Abstract
The great majority of mitochondrial proteins are synthesized by cytosolic ribosomes and then imported into the organelle post-translationally. The translocase of the outer membrane (TOM) is a proteinaceous machinery that contains surface receptors for preprotein recognition and also serves as the main entry gateway into mitochondria. Mitochondrial targeting requires various cytosolic factors, in particular the molecular chaperones Hsc70/Hsp70 and Hsp90. The chaperone activity of Hsc70/Hsp70 and Hsp90 occurs in coordinated cycles of ATP hydrolysis and substrate binding, and is regulated by a number of co-chaperone proteins. The import receptor Tom70 is a member of the tetratricopeptide repeat (TPR) co-chaperone family and contains a conserved TPR clamp domain for interaction with Hsc70 and Hsp90. Such interaction is essential for the initiation of the import process. This review will discuss the roles of Hsc70 and Hsp90 in mitochondrial import and summarize recent progress in understanding these pathways.
Keywords: Mitochondria, import, chaperone, Tom70, Hsc70, Hsp90
INTRODUCTION
Mitochondria are double membrane-bound organelles and contain 4 distinctive sub-compartments – the outer membrane (OM), the inner membrane (IM) with folded cristae, the intermembrane space (IMS) and the matrix. The respiratory chain is located in the IM, and is responsible for the generation of ATP using an electrochemical gradient. Mitochondria are also key sites of lipid metabolism, iron homeostasis and regulation of apoptosis [1–3]. Recent proteomic studies suggested that there are as many as 1000 proteins in mitochondria from budding yeast Saccharomyces cerevisiae [4, 5], and 1500 mitochondrial proteins in mammals [6, 7]. Except for a handful of proteins encoded by the mitochondrial genomic DNA and translated by matrix ribosomes, mitochondrial proteins are encoded by the nuclear genome and translated by cytosolic ribosomes. These polypeptides are post-translationally targeted to the mitochondria, imported and subsequently sorted to their destined sub-compartments within the organelle Fig. (1). Therefore, the mitochondrial import and sorting machineries are of outmost importance in maintaining proper mitochondrial functions. The protein-based mechanisms for polypeptide sorting within mitochondria are the subject of much research over the last 20 years (for reviews, see [8–10]). It has long been known that mitochondrial preproteins have various requirements for cytosolic factors prior to import. In particular, the role of molecular chaperones, which generally assist protein folding, has become one aspect of recent interest. Here, a brief overview on the classical mitochondrial import pathways and the cytosolic chaperones, Hsc70 and Hsp90, will be provided. The roles of the chaperones in mitochondrial import will be discussed, and recent findings on the known chaperone-dependent import mechanism will be highlighted.
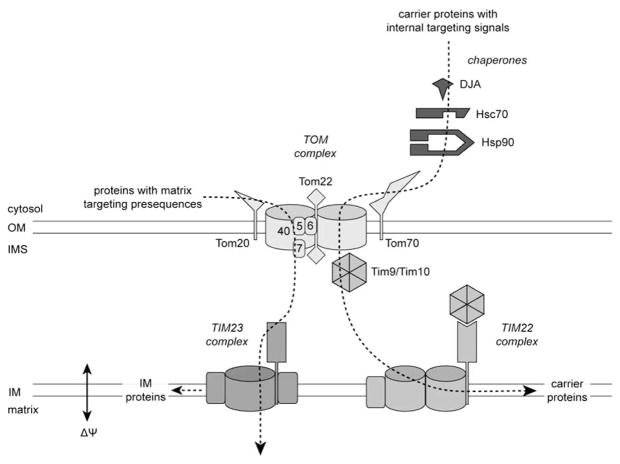
Figure 1.
Mitochondrial import pathways. Proteins pass through a common TOM complex in the outer membrane (OM), consisting of Tom40, Tom22, Tom5, Tom6 and Tom7, and the import receptors Tom20 and Tom70. Proteins with matrix targeting presequences are recognized by Tom20 and directed to the matrix or released into the inner membrane (IM) or intermembrane space (IMS) by the TIM23 complex. Metabolite carrier proteins are recognized by Tom70 and inserted in the IM by the TIM22 complex. Translocation across the IM via the TIM23 or TIM22 complexes requires the (positive/negative) membrane potential (ΔΨ).
MITOCHONDRIAL IMPORT PATHWAYS
The translocase of outer membrane (TOM) machinery serves as the general entry gateway into the mitochondria Fig. (1). The major component of the TOM machinery is the channel-forming protein Tom40 (numbering of mitochondrial import components refers to approximate molecular weight). Tom40, along with an organizing receptor Tom22, and small Tom proteins Tom5, Tom6 and Tom7, constitutes the general import pore (GIP) on the OM. The majority of the mitochondrial precursor proteins are synthesized with classical N-terminal amphipathic presequences. They are recognized by the Tom20 receptor on the OM, translocated through the GIP and subsequently sorted to the matrix or IM by the translocase of inner membrane (TIM) complex TIM23. Another receptor, Tom70, recognizes preproteins with targeting signals embedded in their internal sequences. These preproteins usually contain multiple membrane-spanning domains and many of them are metabolite carriers of the IM. Due to their hydrophobic nature, molecular chaperones are required to prevent aggregation in the cytosol and the import of these preproteins are also chaperone-dependent. After translocation across the OM, they are guided by a heterohexameric complex of Tim9-Tim10 in the IMS to the TIM22 complex, which facilitates the insertion into the IM. Translocation through both the TIM22 and TIM23 complexes requires an active electrochemical potential across the IM [8–10]. The post-translational nature of import implies that precursors are exposed to the cytosolic environment before recognition by the Tom20 or Tom70 receptors. The involvement of cytosolic chaperones in targeting to these outer membrane receptors has been studied. The Tom70-mediated import pathway for metabolite carriers is the most clearly dependent on chaperones, as it requires a specific interaction between Tom70 and the chaperones Hsc70/Hsp70 and Hsp90.
HSC70
The Hsp70 family is a class of abundant chaperones that perform versatile functions ranging from folding and preventing aggregation of nascent polypeptide chains to post-translation targeting [11, 12]. Two members of the Hsp70 family are found in the human cytosol – the stress-inducible Hsp70 (70-kDa heat shock protein, human HSPA1) and its constitutively expressed homologue Hsc70 (70-kDa heat shock cognate protein, HSPA8). Hsc70 handles a broad range of client proteins due to its substrate specificity for short, hydrophobic peptides [13]. It was shown to bind a large number of nascent polypeptide chains co-translationally to prevent aggregation and assist in de novo folding [14, 15]. All homologues of the Hsp70 family have the same domain architecture, with a 44-kDa N-terminal ATPase domain and a 27-kDa C-terminal substrate binding domain joined by a well-conserved linker [16, 17]. The AT-Pase domain showed structural similarities to those of actin and hexokinases [16]. Consistent with the chaperone’s preference for short, hydrophobic peptides, the substrate binding domain of DnaK (bacterial Hsp70) binds a substrate peptide in an extended form via hydrophobic interactions [17]. Substrate binding and release are strictly dependent on the reaction cycles of ATP hydrolysis.
The co-chaperone family related to bacterial DnaJ is conserved in their J-domains, a 70-residue domain with a His-Pro-Asp (HPD) motif [18, 19], essential for stimulating the ATPase activity of Hsp70 [20]. In eukaryotes, there are three types of J-domain co-chaperones. Type 1 co-chaperones, such as DJA1 (Hdj2/DNAJA1), DJA2 (DNAJA2) and DJA4 (DNAJA4) in human and Ydj1 in yeast, are most homologous to bacterial DnaJ. Following the J-domain in the N-terminus, DnaJ contains a Gly/Phe rich region, a central region with two zinc-finger motifs and a C-terminal dimerization domain. The eukaryotic DnaJ homologues also contain a conserved cysteine in the C-termini for farnesylation. Type 2 co-chaperones, such as human DJB1 (Hsp40/Hdj1/DnaJB1) and yeast Sis1, contain the J-domain and the G/F rich region but are divergent in the central and C-terminal domains, whereas type 3 co-chaperones are common in their J-domains only [21–23]. Type 1 members have chaperone-like activity since they can directly interact with substrate and prevent their aggregation [24, 25]. The central regions of Type 1 members were identified to be the substrate binding site and dimerization is essential for the chaperone activity [26]. In mammals, DJA1, DJA2 and DJA4 are constitutively expressed and co-localized with Hsc70 [27, 28].
Nucleotide exchange factors (NEF) also regulate the reaction cycle of Hsp70 chaperones by accelerating the release of ADP in exchange for ATP, stimulating the ATPase activity together with a DnaJ co-chaperone. In the eukaryotic cytosol, the first nucleotide exchange factor identified was Bag-1, which is structurally unrelated to the bacterial NEF GrpE but induces a similar conformational change in the ATPase domain [29, 30]. The Bag co-chaperone family is characterized by the Bag domain which can recognize the ATPase domain of Hsc70 [31]. Other nucleotide exchange factors in human cells include HspBP1 and Hsp110, which are again structurally unrelated to Bag-1 or GrpE but also interact with the Hsc70 ATPase domain [32–34].
The ATPase cycle of Hsp70 chaperones is thought to be universally conserved with DnaK Fig. (2) [11]. DnaK binds ATP with a high affinity, but the chaperone has low affinity for substrate peptide in the ATP-bound form, and is in a rapid exchange between substrate-bound and substrate-free states. DnaJ can form a ternary complex with DnaK and the substrate, and stimulate ATP hydrolysis, thus stabilizing the interaction between the substrate and the DnaK. Following ATP hydrolysis, the NEF mediates the release of ADP from DnaK. Subsequent ATP binding to DnaK can trigger the release of the substrate [18, 35–38]. The substrate could either be released in its folded native state, or re-enter the Hsp70 ATPase cycle. In the human Hsc70 cycle, released substrate could also be delivered to Hsp90 for folding.
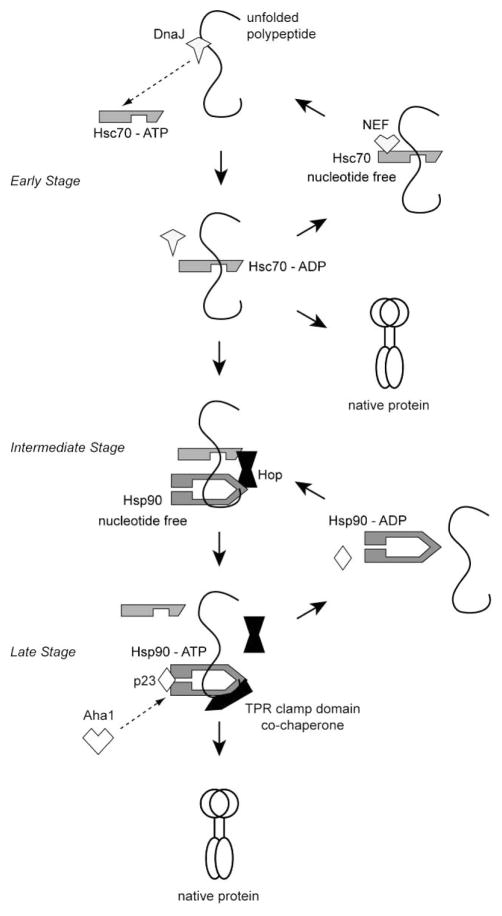
Figure 2.
The Hsc70-Hsp90 multichaperone system. DnaJ co-chaperones can bind substrate and activate Hsc70 to hydrolyze ATP and bind the substrate (early stage). Nucleotide exchange factors (NEF) allow Hsc70 to return to the ATP state. Released substrates may be folded or transferred to Hsp90 in complexes containing the TPR clamp domain co-chaperone Hop (intermediate stage). Hsp90 in the ATP state stably binds substrate. Other TPR co-chaperones can replace Hop on Hsp90, and the co-chaperones p23 and Aha1 regulate the Hsp90 ATPase cycle for substrate folding (late stage). ATP hydrolysis by Hsp90 releases substrate.
HSP90
The 90-kDa heat shock proteins (Hsp90) are a family of chaperones including Hsp90α and Hsp90β in the human cell cytosol. In eukaryotes, Hsp90 is highly abundant and is essential for viability [39, 40]. Unlike Hsc70, Hsp90 preferentially recognizes more structured folding intermediates [41, 42]. Hsp90 functions in vivo include the maturation of a specific set of substrates, with the best characterized examples being steroid hormone receptors and protein kinases [40, 43]. The Hsp90 chaperones are homodimers and dependent on ATP. Each subunit contains an N-terminal ATPase domain, followed by a linker region, a middle domain and a C-terminal homodimerization domain [44, 45]. The crystal structure of the 25-kDa N-terminal domain of yeast Hsp90 bound ATP/ADP and displayed structural similarities with the ATPase domains from DNA gyrase B (GyrB) [44]. Mutagenesis and biochemical assays confirmed that Hsp90 function is dependent on its ATPase activity [46, 47]. The 33-kDa middle domain contains a catalytic loop which contacts the N-terminal domain to activate the hydrolysis of ATP, and a hydrophobic patch implicated in substrate binding [48]. The middle domain is also the binding site for co-chaperone, Aha1, which stimulates the ATPase activity of Hsp90 [49–51]. The 12-kDa C-terminal domain is essential for dimerization, and the intersubunit interface is formed by a pair of α-helices from each subunit [52]. The full-length structure of yeast Hsp90 [53] depicts a homodimer of Hsp90 in the nucleotide-bound state, complexed with co-chaperone, p23, which was known to regulate the ATPase cycle of Hsp90 [54, 55]. The subunits are in a parallel arrangement with extensive intersubunit contacts between the N-terminal domains [53].
Hsp90 cooperates with Hsc70 in a multi-chaperone system [43] and Hop (Hsc70/Hsp90-organizing protein) is the functional link between the two molecular chaperones Fig. (2). Hop can simultaneously bind both Hsc70 and Hsp90 and acts as an adaptor protein between them [56]. The reaction cycle of Hsp90 is best characterized with steroid hormone receptors as substrates. Hop recognizes Hsc70 in the substrate-binding ADP-bound state and transfers the bound substrate to Hsp90, which is in the nucleotide-free state. Upon ATP binding by Hsp90, Hop and Hsc70 are released from the complex. In addition, the N-terminal ATPase domains of Hsp90 transiently dimerize when in ATP-bound state. This conformational change is required for the binding of p23, which stabilizes the substrate-Hsp90 complex and promotes substrate release after ATP hydrolysis [55, 57–59].
The chaperone-binding domains of Hop each have three tetratricopeptide repeat (TPR) motifs – the TPR1 domain binds Hsc70 and TPR2A binds Hsp90 [60]. TPR motifs are loosely conserved 34-residue sequences. The C-termini of both Hsc70 and Hsp90 contain a conserved EEVD motif that is required for interaction with Hop. Crystal structures of the TPR1 and TPR2A domains in the presence of chaperone-derived peptides showed that three TPR motifs are arranged in a superhelical structure, providing a cradle-shaped groove that anchors the EEVD motifs. Interactions with the terminal Asp residue of the chaperones are particularly important, thus they are named “TPR dicarboxylate clamp domains”. Additional hydrophobic interactions determine the specificity of chaperone binding [60, 61]. The TPR clamp interactions are conserved in a number of other co-chaperones, including Tom70.
CYTOSOLIC FACTORS IN IMPORT
Mitochondrial preproteins have various import requirements for cytosolic molecular chaperones. In vivo depletion of the Hsc70 homologue Ssa1 in yeast led to accumulation of Tom20-dependent preprotein F1-ATP synthase subunit β (F1β) [62]. Studies in vitro with a Tom20-dependent precursor of ornithine transcarbamylase (OTC) showed the importance of Hsc70 in preventing aggregation of preproteins and maintaining them in an import-competent state [63, 64]. The presence of Hsc70 during preprotein translation is a requirement for import competence, since immunodepletion of the chaperone from rabbit reticulocyte lysate led to disruption of preprotein import, which could not be restored by re-addition of purified Hsc70 after translation [64]. Similarly, studies with another preprotein, precursor of the mitochondrial aspartate aminotransferase (pmAAT), showed that Hsc70 interacts with the preprotein during a very early stage of translation, and that such interaction is important for distinguishing the preprotein from its cytosolic isozyme for targeting into mitochondria [65, 66].
The roles of DnaJ co-chaperones in mitochondrial import have also been investigated. Immunodepletion and re-addition studies provided evidence that DJA1 and DJA2 are required for OTC import into mitochondria [28, 67, 68]. Hsc70 and Ydj1 were shown to form complexes with purified preprotein pmAAT, and the complexes were in a competent form for import in the presence of rabbit reticulocyte [69]. Type 1 and some Type 2 DnaJ co-chaperones are farnesylated in the C-terminus, but it is questionable whether or not farnesylation is important for mitochondrial targeting and import. A yeast strain with a Ydj1 mutant which cannot be farnesylated showed accumulation of F1β in vivo under restrictive temperature [70]. The import defect of OTC caused by immunodepletion of DJA1, however, could be restored by re-addition of bacterial DnaJ homologue, which lacks the C-terminal farnesylation [68]. The dependence on Hsc70 and DnaJ co-chaperones may vary upon the preprotein concerned. Various preproteins could be imported efficiently when translated in the Hsc70-immunodepleted rabbit reticulocyte lysate or in a wheat germ-based translation extract, which does not contain sufficient amounts of Hsc70 [71, 72].
THE TOM70 IMPORT PATHWAY
The import receptor Tom70 Fig. (3) exhibits substrate specificity for large, hydrophobic proteins with multiple internal targeting sequences. Mitochondrial carrier proteins, such as the inorganic phosphate carrier (PiC) and adenine nucleotide transporter (ANT), the mammalian homologue of yeast ADP/ATP carrier (AAC), are a family of proteins that shuttle metabolites across the IM [73], and typically require Tom70 for import. Tom70 is anchored by an N-terminal transmembrane domain and exposes a ~60-kDa domain in the cytosol. It was first identified as a receptor for AAC [74, 75]. The first mammalian homologue was characterized in rat with 20% and 28% sequence identity to the N. crassa and S. cerevisiae Tom70, and a similar function [76]. Limited proteolysis of yeast Tom70 identified a 25-kDa middle region which showed comparable carrier preprotein binding as the full cytosolic domain, suggesting that it constitutes part of the preprotein binding site [77]. Tom70 was thought to exist as a homodimer due to its slow mobility on Blue Native PAGE (BNP) and its ability to generate cross-linked dimer on isolated yeast mitochondria [78, 79]. Cross-linking the arrested preprotein AAC produced adducts with molecular weight consistent with a preprotein binding to Tom70 dimers [80, 81]. Analyses of the translocation intermediates on BNP revealed a high molecular weight complex, suggesting that each of the three targeting signals in AAC can recruit Tom70 [81]. It was proposed that the binding of multiple dimers of Tom70 to a single AAC molecule may have chaperone-like effect in preventing preprotein aggregation, although direct experimental evidence for the chaperone activity was lacking.
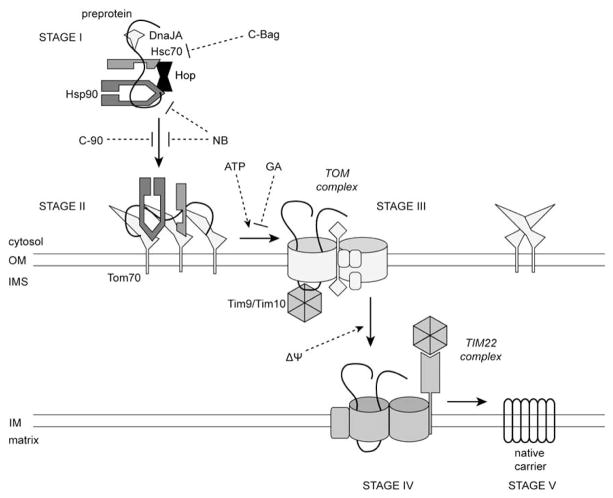
Figure 3.
Stages of Tom70-mediated import. In Stage I, carrier preprotein is bound by the cytosolic chaperone complex. Inhibition of Hsp90 by novobiocin (NB), or of Hsc70 by the Bag-1 NEF (C-Bag) disrupts targeting. In Stage II, chaperones dock onto Tom70 which recognizes targeting signals in the preprotein. Tom70 competition with an Hsp90 fragment (C-90) blocks docking. ATP cycling by the chaperones releases preprotein to the TOM translocation pore, Stage III. Depletion of ATP, or geldanamycin (GA) inhibition of Hsp90, impairs progression to Stage III. The TIM22 complex inserts the carrier in the IM in Stage IV, dependent on the membrane potential. In Stage V, the carrier is folded and assembled into its native state.
Instead, it was found that Tom70 directly interacts with Hsc70, but also with Hsp90, which had previously been unknown to function in import [82]. The mammalian carrier preproteins including PiC are bound by cytosolic chaperones, Hsc70/Hsp70 and Hsp90, prior to import. Sequence alignments suggested that Tom70 has a TPR clamp domain in its N-terminal region, and interaction with Hsc70 and Hsp90 was confirmed with purified Tom70 cytosolic fragment. Thus, the TPR clamp domain of Tom70 serves as a docking point for either of these chaperones before preproteins are delivered to the receptor for direct interaction. Competition for the Tom70 TPR clamp domain with a purified fragment of Hsp90 blocked import in vitro, and mutation of the chaperone docking site impaired Tom70 function in vitro and in live yeast. Crosslinking of preproteins showed that they were bound by Tom70 after the docking step. Yeast Tom70 only recognizes Ssa1 (yeast Hsc70) but not Hsp82 (yeast Hsp90), although the chaperone-docking step remains a requirement for preprotein interaction [82].
The Tom70-dependent import of carrier proteins is conceptually divided into 5 stages Fig. (3), most of which could be characterized by BNP [78, 83]. Stage I describes the preprotein in the cytosolic multi-chaperone complex. The cytosolic complex elutes in a 500 kDa peak on size-exclusion chromatography, characteristic of Hsp90 and Hsp90-bound substrate due to the extended structure of the chaperone [84, 85]. In stage II, the preprotein is in the receptor-bound state on the OM. Upon depletion of ATP, stage II could be characterized as a complex in a broad molecular weight range of 400–500 kDa on BNP. A stage III translocation intermediate could be arrested at the TOM complex by dissipation of the IM electrochemical potential to prevent TIM22 docking, and observed in the low molecular weight range below 60 kDa. This intermediate is protected from protease digestion, and is suggested to be preprotein partially translocated through the GIP and bound by the Tim9-Tim10 complex in the IMS [78]. If the membrane potential is only partially dissipated, a stage IV complex could be observed at around 350 kDa, which corresponds to the combined molecular weights of the TIM22 complex and a preprotein [86]. Finally, stage V describes the mature carrier that is properly integrated in the inner membrane as homodimers migrating at around 100 kDa [78, 81].
CHAPERONE-TOM70 MECHANISMS
A recent study investigated if the chaperones have an active role in translocation, other than simply docking on the import receptor. Among the chaperones, Hsp90 is unique in having small molecule inhibitors that target it. Two inhibitors with different mechanisms, geldanamycin (GA) and novobiocin (NB), were used. GA is a competitive inhibitor that targets the N-terminal ATP binding domain, whereas the targeting site of NB was localized to a C-terminal region [45, 87]. Indeed, the inhibitors affected the import pathway at different steps. NB acted in earlier steps by dual mechanisms – it decreased preprotein-chaperone complex (stage I) formation and also reduced the efficiency of chaperone docking (stage II). The combined effect led to inhibition of Tom70-dependent import down to basal level. On the other hand, GA acted at a later step and decreased the formation of translocation intermediate (stage III) [84]. Thus, we conclude that Hsp90 remains stably bound to the receptor-substrate complex on the outer membrane, and that progression to the GIP requires ATP cycling by Hsp90.
The co-chaperones interacting with a Tom70-dependent preprotein in the cytosol before import have been investigated. Import-competent complexes of chaperones with purified ANT, a Tom70-dependent metabolite carrier, were analyzed by mass spectrometry. ANT formed a complex with Hsc70, Type 1 DnaJ co-chaperones DJA1, DJA2 and DJA4, the Type 3 J-domain and TPR clamp domain co-chaperone Tpr2 (DnaJC7), as well as Hsp90, Hop and p23. The three Type 1 DJA proteins were functional in import based on dominant mutant inhibition in vitro. Nonetheless, the three co-chaperones differed in their abilities in preprotein binding in vitro and in increasing import efficiency in cultured cells [88]. In addition to ANT, the DnaJ co-chaperones could also bind to other Tom70-dependent preproteins, including PiC, citrate carrier (CiC) and oxoglutarate carrier (OGC), with differential affinities [89]. Another study compared the function of the cleaved presequences of PiC and CiC, which are unrelated to matrix targeting sequences: the PiC presequence lacks the amphipathic pattern and the CiC presequence is polar but short. Both presequences were not essential for import but increased efficiency by different means – the PiC sequence provided a binding site for Hsc70, while the CiC sequence increased the physical solubility [90].
Tom70 was implicated in the import of other hydrophobic mitochondrial proteins outside of the carrier family. Import of Tim54, an IM protein, requires Tom70 [91], as does the import of some matrix proteins such as mitochondrial alcohol dehydrogenase isozyme III and citrate synthase [92, 93]. Tom70 also plays a role in the insertion of OM proteins, including Tom40, which follows the same Hsp90- and Hsc70-mediated targeting pathway as the inner membrane carrier proteins [94, 95]. The Bcl-2 homology protein Mcl-1 has been reported to target to the OM for insertion of its tail anchor via Tom70, but through a direct interaction with the TPR clamp domain [96]. Recently, Tom70 has been implicated in the import of OM proteins with multiple α-helical transmembrane segments, particularly the cholesterol translocator protein (TSPO) and the mitofusin protein Mfn2. The involvement of Hsp90 in TSPO import was demonstrated by Tom70 competition and drug inhibition of Hsp90, and its import also involved the OM protein Metaxin1 as well as the TOM complex [97, 98].
While a direct involvement of Tom70 has not been shown, recent studies demonstrated that Hsp90 and its co-chaperone Cdc37 regulate the subcellular distribution of the PTEN-induced putative kinase, Pink1, mutations of which are linked to autosomal recessive Parkinson’s disease [99–101]. Pink1 contains a classical N-terminal presequence for translocation to mitochondria, where it is proteolytically processed from its full-length precursor form (66 kDa) to two smaller forms (55 kDa and 46 kDa) [99, 102]. Intriguingly, both full-length and mature forms of Pink1 exhibit dual localization in the cytosol and mitochondria [101, 103]. In addition to its function in stabilizing Pink1, Hsp90 was also shown to cooperate with its co-chaperone, Cdc37, and bind different forms of Pink1 to regulate their subcellular distributions between cytosol and mitochondria [99, 101]. Another Hsp90 co-chaperone AIP (arylhydrocarbon receptor-interacting protein), was reported to mediate the Tom20-dependent import of OTC, and ternary complexes of AIP-OTC-Tom20 were also observed [104]. Interestingly, AIP contains a TPR clamp domain for chaperone interaction with Hsc70 and Hsp90, and binds the C-terminal DDVE sequence of Tom20 at the same site [104, 105].
TOM70 STRUCTURE AND FUNCTION
The crystal structure of the cytosolic domain of yeast Tom70 depicts the receptor as a homodimer of two compact subunits with a relatively flat dimerization interface. Each subunit contains a chaperone-binding TPR clamp domain in the N-terminal region, followed by a disordered linker, and a large C-terminal α-helical domain that forms a conserved, somewhat hydrophobic cavity and was proposed to be the preprotein binding pocket [106]. However, the putative preprotein binding cavities cannot cooperate in binding the multiple targeting signals of a preprotein because the subunits depicted in the homodimer are in opposite orientation. Chaperone docking is also unfavorable due to steric hindrance from the C-terminal region helices, which fold down to form the dimerization interface with helices from the N-terminal region and occlude parts of the TPR clamp domain. A similar cytosolic fragment of yeast Tom70 was reported to adopt an extended, monomeric conformation in analytical ultracentrifugation experiments, and measurements of unfolding pathways provided evidence for a monomer with domains relatively independent of each other [107, 108]. Small-angle X-ray scattering experiments generated molecular envelopes of yeast Tom70 alone or with a bound chaperone-derived peptide. The envelopes portray an elongated “club” shape that fit well with a monomeric Tom70 with an end-to-end arrangement of the domains [109]. These conflicting data suggest different molecular mechanisms of the receptor. If Tom70 exists as a homodimer as depicted in the crystal structure [106], the receptor would need to undergo substantial domain rearrangement or even partial unfolding for chaperone docking to occur. The conformational change involved would be substantial and therefore might require chaperone activity. However, the formation of the receptor-bound intermediate (stage II) does not require ATP, suggesting that the role of the chaperones may not be related to a conformational switch of Tom70. Rather, the chaperone activity occurs after docking, as evidenced by the effect of GA in blocking progression of the preprotein to the GIP [84]. On the other hand, in the monomeric model, both the TPR clamp and the putative preprotein binding domain are exposed and compatible for chaperone docking and preprotein interaction, which seems more consistent with the ATP-independent docking step. Studies with yeast Tom71, a paralogue of yeast Tom70, provides a model in which Tom70 exists in equilibrium of the two states [110].
These ideas were investigated with human Tom70, also in light of earlier cross-linking and BNP experiments which provided evidence for homodimeric yeast Tom70. Analytical ultracentrifugation experiments showed that a similar cytosolic fragment of human Tom70 exists in equilibrium between monomer and dimer. The equilibrium could be shifted to predominantly monomer by point mutations located on the predicted dimerization interface. Notably, preprotein binding was more efficient with the mutant Tom70, and Tom70 on human mitochondrial OM was found not to dimerize. Taken together with the structural work, the functional state of Tom70 was confirmed to be the monomer [111].
OUTLOOK
A number of important questions remain on the mechanisms of Tom70-mediated import and on the role of molecular chaperones in mitochondrial targeting. For example, a molecular understanding is lacking of how Tom70 receives the preprotein from the chaperones and subsequently delivers it to the GIP for translocation. The insertion mechanism for many OM proteins (except for β barrel proteins such as Tom40 and porin/VDAC, which utilize the sorting and assembly machinery SAM) is still poorly characterized. How does Tom70 facilitate the import of the tail-anchored Mcl-1 and the multi-spanning TSPO? In particular, elucidation of the mechanism by which Bcl-2 family members are targeted to the OM present a challenge. Insertion of Bak and Bax was independent of TOM components and cytosolic factors, but may involve the OM porin VDAC [112, 113]. Targeting of Bcl-2 was reported to involve FKBP38, a transmembrane immunophilin that also contains a TPR clamp domain [114, 115], but the role of chaperones is still unclear. On the other hand, Hsp90 and Cdc37 are required for mitochondrial targeting of Pink1, yet the involvement of Tom70 remains to be confirmed. Finally, the possible function of chaperones in the regulation and degradation of OM proteins is still to be explored.
Acknowledgments
The authors are supported by the Canadian Institutes of Health Research, and Fonds de la recherche en santé du Québec. JCY holds a Canada Research Chair, Tier II.
LIST OF ABBREVIATIONS
- AAC
ADP/ATP carrier
- (pm)AAT
(Precursor of the mitochondrial) aspartate aminotransferase
- AIP
Arylhydrocarbon receptor-interacting protein
- ANT
Adenine nucleotide transporter
- BNP
Blue native polyacrylamide gel electrophoresis
- CiC
Citrate carrier
- F1β
F1-ATP synthase subunit β
- GA
Geldanamycin
- GIP
General import pore
- Hop
Hsc70/Hsp90-organizing protein
- Hsc70
70-kDa heat shock cognate protein
- Hsp70
70-kDa heat shock protein
- Hsp90
90-kDa heat shock protein
- IM
Inner membrane
- IMS
Intermembrane space
- NB
Novobiocin
- NEF
Nucleotide exchange factor
- OGC
2-Oxoglutarate carrier
- OM
Outer membrane
- OTC
Ornithine transcarbamylase
- PiC
Inorganic phosphate carrier
- Pink1
PTEN-induced putative kinase 1
- TIM
Translocase of the inner membrane
- TPR
Tetratricopeptide repeat
- TOM
Translocase of the outer membrane
- TSPO
Cholesterol translocator protein
- VDAC
Voltage-dependent anion channel
References
- 1.DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N Engl J Med. 2003;348(26):2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- 2.Lill R, Muhlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu Rev Biochem. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- 3.Newmeyer DD, Ferguson-Miller S. Mitochondria: releasing power for life and unleashing the machineries of death. Cell. 2003;112(4):481–490. doi: 10.1016/s0092-8674(03)00116-8. [DOI] [PubMed] [Google Scholar]
- 4.Reinders J, Zahedi RP, Pfanner N, Meisinger C, Sickmann A. Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J Proteome Res. 2006;5(7):1543–1554. doi: 10.1021/pr050477f. [DOI] [PubMed] [Google Scholar]
- 5.Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schonfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C. The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci US A. 2003;100(23):13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134(1):112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor SW, Fahy E, Zhang B, Glenn GM, Warnock DE, Wiley S, Murphy AN, Gaucher SP, Capaldi RA, Gibson BW, Ghosh SS. Characterization of the human heart mitochondrial proteome. Nat Biotechnol. 2003;21(3):281–286. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]
- 8.Bolender N, Sickmann A, Wagner R, Meisinger C, Pfanner N. Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 2008;9(1):42–49. doi: 10.1038/sj.embor.7401126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138(4):628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 11.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62(6):670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Young JC, Barral JM, Ulrich Hartl F. More than folding: localized functions of cytosolic chaperones. Trends Biochem Sci. 2003;28(10):541–547. doi: 10.1016/j.tibs.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Rudiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 1997;16(7):1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beckmann RP, Mizzen LE, Welch WJ. Interaction of Hsp 70 with newly synthesized proteins: implications for protein folding and assembly. Science. 1990;248(4957):850–854. doi: 10.1126/science.2188360. [DOI] [PubMed] [Google Scholar]
- 15.Frydman J, Nimmesgern E, Ohtsuka K, Hartl FU. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature. 1994;370(6485):111–117. doi: 10.1038/370111a0. [DOI] [PubMed] [Google Scholar]
- 16.Flaherty KM, DeLuca-Flaherty C, McKay DB. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature. 1990;346(6285):623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- 17.Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272(5268):1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci US A. 1991;88(7):2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qian YQ, Patel D, Hartl FU, McColl DJ. Nuclear magnetic resonance solution structure of the human Hsp40 (HDJ-1) J-domain. J Mol Biol. 1996;260(2):224–235. doi: 10.1006/jmbi.1996.0394. [DOI] [PubMed] [Google Scholar]
- 20.Wall D, Zylicz M, Georgopoulos C. The NH2-terminal 108 amino acids of the Escherichia coli DnaJ protein stimulate the AT-Pase activity of DnaK and are sufficient for lambda replication. J Biol Chem. 1994;269(7):5446–5451. [PubMed] [Google Scholar]
- 21.Craig EA, Huang P, Aron R, Andrew A. The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev Physiol Biochem Pharmacol. 2006;156:1–21. doi: 10.1007/s10254-005-0001-0. [DOI] [PubMed] [Google Scholar]
- 22.Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci. 2006;63(22):2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramos CH, Oliveira CL, Fan CY, Torriani IL, Cyr DM. Conserved central domains control the quaternary structure of type I and type II Hsp40 molecular chaperones. J Mol Biol. 2008;383(1):155–166. doi: 10.1016/j.jmb.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langer T, Lu C, Echols H, Flanagan J, Hayer MK, Hartl FU. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature. 1992;356(6371):683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- 25.Szabo A, Korszun R, Hartl FU, Flanagan J. A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. EMBO J. 1996;15(2):408–417. [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Qian X, Sha B. The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate. Structure. 2003;11(12):1475–1483. doi: 10.1016/j.str.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 27.Abdul KM, Terada K, Gotoh T, Hafizur RM, Mori M. Characterization and functional analysis of a heart-enriched DnaJ/Hsp40 homolog dj4/DjA4. Cell Stress Chaperones. 2002;7(2):156–166. doi: 10.1379/1466-1268(2002)007<0156:cafaoa>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terada K, Mori M. Human DnaJ homologs dj2 and dj3, and bag-1 are positive cochaperones of hsc70. J Biol Chem. 2000;275(32):24728–24734. doi: 10.1074/jbc.M002021200. [DOI] [PubMed] [Google Scholar]
- 29.Hohfeld J, Jentsch S. GrpE-like regulation of the hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J. 1997;16(20):6209–6216. doi: 10.1093/emboj/16.20.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sondermann H, Scheufler C, Schneider C, Hohfeld J, Hartl FU, Moarefi I. Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science. 2001;291(5508):1553–1557. doi: 10.1126/science.1057268. [DOI] [PubMed] [Google Scholar]
- 31.Takayama S, Bimston DN, Matsuzawa S, Freeman BC, Aime-Sempe C, Xie Z, Morimoto RI, Reed JC. BAG-1 modulates the chaperone activity of Hsp70/Hsc70. EMBO J. 1997;16(16):4887–4896. doi: 10.1093/emboj/16.16.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25(11):2519–2528. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabani M, McLellan C, Raynes DA, Guerriero V, Brodsky JL. HspBP1, a homologue of the yeast Fes1 and Sls1 proteins, is an Hsc70 nucleotide exchange factor. FEBS Lett. 2002;531(2):339–342. doi: 10.1016/s0014-5793(02)03570-6. [DOI] [PubMed] [Google Scholar]
- 34.Raviol H, Sadlish H, Rodriguez F, Mayer MP, Bukau B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 2006;25(11):2510–2518. doi: 10.1038/sj.emboj.7601139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flynn GC, Chappell TG, Rothman JE. Peptide binding and release by proteins implicated as catalysts of protein assembly. Science. 1989;245(4916):385–390. doi: 10.1126/science.2756425. [DOI] [PubMed] [Google Scholar]
- 36.McCarty JS, Buchberger A, Reinstein J, Bukau B. The role of ATP in the functional cycle of the DnaK chaperone system. J Mol Biol. 1995;249(1):126–137. doi: 10.1006/jmbi.1995.0284. [DOI] [PubMed] [Google Scholar]
- 37.Palleros DR, Reid KL, Shi L, Welch WJ, Fink AL. ATP-induced protein-Hsp70 complex dissociation requires K+ but not ATP hydrolysis. Nature. 1993;365(6447):664–666. doi: 10.1038/365664a0. [DOI] [PubMed] [Google Scholar]
- 38.Szabo A, Langer T, Schroder H, Flanagan J, Bukau B, Hartl FU. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc Natl Acad Sci US A. 1994;91(22):10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borkovich KA, Farrelly FW, Finkelstein DB, Taulien J, Lindquist S. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol Cell Biol. 1989;9(9):3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pearl LH, Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- 41.Freeman BC, Morimoto RI. The human cytosolic molecular chaperones hsp90, hsp70 (hsc70) and hdj-1 have distinct roles in recognition of a non-native protein and protein refolding. EMBO J. 1996;15(12):2969–2979. [PMC free article] [PubMed] [Google Scholar]
- 42.Jakob U, Lilie H, Meyer I, Buchner J. Transient interaction of Hsp90 with early unfolding intermediates of citrate synthase. Implications for heat shock in vivo. J Biol Chem. 1995;270(13):7288–7294. doi: 10.1074/jbc.270.13.7288. [DOI] [PubMed] [Google Scholar]
- 43.Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med (Maywood) 2003;228(2):111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 44.Prodromou C, Roe SM, O’Brien R, Ladbury JE, Piper PW, Pearl LH. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell. 1997;90(1):65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 45.Stebbins CE, Russo AA, Schneider C, Rosen N, Hartl FU, Pavletich NP. Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell. 1997;89(2):239–250. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 46.Obermann WM, Sondermann H, Russo AA, Pavletich NP, Hartl FU. In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J Cell Biol. 1998;143(4):901–910. doi: 10.1083/jcb.143.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Panaretou B, Prodromou C, Roe SM, O’Brien R, Ladbury JE, Piper PW, Pearl LH. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 1998;17(16):4829–4836. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meyer P, Prodromou C, Hu B, Vaughan C, Roe SM, Panaretou B, Piper PW, Pearl LH. Structural and functional analysis of the middle segment of hsp90: implications for ATP hydrolysis and client protein and cochaperone interactions. Mol Cell. 2003;11(3):647–658. doi: 10.1016/s1097-2765(03)00065-0. [DOI] [PubMed] [Google Scholar]
- 49.Lotz GP, Lin H, Harst A, Obermann WM. Aha1 binds to the middle domain of Hsp90, contributes to client protein activation, and stimulates the ATPase activity of the molecular chaperone. J Biol Chem. 2003;278(19):17228–17235. doi: 10.1074/jbc.M212761200. [DOI] [PubMed] [Google Scholar]
- 50.Meyer P, Prodromou C, Liao C, Hu B, Roe SM, Vaughan CK, Vlasic I, Panaretou B, Piper PW, Pearl LH. Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. EMBO J. 2004;23(6):1402–1410. doi: 10.1038/sj.emboj.7600141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Panaretou B, Siligardi G, Meyer P, Maloney A, Sullivan JK, Singh S, Millson SH, Clarke PA, Naaby-Hansen S, Stein R, Cramer R, Mollapour M, Workman P, Piper PW, Pearl LH, Prodromou C. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol Cell. 2002;10(6):1307–1318. doi: 10.1016/s1097-2765(02)00785-2. [DOI] [PubMed] [Google Scholar]
- 52.Harris SF, Shiau AK, Agard DA. The crystal structure of the carboxy-terminal dimerization domain of htpG, the Escherichia coli Hsp90, reveals a potential substrate binding site. Structure. 2004;12(6):1087–1097. doi: 10.1016/j.str.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 53.Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440(7087):1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McLaughlin SH, Smith HW, Jackson SE. Stimulation of the weak ATPase activity of human hsp90 by a client protein. J Mol Biol. 2002;315(4):787–798. doi: 10.1006/jmbi.2001.5245. [DOI] [PubMed] [Google Scholar]
- 55.Young JC, Hartl FU. Polypeptide release by Hsp90 involves ATP hydrolysis and is enhanced by the co-chaperone p23. EMBO J. 2000;19(21):5930–5940. doi: 10.1093/emboj/19.21.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen S, Prapapanich V, Rimerman RA, Honore B, Smith DF. Interactions of p60, a mediator of progesterone receptor assembly, with heat shock proteins hsp90 and hsp70. Mol Endocrinol. 1996;10(6):682–693. doi: 10.1210/mend.10.6.8776728. [DOI] [PubMed] [Google Scholar]
- 57.Kanelakis KC, Shewach DS, Pratt WB. Nucleotide binding states of hsp70 and hsp90 during sequential steps in the process of glucocorticoid receptor.hsp90 heterocomplex assembly. J Biol Chem. 2002;277(37):33698–33703. doi: 10.1074/jbc.M204164200. [DOI] [PubMed] [Google Scholar]
- 58.Morishima Y, Murphy PJ, Li DP, Sanchez ER, Pratt WB. Stepwise assembly of a glucocorticoid receptor.hsp90 heterocomplex resolves two sequential ATP-dependent events involving first hsp70 and then hsp90 in opening of the steroid binding pocket. J Biol Chem. 2000;275(24):18054–18060. doi: 10.1074/jbc.M000434200. [DOI] [PubMed] [Google Scholar]
- 59.Prodromou C, Panaretou B, Chohan S, Siligardi G, O’Brien R, Ladbury JE, Roe SM, Piper PW, Pearl LH. The AT-Pase cycle of Hsp90 drives a molecular ‘clamp’ via transient dimerization of the N-terminal domains. EMBO J. 2000;19(16):4383–4392. doi: 10.1093/emboj/19.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101(2):199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 61.Brinker A, Scheufler C, Von Der Mulbe F, Fleckenstein B, Herrmann C, Jung G, Moarefi I, Hartl FU. Ligand discrimination by TPR domains. Relevance and selectivity of EEVD-recognition in Hsp70 x Hop x Hsp90 complexes. J Biol Chem. 2002;277(22):19265–19275. doi: 10.1074/jbc.M109002200. [DOI] [PubMed] [Google Scholar]
- 62.Deshaies RJ, Koch BD, Werner-Washburne M, Craig EA, Schekman R. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature. 1988;332(6167):800–805. doi: 10.1038/332800a0. [DOI] [PubMed] [Google Scholar]
- 63.Sheffield WP, Shore GC, Randall SK. Mitochondrial precursor protein. Effects of 70-kilodalton heat shock protein on poly-peptide folding, aggregation, and import competence. J Biol Chem. 1990;265(19):11069–11076. [PubMed] [Google Scholar]
- 64.Terada K, Ohtsuka K, Imamoto N, Yoneda Y, Mori M. Role of heat shock cognate 70 protein in import of ornithine transcarbamylase precursor into mammalian mitochondria. Mol Cell Biol. 1995;15(7):3708–3713. doi: 10.1128/mcb.15.7.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lain B, Iriarte A, Martinez-Carrion M. Dependence of the folding and import of the precursor to mitochondrial aspartate aminotransferase on the nature of the cell-free translation system. J Biol Chem. 1994;269(22):15588–15596. [PubMed] [Google Scholar]
- 66.Lain B, Iriarte A, Mattingly JR, Jr, Moreno JI, Martinez-Carrion M. Structural features of the precursor to mitochondrial aspartate aminotransferase responsible for binding to hsp70. J Biol Chem. 1995;270(42):24732–24739. doi: 10.1074/jbc.270.42.24732. [DOI] [PubMed] [Google Scholar]
- 67.Kanazawa M, Terada K, Kato S, Mori M. HSDJ, a human homolog of DnaJ, is farnesylated and is involved in protein import into mitochondria. J Biochem. 1997;121(5):890–895. doi: 10.1093/oxfordjournals.jbchem.a021670. [DOI] [PubMed] [Google Scholar]
- 68.Terada K, Kanazawa M, Bukau B, Mori M. The human DnaJ homologue dj2 facilitates mitochondrial protein import and luciferase refolding. J Cell Biol. 1997;139(5):1089–1095. doi: 10.1083/jcb.139.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Artigues A, Iriarte A, Martinez-Carrion M. Binding to chaperones allows import of a purified mitochondrial precursor into mitochondria. J Biol Chem. 2002;277(28):25047–25055. doi: 10.1074/jbc.M203474200. [DOI] [PubMed] [Google Scholar]
- 70.Caplan AJ, Cyr DM, Douglas MG. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell. 1992;71(7):1143–1155. doi: 10.1016/s0092-8674(05)80063-7. [DOI] [PubMed] [Google Scholar]
- 71.Miller BR, Cumsky MG. Intramitochondrial sorting of the precursor to yeast cytochrome c oxidase subunit Va. J Cell Biol. 1993;121(5):1021–1029. doi: 10.1083/jcb.121.5.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Terada K, Ueda I, Ohtsuka K, Oda T, Ichiyama A, Mori M. The requirement of heat shock cognate 70 protein for mitochondrial import varies among precursor proteins and depends on precursor length. Mol Cell Biol. 1996;16(11):6103–6109. doi: 10.1128/mcb.16.11.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arco AD, Satrustegui J. New mitochondrial carriers: an overview. Cell Mol Life Sci. 2005;62(19–20):2204–2227. doi: 10.1007/s00018-005-5197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hines V, Brandt A, Griffiths G, Horstmann H, Brutsch H, Schatz G. Protein import into yeast mitochondria is accelerated by the outer membrane protein MAS70. EMBO J. 1990;9(10):3191–3200. doi: 10.1002/j.1460-2075.1990.tb07517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sollner T, Pfaller R, Griffiths G, Pfanner N, Neupert W. A mitochondrial import receptor for the ADP/ATP carrier. Cell. 1990;62(1):107–115. doi: 10.1016/0092-8674(90)90244-9. [DOI] [PubMed] [Google Scholar]
- 76.Suzuki H, Maeda M, Mihara K. Characterization of rat TOM70 as a receptor of the preprotein translocase of the mitochondrial outer membrane. J Cell Sci. 2002;115(Pt 9):1895–1905. doi: 10.1242/jcs.115.9.1895. [DOI] [PubMed] [Google Scholar]
- 77.Brix J, Ziegler GA, Dietmeier K, Schneider-Mergener J, Schulz GE, Pfanner N. The mitochondrial import receptor Tom70: identification of a 25 kDa core domain with a specific binding site for preproteins. J Mol Biol. 2000;303(4):479–488. doi: 10.1006/jmbi.2000.4120. [DOI] [PubMed] [Google Scholar]
- 78.Ryan MT, Muller H, Pfanner N. Functional staging of ADP/ATP carrier translocation across the outer mitochondrial membrane. J Biol Chem. 1999;274(29):20619–20627. doi: 10.1074/jbc.274.29.20619. [DOI] [PubMed] [Google Scholar]
- 79.Sollner T, Rassow J, Wiedmann M, Schlossmann J, Keil P, Neupert W, Pfanner N. Mapping of the protein import machinery in the mitochondrial outer membrane by crosslinking of translocation intermediates. Nature. 1992;355(6355):84–87. doi: 10.1038/355084a0. [DOI] [PubMed] [Google Scholar]
- 80.Truscott KN, Wiedemann N, Rehling P, Muller H, Meisinger C, Pfanner N, Guiard B. Mitochondrial import of the ADP/ATP carrier: the essential TIM complex of the intermembrane space is required for precursor release from the TOM complex. Mol Cell Biol. 2002;22(22):7780–7789. doi: 10.1128/MCB.22.22.7780-7789.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wiedemann N, Pfanner N, Ryan MT. The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J. 2001;20(5):951–960. doi: 10.1093/emboj/20.5.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Young JC, Hoogenraad NJ, Hartl FU. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 2003;112(1):41–50. doi: 10.1016/s0092-8674(02)01250-3. [DOI] [PubMed] [Google Scholar]
- 83.Pfanner N, Neupert W. Distinct steps in the import of ADP/ATP carrier into mitochondria. J Biol Chem. 1987;262(16):7528–7536. [PubMed] [Google Scholar]
- 84.Fan AC, Bhangoo MK, Young JC. Hsp90 functions in the targeting and outer membrane translocation steps of Tom70-mediated mitochondrial import. J Biol Chem. 2006;281(44):33313–33324. doi: 10.1074/jbc.M605250200. [DOI] [PubMed] [Google Scholar]
- 85.Richter K, Buchner J. Hsp90: chaperoning signal transduction. J Cell Physiol. 2001;188(3):281–290. doi: 10.1002/jcp.1131. [DOI] [PubMed] [Google Scholar]
- 86.Rehling P, Model K, Brandner K, Kovermann P, Sickmann A, Meyer HE, Kuhlbrandt W, Wagner R, Truscott KN, Pfanner N. Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science. 2003;299(5613):1747–1751. doi: 10.1126/science.1080945. [DOI] [PubMed] [Google Scholar]
- 87.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5(10):761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 88.Bhangoo MK, Tzankov S, Fan AC, Dejgaard K, Thomas DY, Young JC. Multiple 40-kDa heat-shock protein chaperones function in Tom70-dependent mitochondrial import. Mol Biol Cell. 2007;18(9):3414–3428. doi: 10.1091/mbc.E07-01-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tzankov S, Wong MJ, Shi K, Nassif C, Young JC. Functional divergence between co-chaperones of Hsc70. J Biol Chem. 2008;283(40):27100–27109. doi: 10.1074/jbc.M803923200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zara V, Ferramosca A, Robitaille-Foucher P, Palmieri F, Young JC. Mitochondrial carrier protein biogenesis: role of the chaperones Hsc70 and Hsp90. Biochem J. 2009;419(2):369–375. doi: 10.1042/BJ20082270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kurz M, Martin H, Rassow J, Pfanner N, Ryan MT. Biogenesis of Tim proteins of the mitochondrial carrier import pathway: differential targeting mechanisms and crossing over with the main import pathway. Mol Biol Cell. 1999;10(7):2461–2474. doi: 10.1091/mbc.10.7.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chan NC, Likic VA, Waller RF, Mulhern TD, Lithgow T. The C-terminal TPR domain of Tom70 defines a family of mitochondrial protein import receptors found only in animals and fungi. J Mol Biol. 2006;358(4):1010–1022. doi: 10.1016/j.jmb.2006.02.062. [DOI] [PubMed] [Google Scholar]
- 93.Yamamoto H, Fukui K, Takahashi H, Kitamura S, Shiota T, Terao K, Uchida M, Esaki M, Nishikawa S, Yoshihisa T, Yamano K, Endo T. Roles of Tom70 in import of presequence-containing mitochondrial proteins. J Biol Chem. 2009;284(46):31635–31646. doi: 10.1074/jbc.M109.041756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Humphries AD, Streimann IC, Stojanovski D, Johnston AJ, Yano M, Hoogenraad NJ, Ryan MT. Dissection of the mitochondrial import and assembly pathway for human Tom40. J Biol Chem. 2005;280(12):11535–11543. doi: 10.1074/jbc.M413816200. [DOI] [PubMed] [Google Scholar]
- 95.Keil P, Weinzierl A, Kiebler M, Dietmeier K, Sollner T, Pfanner N. Biogenesis of the mitochondrial receptor complex. Two receptors are required for binding of MOM38 to the outer membrane surface. J Biol Chem. 1993;268(26):19177–19180. [PubMed] [Google Scholar]
- 96.Chou CH, Lee RS, Yang-Yen HF. An internal EELD domain facilitates mitochondrial targeting of Mcl-1 via a Tom70-dependent pathway. Mol Biol Cell. 2006;17(9):3952–3963. doi: 10.1091/mbc.E06-04-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Otera H, Taira Y, Horie C, Suzuki Y, Suzuki H, Setoguchi K, Kato H, Oka T, Mihara K. A novel insertion pathway of mitochondrial outer membrane proteins with multiple transmembrane segments. J Cell Biol. 2007;179(7):1355–1363. doi: 10.1083/jcb.200702143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rone MB, Liu J, Blonder J, Ye X, Veenstra TD, Young JC, Papadopoulos V. Targeting and insertion of the cholesterol-binding translocator protein into the outer mitochondrial membrane. Biochemistry. 2009;48(29):6909–6920. doi: 10.1021/bi900854z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lin W, Kang UJ. Characterization of PINK1 processing, stability, and subcellular localization. J Neurochem. 2008;106(1):464–474. doi: 10.1111/j.1471-4159.2008.05398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, Albanese A, Nussbaum R, Gonzalez-Maldonado R, Deller T, Salvi S, Cortelli P, Gilks WP, Latchman DS, Harvey RJ, Dallapiccola B, Auburger G, Wood NW. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304(5674):1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 101.Weihofen A, Ostaszewski B, Minami Y, Selkoe DJ. Pink1 Parkinson mutations, the Cdc37/Hsp90 chaperones and Parkin all influence the maturation or subcellular distribution of Pink1. Hum Mol Genet. 2008;17(4):602–616. doi: 10.1093/hmg/ddm334. [DOI] [PubMed] [Google Scholar]
- 102.Silvestri L, Caputo V, Bellacchio E, Atorino L, Dallapiccola B, Valente EM, Casari G. Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum Mol Genet. 2005;14(22):3477–3492. doi: 10.1093/hmg/ddi377. [DOI] [PubMed] [Google Scholar]
- 103.Takatori S, Ito G, Iwatsubo T. Cytoplasmic localization and proteasomal degradation of N-terminally cleaved form of PINK1. Neurosci Lett. 2008;430(1):13–17. doi: 10.1016/j.neulet.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 104.Yano M, Terada K, Mori M. AIP is a mitochondrial import mediator that binds to both import receptor Tom20 and preproteins. J Cell Biol. 2003;163(1):45–56. doi: 10.1083/jcb.200305051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bell DR, Poland A. Binding of aryl hydrocarbon receptor (AhR) to AhR-interacting protein. The role of hsp90. J Biol Chem. 2000;275(46):36407–36414. doi: 10.1074/jbc.M004236200. [DOI] [PubMed] [Google Scholar]
- 106.Wu Y, Sha B. Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nat Struct Mol Biol. 2006;13(7):589–593. doi: 10.1038/nsmb1106. [DOI] [PubMed] [Google Scholar]
- 107.Beddoe T, Bushell SR, Perugini MA, Lithgow T, Mulhern TD, Bottomley SP, Rossjohn J. A biophysical analysis of the tetratricopeptide repeat-rich mitochondrial import receptor, Tom70, reveals an elongated monomer that is inherently flexible, unstable, and unfolds via a multistate pathway. J Biol Chem. 2004;279(45):46448–46454. doi: 10.1074/jbc.M405639200. [DOI] [PubMed] [Google Scholar]
- 108.Bushell SR, Bottomley SP, Rossjohn J, Beddoe T. Tracking the unfolding pathway of a multirepeat protein via tryptophan scanning: evidence of localized instability in the mitochondrial import receptor Tom70. J Biol Chem. 2006;281(34):24345–24350. doi: 10.1074/jbc.M602966200. [DOI] [PubMed] [Google Scholar]
- 109.Mills RD, Trewhella J, Qiu TW, Welte T, Ryan TM, Hanley T, Knott RB, Lithgow T, Mulhern TD. Domain organization of the monomeric form of the Tom70 mitochondrial import receptor. J Mol Biol. 2009;388(5):1043–1058. doi: 10.1016/j.jmb.2009.03.070. [DOI] [PubMed] [Google Scholar]
- 110.Li J, Qian X, Hu J, Sha B. Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading. J Biol Chem. 2009;284(35):23852–23859. doi: 10.1074/jbc.M109.023986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fan AC, Gava LM, Ramos CH, Young JC. Human mitochondrial import receptor Tom70 functions as a monomer. Biochem J. 2010;429(3):553–563. doi: 10.1042/BJ20091855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ott M, Norberg E, Zhivotovsky B, Orrenius S. Mitochondrial targeting of tBid/Bax: a role for the TOM complex? Cell Death Differ. 2009;16(8):1075–1082. doi: 10.1038/cdd.2009.61. [DOI] [PubMed] [Google Scholar]
- 113.Setoguchi K, Otera H, Mihara K. Cytosolic factor- and TOM-independent import of C-tail-anchored mitochondrial outer membrane proteins. EMBO J. 2006;25(24):5635–5647. doi: 10.1038/sj.emboj.7601438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Edlich F, Erdmann F, Jarczowski F, Moutty MC, Weiwad M, Fischer G. The Bcl-2 regulator FKBP38-calmodulin-Ca2+ is inhibited by Hsp90. J Biol Chem. 2007;282(21):15341–15348. doi: 10.1074/jbc.M611594200. [DOI] [PubMed] [Google Scholar]
- 115.Shirane M, Nakayama KI. Inherent calcineurin inhibitor FKBP38 targets Bcl-2 to mitochondria and inhibits apoptosis. Nat Cell Biol. 2003;5(1):28–37. doi: 10.1038/ncb894. [DOI] [PubMed] [Google Scholar]