Abstract
PURPOSE
WRN promoter CpG island hypermethylation in colorectal cancer (CRC) has been reported to increase sensitivity to irinotecan-based therapies. We aimed to characterize methylation of the WRN promoter; determine the effect of WRN promoter hypermethylation upon expression; and validate a previous report that WRN promoter hypermethylation predicts improved outcomes for metastatic colorectal cancer (mCRC) patients treated with irinotecan-based therapy.
DESIGN
WRN methylation status was assessed using methylation-specific PCR and bisulfite sequencing assays. WRN expression was determined using qRT-PCR and Western blotting. WRN methylation status was correlated with overall survival (OS) and progression-free survival (PFS) in 183 patients with mCRC. Among these patients 90 received capecitabine monotherapy (CAP) as first line therapy and 93 received capecitabine plus irinotecan (CAPIRI) therapy as part of the CAIRO Phase III clinical trial.
RESULTS
WRN mRNA and WRN protein expression levels were low in CRC cell lines and in primary CRC, and were largely independent of WRN methylation status. Patients with methylated WRN CRC had a shorter OS compared to patients who had unmethylated WRN CRC (hazard ratio [HR]=1.6 (95%CI1.2–2.2), p=0.003). Patients with unmethylated WRN showed a significantly longer PFS when treated with CAPIRI compared to CAP alone (HR=0.48 (95%CI 0.32–0.70), p=0.0001). In contrast, patients did not benefit from adding irinotecan to CAP when WRN was methylated (HR=1.1 (95%CI 0.69–1.77), p=0.7).
CONCLUSION
WRN expression is largely independent of WRN promoter hypermethylation in CRC. Moreover, we could not validate previous finding that WRN promoter hypermethylation predicts improved clinical outcomes of mCRC treated with irinotecan-based therapy and found instead the opposite result.
INTRODUCTION
Colorectal cancer (CRC) is among the most common cancers in the world, with an incidence of over 1.2 million and with nearly 700,000 deaths per year (1). Half of CRC patients have or will develop distant metastases by the time of diagnosis, or shortly thereafter (2). The majority of patients with metastatic disease are not candidates for curative surgical therapy, and thus receive systemic palliative therapy, most often with a fluoropyrimidine together with irinotecan or oxaliplatin (3). The more recent addition of molecularly targeted drugs such as anti-EGFR or anti-VEGF antibodies has further improved survival (4, 5). CRC is a heterogeneous disease at the molecular level, and recurrent genetic and epigenetic alterations may be important drivers of clinical behavior and the response to therapy (6, 7). Despite this, we lack robust tools to select the best therapy for individual patients to reliably improve treatment outcomes.
Promoter region DNA hypermethylation has been associated with loss of expression at many genetic loci (8). Simple, reliable gene-specific assays can detect DNA hypermethylation in clinical specimens, and thus could be used to help guide the selection of therapy for genes whose expression level modulates the response to clinically approved drugs (9). One association of this type was reported in 2006: hypermethylation of the Werner syndrome WRN RECQ helicase gene was linked to transcriptional silencing of WRN in colorectal cancers (10), and WRN silencing was suggested to improve treated outcomes for cancer patients receiving irinotecan therapy (10–12).
WRN is a human RECQ helicase protein that plays critical roles in DNA replication, recombination, repair and telomere maintenance (13, 14). The heritable loss of WRN leads to Werner syndrome, a progeroid syndrome associated with genetic instability, an elevated risk of cancer, and cellular sensitivity to DNA topoisomerase I inhibitors such as camptothecin and irinotecan and several other important classes of chemotherapeutic drugs (15). WRN was recently identified as the top-ranked gene associated with advanced clinical stage CRC by the combined analysis of copy number alterations (CNA), methylation status and expression; i.e. WRN promoter hypermethylation, CNA/loss and decreased expression were all associated with Stage III and IV CRC (16). These provocative and potentially exciting findings suggested that methylated WRN might be a predictive marker for irinotecan sensitivity in advanced stage CRC.
In the present study we determined the methylation and expression status of WRN in CRC cell lines and primary CRC tissue samples. We also examined whether methylated WRN predicted clinical outcomes for patients enrolled in the Dutch CApecitabine, IRinotecan and Oxaliplatin (CAIRO) study (17). We developed and validated assays to determine WRN promoter methylation status, then used these assays to determine whether WRN methylation status correlated with WRN expression at the mRNA or protein levels (10, 12) and predicted survival in CRC patients who received irinotecan therapy (10).
MATERIALS AND METHODS
Experiments were conducted at the University of Washington in Seattle (UWSEA) and the VU University Medical Center in Amsterdam, the Netherlands (VUmc) using cell lines and patient samples. A brief overview of materials and methods is given below, with full sample detail and methods in the Supplementary Information.
Cell lines and tissue samples
Two independent collections of cultured CRC-derived cell lines were investigated. The adenoma cell line AAC1 and CRC cell lines RKO, LoVo, SW480, LS174T, AAC1/SB10, HCT116, SW48, FET, VACO400, VACO411, VACO5 were cultured at UWSEA. The UWSEA lines were authenticated by DNA fingerprint analysis prior to use (IDEXX/Radil Bioresearch; IRB). CRC cell lines Colo205, Colo320, HCT116, HCT15, HT29, LIM1863, LS174T, LS513, RKO, SW480, and SW1398 were cultured at VUmc and authenticated by array comparative genomic hybridization (aCGH, 244 k Agilent oligonucleotide platform) at the VU University Medical Center, Amsterdam, the Netherlands. The patterns of chromosomal changes observed were in concordance to the previously described chromosomal changes in these cell lines (18, 19). Twenty-six fresh frozen (FF) primary CRC tissues with matched FF normal colon tissue, and 21 formalin-fixed paraffin-embedded (FFPE) normal colon tissues from cancer-free patients were collected and studied following IRB approved protocols and in accordance with the ethical regulations of the corresponding institutions (UWSEA and VUmc). The samples used at UWSEA were provided by the Cooperative Human Tissue Network (CHTN). Collection, storage and use of patient-derived tissue and data from VUmc was performed in accordance with the Code for Proper Secondary Use of Human Tissue in The Netherlands (20).
Tissue samples from the CAIRO clinical trial
In the CAIRO study CRC patients with metastatic disease were randomized between sequential treatment (capecitabine (CAP) followed upon disease progression by irinotecan (IRI), then oxaliplatin plus capecitabine (CAPOX)), or combination therapy with irinotecan plus capecitabine (CAPIRI) followed by CAPOX(17). The primary endpoint of the study was overall survival (OS). DNA was isolated from FFPE tissue of surgically resected primary tumors from 183 patients that participated in the CAIRO study. Of these 183 patients, 93 received CAPIRI as first-line therapy while 90 received first-line CAP monotherapy. From the 90 patients that received first-line CAP monotherapy, 52 received more than 2 cycles of second-line IRI. These samples were selected to match stratification factors in the original study for the subgroup of patients that underwent primary tumor resection, i.e. resection status, WHO performance status, predominant localization of metastases, previous adjuvant therapy and serum lactate dehydrogenase levels. Samples were also selected based on a high proportion of tumor cells in sections (at least 70%). A large proportion of these samples overlap with samples described in (21).
WRN methylation analyses
WRN methylation status was assessed by two different methylation-specific PCR (MSP) assays together with bisulfite sequencing (see Supplementary Methods for additional detail). A WRN 5′ region from −31 bp to +128 relative to the transcription start site (TSS), hereafter referred to as Region 1, was analyzed by a gel-based MSP assay. Region 2, located at −410 to −331 bp upstream of the WRN TSS was analyzed with a quantitative MSP assay. Bisulfite sequencing was performed for the region −193 bp to +157bp that encompassed the TSS, and overlapped with the locations of the WRN MSP primer pairs described in Agrelo et al. (10) and an independent set of WRN MSP primers reported by Ogino et al.(22).
WRN expression analyses
RNA expression analyses were performed by real-time quantitative PCR assays using TaqMan® Gene Expression Assays from Applied Biosystems for WRN (Hs00172155_m1), β-2 micoglobulin (B2M, Hs00984230_m1), and β-glucuronidase (GusB, Hs99999908_m1). Protein expression analyses were performed by Western blotting, using monoclonal antibodies for WRN (W0393, Sigma) and beta-actin (13E5, Cell Signaling Technologies).
TCGA data
WRN DNA methylation (Illumina Infinium HM27 bead array; HM27) and mRNA expression (Agilent microarray) data from 223 CRC tumors from The Cancer Genome Atlas (TCGA) Colorectal Cancer project (23) were obtained via cBioPortal (http://www.cbioportal.org; data downloaded on the 2 March 2014)(24). When data from more than one probe per gene is available from the methylation assay, cBioPortal uses methylation data from the probe with the strongest negative correlation between the methylation signal and mRNA gene expression.
Statistical analyses
Student’s T-test was used to compare WRN expression levels in HCT116 and Colo205 before and after 5-aza-2-deoxycytidine (DAC) and/or trichostatin A (TSA) treatment. Pearson correlation analysis was used to measure correlation between WRN methylation and mRNA expression levels.
Progression-free survival (PFS) for first-line treatment was calculated from the date of randomization to the date of first observed disease progression or death after first-line treatment. Overall survival (OS) was measured from the date of randomization to date of death due to cancer. Other causes of death were censored. The prognostic or predictive value of WRN methylation status was assessed by a Kaplan-Meier survival analysis and log-rank test.
A Cox proportional hazard regression model was used to estimate Hazard Ratios (HR) and 95% Confidence Intervals (95%CI). A multivariate Cox regression model was used to assess and adjust for important prognostic variables including age, gender, serum lactate dehydrogenase (LDH), WHO performance status, previous adjuvant therapy and location of metastases. Multivariate Cox regression analysis was also used to assess and adjust for possible prognostic variables Microsatellite Instability (MSI) status, BRAF mutational status and mucinous differentiation, for which information was available on a sub-set of the samples (136 out of 183)(25, 26). Results were considered significant when p-values were ≤ 0.05.
RESULTS
WRN methylation and expression status in colon cancer cell lines
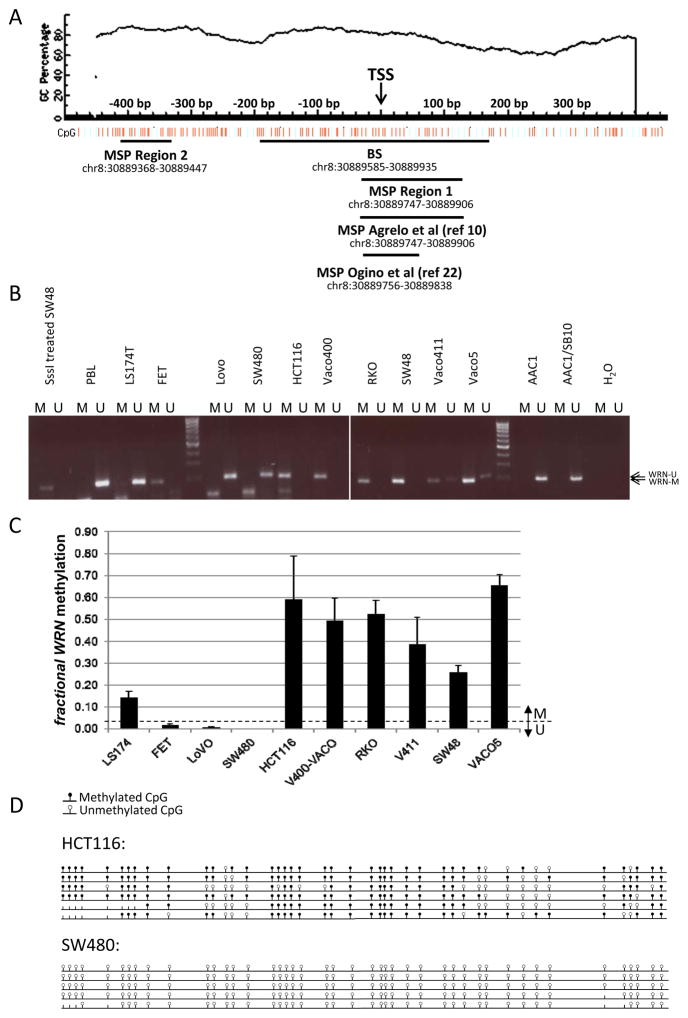
In order to accurately detect and quantify WRN promoter methylation in CRC samples, we independently developed and cross-validated methylation-specific PCR (MSP) primer sets and assays in both labs (UWSEA and VUmc) for two WRN regions adjacent to and overlapping the TSS at base pair position +1: Region 1 (−31 bp to +128 bp) and Region 2 (−410 to −331 bp) (Figure 1A).
Figure 1. WRN promoter region methylation analysis in cell lines.
A. WRN promoter region CpG island and primer locations Genomic coordinates, CpG density and positions are shown in the top panel. Each vertical bar in the lower panel represents the presence of a CpG dinucleotide. Black horizontal bars indicate regions amplified by newly designed and validated MSP primer pairs (Region 1 and Region 2), the region amplified by the original primer pair described by Agrelo et al (10), and the region targeted for bisulfite sequencing (BS). TSS, Transcription Start Site. This figure was created using MethPrimer (39). B. Methylation analysis of Region 1 (see Figure 1) in the adenoma cell line AAC1 and in colon cancer cell lines. DNA from Peripheral Blood Lymphocytes (PBL) was used as an unmethylated control. H2O and DNA from SssI methylase-treated DNA from the colorectal cancer cell line SW48 were used, respectively, as ‘no template’ and ‘methylated template’ controls. C. Quantitative methylation analysis of WRN promoter Region 2 in the same colon cancer cell lines shown in panel A. D. Sodium bisulfite sequencing results of WRN gene promoter on cell lines HCT116 and SW480 in the region depicted in Figure 1. Each row represents an individual cloned allele and each circle indicates a CpG dinucleotide. Black circle=methylated CpG site; white circle=unmethylated CpG site; no circle=not determined.
WRN methylation status in Region 1 was assessed in 11 colon cancer cell lines (SW480, Vaco411, AAC1/SB10, Vaco400 LS174T, LoVo, HCT116, Vaco5, FET, RKO, SW48), and 1 adenoma cell line (AAC1) from UWSEA. Seven of 11 colon cancer cell lines (64%) had Region 1-methylated WRN (Figure 1B), while the adenoma cell line was unmethylated. There was no association between WRN Region 1 methylation and MSI and/or CpG Island Methylator Phenotype (CIMP) (Supplementary M&M and Supplementary Table 1).
WRN methylation status in Region 2 was successfully evaluated in 10 colon cancer cell lines (SW480, Vaco411, Vaco400, LS174T, LoVo, HCT116, Vaco5, FET, RKO, SW48; UWSEA), and was comparable to Region 1 methylation status within a cell line (Figure 1C). Bisulfite sequencing of cells lines with methylated (HCT116) or unmethylated WRN (SW480) was performed to confirm the methylation status of both regions and validate the MSP results using an orthogonal assay (Figure 1D). We assessed WRN Region 2 methylation status in a second, overlapping series of colon cancer cell lines (Colo205, Colo320, HCT116, HCT15, HT29, LIM1863, LS174T, LS513, RKO, SW480, and SW1398, SW48 and Caco2; VUmc). These analyses revealed that 10 of 13 cell lines, or 77%, were WRN Region 2 methylated (Figure 2B).
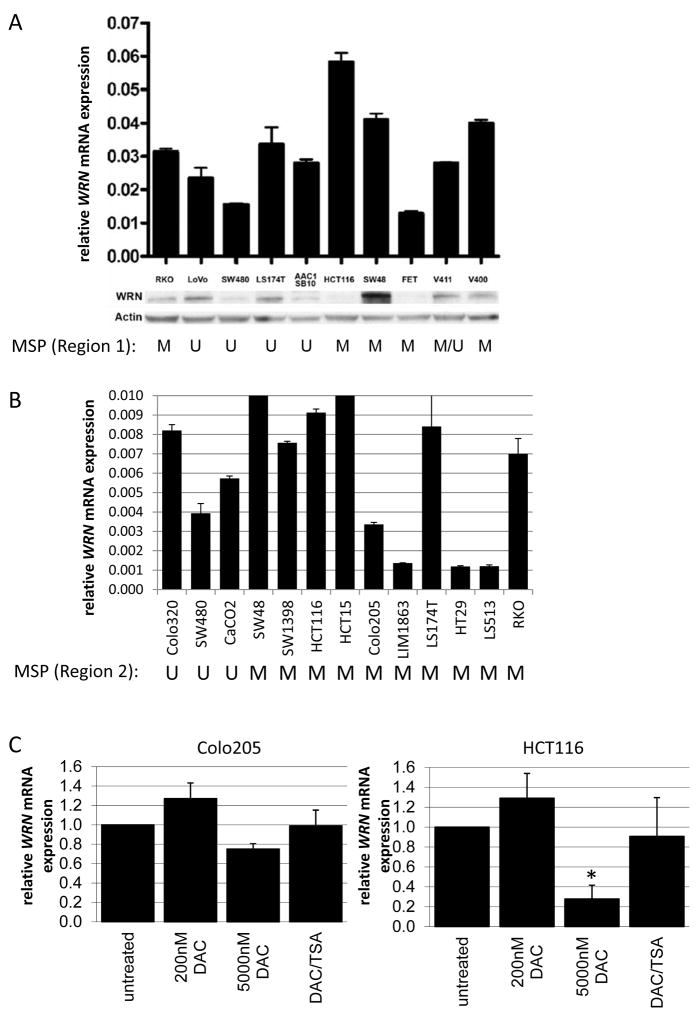
Figure 2. WRN expression analysis in cell lines.
A. WRN mRNA (upper panel) and protein (lower paired panels) expression in CRC cell lines in relation to methylation status in WRN promoter Region 1 (lower panel). Error bars represent standard deviations across triplicate independent experiments, in which WRN mRNA was normalized to mRNA expression of the reference gene GUSB (upper panel) and, for protein expression β-actin (lower panel). Methylation status of WRN promoter Region 1 is indicated below each pair of immunoblots (M = methylated; U = unmethylated). B. WRN mRNA expression level in relation to methylation status of WRN promoter Region 2.Error bars represent standard deviations of mean expression values of two independent experiments. Methylation status of WRN promoter Region 2 is indicated below each cell line designation (M = methylated; U = unmethylated). C. WRN mRNA expression analysis of Colo205 (left) and HCT116 (right) with and without 5-aza-2-deoxycytidine (DAC) or DAC/trichostatin A (TSA) treatment. Bars represent mean in two independent experiments, with error bars represent standard deviations. Expression was quantified relative to mRNA expression levels of B2M. *p=0.001
Cell lines that carried methylated WRN expressed relatively high levels of WRN as assessed by WRN mRNA qRT-PCR (Figure 2A&B). There was either no or a slightly positive correlation between WRN Region 2 methylation and expression level in two different groups of CRC cell lines: SW480, Vaco411, Vaco400 LS174T, LoVo, HCT116, Vaco5, FET, RKO, SW48 (UWSEA; Pearson correlation of 0.32, p=0.3); and Colo205, Colo320, HCT116, HCT15, HT29, LIM1863, LS174T, LS513, RKO, SW480, and SW1398, SW48 and Caco2 (VUmc; Pearson correlation of 0.68, p=0.04). Consistent with these results, treatment of the methylated CRC cell lines HCT116 and Colo205 with the demethylating agent 5-aza-2’-deoxycytidine (DAC) and/or tricostatin A (TSA) either did not change or resulted in decreased WRN mRNA expression (Figure 2C). Western blot analysis of WRN protein expression as a function of Region 1 and 2 promoter methylation in CRC cell lines in the UWSEA collection further emphasized the lack of correlation between WRN promoter hypermethylation and mRNA and protein expression (Figure 2A&B).
WRN methylation and expression status in CRC tissues
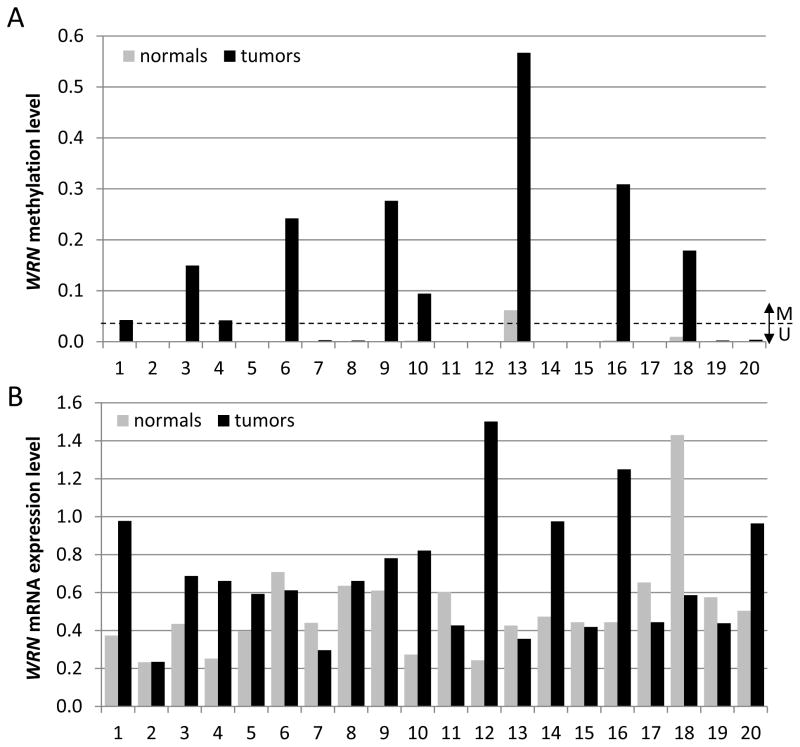
In order to determine whether there was a more consistent relationship between WRN methylation status and expression in primary tumor samples, we analyzed WRN methylation status and expression in primary CRC samples and in adjacent normal colon mucosa. We detected Region 1 methylation in 33% (7 of 21) of primary CRCs, but in none of the paired normal mucosa samples tested (N=12). Region 2 methylation was detected in 45% (9 of 20) of primary CRCs, and in 1 of 20 matched normal mucosa samples (Figure 3A). Methylation status was largely concordant between the two regions: all samples that showed methylation in Region 1 were also Region 2-methylated. Only two cases showed an unmethylated Region 1 and a methylated Region 2. Bisulfite sequencing of a subset of these samples (8 CRCs and 2 normal mucosa samples) confirmed the results of MSP assays (data not shown). A second analysis of Region 2 methylation using an independent series of primary colorectal cancers (N=183 from the CAIRO series, see next section) and normal colon mucosa samples (N=21, VUmc) revealed WRN promoter hypermethylation in 40% (74/183) of the primary CRCs, and very low or absent WRN methylation level in normal colon mucosa.
Figure 3. WRN promoter region methylation and expression analyses in CRC and matched normal colon tissues.
A. WRN methylation levels in CRC tumor tissues (black bars) and matched normal colon tissues (grey bars). Bars represent mean expression of duplicate measurements in one experiment. A sample was considered methylated when the Ct ratio exceeded the threshold of 0.03, which was set based on an analysis of normal colon samples (N=21), which all had values below this threshold. B. WRN mRNA expression versus a GUSB control in the same CRC tumor (black bars) and matched normal colon (grey bars) samples shown in panel A. Bars represent mean expression of triplicate measurements in one experiment.
In our first series of colon tissues, WRN mRNA expression was higher in primary CRC vs matched normal mucosa samples in 10 of 20 patients (50%), lower in 6 samples (6 of 20 or 30%) and equivalent in the remaining 4 samples (20%; Figure 3B). No association was observed between WRN Region 1 or 2 hypermethylation and mRNA expression (Region 2: Pearson correlation 0.14, p=0.4). WRN protein expression could not be detected by Western blot in 10 of 20 (50%) of paired primary CRC/normal mucosa samples (data not shown). An independent assessment of WRN methylation status and mRNA expression in 223 CRCs included in the TCGA Colorectal Cancer Project (see The Cancer Genome Atlas (TCGA) database at www.cBioportal.org; (24)) did not reveal a negative correlation between WRN methylation level and mRNA expression (Pearson correlation of 0.1, p=0.03; Supplementary Figure 1).
Relationship of WRN methylation to clinical outcome
In order to determine if there is a relationship between WRN promoter hypermethylation and treatment outcomes, we assessed the correlation between WRN promoter methylation status and survival in patients who participated in the CAIRO study (17). OS did not differ between the two treatment arms in the original study population, or in the subset included in this analysis. Patient characteristics such as age, sex, performance status, predominant localization of metastases, previous adjuvant therapy and serum lactate dehydrogenase level (LDH) were comparable between the two treatment arms in the subset included in this analysis (Supplementary Table 2). Thus we pooled patients from the two treatment arms to evaluate the association of WRN promoter methylation status and OS.
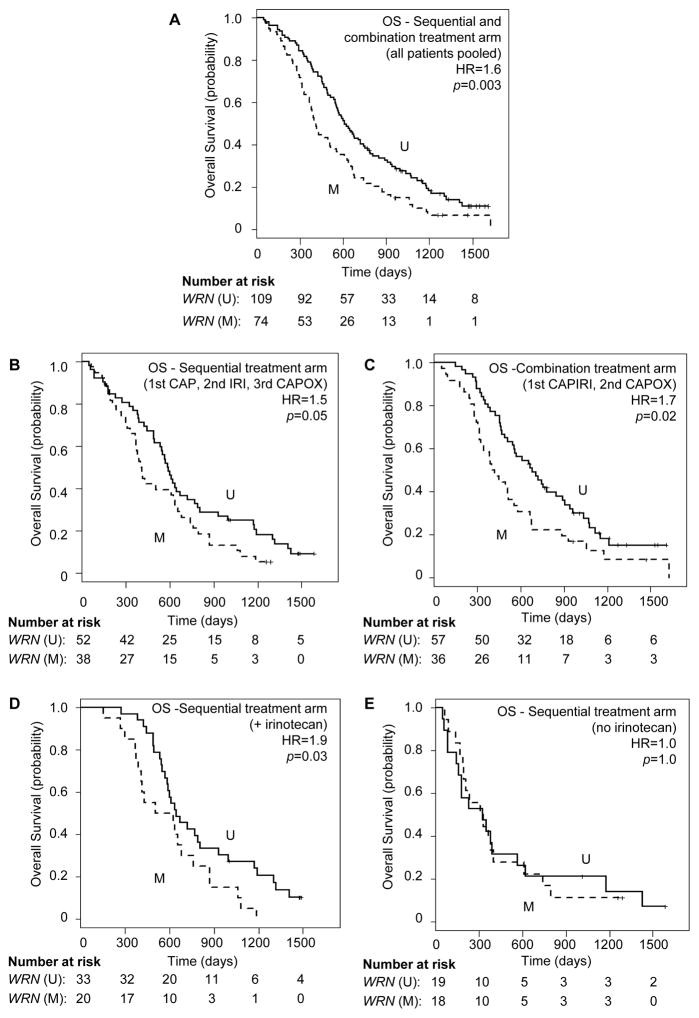
The cohort of 183 patients included a total of 160 death events. The group of 109 patients with unmethylated WRN had 91 death events and the group of 74 patients with methylated WRN had 69 death events. Patients with methylated WRN CRC had shorter OS compared to patients with unmethylated WRN (median OS of 407 vs 610 days for methylated vs unmethylated WRN, respectively (HR = 1.6 (95%CI 1.2–2.2), p = 0.003; Figure 4A). This was observed for patients in the sequential treatment arm (median OS of 405 vs 589 days; HR = 1.5 (95%CI 1.0–2.4), p=0.05), as well as in the combination treatment arm (median OS of 410 vs 680 days for methylated vs unmethylated WRN, respectively; HR = 1.7 (95%CI 1.1–2.7), p=0.02; compare Figure 4B, 4C). However, in the sequential treatment arm, a negative effect of WRN promoter hypermethylation on outcome was observed only for patients who received irinotecan during their treatment course (n=55; median OS of 567 vs 646 days for methylated vs unmethylated WRN, respectively; HR = 1.9 (95%CI 1.1–3.5), p=0.03; Figure 4D). This effect was not observed in patients who did not receive irinotecan (n=37; median OS of 320 vs 326 days for methylated vs unmethylated WRN, respectively; HR = 1.0 (95%CI 0.5–2.0), p=1.0; Figure 4E).
Figure 4. Overall survival in metastatic CRC patients with unmethylated or methylated WRN promoter regions.
Overall survival (OS) of CRC patients with unmethylated (solid lines, U) or methylated (dashed lines, M) WRN promoter regions in response to (A) sequential and combination treatment arms combined (sequential or combined capecitabine (CAP) and Irinotecan (IRI), followed by capecitabine + oxaliplatin (CAPOX)); (B) in the sequential treatment arm alone (1st line capecitabine (CAP), 2nd line Irinotecan (IRI), 3rd line capecitabine + oxaliplatin (CAPOX)); (C) in the combination treatment arm alone (1st line capecitabine + irinotecan (CAPIRI), 2nd line capecitabine + oxaliplatin (CAPOX)); in the subset of patients who received (D) or did not receive (E) irinotecan (IRI) in the sequential treatment arm. HR = Hazard Ratio (Methylated WRN vs unmethylated WRN).
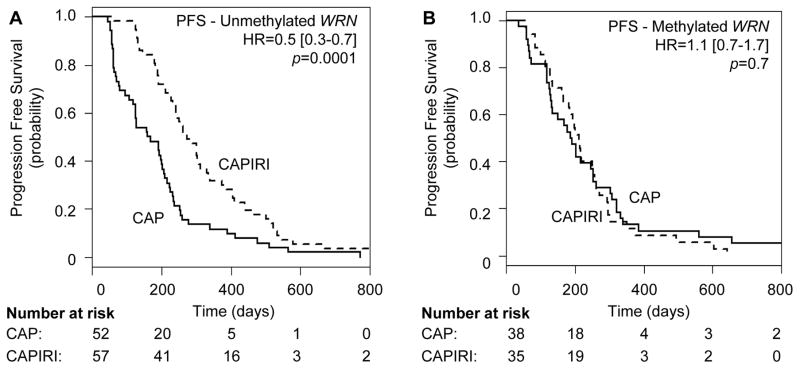
We next determined whether WRN methylation status had predictive value for irinotecan-treated outcomes by assessing the relationship between WRN methylation status and response to CAPIRI. Patients with unmethylated WRN showed significantly longer PFS when treated with CAPIRI compared to CAP alone, as was expected from the results of the original CAIRO trial (17) (median PFS of 272 vs 164 days for CAPIRI vs CAP, respectively; HR=0.48 (95%CI 0.32–0.70), p=0.0001; Figure 5A). However, patients with methylated WRN did not benefit from CAPIRI therapy (median PFS of 211 vs 190 days for CAPIRI vs CAP, respectively; HR=1.1(95%CI 0.69–1.77), p=0.7; Figure 5B). The same trend was observed for patients receiving second-line irinotecan monotherapy in the sequential treatment arm, though the number of patients was small (Supplementary Figure 2).
Figure 5. Progression-free survival in metastatic CRC patients treated with CAP (solid lines) or CAPIRI (dashed lines) as a function of WRN promoter region methylation.
PFS is shown for CRCs with unmethylated (panel A) or methylated (panel B) WRN promoter regions. HR = Hazard Ratio (CAPIRI vs CAP).
Multivariate Cox regression analysis showed significant interaction effects between treatment arm and WRN methylation status, even after adjusting for potentially confounding factors including age, gender, serum LDH, WHO performance status, previous adjuvant therapy, predominant location of metastasis, MSI status, BRAF mutational status and mucinous differentiation (Table 1 and Supplementary Table 4).
Table 1.
Multivariate Cox regression analysis showing the relationship between different covariates, including an interaction between WRN methylation and treatment arm, and progression free survival
| Covariate | HR | 95% CI | p-value* |
|---|---|---|---|
| treatment arm | 0.47 | 0.32–0.70 | 0.0002 |
| WRN methylation status | 0.36 | 0.14–0.98 | 0.05 |
| previous adjuvant therapy | 1.65 | 1.09–2.50 | 0.02 |
| serum LDH | 1.52 | 1.07–2.15 | 0.02 |
| WHO performance status | 1.18 | 0.89–1.56 | 0.26 |
| gender | 0.74 | 0.52–1.04 | 0.09 |
| age | 0.99 | 0.98–1.01 | 0.51 |
| location of metastases | 1.10 | 0.85–1.43 | 0.45 |
| interaction of treatment arm and WRN methylation status | 2.11 | 1.13–3.92 | 0.02 |
Analysis based on 182 samples, of which 179 events (1 observation deleted due to missingness)
CI, confidence interval; HR, hazard ratio; WRN, Werner gene; LDH, Lactate dehydrogenase; WHO, World Health Organisation
Wald test
DISCUSSION
DNA repair proteins such as the RECQ helicase WRN are promising biomarkers for predicting the response to genotoxic chemotherapy. In this study, we aimed to validate the reported association between WRN promoter hypermethylation and transcriptional silencing, and determine the predictive value of WRN promoter hypermethylation for increased sensitivity to IRI-based therapy in CRC patients (10).
We developed and used two new sets of MSP PCR primers to reliably assess WRN methylation status in both CRC and normal colon tissue. Methylation status was also analyzed by bisulfite sequencing (BS) of a region overlapping the WRN TSS. Our new MSP primer pairs and BS assay covered the regions analyzed in previous reports (10, 22) (Figure 1A), and proved more reliable in our hands than the originally reported primer pair for WRN MSP assays (10). Despite using these newly developed and well-validated methylation-specific reagents, we found no consistent association between WRN promoter hypermethylation and WRN expression at the mRNA or protein level. Moreover, we found that WRN promoter hypermethylation was associated with reduced, as opposed to the previously reported increased, OS in CRC patients with metastases who received irinotecan (10). Progression free survival (PFS) improved only when irinotecan was added to CAP in the presence of unmethylated WRN, which was not expected from the results of the original CAIRO trial (17).
One explanation for the differing results between our study and a previous report (10) could be the use of different methylation assays. However, this is unlikely: we designed and validated new primer sets for overlapping MSP and bisulfite sequencing assays that worked reliably, and covered a 567 bp region that encompassed the TSS. These reagents reliably and accurately detected WRN promoter methylation status in both cell lines and primary tumor samples across the locations of both the originally reported (10) and an additional reported overlapping primer pair (22) (Figure 1A). Other possible reasons for the contrasting results in the current and previous report (10) encompass the lack of robust analytical tools in the previous report (10), together with the limited number of cell lines and the small size and nature of the clinical samples analyzed (10). Of note, The clinicopathological details of the 88 patients reported in (10) were not described in the original report or in the reference to this cohort included in the initial report (10). Hence, selection bias cannot be excluded.
We further corroborated our finding of no consistent relationship between WRN promoter methylation level and gene expression using data on 223 CRC samples included in the TCGA Colorectal Cancer Project, where again no correlation could be identified between WRN hypermethylation and WRN transcriptional silencing (23, 24).
In order to test the association between WRN methylation status and clinical outcomes, we used material from patients enrolled in the CAIRO study (the Dutch CApecitabine, IRinotecan and Oxaliplatin (CAIRO) study (17). Our CAIRO study cohort (n=183) was larger than the initial cohort (n=183 vs 88) and has been described in detail. The CAIRO study provided high quality clinical data, which are essential to evaluate predictive biomarkers (27, 28) and to test the association between WRN methylation status and clinical outcomes. The CAIRO cohort also offered the opportunity to compare first-line CAP monotherapy versus CAPIRI therapy.
Despite our larger well-characterized study population, we were not able to confirm the initial observation that WRN promoter hypermethylation was associated with improved outcome in irinotecan-treated metastatic CRC patients (10). In contrast, we observed a significantly worse outcome for irinotecan-treated colorectal cancer patients with WRN-methylated tumors. This is similar to the outcome observed in an independent, well-described study (22) that used primer pairs targeting the same WRN region as the initial report (10) (see Figure 1A). These observations indicate that WRN promoter hypermethylation may be useful as a biomarker, to predict a worse response to irinotecan treatment.
This effect is likely to reflect as-yet unidentified co-variables, as WRN promoter hypermethylation does not consistently alter WRN expression. WRN is a housekeeping gene that is expressed at comparatively low copy number (≤1000 to 10,000 copies/cell) in many cell types (29–32). The WRN promoter region includes Sp1, RCE (retinoblastoma/TP53), AP2 and MYC E-box binding sites, and there are experimental data showing that these binding sites and/or transcription factors can alter WRN transcription (33, 34). WRN expression is also known to be cell cycle-responsive, and upregulated by cellular oncogenic transformation (31), though none of these mechanisms has been shown thus far to be WRN DNA methylation-dependent or modulated.
Alternatively, WRN promoter hypermethylation has been associated to microsatellite instability, CpG island methylator phenotype, BRAF mutations and mucinous differentiation, which themselves are associated to clinical outcome in colon cancer (11, 22, 35). Information on MSI, BRAF and mucinous differentiation available on a sub-set of our sample set revealed that those variables did not explain the association between WRN promoter hypermethylation and clinical outcome after treatment with irinotecan-based therapy. However, the number of samples with MSI status and/or BRAF mutation was very low (n=6 and n=11, respectively), hence no hard conclusions can be drawn from these results. Future functional analyses and validation studies in large, independent and well-annotated cohorts are needed to shed light on the role of WRN promoter hypermethylation as a determinant of the response to irinotecan-based therapy.
Our study has the following limitations. First, measurements were performed on the primary tumors, while patients were treated for their metastases, which raises the question whether intra tumor heterogeneity could play a role. Although metastases can acquire additional genomic alterations, they keep most alterations present in the primary tumor (36, 37). Furthermore, DNA methylation is usually an early event in colorectal carcinogenesis, which we suspect is true for WRN methylation as well (38).
Second, we were not able to independently analyze all cell lines at both participating institutions, though note that the subset of cells analyzed by both groups gave concordant results. This strengthens our conclusion that previous findings on the negative relationship between WRN promoter methylation level and gene expression at the mRNA or protein level could not be validated.
A final limitation of the current study was the use of DNA from 183 patients and tumor tissue which represented a subset of all patients in the CAIRO trial (17). However, this selection was representative for the subgroup of patients that underwent resection of the primary tumor in terms of clinical characteristics and survival outcome (see also ref (21)). Furthermore, the current cohort is larger than the cohort as described in (10) (n=183 vs n=88) and was large enough to have statistical power.
In summary, we found that the methylation status of the WRN promoter region can be reliably assessed in both CRC and normal colorectal tissue using newly developed methylation-specific PCR and bisulfite sequencing assays. However, there was no consistent association between WRN promoter hypermethylation and loss of WRN expression at the mRNA or protein level in CRC cell lines or tumors. Moreover, we could not validate findings from a previous study that WRN promoter hypermethylation was associated with a better response to irinotecan-based therapy and found, instead, that WRN promoter hypermethylation was associated with reduced OS and PFS in our well-characterized CRC patient cohort who received irinotecan-based therapy. Despite growing evidence for a role for WRN genomic alterations in CRC disease progression (16), our results indicate that WRN promoter hypermethylation does not reliably predict WRN gene expression or, as originally reported (10), improved clinical outcomes in CRC patients treated with irinotecan-based chemotherapy regimens.
Supplementary Material
Significance of the study.
The current care for metastatic colorectal cancer includes, if clinically indicated, surgical resection of the primary tumor and/or liver metastases, together with chemotherapy (5-fluoruracil and oxaliplatin or irinotecan) and in some patients targeted therapy (anti-EGFR antibodies or anti-VEGF therapy). The clinical response to this regimen is variable, and it is difficult to predict who will benefit from treatment. Moreover, for most therapies, we lack accurate biomarkers to identify the optimal treatment for individual patients. DNA repair proteins such as the Werner syndrome RECQ helicase, WRN, are promising biomarkers for predicting the response to genotoxic chemotherapy. We attempted to validate previous studies that showed WRN promoter hypermethylation predicted the response to irinotecan using an independent sample set. We did not find a clear association between aberrant WRN promoter hypermethylation and reduced WRN expression. Moreover, in contrast to earlier studies we found an inverse correlation of WRN promoter hypermethylation with survival in metastatic colorectal cancer patients treated with irinotecan. Our results highlight the need for further studies to identify biomarkers that can predict the response of colorectal cancer to standard-of-care chemotherapeutic agents including irinotecan, oxaliplatin and 5-fluorouracil.
Acknowledgments
Grant support:
Support for these studies was provided by the NIH (RO1CA115513, P30CA15704, UO1CA152756, U54CA143862, and P01CA077852), and a Burroughs Wellcome Fund Translational Research Award for Clinician Scientist (WMG). VVL was supported by ACS fellowship PF-11–086–01-TBG; 2T32DK007742–16; ASCRS GSRRIG; and NIH NCI F32CA1591555–01 (VVL). LJWB was supported by Dutch Cancer Society (KWF Fellowship VU 2013–5885). Support for these studies was also provided by National Natural Science Foundation of China (81201920,81472257, YL). Parts of this study were performed within the framework of CTMM, the Center for Translational Molecular Medicine, DeCoDe project (grant 03O-101) and the Dutch Colorectal Cancer Group (DCCG).
The authors thank dr. H. van Tinteren for his critical review of the manuscript and his valuable suggestions.
Non-standard abbreviations
- mCRC
metastatic Colorectal Cancer
- WRN
Werner syndrome
- MSP
Methylation Specific PCR
- BS
Bisulfite Sequencing
- OS
Overall Survival
- PFS
Progression Free Survival
- CAIRO trial
Capecitabine Irinotecan and Oxaliplatin trial
- CAP
capecitabine
- IRI
irinotecan
- CAPIRI
combination of capecitabine and irinotecan
- CAPOX
combination of capecitabine and oxaliplatin
- Ct
Cycle threshold
- DAC
5-aza-2’-deoxycytidine
- TSA
trichostatin A
- TSS
Transcription Start Site
Footnotes
Disclosure: WvC is an employee of and holds stock options in MDxHealth. GAM and JGH are consultants to MDxHealth and GAM receives MDxHealth research funding. All other authors have nothing to disclose.
Author contributions:
Study concept and design: LJWB, WvC, GAM, BC, RJMJr and WMG; Data acquisition: LJWB, YL,VVL, PS, SM, GT, IL, SM, WT and PW; Data analysis and interpretation: LJWB, YL, VVL, PS, GT, JH, WvC, GAM, RJMJr, BC and WMG; Manuscript drafting: LJWB, YL, RJMJr, GAM, BC and WMG; Critical revision of the manuscript: all authors; Obtained funding: GAM, RJMJr and WMG; Provided study materials: MK, IN, CJAP.
References
- 1.American Cancer Society. Global Cancer Facts & Figures. 2. Atlanta: American Cancer Society; 2011. pp. 13–5. [Google Scholar]
- 2.DeVita, Hellman, Rosenberg . Cancer: Principles and Practice of Oncology. 10. 2015. [Google Scholar]
- 3.Koopman M, Punt CJ. Chemotherapy, which drugs and when. Eur J Cancer. 2009;45(Suppl 1):50–6. doi: 10.1016/S0959-8049(09)70016-4. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, et al. Colorectal cancer. Lancet. 2010;375:1030–47. doi: 10.1016/S0140-6736(10)60353-4. [DOI] [PubMed] [Google Scholar]
- 5.Tol J, Punt CJ. Monoclonal antibodies in the treatment of metastatic colorectal cancer: a review. Clin Ther. 2010;32:437–53. doi: 10.1016/j.clinthera.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 6.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011;8:686–700. doi: 10.1038/nrgastro.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–22. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paz MF, Yaya-Tur R, Rojas-Marcos I, Reynes G, Pollan M, Aguirre-Cruz L, et al. CpG island hypermethylation of the DNA repair enzyme methyltransferase predicts response to temozolomide in primary gliomas. Clin Cancer Res. 2004;10:4933–8. doi: 10.1158/1078-0432.CCR-04-0392. [DOI] [PubMed] [Google Scholar]
- 10.Agrelo R, Cheng WH, Setien F, Ropero S, Espada J, Fraga MF, et al. Epigenetic inactivation of the premature aging Werner syndrome gene in human cancer. Proc Natl Acad Sci USA. 2006;103:8822–7. doi: 10.1073/pnas.0600645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawasaki T, Ohnishi M, Suemoto Y, Kirkner GJ, Liu Z, Yamamoto H, et al. WRN promoter methylation possibly connects mucinous differentiation, microsatellite instability and CpG island methylator phenotype in colorectal cancer. Mod Pathol. 2008;21:150–8. doi: 10.1038/modpathol.3800996. [DOI] [PubMed] [Google Scholar]
- 12.Masuda K, Banno K, Yanokura M, Tsuji K, Kobayashi Y, Kisu I, et al. Association of epigenetic inactivation of the WRN gene with anticancer drug sensitivity in cervical cancer cells. Oncol Rep. 2012;28:1146–52. doi: 10.3892/or.2012.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Croteau DL, Popuri V, Opresko PL, Bohr VA. Human RecQ helicases in DNA repair, recombination, and replication. Annu Rev Biochem. 2014;83:519–52. doi: 10.1146/annurev-biochem-060713-035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sidorova JM, Monnat RJ., Jr Human RECQ helicases: roles in cancer, aging, and inherited disease. Advances in Genomics and Genetics. 2015;5:19–33. [Google Scholar]
- 15.Mao FJ, Sidorova JM, Lauper JM, Emond MJ, Monnat RJ. The human WRN and BLM RecQ helicases differentially regulate cell proliferation and survival after chemotherapeutic DNA damage. Cancer Res. 2010;70:6548–55. doi: 10.1158/0008-5472.CAN-10-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee H, Flaherty P, Ji HP. Systematic genomic identification of colorectal cancer genes delineating advanced from early clinical stage and metastasis. BMC Med Genomics. 2013;6:54. doi: 10.1186/1755-8794-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koopman M, Antonini NF, Douma J, Wals J, Honkoop AH, Erdkamp FL, et al. Sequential versus combination chemotherapy with capecitabine, irinotecan, and oxaliplatin in advanced colorectal cancer (CAIRO): a phase III randomised controlled trial. Lancet. 2007;370:135–42. doi: 10.1016/S0140-6736(07)61086-1. [DOI] [PubMed] [Google Scholar]
- 18.The Wellcome Trust Sanger Institute Cancer Genome Project web site. The Wellcome Trust Sanger Institute Cancer Genome Project web site. 2011 cited; Available from: http://www.sanger.ac.uk/genetics/CGP.
- 19.Hermsen M, Snijders A, Guervos MA, Taenzer S, Koerner U, Baak J, et al. Centromeric chromosomal translocations show tissue-specific differences between squamous cell carcinomas and adenocarcinomas. Oncogene. 2005;24:1571–9. doi: 10.1038/sj.onc.1208294. [DOI] [PubMed] [Google Scholar]
- 20.Dutch Federation of Biomedical Scientific Societies. Code for Proper Secondary Use of Human Tissue in the Netherlands. 2011. [Google Scholar]
- 21.Haan JC, Labots M, Rausch C, Koopman M, Tol J, Mekenkamp LJ, et al. Genomic landscape of metastatic colorectal cancer. Nat Commun. 2014;5:5457. doi: 10.1038/ncomms6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogino S, Meyerhardt JA, Kawasaki T, Clark JW, Ryan DP, Kulke MH, et al. CpG island methylation, response to combination chemotherapy, and patient survival in advanced microsatellite stable colorectal carcinoma. Virchows Arch. 2007;450:529–37. doi: 10.1007/s00428-007-0398-3. [DOI] [PubMed] [Google Scholar]
- 23.The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mekenkamp LJ, Heesterbeek KJ, Koopman M, Tol J, Teerenstra S, Venderbosch S, et al. Mucinous adenocarcinomas: poor prognosis in metastatic colorectal cancer. European journal of cancer. 2012;48:501–9. doi: 10.1016/j.ejca.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 26.Venderbosch S, Nagtegaal ID, Maughan TS, Smith CG, Cheadle JP, Fisher D, et al. Mismatch repair status and BRAF mutation status in metastatic colorectal cancer patients: a pooled analysis of the CAIRO, CAIRO2, COIN, and FOCUS studies. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:5322–30. doi: 10.1158/1078-0432.CCR-14-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koopman M, Venderbosch S, Nagtegaal ID, van Krieken JH, Punt CJ. A review on the use of molecular markers of cytotoxic therapy for colorectal cancer, what have we learned? EurJCancer. 2009;45:1935–49. doi: 10.1016/j.ejca.2009.04.023. [DOI] [PubMed] [Google Scholar]
- 28.Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst. 2009;101:1446–52. doi: 10.1093/jnci/djp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, et al. The quantitative proteome of a human cell line. Molecular systems biology. 2011;7:549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, et al. Deep proteome and transcriptome mapping of a human cancer cell line. Molecular systems biology. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawabe T, Tsuyama N, Kitao S, Nishikawa K, Shimamoto A, Shiratori M, et al. Differential regulation of human RecQ family helicases in cell transformation and cell cycle. Oncogene. 2000;19:4764–72. doi: 10.1038/sj.onc.1203841. [DOI] [PubMed] [Google Scholar]
- 32.Moser MJ, Kamath-Loeb AS, Jacob JE, Bennett SE, Oshima J, Monnat RJ., Jr WRN helicase expression in Werner syndrome cell lines. Nucleic acids research. 2000;28:648–54. doi: 10.1093/nar/28.2.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamabe Y, Shimamoto A, Goto M, Yokota J, Sugawara M, Furuichi Y. Sp1-mediated transcription of the Werner helicase gene is modulated by Rb and p53. Molecular and cellular biology. 1998;18:6191–200. doi: 10.1128/mcb.18.11.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grandori C, Wu KJ, Fernandez P, Ngouenet C, Grim J, Clurman BE, et al. Werner syndrome protein limits MYC-induced cellular senescence. Genes & development. 2003;17:1569–74. doi: 10.1101/gad.1100303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–6. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knijn N, Mekenkamp LJ, Klomp M, Vink-Borger ME, Tol J, Teerenstra S, et al. KRAS mutation analysis: a comparison between primary tumours and matched liver metastases in 305 colorectal cancer patients. Br J Cancer. 2011;104:1020–6. doi: 10.1038/bjc.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mekenkamp LJ, Haan JC, Israeli D, van Essen HF, Dijkstra JR, van CP, et al. Chromosomal copy number aberrations in colorectal metastases resemble their primary counterparts and differences are typically non-recurrent. PLoS One. 2014;9:e86833. doi: 10.1371/journal.pone.0086833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Derks S, Postma C, Moerkerk PT, van den Bosch SM, Carvalho B, Hermsen MA, et al. Promoter methylation precedes chromosomal alterations in colorectal cancer development. Cell Oncol. 2006;28:247–57. doi: 10.1155/2006/846251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–31. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.