka S, Kato K, Nishida M, Mio K, Numaga T, Sawaguchi Y, et al. Selective and direct inhibition of TRPC3 channels underlies biological activities of a pyrazole compound. Proc Natl Acad Sci U S A. 2009;106:5400–5. doi: 10.1073/pnas.0808793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Proksch P, Giaisi M, Treiber MK, Palfi K, Merling A, Spring H, et al. Rocaglamide derivatives are immunosuppressive phytochemicals that target NF-AT activity in T cells. J Immunol. 2005;174:7075–84. doi: 10.4049/jimmunol.174.11.7075. [DOI] [PubMed] [Google Scholar]
- 49.Umehara H, Asai A. Tributylhexadecylphosphonium bromide, a novel nuclear factor of activated T cells signaling inhibitor, blocks interleukin-2 induction associated with inhibition of p70 ribosomal protein S6 kinase phosphorylation. Biol Pharm Bull. 2012;35:805–9. doi: 10.1248/bpb.35.805. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.