Abstract
In his 1875 monograph on insectivorous plants, Darwin described the feeding reactions of Drosera flypaper traps and predicted that their secretions contained a “ferment” similar to mammalian pepsin, an aspartic protease. Here we report a high-quality draft genome sequence for the cape sundew, Drosera capensis, the first genome of a carnivorous plant from order Caryophyllales, which also includes the Venus flytrap (Dionaea) and the tropical pitcher plants (Nepenthes). This species was selected in part for its hardiness and ease of cultivation, making it an excellent model organism for further investigations of plant carnivory. Analysis of predicted protein sequences yields genes encoding proteases homologous to those found in other plants, some of which display sequence and structural features that suggest novel functionalities. Because the sequence similarity to proteins of known structure is in most cases too low for traditional homology modeling, 3D structures of representative proteases are predicted using comparative modeling with all-atom refinement. Although the overall folds and active residues for these proteins are conserved, we find structural and sequence differences consistent with a diversity of substrate recognition patterns. Finally, we predict differences in substrate specificities using in silico experiments, providing targets for structure/function studies of novel enzymes with biological and technological significance.
Keywords: genome sequence, protein structure prediction, aspartic protease, cysteine protease, carnivorous plant, enzyme substrate specificity, molecular docking, Rosetta, plant-specific insert, digestive enzyme
Introduction
The digestive enzymes of carnivorous plants have been a topic of biological interest at least since Darwin's 1875 monograph on insectivorous plants [1], where he noted that the mucilage secretions of plants in the genus Drosera appeared to contain a “ferment” that he conjectured to be similar to mammalian pepsin (now known to be an aspartic protease). Despite the tremendous advances in our understanding of biochemistry since Darwin's era, researchers are only beginning to characterize the carnivorous plant digestive enzymes whose existence he and others posited. To date, only two carnivorous plant genomes, those of Genlisea aurea [2] and Utricularia gibba [3], both members of the asterid order Lamiales, have been sequenced. The focus of these studies was on the genomes themselves, both of which are remarkably small due to their unusually low non-coding DNA content. Both Genlisea and Utricularia feed on small, often microscopic, prey and perform their digestive functions in closed traps in a relatively thermostable environment (underground or under water); thus they are less subject to the environmental constraints faced by carnivorous plants that perform their prey capture in exposed environments. These plants, by contrast, require stable, highly active digestive enzymes that permit the processing of animal prey tissues over relatively long time spans and usually under milder chemical conditions than those of their animal counterparts. The digestive process must occur without mastication or other mechanical disruption of the prey tissue, and in competition with bacterial and fungal growth. The unique proteomic challenges faced by these plants make them attractive targets for enzyme discovery.
Carnivorous plant digestive enzymes constitute a rich resource for chemical biology and biotechnology applications: their stability, substrate specificities, cleavage patterns, and ability to function over different pH ranges are potentially useful in a variety of laboratory applications. Furthermore, the evolution of plant carnivory, which has happened independently in several lineages [4] presents an opportunity to understand how complex signaling mechanisms and mechanical structures can be modified to serve different purposes. For our present genome sequencing and enzyme discovery study, we chose the Cape sundew (Drosera capensis), which is native to the Cape region of South Africa and belongs to the order Caryophyllales. D. capensis represents an excellent model organism for the study of carnivory in plants; it is easily cultivated, is capable of self-pollination, matures quickly, requires no period of dormancy, and is large and robust, facilitating tissue collection for multiple experiments from the same specimen.
The Drosera belong to the order Caryophyllales, which includes some of the most specialized and charismatic examples of plant carnivory; the order contains the tropical pitcher plants (Nepenthes sp.), the Venus flytrap (Dionaea muscipula), the waterwheel plant (Aldrovanda vesiculosa), the dewy pine (Drosophyllum luscitanica), and the sundews (Drosera sp.). Proteomic analysis of the trap fluid in Nepenthes established the description of the nepenthesins as a distinct aspartic protease family [5]. Nepenthesins from several species have been cloned and expressed [6, 7], enabling characterization of their substrate specificities, resistance to thermal and chemical denaturation, and pH dependence. More recently, a combination of in vitro and in vivo approaches have determined that nepenthesins account for most of the protease activity in Nepenthes pitchers, and that prey capture induces both nepenthesin upregulation and a decrease in pitcher fluid pH [8]. The prey capture and digestive processes are regulated by the jasmonate signaling pathway in Nepenthes mirabilis [8], D. muscipula [9] and D. capensis [10]. Because the jasmonate phytohormones are also involved in upregulation of genes required for defensive responses to wounding and chemical cues from herbivorous insects, this supports the hypothesis that carnivory in the Caryophyllales evolved from defensive responses [11]. Despite the intense interest in these plants and their prey capture systems and associated proteins, no genome of a carnivorous plant from order Caryophyllales has previously been sequenced, limiting both the investigation of their evolutionary relationships and the discovery of novel enzymes from these sources.
Proteases are a ubiquitous class of enzymes that catalyze the breaking of peptide bonds, an essential function in both normal protein turnover and in the digestion of prey. Despite differences in evolutionary history and substrate specificity, proteases can be broadly classified by catalytic mechanism based on the primary functional group used to perform peptide bond hydrolysis, as tabulated in the MEROPS protease database [12]. The active moiety can be the thiol group of a cysteine, the hydroxyl group of a serine or threonine, the carboxylic acid of a glutamic acid or aspartic acid, or a metal ion bound to the active site. In addition to the widely conserved plant proteases that perform cellular functions related to protein turnover, fruit ripening, and programmed cell death, carnivorous plants must also produce digestive enzymes; thus, their proteases are expected to provide a rich source of biochemical and structural targets.
The genes coding for cysteine proteases and aspartic proteases in the D. capensis genome have only moderate sequence identity to known proteases, although important functional features are conserved. This presents a problem for traditional homology modeling, which is performed by superimposing the primary sequence of the unknown protein over the structure of a close homolog and refining via energy minimization. Because traditional homology models rely on a high degree of sequence identity (typically 60% or more) to a protein of known structure, we use Rosetta [13] via the Robetta server [14] to perform comparative modeling with all-atom refinement. This approach uses a combination of fragment homology and de novo structure prediction, allowing accurate prediction of tertiary (and in some cases quaternary) structure from sequence information. Rosetta has been productively applied to understanding the determinants of substrate binding and enzyme activity [15], active site design [16], and guiding molecular replacement for protein structure determination [17]. Structure prediction using Robetta is regularly evaluated both internally and as part of the biennial CASP competition, in which molecular modeling programs are tested against new, unpublished structures [18, 19]. Here we apply this strategy to predicting enzyme functionality based on structural homology in the absence of close relatives of known structure, illustrating how comparative modeling with all-atom refinement can be used to guide target selection for a focused structural genomics study. The predicted structures enable selection of proteins for resource-intensive biochemical characterization and structure determination that are likely to be structurally and functionally interesting. Although the proteins here can be understood in terms of comparison to known structures, in all three enzyme classes, examples are found where significant structural differences are predicted by the models, suggesting promising targets for structure determination and biochemical characterization.
Here we report the sequencing and assembly of a high-quality draft genome for D. capensis and a comparative analysis of the sequences and predicted structures of representative members of three classes of proteases. Four cysteine proteases and seven aspartic proteases were identified from genomic DNA based on phylogenetic analysis and selected for further study; homology to cDNA from the previously reported transcriptome of D. muscipula [20] as well as data from model organisms is employed to confirm similarity to expressed proteins. We use comparative modeling with all-atom refinement to predict three-dimensional structures for these proteins as well as functional subsequences thereof, finding putative global and active site structures that are a close match to those of other known members of these protein families, despite limited sequence identity. Differences in protease substrate affinity are predicted in silico via substrate docking experiments. The conservation of the active site residues suggests that these proteins are functional, while their sequence differences from known homologs and variation in substrate affinities make them excellent candidates for further biochemical and structural characterization. The predicted structures and substrate docking studies provide many opportunities for testing hypotheses about structure-function relationships in these aggressively selected enzymes. In particular, we identify one group of aspartic proteases—which we refer to as droserasins—whose relative similarity to pepsin make them natural candidates for the “ferment” identified by Darwin in his 1875 monograph.
Methods
Approximately 1 g (wet mass) of leaf and petiole material was taken from a mature (approximately 16 months in age) specimen of D. capensis. Genomic DNA was isolated using a protocol developed for recalcitrant plants [21].
Genomic DNA Sequencing
Using a Covaris S2, gDNA was sheared to generate 300 - 500bp fragments. The Bioo Scientific NEXTflex Rapid DNA-Seq kit for the Illumina platform was used. The ends of the sheared DNA were repaired and then adenylated on the 3′ end, after which the fragments were ligated with NEXTflex DNA-Seq adapters and enriched by PCR. The libraries were validated by qPCR and sized by Agilent Bioanalyzer DNA high-sensitivity chip. The libraries were clustered on the flow-cell at 12pM and sequenced on an Illumina HiSeq 2500 high output lane using paired end 100 cycles. The version of HiSeq control software was HCS 2.2.38 with real time analysis software, RTA 1.18.64. Reads were prepared as 100bp paired-end libraries, with an average insert size of approximately 284bp. This process resulted in approximately 133 million usable reads.
Genome Assembly and Validation
Paired-end reads were assembled using the MaSuRCA pipeline (v2.3.2) [22], which employs a hybrid algorithm combining de Bruijn graph and overlap-layout-consensus approaches. As MaSuRCA utilizes an internal error-correction algorithm (QuorUM [23]), reads were not quality-trimmed prior to processing (although adaptor sequences were removed). (Alternate assemblies constructed using SOAPdenovo2 (v2.04) [24] with a variety of pre-processing steps also showed no benefit to quality trimming, removal of low-entropy sequences, or related measures.) Following initial assembly, scaffolds were screened for organelle DNA by BLAST searches against known organelle genomes from related organisms; specifically, the Fagopyrum esculentum subsp. ancestrale (NC 010776) chloroplast genome and the mitochondrial genomes for Silene latifolia (NC 014487.1), Beta macrocarpa (NC 015994.1), and Beta vulgaris subsp. maritima (FP 885845.1) were employed as references. Scaffolds returning any hits at ≥80% identity with an e-value of 1e-8 versus any reference were tagged as containing non-genomic DNA and were removed from the assembly. Additional screening for vectors, human DNA, or other contaminants was performed using the NCBI VecScreen service (with any problematic scaffolds removed). The final assembly consists of 18,637 contigs in 13,142 scaffolds, spanning a total of 264Mbp. This covers approximately 90% of the estimated 293Mbp [25] in the D. capensis genome. The assembly was successfully submitted to the NCBI WGS repository (project PRJNA291419, accession LIEC00000000).
Initial evaluation of the assembled genome was performed using QUAST (v2.3) [26]. Exclusion of all contigs of length <500bp leads to an N50 of 82,651bp, with a maximum scaffold size of approximately 405Kbp (longest contig apx 242Kbp, mean 19Kpb). This compares favorably with other published assemblies using Illumina HiSeq reads (e.g., [2]). QUAST's default (GlimmerHMM) eukaryotic gene finder predicts approximately 62,000 unique genes, which is also comparable with other published carnivorous plant assemblies [2, 3]. Proteome coverage was assessed using CEGMA pipeline [27] by examining the number of Core Eukaryotic Genes contained within the assembly; of the 248 genes in the CEGMA core set, complete matches were found for approximately 90% (223/248) and partial matches for essentially all (99%, or 245/248). Our assembly thus appears to cover the overwhelming majority of the coding region of the D. capensis genome.
Annotation
De novo gene annotation was performed using the MAKER-P (v2.31.8) pipeline [28], following the protocol of Campbell et al. [29]. The cDNA library from the D. muscipula transcriptome [20] was employed as a source of EST evidence from a closely related species; the set of all proteins in the UniProt database from plants in order Caryophyllales with evidence at the transcript or protein level was employed as an additional source of information on homologous proteins. The Augustus [30] “tomato” model was employed for the initial annotation (as the closest relative with an available trained model). Per the Campbell et al. protocol, SNAP [31] was also employed via a five-step training cycle: an initial MAKER annotation cycle was performed with SNAP untrained; SNAP was trained on the initial MAKER output; a second MAKER annotation cycle was performed using the trained SNAP model; SNAP was trained on the second round of MAKER output; and, finally, a third MAKER annotation cycle was performed using the retrained SNAP model. Annotations from this cycle were retained for subsequent analysis.
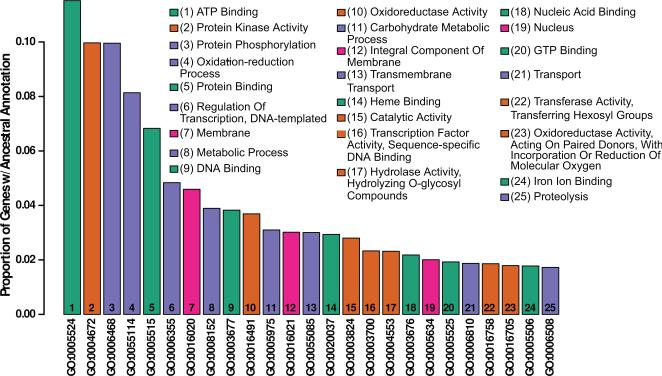
Following identification, putative genes were assigned functional annotations using BLAST against SwissProt (downloaded 8/30/15) and InterProScan [32] (Campbell et al. support protocol 3, basic protocol 5). 8,120 genes were identified by the annotation pipeline, with 92% having AED scores less than 0.5 and 91% having a recognized Pfam domain. Given the substantially higher estimated gene count from QUAST, we regard this as an extremely conservative lower bound on the gene content of the D. capensis genome. InterProScan was used to assign ontology codes [33] to each putative gene; for each identified GO code, all codes ancestral to it in the gene ontology network (under the SUBCLASS relation) were identified using OWLtools [34], yielding a total of 1,761 unique annotations. For display in Figure 1, GO codes were clustered based on their geodesic distances in the symmetrized SUBCLASS network using the ward.D hierarchical clustering method in the R statistical computing platform [35] (network analysis performed using the network and sna libraries from the statnet library [36, 37, 38]).
Figure 1.
D. capensis genome annotation. The 25 most common functional annotations either directly associated or ancestral to directly associated annotations on MAKER-identified genes in D. capensis. The fraction of identified genes associated directly or indirectly with each annotation code is shown on the vertical axis, with annotations shown in descending order of frequency. The description field of each GO code is shown (inset) by rank order. Hierarchical clustering of intercode distances within the ontology network was used to group the 25 most common functional annotations into four classes (denoted by color): ligand binding (aqua); membrane or nuclear location (magenta); and two groups of enzymatic functions (orange and violet).
Sequence Alignment and Prediction of Putative Protein Properties
Sequence alignments were performed using ClustalOmega [39], with settings for gap open penalty = 10.0 and gap extension penalty = 0.05, hydrophilic residues = GPSNDQERK, and the BLOSUM weight matrix. The presence and position of a signal sequence flagging the protein for secretion was predicted using the program SignalP 4.1 server [40]. Secondary structure prediction was performed using the PsiPred server [41, 42] and protein domains were predicted using GenTHREADER and pDomTHREADER [43, 44]. Structures were predicted using the Robetta server [14]. The PDB files generated by Rosetta for all the proteins discussed in this manuscript are available in the Supplementary Information; the available files are tabulated in Supplementary Tables S1 and S2.
Substrate Docking and Predicted Affinity Comparisons
Differences in protease substrate affinity were predicted in silico via the following procedure. First, hexameric test peptides were constructed using PyMol and prepared as flexible ligands using Autodock Tools [45]. Each test peptide was then docked to the target protease using Autodock Vina (v1.1.2) [46]—which has been found to show favorable performance with this protein family [47]—with a search space centered on the active site. Following the recommendations of [48], a linear dimension of 2.857 times the peptide radius of gyration was employed, and a maximum of 20 poses were extracted per peptide/protease combination. This process was repeated multiple times to obtain a larger pose set for subsequent analysis, as described below.
The complete set of poses was employed to construct affinity weights, based on a simplified Boltzmann-like model. For each peptide, the total set of observed poses is treated as an approximate ensemble of pose microstates for that peptide. We approximate the probability of finding the system in microstate i by exp(−Ei/(RT))/Z(T), with Ei being the Vina affinity score (converted to appropriate units), T being the system temperature, R being the gas constant, and Z being the partition function (here, the sum of the numerator over all observed poses). We employ 300K as the effective temperature for purposes of analysis; results were not found to be sensitive to this parameter over a physically reasonable range. For each pose, we examine the minimum distance from each backbone C=O bond to the active site oxygens (the “active site distance”); the C-N bond following the closest C=O bond is taken to be the potential scissile bond for that pose. As the hydrolysis of the C-N bond is initiated by a nucleophilic attack on the C=O carbon, successful cleavage depends on the carbonyl being within an appropriate distance of the active site. Here, we label poses with an active site distance within a specified range (calibration described below) as “viable,” while poses with active site distances outside this range are referred to as “non-viable;” non-viable poses are not expected to contribute to cleavage, but will compete with viable poses for the position of the ligand in the active site. Given this model, we define the affinity weight of a given bond as the log of the total probability of finding the system in a viable pose with that bond as the scissile bond at a random time. Intuitively, high-weight bonds are those that have a high total affinity without also having many non-viable poses that compete with local viable ones–both factors can conspire to render a bond low-weight. Given that a peptide is presented to the active site, the high-affinity bonds are predicted to be those with the most opportunities to be severed, and hence those that will be cleaved at the highest rates.
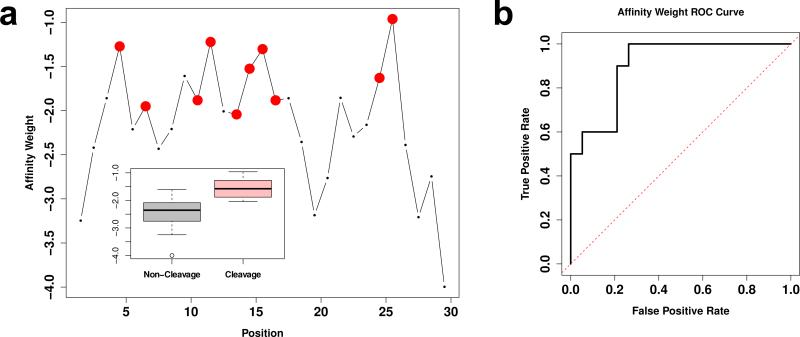
Validation of this approach was perfomed using experimental data from Athauda et al. [49] on cleavage sites for Nepenthesin 1 versus the B chain of human insulin (UNIPROT P01308, residues 25-54). Docking was performed on each sequential insulin hexamer 50 times, for a total of approximately 1000 poses per hexamer (approximately 25,000 poses total); affinity weights were calculated for each bond using the above procedure, with weights for each bond combined by logarithmic summation across all hexamers in which it appeared. The range of viable active site distances was found by simulated annealing with the affinity weight/cleavage indicator correlation as the objective function; the resulting range was found to be 5.11–5.74 Å. The resulting affinity weights are strong cleavage site predictors, as shown in Figure 2, with an affinity weight/cleavage correlation of 0.64 (p < 0.001).
Figure 2.
Docking procedure validation for Nepenthesin 1 on human insulin. a. Affinity weights by sequence position for human insulin B chain; larger values indicate higher predicted affinity. Cleavage sites reported by Athauda et al. [49] are shown in red. Higher affinities are significantly associated with cleavage sites (correlation 0.64, p < 0.001), as revealed by the marginal distributions of affinity weights for cleavage versus non-cleavage sites (inset). b. Receiver operating characteristic (ROC) curve for cleavage/non-cleavage site classification in insulin using affinity weights. Affinity weight is a strong predictor of cleavage sites, with high true positive rates attainable at fairly low false positive rates.
Our approach was employed to predict differences in substrate affinity across aspartic proteases, as follows. Test peptides were constructed from Drosophila melanogaster myosin (heavy chain, muscle; Uniprot P05661) by dividing the complete protein into 327 sequential non-overlapping hexameric segments; each hexapeptide was constructed and docked using the above procedure, with a minimum of 100 poses per peptide per protease (≥32,700 poses per protease total). Each pose set was employed to construct affinity weights as specifed above, with pose viability determined via the previously calibrated 5.11–5.74 Å active site distance range. This resulted in a total of 1635 affinity weights per protein (5/peptide). For purposes of comparison, the relative affinity weights of each potential cleavage site were rank ordered for each protease, and the summed absolute differences in ranks (i.e., Manhattan metric or L1 norm) were computed for every pair of proteases. To ensure that sufficient pose coverage was obtained, split-half reliability was calculated for the total distance matrix (with splits being taken over the set of affinity weights); the mean reliability (i.e., product-moment correlation between off-diagonal distances) over 1,000 random splits was >0.96, indicating a high level of stability.
Results and Discussion
The D. capensis Genome Contains the Expected Complement of Genes
The D. capensis genome assembly consists of 18,637 contigs in 13,142 scaffolds, spanning a total of 264Mbp. This assembly has been submitted to the NCBI WGS repository (project PRJNA291419, accession LIEC00000000). Putative genes from the D. capensis genome were identified using the MAKER-P [28] pipeline. Gene ontology annotations [33] were assigned to putative genes using InterProScan [32]; for each identified GO code, all codes ancestral to it in the gene ontology network (under the SUBCLASS relation) were identified using OWLtools [34], yielding a total of 1,761 unique annotations (Fig. 1). The high diversity of identified functions is indicated by the long-tailed distribution of frequencies. Sequence similarity searches to the D. capensis genomic DNA and the recently sequenced D. muscipula transcriptome [20] reveal many predicted cysteine proteases and aspartic proteases, most with only moderate (30%-40%) sequence identity to their nearest matches from model organisms. Although the genome annotation indicates that D. capensis contains a full complement of digestive enzymes, including nucleases, chitinases, lipases, and amylases, here we focus on we focus on comparative analysis of three classes of proteases: the cysteine proteases of MEROPS family C1, and the aspartic proteases of MEROPS families A1A and A1B [12].
Key Residues Are Conserved in D. capensis Cysteine Proteases
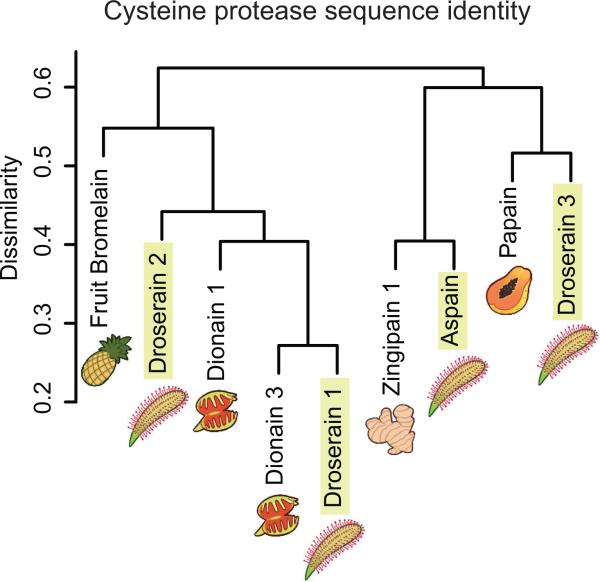
Clustering of representative sequences with previously characterized cysteine proteases (Fig. 3) indicates that a diverse array of protease types is found in both D. capensis and D. muscipula. Each of the recently identified dionains 1 and 3 has a close relative in D. capensis. A sequence alignment comparing four putative cysteine proteases from D. capensis (Supplementary Fig. S1) to the well-characterized enzymes papain (Carica papaya, UniProt P00784), and pineapple fruit bromelain (Ananas comosus, UniProt O23791) shows that these proteins contain the conserved Cys and His residues comprising the cysteine protease catalytic dyad as well as the pro-sequences that would be expected for expected for functional proteases. All but aspain are predicted to have an N-terminal signal sequence targeting the protein for secretion (Supplementary Figure S1).
Figure 3.
Cluster analysis of cysteine protease sequences identified from D. capensis, which are homologous to a variety of known plant cysteine proteases, including dionain 1 and dionain 3 from the Venus flytrap, Dionaea muscipula.
Included for comparison are related plant cysteine proteases that have been previously identified but not yet extensively characterized; zingipain 1 from Zingiber officianale (UniProt P82473), and the dionains 1 and 3 from the related Dionaea muscipula (UniProt A0A0E3GLN3, and A0A0E3M338, respectively). The sequences are annotated to indicate both general amino acid properties and specific functional aspects of C1 family proteases, as described in the S.I. For the previously uncharacterized cysteine proteases, SignalP 4.1 [40] was used to predict the signal sequences (Supplementary Figs. S2-S3), while the pro-sequences were predicted by similarity to papain and fruit bromelain. Because the zingipain-1 sequence was identified via protein sequencing of the mature enzyme via mass spectrometry, this sequence lacks the signal peptide and pro-sequence [50]. As-pain and dionain 3 appear to lack signal sequences, although each contains a pro-sequence.
Molecular Modeling of Cysteine Proteases Reveals Similarities and Differences to Known Structures
Papain-type enzymes (MEROPS C1) are common in plants, both as vacuolar proteins and in the flesh of many fruits, where they deter insects and cleave endogenous proteins as part of the ripening process. This family includes endopeptidases, dipeptidyl peptidases, and aminopeptidases [51]. Some are secreted, while others are active in the vacuole, serving functions analogous to those of lysozomal cathepsins in animals [52]. The same species may have multiple paralogs with different substrate specificities; the tobacco plant (Nicotiana tabacum) has at least 60 different cysteine protease genes [53]. This sequence diversity enables the organism to produce a broad portfolio of cleavage activities, as demonstrated for the ervatamins, where subtle structural differences in the substrate-binding pocket result in variable substrate preferences [54].
C1 cysteine proteases necessarily contain a catalytic dyad made up of Cys and His residues; in many examples, an asparagine residue also helps to position the catalytic His in the correct orientation to deprotonate the Cys [55]. Like many other proteases, the papain-family proteins are protected during processing by a pro-sequence that must be cleaved for the enzyme to become active. The pro-sequences of C1 proteases in plants can inhibit exogenous cysteine proteases, inhibiting the feeding of insect [56], nematode [57], and spider mite pests [58]. The inhibitory effect of the pro-sequence can be used to provide protection in transgenic plants, conferring resistance to crop varieties otherwise lacking the relevant cysteine proteases [59] and protecting against Bt-resistant pests [60]. Despite some variation in the lengths of the C-terminal and N-terminal regions, all the cysteine proteases investigated here show substantial similarity in the pro-sequences; in particular, the ERFNIN motif often found in the pro-sequence of C1 proteases [61] is conserved. The diversity of cleavage modes and substrate activities found in the C1 protease family are particularly interesting in the context of carnivorous plants, which need a variety of cleavage activities to effectively digest the proteins from their prey. The presence of cysteine proteases in the digestive fluids of D. indica has previously been inferred from biochemical activity assays [62], but the proteins themselves have not yet been characterized.
In this study, four cysteine proteases with moderate sequence homology to proteins of known structure have been identified from the genome of D. capensis. Sequence analysis (Supplementary Figure S1) and molecular modeling (Supplementary Table 1) predict that all four are similar in overall fold to papain and related proteases. Because of their limited sequence similarity to targets of known structure, we employ a strategy of comparative modeling with all-atom refinement using Rosetta to predict the protease structures. The Robetta server employs a combined strategy of comparative and de novo modeling, first attempting to find templates from homologues in the protein data bank and then moving to de novo prediction in the case where no close homologues exist [13]. For all of the D. capensis proteases examined here, structural templates with varying degrees of homology were available in the PDB; the modeling was therefore done using iterative comparative modeling in all cases. In the Rosetta refine-and-rebuild protocol [63], the target sequence is aligned to the parent seqence(s), followed by all-atom refinement runs, generating multiple models from which the lowest energy models are selected. This process is repeated iteratively, using more conservative alignment to fewer templates in the case of high homology and a broader search of the available conformational space, making use of more template sequences, in the case of lower homology. Loops and insertions present in the target but not the template are handled as separate fragment structures [64]. All PDB structures used to generate the domain predictions for two representative cysteine proteases, Dionain 1 and Aspain, are tabulated in Supplementary Tables 2 and 3, respectively. The enzymes used in these structure predictions include plant cysteine proteases, e.g. papain and variants thereof, as well as mammalian capthepsins.
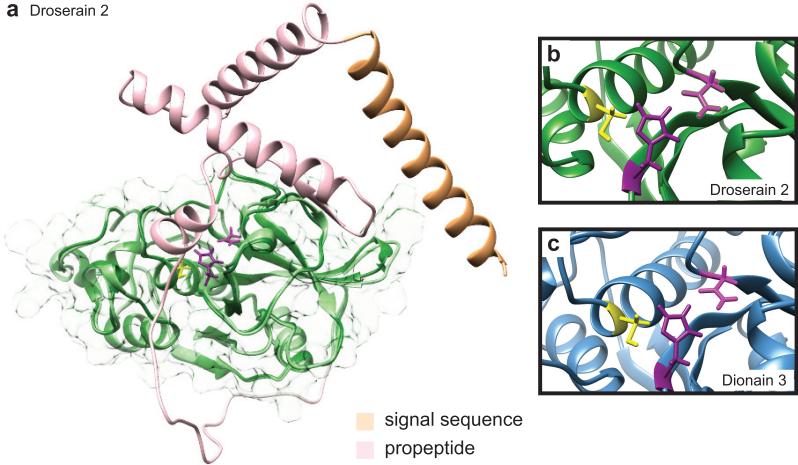
We separately calculated predicted structures for both the proenzymes and the predicted mature sequences for the D. capensis enzymes, as well as the dionains and zingipain 1. Interestingly, the structure selected for use as the primary template for the predicted structure is not necessarily that with the greatest sequence identity to the target. The parent structures for all cysteine proteases described in this study are reported in Supplementary Table 3. The Cys and His residues of the catalytic dyad are conserved in all cases, and adopt the same conformation within our predicted structures as in the crystal structure of papain [65]. Figure 4a shows the predicted structures of the full and mature sequences overlaid for droserain 2. Details of the active site regions with the catalytically important residues are highlighted for droserain 2 (Fig. 4b) and dionain 3 (Fig. 3c). With one exception (discussed below), in all of the cysteine proteases described here the active site residues are identical to those in papain: a Cys-His dyad with a stablizing Asn residue.
Figure 4.
Cysteine protease structure predictions. Rosetta was used to predict the structures of the D. capensis and D. muscipula cysteine proteases. a Two structures of each enzyme were calculated: the full-length chain, and the subsequence believed to represent the mature enzyme (dark green, transparent surface). This is illustrated here for droserain 2, with the full-length protein in light colors overlaid on the mature enzyme in dark green. Like all the droserains and dionains investigated, droserain 2 has a two-domain architecture with the active residues located in the interdomain cleft, similar to papain. The signal sequence (light orange) and the pro-sequence (pink) are cleaved during maturation. The PDB files for the predicted structures corresponding to the full-length and mature sequences of all the enzymes investigated are available in the Supporting Information. b, c The active sites of all the cysteine proteases investigated contain the active Cys and His residues in the expected configuration. Most also have the Asn residue that generally stabilizes the protonated His in the active configuration, shown here for droserain 2 and dionain 3.
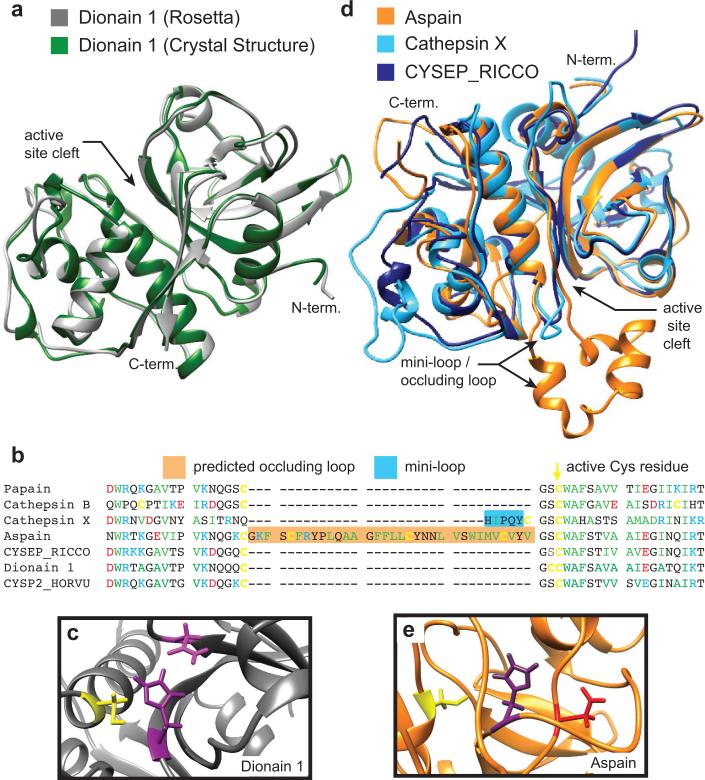
Comparison between our predicted structure (calculated on Sept. 15, 2015), and the crystal structure of Dionain 1 [66] (submitted to the PDB on Dec. 9, 2015), provides a serendipitous opportunity to validate our modeling approach. Because the Dionain 1 structure was submitted to the PDB after the calculation of our predicted structure, it was not in the training set available to Rosetta. This structure therefore serves as a useful validation against an out-of-sample observation. Figure 5a shows the predicted structure from Rosetta (gray) overlaid with the crystal structure (PDB ID 5A24, green). The agreement between the prediction and experimental results is excellent, with overlap of all major secondary structural elements and only minor deviations in the loop regions, which are expected to be more flexible. Figure 5b shows a partial sequence alignment of Dionain 1 with other cysteine proteases, including the parent of the predicted structure, which is cysteine endopeptidease B2 from Hordeum vulgare (PDB ID 2FO5) [67]. Comparison of these sequences reveals substantial conservation of the residues immediately surrounding the active cysteine. Figure 5c shows the active site residues of the predicted structure for Dionain 1, which are consistent with this enzyme being a functional cysteine protease. The excellent agreement between the molecular model and the experimentally deterimined structure indicate that the comparative modeling with all-atom refinement approach is capable of generating reasonable structural models, even in the case of only moderate sequence homology to the template molecules.
Figure 5.
a The predicted Rosetta structure of Dionain 1 (gray) overlaid with the subsequently solved crystal structure (PDBID 1EF7, green) [66]. b Structural comparison among the predicted structure of Aspain (orange), the structure of Ricinus communis CysEP (CYSEP_RICCO) used by Rosetta as the parent structure of Aspain (PDBID 1S4V, dark blue) [70], and Cathepsin X (PDBID 1EF7, light blue) [69]. Structure prediction suggests that aspain has an occluding loop located near the active site, in the same position as the mini-loop of Cathepsin X. c A sequence alignment of the residues near the active sites showing the position of the insertions representing the Cathepsin X mini-loop and Aspain occluding loop relative to the active cysteine residues. d The predicted active site of Dionain 1 with the active residues highlighted. e The active site of aspain contains the active Cys and His residues in the expected configuration, but the His appears to be stabilized by an Asp instead of an Asn.
In contrast to Dionain 1, which appears to be a relatively typical plant cysteine protease, Aspain contains some structural features not seen in known plant cysteine proteases, but observed in proteins from other organisms. An alignment of all sequences used in predicting the structure of this enzyme is shown in Supplementary Figure S5. The essential catalytic residues and disulfide bonds are conserved, however this protein also has a potential occluding loop, which may confer enhanced substrate specificity. This loop resembles the occluding loops that control substrate access, sometimes in a pH-dependent manner, in cysteine proteases from other organisms [51]. For example, the mini-loop of the human lysozomal protein Cathepsin X confers carboxypeptidase specificity [68, 69]. The predicted occluding loop appears immediately preceding the active Cys residue, in the same sequence position as the Cathepsin X mini-loop; however the corresponding insertion in Aspain is significantly longer (Figure 5b). Figure 5d shows structural comparisons of Aspain (orange), its parent structure, which is a cysteine endopeptidase (CysEP) from Ricinus communis (PDB ID 1S4V, dark blue) [70], and its structural homolog Cathepsin X (PDB ID 1EF7, light blue) [69]. Examination of the sequence alignment indicates that the Ricinus CysEP does not contain any insertions at the position of the mini-loop; the template structures used by Rosetta are chosen for overall structural homology while the structures of loop and linker regions are calculated from small fragments of the template structures [64].
Aspain also displays an unusual active site architecture, shown in Figure 5e. The stabilizing Asn residue is absent, but an Asp residue is located close enough to the protonated His to play this role (4.5 Å). An example of a similar active site in a papain-like protease is observed in the foot-and-mouth disease virus leader proteinase, where an Asp residue located 4.5 Å from the active His is required for full activity [71]. This enzyme has a highly specific substrate requirement, cleaving Lys-Leu-Lys*Gly-Ala-Gly to remove its own pro-sequence and Asn-Leu-Gly*Arg-Thr-Thr to disable the 5′ cap of the host ribosome [72]. Although it is impossible to determine the substrate preferences of this enzyme based on the predicted structure, it does provide an example of how molecular modeling can identify enzymes with unusual features that may represent novel activities as candidates for further experimental characterization.
D. capensis Aspartic Proteases Cluster Into Two Distinct Families
Aspartic proteases are acid-activated endopeptidases found in a diverse array of organisms, including animals, plants, fungi, eubacteria, and archaea. In most plants, aspartic proteases play different roles ranging from protein processing [73] to senescence and programmed cell death [74]. Some are constituitively expressed in many types of plant tissue [75], while others are involved in responses to stressors such as drought [76] and pathogen infection [77, 78, 79]. Here we focus on examples from MEROPS families A1A (pepsin, cathepsins) and A1B (nepenthesins), because these are known to function as digestive enzymes and are therefore of particular interest in carnivorous plants. Like the cysteine proteases, the aspartic proteases are expressed as zymogens and then cleaved post-translationally to yield the mature enzyme.
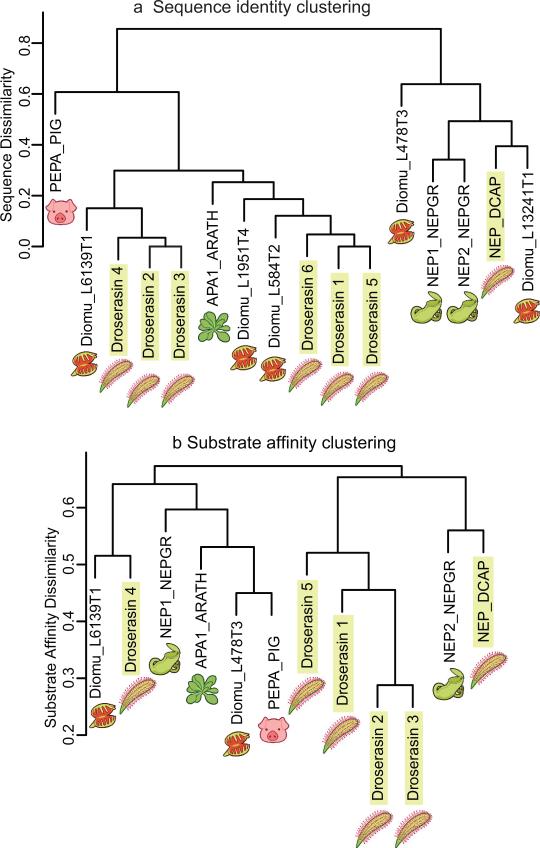
The aspartic proteases found in D. capensis and their homologs identified from the transcriptome of D. muscipula cluster into two distinct families; the nepenthesins, which were first discovered in the tropical pitcher plants (Nepenthes sp.), and the droserasins, which are related to pepsin. The cluster in Figure 6a shows the relationships among these carnivorous plant enzymes and the previously-annotated enzymes porcine pepsin and aspartic protease 1 from Arabidopsis thaliana (APA1_ARATH, a housekeeping protease). We find droserasins and nepenthesins both D. capensis and D. muscipula. A protein sequence alignment (Supplementary Fig. S6) comparing aspartic proteases from D. capensis and D. muscipula with some of their counterparts from N. gracilis and selected model organisms reveals well-defined sequence features differentiating the droserasins from the nepenthesins. A comparison among the six droserasins identified from D. capensis is shown in Supplementary Fig. S7. In addition to the full sequence alignments, we also compared the sequences of droserasins 1 and 2 to an 80-amino acid aspartic protease fragment previously identified from D. capensis (UniProt E9RJM0_DROCA) (Supplementary Fig. S8). Although the active residues are not present, this fragment clearly represents part of a droserasin: 74/80 residues are identical to droserasin 1 and 64/80 are identical to droserasin 2. Examination of the template structures used to model the droserasins and nepenthesins (Suppementary Tables 6-8 and Suppementary Figures S10-S12) shows that structures of porcine pepsin, porcine pepsinogen, as well as other mammalian aspartic proteases such as renin, represent a signficant fraction of the structures used. However, the nepenthesins and the droserasins are each characterized by features not found in mammalian pepsin that confer additional functionality.
Figure 6.
Cluster analysis of aspartic proteases from Caryophyllales carnivorous plants and model organisms. a The aspartic proteases found in D. capansis and D. muscipula. belong to two distinct groups, the pepsin family and the nepenthesin family. b Cluster analysis of aspartic proteases from Caryophyllales carnivorous plants and model organisms based on predicted substrate affinity. Here, porcine pepsin shows a similar substrate affinity pattern to Diomu_L478T3 (to which it shows little sequence similarity). The nepenthesins have diverse substrate preferences; Nepenthesin 1 clusters with droserasin 4, porcine pepsin, and the Arabadopsis protease APA1_ARATH), while Nepenthesin 2 and NAP_DCAP cluster separately. Droserasins 1-3 and 5 form a distinct group with similar substrate specificities, while droserasin 4 and Diomu_L6139T1 are similar to each other.
Aspartic Protease Models Reveal Diversity in Structural Features
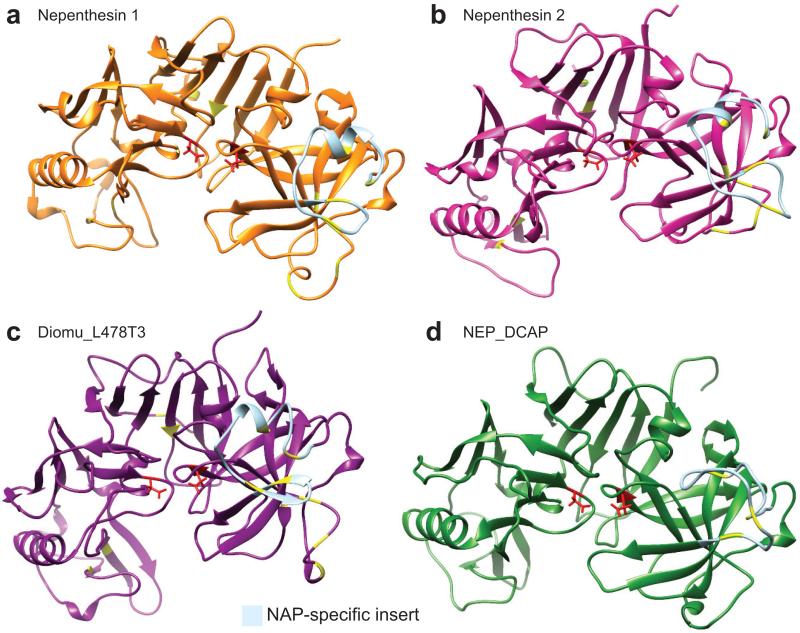
Signal sequences were predicted using the SignalP 4.1 server [40] for droserasins 1 and 2 and the putative D. capensis nepenthesin (Supplementary Fig. S9), as reported for nepenthesins 1 and 2 from N. gracilis [8]. Structural models of the subsequences believed to represent the mature nepenthesins (MEROPS family A1B) are shown in Fig. 7. Nepenthesin 1 (Fig. 7a) and nepenthesin 2 (Fig. 7b) from N. gracilis have been biochemically characterized, but no structures are yet available; we thus predict their three-dimensional structures using Rosetta. A list of the molecular models available can be found in Supplementary Table 5. The structures used in modeling a representative nepenthesin (Nepenthesin 2) are presented in Supplementary Table 6 and Supplementary Figures S10-S11. The nepenthesin homologs from D. muscipula (Fig. 7c, Diomu_L478T3) and D. capensis (Fig. 7d, NEP_DCAP) share with nepenthesins 1 and 2 a two-domain architecture, with the active Asp residues (red) located in a cleft between the domains. Their most distinct common feature is the nepenthesin aspartic protease (NAP)-specific insert (light blue). The nepenthesins contain the nepenthesin aspartic protease (NAP)-specific insert, which is a structured region containing three additional disulfide bonds not found in pepsin [7, 5]. These disulfide bonds are thought to account for the unusually high stability of nepenthesins. Overall, the nepenthesins have six additional cysteines, 4 in the NAP-specific insert and two preceding it, that are not present in mature pepsin-like proteases.
Figure 7.
Nepenthesin structure predictions. Three-dimensional structure prediction of nepenthesins 1 and 2 from N. gracilis (a and b), Diomu_L478T3 from D. muscipula (c), and NEP_DCAP from D. capensis (d). The active site Asp residues are highlighted in red, while the cysteines believed to be involved in disulfide bonds (by homology to nepenthesin 1) are shown in yellow. The NAP-specific insert, which is not removed during processing and contributes to the enhanced stability of the nepenthesins, is colored light blue.
The high stability and unique cleavage patterns of nepenthesins 1 and 2 have recently been used in mass spectrometry-based proteolysis studies [80, 81]. The molecular models shown in Fig. 7 predict similar structures for nepenthesins from different carnivorous plant species; however close examination of the models reveals some differences that may account for the diversity of their substrate preferences. Experimental structure determination of these enzymes and their complexes with substrates and inhibitors, will be critical for understanding their functional differences.
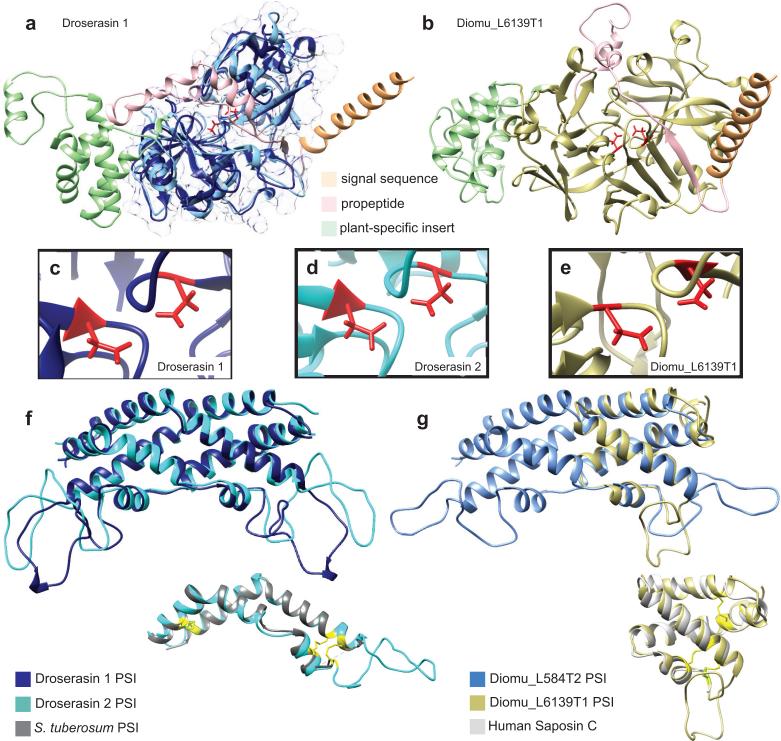
The droserasins (MEROPS A1A), are in many ways more straightforwardly similar to pepsin; however, enough subtle differences exist that the use of molecular modeling is required to identify the mature form of the enzyme for use in docking studies. The sequences used in the structure prediction of Droserasin 2 are presented in Supplementary Table 7 and Supplementary Figure S12, while the full list of parent sequences for all the aspartic proteases is given in Supplementary Table 8. As for the nepenthesins, mammalian aspartic proteases such as pepsin and renin appear frequently, as do the plant enzymes prophytepsin and edgp. Although the droserasins share the same overall architecture as the nepenthesins, they lack the NAP-specific insert and instead contain an interesting feature not found in mammalian enzymes or nepenthesins, the plant-specific insert (PSI). The PSI is a domain of 50-100 amino acids with moderate sequence similarity to mammalian saposins that is removed during post-translational processing to form a separate functional protein. Figure 9a shows both the subsequence predicted to represent the mature enzyme for droserasin 1 (dark blue) and the full sequence (overlaid in lighter colors representing different sequence regions).
Figure 9.
Structures of both pepsin-type aspartic proteases from D. capensis and D. muscipula predicted using Rosetta. a Structures were separately predicted for the full-length and mature sequences, illustrated here for droserasin 1. The mature sequence is shown in dark blue with a transparent surface, while the full sequence is overlaid in light colors. The signal sequence (light orange), pro-sequence (pink) and PSI (light green) are cleaved during maturation. b The full-length sequence for Diomu_L6139T1, with cleaved sequence regions indicated using the same annotations as in part a. c, d, e The active sites of droserasin 1, droserasin 2, and Diomu_6139T1, showing the active Asp residues in red. f Overlay of the PSIs for droserasins 1 (dark blue) and 2 (cyan). Inset: monomer of the droserasin 2 PSI overlaid with the crystal structure (PDBID 3RFI) of the PSI from a Solanum tuberosum aspartic protease (gray). The compact structure is held together by three disulfide bonds (yellow, Cys positions shown for the S. tuberosum PSI). g Overlay of the PSIs from Diomu_L584T2 and Diomu_L6139T1. Inset: the Diomu_L6139T1 PSI overlaid with a crystal structure of human saposin C (PDBID 2GTG). Disulfide bonds are shown in yellow.
A structural model of the full-length sequence of Diomu_L6139T1, a droserasin from D. muscipula is shown in Figure 9b. Diomu_L6139T1 has all the features of a functional droserasin, including the signal sequence, the pro-sequence, the active site, and the PSI. For each protein, the structures of the zymogen and the mature sequence were predicted independently using Rosetta; the structures are nearly superimposable over the sequence region where they coincide. The active sites of all the aspartic protease models examined have two Asp residues pointed toward each other as expected from the crystal structures of pepsin, as shown for droserasin 1 (Fig. 9c) and droserasin 2 (Fig. 9d, and Diomu_L6139T1 ((Fig. 9e). As expected, the secretion signal sequence (light orange) is a long helix, while the pro-sequence (pink) consists of helical and loop regions and blocks the active site.
Because the PSI is biologically active in its own right, the droserasin PSIs were modeled independently from the full-length protein. The template sequences used for the Droserasin 2 PSI are presented in Supplementary Table 7 and Supplementary Figure 13. The PSI domain was first observed in the crystal structure of a barley (Hordeum vulgare) aspartic protease, prophytepsin [82]. Known PSI proteins form membrane-associated dimers, and act to suppress the growth of fungal pathogens affecting both plants and humans [83]. After cleavage from its parent aspartic protease, the PSI acts as a pH-dependent fusogenic enzyme, disrupting membranes and promoting fusion in a similar manner to mammalian saposins and viral hemagglutinins, both of which it resembles in sequence and 3D structure. Interestingly, although the full-length enzymes are modeled using different structures, all the droserasin PSIs examined here are modeled based on the crystal structure of the S. tuberosum PSI (PDBID 3RFI) [84]. Except for that of Diomu_L6139T1, the droserasin PSIs are predicted to adopt a kinked structure composed of four short helices, which then assembles into a domain-swapped dimer (Fig. 9f and g. The PSI of Diomu_L6139T1 is predicted to form a more compact four-helix bundle, similar to structures observed for human saposins C and D. Upon closer inspection of the two different PSI structures from D. muscipula, the saposin fold can be overlaid over one half of the more extended dimeric structure, showing a clear relationship between these apparently disparate folds. Both types of PSI structure predicted here were observed in a recent modeling study of the S. tuberosum PSI [85] at different pH values, suggesting that the conformation of this domain is strongly dependent on solution conditions and providing a basis for future experimental studies. The question of pH-dependent structural changes and differences in activity are highly relevant to comparative studies of carnivorous plants, as the pH of Nepenthes pitcher fluid is about 2, while that of Drosera capensis mucilage is approximately 5. This suggests that the D. capensis digestive enzymes may be particularly useful in a laboratory context, because they are active under milder solution conditions.
Molecular Docking Predicts Differences in Aspartic Protease Substrate Affinities
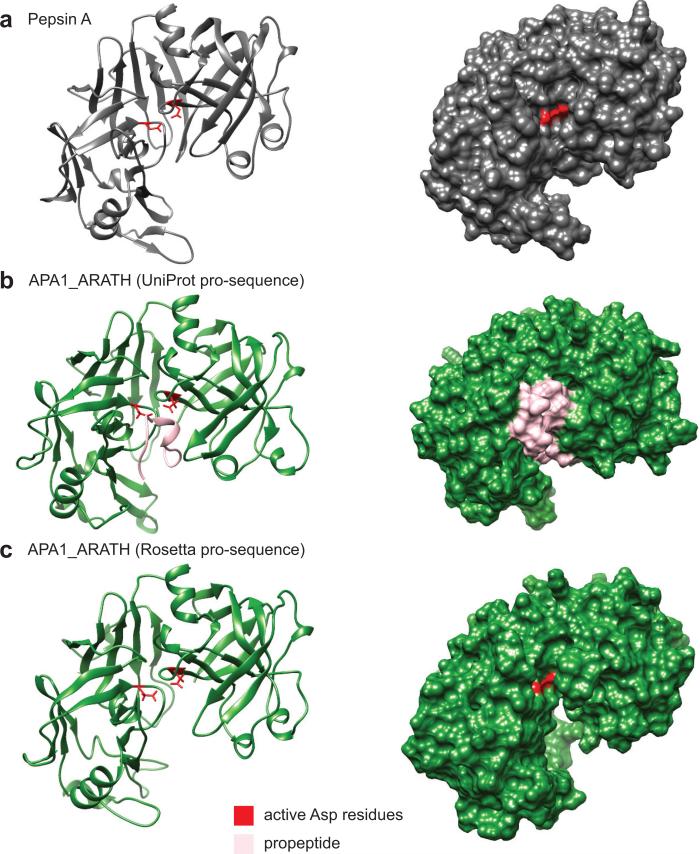
The cleavage sites for the pro-sequences of the droserasins were initially predicted using the UniProt annotations for a pepsin-family aspartic protease from Arabidopsis thaliana (UniProt O65390) for which no structures yet exist; the predicted cut site was based on sequence homology to pig pepsin A (Fig. 8a). However, the Rosetta structures revealed that some of the pro-sequence remained, blocking the active site (Fig. 8b). A new cleavage site was then predicted, based on structural rather than sequence homology to pepsin A (Fig. 8c). This approach resulted in structures with exposed active sites for all the proteins examined here, enabling the docking studies (Supplementary Fig. S13).
Figure 8.
Predictions of the pro-sequence cut site in a model aspartic protease from A. thaliana. a The 3D structure of the mature form of pig pepsin (PDBID: 4PEP). b The pro-sequence of APA1_ARATH was removed using sequence homology to pig pepsin. This approach leaves a residual part of the pro-sequence (pink), blocking the active site. c Removing the pro-sequence of APA1_ARATH according to structural homology to pig pepsin, using structures predicted with Rosetta, results in exposure of the active site residues (red).
Differences in protease substrate affinity were predicted in silico via docking experiments using Autodock Vina (v1.1.2) [46]. 327 test peptides six residues in length were constructed from Drosophila melanogaster myosin (heavy chain, muscle; Uniprot P05661) and docked to each predicted aspartic protease structure. Cleavage propensity for each peptide bond was predicted using affinity weights (see methods), and was ranked (over all 1635 bonds) for each protease; summed absolute differences in ranks were employed as a measure of difference in substrate affinity. A complete link hierarchical clustering of the resulting distances (rescaled against the maximum possible distance) is shown in Fig. 6b. Proteases clustered together show relatively similar affinity patterns across the test peptides. The pattern of similarity in substrate affinity has some resemblance to sequence similarity (Fig. 6a), but is in many respects distinct. For instance, porcine pepsin shows substantial differences in amino acid sequence from most of the other proteins examined, but shows a similar substrate affinity pattern to Diomu_L478T3 (to which it shows little sequence similarity). The nepenthesins appear to have diverse substrate preferences (with NEP_DCAP and NEP2 being closer to to each other than to the other proteins, and both being far from NEP1), with droserasins 1-3 and 5 having more consistent (and distinct) behavior. Droserasin 4 appears to behave most similarly to Diomu_L6139T1, with both being fairly dissimilar to their closest neighbor (NEP1); the Arabadopsis protease APA1_ARATH is somewhat intermediate between NEP1 and the DiomuŁ478T3/porcine pepsin cluster.
Conclusion
In summary, we report here the first genome of a carnivorous plant from the order Caryophyllales, Drosera capensis. Several new proteases were identified directly from the genomic DNA of D. capensis, and compared to their homologs found in the cDNA of Dionaea muscipula and in previously studied model organisms. Molecular modeling of these proteins suggests that they are functional and may display useful variation in their substrate specificity and cleavage patterns, making them promising targets for biochemical studies. The aspartic proteases identified here likely constitute the “ferment” conjectured in early observations of Drosera digestion. Although our focus here is on digestive enzymes, the prey capture mode of D. capensis suggests the presence of potentially novel transporters, glycosyltransferases, and other functional proteins. We expect that the D. capensis genome will be a rich resource for these and other potentially useful enzymes, and that molecular modeling techniques will be invaluable in identifying promising structural targets with biological relevance.
Supplementary Material
Acknowledgments
This work was made possible, in part, through access to the Genomic High Throughput Facility Shared Resource of the Cancer Center Support Grant (CA-62203) at the University of California, Irvine and NIH shared instrumentation grants 1S10RR025496-01 and 1S10OD010794-01; this research was also supported by NSF award DMS-1361425. The authors acknowledge Melanie Oakes, Jered Haun, Seung-Ah Chung, and Valentina Ciobanu at the UCI Genomics High-Throughput Facility for assistance with DNA sequencing, the Martin group for assistance with molecular modeling, and Veronika César for assistance with artwork in Figs. 3 and 6.
Footnotes
Author Contributions
C.T.B. grew the plant specimen, assembled and annotated the genome, and performed and analyzed the docking experiments. J.C.B. prepared samples and analyzed data. R.W.M. extracted the gDNA and performed protein sequence and structure analysis. All authors contributed to writing the manuscript.
References
- 1.Darwin C. Insectivorous Plants. John Murray; London: 1875. available: http://darwin-online.org.uk/converted/published/1875_insectivorous_f1217/1875_insect_f1217.html edition. [Google Scholar]
- 2.Leushkin EV, Sutormin RA, Nabieva ER, Penin AA, Kondrashov AS, Logacheva MD. The miniature genome of a carnivorous plant Genlisea aurea contains a low number of genes and short non-coding sequences. BMC Genomics. 2013;14 doi: 10.1186/1471-2164-14-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ibarra-Laclette E, Lyons E, Hernandez-Guzman G, Perez-Torres CA, Carretero-Paulet L, Chang T-H, Lan T, Welch AJ, Juarez MJA, Simpson J, Fernandez-Cortes A, Arteaga-Vazquez M, Gongora-Castillo E, Acevedo-Hernandez G, Schuster SC, Himmelbauer H, Minoche AE, Xu S, Lynch M, Oropeza-Aburto A, Cervantes-Perez SA, de Jesus Ortega-Estrada M, Cervantes-Luevano JI, Michael TP, Mock-ler T, Bryant D, Herrera-Estrella A, Albert VA, Herrera-Estrella L. Architecture and evolution of a minute plant genome. Nature. 2013;498:94–98. doi: 10.1038/nature12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellison AM, Gotelli NJ. Energetics and the evolution of carnivorous plants—Darwin's ’most wonderful plants in the world’. Journal of Experimental Botany. 2009;60:19–42. doi: 10.1093/jxb/ern179. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K, Athauda S, Matsumoto K, Rajapakshe S, Kuribayashi M, Kojima M, Kubomura-Yoshida N, Iwamatsu A, Shibata C, Inoue H. Nepenthesin, a unique member of a novel subfamily of aspartic proteinases: enzymatic and structural characteristics. Current Protein and Peptide Science. 2005;6:513–525. doi: 10.2174/138920305774933259. [DOI] [PubMed] [Google Scholar]
- 6.An CL, Fukusaki E, Kobayashi A. Aspartic proteinases are expressed in pitchers of the carnivorous plant nepenthes alata blanco. Planta. 2002;214:661–667. doi: 10.1007/s004250100665. [DOI] [PubMed] [Google Scholar]
- 7.Athauda S, Matsumoto K, Rajapakshe S, Kuribayashi M, Kojima N, and Kubomura-Yoshida M, Iwamatsu A, Shibata C, Inoue H, Takahashi K. Enzymic and structural characterization of nepenthesin, a unique member of a novel subfamily of aspartic proteinases. Biochemical Journal. 2004;381:295–306. doi: 10.1042/BJ20031575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buch F, Kaman WE, Bikker FJ, Yilamujiang A, Mithöfe A. Nepenthesin protease activity indicates digestive fluid dynamics in carnivorous Nepenthes plants. PLoS ONE. 2015;10:e0118853. doi: 10.1371/journal.pone.0118853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Libiaková M, Floková K, Novák O, Slováková L, Pavlovič A. Abundance of cysteine endopeptidase dionain in digestive fluid of Venus flytrap (dionaea muscipula ellis) is regulated by different stimuli from prey through jasmonates. PLoS ONE. 2014;9:e104424. doi: 10.1371/journal.pone.0104424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakamura Y, Reichelt M, Mayer VE, Mithöfer A. Jasmonates trigger prey-induced formation of outer stomach in carnivorous sundew plants. Proceedings of the Royal Society B. 2013;280 doi: 10.1098/rspb.2013.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bemm F, Becker D, Larisch C, Kreuzer I, Escalante-Perez M, Schulze WX, Ankenbrand M, Van de Weyer A-L, Krol E, Al-Rasheid KA, Mithöfer A, Weber AP, Schultz J, Hedrich R. Venus flytrap carnivorous lifestyle builds on herbivore defense strategies. Genome Research. 2016 doi: 10.1101/gr.202200.115. in press doi: 10.1101/gr.202200.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rawlings N, Waller M, Barrett A, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Research. 2014;42:D503–D509. doi: 10.1093/nar/gkt953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raman S, Vernon R, Thompson J, Tyka M, Sadreyev R, Pei J, Kim D, Kellogg E, DiMaio F, Lange O, Kinch L, Sheffler W, Kim B-H, Das R, Grishin NV, Baker D. Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins. 2009;77:89–99. doi: 10.1002/prot.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim D, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Research. 2004;32:W526–31. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thyme SB, Jarjour J, Takeuchi R, Havranek JJ, Ashworth J, Scharenberg AM, Stoddard BL, Baker D. Exploitation of binding energy for catalysis and design. Nature. 2009;461:1300–1304. doi: 10.1038/nature08508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajagopalan S, Wang C, Yu K, Kuzin AP, Richter F, Lew S, Miklos AE, Matthews ML, Seetharaman J, Su M, Hunt JF, Cravatt BF, Baker D. Design of activated serine–containing catalytic triads with atomic-level accuracy. Nature Chemical Biology. 2014;10:386–391. doi: 10.1038/nchembio.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M, DiMaio F, Zhou D, Gustchina A, Lubkowski J, Dauter Z, Baker D, Wlodawer A. Crystal structure of XMRV protease differs from the structures of other retropepsins. Nature Structural and Molecular BIology. 2011;18:207–209. doi: 10.1038/nsmb.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cozzetto D, Kryshtafovych A, Fidelis K, Moult J, Rost B, Tramontano A. Evaluation of template-based models in CASP8 with standard measures. Proteins: Structure, Function and Bioinformatics. 2009;77:18–28. doi: 10.1002/prot.22561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kryshtafovych A, Krysko O, Daniluk P, Dmytriv Z, Fidelis K. Protein structure prediction center in casp8, Proteins: Structure. Function and Bioinformatics. 2009;77:5–9. doi: 10.1002/prot.22517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen MK, Vogt JK, Bressendorff SA, Seguin-Orlando A, Petersen M, Sicheritz-Pontén T, Mundy J. Transcriptome and genome size analysis of the venus flytrap. PLoS ONE. 2015;10:e0123887. doi: 10.1371/journal.pone.0123887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Healey A, Furtado A, Cooper T, Henry RJ. Protocol: a simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods. 2014;10:21. doi: 10.1186/1746-4811-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimin A, Marçais G, Puiu D, Roberts M, Salzberg SL, Yorke JA. The MaSuRCA genome assembler. Bioinformatics. 2013;29 doi: 10.1093/bioinformatics/btt476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marçais G, Yorke JA, Zimin A. QuorUM: An error corrector for Illumina reads. PLoS ONE. 2015;10:e0130821. doi: 10.1371/journal.pone.0130821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo R, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1 doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett MD, Smith JB. Nuclear DNA amounts in angiosperms. Philosophical Transactions of the Royal Society of London B. 1976;274:227–274. doi: 10.1098/rstb.1976.0044. [DOI] [PubMed] [Google Scholar]
- 26.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parra G, Bradnam K, Korf I. CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 28.Campbell M, Law M, Holt C, Stein J, Moghe G, Hufnagel D, Lei J, Achawanantakun R, Jiao D, Lawrence CJ, Ware D, Shiu SH, Childs KL, Sun Y, Jiang N, Yandell M. MAKER-P: A tool-kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiology. 2013;164:513–524. doi: 10.1104/pp.113.230144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campbell MS, Holt C, Moore B, Yandell M. Genome annotation and curation using MAKER and MAKER-P. Current Protocols in Bioinformatics. 2014;48:4.11.1–4.11.39. doi: 10.1002/0471250953.bi0411s48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanke M, Keller O, Gunduz I, Hayes A, Waack S, Morgenstern B. AUGUSTUS: Ab initio prediction of alternative transcripts. Nucleic Acids Research. 2006;34:W435–W439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. InterProScan: Protein domains identifier. Nucleic Acids Research. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.OWLTools Development Team Owltools, software package. 2015 [Google Scholar]
- 35.R Core Team R. A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2015. [Google Scholar]
- 36.Butts CT. network: a package for managing relational data in R. Journal of Statistical Software. 2008;24 [Google Scholar]
- 37.Butts CT. Social network analysis with sna. Journal of Statistical Software. 2008;24 doi: 10.18637/jss.v024.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Handcock MS, Hunter DR, Butts CT, Goodreau SM, Morris M. statnet: Software tools for the representation, visualization, analysis and simulation of network data. Journal of Statistical Software. 2008;24:1–11. doi: 10.18637/jss.v024.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology. 2011;7:539–539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petersen T, Brunak S, von Heijne G, Henrik Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 41.Jones D. Protein secondary structure prediction based on position-specific scoring matrices. Journal of Molecular Biology. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 42.Buchan D, Minneci F, Nugent T, Bryson K, Jones D. Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Research. 2013;41:W340–W348. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones D. GenTHREADER: an efficient and reliable protein fold recognition method for genomic sequences. Journal of Molecular Biology. 1999;287:797–815. doi: 10.1006/jmbi.1999.2583. [DOI] [PubMed] [Google Scholar]
- 44.Lobley A, Sadowski M, Jones D. pGenTHREADER and pDomTHREADER: New methods for improved protein fold recognition and superfamily ddiscrimination. Bioinformatics. 2009;25:1761–1767. doi: 10.1093/bioinformatics/btp302. [DOI] [PubMed] [Google Scholar]
- 45.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity. Journal of Computational Chemistry. 2009;16:2785–91. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. Journal of Computational Chemistry. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z, Sun H, Yao X, Li D, Xu L, Li Y, Tian S, Hou T. Comprehensive evaluation of ten docking programs on a diverse set of protein-ligand complexes: the prediction accuracy of sampling power and scoring power. Phys. Chem. Chem. Phys. 2016;18:12964–12975. doi: 10.1039/c6cp01555g. [DOI] [PubMed] [Google Scholar]
- 48.F. W. P., Brylinski M. Calculating an optimal box size for ligand docking and virtual screening against experimental and predicted binding pockets. Journal of Cheminformatics. 2015;7:7–18. doi: 10.1186/s13321-015-0067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Athauda SBP, Matsumoto K, Rajapakshe S, Kuribayashi M, Kojima M, Kubomura-Yoshida N, Iwamatsu A, Shibata C, Inoue H, Takahashi K. Enzymic and structural characterization of nepenthesin, a unique member of a novel subfamily of aspartic proteinases. Biochemical Journal. 2004;381:295–306. doi: 10.1042/BJ20031575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi K, Laursen R. Amino-acid sequence and glycan structures of cysteine proteases with proline specificity from ginger rhizome Zingiber officinale. European Journal of Biochemistry. 2000;267:1516–1526. doi: 10.1046/j.1432-1327.2000.01152.x. [DOI] [PubMed] [Google Scholar]
- 51.Novinec M, Lenarčič B. Papain-like peptidases: structure, function, and evolution. Biomolecular Concepts. 2013;4:287–308. doi: 10.1515/bmc-2012-0054. [DOI] [PubMed] [Google Scholar]
- 52.Turk V, Stoka V, Vasiljeva O, Renko M, Sun T, Turk B, Turk D. Cysteine cathepsins: From structure, function and regulation to new frontiers. Biochimica et Biophysica Acta. 2012;1824:68–88. doi: 10.1016/j.bbapap.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duwadi K, Chen L, Menassa R, Dhaubhadel S. Identification, characterization and down-regulation of cysteine protease genes in tobacco for use in recombinant protein production. PLoS ONE. 2015;10:e0130556. doi: 10.1371/journal.pone.0130556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghosh R, Chakraborty S, Chakrabarti C, Dattagupta JK, Biswas S. Structural insights into the substrate specificity and activity of ervatamins, the papain-like cysteine proteases from a tropical plant, Ervatamia coronaria. FEBS Journal. 2008;275 doi: 10.1111/j.1742-4658.2007.06211.x. [DOI] [PubMed] [Google Scholar]
- 55.Vernet T, Tessier DC, Chatellier J, Plouffe C, Lee TS, Thomas D, Storer R, M'enard AC. Structural and functional roles of asparagine 175 in the cysteine protease papain. The Journal of Biological Chemistry. 1995;270:16645–16652. doi: 10.1074/jbc.270.28.16645. [DOI] [PubMed] [Google Scholar]
- 56.Visal S, Taylor M, Michaud D. The proregion of papaya proteinase IV inhibits Colorado potato beetle digestive cysteine proteinases. FEBS Letters. 1998;434:401–405. doi: 10.1016/s0014-5793(98)01018-7. PMID: 9742962. [DOI] [PubMed] [Google Scholar]
- 57.Silva F, Batista J, Marra B, Fragoso R, Monteiro A, Figueira E, Grossi-de M. Sá. Prodomain peptide of HGCP-iv cysteine proteinase inhibits nematode cysteine proteinases. Genetics and Molecular Research. 2004;3:342–355. [PubMed] [Google Scholar]
- 58.Santamaria ME, Arnaiz A, Diaz-Mendoza M, Martinez I, andDiaz M. Inhibitory properties of cysteine protease pro-peptides from barley confer resistance to spider mite feeding. PLoS ONE. 2015;10:e0128323. doi: 10.1371/journal.pone.0128323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marra B, Souza D, Aguiar J, Firmino A, Sarto R, Silva F, Almeida C, Cares J, Continho M, Martins-de-Sá C, Franco O, Grossi-de-Sá M. Protective effects of a cysteine proteinase propeptide expressed in transgenic soybean roots. Peptides. 2009;30:825–831. doi: 10.1016/j.peptides.2009.01.022. [DOI] [PubMed] [Google Scholar]
- 60.Rovenska G, Zemek R, Schmidt J, Hilbeck A. Altered host plant preference of Tetranychus urticae and prey preference of its predator Phytoseiulus persimilis (Acari: Tetranychidae, Phytoseiidae) on transgenic Cry3Bbeggplants. Biological Control. 2005;33:293–300. PMID: 15781137. [Google Scholar]
- 61.Karrer K, Peiffer S, DiTomas M. Two distinct gene subfamilies within the family of cysteine protease genes. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:3063–3067. doi: 10.1073/pnas.90.7.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takahashi K, Nishii W, Shibata C. The digestive fluid of drosera indica contains a cysteine endopeptidase (“droserain”) similar to dionain from Dionaea muscipula. Carnivorous Plant Newsletter. 2012;41:132–134. [Google Scholar]
- 63.Qian B, Raman S, Das R, Bradley P, McCoy A, Read R, Baker D. High-resolution structure prediction and the crystallographic phase problem. Nature. 2007;450:259–264. doi: 10.1038/nature06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang C, Schueler-Furman O, Andre I, London N, Fleishman S, Bradley P, Qian B, Baker D. RosettaDock in CAPRI rounds 6–12. Proteins. 2007;69:758–763. doi: 10.1002/prot.21684. [DOI] [PubMed] [Google Scholar]
- 65.Kamphuis I, Kalk K, Swarte M, Drenth J. Structure of papain refined at 1.65å resolution. Journal of Molecular Biology. 1984;2:233–256. doi: 10.1016/0022-2836(84)90467-4. [DOI] [PubMed] [Google Scholar]
- 66.Risør MW, Thomsen LR, Sanggaard KW, Nielsen TA, Thøgersen IB, Lukassen MV, Rossen L, Garcia-Ferrer I, Guevara T, Scavenius C, Meinjohanns E, Gomis-Rüth FX, Enghild JJ. Enzymatic and structural characterization of the major endopeptidase in the Venus fly-trap digestion fluid. The Journal of Biological Chemistry. 2016;291:2271–2287. doi: 10.1074/jbc.M115.672550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bethune M, Strop P, Tang Y, Sollid L, Khosla C. Heterologous expression, purification, refolding, and structural-functional characterization of EP-B2, a self-activating barley cysteine endoprotease. Chemistry & Biology. 2006;13:637–647. doi: 10.1016/j.chembiol.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 68.Nägler D, Zhang R, Tam W, Sulea T, Purisima E, Ménard R. Human cathepsin X: A cysteine protease with unique carboxypeptidase activity. Biochemistry. 1999;38:12648–12654. doi: 10.1021/bi991371z. [DOI] [PubMed] [Google Scholar]
- 69.Guncar G, Klemencic I, Turk B, Turk V, Karaoglanovic-Carmona A, Juliano L, Turk D. Crystal structure of cathepsin X: a flip-flop of the ring of His23 allows carboxy-monopeptidase and carboxy-dipeptidase activity of the protease. Structure: Folding and Design. 2000;8:305–313. doi: 10.1016/s0969-2126(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 70.Than ME, Helm M, Simpson DJ, Lottspeich F, Huber R, Gietl C. The 2.0 ǎcrystal structure and substrate specificity of the KDEL-tailed cysteine endopeptidase functioning in programmed cell death of Ricinus communis endosperm. Journal of Molecular Biology. 2004;336:1103–1116. doi: 10.1016/j.jmb.2003.12.075. [DOI] [PubMed] [Google Scholar]
- 71.Kronovetr J, Skern T. Foot-and-mouth disease virus leader proteinase: a papain-like enzyme requiring an acidic environment in the active site. FEBS Letters. 2002;528:58–62. doi: 10.1016/s0014-5793(02)03237-4. [DOI] [PubMed] [Google Scholar]
- 72.Glaser W, Cencic R, Skern T. Foot-and-mouth disease virus leader proteinase: Involvement of C-terminal of residues in self-processing and cleavage of eIF4GI. Journal of Biological Chemistry. 2001;276:35473–35481. doi: 10.1074/jbc.M104192200. [DOI] [PubMed] [Google Scholar]
- 73.Simoes I, Faro C. Structure and function of plant aspartic proteinases. European Journal of Biochemistry. 2004;271:2067–2075. doi: 10.1111/j.1432-1033.2004.04136.x. [DOI] [PubMed] [Google Scholar]
- 74.García-Lorenzo M, Sjödin A, Jansson S, Funk C. Protease gene families in populus and arabidopsis. BMC Plant Biology. 2006;6:30. doi: 10.1186/1471-2229-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takahashi K, Niwa H, Yokota N, Kubota K, Inoue H. Widespread tissue expression of nepenthesin-like aspartic protease genes in Arabidopsis thaliana. Plant Physiology and Biochemistry. 2008;46:724–729. doi: 10.1016/j.plaphy.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 76.Yao X, Xiong W, Ye T, Wu Y. Overexpression of the aspartic protease ASPG1 gene confers drought avoidance in Arabidopsis. Journal of Experimental Botany. 2012;63:2579–2593. doi: 10.1093/jxb/err433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guo R, Xu C, Bassett X, Li M, Gao X, Zheng Y, Wang X. Genome-wide identification, evolutionary and expression analysis of the aspartic protease gene superfamily in grape. BMC Genomics. 2013;14:554. doi: 10.1186/1471-2164-14-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vicente Ramírez V, López A, Brigitte Mauch-Mani B, Ma José Gil MJ, Vera P. An extracellular subtilase switch for immune priming in Arabidopsis. PLoS Pathogens. 2013;9:e1003445. doi: 10.1371/journal.ppat.1003445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fernandez MB, Daleo GR, Guevara MG. Isolation and characterization of a Solanum tuberosum subtilisin-like protein with caspase-3 activity (StSBTc-3) Plant Physiology and Biochemistry. 2015;86:137–146. doi: 10.1016/j.plaphy.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 80.Kadek A, Mrazek H, Halada P, Rey M, Schriemer DC, Man P. Aspartic protease nepenthesin-1 as a tool for digestion in hydrogen/deuterium exchange mass spectrometry. Analytical Chemistry. 2014;86:4287–4294. doi: 10.1021/ac404076j. [DOI] [PubMed] [Google Scholar]
- 81.Yang M, Hoeppner M, Rey M, Kadek A, Man P, Schriemer DC. Recombinant nepenthesin II for hydrogen/deuterium exchange mass spectrometry. Analytical Chemistry. 2015;87:6681–6687. doi: 10.1021/acs.analchem.5b00831. [DOI] [PubMed] [Google Scholar]
- 82.Kervinen G, Tobin J, Costa J, Waugh D, Wlodawer A, Zdanov A. Crystal structure of plant aspartic proteinase prophytepsin: inactivation and vacuolar targeting. EMBO Journal. 1999;18:3947–3955. doi: 10.1093/emboj/18.14.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guevara M, Verissimo P, Pires E, Faro C, Daleo D. Potato aspartic proteases: induction, antimicrobial activity and substrate specificity. Journal of Plant Pathology. 2004;86:233–238. [Google Scholar]
- 84.Bryksa B, Bhaumik P, Magracheva E, De Moura D, Kurylowicz M, Zdanov A, Dutcher J, Wlodawer A, Yada R. Structure and mechanism of the saposin-like domain of a plant aspartic protease. Journal of Biological Chemistry. 2011;286:28265–28275. doi: 10.1074/jbc.M111.252619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.De Moura DC, Bryksa BC, Yada RY. In silico insights into protein-protein interactions and folding dynamics of the saposin-like domain of Solanum tuberosum aspartic protease. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.