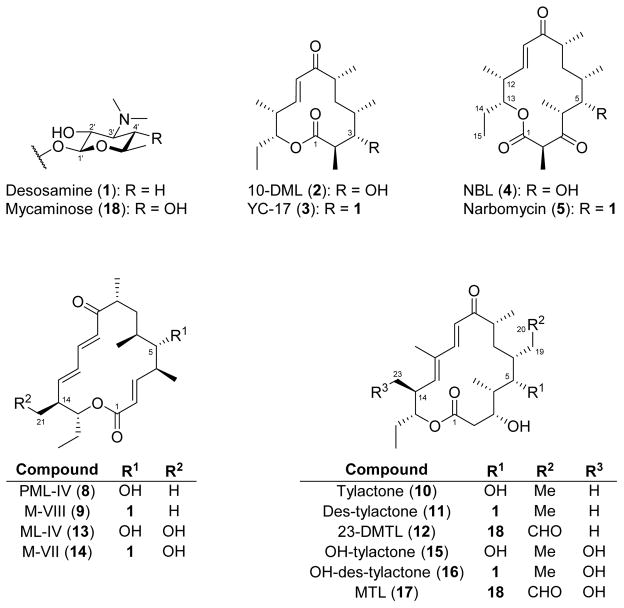
Abstract
Cytochrome P450 monooxygenases (P450s) are some of nature’s most ubiquitous and versatile enzymes for performing oxidative metabolic transformations. Their unmatched ability to selectively functionalize inert C-H bonds has led to their increasing employment in academic and industrial settings for the production of fine and commodity chemicals. Many of the most interesting and potentially biocatalytically useful P450s come from microorganisms, where they catalyze key tailoring reactions in natural product biosynthetic pathways. While most of these enzymes act on structurally complex pathway intermediates with high selectivity, they often exhibit narrow substrate scope, thus limiting their broader application. In the present study, we investigated the reactivity of the P450 MycCI from the mycinamicin biosynthetic pathway toward a variety of macrocyclic compounds and discovered that the enzyme exhibits appreciable activity on several 16-membered ring macrolactones independent of their glycosylation state. These results were corroborated by performing equilibrium substrate binding experiments, steady-state kinetics studies, and x-ray crystallographic analysis of MycCI bound to its native substrate mycinamicin VIII. We also characterized TylHI, a homologous P450 from the tylosin pathway, and showed that its substrate scope is severely restricted compared to MycCI. Thus, the ability of the latter to hydroxylate both macrocyclic aglycones and macrolides sets it apart from related biosynthetic P450s and highlights its potential for developing novel P450 biocatalysts with broad substrate scope and high regioselectivity.
Cytochromes P450 (P450s) comprise one of the most widely distributed and versatile groups of enzymes found in nature, catalyzing a broad range of physiologically important oxidative transformations in organisms across all domains of life.1–4 To date, more than 21,000 individual sequences of these ubiquitous heme-thiolate proteins have been identified.5 Despite their vast diversity, P450s share a conserved three-dimensional fold as well as a general catalytic cycle for dioxygen activation and transfer.6–8 Although P450s most commonly catalyze hydroxylation, epoxidation, heteroatom (e.g., N or S) oxygenation, and dealkylation reactions, their catalytic repertoire extends far beyond these well-documented transformations.4,9,10
With P450s, nature has provided an impressive class of oxidation catalysts that can functionalize a diverse array of molecular scaffolds with high degrees of selectivity. Accordingly, they have garnered increasing attention in recent years as potential biocatalysts for the oxidative tailoring of molecules ranging from simple hydrocarbons to complex natural products.11–20 In addition, since they operate under mild reaction conditions and employ abundant Fe as a cofactor, P450s have the potential to contribute to the realization of environmentally sustainable approaches toward the production of fine and commodity chemicals.21 Microbes offer a particularly rich source of novel P450s with potential broad applicability in various biocatalytic processes.20,22,23 In many cases, these enzymes are found in gene clusters for the biosynthesis of secondary metabolites, where they are responsible for catalyzing a diverse array of tailoring reactions including C-H bond hydroxylation, oxidative phenolic coupling, and C-C bond formation/cleavage among many others.24,25
Actinomycetes, and Streptomyces spp. in particular, are prolific producers of biologically active natural products, many of which have been developed into important pharmaceuticals.25,26 Accordingly, many of the secondary metabolic (biosynthetic) P450s that have hitherto been structurally and functionally characterized originate from these organisms.17 Whereas mammalian P450s involved in xenobiotic metabolism are known to act on a variety of different substrates, the bacterial biosynthetic enzymes typically possess limited substrate scope and catalyze oxidative transformations with high degrees of regio- and stereoselectivity. For example, EryF, which is responsible for hydroxylating the aglycone precursor to erythromycin (6-deoxyerythronolide B), exhibits an extremely narrow substrate scope and cannot tolerate even minor changes to the structure of its native substrate.27 Conversely, a few biosynthetic P450s inherently possess some degree of substrate flexibility. PikC, a well-studied P450 from the methymycin/pikromycin pathway in Streptomyces venezuelae, is capable of hydroxylating both 12- and 14-membered ring macrolides at either of two positions, leading to the production of up to six unique macrolide antibiotics from a single biosynthetic pathway.28–30 Because biosynthetic P450s often catalyze oxidation reactions on complex and densely functionalized substrates, they provide an important and underexplored starting point for the development of new biocatalysts.20,22 Consequently, much of our recent work has focused on the discovery and biochemical/structural characterization of new P450s involved in the biosynthesis of bacterial natural products.31–35 As we have demonstrated with PikC,36–38 these fundamental studies serve as a key starting point to guide future efforts to modulate the substrate scope and selectivity properties of P450 enzymes. Engineered biosynthetic P450s may ultimately be employed in vitro or in vivo as biocatalysts for the efficient production of novel biologically active compounds. For example, new sites of oxidation on macrolide scaffolds could enable further diversification of these molecules via chemoenzymatic synthesis, potentially leading to new and more effective antimicrobial agents.
In previous work, we demonstrated that the desosamine (1) moiety facilitates recognition and anchoring of the macrolide substrates YC-17 (3) and narbomycin (5) in the active site of PikC.31,39 Specifically, the protonated N,N-dimethylamino group on 1 participates in a salt bridge interaction with either of two proximal glutamate residues in the enzyme active site. We subsequently demonstrated that the presence of 1 on a series of unfunctionalized carbocycles was sufficient for PikC to bind and hydroxylate these unnatural substrates.36 More recently, we have shown that PikC can accept a variety of other substrates attached to structurally diverse non-sugar anchors containing the N,N-dimethylamino moiety.37,38 Although such an explicit anchoring mechanism has not been clearly established for other P450s that act on macrolide substrates, the presence of one or more sugar moieties is typically required for substrate recognition, leading in turn to effective binding and catalysis. Conversely, other P450s involved in macrolide biosynthetic pathways (e.g., EryF) strictly act on aglycone intermediates prior to glycosylation. Here, we report that the P450 MycCI from the mycinamicin (6) pathway in Micromonospora griseorubida (Scheme 1a) is able to effectively accept 16-membered ring macrolactones as substrates regardless of their glycosylation state. Our findings are further clarified on the basis of equilibrium substrate binding experiments, steady-state kinetics studies, and x-ray crystallographic analysis of the enzyme in complex with its native substrate. Comparison with TylHI, a homologous P450 from the tylosin (7) biosynthetic pathway (Scheme 1b), further reveals that MycCI is unique in exhibiting such unusually broad substrate tolerance.
RESULTS AND DISCUSSION
MycCI is capable of hydroxylating 16-membered ring macrolides and aglycones
To test the substrate scope of MycCI and other P450s involved in macrolide biosynthetic pathways, we envisioned generating a set of macrocyclic compounds of varying ring sizes and glycosylation states. Using a biotransformation method we previously developed for producing the macrolide antibiotics methymycin and pikromycin directly from the corresponding aglycones 10-deoxymethynolide (10-DML, 2) and narbonolide (NBL, 4), respectively,40 we were able to efficiently append desosamine (1) to various 12-, 14-, and 16-membered ring macrolactone aglycones (Figure 1). Consistent with prior reports detailing the extremely broad substrate specificity of DesVII (the native glycosyltransferase in the methymycin/pikromycin pathway in S. venezuelae) acting in conjunction with its effector protein DesVIII,41,42 tylactone (10) was completely converted to 5-O-desosaminyl-tylactone (des-tylactone, 11), and the majority (> 90%) of protomycinolide IV (PML-IV, 8) was converted to mycinamicin VIII (M-VIII, 9) following a 24 h incubation period. An additional potential substrate, 23-deoxy-5-O-mycaminosyl-tylonolide (23-DMTL, 12), was acquired through fermentation of a Streptomyces fradiae mutant strain that had previously been generated via chemical mutagenesis.43 In each case, sufficient quantities of pure substrate to be used for enzymatic assays were obtained by semi-preparative HPLC purification of crude extracts.
Figure 1.
Structures of compounds investigated in this study.
In the mycinamicin biosynthetic gene cluster, mycCI is located adjacent to mycCII, which encodes a [3Fe-4S]-type ferredoxin. Our previous work demonstrated that MycCI converts 9 to the monohydroxylated product mycinamicin VII (M-VII, 14) more effectively when it is paired with MycCII compared to spinach ferredoxin.33 Therefore, we purified MycCI and MycCII expressed separately as N-terminal His-tagged constructs and used them in conjunction with purified N-terminal MBP-tagged spinach ferredoxin reductase (MBP-FdR) in small-scale in vitro reactions. MycCI exhibited equivalent activity on all three of the 16-membered ring macrolides tested (9, 11, and 12; see Table 1), demonstrating its inherent ability to accept large substrates with slight differences in overall architecture. More surprising, however, was the ability of MycCI to catalyze hydroxylation of the corresponding 16-membered ring aglycones (8 and 10) with only relatively minor decreases in total turnover number (TTN) (Table 1). To the best of our knowledge, such tolerance with respect to the glycosylation state of the substrate has not been reported for related P450s involved in macrolide biosynthetic pathways or for those that play roles in the biosynthesis of other glycosylated natural products (e.g., glycopeptides). Moreover, experiments that we performed with a suite of purified P450s from the pikromycin, erythromycin, and tylosin (vide infra) pathways using the substrates presented here failed to identify another P450 enzyme besides MycCI that could accept both a given glycosylated compound and its corresponding aglycone. In addition to the 16-membered ring substrates, MycCI displayed low-level ability to catalyze oxidation of the 14-membered ring macrolide narbomycin (5) (Tables 1 and S5). In contrast to the other compounds tested, which yielded single monohydroxylated products in each case, LC-MS analysis of reaction mixtures containing 5 revealed the formation of two distinct monohydroxylated species as well as a potential dihydroxylated molecule, all in roughly equal abundance (Figure S4). Interestingly, MycCI was unable to catalyze C-H oxidation of the 14-membered ring aglycone 4 or the smaller 12-membered ring compounds YC-17 (3) and 2.
Table 1.
Total Turnover Numbers (TTNs) for Selected Enzymatic Reactionsa
| Catalytic system | Substrate
|
|||||
|---|---|---|---|---|---|---|
| 5 | 8 | 9 | 10 | 11 | 12 | |
| MycCI/MycCIIb | 6.8 ± 0.4 | 65 ± 1 | 85 ± 2 | 72 ± 1 | 87 ± 4 | 89 ± 2 |
| MycCI-RhFRED | 0.6 ± 0.1 | 18.8 ± 0.8 | 167 ± 13 | 49.3 ± 0.7 | 177 ± 7 | 114 ± 4 |
| TylHI/TylHIIb | 0 | 0 | 0 | 0 | 1.0 ± 0.2 | 18.2 ± 0.8 |
| TylHI/MycCIIb | 0 | 0 | 0 | 0 | 0.2 ± 0.1 | 69 ± 2 |
| TylHI-RhFRED | 0 | 0 | 0 | 0 | 0.8 ± 0.1 | 162 ± 6 |
| MycCIS172A/MycCII b | 2.0 ± 0.1 | 74 ± 3 | 71 ± 2 | 77 ± 1 | 61 ± 4 | 66 ± 4 |
TTN = mol product/mol P450. Reported errors are standard deviations calculated from experiments performed in triplicate.
Includes MBP-FdR.
Over the course of optimizing reaction conditions, it became evident that the amount of MycCII present in reaction mixtures was an important factor in determining reaction outcome as assessed by TTN. With the concentration of MycCI kept relatively low (0.5 μM), increased concentrations of MycCII (e.g., 10–100 μM) correlated with higher TTN values for all substrates tested. In contrast, increasing the amount of MBP-FdR (e.g., 1–10 μM) by itself had little to no effect on this parameter. However, when the levels of both MycCII and MBP-FdR were increased, the positive effect on TTN was synergistic (Figure S6). MycCII interacts directly with MycCI to deliver the requisite electrons for P450 Compound I formation, and we have previously reported an apparent dissociation constant (Kd) of 7 μM for N-terminal His-tagged MycCII binding to MycCI.33 In light of this information, it is not surprising that further increases in MycCII concentration beyond values close to its Kd with respect to MycCI led to enhanced substrate turnover.
Engineering a catalytically self-sufficient MycCI biocatalyst
Although MycCI exhibited reasonable activity on 8–12, we wished to further improve its efficiency since optimal turnover was dependent on relatively high levels of MycCII and MBP-FdR. Therefore, we decided to create a self-sufficient biocatalytic system to facilitate reaction scale-up, product isolation, and structural verification. Our laboratory has previously succeeded in generating self-sufficient P450 C-H oxidation catalysts via genetic fusion to the FMN/[2Fe-2S]-containing reductase domain of P450RhF (RhFRED) from Rhodococcus sp. NCIMB 9784.34,44,45 Typically, these chimeric enzymes exhibit improved activity, especially when compared to three-component systems comprising the P450 and non-native redox partners (e.g., spinach ferredoxin and spinach ferredoxin reductase). However, as the native ferredoxin partner for MycCI was known and had been demonstrated to support catalytic activity of its associated P450, we were unsure whether fusion to RhFRED (containing both the ferredoxin and reductase domains) would indeed yield a more active biocatalyst. Thus, we were pleased to verify that purified MycCI-RhFRED exhibited improved activity relative to MycCI in conjunction with MycCII and MBP-FdR on each of the three 16-membered ring macrolides tested. Specifically, TTN values were doubled for 9 and 11 while the improvement was more modest for 12 (Table 1). Although the fusion protein could still accept the two 16-membered ring aglycones (8 and 10) as substrates, TTN values were lower than those reported for the stand-alone MycCI protein with ferredoxin and ferredoxin reductase partners acting in trans. The decrease in activity was more pronounced for 8, with a TTN value of 19 compared to 65 for the three-component system. MycCI-RhFRED also showed an almost complete lack of activity on the 14-membered ring macrolide 5. As observed for the MycCI/MycCII/MBP-FdR system, the MycCI-RhFRED fusion enzyme did not accept 2–4 as substrates. Interestingly, TTN values for MycCI-RhFRED were increased about two-fold across 8–12 when the enzyme was prepared using a faster, less rigorous purification procedure, yielding “semi-pure” P450 enzyme that co-purified with a number of additional protein impurities (see Supporting Information for further discussion).
Product isolation and structure elucidation
Our earlier studies on the mycinamicin biosynthetic pathway established that MycCI is responsible for installing a hydroxyl group on C21 of 9 to produce the intermediate 14, which is then primed for subsequent glycosylation and further tailoring by two methyltransferases and the multifunctional P450 MycG (Scheme 1a).33,46 Comparing the structure of 9 to those of 11 and 12, we surmised that MycCI would likely introduce a hydroxyl group at C23 of the latter two compounds. In order to rigorously assess the regioselectivity of MycCI when acting upon these macrolides and the corresponding aglycones, we scaled up the reactions so that adequate product could be isolated for full NMR characterization in each case. MycCI-RhFRED was employed in large-scale reactions with macrolide substrates, which showed conversions averaging around 60% based on analytical HPLC. Because the fusion enzyme exhibited lower activity on aglycone substrates compared to wild-type MycCI in conjunction with MycCII and MBP-FdR, we used the latter biocatalytic system in scale-up reactions involving 8 and 10; conversions for these two compounds were ~30% and ~50%, respectively.
Following isolation of products by semi-preparative HPLC, we used 1D and 2D NMR to confirm that 8 and 9 were both hydroxylated at C21 to produce mycinolide IV (ML-IV, 13) and 14, respectively, while 10, 11, and 12 were each hydroxylated at the corresponding C23 position, yielding 23-hydroxy-tylactone (OH-tylactone, 15), 23-hydroxy-5-O-desosaminyl-tylactone (OH-des-tylactone, 16), and 5-O-mycaminosyl-tylonolide (MTL, 17), respectively (Figure 1; see Supporting Information for full spectroscopic characterization of all starting materials and products). Despite the low activity of MycCI on 5, we attempted to scale up the reaction so that we could determine the regioselectivity of the enzyme on this substrate. Using the same conditions as described for the aglycone substrates, 5 was converted to two monohydroxylated products, with one representing ~2% and the other slightly more polar compound making up ~0.5% of the reaction mixture (Figure S3). Unfortunately, we were unable to isolate enough material for NMR characterization, but comparison to the products of the same reaction carried out on an analytical scale using PikCD50N-RhFRED36,37 revealed that hydroxylation by MycCI does not occur at C12 of 5 to produce pikromycin (Figure S5). In the LC-MS trace, both products elute earlier than pikromycin and neopikromycin, suggesting that the analogs are more polar. Comparing the structure of 5 to those of the 16-membered ring substrates, MycCI may catalyze hydroxylation at C14 and/or at the adjacent C15 position (Figure 1).
Binding affinities of aglycone and macrolide substrates to MycCI
In order to probe the biochemical basis for the differences in reactivity observed in the activity assays described above, we conducted substrate binding experiments on both MycCI and MycCI-RhFRED. During the P450 catalytic cycle, binding of substrate displaces the heme axial water ligand, which shifts the spin-state equilibrium of the heme iron toward high spin, resulting in a blue-shifted Soret band that can be quantified spectrophotometrically.7,47 The established substrate titration methodology associated with this principle was used to determine the dissociation constants for each of the substrates (Table 2; Figure S7). Narbomycin (5) and PML-IV (8) bound to MycCI with Kd values of 17 μM and 19 μM, respectively, while tylactone (10) bound more tightly with a Kd value of about 3 μM. These binding affinities are readily comparable to those reported for many P450s and their natural substrates. Unexpectedly, when initial titration experiments were conducted with the 16-membered ring macrolide compounds (9, 11, and 12) under the same conditions, saturation was observed after the addition of about one molar equivalent of substrate. When sub-stoichiometric concentrations of substrate were titrated into the enzyme solution, a linear increase in signal was observed up until the equivalence point, after which no further increase in signal was detected. At sufficiently low concentrations of enzyme, dissociation constants could be estimated by fitting the data to a quadratic tight-binding equation. M-VIII (9) bound with an estimated Kd of 1 nM while des-tylactone (11) and 23-DMTL (12) bound with Kd values of 6 nM and 45 nM, respectively, indicating extremely tight binding of the large macrolide substrates. These dissociation constants rival those found for the binding of azole inhibitors to various human and bacterial P450s. For example, ketoconazole binds to OleP48 with Kd = 5 nM and clotrimazole binds to EryK49 with Kd < 10 nM. Interestingly, the 12-membered ring macrolide YC-17 (3) bound to MycCI more weakly (Kd = 40 μM), but the affinity was still comparable to that observed for other P450s and their substrates. However, as previously noted, MycCI did not exhibit catalytic activity toward this smaller molecule. The aglycones 10-DML (2) and NBL (4) did not induce a concentration-dependent change in the spin state of the enzyme, which explains why MycCI failed to hydroxylate these molecules. Consistent with prior findings,44,50 fusion to the RhFRED domain did not substantially impact the binding affinities of the various substrates tested toward MycCI (Table 2).
Table 2.
Kd Values from Equilibrium Substrate Binding Assaysa
| Substrate | Enzyme
|
||||
|---|---|---|---|---|---|
| MycCI | MycCI-RhFRED | TylHI | TylHI-RhFRED | MycCIS172A | |
| 3 | 40 ± 4 | 40 ± 2 | N.B.b | N.B. | 63 ± 5 |
| 5 | 17 ± 1 | 15.0 ± 0.8 | N.B. | N.B. | 23 ± 3 |
| 8 | 19 ± 2 | 15 ± 2 | N.B. | 1524 ± 546 | 9.2 ± 1.0 |
| 9 | 0.001 ± 0.001 | 0.005 ± 0.001 | 1291 ± 59 | 1230 ± 134 | 0.003 ± 0.004 |
| 10 | 3.3 ± 0.3 | 3.0 ± 0.3 | 433 ± 40 | 469 ± 39 | 1.35 ± 0.09 |
| 11 | 0.006 ± 0.004 | 0.010 ± 0.003 | 275 ± 22 | 205 ± 20 | 0.04 ± 0.01 |
| 12 | 0.045 ± 0.006 | 0.038 ± 0.005 | 0.60 ± 0.03 | 0.66 ± 0.05 | 0.12 ± 0.04 |
Kd values are in μM. Reported errors are those obtained from fitting data averaged from experiments performed at least in triplicate.
N.B. = no binding detected.
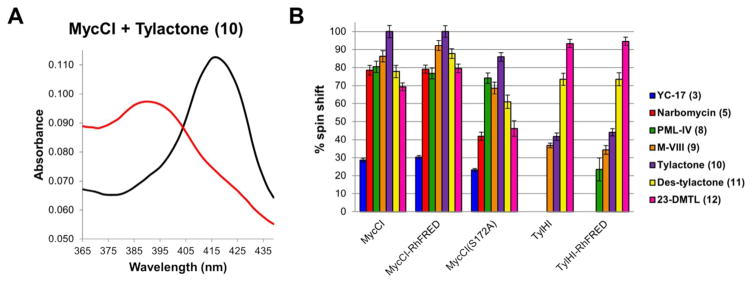
In most cases, the binding affinities of the compounds correlated well with their ability to be accepted as substrates for enzymatic oxidation. However, despite the fact that 5 bound to MycCI with about the same affinity as 8, the former was a considerably poorer substrate. Moreover, while 3 bound to MycCI with modest affinity, no substrate turnover was observed. In order to rationalize these results, we examined the degree to which each substrate was able to induce a shift of the heme iron from low spin to high spin. Normalizing the maximal change in absorbance acquired from non-linear regression analysis of the raw data to the enzyme concentration, we found that 10 induced the largest change in the spin state for both MycCI and MycCI-RhFRED (Table S9). The percentage of each enzyme in the high-spin state at saturating levels of a given substrate was estimated by assuming that each was completely converted to high spin when saturated with 10 (Figure 2; Table S9). Almost all substrates bound very effectively as demonstrated by their ability to elicit particularly high changes in the spin state of the enzyme (> 75%) (Figures 2 and S8). In contrast, saturation with 3 resulted in only ~30% high spin enzyme, which provides a potential explanation for the lack of activity with this substrate. Curiously, 5 and 8 induced the same shift in spin state (~80%), indicating that differences in substrate binding could not be used to rationalize the differences in reactivity observed for these two compounds. The α,β,γ,δ-unsaturated ketone moiety present in 8 and the other 16-membered ring macrolactones may play a key role in rigidifying the structures of these molecules such that the appropriate primary C-H bond in each case is presented to the iron-oxo species. The more flexible scaffold of 5 may hinder its ability to adopt one particular orientation within the enzyme active site, resulting in fewer turnovers and multiple oxidized products (Table S5).
Figure 2.
Equilibrium substrate binding analysis. (A) Absolute spectra of MycCI (0.5 μM) in the absence of substrate (black trace) or in the presence of 128 μM 10 (red trace). (B) Percent spin shift of each enzyme at saturating levels of different substrates. Values were calculated as described in Supporting Information.
Increased mobility of a substrate in the P450 active site makes it more likely that an inherently weaker C-H bond will present itself to the active iron-oxo species, leading to preferential oxidation of this bond over others that have stronger bond-dissociation energies. A variety of P450 ω-hydroxylases have been identified, which are highly selective for oxidizing primary unactivated C-H bonds in linear hydrocarbons (e.g., members of the CYP153 family in bacteria and the CYP52 family in fungi), linear and branched fatty acids (e.g., members of the mammalian CYP4 family), and cholesterol (e.g., CYP124A1 and CYP125A1 from Mycobacterium tuberculosis).51 These remarkable enzymes are able to control their regioselectivity through specific interactions with their substrates, which typically bind tightly and whose movement in the active site is therefore severely restricted. This tight control of the bound substrate is required for ω-hydroxylation to take place over hydroxylation of more activated secondary, tertiary, or allylic carbons. In the 16-membered ring aglycone and macrolide substrates tested here, the primary C-H bond subject to oxidation is located adjacent to a much weaker tertiary allylic C-H bond. Given the great deal of control over substrate orientation that must be required to completely restrict access of more labile C-H bonds to the iron-oxo species, it is not surprising that these substrates bind MycCI with such high affinities.
Steady-state kinetic analysis of MycCI-catalyzed substrate hydroxylation
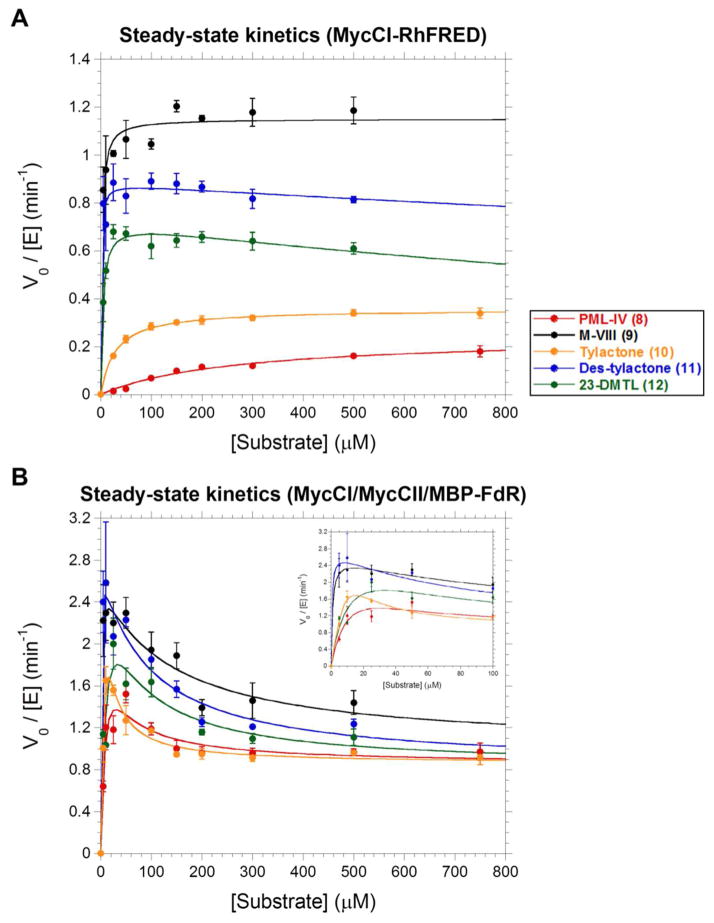
Steady-state kinetics experiments were conducted for both MycCI/MycCII/MBP-FdR and MycCI-RhFRED using 8–12. First, we evaluated the kinetics of product formation using the latter self-sufficient enzyme. Generally, the rates were rather slow, with kcat values ranging from 1.15 ± 0.02 min−1 for 9 to 0.24 ± 0.02 min−1 for 8 (Figure 3a; Table 3). Although the latter value is lower than that observed for many other P450/substrate combinations, it is close to that reported for CYP107W1, which hydroxylates a tertiary carbon atom in the 26-membered ring macrolide oligomycin C.52 In addition, all of the rates are higher than that determined for TamI-RhFRED acting on tirandamycin A (kcat = 0.11 ± 0.01 min−1),34 which is one of the few other examples of a biosynthetic P450 that hydroxylates an unactivated primary carbon atom. The slow rates may in part be explained by the higher activation energy required to oxidize a primary C-H bond, although examples of engineered P450s that perform similar transformations more rapidly have been reported.53,54 Consistent with the extremely tight binding of the macrolides, the Km values for these substrates were low and thus difficult to determine accurately. Despite much lower catalytic rates, kcat/Km values (specificity constants) were comparable to those for PikC and PikC-RhFRED acting on native substrates 3 and 5.44 Relative to the macrolides, the specificity constants for the aglycones were considerably reduced, largely due to higher Km values. The value for 10 (0.0127 ± 0.0006 μM−1min−1) is similar to that for CYP107W1 acting on its native substrate (0.012 μM−1min−1).52 The specificity constant is ~14-fold lower for 8 (0.0009 ± 0.0002 μM−1min−1), but it is still higher than that for the oxidation of tirandamycin A by TamI-RhFRED (0.00058 μM−1min−1).34 Notably, the results from these kinetics experiments are consistent with those obtained from the TTN assays, which indicate a clear preference for MycCI-RhFRED toward substrates bearing a deoxyamino sugar.
Figure 3.
Steady-state kinetics profiles for (A) MycCI-RhFRED and (B) MycCI in conjunction with MycCII and MBP-FdR. Plots are of initial product formation rate (V0) as assessed by HPLC normalized to the concentration of P450 (0.5 μM) used in the kinetics assays. Mean values and standard deviations (error bars) associated with each data point were calculated from the results of experiments performed at least in triplicate. The data were fit to the Michaelis-Menten equation (MycCI-RhFRED + 8–10) or to equations describing single-site (MycCI-RhFRED + 11, 12) or two-site (MycCI/MycCII/MBP-FdR + 8–12) substrate inhibition as described in Supporting Information. The inset in the bottom graph shows a close-up version of the data at low substrate concentrations (0–100 μM).
Table 3.
Steady-state Kinetic Parameters for MycCI-RhFREDa
| Substrate | Product formation | NADPH consumption | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Kcat (min−1) | Km (μM) | Kcat/Km (μM−1min−1) | Kcat (min−1) | Km (μM) | Kcat/Km (μM−1min−1) | |
| 8 | 0.24 ± 0.02 | 264 ± 50 | 0.0009 ± 0.0002 | 5.4 ± 0.2 | 99 ± 8 | 0.055 ± 0.005 |
| 9 | 1.15 ± 0.02 | 2.1 ± 0.4 | 0.6 ± 0.1 | 8.0 ± 0.3 | 1.5 ± 0.4 | 5 ± 1 |
| 10 | 0.355 ± 0.003 | 28 ± 1 | 0.0127 ± 0.0006 | 6.38 ± 0.08 | 14.9 ± 0.9 | 0.43 ± 0.03 |
| 11 | 0.88 ± 0.04 | 0.9 ± 0.5 | 1.0 ± 0.5 | 9.1 ± 0.3 | 1.5 ± 0.3 | 6 ± 1 |
| 12 | 0.72 ± 0.03 | 3.9 ± 0.8 | 0.19 ± 0.04 | 8.2 ± 0.1 | 1.0 ± 0.1 | 8 ± 1 |
Reported errors are those obtained from fitting data averaged from experiments performed at least in triplicate.
The same set of experiments was then conducted using MycCI in combination with MycCII and MBP-FdR. For all of the compounds tested, plots of initial rate vs. substrate concentration showed a high degree of partial substrate inhibition (Figure 3b). The data revealed relatively “high” turnover rates at low substrate concentrations (e.g., 2.3 ± 0.3 min−1 at 10 μM 9) that quickly diminished to ~1 min−1 in all cases as concentrations of substrate were increased. Partial substrate inhibition is a commonly observed phenomenon in P450-catalyzed reactions, although this situation is most well documented for human drug-metabolizing P450s (e.g., CYP3A4). Such inhibition occurs when saturating levels of substrate lead to incomplete inhibition of P450 activity,55 and it is thought to occur as a consequence of substrate binding to a second site located either proximal or distal to the catalytically productive binding site.56,57 The resulting ternary complex of enzyme bound simultaneously to two substrate molecules exhibits reduced competence with respect to product formation (see Supporting Information for further discussion). Although the data obtained for MycCI/MycCII/MBP-FdR could be fit to a defined rate equation for an enzyme with two substrate binding sites,56 the resulting values for kcat and Km were associated with large errors (Table S11), rendering useful interpretation of their significance difficult. Since the physiological concentration of the intermediate 9 in wild-type M. griseorubida is likely very low,58 substrate inhibition of MycCI may either be irrelevant in vivo, or it could represent a mechanism by which flux through the mycinamicin biosynthetic pathway is regulated.
The rates determined for product formation in the experiments described above were substantially lower than those previously reported for MycCI (e.g., kcat = 104 ± 2 min−1 for 9).33 Since the previous data were determined by monitoring NADPH consumption, it was possible that cofactor and substrate oxidation were highly uncoupled. In order to verify this hypothesis, steady-state kinetics experiments were repeated by spectrophotometrically monitoring rates of NADPH consumption at various concentrations of substrate. For MycCI-RhFRED, the derived turnover rates were indeed significantly higher than those determined by monitoring product formation, indicating poor coupling efficiency (≤ 12% in all cases) (Tables 3, S15, and S16). The ability to test lower substrate concentrations in the NADPH consumption assays also facilitated more accurate determination of Km values, which were generally in agreement with those derived from product formation experiments (Table 3). Due to relatively fast NADPH oxidation rates, however, kcat/Km values were much higher.
Prohibitively high rates of NADPH consumption in the absence of substrate were observed when MycCI was tested in combination with MycCII and MBP-FdR (Table S13), further precluding determination of relevant kinetic parameters for this tripartite biocatalytic system. Nonetheless, we were able to compare turnover frequency (TOF, determined at 500 μM substrate) values to those for MycCI-RhFRED (Table 4). The TOF was 1.4 ± 0.1 min−1 for the tripartite system with 9 acting as a substrate; this value was only slightly reduced for the other substrates, with the lowest being ~1 min−1 for each of the aglycones. MycCI-RhFRED had a similar TOF for 9 (1.19 ± 0.06 min−1), but the rates were more substantially decreased for the other substrates. In particular, the rates for formation of 15 and 13 were about 3-fold and 6-fold lower, respectively, compared to the rates determined using the tripartite MycCI system. These results indicate that while electron transfer may not be rate-limiting for MycCI/MycCII/MBP-FdR, it likely is for MycCI-RhFRED. Interestingly, despite the higher TTN values obtained with MycCI-RhFRED acting on macrolide substrates 9, 11, and 12, the corresponding TOF values were all lower than those observed with MycCI/MycCII/MBP-FdR. The most likely explanation for this result is increased coupling efficiency of the self-sufficient system compared to the tripartite system, leading in turn to attenuated production of reactive oxygen species (ROS) that are involved in the oxidative degradation of the enzyme.59 Thus, despite lower rates of catalysis, MycCI-RhFRED remains active in the reaction mixture for a longer amount of time and is thereby able to turn over more substrate molecules.
Table 4.
Turnover Frequencies (TOFs) for Selected Enzymatic Reactionsa
| Catalytic system | Substrate
|
||||
|---|---|---|---|---|---|
| 8 | 9 | 10 | 11 | 12 | |
| MycCI/MycCIIb | 0.97 ± 0.03 | 1.4 ± 0.1 | 0.96 ± 0.01 | 1.23 ± 0.05 | 1.11 ± 0.08 |
| MycCI-RhFRED | 0.161 ± 0.008 | 1.19 ± 0.06 | 0.34 ± 0.01 | 0.81 ± 0.01 | 0.61 ± 0.02 |
| TylHI/TylHIIb | N.D.c | N.D. | N.D. | N.D. | 0.14 ± 0.03 |
| TylHI/MycCIIb | N.D. | N.D. | N.D. | N.D. | 1.63 ± 0.03 |
| TylHI-RhFRED | N.D. | N.D. | N.D. | N.D. | 0.76 ± 0.02 |
| MycCIS172A/MycCIIb | 1.15 ± 0.05 | 1.09 ± 0.05 | 0.96 ± 0.02 | 0.90 ± 0.03 | 0.86 ± 0.06 |
TOF = mol product/mol P450 per min. Product formation was monitored by HPLC over the first 30 min of the reaction (initial rates). Reported errors are standard deviations calculated from experiments performed in triplicate.
Includes MBP-FdR.
N.D. = value not determined.
Comparative analysis of the MycCI homolog TylHI
MycCI (CYP105L2) is a member of the CYP105 family of P450 enzymes. Homologs of CYP105 have been identified in all species of Streptomyces hitherto characterized, where they are chiefly involved in xenobiotic metabolism and in the biosynthesis of secondary metabolites.60 In turn, many of the latter variety are involved in polyketide biosynthesis, particularly polyene antifungals (e.g., amphotericin, pimaricin, nystatin, filipin, and candicidin) and macrolide antibiotics (e.g., tylosin, mycinamicin, and geldanamycin). MycCI has close homologs in the biosynthetic pathways for the production of the 16-membered ring macrolides tylosin (7) in Streptomyces fradiae (TylHI)61,62 and chalcomycin in Streptomyces bikiniensis (ChmHI).63 The pathways associated with the production of these molecules are highly homologous to each other and to that involved in the assembly of the mycinamicins in M. griseorubida (Scheme 1). Accordingly, the chemical structures of the final compounds are very similar, all consisting of a 16-membered ring macrolactone core containing a characteristic unsaturated ketone moiety to which at least two different sugars are appended.
Like the mycinamicin biosynthetic gene cluster, the tylosin cluster contains two P450 enzymes (TylI and TylHI) whose functions have been provisionally assigned based on early co-fermentation and biotransformation studies with mutants of S. fradiae blocked in tylosin biosynthesis43,64,65 as well as phylogenetic comparison with homologous P450s from other pathways. The preferred order of oxidation events was also demonstrated via experiments that employed S. fradiae crude cell lysate for bioconversion of tylosin biosynthetic intermediates.66 Based on these collective efforts, it was proposed that TylI installs the aldehyde at C20 of 5-O-mycaminosyl-tylactone just prior to hydroxylation of C23 by TylHI to produce the key intermediate 17 (Scheme 1b). Until now, the lack of a robust in vitro system to explore these two P450 enzymes has resulted in persistent ambiguity with respect to their substrate selectivity. The primary sequence of TylHI (CYP105L1) is 55% identical to that of MycCI; hence, they are classified as members of the same P450 subfamily. In addition, a small gene encoding a [3Fe-4S]-type ferredoxin, designated tylHII, is located adjacent to tylHI in the gene cluster.61,62 The same juxtaposition of P450 and ferredoxin genes encoding MycCI and MycCII, respectively, occurs in the mycinamicin gene cluster,67 suggesting that TylHI acts in conjunction with TylHII during C23 hydroxylation of its proposed native substrate 12.
Given its high homology to MycCI, we decided to probe the substrate scope of TylHI. Although previous studies seemed to suggest that this enzyme could only act upon glycosylated substrates with an adjacent ethyl group that had already been fully oxidized to an aldehyde,66 we predicted that the purified enzyme might have a broader substrate scope based on its close association with MycCI. Following the cloning, overexpression, and purification of TylHI and its proposed ferredoxin partner TylHII, we tested their activity in conjunction with MBP-FdR toward all of the compounds previously employed to assess the substrate scope of MycCI. The results demonstrated that TylHI partnered with TylHII converted 12 to a monohydroxylated product with the same retention time as 17, but the catalytic system only supported about 18 substrate turnovers (Table 1). In addition, TylHI could hydroxylate 11 at the same relative position to produce 16, but only a single turnover was observed. The enzyme exhibited no activity on any of the other substrates tested, even when the concentration of TylHI in the reaction mixture was increased ten-fold to 5 μM (1.0 mol %). Although the gene encoding TylHII was codon optimized for expression in E. coli, the protein expressed much more poorly than MycCII, even when GroEL/GroES were co-expressed. Furthermore, spectrophotometric analysis of the purified protein indicated diminished absorbance at 410 nm compared to MycCII at a given concentration (Figure S1), which could indicate the presence of higher amounts of apo protein. Accordingly, we tested TylHI in combination with MycCII to determine if the relative lack of activity observed for this enzyme could be attributed to a suboptimal ferredoxin partner. While the TTN for 12 nearly quadrupled, that for 11 was essentially naught (Table 1). Moreover, the enzyme remained inactive on all of the other substrates, even when it was present at high concentrations (5 μM).
Since MycCI-RhFRED exhibited improved activity compared to MycCI/MycCII/MBP-FdR, we opted to construct the corresponding self-sufficient version of TylHI. Although activity increased to around 160 turnovers for the native substrate 12, no improvement was observed for 11, and the enzyme remained inactive toward all other substrates (Table 1). These data unambiguously demonstrate that the substrate scope for TylHI is extremely narrow, especially compared to MycCI. While this finding is surprising given the high homology of the two enzymes, it is consistent with the results of previous studies.66
Subsequently, we conducted substrate binding and steady-state kinetics studies with TylHI and TylHI-RhFRED, which provided some insights into the results of the activity assays. TylHI bound 12 with very high affinity (Kd = 0.60 ± 0.03 μM), but its affinity toward 11 was considerably diminished (Kd = 275 ± 22 μM) (Table 2; Figure S7). Curiously, these two compounds differ only in the identity of the deoxyamino sugar (1 vs. 18) and the oxidation state of C20 (methyl vs. formyl) (Figure 1). Therefore, one or both of these features play key roles in substrate recognition. Although none of the other substrates tested were converted to monohydroxylated products by TylHI, some were able to induce a shift in the spin state of the enzyme, including aglycone 10 (Table 2; Figures 2b and S8).
Steady-state kinetic analysis of the reaction between TylHI-RhFRED and 12 revealed little to no substrate inhibition and parameters for product formation that were about the same compared to MycCI-RhFRED acting on the same substrate (Figure S9a). However, TylHI-RhFRED oxidized NADPH at a slightly faster rate than MycCI-RhFRED when saturated with 12 (Figure S10c). As for the tripartite MycCI system, partial substrate inhibition was observed for TylHI/TylHII/MBP-FdR acting on 12 (Figure S9b), but the rate of product formation was nearly an order of magnitude lower (TOF = 0.14 ± 0.03 min−1) (Table 4). When MycCII was employed as surrogate ferredoxin, the TOF increased 12-fold to about 1.6 min−1, which was higher than that observed for MycCI/MycCII/MBP-FdR.
The highly limited substrate scope of TylHI is in stark contrast to the flexibility that MycCI exhibits toward different types of 16-membered ring macrolactones. Whereas the deoxyamino sugar is a dispensable feature of MycCI substrates, it is absolutely required for TylHI catalysis. Additionally, the identity of the sugar and/or the nature of the carbon side chains comprising the macrolactone core plays a significant role in substrate binding. Based on the results of our experiments, it remains unclear what specific aspects of 12 make it a far superior substrate for TylHI than 11. The aldehyde and/or the additional hydroxyl group on the sugar appears to be critical for TylHI-catalyzed hydroxylation of 12. Work is ongoing to solve the x-ray crystal structure of TylHI in complex with its native substrate, which will provide important insights into the specific interactions that are responsible for the enzyme’s high substrate specificity. Despite the current lack of a TylHI structure, alignment with MycCI revealed that while differences are fairly evenly distributed throughout the sequence, nearly half of the residues that are located within 4 Å of 9 in the MycCI co-crystal structure (vide infra) are different in the TylHI sequence. Moreover, most of these changes appear in residues that are proximal to the desosamine sugar moiety, including those found in and around the BC loop (substrate recognition site 1) and FG loop (substrate recognition sites 2–3) regions.68 These findings are consistent with the differences observed between MycCI and TylHI with respect to the importance of the sugar moiety in substrate binding and catalysis.
Another interesting example of highly homologous P450s that nonetheless exhibit significant differences in their substrate scope can be found in glycopeptide antibiotic biosynthesis. Recent work carried out by Cryle and coworkers has demonstrated the importance of the non-ribosomal peptide synthetase X-domain in helping to coordinate the P450-mediated oxidative cascade that leads to fully cyclized aglycones in the biosynthesis of vancomyin, teicoplanin, and other glycopeptides.69–71 While the P450 OxyB from the vancomycin pathway (OxyBvan) has long been recognized as a versatile biocatalyst with the ability to accept a wide range of peptidyl carrier protein (PCP)-bound substrates,72–75 the homolog from the teicoplanin pathway (OxyBtei, 74% sequence identity) exhibits a much narrower substrate scope when acting on PCP-bound peptides in the absence of the X-domain.76 However, OxyBtei and homologs from other pathways are able to act on a broader range of substrates when the X-domain is present, although they tend to exhibit lower overall activity compared to OxyBvan.69,70 Along with these fascinating studies, our results with MycCI underscore the importance of empirically testing and comparing P450 homologs from related biosynthetic pathways in order to identify those that might prove most useful in the development of novel biocatalysts.
Structural analysis of MycCI bound to M-VIII
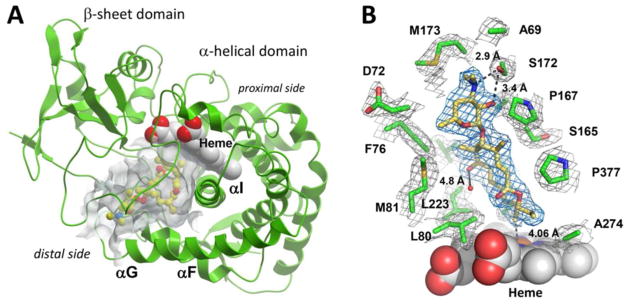
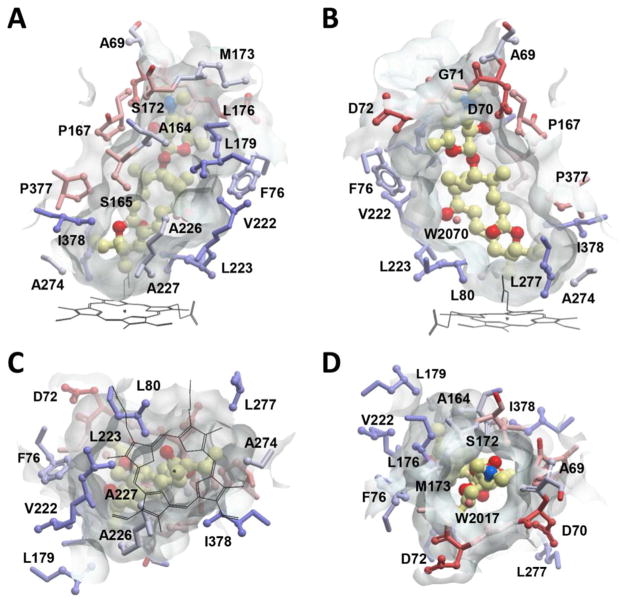
In order to gain further insight into the unique reactivity exhibited by MycCI, we determined the crystal structure of the enzyme in complex with its native substrate M-VIII (9) to a resolution of 2.21 Å. The asymmetric unit of the crystal lattice contains two molecules of MycCI, with no conformational ambiguity observed between chains A and B, both of which have substrate bound in the active site. MycCI has an overall tertiary structure typical of cytochrome P450 enzymes (Figures 4a and S11).25 The 7000 Å3 substrate binding pocket at the “distal” face of the heme macrocycle accommodates 9 bound in an orientation tilted relative to the plane of the macrocycle (Figure 4b). The 16-membered ring polyketide macrolactone and desosamine sugar (1) moieties of the substrate are both clearly defined in the electron density map. The primary methyl group (C21) is exposed to the oxygen scission site within 4.06 Å of the iron center (distance averaged between the two MycCI molecules constituting an asymmetric unit). The proximity of C21 and the orientation of the C14–C21 bond relative to the heme iron are consistent with hydroxylation of 9 exclusively at the C21 position. A few water molecules are observed in the active site, but only one overlaps between chains A and B (Figure 4b).
Figure 4.
Crystal structure of MycCI with 9 bound in the active site. (A) The protein scaffold is represented by green ribbons. 9 (ball-and-stick mode: yellow = carbon, red = oxygen, blue = nitrogen) occupies the 7000 Å3 pocket (semi-transparent grey surface) of the α-helical domain interfacing the β-sheet domain. Image was generated using the MOLSOFT ICM-Browser.77 (B) A fragment of the 2Fo−Fc electron density map contoured at 1 σ delineates 9 (blue mesh) and a set of amino acid residues (green) within 4 Å of 9 (grey mesh). Distances reported are in Ångströms. The heme cofactor is depicted as van der Waals spheres (grey = carbon, red = oxygen, blue = nitrogen, ochre = iron). The water molecule present in both chains A and B of the asymmetric unit is shown as a small red sphere. Image was prepared using PyMOL.78
The most hydrophobic edge of the macrolactone portion of 9 is tightly packed against hydrophobic amino acid side chains emanating from the F-, G-, and I helices (shown in Figure 5a in different shades of blue). The opposite side of the binding site interfacing the β-sheet domain is more polar and loosely constructed (Figure 5b). Six hydrophobic residues (L80, L223, A227, A274, L277, and I378) form a rim adjacent to the porphyrin ring that tightly encloses the end of the macrolactone opposite desosamine (Figure 5c), serving to position it in an orientation that favors hydroxylation at C21. The binding pocket that accommodates desosamine is built by more polar residues, including S172, D70, and D72 (Figure 5d). However, the only potential specific polar contact between enzyme and substrate that can be discerned occurs between the side chain hydroxyl of S172 and the 3′-nitrogen as well as the adjacent 2′-hydroxyl of desosamine (Figure 4b). In order to probe the importance of these potential interactions, the MycCIS172A mutant was generated and tested in reactions with the series of substrates previously described. Only moderate decreases in TTN were observed for the macrolide substrates (9, 11, and 12) while, surprisingly, turnover of aglycone substrates (8 and 10) was slightly improved for the mutant relative to the wild-type enzyme (Table 1). Consistent with these data, the mutant bound the 16-membered ring macrolide compounds with slightly weaker affinity, but the dissociation constants were still in the nanomolar range; in contrast, 8 and 10 bound about twice as tightly to the mutant (Table 2). Despite the marginal impact that the S172A mutation had on substrate binding affinity, a lower percentage of the mutant was shifted to the high-spin state at saturating levels of each substrate compared to the wild-type enzyme (Figure 2b; Table S9). This decrease in percent spin shift was largest for 5, which shifted only ~40% of the mutant to high spin at saturating levels compared to ~80% of the wild-type. Finally, kinetic analysis revealed modest (~25%) reductions in TOF for each of the macrolide substrates while rates either improved (8) or remained unchanged (10) for the aglycones (Table 4). Taken together, these data demonstrate that S172 plays a minor role in facilitating substrate binding and catalysis for MycCI. However, this result is not surprising given that we have already shown the entire sugar moiety to be an unnecessary feature of suitable MycCI substrates. In contrast to what we have observed for PikC, the tertiary amine of desosamine, which carries a positive charge at neutral pH, does not interact with the negatively charged carboxylate-containing residues of MycCI. As previously noted, salt-bridge interactions between the N,N-dimethylamino group of desosamine and the side chains of acidic residues (E85 and E94) play a critical role in positioning both native and semi-synthetic substrates in the active site of PikC.31,36,37,39 In the MycCI crystal structure, the carboxylate groups of both D70 and D72 actually point away from desosamine. Furthermore, the negative charge of the D70 side chain is neutralized by forming a salt-bridge with the positively charged guanidinium side chain of R278. Thus, substrate recognition and binding operate by a completely different mechanism in MycCI, mostly mediated by hydrophobic interactions between the substrate and the plethora of nonpolar amino acid side chains that line the binding pocket.
Figure 5.
The binding site of 9 in MycCI, represented by a semi-transparent surface surrounding the substrate molecule (ball-and-stick mode: yellow = carbon, red = oxygen, blue = nitrogen). Amino acid residues are in ball-and-stick mode and are colored by hydrophobicity in a color gradient ranging from ice-blue (hydrophobic) to red (hydrophilic). Water molecules are shown as small spheres. Images were generated using the MOLSOFT ICM-Browser.77 (A) Side view of the binding site showing the hydrophobic wall built by lipophilic amino acids. (B) The opposite side of the binding pocket facing the β-sheet domain is more polar and loosely built. (C) Bottom view of the binding site with the heme cofactor (black lines) projected onto the semi-transparent surface. Six lipophilic residues along the edge of the heme macrocycle (L80, L223, A227, A274, L277, and I378) form a pocket that surrounds the end of the M-VIII macrolactone opposite desosamine, thus positioning the C21 methyl group above the iron center. (D) Top view along the axis of 9 with the desosamine moiety visible through the surface opening.
Our previous studies had initially led us to hypothesize that MycCI employs a mechanism analogous to that used by PikC for binding its natural desosaminylated substrate 9, as we did not observe conversion of the corresponding aglycone 8 to a monohydroxylated product.33 The reason for the discrepancy between these prior results and our current findings remains unclear. In our initial report, the MycCI construct employed in analytical-scale reactions had a C-terminal His-tag, which was reported to show activity toward 9 comparable to that exhibited by the N-terminal His-tagged enzyme. However, the relative activity of the C-terminal His-tagged construct on 8 may have been diminished to the extent that no product could be detected at all. Moreover, the concentration of MycCII in the reported reactions was significantly lower (3.5 μM compared to 100 μM in the present work), and a NADPH regeneration system was not employed in these prior studies. As noted before, the extent of substrate turnover was highly dependent on the amount of MycCII present in the reaction mixture. It is likely that suboptimal concentrations of this important redox partner along with the lack of continual in situ NADPH regeneration would preclude formation of detectable amounts of monohydroxylated 8. Indeed, we used similar unoptimized conditions when we made the initial discovery that MycCI could hydroxylate 10, observing only a few percent total conversion of this substrate.
Conclusions
We have demonstrated that MycCI, a P450 monooxygenase from the mycinamicin biosynthetic pathway, is capable of catalyzing the hydroxylation of unactivated primary carbons in diverse aglycone and macrolide substrates. This ability sets it apart from related P450s involved in macrolide biosynthesis and highlights its potential to serve as a basis for developing novel hydroxylation biocatalysts with broad substrate scope and high regioselectivity. The crystal structure of MycCI in complex with its native 16-membered ring macrolide substrate 9 provides some insight into the mechanism of substrate binding, revealing the dominance of hydrophobic interactions in properly positioning it to undergo the energetically unfavorable oxidation of a primary C-H bond. Our work further underscores the importance of exploratory efforts in discovering new P450s from diverse organisms that exhibit unexpected and biocatalytically useful properties.
Supplementary Material
Table 5.
Crystallographic Data Summary
| Protein | MycCI |
|---|---|
|
| |
| Ligand | Mycinamicin VIII |
|
| |
| PDB ID | 5FOI |
| Data collection | |
| Space group | P1 |
| Cell dimensions | |
| a, b, c (Å) | 54.8, 59.5, 74.6 |
| α, β, γ (°) | 83.2, 72.2, 62.6 |
| Molecules in AU | 2 |
| Wavelength | 1.11587 |
| Resolution (Å) | 2.21 |
| Rsym (%) | 24.4 |
| I/σI | 3.8 (1.3)a |
| Completeness (%) | 91.1 (71.2) |
| Redundancy | 2.0 (1.9) |
| Refinement | |
| No. reflections | 34383 |
| Rwork/Rfree (%) | 18.2/25.2 |
| No. atoms | |
| Protein | 6013 |
| Heme | 86 |
| Substrate | 72 |
| Solvent | 147 |
| Mean B value | 27.2 |
| B-factors | |
| Protein | 27.9 |
| Heme | 17.0 |
| Substrate | 21.0 |
| Solvent | 28.6 |
| RMSD | |
| Bond lengths (Å) | 0.014 |
| Bond angles (°) | 1.907 |
Values in parentheses are for the highest resolution shell.
Acknowledgments
The plasmid for the expression of MBP-FdR was a kind gift from G. Zanetti (University of Milan). We thank Y. Anzai (Toho University) for the generous gifts of protomycinolide IV and mycinolide IV. We also thank K. C. Chiou (University of Michigan) for assistance with cloning TylHI constructs, V. Shende (University of Michigan) for assistance with fermentation and purification of tylactone, A. Tripathi (University of Michigan) for assistance with NMR data analysis, and D. P. Ballou (University of Michigan) for helpful discussions related to P450s and steady-state kinetics. We are grateful to the National Science Foundation under the CCI Center for Selective C-H Functionalization (CHE-1205646), the National Institutes of Health for financial support (GM078553 and GM118101), and the Hans W. Vahlteich Professorship (to D.H.S.). M.D.D. II acknowledges support from the University of Michigan Cellular Biotechnology Training Program (NIH grant T32-GM008353) and a University of Michigan Rackham Predoctoral Fellowship.
Footnotes
Accession codes.
Atomic coordinates and structure factors for MycCI/M-VIII have been deposited in the Protein Data Bank under PDB accession code 5FOI.
Notes.
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Detailed experimental materials and methods, supplemental tables, schemes, figures, and discussions, and NMR data for compounds 8–17. This material is available free of charge via the Internet.
References
- 1.Ortiz de Montellano PR, editor. Cytochrome P450: structure, mechanism, and biochemistry. 3. Kluwer Academic/Plenum Publishers; New York: 2005. [Google Scholar]
- 2.McLean KJ, Girvan HM, Mason AE, Dunford AJ, Munro AW. Structure, mechanism and function of cytochrome P450 enzymes. In: de Visser SP, Kumar D, editors. Iron-containing enzymes: versatile catalysts of hydroxylation reactions in nature. Royal Society of Chemistry; Cambridge: 2011. pp. 255–280. [Google Scholar]
- 3.Munro AW, Girvan HM, Mason AE, Dunford AJ, McLean KJ. What makes a P450 tick? Trends Biochem Sci. 2013;38:140–150. doi: 10.1016/j.tibs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Guengerich FP. Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem Res Toxicol. 2001;14:611–650. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- 5.Nelson DR. The Cytochrome P450 Homepage. Hum Genomics. 2009;4:59–65. doi: 10.1186/1479-7364-4-1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meunier B, de Visser SP, Shaik S. Mechanism of oxidation reactions catalyzed by cytochrome P450 enzymes. Chem Rev. 2004;104:3947–3980. doi: 10.1021/cr020443g. [DOI] [PubMed] [Google Scholar]
- 7.Denisov IG, Makris TM, Sligar SG, Schlichting I. Structure and chemistry of cytochrome P450. Chem Rev. 2005;105:2253–2277. doi: 10.1021/cr0307143. [DOI] [PubMed] [Google Scholar]
- 8.Ortiz de Montellano PR. Hydrocarbon hydroxylation by cytochrome P450 enzymes. Chem Rev. 2010;110:932–948. doi: 10.1021/cr9002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Isin EM, Guengerich FP. Complex reactions catalyzed by cytochrome P450 enzymes. Biochim Biophys Acta. 2007;1770:314–329. doi: 10.1016/j.bbagen.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Guengerich FP, Munro AW. Unusual cytochrome P450 enzymes and reactions. J Biol Chem. 2013;288:17065–17073. doi: 10.1074/jbc.R113.462275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernhardt R. Cytochromes P450 as versatile biocatalysts. J Biotechnol. 2006;124:128–145. doi: 10.1016/j.jbiotec.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 12.Urlacher VB, Eiben S. Cytochrome P450 monooxygenases: perspectives for synthetic application. Trends Biotechnol. 2006;24:324–330. doi: 10.1016/j.tibtech.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Munro AW, Girvan HM, McLean KJ. Variations on a (t)heme—novel mechanisms, redox partners and catalytic functions in the cytochrome P450 superfamily. Nat Prod Rep. 2007;24:585–609. doi: 10.1039/b604190f. [DOI] [PubMed] [Google Scholar]
- 14.O’Reilly E, Köhler V, Flitsch SL, Turner NJ. Cytochromes P450 as useful biocatalysts: addressing the limitations. Chem Commun. 2011;47:2490–2501. doi: 10.1039/c0cc03165h. [DOI] [PubMed] [Google Scholar]
- 15.Grogan G. Cytochromes P450: exploiting diversity and enabling application as biocatalysts. Curr Opin Chem Biol. 2011;15:241–248. doi: 10.1016/j.cbpa.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 16.Fasan R. Tuning P450 enzymes as oxidation catalysts. ACS Catal. 2012;2:647–666. [Google Scholar]
- 17.Urlacher VB, Girhard M. Cytochrome P450 monooxygenases: an update on perspectives for synthetic application. Trends Biotechnol. 2012;30:26–36. doi: 10.1016/j.tibtech.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 18.Caswell JM, O’Neill M, Taylor SJC, Moody TS. Engineering and application of P450 monooxygenases in pharmaceutical and metabolite synthesis. Curr Opin Chem Biol. 2013;17:271–275. doi: 10.1016/j.cbpa.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 19.Bernhardt R, Urlacher VB. Cytochromes P450 as promising catalysts for biotechnological application: chances and limitations. Appl Microbiol Biotechnol. 2014;98:6185–6203. doi: 10.1007/s00253-014-5767-7. [DOI] [PubMed] [Google Scholar]
- 20.Behrendorff JBYH, Huang W, Gillam EMJ. Directed evolution of cytochrome P450 enzymes for biocatalysis: exploiting the catalytic versatility of enzymes with relaxed substrate specificity. Biochem J. 2015;467:1–15. doi: 10.1042/BJ20141493. [DOI] [PubMed] [Google Scholar]
- 21.Guengerich FP. Cytochrome P450 enzymes in the generation of commercial products. Nat Rev Drug Discov. 2002;1:359–366. doi: 10.1038/nrd792. [DOI] [PubMed] [Google Scholar]
- 22.Kelly SL, Lamb DC, Jackson CJ, Warrilow AGS, Kelly DE. The biodiversity of microbial cytochromes P450. In: Poole RK, editor. Advances in Microbial Physiology. Vol. 47. Academic Press; London: 2003. pp. 131–186. [DOI] [PubMed] [Google Scholar]
- 23.Kelly SL, Kelly DE. Microbial cytochromes P450: biodiversity and biotechnology. Where do cytochromes P450 come from, what do they do and what can they do for us? Phil Trans R Soc B. 2013:368. doi: 10.1098/rstb.2012.0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cryle MJ, Stok JE, De Voss JJ. Reactions catalyzed by bacterial cytochromes P450. Aust J Chem. 2003;56:749–762. [Google Scholar]
- 25.Podust LM, Sherman DH. Diversity of P450 enzymes in the biosynthesis of natural products. Nat Prod Rep. 2012;29:1251–1266. doi: 10.1039/c2np20020a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newman DJ, Cragg GM. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod. 2016;79:629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 27.Andersen JF, Tatsuta K, Gunji H, Ishiyama T, Hutchinson CR. Substrate specificity of 6-deoxyerythronolide B hydroxylase, a bacterial cytochrome P450 of erythromycin A biosynthesis. Biochemistry. 1993;32:1905–1913. doi: 10.1021/bi00059a004. [DOI] [PubMed] [Google Scholar]
- 28.Xue Y, Wilson D, Zhao L, Liu H-w, Sherman DH. Hydroxylation of macrolactones YC-17 and narbomycin is mediated by the pikC-encoded cytochrome P450 in Streptomyces venezuelae. Chem Biol. 1998;5:661–667. doi: 10.1016/s1074-5521(98)90293-9. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Q, Sherman DH. Isolation and structure determination of novamethymycin, a new bioactive metabolite of the methymycin biosynthetic pathway in Streptomyces venezuelae. J Nat Prod. 2001;64:1447–1450. doi: 10.1021/np010146r. [DOI] [PubMed] [Google Scholar]
- 30.Lee SK, Park JW, Kim JW, Jung WS, Park SR, Choi CY, Kim ES, Kim BS, Ahn JS, Sherman DH, Yoon YJ. Neopikromycin and novapikromycin from the pikromycin biosynthetic pathway of Streptomyces venezuelae. J Nat Prod. 2006;69:847–849. doi: 10.1021/np060026p. [DOI] [PubMed] [Google Scholar]
- 31.Sherman DH, Li S, Yermalitskaya LV, Kim Y, Smith JA, Waterman MR, Podust LM. The structural basis for substrate anchoring, active site selectivity, and product formation by P450 PikC from Streptomyces venezuelae. J Biol Chem. 2006;281:26289–26297. doi: 10.1074/jbc.M605478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding Y, Seufert WH, Beck ZQ, Sherman DH. Analysis of the cryptophycin P450 epoxidase reveals substrate tolerance and cooperativity. J Am Chem Soc. 2008;130:5492–5498. doi: 10.1021/ja710520q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anzai Y, Li S, Chaulagain MR, Kinoshita K, Kato F, Montgomery J, Sherman DH. Functional analysis of MycCI and MycG, cytochrome P450 enzymes involved in biosynthesis of mycinamicin macrolide antibiotics. Chem Biol. 2008;15:950–959. doi: 10.1016/j.chembiol.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carlson JC, Li S, Gunatilleke SS, Anzai Y, Burr DA, Podust LM, Sherman DH. Tirandamycin biosynthesis is mediated by co-dependent oxidative enzymes. Nat Chem. 2011;3:628–633. doi: 10.1038/nchem.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li S, Tietz DR, Rutaganira FU, Kells PM, Anzai Y, Kato F, Pochapsky TC, Sherman DH, Podust LM. Substrate recognition by the multifunctional cytochrome P450 MycG in mycinamicin hydroxylation and epoxidation reactions. J Biol Chem. 2012;287:37880–37890. doi: 10.1074/jbc.M112.410340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li S, Chaulagain MR, Knauff AR, Podust LM, Montgomery J, Sherman DH. Selective oxidation of carbolide C–H bonds by an engineered macrolide P450 mono-oxygenase. Proc Natl Acad Sci USA. 2009;106:18463–18468. doi: 10.1073/pnas.0907203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Negretti S, Narayan ARH, Chiou KC, Kells PM, Stachowski JL, Hansen DA, Podust LM, Montgomery J, Sherman DH. Directing group-controlled regioselectivity in an enzymatic C–H bond oxygenation. J Am Chem Soc. 2014;136:4901–4904. doi: 10.1021/ja5016052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Narayan ARH, Jiménez-Osés G, Liu P, Negretti S, Zhao W, Gilbert MM, Ramabhadran RO, Yang YF, Furan LR, Li Z, Podust LM, Montgomery J, Houk KN, Sherman DH. Enzymatic hydroxylation of an unactivated methylene C–H bond guided by molecular dynamics simulations. Nat Chem. 2015;7:653–660. doi: 10.1038/nchem.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li S, Ouellet H, Sherman DH, Podust LM. Analysis of transient and catalytic desosamine-binding pockets in cytochrome P-450 PikC from Streptomyces venezuelae. J Biol Chem. 2009;284:5723–5730. doi: 10.1074/jbc.M807592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansen DA, Rath CM, Eisman EB, Narayan ARH, Kittendorf JD, Mortison JD, Yoon YJ, Sherman DH. Biocatalytic synthesis of pikromycin, methymycin, neomethymycin, novamethymycin, and ketomethymycin. J Am Chem Soc. 2013;135:11232–11238. doi: 10.1021/ja404134f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borisova SA, Zhang C, Takahashi H, Zhang H, Wong AW, Thorson JS, Liu H-w. Substrate specificity of the macrolide-glycosylating enzyme pair DesVII/DesVIII: opportunities, limitations, and mechanistic hypotheses. Angew Chem Int Ed. 2006;45:2748–2753. doi: 10.1002/anie.200503195. [DOI] [PubMed] [Google Scholar]
- 42.Borisova SA, Kim HJ, Pu X, Liu H-w. Glycosylation of acyclic and cyclic aglycone substrates by macrolide glycosyltransferase DesVII/DesVIII: analysis and implications. ChemBioChem. 2008;9:1554–1558. doi: 10.1002/cbic.200800155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baltz RH, Seno ET. Properties of Streptomyces fradiae mutants blocked in biosynthesis of the macrolide antibiotic tylosin. Antimicrob Agents Chemother. 1981;20:214–225. doi: 10.1128/aac.20.2.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li S, Podust LM, Sherman DH. Engineering and analysis of a self-sufficient biosynthetic cytochrome P450 PikC fused to the RhFRED reductase domain. J Am Chem Soc. 2007;129:12940–12941. doi: 10.1021/ja075842d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang W, Liu Y, Yan J, Cao S, Bai F, Yang Y, Huang S, Yao L, Anzai Y, Kato F, Podust LM, Sherman DH, Li S. New reactions and products resulting from alternative interactions between the P450 enzyme and redox partners. J Am Chem Soc. 2014;136:3640–3646. doi: 10.1021/ja4130302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anzai Y, Tsukada S, Sakai A, Masuda R, Harada C, Domeki A, Li S, Kinoshita K, Sherman DH, Kato F. Function of cytochrome P450 enzymes MycCI and MycG in Micromonospora griseorubida, a producer of the macrolide antibiotic mycinamicin. Antimicrob Agents Chemother. 2012;56:3648–3656. doi: 10.1128/AAC.06063-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luthra A, Denisov IG, Sligar SG. Spectroscopic features of cytochrome P450 reaction intermediates. Arch Biochem Biophys. 2011;507:26–35. doi: 10.1016/j.abb.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Montemiglio LC, Parisi G, Scaglione A, Sciara G, Savino C, Vallone B. Functional analysis and crystallographic structure of clotrimazole bound OleP, a cytochrome P450 epoxidase from Streptomyces antibioticus involved in oleandomycin biosynthesis. Biochim Biophys Acta. 2016;1860:465–475. doi: 10.1016/j.bbagen.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 49.Montemiglio LC, Gianni S, Vallone B, Savino C. Azole drugs trap cytochrome P450 EryK in alternative conformational states. Biochemistry. 2010;49:9199–9206. doi: 10.1021/bi101062v. [DOI] [PubMed] [Google Scholar]
- 50.Robin A, Roberts GA, Kisch J, Sabbadin F, Grogan G, Bruce N, Turner NJ, Flitsch SL. Engineering and improvement of the efficiency of a chimeric [P450cam-RhFRed reductase domain] enzyme. Chem Commun. 2009:2478–2480. doi: 10.1039/b901716j. [DOI] [PubMed] [Google Scholar]
- 51.Johnston JB, Ouellet H, Podust LM, Ortiz de Montellano PR. Structural control of cytochrome P450-catalyzed ω-hydroxylation. Arch Biochem Biophys. 2011;507:86–94. doi: 10.1016/j.abb.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han S, Pham TV, Kim JH, Lim YR, Park HG, Cha GS, Yun CH, Chun YJ, Kang LW, Kim D. Functional characterization of CYP107W1 from Streptomyces avermitilis and biosynthesis of macrolide oligomycin A. Arch Biochem Biophys. 2015;575:1–7. doi: 10.1016/j.abb.2015.03.025. [DOI] [PubMed] [Google Scholar]
- 53.Meinhold P, Peters MW, Hartwick A, Hernandez AR, Arnold FH. Engineering cytochrome P450 BM3 for terminal alkane hydroxylation. Adv Synth Catal. 2006;348:763–772. [Google Scholar]
- 54.Zhang K, Shafer BM, Demars MD, II, Stern HA, Fasan R. Controlled oxidation of remote sp3 C–H bonds in artemisinin via P450 catalysts with fine-tuned regio- and stereoselectivity. J Am Chem Soc. 2012;134:18695–18704. doi: 10.1021/ja3073462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hutzler JM, Tracy TS. Atypical kinetic profiles in drug metabolism reactions. Drug Metab Dispos. 2002;30:355–362. doi: 10.1124/dmd.30.4.355. [DOI] [PubMed] [Google Scholar]
- 56.Korzekwa KR, Krishnamachary N, Shou M, Ogai A, Parise RA, Rettie AE, Gonzalez FJ, Tracy TS. Evaluation of atypical cytochrome P450 kinetics with two-substrate models: evidence that multiple substrates can simultaneously bind to cytochrome P450 active sites. Biochemistry. 1998;37:4137–4147. doi: 10.1021/bi9715627. [DOI] [PubMed] [Google Scholar]
- 57.Lin Y, Lu P, Tang C, Mei Q, Sandig G, Rodrigues AD, Rushmore TH, Shou M. Substrate inhibition kinetics for cytochrome P450-catalyzed reactions. Drug Metab Dispos. 2001;29:368–374. [PubMed] [Google Scholar]
- 58.Kinoshita K, Imura Y, Takenaka S, Hayashi M. Mycinamicins, new macrolide antibiotics. XI Isolation and structure elucidation of a key intermediate in the biosynthesis of the mycinamicins, mycinamicin VIII. J Antibiot. 1989;42:1869–1872. doi: 10.7164/antibiotics.42.1869. [DOI] [PubMed] [Google Scholar]
- 59.Loida PJ, Sligar SG. Molecular recognition in cytochrome P-450: mechanism for the control of uncoupling reactions. Biochemistry. 1993;32:11530–11538. doi: 10.1021/bi00094a009. [DOI] [PubMed] [Google Scholar]
- 60.Moody SC, Loveridge EJ. CYP105—diverse structures, functions and roles in an intriguing family of enzymes in Streptomyces. J Appl Microbiol. 2014;117:1549–1563. doi: 10.1111/jam.12662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fouces R, Mellado E, Díez B, Barredo JL. The tylosin biosynthetic cluster from Streptomyces fradiae: genetic organization of the left region. Microbiology. 1999;145:855–868. doi: 10.1099/13500872-145-4-855. [DOI] [PubMed] [Google Scholar]
- 62.Bate N, Cundliffe E. The mycinose-biosynthetic genes of Streptomyces fradiae, producer of tylosin. J Ind Microbiol Biotechnol. 1999;23:118–122. doi: 10.1038/sj.jim.2900707. [DOI] [PubMed] [Google Scholar]
- 63.Ward SL, Hu Z, Schirmer A, Reid R, Revill WP, Reeves CD, Petrakovsky OV, Dong SD, Katz L. Chalcomycin biosynthesis gene cluster from Streptomyces bikiniensis: novel features of an unusual ketolide produced through expression of the chm polyketide synthase in Streptomyces fradiae. Antimicrob Agents Chemother. 2004;48:4703–4712. doi: 10.1128/AAC.48.12.4703-4712.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Omura S, Sadakane N, Matsubara H. Bioconversion and biosynthesis of 16-membered macrolide antibiotics. XXII Biosynthesis of tylosin after protylonolide formation. Chem Pharm Bull. 1982;30:223–229. [Google Scholar]
- 65.Baltz RH, Seno ET, Stonesifer J, Wild GM. Biosynthesis of the macrolide antibiotic tylosin: a preferred pathway from tylactone to tylosin. J Antibiot. 1983;36:131–141. doi: 10.7164/antibiotics.36.131. [DOI] [PubMed] [Google Scholar]
- 66.Omura S, Tanaka H, Tsukui M. Biosynthesis of tylosin: oxidations of 5-O-mycaminosylprotylonolide at C-20 and C-23 with a cell-free extract from Streptomyces fradiae. Biochem Biophys Res Commun. 1982;107:554–560. doi: 10.1016/0006-291x(82)91527-3. [DOI] [PubMed] [Google Scholar]
- 67.Anzai Y, Saito N, Tanaka M, Kinoshita K, Koyama Y, Kato F. Organization of the biosynthetic gene cluster for the polyketide macrolide mycinamicin in Micromonospora griseorubida. FEMS Microbiol Lett. 2003;218:135–141. doi: 10.1111/j.1574-6968.2003.tb11509.x. [DOI] [PubMed] [Google Scholar]
- 68.Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- 69.Haslinger K, Peschke M, Brieke C, Maximowitsch E, Cryle MJ. X-domain of peptide synthetases recruits oxygenases crucial for glycopeptide biosynthesis. Nature. 2015;521:105–109. doi: 10.1038/nature14141. [DOI] [PubMed] [Google Scholar]
- 70.Brieke C, Peschke M, Haslinger K, Cryle MJ. Sequential in vitro cyclization by cytochrome P450 enzymes of glycopeptide antibiotic precursors bearing the X-domain from nonribosomal peptide biosynthesis. Angew Chem Int Ed. 2015;54:15715–15719. doi: 10.1002/anie.201507533. [DOI] [PubMed] [Google Scholar]
- 71.Peschke M, Haslinger K, Brieke C, Reinstein J, Cryle MJ. Regulation of the P450 oxygenation cascade involved in glycopeptide antibiotic biosynthesis. J Am Chem Soc. 2016;138:6746–6753. doi: 10.1021/jacs.6b00307. [DOI] [PubMed] [Google Scholar]
- 72.Woithe K, Geib N, Zerbe K, Li DB, Heck M, Fournier-Rousset S, Meyer O, Vitali F, Matoba N, Abou-Hadeed K, Robinson JA. Oxidative phenol coupling reactions catalyzed by OxyB: a cytochrome P450 from the vancomycin producing organism. Implications for vancomycin biosynthesis. J Am Chem Soc. 2007;129:6887–6895. doi: 10.1021/ja071038f. [DOI] [PubMed] [Google Scholar]
- 73.Woithe K, Geib N, Meyer O, Wörtz T, Zerbe K, Robinson JA. Exploring the substrate specificity of OxyB, a phenol coupling P450 enzyme involved in vancomycin biosynthesis. Org Biomol Chem. 2008;6:2861–2867. doi: 10.1039/b805956j. [DOI] [PubMed] [Google Scholar]
- 74.Schmartz PC, Wölfel K, Zerbe K, Gad E, El Tamany ES, Ibrahim HK, Abou-Hadeed K, Robinson JA. Substituent effects on the phenol coupling reaction catalyzed by the vancomycin biosynthetic P450 enzyme OxyB. Angew Chem Int Ed. 2012;51:11468–11472. doi: 10.1002/anie.201204458. [DOI] [PubMed] [Google Scholar]
- 75.Brieke C, Kratzig V, Haslinger K, Winkler A, Cryle MJ. Rapid access to glycopeptide antibiotic precursor peptides coupled with cytochrome P450-mediated catalysis: towards a biomimetic synthesis of glycopeptide antibiotics. Org Biomol Chem. 2015;13:2012–2021. doi: 10.1039/c4ob02452d. [DOI] [PubMed] [Google Scholar]
- 76.Haslinger K, Maximowitsch E, Brieke C, Koch A, Cryle MJ. Cytochrome P450 OxyBtei catalyzes the first phenolic coupling step in teicoplanin biosynthesis. ChemBioChem. 2014;15:2719–2728. doi: 10.1002/cbic.201402441. [DOI] [PubMed] [Google Scholar]
- 77.Abagyan R. Internal Coordinate Mechanics (ICM) version 3.8-4a. doi: 10.1021/ct501138c. http://www.molsoft.com/icm_pro.html. [DOI] [PMC free article] [PubMed]
- 78.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.