Abstract
Purpose
To test the efficacy of 7T MRS for in-vivo detection of 2-hydroxyglutarate (2HG) in brain tumors.
Methods
The subecho times of point-resolved spectroscopy (PRESS) were optimized at 7T, with density-matrix simulations and phantom validation, for improving the 2HG signal selectivity with respect to the neighboring resonances of GABA, glutamate (Glu), and glutamine (Gln). MRS data were acquired from 12 subjects with gliomas in vivo and analyzed with LCModel using calculated basis spectra. Metabolite levels were quantified using unsuppressed short-TE water as reference.
Results
The PRESS TE was optimized as TE=78 ms (TE1=58 ms and TE2=20 ms), at which the 2HG 2.25 ppm resonance appeared as a temporally-maximum inverted narrow peak while the GABA, Glu and Gln resonances between 2.2 and 2.5 ppm were all positive peaks. The PRESS TE=78 ms method offered improved discrimination of 2HG from Glu, Gln and GABA when compared with short-TE MRS. 2HG was detected in all patients enrolled in the study, the estimated 2HG concentrations ranging from 1.0 to 6.2 mM, with percentage standard deviation of 2 - 7%.
Conclusion
Data indicate that the optimized MRS provides good selectivity of 2HG from other metabolite signals and may confer reliable in-vivo detection of 2HG at relatively low concentrations.
Keywords: 2-Hydroxyglutarate (2HG), 1H MRS, 7T, PRESS (point-resolved spectroscopy), human brain tumors, gliomas, IDH (isocitrate dehydrogenase)
INTRODUCTION
Mutations in the isocitrate dehydrogenase (IDH) 1 and 2 family of enzymes in the tricarboxylic acid cycle cause NADPH-dependent reduction of α-ketoglutarate to 2-Hydroxyglutarate (2HG), resulting in a two-to-three order-of-magnitude increase in the cellular 2HG concentration, which otherwise is present at very low levels in normal conditions (1-3) . This coincides with loss of catalytic activities of mutated IDH to convert isocitrate to α-ketoglutarate (1,4). These modifications of metabolic activities have become a hallmark of cancer and are strongly implicated in tumorigenesis (5). Moreover, IDH mutations, which occur in the majority of World Health Organization grade II and III gliomas and secondary glioblastomas, are associated with longer overall survival compared to IDH wild-type tumors (6,7). Noninvasive in vivo detection of 2HG may therefore provide new insights into tumor transformation and for development of therapeutic targets.
The 2HG molecule has five non-exchangeable protons, which resonate at 4.02, 2.27, 2.22, 1.98 and 1.83 ppm (see Fig. 1a) (8). These protons are J coupled and give rise to multiplets at 4.02 ppm, 2.25 ppm, and ~1.90 ppm in in-vivo situations. Precise measurement of 2HG by standard short-TE MRS is challenging due to extensive spectral overlaps. The 2HG 4.02 ppm signal is obscured by the creatine (Cr) 3.92 ppm and myo-inositol (mI) 4.06 ppm resonances. Detection of the 2HG 1.9 ppm signal is challenging due to its proximity to the N-acetylaspartate (NAA) singlet at 2.01 ppm. The 2HG signal at 2.25-ppm, which is larger than the other 2HG signals, is overlapped with the resonances of glutamate (Glu), glutamine (Gln) and γ-aminobutyric acid (GABA). Several studies have recently reported in vivo detection of 2HG in patients with gliomas at 3T, achieved by means of optimized-TE point-resolved spectroscopy (PRESS) (9,10), J-difference editing (9,11), correlation spectroscopy (11), and standard short-TE MRS (12). Given the role of 2HG as a diagnostic and prognostic imaging biomarker, in-vivo measurement of 2HG is of high significance, but accurate evaluation of 2HG in vivo remains challenging, especially when 2HG elevation is moderate.
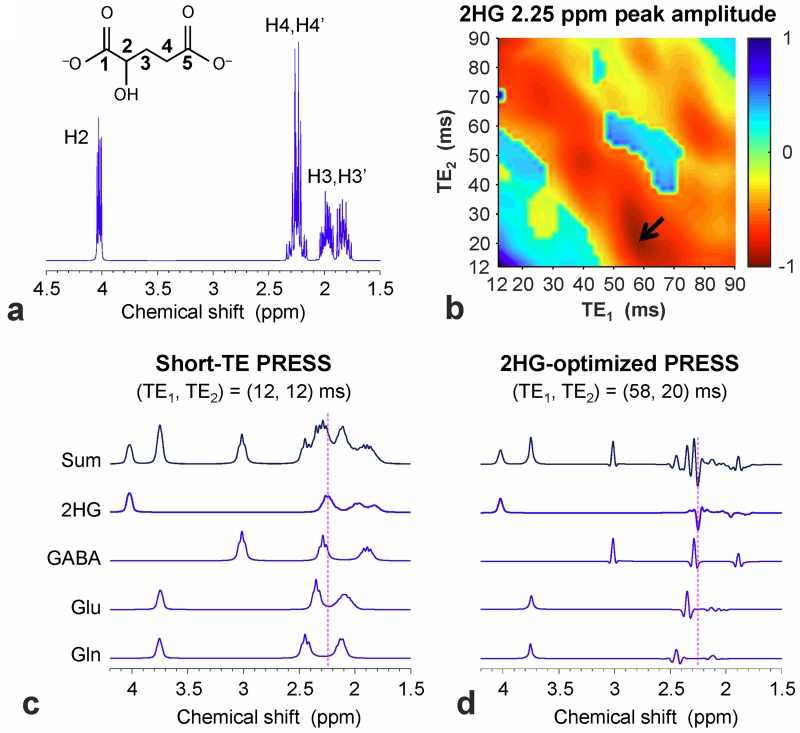
Figure 1.
(a) A 1H MR absorption spectrum of 2HG, numerically calculated for single-pulse acquisition at 7T, is shown with the H2, H3 and H4 proton labels. The line broadening corresponds to a singlet linewidth of 1 Hz. (b) The 2HG 2.25 ppm peak amplitude, obtained from calculated PRESS spectra, was color mapped vs. TE1 and TE2 between 12 and 90 ms, ignoring T2 relaxation effects. An arrow indicates the subecho time set used for 2HG detection in this study. (c, d) Calculated PRESS spectra of 2HG, GABA, Glu and Gln at equal concentrations for (TE1, TE2) = (12, 12) and (58, 20) ms. A vertical dotted line is drawn at 2.25 ppm. In (b), (c) and (d), the spectra were broadened to a singlet linewidth of 9 Hz.
High-field MRS benefits from enhancement of spectral resolution and signal gain. At 7T, which is increasingly available for patient studies, short-TE MRS is widely used with the advantage of minimal T2 signal loss. However, the spectral analysis for 2HG may be complicated by the presence of the broad complex signals from macromolecules (MM) at short TE. Since the MM signals diminish rapidly with TE, a long-TE approach can be employed as an alternative, with the additional advantage that TE optimization may improve discrimination between J-coupled spin resonances. Specifically, as shown in prior 7T MRS studies (13-15), the spectral resolution of the GABA, Glu and Gln resonances between 2.2 and 2.5 ppm can be improved at carefully-chosen long echo times, largely due to the effects of multiplet narrowing and in part due to the spectral simplification arising from the attenuated MM signals. Preliminary studies of 2HG detection using long TEs (78 - 110 ms) at 7T were recently reported (16-19). Here we report a 1H MRS method for detecting 2HG in gliomas at 7T, which provides improved selectivity of 2HG against neighboring resonances, compared to short-TE approaches. A long PRESS echo time is developed for good differentiation of the 2HG 2.25 ppm resonance from the Glu, Gln and GABA signals. Preliminary in-vivo data from 12 glioma patients are presented.
METHODS
To improve the detectability of 2HG signal at 2.25 ppm with respect to the neighboring resonances of GABA, Glu, and Gln, the echo time of PRESS was optimized with density-matrix simulations that incorporated experimental RF and gradient pulse waveforms. 2D volume-localized spectra were calculated for the first and second subecho times of PRESS, TE1 and TE2, in the range of 12 - 90 ms (with 1 ms increments), using a product-operator based transformation-matrix algorithm described in a prior study (9). Volume localization was obtained with an 8.8 ms 90° RF pulse (bandwidth = 4.7 kHz) and two 12 ms 180° RF pulses (bandwidth = 1.4 kHz) at RF field intensity (B1) of 15 µT. Published chemical shift and J coupling constants were used for the simulations (8,20,21). The computer simulation was programmed with MATLAB® (The MathWorks, Inc., Natick, MA).
Twelve patients with brain tumors were recruited for the study, of which 6 patients had post-scan surgical biopsies and 6 had mass lesions clinically consistent with a grade 2 or 3 glioma. The tumor grades and types were determined by the histopathological analysis of biopsied tissue, according to the World Health Organization (WHO) criteria. IDH mutational status was obtained with immunohistochemical analysis and genomic sequencing. Written informed consent was obtained prior to the MR scans. The study was approved by the Institutional Review Board at University of Texas Southwestern Medical Center.
MR experiments were carried out on a human whole-body 7T MR scanner (Philips Medical Systems, Cleveland, OH) using a quadrature transmit (4 kW RF amplifier) and 16-channel receive head coil (Nova Medical, Wilmington, MA). The optimized PRESS sequence was tested in two spherical phantoms (6 cm diameter; pH = 7.0); one with 2HG (12 mM) and glycine (Gly) (20 mM), and the other with 2HG (5 mM), GABA (1 mM), Glu (5 mM), Gln (5 mM), and Cr (10 mM). Data were acquired from a 20 × 20 × 20 mm3 voxel with TE = 78, TR = 9 s, and number of signal averages (NSA) = 64. For in vivo experiments, T2-weighted fluid attenuated inversion recovery images (T2w-FLAIR; TR/TE/TI = 11,000/93/2800 ms; field of view (FOV) = 230 × 230 mm2; slice thickness of 5 mm; 20 transverse slices) were acquired to identify tumors. Water-suppressed PRESS TE = 78 ms single-voxel data were obtained from a voxel (size 4.5 - 15.6 mL) within the tumor mass, with NSA from 64 to 256. Unsuppressed water was acquired with the PRESS TE = 78 ms sequence for eddy current compensation. The scan parameters included a TR of 2.5 sec, a spectral width of 5000 Hz, and 4096 complex points. The carrier frequency of the PRESS RF pulses was set to 2.5 ppm. A vendor-supplied four-pulse scheme was used for water suppression. An unsuppressed STEAM (stimulated-echo acquisition mode) water signal was acquired, with TE = 13 ms and TR = 2.5 s, from the same voxel for use as reference in metabolite quantification. The target B1 (15 μT) was obtained for the PRESS localized volumes using a vendor-supplied B1-calibration method (22). Surgical cavities, areas of intratumoral hemorrhage, cystic changes, and necrosis, identified on T2w-FLAIR anatomical images, were excluded from the tumor voxels.
The 16-channel data were combined using a scanner built-in function in 7 patients and using an in-house Matlab script in 5 patients which was developed according to a published algorithm (23). Free-induction decay (FID) signals were multiplied by a 1-Hz exponential function to prior to Fourier transformation. Spectral fitting was undertaken with LCModel (Ver 6.3; Stephen Provencher, Oakville, Canada) (24), using numerically-calculated basis spectra of 20 metabolites; 2HG, GABA, Glu, Gln, NAA, N-acetylaspartylglutamate (NAAG), tCr (creatine + phosphocreatine), tCho (glycerophosphocholine + phosphocholine), glutathione, myo-inositol, glycine, lactate, taurine, scyllo-inositol, aspartate, phosphoethanolamine, ethanolamine, citrate, succinate, and threonine. The spectral fitting was conducted between 0.5 and 4.1 ppm. For validation purpose, additional LCModel spectral fitting was performed without 2HG in the basis set. Cramér–Rao lower bounds (CRLB) and individual metabolite fits were obtained from LCModel. For evaluation of the Glu, tNAA, tCr, and tCho concentrations, the transverse relaxation effects were corrected using published healthy-brain T2 values of 120, 150, 110 and 140 ms, respectively (14,25). The T2 values of 2HG, GABA, and Gln were assumed to be equal to the Glu T2. Metabolite concentrations were calculated with reference to water at 43 M, whose signal strength was evaluated from LCModel fitting using a STEAM water basis set. Statistical tests were performed for potential correlation of 2HG levels with Glu, Gln, GABA, tCho, and tNAA concentrations, using Minitab® (Minitab, Inc., State College, Pennsylvania, USA). Statistical significance was declared at p = 0.05. Data are presented as mean ± standard deviation.
RESULTS
The PRESS volume-localized density-matrix simulation indicated that the 2HG 2.25 ppm signal from the strongly-coupled C4-proton spins depends on the PRESS subecho times, TE1 and TE2, extensively (Fig. 1b). The 2.25 ppm signal, which was positive at short TE (= TE1 + TE2 < 40 ms), shrank substantially at a TE value of ~60 ms, then grew larger but negative with increasing TE (Sup. Fig. S1). The dependence of the 2HG signal intensity and pattern on TE1 and TE2 was asymmetric with respect to the TE1 = TE2 line. At PRESS (TE1, TE2) = (58, 20) ms, the 2HG 2.25 ppm signal was temporally maximum with negative polarity. For an identical volume, ignoring T2 relaxation effects, the 2HG 2.25-ppm signal amplitude at TE = 78 ms was 56% with respect to a 90° pulse-and-acquire sequence (data not shown) and 92% compared to PRESS TE = 24 ms (Fig. 1b-d), which was the shortest possible TE in the simulation. Most importantly, using the TE 78ms PRESS sequence resulted in apparent line narrowing of the 2HG multiplet. For broadening to a singlet linewidth of 9 Hz, the width of the 2HG 2.25 ppm multiplet using the TE 78ms PRESS sequence was 8.2 Hz, much smaller than those of pulse-acquisition (23.7 Hz) and PRESS TE = 24 ms (31.2 Hz). Apparent multiplet narrowing also occurred to the signals of GABA, Glu and Gln between 2.2 and 2.5 ppm, assuring improved spectral resolution between 2HG, GABA, Glu and Gln (Fig. 1c,d). The 2HG and GABA signals, which are very proximate to each other, were predicted to be separable even when the peaks are broadened (Sup. Fig. S2). Taken together, the PRESS echo time was optimized as TE = 78 ms (TE1 = 58 ms and TE2 =20 ms) for 2HG detection in the present study.
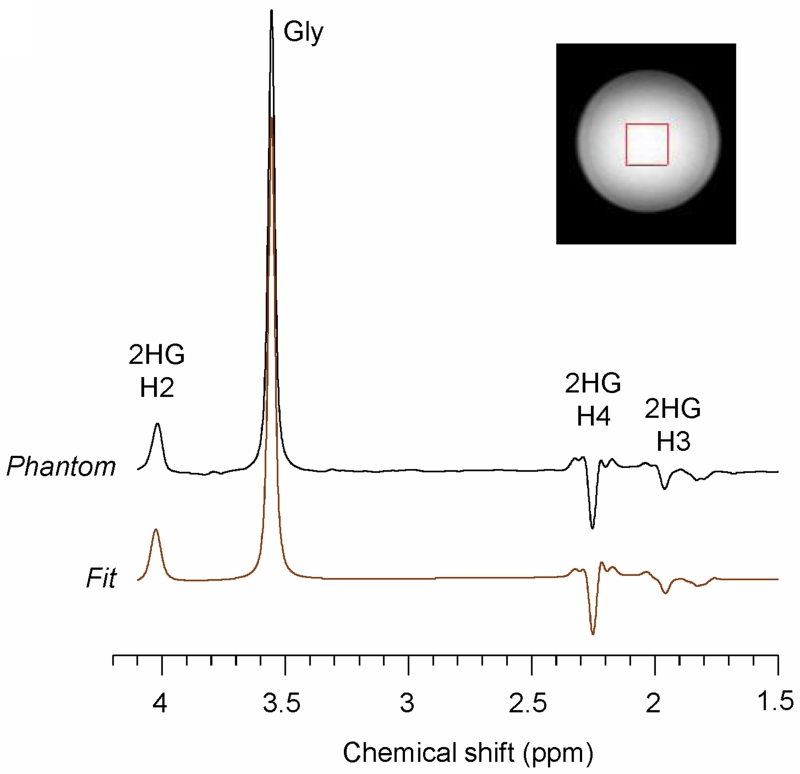
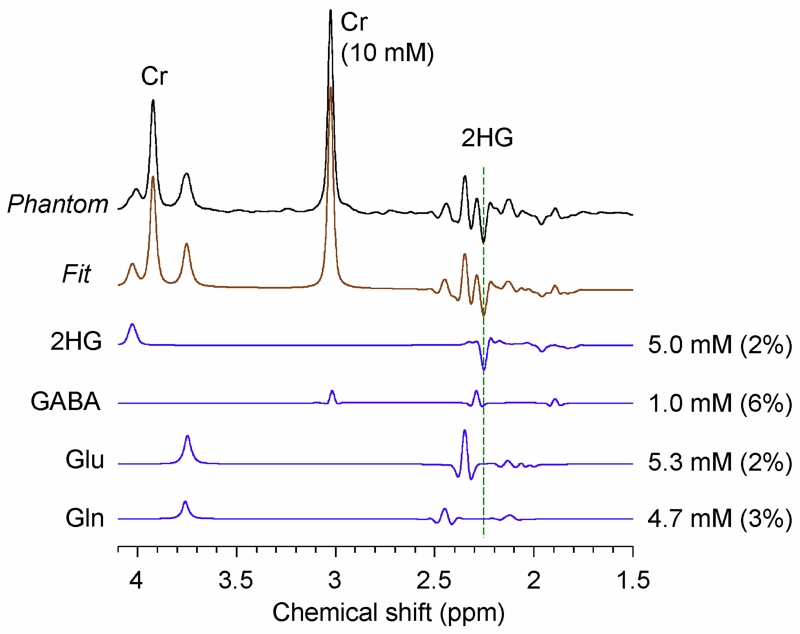
The 2HG-optimized PRESS TE = 78 ms method was tested in a phantom solution with 2HG and Gly (Fig. 2). The signal strengths and patterns of the 2HG resonances were all in good agreement between calculation and experiment. The phantom data showed an inverted peak of 2HG at 2.25 ppm, as predicted by the simulation. The prepared 2HG-to-Gly concentration ratio of 0.6 (=12/20) was reproduced within experimental errors (~5%), following the correction for the transverse relaxation effects in the solution using experimentally-measured phantom T2 values (Gly T2 = 1500 ms and 2HG T2 = 650 ms). Figure 3 shows a spectrum from a composite phantom with 2HG, GABA, Glu and Gln, together with the LCModel fit. The phantom spectrum and the fit agreed closely. The prepared concentrations were well reproduced by the spectral fitting, which resulted in concentrations of 2HG, GABA, Glu, and Gln (with respect to Cr at 10 mM) as 5.0, 1.0, 5.3 and 4.7 mM, respectively, after correction for the T2 relaxation effects in 2HG, GABA, Glu and Glu using the phantom 2HG T2 value. Also, we tested the 2HG-optimized PRESS sequence in a healthy brain. The data from three regions (frontal, occipital and mid-brain) showed the expected metabolite signals without evidence of 2HG (Sup. Fig. S3).
Figure 2.
An in vitro spectrum from 2HG (12 mM) and glycine (20 mM), obtained with PRESS at TE = 78 ms (TE1 = 58 ms, TE2 = 20), is shown together with an LCModel fit. Data were obtained from a 20 × 20 × 20 mm3 voxel, with TR = 9 s and NSA = 64. The spectrum was broadened to glycine linewidth of 9 Hz prior to the spectral fitting. A vertical dotted line is drawn at 2.25 ppm.
Figure 3.
A PRESS TE = 78 ms spectrum from a composite phantom solution, containing 2HG (5 mM), GABA (1 mM), Glu (5 mM), Gln (5 mM), and Cr (10 mM), is shown together with LCModel outputs. LCModel-returned signals of 2HG, GABA, Glu and Gln are shown together with the estimated concentrations and CRLB values. Data were obtained from a 20 × 20 × 20 mm3 voxel, with TR = 9 s and NSA = 64. The spectrum was broadened to Cr linewidth of 9 Hz prior to the spectral fitting. A vertical dotted line is drawn at 2.25 ppm.
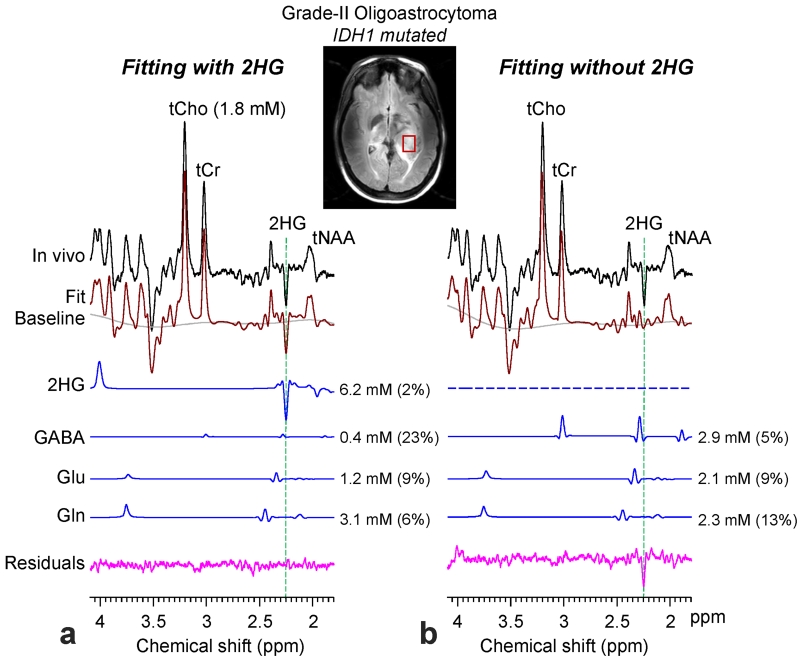
Figure 4 presents an in-vivo spectrum, obtained with the 2HG-optimized PRESS TE = 78 ms method, from a subject with IDH1-mutated oligoastrocytoma, in which T2w-FLAIR imaging identified a left mesial temporal lobe mass. The spectrum showed a large inverted peak at 2.25 ppm, which was closely reproduced by the LCModel fit when using a basis set with 2HG (Fig. 4a). The 2HG level was estimated to be 6.2 mM, with high precision (CRLB = 2%). The estimates of GABA, Glu, and Gln were 0.4, 1.2 and 3.1 mM, with CRLB at 23%, 9% and 6%, respectively. The concentration of tCho, whose level in healthy brain may be 1 - 1.5 mM (14,26), was moderately increased in this tumor (1.8 mM). When 2HG was excluded from the basis set, the resulting fit was substantially different from the in-vivo spectrum at ~2.25 and ~4.0 ppm (Fig. 4b). The fitting resulted in large residual signals at 2.25 and ~4.0 ppm and some change in the LCModel baseline at > 3.5 ppm, indicating that the signals at the two spectral locations were primarily attributed to the 2HG H4 and H2 resonances. The absence of 2HG in the basis set influenced the estimation of other metabolites, such as GABA and Glu whose major signals are close to 2.25 ppm.
Figure 4.
An in-vivo spectrum from an IDH1 mutated grade-II oligoastrocytoma is shown together with LCModel fits, baseline and residuals from fitting with 2HG (left) or without 2HG (right) in the basis set. The signals of 2HG, GABA, Glu and Gln are shown with the estimated concentrations and CRLBs in parenthesis. The voxel size was 2.0 × 1.5 × 1.5 cm3 (TR = 2.5 s, NSA = 128). Vertical dashed lines are drawn at 2.25 ppm to indicate the location of the 2HG C4-proton resonance.
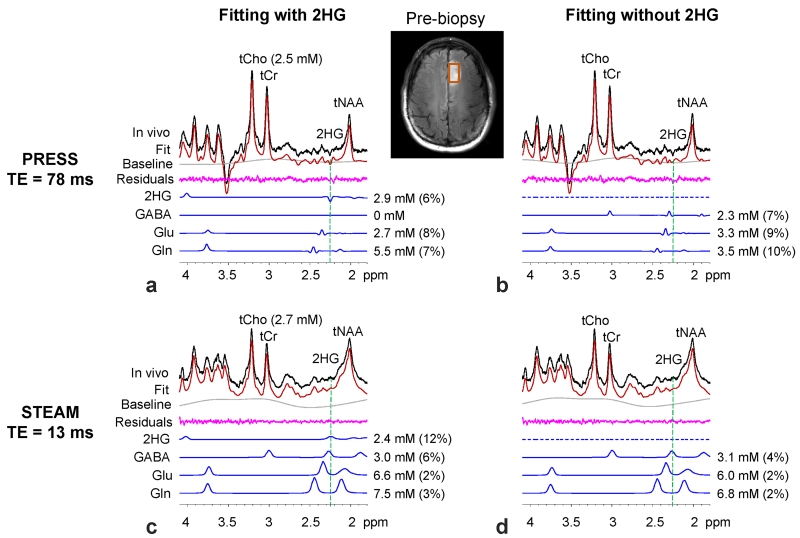
The 2HG detectability of the PRESS TE = 78 ms sequence was compared with short-TE MRS. Figure 5 presents in-vivo spectra from a radiography-suggested glioma patient, obtained with the PRESS and STEAM TE = 13 ms methods. The PRESS spectrum showed a relatively small but clearly discernible negative peak at 2.25 ppm while the STEAM data had a smoothly-varying signal pattern between 2.1 - 2.4 ppm, most likely due to the overlaps of the broad peaks from several metabolites, macromolecule and lipids. In the spectral fitting with 2HG in the basis set, the PRESS and STEAM spectra were both well reproduced by LCModel fits, without showing large residuals at 2.25 ppm (Fig 5a,c). Following correction for T2 relaxation effects, the estimated 2HG concentration was slightly different between PRESS and STEAM (2.9 vs. 2.4 mM) while the tCho estimates were about the same (2.5 vs. 2.7 mM). The CRLB of 2HG was considerably smaller in PRESS than in short-TE STEAM (6% vs. 12%). When 2HG was removed from the basis set, the fitting outputs from the PRESS and STEAM data were quite different. The without-2HG fitting of the PRESS data left an inverted peak at 2.25 ppm in the residual spectrum. In contrast, in the STEAM data, the residuals at ~2.25 ppm were featureless for both the with-2HG and without-2HG fittings, indicating that the 2HG signal from the with-2HG fitting was not very selective. In addition, estimates of GABA, Glu and Gln were influenced by the removal of 2HG from the basis set in both PRESS and STEAM.
Figure 5.
In-vivo spectra from a pre-biopsy patient, obtained with PRESS TE = 78 ms (upper panel) and with STEAM (TE, TM) = (13, 17) ms (lower panel), are shown together with LCModel outputs from with-2HG (left) and without-2HG (right) fittings. The voxel size was 2.0 × 1.5 × 1.5 cm3, with TR = 2.5 s and NSA = 128 (PRESS) and 256 (STEAM). Estimated metabolite concentrations are shown with CRLBs in parenthesis.
Table 1 presents the estimated concentrations and CRLBs of 2HG, GABA, Glu, Gln, tCho and tNAA for the 12 tumor patients enrolled in the study (6 IDH mutated and 6 IDH status unknown). For these 12 patients, the mean concentration of 2HG was 3.1 ± 1.7 mM with mean CRLB of 5 ± 2%. For the 6 patients with biopsy-proven IDH mutation, the mean 2HG level was 3.3 ± 2.0 mM, ranging from 1.0 to 6.2 mM. Similar 2HG concentrations were obtained in the 6 pre-biopsy patients, with a mean 2HG level at 2.9 ± 1.5 mM, ranging from 1.0 to 4.4 mM. In addition, we evaluated potential correlation of 2HG levels with tumor grade and the concentrations of tCho, tNAA, Glu, Gln, and GABA. Significant correlation was observed between 2HG and Gln with p = 0.03.
Table 1.
For the 12 glioma patients (6 IDH mutated and 6 no biopsy) enrolled in the study, the in-vivo estimated concentrations (mM) and CRLBs (% in parenthesis) of 2HG and other metabolites are tabulated together with the voxel size. G2 and G3 denote grade-II and grade-III gliomas respectively. IDH1 and IDH2 denote IDH1-mutated and IDH2-mutated gliomas respectively. O, oligodendroglioma; OA, oligoastrocytoma; A, astrocytoma; AA, anaplastic astrocytoma; AOA, anaplastic oligoastrocytoma.
| Voxel (mL) |
2HG | GABA | Glu | Gln | tCho | tNAA | |
|---|---|---|---|---|---|---|---|
|
|
|||||||
| G2, OA, IDH1 | 4.5 | 6.2 (2) | 0.4 (23) | 1.2 (9) | 3.1 (6) | 1.8 (1) | 1.2 (4) |
| G2, O, IDH2 | 6.4 | 4.6 (3) | 0 | 1.5 (9) | 2.1 (11) | 1.5 (1) | 2.4 (2) |
| G2, A, IDH1 | 15.6 | 1.2 (7) | 0.3 (29) | 2.1 (5) | 0.5 (36) | 2.4 (1) | 3.1 (2) |
| G2, A, IDH1 | 8 | 1.0 (6) | 0.2 (25) | 1.0 (7) | 1.3 (9) | 1.3 (1) | 1.7 (2) |
| G3, AA, IDH1 | 8 | 3.8 (5) | 0 | 2.1 (5) | 3.5 (4) | 2.6 (1) | 5.2 (2) |
| G3, AOA, IDH1 | 8 | 2.7 (7) | 0.5 (46) | 1.7 (15) | 1.0 (43) | 3.1 (1) | 1.3 (3) |
| No biopsy | 5.2 | 4.4 (3) | 0 | 1.6 (9) | 3.6 (6) | 3.2 (0) | 1.5 (3) |
| No biopsy | 8 | 4.1 (6) | 0 | 1.9 (6) | 1.5 (9) | 1.2 (1) | 1.7 (2) |
| No biopsy | 12.5 | 3.5 (5) | 0 | 4.2 (5) | 2.8 (10) | 7.4 (0) | 4.0 (2) |
| No biopsy | 4.5 | 2.9 (6) | 0 | 2.7 (8) | 5.5 (7) | 2.5 (1) | 4.5 (2) |
| No biopsy | 6 | 1.2 (5) | 0 | 1.5 (5) | 1.5 (8) | 1.0 (1) | 1.3 (3) |
| No biopsy | 4.5 | 1.0 (5) | 0 | 1.0 (6) | 1.0 (10) | 0.8 (1) | 0.4 (4) |
DISCUSSION AND CONCLUSION
The current paper reports in vivo measurement of the onco-metabolite 2HG at 7T. Given the fact that the 2HG 2.25 ppm resonance, which gives the most pronounced signal in 2HG, is extensively overlapped with the neighboring resonances of GABA, Glu and Gln at short TE (see Figs. 1c and 5c), a long TE approach has been developed for detection of 2HG and tested in glioma patients in vivo. The in-vitro and in-vivo data show clearly that the 2HG-optimized PRESS TE = 78 ms method brings about apparent multiplet narrowing in 2HG as well as GABA, Glu and Gln, as predicted by the numerical simulation, and thus provides improved selectivity of 2HG with respect to the neighboring resonances, compared to short-TE MRS. Moreover, the 2HG 2.25 ppm peak polarity is inverted at the optimized PRESS TE, giving further possibility of 2HG signal differentiation from the adjacent positive signals of GABA and Glu. Using the optimized-TE MRS at 7T, 2HG was measurable in all of the 12 IDH-mutated and/or radiography-suggested glioma patients, of which four tumors had relatively low 2HG concentrations (< 2 mM) that were obtained with acceptable precision (CRLB ≤ 7%).
Despite the similarity in molecular structure, the J evolution of the 2HG resonances appears to be quite different than those of the Glu and Gln spins at 7T, resulting in the negative polarity of the 2.25 ppm multiplet of 2HG at TE = 60 - 100 ms while the polarity of the Glu and Gln multiplets between 2.3 and 2.5 ppm remains positive for the TEs studied. For each of 2HG, Glu and Gln, the five non-exchangeable protons (i.e., H2, H3, H3′, H4 and H4′) may be modeled as AMNPQ, of which the C4-proton spins (H4 and H4′), P and Q, that give signals between 2.2 and 2.5 ppm, are strongly coupled with the C3 protons. Following the 90° excitation, the inphase coherences, –Py and –Qy, evolve to antiphase coherences over time. The strength and pattern of the C4-proton multiplet may then be determined by the 4 inphase coherences (i.e., Px, Py, Qx and Qy) and 28 antiphase coherences, which include 14 terms of P-spin magnetization in antiphase with respect to Q, M and/or N spins (e.g., 2PxQz, 4PyQzMz, 8PxQzMzNz, etc.), and 14 terms of Q-spin magnetization in antiphase with respect to P, M and/or N spins (e.g., 2QyPz, 4QxPzMz, 8QyPzMzNz, etc.). A PRESS volume-localized numerical simulation indicates that, when the absorption signal of an uncoupled spin I is given by –Iy, the C4-proton signals of Glu and Gln at the TE = 78 ms are mainly attributed to 4PyMzNz and 4QyMzNz coherences (~90%), with coefficients at 0.54 and 0.58 for Glu and 0.3 and 0.31 for Gln, respectively. In contrast, the 2HG 2.25 ppm signal at the PRESS TE = 78 ms has contributions from several coherences. About 35% of the 2HG signal is attributed to 8PxQzMzNz and 8QxPzMzNz (coefficients of approximately −0.3 for both), about 20% to 4PyMzNz and 4QyMzNz (coefficients of approximately −0.2 for both), about 20% to 2PxMz, 2PxNz, 2QxMz and 2QxNz (coefficients of approximately −0.1 for all), and the remaining to other coherences. The coherence attribute of the GABA 2.29 ppm resonance is similar to those of the Glu and Gln C4-proton resonances at the PRESS TE = 78 ms. When two of the 2.29 ppm spins and the J coupling partners, two of the 1.9 ppm spins, are modeled as A2M2, the GABA 2.29 ppm signal is primarily attributed to 4A1yM1zM2z and 4A2yM1zM2z (coefficient = 0.62 for both).
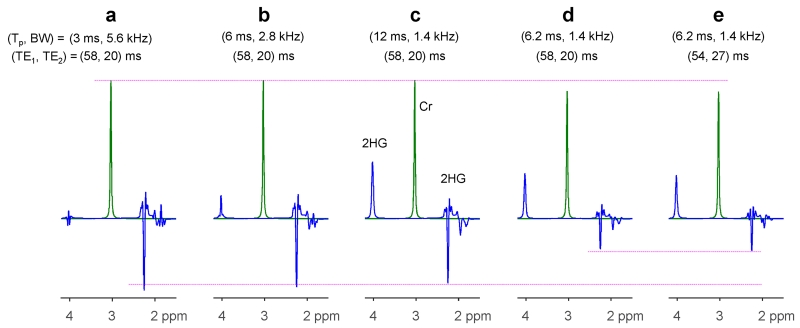
The 2HG signal strength and pattern depends on the PRESS 180° RF pulses. For the 180° pulse envelope used in the present study (see Fig. 1d of reference 10), the pattern of the C4-proton multiplet is not sensitive to the bandwidth of PRESS 180° RF pulses due to the relatively small spectral distance between the C4 and C3 protons (< 0.5 ppm), but the peak amplitude is somewhat dependent on the bandwidth. A computer simulation indicates that, for bandwidth of 1.4, 2.8 and 5.6 kHz (with durations of 12, 6 and 3 ms), the 2HG 2.25 ppm signal amplitude for the TE 78ms PRESS sequence relative to a 90° pulse-and-acquire sequence was 56, 60 and 64% for an identical volume, respectively (Fig. 6a-c). Unlike the C4-proton resonances, the intensity and pattern of the C3- and C2-proton multiplets of 2HG both depend on bandwidth due to the large spectral distance between the coupling partners. In addition, for a 180° RF pulse with a different envelope (see Fig. 1b of reference 10), whose bandwidth is, with duration of 6.2 ms, the same as that of the 180° pulse of the present study (1.4 kHz), the 2HG 2.25 ppm signal at PRESS (TE1, TE2) = (58, 20) ms was quite small (27% with respect to 90°-acquisition) (Fig. 6d). For this pulse, the 2HG 2.25 ppm signal was maximized at (TE1, TE2) = (54, 27) ms, but the amplitude was only 31% relative to 90°-acquisition (Fig. 6e). Taken together, the 2HG 2.25 ppm multiplet from the strongly-coupled C4-proton spins depends on the 180° RF pulse bandwidth, duration and envelope. Since the spectral pattern and signal yield depend on the RF pulses, basis spectra generated with actual RF pulse envelopes may be required for spectral fitting.
Figure 6.
Numerically-calculated PRESS spectra of 2HG for various durations and bandwidths of 180° RF pulses and various subecho time sets. The envelope of the 180° pulse in (a) - (c) was the same as that used for in-vivo tests in the present study (see Fig. 1d of reference 10). The RF pulse for (d-e) had a different envelope (see Fig. 1b of reference 10). Spectra were broadened to Cr singlet width of 9 Hz. The singlet strength in (d) - (e) is smaller by ~10% compared to (a) - (c) due to the difference in ripple size in the refocusing profiles.
The detectability of 2HG may depend on linewidth and signal-to-noise ratio (SNR). The mean in vivo linewidth (FWHM) of the tCho singlet in our data from 12 patients was 11±2.0 Hz, similar as reported in our prior study (14). To evaluate the influence of linewidth on 2HG detection, we performed LCModel analyses on simulated spectra with random noise for singlet linewidths of 5 - 20 Hz. The spectra included 2HG, GABA, Glu, Gln and Cr at a concentration ratio of 5:1:5:5:5, with 2HG SNR (i.e., peak amplitude to noise standard deviation ratio) at 10 for the various linewidths. The result indicated that the 2HG-to-Cr concentration ratio of unity was closely reproduced up to singlet linewidth of ~15 Hz, much larger than the mean in-vivo linewidth of this study. Also, LCModel fitting was performed on similar synthetic data for evaluating SNR dependence of 2HG detectability. For a singlet linewidth of 11 Hz, the 2HG-to-Cr ratio of unity was reproduced in spectral fitting when 2HG SNR ≥ 2. In the in-vivo data of the present study, the mean 2HG SNR was 8.9 ± 5.9, much higher than the theoretical SNR limit for proper 2HG detection. The results from the analyses of the synthetic data may support the reliable detection of 2HG at low concentration (< 2 mM) by our proposed MRS method.
Signal reduction due to T2 relaxation effects may be a major pitfall of the proposed 2HG MRS. The 2HG signal could decay by ~50% during the 78 ms TE for T2 = 120 ms, which was used for T2 signal loss correction in this study. The disadvantage may be outweighed by improved resolution of 2HG signal, achieved by the optimized long TE. Suppression of MM signals at the long TE may be another benefit for enhancing inter-metabolite discrimination. Although a long-TE acquisition may provide enhanced resolution, a caveat with respect to 2HG quantification is that the measured signal intensity depends on the accuracy of 2HG T2. In addition, the B1 field is relatively inhomogeneous at 7T and thus extension of the proposed PRESS method to spectroscopic imaging of 2HG may not be straightforward. Although the B1 variation within a small volume may not be extensive (see Sup. Fig. S4), imaging of metabolites in large volumes often suffers from B1 inhomogeneity. Reliable spectroscopic imaging of 2HG may require B1 shimming with multichannel RF transmission (27) and/or use of adiabatic RF pulses for obtaining uniform flip angles across a large volume of interest (19).
Several sources of errors may be present in the present study. First, the use of the healthy brain Glu T2 value for correcting for the T2 relaxation effects of 2HG, GABA, Glu and Gln in tumors, may introduce errors in the quantification, depending on the differences of their T2 values from the Glu T2. Also, potential differences of T2 between healthy brain and tumors will cause errors in metabolite estimation, but we believe these errors may not be substantial since metabolite T2 differences between tumors and healthy brain are not extensive (28). Second, differences in T1 saturation effects on metabolites and water signals, which were ignored in this study, could introduce error in metabolite quantification.
Third, some errors may be present due to the effects of chemical-shift misregistration. The slice displacements due to the limited bandwidth (1.4 kHz) of the 180° RF pulse was quite large at 7T (~20% of the slice thickness per ppm), preventing acquisition of multiple resonances from an identical volume. The metabolite concentrations may be inhomogeneous in heterogeneous tumors. Our estimates of metabolites represent the concentrations in voxels shifted according to the spectral distances of the metabolite resonances from the RF carrier frequency (2.5 ppm). Fourth, use of STEAM water for quantifying PRESS-acquired metabolite signals may cause errors because the frequency profiles of the 90° and 180° RF pulses used were not identical. However, since the difference in the profiles was not substantial (i.e., transition width / bandwidth ≅ 10% and 12% respectively (14)) and the water T2 is different between tumors (28), the errors due to the profile discrepancy may be smaller than the errors that can be introduced when PRESS water signals at the shortest possible TE (~45 ms in the PRESS sequence used) are used as reference. Lastly, use of a constant water concentration (43 M) for metabolite quantification can cause errors when the water concentration differs between tumors.
Comparison of 2HG detectability between 7T vs. 3T may be of interest. When comparing the present 7T data with 3T data, obtained with our previously-reported TE=97 ms PRESS method (9,10), 7T appears to be advantageous over 3T for 2HG detection. For five patients who were scanned at both 7T and 3T with time intervals of < 1 week, the mean estimates of 2HG and tCho were similar between 7T and 3T (2.9 vs. 3.1 mM for 2HG, and 3.1 vs. 3.2 mM for tCho). The mean 2HG CRLB was, however, quite different between 7T and 3T (5% and 8% respectively), which is likely due to SNR difference (see Sup. Fig. S5). When taking into account the differences in voxel size and scan durations, the mean SNR of tCho was higher by ~1.4 fold at 7T than at 3T although signal reduction due to T2 relaxation effect is more extensive at 7T than at 3T (e.g., tCho T2 ≅ 140 vs. 230 ms respectively). The higher SNR at 7T than at 3T may be due to signal gain at high field and difference in the signal reception coils (16-vs. 8-channel). Most importantly, the mutual dependence of 2HG and GABA signal estimation was markedly lower at 7T than at 3T, as indicated by the 2HG-GABA correlation coefficients returned by LCModel: maximum and minimum values over the five patients were −0.18 and 0.05 at 7T vs. −0.61 and −0.34 at 3T. This result demonstrates that the opposite polarity and apparent narrowing of 2HG and GABA signals, achieved using long TE at 7T, provide improved signal separation compared to 3T long-TE MRS that gives only line narrowing with respect to short TE (10).
In conclusion, noninvasive in vivo detection of elevated 2HG in IDH mutant gliomas has been largely demonstrated at low field strengths (3T) to date, where the SNR and spectral resolution of 2HG from GABA and Glu may be limited. Our proposed MRS method at 7T provides a new tool for reliable measurement of 2HG, with excellent separation of 2HG from neighboring resonances. With this 7T MRS, detection of 2HG in small tumor mass with low cellularity may be possible, which is challenging at low field strengths. The role of 2HG in IDH mutated gliomas is well established as a diagnostic and prognostic tool (1,6,7). Our pre-biopsy 2HG measurement with good precision may provide great potential for noninvasive diagnosis of brain lesions. When used in conjunction with other imaging methods such as magnetic resonance angiography, susceptibility-weighted imaging, both of which show significant increase in resolution at 7T (29,30), 2HG MRS may provide new information on tumor metabolism and microenvironment. Several therapeutic studies are currently underway, targeting mutated forms of IDH1 and IDH2 (31,32). Noninvasive measurement of this unprecedented MRS biomarker 2HG may have great potential for drug development as well as for monitoring tumor progression and response to treatment (33).
Supplementary Material
Acknowledgments
This work was supported by Cancer Prevention Research Institute of Texas grant RP140021-P04 and RP130427 and a US National Institutes of Health grant CA184584. We thank Dr. Ivan Dimitrov for technical assistance.
REFERENCES
- 1.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Losman JA, Kaelin WG., Jr. What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes & development. 2013;27:836–852. doi: 10.1101/gad.217406.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parker SJ, Metallo CM. Metabolic consequences of oncogenic IDH mutations. Pharmacology & therapeutics. 2015;152:54–62. doi: 10.1016/j.pharmthera.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, Rabinowitz JD, Carroll M, Su SM, Sharp KA, Levine RL, Thompson CB. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Jr., Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bal D, Gryff-Keller A. 1H and 13C NMR study of 2-hydroxyglutaric acid and its lactone. Magn Reson Chem. 2002;40:533–536. [Google Scholar]
- 9.Choi C, Ganji SK, Deberardinis RJ, Hatanpaa KJ, Rakheja D, Kovacs Z, Yang XL, Mashimo T, Raisanen JM, Marin-Valencia I, Pascual JM, Madden CJ, Mickey BE, Malloy CR, Bachoo RM, Maher EA. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med. 2012;18:624–629. doi: 10.1038/nm.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi C, Ganji S, Hulsey K, Madan A, Kovacs Z, Dimitrov I, Zhang S, Pichumani K, Mendelsohn D, Mickey B, Malloy C, Bachoo R, Deberardinis R, Maher E. A comparative study of short- and long-TE 1H MRS at 3 T for in vivo detection of 2-hydroxyglutarate in brain tumors. NMR Biomed. 2013;26:1242–1250. doi: 10.1002/nbm.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andronesi OC, Kim GS, Gerstner E, Batchelor T, Tzika AA, Fantin VR, Vander Heiden MG, Sorensen AG. Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci Transl Med. 2012;4:116ra114. doi: 10.1126/scitranslmed.3002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pope WB, Prins RM, Albert Thomas M, Nagarajan R, Yen KE, Bittinger MA, Salamon N, Chou AP, Yong WH, Soto H, Wilson N, Driggers E, Jang HG, Su SM, Schenkein DP, Lai A, Cloughesy TF, Kornblum HI, Wu H, Fantin VR, Liau LM. Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J Neurooncol. 2012;107:197–205. doi: 10.1007/s11060-011-0737-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi C, Dimitrov IE, Douglas D, Patel A, Kaiser LG, Amezcua CA, Maher EA. Improvement of resolution for brain coupled metabolites by optimized 1H MRS at 7T. NMR Biomed. 2010;23:1044–1052. doi: 10.1002/nbm.1529. [DOI] [PubMed] [Google Scholar]
- 14.Ganji SK, An Z, Banerjee A, Madan A, Hulsey KM, Choi C. Measurement of regional variation of GABA in the human brain by optimized point-resolved spectroscopy at 7 T in vivo. NMR Biomed. 2014;27:1167–1175. doi: 10.1002/nbm.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.An L, Li S, Murdoch JB, Araneta MF, Johnson C, Shen J. Detection of glutamate, glutamine, and glutathione by radiofrequency suppression and echo time optimization at 7 tesla. Magn Reson Med. 2015;73:451–458. doi: 10.1002/mrm.25150. [DOI] [PubMed] [Google Scholar]
- 16.Choi C, Ganji S, Banerjee A, Dimitrov I, DeBerardinis R, Malloy C, Mickey B, Bachoo R, Maher E. Noninvasive detection of 2-hydroxyglutarate in gliomas by 1H MR spectroscopy at 7.0 T in vivo. In Proceedings of the 20th Annual Meeting of ISMRM, Melbourne, Australia. 2012:1813. [Google Scholar]
- 17.Ganji S, Hulsey K, Maher E, Choi C. In vivo spectroscopic imaging of 2-hydroxyglutarate in human gliomas at 7.0 T. In Proceedings of the 21st Annual Meeting of ISMRM, Salt Lake City, USA. 2013:214. [Google Scholar]
- 18.Emir UE, Larkin SJ, de Pennington N, Voets N, Plaha P, Stacey R, McCullagh J, Clare S, Jezzard P, Schofield CJ, Ansorge O, Cadoux-Hudson T. The improved detection of 2-hydroxyglutarate in gliomas at 7T using high-bandwidth adiabatic refocusing pulses. In Proceedings of the 23rd Annual Meeting of ISMRM, Toronto, Canada. 2013:2236. [Google Scholar]
- 19.Emir UE, Larkin SJ, de Pennington N, Voets N, Plaha P, Stacey R, Al-Qahtani K, McCullagh J, Schofield CJ, Clare S, Jezzard P, Cadoux-Hudson T, Ansorge O. Noninvasive Quantification of 2-Hydroxyglutarate in Human Gliomas with IDH1 and IDH2 Mutations. Cancer Res. 2016;76:43–49. doi: 10.1158/0008-5472.CAN-15-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. 2000;13:129–153. doi: 10.1002/1099-1492(200005)13:3<129::aid-nbm619>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 21.Kaiser LG, Young K, Meyerhoff DJ, Mueller SG, Matson GB. A detailed analysis of localized J-difference GABA editing: theoretical and experimental study at 4 T. NMR Biomed. 2008;21:22–32. doi: 10.1002/nbm.1150. [DOI] [PubMed] [Google Scholar]
- 22.Versluis MJ, Kan HE, van Buchem MA, Webb AG. Improved signal to noise in proton spectroscopy of the human calf muscle at 7 T using localized B1 calibration. Magn Reson Med. 2010;63:207–211. doi: 10.1002/mrm.22195. [DOI] [PubMed] [Google Scholar]
- 23.Hall EL, Stephenson MC, Price D, Morris PG. Methodology for improved detection of low concentration metabolites in MRS: optimised combination of signals from multi-element coil arrays. Neuroimage. 2014;86:35–42. doi: 10.1016/j.neuroimage.2013.04.077. [DOI] [PubMed] [Google Scholar]
- 24.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30:672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- 25.Michaeli S, Garwood M, Zhu XH, DelaBarre L, Andersen P, Adriany G, Merkle H, Ugurbil K, Chen W. Proton T2 relaxation study of water, N-acetylaspartate, and creatine in human brain using Hahn and Carr-Purcell spin echoes at 4T and 7T. Magn Reson Med. 2002;47:629–633. doi: 10.1002/mrm.10135. [DOI] [PubMed] [Google Scholar]
- 26.Tkac I, Oz G, Adriany G, Ugurbil K, Gruetter R. In vivo 1H NMR spectroscopy of the human brain at high magnetic fields: metabolite quantification at 4T vs. 7T. Magn Reson Med. 2009;62:868–879. doi: 10.1002/mrm.22086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hetherington HP, Avdievich NI, Kuznetsov AM, Pan JW. RF shimming for spectroscopic localization in the human brain at 7 T. Magn Reson Med. 2010;63:9–19. doi: 10.1002/mrm.22182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madan A, Ganji SK, An Z, Choe KS, Pinho MC, Bachoo RM, Maher EM, Choi C. Proton T2 measurement and quantification of lactate in brain tumors by MRS at 3 Tesla in vivo. Magn Reson Med. 2015;73:2094–2099. doi: 10.1002/mrm.25352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lupo JM, Li Y, Hess CP, Nelson SJ. Advances in ultra-high field MRI for the clinical management of patients with brain tumors. Current opinion in neurology. 2011;24:605–615. doi: 10.1097/WCO.0b013e32834cd495. [DOI] [PubMed] [Google Scholar]
- 30.Radbruch A, Eidel O, Wiestler B, Paech D, Burth S, Kickingereder P, Nowosielski M, Baumer P, Wick W, Schlemmer HP, Bendszus M, Ladd M, Nagel AM, Heiland S. Quantification of tumor vessels in glioblastoma patients using time-of-flight angiography at 7 Tesla: a feasibility study. PLoS One. 2014;9:e110727. doi: 10.1371/journal.pone.0110727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, Tsoi J, Clark O, Oldrini B, Komisopoulou E, Kunii K, Pedraza A, Schalm S, Silverman L, Miller A, Wang F, Yang H, Chen Y, Kernytsky A, Rosenblum MK, Liu W, Biller SA, Su SM, Brennan CW, Chan TA, Graeber TG, Yen KE, Mellinghoff IK. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang F, Travins J, DeLaBarre B, Penard-Lacronique V, Schalm S, Hansen E, Straley K, Kernytsky A, Liu W, Gliser C, Yang H, Gross S, Artin E, Saada V, Mylonas E, Quivoron C, Popovici-Muller J, Saunders JO, Salituro FG, Yan S, Murray S, Wei W, Gao Y, Dang L, Dorsch M, Agresta S, Schenkein DP, Biller SA, Su SM, de Botton S, Yen KE. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340:622–626. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 33.Choi C, Ganji S, Madan A, DeBerardinis R, Mickey B, Malloy C, Bachoo R, Maher E. Clinical utility of in-vivo MRS measurements of 2-hydroxyglutarate in IDH-mutated gliomas. In Proceedings of the 21st Annual Meeting of ISMRM, Salt Lake City, USA. 2013:509. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.