Abstract
Cancer cells have high rates of glycolysis and lactic acid fermentation in order to fuel accelerated rates of cell division (Warburg effect). Here, we present a strategy for merging cancer and yeast metabolism to remove pyruvate, a key intermediate of cancer cell metabolism, and produce the toxic compound acetaldehyde. This approach was achieved by administering the yeast enzyme pyruvate decarboxylase to triple negative breast cancer cells. To overcome the challenges of protein delivery, a nanoparticle-based system consisting of cationic lipids and porous silicon was employed to obtain efficient intracellular uptake. The results demonstrate that the enzyme therapy decreases cancer cell viability through production of acetaldehyde and reduction of lactic acid fermentation.
Keywords: Cancer metabolism, multistage vector, nanotechnology, pyruvate decarboxylase, Warburg effect
Graphical abstract
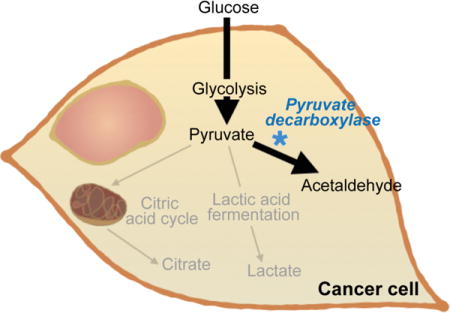
1. Introduction
Approximately half a million people are annually expected to die of cancer in the United States, making this disease the second leading cause of death [1]. Although cancer is an umbrella term for over one hundred diseases, there exist common characteristics shared by all cancers, often referred to as the hallmarks of cancer [2, 3]. These hallmarks include self-sustained growth, evasion of apoptosis, unlimited replication, invasion, angiogenesis, abnormal metabolism, evasion of the immune system, genomic instability, and inflammation. Many strategies have been employed to target these hallmarks, including cytotoxic drugs, natural compounds, hormonal therapies, antibodies, oncolytic viruses, immune system modulators, photothermal therapies, and small interfering RNA (siRNA) [4–16]. The majority of these cancer therapeutics aims to halt cancer progression by initiating programmed cell death or preventing cell division. The major challenge in developing therapeutics lies in finding ways to specifically target cancer cells without detrimentally affecting normal cells. Moreover, the prevalence of drug resistance necessitates the development of new therapeutics and strategies so that patients may continue to receive efficacious therapies [4]. A promising but less commonly used strategy for suppressing cancer growth involves targeting abnormal metabolic pathways.
Here, we have devised a strategy for merging metabolic pathways in cancer and yeast cells for therapeutic purposes. Namely, the yeast enzyme pyruvate decarboxylase has been used to interfere with cancer cell metabolism through the consumption of pyruvate and the production of acetaldehyde (Figure 1), a reaction that does not typically occur in mammals [17]. Pyruvate is the substrate for both the citric acid cycle (aerobic metabolism) and lactic acid fermentation (anaerobic metabolism). These respiratory pathways are essential for cell survival as they produce energy in the form of adenosine triphosphate (ATP) and precursors for biomolecules utilized to generate new cells [18]. Despite lower energy yields, cancer cells have been shown to favor high rates of lactic acid fermentation even in the presence of oxygen [19]. This phenomenon, termed the Warburg effect, is thought to occur because cancer cells are less dependent on efficient energy production than the generation of biological precursors that can serve as building blocks for new cells [20]. In fact, the process of converting pyruvate to lactate produces a supply of intermediate metabolites, which can be used to construct lipids, nucleic acids, and proteins that are essential materials for cells. The Warburg effect also entails higher levels of glycolysis, resulting in increased production of the glycolytic metabolite pyruvate [20, 21]. Since pyruvate decarboxylase specifically targets pyruvate, which is produced in high amounts in cancer cells, the resulting therapeutic effects are likely to be more pronounced in malignant cells. In addition to suppressing lactic acid fermentation, the conversion of pyruvate to acetaldehyde could also have detrimental effects on cancer cells due to the toxic effects of acetaldehyde, which include increased membrane permeability and reduced cell viability [22]. In particular, exposure to acetaldehyde has been shown to cause polymerization of microtubules, lipid peroxidation, collagen production, and adverse mitochondrial effects [23].
Figure 1.
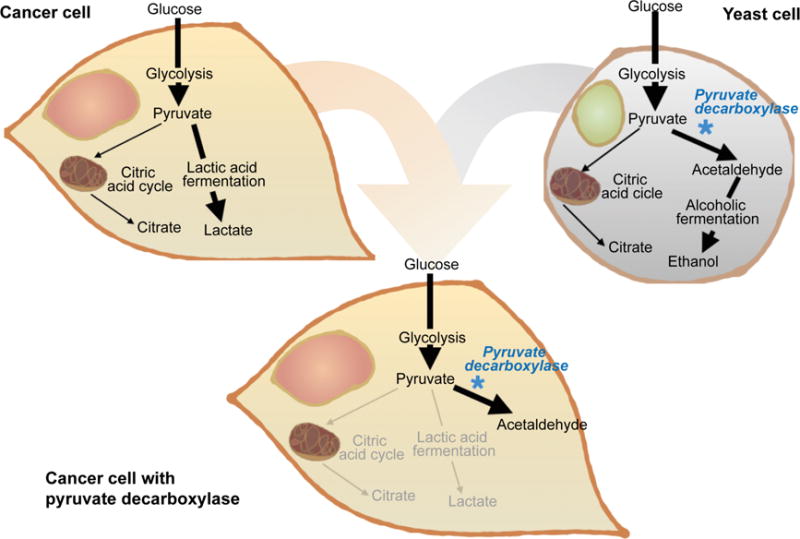
Schematic representation of select metabolic pathways in cancer cells and yeast cells. The figure demonstrates how the enzyme pyruvate decarboxylase can be used to converge cancer and yeast metabolic pathways. The exposure of cancer cells to this enzyme prevents pyruvate from efficiently being utilized for lactic acid fermentation. In addition, pyruvate decarboxylase produces the toxic compound acetaldehyde. This therapeutic strategy aims to reduce cancer cell viability through a dual approach: the suppression of critical metabolic pathways and the generation of a toxic substance.
Pyruvate decarboxylase has not been used in cancer treatment since 1966 when it was proven to successfully inhibit tumor cell glycolysis by competitive enzyme inhibition, resulting in significant reduction of tumor growth [24]. Despite the study’s promising results, it is likely that the therapeutic effects were drastically limited as the authors lacked strategies for intracellular delivery of the enzyme into cancer cells. Notably, the cellular internalization of therapeutic peptides and proteins is an area of great interest in drug delivery research today [25]. Moreover, even if proteins are able to enter the cells through receptor-mediated endocytosis, entry through endosomal uptake usually concludes with enzymatic degradation in the lysosome [26]. Protein delivery is now possible with current technologies, and studies have demonstrated varying success with liposomal vehicles [27, 28] and cell-penetrating peptides [29–31]. In fact, nanomedicine has shown great promise in recent years and several nanodrugs have received clinical approval, including various liposomal and polymer-based carriers [32, 33]. Nanoparticles offer an approach to overcome transport obstacles in the body, consequently increasing therapeutic efficacy and reducing side effects. In particular, multistage delivery platforms that have several components are well-equipped to navigate biological barriers [34–36].
In this study, the commercial cationic lipid-based protein delivery vehicle PULSin was used in combination with a multistage vector (MSV) system to facilitate cellular internalization of pyruvate decarboxylase. PULSin works by forming a protein-lipid complex that can enter cells through endocytosis and then escape from the lysosome into the cytosol [37]. PULSin consists of the cationic lipid dioctadecylamido-glycylspermine (DOGS) [37]. Since DOGS and other cationic lipids frequently display charge-induced toxicity [38], PULSin was assembled with the MSV delivery system to increase biocompatibility. The MSV is a porous silicon-based microparticle that can be loaded with a wide variety of nanoparticles [39–48]. Cell culture studies have shown that MSV particles are internalized through endocytosis [39, 49–51]. Additionally, multiple animal studies have demonstrated that the MSV carrier is non-toxic [39, 52–55]. Previously, we have also demonstrated that the MSV can eliminate the toxicity of positively-charged polymeric nanoparticles [39]. In addition to increasing the safety of nanotherapeutics, the MSV protects the cargo from degradation, preferentially binds to tumor vasculature, and enables sustained release of cargo [35]. Previous studies investigating the degradation of the MSV have demonstrated that the particles disintegrate within 48 h, leading to the gradual release of cargo [56–59]. In particular, studies have shown that the MSVs degrade faster in cell culture media (24 h) than in saline (48 h) [57]. Taken together, these characteristics make the MSV carrier an ideal candidate for protein delivery.
2. Materials and methods
2.1. Materials
Materials were acquired from the following sources: penicillin-streptomycin solution, M-PER mammalian protein extraction reagent, phosphate buffered saline (PBS; Hyclone), and fetal bovine serum (FBS; Hyclone) from Thermo Fisher Scientific; Dulbecco’s modified Eagle’s Medium (DMEM; 4 mM L-glutamine, 4.5 g/L glucose, sodium pyruvate; Hyclone) from GE Healthcare Life Sciences; PULSin protein delivery kit (with R-phycoerythrin and HEPES buffer) from Polyplus-transfection; pyruvate decarboxylase from baker’s yeast (S. cerevisiae) from Sigma Aldrich; MDA-MB-468 and MDA-MB-231 human breast cancer cells from ATCC; L-lactate assay kit (colorimetric) and aldehyde quantification assay kit (colorimetric) from Abcam; CellTiter 96 AQueous non-radioactive cell proliferation assay from Promega.
2.2. Particle assembly and characterization
A PULSin/protein complex was formed according to the manufacturer’s instruction. Briefly, pyruvate decarboxylase or R-phycoerythrin were diluted in 20 mM Hepes buffer at a concentration of 15 μg/ml and mixed with PULSin at a concentration of 4 μl/μg protein. The PULSin/protein complex was separated from free protein through centrifugation (21,100 × g, 1 h). The supernatant was analyzed with dynamic light scattering (DLS) to ensure that the PULSin/protein complex was pelleted. The protein encapsulation efficiency was determined by measuring the fluorescence intensity of R-phycoerythrin in the supernatant using a Synergy H4 Hybrid microplate reader (BioTek, Ex 488 nm/Em 575 nm). The size of pyruvate decarboxylase, PULSin, and PULSin/pyruvate decarboxylase in HEPES buffer was measured with DLS using a Zetasizer Nano ZS (ZEN 3600) from Malvern Instruments.
MSV particles were fabricated using electrochemical etching and photolitography as previously reported [60]. The particles were oxidized in a 30% solution of hydrogen peroxide at 95 °C for 2 h, washed, and counted following previously established protocols [41]. Scanning electron microscopy (SEM) images of the MSVs were obtained as previously described [57]. For MSV loading, the PULSin/protein complex was mixed with MSV particles (7 × 104 – 1.06 × 106 MSV particles/μg protein) using a benchtop shaker (750 rpm, 30 min, room temperature). The MSV particle/PULSin/protein complex was separated from free PULSin and protein through centrifugation (900 × g, 3 min). The supernatant was analyzed with DLS to ensure that the PULSin/protein complex remained in the supernatant, while the MSV particle/PULSin/protein complex was pelleted. The PULSin loading efficiency in the MSV was determined by measuring the fluorescence intensity of R-phycoerythrin (Ex 488 nm/Em 575 nm) in the supernatant as described above and accounting for the protein encapsulation efficiency in PULSin. For further studies, a ratio of 1.3 × 105 MSV particles/μg protein was used. The zeta potential of pyruvate decarboxylase, PULSin, PULSin/pyruvate decarboxylase, MSV particles, MSV particle/PULSin, and MSV particle/PULSin/pyruvate decarboxylase in distilled water was measured by laser doppler microelectrophoresis (Smoluchowski’s theory) using a Zetasizer Nano ZS (ZEN 3600).
2.3. Cell culture
MDA-MB-231 and MDA-MB-468 cells were cultured in DMEM supplemented with 10% fetal bovine serum, 100 units/mL penicillin, and 100 μg/mL streptomycin. Cells were maintained in a humidified incubator at 37°C and 5% CO2.
2.4. Cellular uptake efficiency
Protein delivery with the PULSin protein delivery kit was performed as per manufacturer’s directions. MDA-MB-468 cells were seeded overnight in 96-well plates at a density of 4 × 103 cells/well. The cells were exposed to PULSin (1.2 μl/well) or PULSin/R-phycoerythrin (0.3 μg protein/1.2 μl PULSin/well) in serum-free media for 4 h, after which the media was replaced with complete growth media. The fluorescence intensity of the cells was measured after 24 h and 72 h using a Synergy H4 Hybrid microplate reader (BioTek, Ex 488 nm/Em 575 nm). For studies of R-phycoerythrin uptake and MSV particle/PULSin/R-phycoerythrin uptake, MDA-MB-468 cells were seeded as described above and particles were added to complete media and incubated with cells for 24 h, 48 h, or 72 h (0.3 μg protein/well). The fluorescence intensity was measured as described above. The cellular uptake of R-phycoerythrin delivered with the MSV/PULSin complex was also visualized in live cells using an Eclipse Ti inverted fluorescence microscope (Nikon).
2.5. Cell viability studies
MDA-MB-468 cells were seeded in 96-well plates overnight at a density of 4 × 103 cells/well. The cells were exposed to PULSin (1.2 μl/well), pyruvate decarboxylase (0.3 μg/well), or PULSin/pyruvate decarboxylase (0.3 μg protein/1.2 μl PULSin/well) in serum-free media for 4 h, after which the media was replaced with complete media. Cell viability was measured with the CellTiter 96AQueous non-radioactive cell proliferation assay according to the manufacturer’s instructions. Absorbance was measured with a Synergy H4 Hybrid microplate reader (BioTek, λ = 490 nm). Results were normalized to control cells. For studies of cell viability upon exposure to MSV particle/PULSin, MDA-MB-468 and MDA-MB-231 cells were seeded as described above. The cells were then treated with MSV particles (5.6 × 104 particles/well), PULSin (1.2 μl/well), or MSV particles/PULSin (1.2 μl PULSin/5.6 × 104 particles/well) for 24 h, 48 h, or 72 h. MDA-MB-468 cells were also exposed to MSV particles/PULSin/pyruvate decarboxylase (0.3 μg protein/well). Cell viability was measured as described above.
2.6. Measurements of lactate and acetaldehyde
For measurements of lactate concentration, MDA-MB-468 cells were seeded overnight in 96-well plates at a density of 6.6 × 103 cells/well. The cells were exposed to PULSin (1.2 μl/well), pyruvate decarboxylase (0.3 μg/well), or PULSin/pyruvate decarboxylase (0.3 μg protein/1.2 μl PULSin/well) in serum-free media for 4 h, after which the media was replaced with media containing 1% FBS. After 24, the cell culture media was analyzed using the colorimetric L-lactate assay kit according to the manufacturer’s instructions. The absorbance was read with a Synergy H4 Hybrid microplate reader (BioTek, λ = 450 nm). For measurements of acetaldehyde concentration, cells were seeded and treated as described above. Following treatment, the media was replaced with complete media. After 24 h, cells were lysed with the M-PER mammalian protein extraction reagent and the lysates were analyzed using the colorimetric aldehyde quantification assay kit according to the manufacturer’s instructions. The absorbance of the lysates was measured at various time points using a Synergy H4 Hybrid microplate reader (BioTek, λ = 405 nm).
2.7. Statistical analysis
T-test comparisons (two-tailed, unpaired) were conducted to evaluate statistical significance between two groups.
3. Results and discussion
3.1. PULSin assembly and characterization
A crucial determinant of the success of pyruvate decarboxylase therapy is successful delivery of this enzyme. Notably, protein delivery is usually challenging due to the size and instability of these biomolecules [25]. Enzymes are especially vulnerable, since minor alterations in protein structure can lead to loss of activity. PULSin was selected as a delivery vehicle, since this lipid formulation has previously been shown to form a complex with proteins without affecting protein activity [37]. Moreover, while most protein delivery strategies require complex conjugation procedures, a protein-PULSin complex can be formed with a simple mixing step. Additionally, PULSin has been shown to permit cellular internalization and endosomal escape of proteins [37]. In this study, pyruvate decarboxylase was complexed with PULSin in order to enable intracellular delivery. The size and zeta potential of the free enzyme, PULSin, and the PULSin/enzyme complex were measured with DLS and laser doppler microelectrophoresis. The results reveal that PULSin nanoparticles (64 ± 14 nm) undergo a slight increase in size upon forming a complex with the enzyme (70 ± 28 nm) (Figure 2A). Moreover, the zeta potential of PULSin decreases from +53.2 mV (± 0.8 mV) to +27.2 mV (± 0.5 mV) after protein complexation (Figure 2B). Analysis of the protein encapsulation efficiency demonstrated that 70.7% of the added protein amount was complexed with the PULSin carrier (Figure 2C). The corresponding loading capacity was 8.8%
Figure 2.
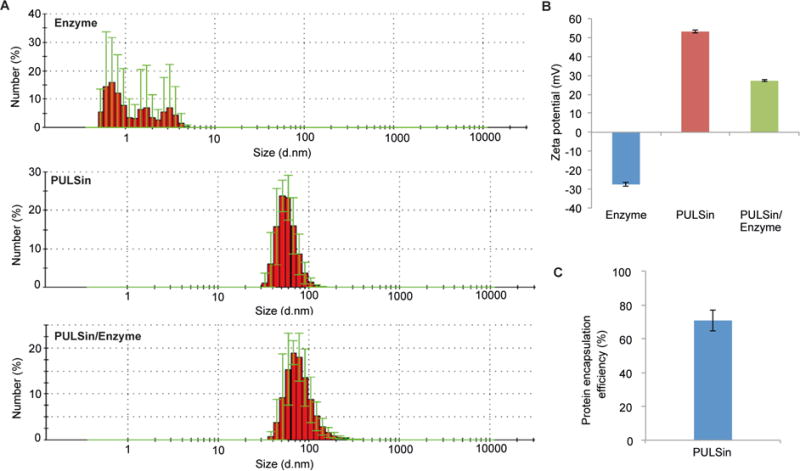
Characterization of the enzyme delivery system. A) The size of the enzyme (pyruvate decarboxylase), PULSin, and the PULSin/enzyme complex were determined using dynamic light scattering. Data is presented as mean ± s.d. of five measurements (10 runs each). B) The zeta potential was measured with laser doppler micro-electrophoresis. Data is presented as mean ± s.d. of five measurements (10 runs each). C) Protein encapsulation efficiency was determined utilizing the fluorescent protein R-phycoerythrin. Data is presented as mean ± s.d. of triplicates.
3.2. Enzyme delivery
The treatment strategy of merging cancer and yeast metabolism is particularly well suited for triple-negative breast cancer (TNBC), since this type of cancer displays increased levels of glucose uptake [61, 62] and glycolysis [63], the metabolic pathway that converts glucose to pyruvate. Moreover, the treatment options for TNBC are limited and the disease usually has worse outcomes compared to other types of breast cancer, necessitating the development of new therapeutic strategies [64]. Here, the anticancer effect of pyruvate decarboxylase therapy was assessed in the TNBC cell line MDA-MB-468, since this cell line was previously reported to have the highest rates of glucose consumption and lactate production in a panel of six breast cancer cell lines [65]. To assess the cellular uptake efficiency of the PULSin complex, the fluorescent protein R-phycoerythrin was used as a model protein. The results indicate that intracellular protein delivery in MDA-MB-468 cells was effective using the PULSin transfection method. In particular, the fluorescence intensity of cells exposed to the PULSin/protein complex was significantly higher than the background fluorescence of cells (Figure 3A).
Figure 3.
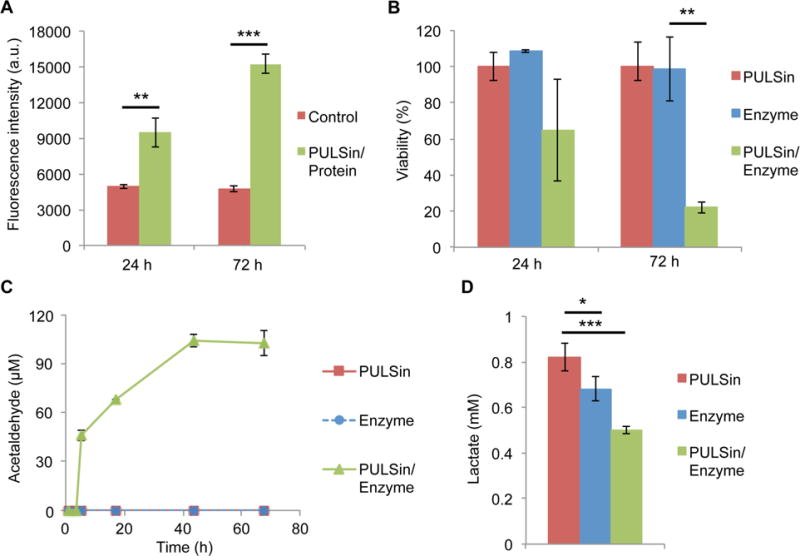
Biological effects of the PULSin/pyruvate decarboxylase complex in MDA-MB-468 breast cancer cells. A) Cellular uptake of PULSin/protein (R-phycoerythrin). Untreated cells were used as a control. B) Cell viability after 24 h and 72 h. Values were normalized to those of cells exposed to PULSin. C) Measurements of acetaldehyde concentration in cell culture lysates after 24 h. Time points represent the reaction time after initiating the assay. D) Measurements of lactate concentration in cell culture media after 24 h. Data is presented as mean ± s.d. of triplicates. **P < 0.01; ***P < 0.001.
3.3. Anticancer effects of pyruvate decarboxylase
Upon confirming that the PULSin carrier could be used to deliver proteins to MDA-MB-468 cells, a cell viability assay was performed to assess the anticancer activity of pyruvate decarboxylase. The results demonstrate that the enzyme caused an ~80% reduction in cell viability after 72 h, while no statistically significant difference between control cells and treated cells could be seen after 24 h (Figure 3B). Notably, cells treated with the free enzyme, did not display any loss of viability at 24 h or 72 h, indicating that intracellular delivery of the protein is necessary for therapeutic activity (Figure 3B). To confirm that enzyme activity was maintained following intracellular delivery the levels of acetaldehyde, the product in the enzyme reaction, was measured in protein lysates of cells exposed to the PULSin/pyruvate decarboxylase complex. Since the enzyme caused the majority of cells to undergo cell death in response to treatment after 72 h, the levels of acetaldehyde were measured after 24 h. Analysis of lysates from cells treated with PUSLin/pyruvate decarboxylase showed that the levels of acetaldehyde increased for 44 h, after which a plateau was observed, potentially due to depletion of pyruvate (Figure 3C). In fact, no detectable levels of acetaldehyde could be observed in the samples during the first 4 h of analysis, indicating that the production of acetaldehyde was initiated after cell lysis when samples were brought to room temperature. Therefore, it is plausible that cells use efflux pumps to rapidly expel acetaldehyde, a phenomenon that has previously been proposed to take place in yeast cells [66]. These results provide evidence that complexation with PULSin and exposure to an intracellular environment does not impair enzyme function.
Moreover, since glycolysis-derived pyruvate is primarily diverted to lactate fermentation in cancer cells, the levels of lactate in MDA-MB-468 cells were measured. Exposure to the PULSin/pyruvate decarboxylase complex caused the lactate levels in the cell culture media to decrease from 0.82 mM to 0.5 mM after 24 h (Figure 3D). Interestingly, the free enzyme also caused a slight decrease in lactate levels (Figure 3D), which could be due to depletion of pyruvate from the cell culture media.
3.4. MSV delivery
The MSV platform has previously been used for the delivery of liposomes [44, 47, 52, 53, 67, 68], micelles [69], and various polymeric nanoparticles [39, 41, 54, 70]. Here, the PULSin/pyruvate decarboxylase complex was assembled with the MSV in order to facilitate delivery. SEM revealed that the discoidal MSV had a diameter of 1 μm and a height of 400 nm (Figure 4A). The zeta potential of the oxidized MSV, the MSV/PULSin complex, and the MSV/PULSin/enzyme complex were −22.5 mV (± 5.6 mV), +35.6 mV (± 2.4 mV), and +20.8 mV (± 0.6 mV), respectively (Figure 4B). The PULSin loading efficiency in the MSV was assessed at various MSV particle/μg protein ratios. At 7 × 104 particles/μg protein the PULSin loading efficiency was 46.6% (Figure 4C). Upon increasing the particle amount to 1.3 × 105, the loading efficiency increased to 70.4% (Figure 4C). Further increasing the number of particles resulted in marginal improvements in loading efficiency. Therefore, a ratio of 1.3 × 105 MSV sparticles/μg protein was used for experimental studies. It is worth noting that the reported increase in loading efficiency does not indicate that the amount of encapsulated PULSin in a single particle increases.
Figure 4.
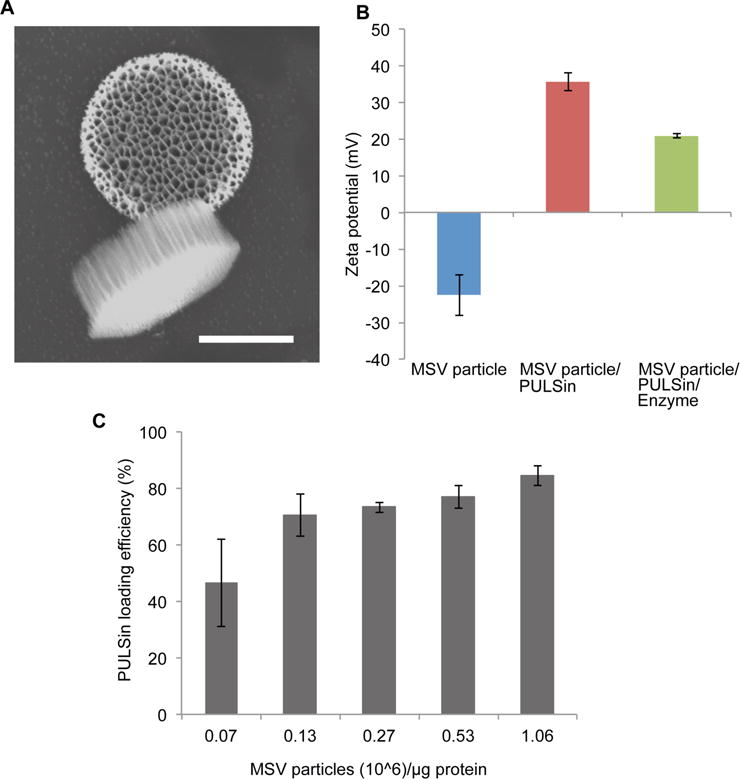
Characterization of the multistage vector (MSV) particle/PULSin/enzyme system. A) Scanning electron microscopy image of the MSV particle. Scale bar, 500 nm. B) Zeta potential measurements of the MSV particle, MSV particle/PULSin, MSV particle/PULSin/enzyme. Data is presented as mean ± s.d. of five measurements (10 runs each). C) PULSin loading efficiency inside MSV particles. Data is presented as mean ± s.d. of triplicates.
Since cationic particles frequently display toxicity, the effects of the empty PULSin carrier on MDA-MB-468 cells were assessed. The results demonstrate that PULSin did not reduce cell viability following a 24 h, 48 h, or 72 h incubation period (Figure 5A), suggesting that the anticancer effects observed with the PULSin/puruvate decrboxylase complex were solely due to enzyme activity. As responses to nanoparticles often vary among cell lines [71], the potential toxicity of the PULSin carrier was also evaluated in MDA-MB-231 human breast cancer cells. On the contrary to the MDA-MB-468 cells, MDA-MB-231 cells displayed a ~ 40% reduction in cell viability in response to PULSin (Figure 5B). Notably, one of the many benefits of the MSV delivery system, is its capability to reduce the toxicity of cationic nanoparticles, as previously demonstrated [39]. Here, we evaluated whether MSV loading could lower the toxicity of PULSin in MDA-MB-231 cells. Indeed, the results demonstrate that the MSV completely eliminated the toxicity of PULSin after 24 h and 48 h, while toxicity was reduced after 72 h (Figure 5B). To confirm that the MSV-mediated improvement in cell viability was not a result of lower levels of PULSin uptake, the cellular internalization of the fluorescent control protein R-phycoerythrin was assessed. Accordingly, levels of protein uptake were similar upon delivery with PULSin or MSV/PULSin (Figure 5C). It is worth noting that since these uptake studies were performed in complete media, protein internalization was substantially lower than that observed in serum-free media (Figure 3A).
Figure 5.
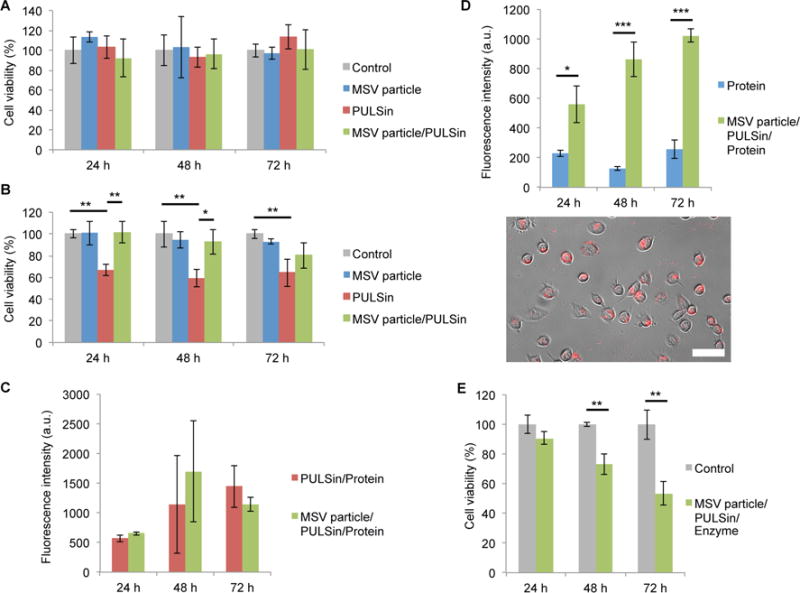
Biological effects of the MSV particle/PULSin/enzyme platform. A,B) Cell viability in response to the MSV particles, PUSLin, and the MSV particle/PULSin complex in MDA-MB-468 cells (A) and MDA-MB-231 cells (B). C) Uptake of PULSin/protein (R-phycoeythrin) and MSV particle/PULSin/protein (R-phycoeythrin) in MDA-MB-231 cells. Fluorescence intensity was normalized to that of control cells. D) Uptake of protein (R-phycoeythrin) and MSV particle/PULSin/protein (R-phycoeythrin) in MDA-MB-468 cells. Fluorescence intensity was normalized to that of control cells. Representative image of R-phycoerythrin uptake was captured with a fluorescent microscope following MSV/PULSin delivery. Scale bar, 50 μm. C) Cell viability in response to the MSV particle/PULSin/enzyme platform in MDA-MB-468 cells. Data is presented as mean ± s.d. of triplicates. *P < 0.05; **P < 0.01; ***P < 0.001.
Next, the uptake levels of free protein and protein delivered with MSV/PULSin were measured in MDA-MB-468 cells. While the free enzyme displayed low levels of cellular internalization, MSV delivery substantially increased protein uptake (Figure 5D). Furthermore, fluorescent microscopy confirmed that the protein accumulated inside cells as a result of MSV/PULSin delivery (Figure 5D). Moreover, MSV/PULSin delivery of puruvate decarboxylase caused a ~30% and ~50% reduction in cell viability after 48 h and 72 h, respectively (Figure 5E). This observation suggests that enzyme function is maintained following MSV delivery.
4. Conclusion
The majority of cancer drugs are designed to prevent replicative processes, thus focusing outcomes due to drug resistance and toxic side effects, different strategies for suppressing tumor growth could prove highly beneficial. Indeed, therapeutic agents that target invasion, angiogenesis, abnormal metabolism, immune surveillance, genomic instability, and inflammation could provide new solutions for reducing the cancer death toll. Here, we present a novel approach designed to exploit cancer metabolism. In particular, cancer cells produce increased levels of pyruvate, which is shunted into the lactic acid fermentation pathway in order to generate precursors for materials that can be used to build new cells. The proposed therapeutic strategy is to use the yeast enzyme pyruvate decarboxylase to convert pyruvate into acetaldehyde. The benefits from this approach are twofold, since the consumption of pyruvate reduces the production of biomolecules and the generation of acetaldehyde causes cancer cell death.
Notably, enzyme delivery is usually challenging, as proteins do not readily cross the cell membrane. In this study, a multistage nanodelivery platform consisting of porous silicon and cationic lipids was used to successfully obtain enzyme uptake in cancer cells. The results demonstrated that cancer cell viability was reduced in response to enzyme therapy. Measurements of acetaldehyde in cell lysates confirmed that enzyme function was preserved in the intracellular environment following nanoparticle uptake. Moreover, the levels of lactate, the end product of the lactic acid fermentation pathway, were decreased in response to enzyme exposure. The initial investigations described in this study support the anticancer potential of nanoparticle-mediated delivery of pyruvate decarboxylase.
Acknowledgments
This work was funded by the Houston Methodist Research Institute. Partial funds were acquired from: the Ernest Cockrell Jr. Distinguished Endowed Chair (M.F.), the US Department of Defense (W81XWH-09-1-0212, W81XWH-12-1-0414) (M.F.), the National Institutes of Health (U54CA143837, U54CA151668) (M.F.), Nylands nation Finland (J.W.), Victoriastiftelsen Finland (J.W.), and the Cancer Prevention Research Institute of Texas (RP121071) (M.F. and H.S.).
References
- 1.Cancer Facts & Figures 2015. American Cancer Society, Inc, American Cancer Society; 2015. [Google Scholar]
- 2.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Awada G, Kourie HR, Awada AH. Novel mechanisms and approaches in the medical therapy of solid cancers. Discov Med. 2015;20:33–41. [PubMed] [Google Scholar]
- 5.Wen Y, Meng WS. Recent In Vivo Evidences of Particle-Based Delivery of Small-Interfering RNA (siRNA) into Solid Tumors. J Pharm Innov. 2014;9:158–173. doi: 10.1007/s12247-014-9183-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou M, Zhao J, Tian M, Song S, Zhang R, Gupta S, Tan D, Shen H, Ferrari M, Li C. Radio-photothermal therapy mediated by a single compartment nanoplatform depletes tumor initiating cells and reduces lung metastasis in the orthotopic 4T1 breast tumor model. Nanoscale. 2015;7:19438–19447. doi: 10.1039/c5nr04587h. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Shen J, Kim HC, Mu C, Gentile E, Mai J, Wolfram J, Ji LN, Ferrari M, Mao ZW, Shen H. Multifunctional gold nanorods for siRNA gene silencing and photothermal therapy. Adv Healthc Mater. 2014;3:1629–1637. doi: 10.1002/adhm.201400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y, Wolfram J, Boom K, Fang X, Shen H, Ferrari M. Hesperetin impairs glucose uptake and inhibits proliferation of breast cancer cells. Cell Biochem Funct. 2013;31:374–379. doi: 10.1002/cbf.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen H, Mittal V, Ferrari M, Chang J. Delivery of gene silencing agents for breast cancer therapy. Breast Cancer Res. 2013;15:205. doi: 10.1186/bcr3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen H, Sun T, Ferrari M. Nanovector delivery of siRNA for cancer therapy. Cancer Gene Ther. 2012;19:367–373. doi: 10.1038/cgt.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Celia C, Trapasso E, Locatelli M, Navarra M, Ventura CA, Wolfram J, Carafa M, Morittu VM, Britti D, Di Marzio L, Paolino D. Anticancer activity of liposomal bergamot essential oil (BEO) on human neuroblastoma cells. Colloids Surf B Biointerfaces. 2013:548–553. doi: 10.1016/j.colsurfb.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 12.Wolfram J, Suri K, Huang Y, Molinaro R, Borsoi C, Scott B, Boom K, Paolino D, Fresta M, Wang J, Ferrari M, Celia C, Shen H. Evaluation of anticancer activity of celastrol liposomes in prostate cancer cells. J Microencaps. 2014;31:501–507. doi: 10.3109/02652048.2013.879932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolfram J, Scott B, Boom K, Shen J, Borsoi C, Suri K, Grande R, Fresta M, Celia C, Zhao Y, Shen H, Ferrari M. Hesperetin liposomes for cancer therapy. Curr Drug Deliv. 2015 doi: 10.2174/1567201812666151027142412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, Wolfram J, Fang X, Shen H, Ferrari M. Polyarginine Induces an Antitumor Immune Response through Binding to Toll-Like Receptor 4. Small. 2014;10:1250–1254. doi: 10.1002/smll.201302887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen J, Kim HC, Su H, Wang F, Wolfram J, Kirui D, Mai J, Mu C, Mao Z, Shen H. Cyclodextrin and polyethylenimine functionalized mesoporous silica nanoparticles for delivery of siRNA cancer therapeutics. Theranostics. 2014;4:487–497. doi: 10.7150/thno.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molinaro R, Wolfram J, Federico C, Cilurzo F, Di Marzio L, Ventura CA, Carafa M, Celia C, Fresta M. Polyethylenimine and chitosan carriers for the delivery of RNA interference effectors. Expert opinion on drug delivery. 2013;10:1653–1668. doi: 10.1517/17425247.2013.840286. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Dong H, Li S, He H. Theoretical study toward understanding the catalytic mechanism of pyruvate decarboxylase. J Phys Chem B. 2005;109:18664–18672. doi: 10.1021/jp052802s. [DOI] [PubMed] [Google Scholar]
- 18.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nature reviews Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 20.Lu J, Tan M, Cai Q. The Warburg effect in tumor progression: mitochondrial oxidative metabolism as an anti-metastasis mechanism. Cancer Lett. 2015;356:156–164. doi: 10.1016/j.canlet.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altenberg B, Greulich KO. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics. 2004;84:1014–1020. doi: 10.1016/j.ygeno.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 22.Henzel K, Thorborg C, Hofmann M, Zimmer G, Leuschner U. Toxicity of ethanol and acetaldehyde in hepatocytes treated with ursodeoxycholic or tauroursodeoxycholic acid. Biochimica et biophysica acta. 2004;1644:37–45. doi: 10.1016/j.bbamcr.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 23.Lieber CS. Metabolic effects of acetaldehyde. Biochem Soc Trans. 1988;16:241–247. doi: 10.1042/bst0160241. [DOI] [PubMed] [Google Scholar]
- 24.Rommel AJ. Effect of yeast pyruvate decarboxylase on Ehrlich ascites carcinoma in vivo. Nature. 1966;212:201. doi: 10.1038/212201a0. [DOI] [PubMed] [Google Scholar]
- 25.Torchilin V. Intracellular delivery of protein and peptide therapeutics. Drug Discov Today Technol. 2008;5:e95–e103. doi: 10.1016/j.ddtec.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Varga CM, Wickham TJ, Lauffenburger DA. Receptor-mediated targeting of gene delivery vectors: insights from molecular mechanisms for improved vehicle design. Biotechnol Bioeng. 2000;70:593–605. doi: 10.1002/1097-0290(20001220)70:6<593::aid-bit1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 27.Reynolds GC, Baker HJ, Reynolds RH. Enzyme replacement using liposome carriers in feline Gm1 gangliosidosis fibroblasts. Nature. 1978;275:754–755. doi: 10.1038/275754a0. [DOI] [PubMed] [Google Scholar]
- 28.Das PK, Murray GJ, Zirzow GC, Brady RO, Barranger JA. Lectin-specific targeting of beta-glucocerebrosidase to different liver cells via glycosylated liposomes. Biochem Med. 1985;33:124–131. doi: 10.1016/0006-2944(85)90135-8. [DOI] [PubMed] [Google Scholar]
- 29.Fawell S, Seery J, Daikh Y, Moore C, Chen LL, Pepinsky B, Barsoum J. Tat-mediated delivery of heterologous proteins into cells. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:664–668. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 31.Roeder GE, Parish JL, Stern PL, Gaston K. Herpes simplex virus VP22-human papillomavirus E2 fusion proteins produced in mammalian or bacterial cells enter mammalian cells and induce apoptotic cell death. Biotechnol Appl Biochem. 2004;40:157–165. doi: 10.1042/BA20030172. [DOI] [PubMed] [Google Scholar]
- 32.Rink JS, Plebanek MP, Tripathy S, Thaxton CS. Update on current and potential nanoparticle cancer therapies. Curr Opin Oncol. 2013;25:646–651. doi: 10.1097/CCO.0000000000000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolfram J, Zhu M, Yang Y, Shen J, Gentile E, Paolino D, Fresta M, Nie G, Chen C, Shen H, Ferrari M, Zhao Y. Safety of Nanoparticles in Medicine. Curr Drug Targets. 2015;16:1671–1681. doi: 10.2174/1389450115666140804124808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sengupta S, Eavarone D, Capila I, Zhao G, Watson N, Kiziltepe T, Sasisekharan R. Temporal targeting of tumour cells and neovasculature with a nanoscale delivery system. Nature. 2005;436:568–572. doi: 10.1038/nature03794. [DOI] [PubMed] [Google Scholar]
- 35.Wolfram J, Shen H, Ferrari M. Multistage vector (MSV) therapeutics. J Control Release. 2015;219:406–415. doi: 10.1016/j.jconrel.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Maltzahn G, Park JH, Lin KY, Singh N, Schwoppe C, Mesters R, Berdel WE, Ruoslahti E, Sailor MJ, Bhatia SN. Nanoparticles that communicate in vivo to amplify tumour targeting. Nature materials. 2011;10:545–552. doi: 10.1038/nmat3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weiss A, Neuberg P, Philippot S, Erbacher P, Weill CO. Intracellular peptide delivery using amphiphilic lipid-based formulations. Biotechnol Bioeng. 2011;108:2477–2487. doi: 10.1002/bit.23182. [DOI] [PubMed] [Google Scholar]
- 38.Balazs DA, Godbey W. Liposomes for use in gene delivery. J Drug Deliv. 2011;2011:326497. doi: 10.1155/2011/326497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen J, Xu R, Mai J, Kim HC, Guo X, Qin G, Yang Y, Wolfram J, Mu C, Xia X, Gu J, Liu X, Mao ZW, Ferrari M, Shen H. High capacity nanoporous silicon carrier for systemic delivery of gene silencing therapeutics. ACS nano. 2013;7:9867–9880. doi: 10.1021/nn4035316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scavo MP, Gentile E, Wolfram J, Gu J, Barone M, Evangelopoulos M, Martinez JO, Liu X, Celia C, Tasciotti E, Vilar E, Shen H. Multistage vector delivery of sulindac and silymarin for prevention of colon cancer. Colloids Surf B Biointerfaces. 2015;136:694–703. doi: 10.1016/j.colsurfb.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 41.Shen J, Wu X, Lee Y, Wolfram J, Mao Z, Ferrari M, Shen H. Porous silicon microparticles for delivery of siRNA therapeutics. J Vis Exp. 2015;15:52075. doi: 10.3791/52075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia X, Mai J, Xu R, Perez JE, Guevara ML, Shen Q, Mu C, Tung HY, Corry DB, Evans SE, Liu X, Ferrari M, Zhang Z, Li XC, Wang RF, Shen H. Porous silicon microparticle potentiates anti-tumor immunity by enhancing cross-presentation and inducing type I interferon response. Cell reports. 2015;11:957–966. doi: 10.1016/j.celrep.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen H, You J, Zhang G, Ziemys A, Li Q, Bai L, Deng X, Erm DR, Liu X, Li C, Ferrari M. Cooperative, nanoparticle-enabled thermal therapy of breast cancer. Advanced healthcare materials. 2012;1:84–89. doi: 10.1002/adhm.201100005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen H, Rodriguez-Aguayo C, Xu R, Gonzalez-Villasana V, Mai J, Huang Y, Zhang G, Guo X, Bai L, Qin G, Deng X, Li Q, Erm DR, Aslan B, Liu X, Sakamoto J, Chavez-Reyes A, Han HD, Sood AK, Ferrari M, Lopez-Berestein G. Enhancing chemotherapy response with sustained EphA2 silencing using multistage vector delivery. Clin Cancer Res. 2013;19:1806–1815. doi: 10.1158/1078-0432.CCR-12-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gizzatov A, Stigliano C, Ananta JS, Sethi R, Xu R, Guven A, Ramirez M, Shen H, Sood A, Ferrari M, Wilson LJ, Liu X, Decuzzi P. Geometrical confinement of Gd(DOTA) molecules within mesoporous silicon nanoconstructs for MR imaging of cancer. Cancer Lett. 2014;352:97–101. doi: 10.1016/j.canlet.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dave B, Granados-Principal S, Zhu R, Benz S, Rabizadeh S, Soon-Shiong P, Yu KD, Shao Z, Li X, Gilcrease M, Lai Z, Chen Y, Huang TH, Shen H, Liu X, Ferrari M, Zhan M, Wong ST, Kumaraswami M, Mittal V, Chen X, Gross SS, Chang JC. Targeting RPL39 and MLF2 reduces tumor initiation and metastasis in breast cancer by inhibiting nitric oxide synthase signaling. Proc Natl Acad Sci U S A. 2014;111:8838–8843. doi: 10.1073/pnas.1320769111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen X, Iliopoulos D, Zhang Q, Tang Q, Greenblatt MB, Hatziapostolou M, Lim E, Tam WL, Ni M, Chen Y, Mai J, Shen H, Hu DZ, Adoro S, Hu B, Song M, Tan C, Landis MD, Ferrari M, Shin SJ, Brown M, Chang JC, Liu XS, Glimcher LH. XBP1 promotes triple-negative breast cancer by controlling the HIF1alpha pathway. Nature. 2014;508:103–107. doi: 10.1038/nature13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mi Y, Mu C, Wolfram J, Deng Z, Hu TY, Liu X, Blanco E, Shen H, Ferrari M. A Micro/Nano Composite for Combination Treatment of Melanoma Lung Metastasis. Advanced healthcare materials. 2016 doi: 10.1002/adhm.201500910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Serda RE, Gu J, Bhavane RC, Liu X, Chiappini C, Decuzzi P, Ferrari M. The association of silicon microparticles with endothelial cells in drug delivery to the vasculature. Biomaterials. 2009;30:2440–2448. doi: 10.1016/j.biomaterials.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 50.Ferrati S, Mack A, Chiappini C, Liu X, Bean AJ, Ferrari M, Serda RE. Intracellular trafficking of silicon particles and logic-embedded vectors. Nanoscale. 2010;2:1512–1520. doi: 10.1039/c0nr00227e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martinez JO, Parodi A, Liu X, Kolonin MG, Ferrari M, Tasciotti E. Evaluation of cell function upon nanovector internalization. Small. 2013;9:1696–1702. doi: 10.1002/smll.201202001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanaka T, Mangala LS, Vivas-Mejia PE, Nieves-Alicea R, Mann AP, Mora E, Han HD, Shahzad MM, Liu X, Bhavane R, Gu J, Fakhoury JR, Chiappini C, Lu C, Matsuo K, Godin B, Stone RL, Nick AM, Lopez-Berestein G, Sood AK, Ferrari M. Sustained small interfering RNA delivery by mesoporous silicon particles. Cancer research. 2010;70:3687–3696. doi: 10.1158/0008-5472.CAN-09-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu R, Huang Y, Mai J, Zhang G, Guo X, Xia X, Koay EJ, Qin G, Erm DR, Li Q, Liu X, Ferrari M, Shen H. Multistage vectored siRNA targeting ataxia-telangiectasia mutated for breast cancer therapy. Small. 2013;9:1799–1808. doi: 10.1002/smll.201201510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang M, Xu R, Xia X, Yang Y, Gu J, Qin G, Liu X, Ferrari M, Shen H. Polycation-functionalized nanoporous silicon particles for gene silencing on breast cancer cells. Biomaterials. 2014;35:423–431. doi: 10.1016/j.biomaterials.2013.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tanaka T, Godin B, Bhavane R, Nieves-Alicea R, Gu J, Liu X, Chiappini C, Fakhoury JR, Amra S, Ewing A, Li Q, Fidler IJ, Ferrari M. In vivo evaluation of safety of nanoporous silicon carriers following single and multiple dose intravenous administrations in mice. Int J Pharm. 2010;402:190–197. doi: 10.1016/j.ijpharm.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martinez JO, Chiappini C, Ziemys A, Faust AM, Kojic M, Liu X, Ferrari M, Tasciotti E. Engineering multi-stage nanovectors for controlled degradation and tunable release kinetics. Biomaterials. 2013;34:8469–8477. doi: 10.1016/j.biomaterials.2013.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martinez JO, Evangelopoulos M, Chiappini C, Liu X, Ferrari M, Tasciotti E. Degradation and biocompatibility of multistage nanovectors in physiological systems. J Biomed Mater Res A. 2014;102:3540–3549. doi: 10.1002/jbm.a.35017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Godin B, Gu J, Serda RE, Bhavane R, Tasciotti E, Chiappini C, Liu X, Tanaka T, Decuzzi P, Ferrari M. Tailoring the degradation kinetics of mesoporous silicon structures through PEGylation. J Biomed Mater Res A. 2010;94:1236–1243. doi: 10.1002/jbm.a.32807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Godin B, Gu J, Serda RE, Ferrati S, Liu X, Chiappini C, Tanaka T, Decuzzi P, Ferrari M. Multistage Mesoporous Silicon-based Nanocarriers: Biocompatibility with Immune Cells and Controlled Degradation in Physiological Fluids. Controlled release newsletter/Controlled Release Society. 2008;25:9–11. [PMC free article] [PubMed] [Google Scholar]
- 60.Godin B, Chiappini C, Srinivasan S, Alexander JF, Yokoi K, Ferrari M, Decuzzi P, Liu X. Discoidal Porous Silicon Particles: Fabrication and Biodistribution in Breast Cancer Bearing Mice. Adv Funct Mater. 2012;22:4225–4235. doi: 10.1002/adfm.201200869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Basu S, Chen W, Tchou J, Mavi A, Cermik T, Czerniecki B, Schnall M, Alavi A. Comparison of triple-negative and estrogen receptor-positive/progesterone receptor-positive/HER2-negative breast carcinoma using quantitative fluorine-18 fluorodeoxyglucose/positron emission tomography imaging parameters: a potentially useful method for disease characterization. Cancer. 2008;112:995–1000. doi: 10.1002/cncr.23226. [DOI] [PubMed] [Google Scholar]
- 62.Palaskas N, Larson SM, Schultz N, Komisopoulou E, Wong J, Rohle D, Campos C, Yannuzzi N, Osborne JR, Linkov I, Kastenhuber ER, Taschereau R, Plaisier SB, Tran C, Heguy A, Wu H, Sander C, Phelps ME, Brennan C, Port E, Huse JT, Graeber TG, Mellinghoff IK. 18F-fluorodeoxy-glucose positron emission tomography marks MYC-overexpressing human basal-like breast cancers. Cancer Res. 2011;71:5164–5174. doi: 10.1158/0008-5472.CAN-10-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pelicano H, Zhang W, Liu J, Hammoudi N, Dai J, Xu RH, Pusztai L, Huang P. Mitochondrial dysfunction in some triple-negative breast cancer cell lines: role of mTOR pathway and therapeutic potential. Breast Cancer Res. 2014;16:434. doi: 10.1186/s13058-014-0434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 65.Morais-Santos F, Miranda-Goncalves V, Pinheiro S, Vieira AF, Paredes J, Schmitt FC, Baltazar F, Pinheiro C. Differential sensitivities to lactate transport inhibitors of breast cancer cell lines. Endocr Relat Cancer. 2014;21:27–38. doi: 10.1530/ERC-13-0132. [DOI] [PubMed] [Google Scholar]
- 66.Aranda A, del Olmo ML. Exposure of Saccharomyces cerevisiae to acetaldehyde induces sulfur amino acid metabolism and polyamine transporter genes, which depend on Met4p and Haa1p transcription factors, respectively. Appl Environ Microbiol. 2004;70:1913–1922. doi: 10.1128/AEM.70.4.1913-1922.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mai J, Huang Y, Mu C, Zhang G, Xu R, Guo X, Xia X, Volk DE, Lokesh GL, Thiviyanathan V, Gorenstein DG, Liu X, Ferrari M, Shen H. Bone marrow endothelium-targeted therapeutics for metastatic breast cancer. J Control Release. 2014;187:22–29. doi: 10.1016/j.jconrel.2014.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dave B, Granados-Principal S, Zhu R, Benz S, Rabizadeh S, Soon-Shiong P, Yu KD, Shao ZM, Li XX, Gilcrease M, Lai Z, Cheng YD, Huang THM, Shen HF, Liu XW, Ferrari M, Zhan M, Wong STC, Kumaraswami M, Mittal V, Chen X, Gross SS, Chang JC. Targeting RPL39 and MLF2 reduces tumor initiation and metastasis in breast cancer by inhibiting nitric oxide synthase signaling. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:8838–8843. doi: 10.1073/pnas.1320769111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martinez JO, Evangelopoulos M, Bhavane R, Acciardo S, Salvatore F, Liu X, Ferrari M, Tasciotti E. Multistage Nanovectors Enhance the Delivery of Free and Encapsulated Drugs. Curr Drug Targets. 2014 doi: 10.2174/1389450115666141015113914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blanco E, Sangai T, Hsiao A, Ferrati S, Bai L, Liu X, Meric-Bernstam F, Ferrari M. Multistage delivery of chemotherapeutic nanoparticles for breast cancer treatment. Cancer Lett. 2013;334:245–252. doi: 10.1016/j.canlet.2012.07.027. [DOI] [PubMed] [Google Scholar]
- 71.Xia T, Kovochich M, Liong M, Zink JI, Nel AE. Cationic polystyrene nanosphere toxicity depends on cell-specific endocytic and mitochondrial injury pathways. ACS nano. 2008;2:85–96. doi: 10.1021/nn700256c. [DOI] [PubMed] [Google Scholar]


