Abstract
S-adenosyl-l-methionine (AdoMet) is an essential enzyme cosubstrate in fundamental biology with an expanding range of biocatalytic and therapeutic applications. We report the design, synthesis, and evaluation of stable, functional AdoMet isosteres that are resistant to the primary contributors to AdoMet degradation (depurination, intramolecular cyclization, and sulfonium epimerization). Corresponding biochemical and structural studies demonstrate the AdoMet surrogates to serve as competent enzyme cosubstrates and to bind a prototypical class I model methyltransferase (DnrK) in a manner nearly identical to AdoMet. Given this conservation in function and molecular recognition, the isosteres presented are anticipated to serve as useful surrogates in other AdoMet-dependent processes and may also be resistant to, and/or potentially even inhibit, other therapeutically relevant AdoMet-dependent metabolic transformations (such as the validated drug target AdoMet decarboxylase). This work also highlights the ability of the prototypical class I model methyltransferase DnrK to accept non-native surrogate acceptors as an enabling feature of a new high-throughput methyltransferase assay.
Graphical abstract
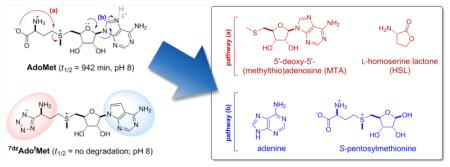
Methyltransferase (MT)-catalyzed S-adenosyl-l-methionine (AdoMet)-dependent methylation is essential to all walks of life where it contributes to modulating the function of a vast array of biomolecules ranging from small metabolites1–5 to macromolecules.5–15 Consistent with this, alterations in methylation-dependent processes are associated with cancer,16 neurodegenerative/neuropsychiatric disorders,17–19 inflammation,20,21 metabolic disorders,22 fundamental development/regenerative medicine,23,24 susceptibility to disease/adverse drug response,25,26 and drug resistance.13–15,27,28 While there have been great advances in methylation-dependent bioinformatics and disease-associated biomarkers,29–31 the study of intracellular MT spatial/temporal resolution, specificity, and/or function remains a challenge.9,14,15,25–28,32–36 Toward this end, the pioneering demonstration of AdoMet analogs, bearing alternative alkyl donor substituents, as cosubstrates for DNA37 or natural product (NP)38 MTs has inspired new tools and strategies to study NP,39–42 protein,43–51 and nucleic acid6,52–57 methylation where the recent development of enzyme-based strategies for the synthesis of differentially alkylated AdoMet analogs has simplified access to these unique cosubstrates.42,49,50,57–59 However, the stability of AdoMet or its corresponding differentially alkylated analogs under physiological conditions limits their utility as reagents or therapeutics by virtue of two fundamental degradative processes: intramolecular cyclization to homoserine lactone and 5′-deoxy-5′-(alkylthio)adenosine (Figure 1, pathway a) and depurination (Figure 1, pathway b).50,60–62 To address this deficiency, herein we report the chemoenzymatic synthesis of AdoMet isosteres designed to circumvent the major AdoMet degradative pathways and demonstrate, via both biochemical and structural studies, these analogs to serve as productive MT cosubstrates for the prototypical class I MT DnrK. This work highlights a unique conceptual strategy to modulate the fundamental properties of AdoMet and corresponding analogs as reagents in epigenetic, proteomic, biocatalytic, and/or therapeutic applications13 and also demonstrates a high-throughput colorimetric assay for DnrK based upon the non-native surrogate substrate 2-chloro-4-nitrophenol (ClNP).
Figure 1.
Utilization and degradation of AdoMet. General scheme highlighting methyltransferase (MT)-catalyzed use of AdoMet in methylation reactions (upper) and major chemical degradation pathways of AdoMet (lower). AdoMet, S-adenosyl-l-methionine (also referred to as SAM); AdoHcy, S-adenosyl-l-homocysteine (also referred to as SAH).
RESULTS AND DISCUSSION
Synthesis of AdoMet Carboxylate Isosteres
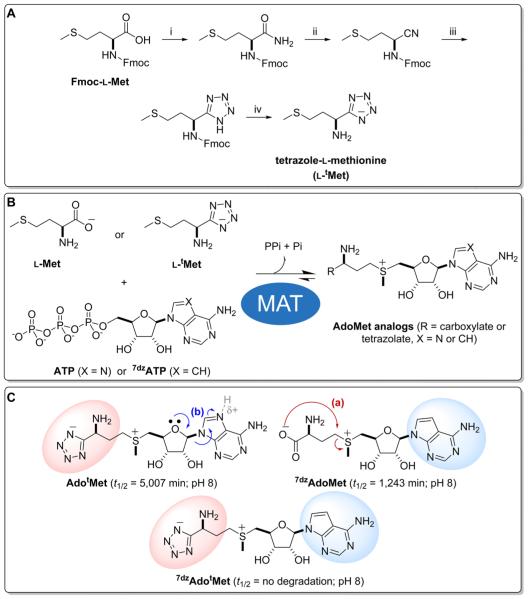
Adenine isosteres, such as 7-deazaadenine (7dzA), have been successfully employed to reduce the propensity for nucleotide depurination (Figure 1, pathway b).63–65 Thus, we focused our attention on addressing the AdoMet degradative intramolecular cyclization to homoserine lactone and 5′-deoxy-5′-(alkylthio)adenosine (Figure 1, pathway a) by replacing the AdoMet carboxylate with a sterically constrained and/or less nucleophilic isosteric functional group. As potential carboxylate isosteres,66,67 tetrazolates display a similar pKa,68 are less nucleophilic,69 and offer improved membrane permeability.70 In addition, MM2 calculations predict the formation of a sterically strained tetrazole-containing bicyclic ring structure via intramolecular cyclization (Figure 1, pathway a) as energetically unfavorable (see Supporting Information). To test this concept, the tetrazole-l-methionine isostere (ltMet) was synthesized from commercially available N-Fmoc-l-methionine in four simple steps (Figure 2, 41% overall yield) via a slight modification of previously reported methods71–78 and subsequently evaluated as a substrate for the permissive methionine adenosyltransferase hMAT2A.42 Importantly, this study revealed the kcat of hMAT2A-catalyzed reactions using native substrate l-methionine (lMet) or isosteric ltMet as nearly identical (lMet, 8.0 ± 0.1 min−1; ltMet, 6.1 ± 0.6 min−1; Table 1, Figure S2) with an 18-fold observed reduction in Km (lMet, 397 ± 27 μM; ltMet, 7,212 ± 1,250 μM) and enabled ~50% conversion to desired AdotMet in 20 h in a standard nonoptimized small scale assay (10 mM ltMet, 2 mM ATP, 10 μM hMAT2A, 25 mM Tris·HCl, pH 8.0, 10 mM MgCl2, 50 mM KCl, 37 °C). Using these parameters as a guide, a subsequent large scale (100 mL) chemoenzymatic reaction followed by preparative HPLC provided 19.9 mg of AdotMet (0.047 mmol, ~25% recovered yield based on the limiting reagent ATP), the purity and composition of which was confirmed via HPLC, NMR, and HRMS (see Supporting Information, Figures S1 and S6).
Figure 2.
Synthesis of AdoMet analogs and corresponding relevant degradation pathways. (A) Synthesis of ltMet: (i) (Boc)2O, pyridine, NH4HCO3, rt, 5 h, 97%; (ii) (TFA)2O/pyridine (1:1), THF, 0 °C, 3 h, 80%; (iii) NaN3, ZnBr2, H2O/2-propanol (2:1), 80 °C, 16 h, 65%; (iv) Et2NH, CH2Cl2, rt, 0.5 h, 82%. (B) General scheme for hMAT2A-catalyzed synthesis of AdoMet, AdotMet, 7dzAdoMet, and 7dzAdotMet. (C) Degradative pathways for AdotMet, 7dzAdoMet, and 7dzAdotMet. Fmoc, fluorenylmethyloxycarbonyl.
Table 1.
Kinetic Parameters and Stability Measurements
| enzyme | substrate | kcat min−1 | Km μmol L−1 | kcat/Km relative | t1/2 mina |
|---|---|---|---|---|---|
| hMAT2A | l-Metb | 8.0 ± 0.1 | 397 ± 29 | 1 | -c |
| l-tMetb | 6.1 ± 0.6 | 7,212 ± 1,250 | 0.04 | -c | |
| DnrK | ClNPd | 0.019 ± 0.001 | 106 ± 10 | N.A.e | -c |
| AdoMetf | 0.017 ± 0.001 | 138 ± 7 | 1 | 942 ± 7 | |
| AdotMetf | 0.031 ± 0.001 | 335 ± 40 | 0.8 | 5007 ± 6 | |
| 7dzAdoMet | – c | – c | – c | 1243 ± 1 | |
| 7dzAdotMet | – c | – c | – c | no degradationg |
Stability studies conducted at 37 °C, pH 8.0.
hMAT2A substrates were assayed in the presence of saturating ATP (2 mM).
Not determined.
DnrK substrate ClNP was assayed in the presence of saturating AdoMet (1.6 mM).
Not applicable.
DnrK substrates AdoMet and AdotMet were assayed in the presence of saturating ClNP (1.5 mM).
No detectable degradation after 3300 min ± standard error (SEM) at 95% confidence interval (CI).
Evaluation of AdoMet Carboxylate Isosteres
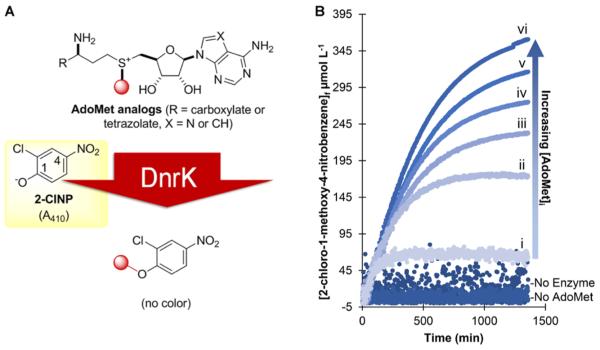
The subsequent comparison of AdoMet and AdotMet stability in 100 mM Tris·HCl, pH 8.0, at 37 °C revealed AdotMet as >5-fold improved in overall stability (AdotMet t1/2 = 5,007 ± 6 min; AdoMet t1/2 = 942 ± 7 min; Table 1, Figures S1 and S5). Consistent with our hypothesis, similar levels of AdoMet and AdotMet depurination (Figure 1, pathway b) were observed while AdotMet enabled complete inhibition of the observed AdoMet degradative intramolecular cyclization reaction (Figure 1, pathway a). It is important to note that while the decay of AdoMet has been reported to follow zero order50 or first order61 kinetics, our results are consistent with the former as is our determined AdoMet t1/2 value consistent with those previously reported.50,56,61 To assess the utility of the stabilized AdotMet as a cosubstrate for MTs, we selected the prototypical class I MT DnrK as a model. DnrK is the carminomycin 4-OMT that catalyzes a culminating step in the biosynthesis of the anticancer agent daunorubicin in Streptomyces peucetius and has also been reported to methylate various modified anthracyclines and flavonoids as surrogate substrates.79–84 Capitalizing on the permissive nature of DnrK and inspired by the recent application of ClNP as a surrogate acceptor for glycosyltransferases,85 we developed a real-time colorimetric assay for DnrK-catalyzed alkylation using ClNP as a surrogate acceptor substrate where alkylation abolishes the classical nitrophenolate color (A410) observed under basic conditions (Figure 3A). While the kinetic parameters for the native DnrK substrate carminomycin have not been reported, the determined ClNP kinetic parameters (kcat = 0.019 ± 0.001 min−1, Km = 106 ± 10 μM with AdoMet as the alkyl-donating cosubstrate) are consistent with ClNP as a sufficient non-native surrogate where clear concentration dependency was observed in realtime assays (Figure 3B). A comparison of the DnrK kinetic parameters for AdoMet (kcat = 0.017 ± 0.001 min−1, Km = 138 ± 7 μM) and AdotMet (kcat = 0.031 ± 0.001 min−1, Km = 335 ± 40 μM) revealed nearly equivalent specificity constants (kcat/ Km; Table 1, Figure S4). As a simple robust high-throughput colorimetric assay for a prototypical class I model MT, this newly validated assay also presents an enabling platform for future class I MT engineering, biochemical studies, and/or tool development.
Figure 3.
DnrK colorimetric assay. (A) General scheme for a high-throughput DnrK colorimetric assay enabled by the surrogate acceptor 2-chloro-4-nitrophenol (ClNP). (B) Representative assay results demonstrating DnrK-catalyzed alkylation of ClNP diminishes classical ClNP color (A410) over time [30 μM DnrK; 500 μM ClNP; AdoMet: (i) 100 μM, (ii) 500 μM, (iii) 800 μM, (iv) 1,000 μM, (v) 1,600 μM, and (vi) 2,000 μM; 25 mM Tris· HCl; pH 8.0].
Elucidation of DnrK/AdotMet/ClNP Ligand-Bound Structures
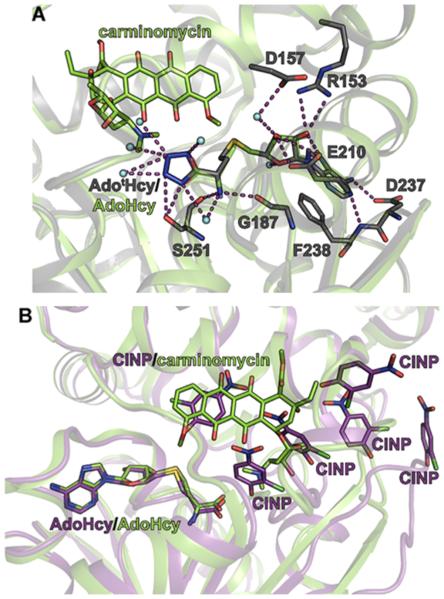
To better understand the molecular recognition of surrogate substrates AdotMet and ClNP by DnrK, the three-dimensional structures of the DnrK/S-adenosyl-tetrazole-lhomocysteine (AdotHcy, the byproduct after methyltransfer) and DnrK-ClNP-AdoHcy were determined at 2.25 and 1.82 Å resolution in space groups C2 and P21, respectively. The average real-space correlation coefficient of all ligands built into these two DnrK structures is over 0.96, and the electron density maps can be found in Figure S7. In both crystal forms, DnrK is identified as a tightly packed dimer with a buried surface area of ~3100 Å2, which is consistent with previously reported DnrK structures.84 As expected, the DnrK-AdotHcy structure reveals AdotHcy and AdoHcy to bind in a nearly identical manner where the AdotHcy tetrazolate forms extra hydrogen bonds to water molecules and the corresponding sulfur resides slightly closer to the donor nucleophile (Figure 4A). This observation is also in agreement with reports of reduced donor–acceptor distances in O-MTs correlating with higher turnover rates,86 as the kcat of DnrK with AdotMet is nearly double the kcat of DnrK with AdoMet. Consistent with the biochemical data, the orientation of one molecule of CINP in the DnrK-ClNP ligandbound structure is poised for alkylation reminiscent of that of the native acceptor carminomycin. Surprisingly, five additional ordered CINPs were also observed in the same structure (Figure 4B), suggesting a wide array of unproductive ClNP binding modes by virtue of key hydrogen bonds and π-stacking interactions.
Figure 4.
DnrK ligand-bound structures. (A) DnrK-AdotHcy (gray) and DnrK-AdoHcy-carminomycin (green) ligand-bound structures. Polar contacts between AdotHcy and nearby residues (stick models) and water molecules (spheres) are highlighted by dashed lines. In this model, the distance between carminomycin O-4 and the AdoHcy or AdotHcy sulfur is 4.2 and 3.8 Å, respectively. (B) DnrK-AdoHcy-ClNP (purple) and DnrK-AdoHcy-carminomycin (green) ligand-bound structures with ligands represented as stick models. In this model, the distance between carminomycin O-4 or ClNP O-1 and the AdoHcy sulfur is the same (4.2 Å).
Synthesis and Evaluation of AdoMet Surrogates Containing Base Isosteres
To confirm that base isosteres prohibit AdoMet depurination and to assess the potential for synergism with carboxylate isosteric replacement, the turnover of 7dzATP by hMAT2A in the presence of lMet and ltMet was subsequently assessed. In hMAT2A-catalyzed 24 h end-point assays, 7dzATP (1 mM) and lMet (10 mM) gave 92% conversion to 7dzAdoMet compared to 57% conversion with 7dzATP (1 mM) and ltMet (10 mM; Figure S3). Stability studies with the purified 7dzAdoMet and 7dzAdotMet confirmed the corresponding 7dzAdoMet as resistant to depurination and 7dzAdotMet as completely resistant to depurination and intramolecular cyclization (7dzAdoMet t1/2 = 1243 ± 1 min; 7dzAdotMet t1/2 = no detectable degradation over 3300 min; Table 1, Figures S1 and S5). Finally, simple coupled ClNPbased hMAT2A-DnrK assays (10 μM hMAT2A, 30 μM DnrK, 2 mM ATP or 7dzATP, 10 mM lMet or ltMet, 1 mM ClNP, 25 mM Tris·HCl, pH 8.0, 10 mM MgCl2, 50 mM KCl, 37 °C, 24 h) revealed all stabilized AdoMet surrogates to serve as suitable MT cosubstrates (ATP/lMet, 52%; ATP/ltMet, 19%; 7dzATP/lMet, 21%; 7dzATP/ltMet, 14%; Figure S1 and Table S1).
Conclusions
In summary, this study highlights the efficient chemoenzymatic synthesis of enzymatically competent AdoMet isosteres that are considerably less prone to the two primary AdoMet physicochemical degradation pathways (intramolecular cyclization and depurination, Figure 1), where the corresponding tetrazole isosteric replacement also restricts the potential for AdoMet carboxylate participation in sulfonium epimerization.15,60 Based on the DnrK-AdotMet biochemical and structural studies presented, AdotMet is able to substitute for AdoMet as a functional enzyme cosubstrate and implicates the isosteric modification as potentially advantageous in the context of other AdoMet-dependent enzymatic processes, as exemplified by AdoMet decarboxylase (AdoMetDC)-catalyzed AdoMet decarboxylation. Notably, AdoMetDC, a central catalyst in polyamine biosynthesis, is a validated anticancer and antiprotozoal drug target where substrate surrogates lacking the requisite AdoMet carboxylate (reminiscent of AdotMet) serve as potent inhibitors.63,87 Thus, the new conceptual framework for engineering AdoMet chemical and metabolic stability put forth presents unique opportunities to advance AdoMet biocatalytic and therapeutic applications including, but not limited to, those involving MTs.
METHODS
Gene Cloning, Protein Expression, and Purification
N-His6-hMAT2A was produced and purified as previously described.42 The dnrK gene encoding carminomycin 4-O-methyltransferase from Streptomyces peucetius was amplified from pWHM90379 using primers (forward, 5′-AGAGCAGTCATATGACCGCTGAACCGACCGTCGCGGCCCGGCCGCAGCAG-3′; reverse, 5′-TACAGTGAATTCTCAGGCGCCGGTGGCCGCGGGGGCAAGGAC-3′) containing NdeI (forward) and EcoRI (reverse) restriction sites. The PCR product was digested and ligated into pET28a to provide an expression plasmid to enable N-His6-DnrK production. The corresponding E. coli BL21(DE3) host was grown in the presence of 50 μg mL−1 of kanamycin at 37 °C to an OD600 of ~0.6, at which point the temperature was lowered to 25 °C and gene expression induced with 0.5 mM IPTG. Cells were allowed to continue to grow at 25 °C for approximately 18 h at 220 rpm. Cells were harvested by centrifugation (30 min, 5000 rpm), resuspended in buffer A (20 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 7.8), and lysed via incubation with 1 mg mL−1 lysozyme (~50 000 U mg–1) for 30 min on ice followed by sonication (VirSonic 475; Virtis, Gardiner, NY; 100 W, 10 × 15 s pulses, ~ 1 min between pulses) on ice. N-His6-DnrK was purified via affnity chromatography (5 mL HiTrap Ni-NTA chelating column, GE Healthcare, Piscataway, NJ) using a standard linear gradient (50 mM NaH2PO4, 300 mM NaCl, pH 8.0 with a linear gradient of imidazole of 10–500 mM) using an AKTA Purifier 10 (GE Healthcare). Concentration of the pooled fractions containing the purified protein was accomplished using an Amicon Ultracel centrifugal filter unit (30 kDa molecular weight cutoff; Merck Millipore Ltd., Tullagreen, Carrigtwohill, Co. Cork, IRL). Buffer exchange of combined and concentrated fractions containing the purified protein was accomplished using a PD-10 column (GE Healthcare) eluted with 25 mM Tris-HCl, pH 8.0. Protein concentrations were determined by Bradford assay (Bio-Rad, Hercules, CA, USA) using BSA as a standard. For this study, all proteins retained the N-terminal-His6 affnity tag.
MAT Assay
To determine the Km and kcat of hMAT2A with varying concentrations of lMet or ltMet, in vitro hMAT2A reactions were conducted in a volume of 50 μL with saturating ATP (2 mM), varied concentrations of lMet or ltMet (20, 50, 100, 200, 500, 1000, 2000, 4000, 6000, 8000, and 10 000 μM), 10 μM purified hMAT2A in 25 mM Tris, pH 8.0, 10 mM MgCl2, and 50 mM KCl. Reactions were incubated for 10 min at 37 °C and subsequently quenched by adding an equal volume of MeOH followed by centrifugation (10 000g, 20 min) to remove the precipitated protein. Product formation for each reaction was subsequently analyzed by RP-HPLC using method A (see Supporting Information, Figure S1). For each reaction, product (AdoMet or AdotMet) concentration was based upon the integration of species at 260 nm and calculated by multiplying the initial concentration of ATP by the quotient of the integrated product (AdoMet plus MTA or AdotMet) HPLC peak area (mAU*sec) over the sum of the integrated peak areas for product and remaining ATP. MTA derives from AdoMet and was thereby also considered as contributing to the total product concentration. Assays were repeated in triplicate where the Michaelis–Menten plots in Figure S2 represent an average of replicates. Controls lacking hMAT2A, lMet, ltMet, and/or ATP led to no product.
To determine if 7dzATP (TriLink Biotechnologies, San Diego, CA) was a viable cosubstrate for hMAT2A in combination with lMet or ltMet, hMAT2A assays containing 10 000 μM lMet or ltMet and variable 7dzATP (500 μM, 1000 μM, 2000 μM, and 4000 μM) were conducted in a similar fashion. Aliquots recovered at defined incubation times (10 min, 30 min, 60 min, 1500 min, and 2940 min) were analyzed via HPLC following an identical protocol as above, and the results are summarized in Figure S3. Controls lacking hMAT2A, lMet, ltMet, and/or 7dzATP led to no product.
DnrK Assay
DnrK reactions were conducted in a volume of 100 μL containing 30 μM purified DnrK in 100 mM Tris, pH 8.0. Acceptor (2-chloro-4-nitrophenol, ClNP) concentrations were saturating (1500 μM) or variable (7.8125, 31.25, 125, 500, 1000, 1500, 1750, or 2000 μM), and donor (AdoMet or AdotMet) concentrations were saturating (1600 μM) or variable (25, 50, 100, 200, 400, 800, 1600, and 3200 μM). Reactions were incubated for 120 min at 37 °C and subsequently quenched and analyzed via HPLC as previously described for the hMAT2A assays using the HPLC method A with detection at 317 nm (Figure S1). Assays were repeated in triplicate where the Michaelis– Menten plots in Figure S4 represent an average of replicates. Corresponding identical plate-based DnrK assays (see Supporting Information) were conducted in clear-bottom 96-well plates where the plate reader limit of quantification required assay dilution at ClNP concentrations >500 μM. The change in absorbance (ΔA410, representing disappearance of ClNP) over time was used as an indirect measure of ClNP methylation where product concentration was inferred by multiplying the percentage change in A410 at t = 0 min and A410 at t = 120 min by the initial ClNP concentration (Figure S4).85 Controls lacking DnrK, AdoMet, AdotMet, and/or ClNP led to no product.
Coupled hMAT2A-DnrK Assay
Coupled hMAT2A-DnrK reactions were conducted in a total volume of 50 μL containing 25 mM Tris·HCl, pH 8.0, 50 mM KCl, 10 mM MgCl2 with 10 μM hMAT2A, 30 μM DnrK, 1000 μM ClNP (saturating), and one of the following combinations of hMAT2A cosubstrates per reaction: lMet (10 000 μM) and ATP (2000 μM), ltMet (10 000 μM) and ATP (2000 μM), lMet (10 000 μM) and 7dzATP (2000 μM), or ltMet (10 000 μM) and 7dzATP (2000 μM). Reactions were incubated at 37 °C in PCR tubes on a thermocycler with a heated lid for 24 h and subsequently quenched and analyzed via HPLC as previously described for DnrK assays using HPLC method A (Figure S1 and Table S1). Controls lacking hMAT2A, DnrK, ClNP, lMet, ltMet, ATP, and/or 7dzATP led to no product.
Chemoenzymatic Synthesis and Purification of AdotMet, 7dzAdoMet, and 7dzAdotMet
Preparative MAT reactions to produce AdotMet (10 mM ltMet, 1 mM 7dzATP), 7dzAdoMet (10 mM lMet, 1 mM 7dzATP), or 7dzAdotMet (10 mM ltMet, 1 mM 7dzATP) were conducted in a volume of 100 mL, 0.8 mL, or 0.8 mL, respectively with 10 μM hMAT2A in 25 mM Tris·HCl, pH 8.0, 50 mM KCl, and 10 mM MgCl2.
The preparative AdotMet reaction was conducted in 2 × 50 mL Falcon tubes incubated at 37 °C for 48 h in a hybridizing oven (Techne Hybridizer HB-1D, Bibby Scientific, Staffordshire, United Kingdom) with rotation. After 48 h, the reactions were combined in a 500 mL round-bottom flask, flash-frozen, and lyophilized to dryness. The mixture was purified via C18 column (3.5 cm × 18 cm) washed with 800 mL of ddH2O (wash) followed by elution with 600 mL of 5:95 CH3CN/ddH2O, 600 mL of 10:90 CH3CN/ddH2O, and 800 mL of 15:85 CH3CN/ddH2O (40 mL fractions collected throughout elution). Analytical HPLC (Method A) revealed fractions 18–50 to contain the desired pure AdotMet, and these product-containing fractions were combined, concentrated in vacuo, flash frozen, and lyophilized to yield 19.9 mg of a brown-orange amorphous solid. Aqueous stock solutions were prepared in 10% EtOH and 5 mM H2SO4 aqueous solution (identical to commercial AdoMet stock preparations) where corresponding concentrations were based upon the molar extinction coefficient for AdoMet (15 400 L mol−1 cm−1 at 254 nm)60 and verified via analytical HPLC (method A) comparison to commercial AdoMet standards.
The preparative 7dzAdoMet/7dzAdotMet reactions were conducted in several PCR tubes (8 × 100 μL for each reaction) and incubated in a thermocycler (with a heated top to prevent evaporation) at 37 °C for 1.5 h (lMet and 7dzATP) or 24 h (ltMet and 7dzATP). Recombinant shrimp alkaline phosphatase (rSAP; New England BioLabs, Ipswich, Massachusetts, USA) was subsequently added to a final concentration of 0.1 U μL−1 and the reactions incubated for an additional 20 min (to convert unreacted 7dzATP to 7dzadenosine). For each target, the cumulative reactions were combined, quenched with an equal volume of MeOH, and centrifuged (10 000g, 20 min, 4 °C) to remove precipitated protein and the supernatant concentrated to ~250 μL in vacuo. The desired product in each case was recovered via HPLC (method B), concentrated, flash frozen, and lyophilized to dryness. Aqueous stock solutions were prepared in 10% EtOH and 5 mM H2SO4 aqueous solution (identical to commercial AdoMet stock preparations) where corresponding concentrations were based upon the molar extinction coefficient for 7-deaza-adenosine (9100 L mol−1 cm−1 at 260 nm)88 and verified via analytical HPLC (method A) comparison to commercial 7dzATP and AdoMet standards.
Determination of t1/2 of AdoMet, AdotMet, 7dzAdoMet, and 7dzAdotMet
AdoMet, AdotMet, 7dzAdoMet, or 7dzAdotMet (1 mM 10% EtOH, 5 mM H2SO4 stock) was diluted to 100 μM in 100 mM Tris buffer at a pH of 8.0 and incubated at 37 °C. Aliquots of each solution were taken at t = 0 min (and at various time points thereafter) and analyzed via HPLC (method A, A254). The concentrations of AdoMet species and corresponding degradation products were determined and graphed on an XY-scatterplot, and linear regression was performed on each data set to obtain degradation rates (Figure S5) as previously described.50,89 The t1/2 of each AdoMet species was determined from the line slope considering zeroth order kinetics, as in the following eq 1:
| (1) |
where t(1/2) is the time (in min) that it takes for one-half of a chemical species (in this example, AdoMet) to degrade, [AdoMet]0 is the starting % concentration of a chemical species, and k is the zero-order rate constant of degradation of a chemical species (in % min−1).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the National Institutes of Health Protein Structure Initiative (U01 GM098248, G.N.P.,J.), National Institutes of Health R37 AI52188 (J.S.T.), National Institutes of Health GM109456 (G.N.P.,J.), the National Center for A dvancing Translational Sciences (UL1TR000117), and the University of Kentucky College of Pharmacy and Markey Cancer Center. We also thank the University of Kentucky Mass Spectrometry Facility (ASTeCC) for HRMS support; the staff at the LS-CAT, SBC-CAT, and GM/CA beamline at the Advanced Photo Source for help in conducting trial attempts and collecting the diffraction data; and K. A. Shaaban (University of Kentucky Center for Pharmaceutical Research and Innovation) for consultation.
Footnotes
Notes
The authors declare the following competing financial interest(s): J.S.T. is a co-founder of Centrose (Madison, WI).
Supporting Information
The Supporting Information is available free of charge on the ACS Publication website at DOI: 10.1021/acschembio.6b00348.
Additional materials and supporting data tables and figures (PDF)
REFERENCES
- (1).Struck A-W, Thompson ML, Wong LS, Micklefield J. S-adenosyl-methionine-dependent methyltransferases: Highly versatile enzymes in biocatalysis, biosynthesis and other biotechnological applications. ChemBioChem. 2012;13:2642–2655. doi: 10.1002/cbic.201200556. [DOI] [PubMed] [Google Scholar]
- (2).Westfall CS, Muehler AM, Jez JM. Enzyme action in the regulation of plant hormone responses. J. Biol. Chem. 2013;288:19304–19311. doi: 10.1074/jbc.R113.475160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Wessjohann LA, Keim J, Weigel B, Dippe M. Alkylating enzymes. Curr. Opin. Chem. Biol. 2013;17:229–235. doi: 10.1016/j.cbpa.2013.02.016. [DOI] [PubMed] [Google Scholar]
- (4).Vance DE. Phospholipid methylation in mammals: From biochemistry to physiological function. Biochim. Biophys. Acta, Biomembr. 2014;1838:1477–1487. doi: 10.1016/j.bbamem.2013.10.018. [DOI] [PubMed] [Google Scholar]
- (5).Liscombe DK, Louie GV, Noel JP. Architectures, mechanisms and molecular evolution of natural product methyltransferases. Nat. Prod. Rep. 2012;29:1238–1250. doi: 10.1039/c2np20029e. [DOI] [PubMed] [Google Scholar]
- (6).Klimašauskas S, Weinhold E. A new tool for biotechnology: AdoMet-dependent methyltransferases. Trends Biotechnol. 2007;25:99–104. doi: 10.1016/j.tibtech.2007.01.006. [DOI] [PubMed] [Google Scholar]
- (7).Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: A new frontier for drug discovery. Nat. Rev. Drug Discovery. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- (8).Chen BF, Chan WY. The de novo DNA methyltransferase DNMT3A in development and cancer. Epigenetics. 2014;9:669–677. doi: 10.4161/epi.28324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Dong H, Fink K, Züst R, Lim SP, Qin CF, Shi PY. Flavivirus RNA methylation. J. Gen. Virol. 2014;95:763–778. doi: 10.1099/vir.0.062208-0. [DOI] [PubMed] [Google Scholar]
- (10).Urbonavičius J, Meškys R, Grosjean H. Biosynthesis of wyosine derivatives in tRNAPhe of Archaea: Role of a remarkable bifunctional tRNAPhe:m1G/imG2 methyltransferase. RNA. 2014;20:747–753. doi: 10.1261/rna.043315.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Lanouette S, Mongeon V, Figeys D, Couture J-F. The functional diversity of protein lysine methylation. Mol. Syst. Biol. 2014;10:724. doi: 10.1002/msb.134974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Yamamoto T, Takano N, Ishiwata K, Ohmura M, Nagahata Y, Matsuura T, Kamata A, Sakamoto K, Nakanishi T, Kubo A, Hishiki T, Suematsu M. Reduced methylation of PFKFB3 in cancer cells shunts glucose towards the pentose phosphate pathway. Nat. Commun. 2014;5:3480. doi: 10.1038/ncomms4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Bottiglieri T. S-Adenosyl-l-methionine (SAMe): From the bench to the bedside - Molecular basis of a pleiotrophic molecule. Am. J. Clin. Nutr. 2002;76:1151S–1157S. doi: 10.1093/ajcn/76/5.1151S. [DOI] [PubMed] [Google Scholar]
- (14).Cacabelos R. Epigenomic networking in drug development: From pathogenic mechanisms to pharmacogenomics. Drug Dev. Res. 2014;75:348–365. doi: 10.1002/ddr.21219. [DOI] [PubMed] [Google Scholar]
- (15).Lu SC, Mato JM. S-adenosylmethionine in liver health, injury, and cancer. Physiol. Rev. 2012;92:1515–1542. doi: 10.1152/physrev.00047.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Campbell RM, Tummino PJ. Cancer epigenetics drug discovery and development: The challenge of hitting the mark. J. Clin. Invest. 2014;124:64–69. doi: 10.1172/JCI71605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Gapp K, Woldemichael BT, Bohacek J, Mansuy IM. Epigenetic regulation in neurodevelopment and neuro-degenerative diseases. Neuroscience. 2014;264:99–111. doi: 10.1016/j.neuroscience.2012.11.040. [DOI] [PubMed] [Google Scholar]
- (18).Tremolizzo L, Rodriguez-Menendez V, Conti E, Zoia C, Cavaletti G, Ferrarese C. Novel therapeutic targets in neuropsychiatric disorders: The neuroepigenome. Curr. Pharm. Des. 2014;20:1831–1839. doi: 10.2174/13816128113199990535. [DOI] [PubMed] [Google Scholar]
- (19).Coşar A, Ipçioğlu OM, Özcan Ö, Gültepe M. Folate and homocysteine metabolisms and their roles in the biochemical basis of neuropsychiatry. Turk. J. Med. Sci. 2014;44:1–9. [PubMed] [Google Scholar]
- (20).Grolleau-Julius A, Ray D, Yung RL. The role of epigenetics in aging and autoimmunity. Clin. Rev. Allergy Immunol. 2010;39:42–50. doi: 10.1007/s12016-009-8169-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Doyle HA, Yang M-L, Raycroft MT, Gee RJ, Mamula MJ. Autoantigens: Novel forms and presentation to the immune system. Autoimmunity. 2014;47:220–233. doi: 10.3109/08916934.2013.850495. [DOI] [PubMed] [Google Scholar]
- (22).Vickers MH. Early life nutrition, epigenetics and programming of later life disease. Nutrients. 2014;6:2165–2178. doi: 10.3390/nu6062165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Wang T, Warren ST, Jin P. Toward pluripotency by reprogramming: Mechanisms and application. Protein Cell. 2013;4:820–832. doi: 10.1007/s13238-013-3074-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Hu K. All roads lead to induced pluripotent stem cells: The technologies of iPSC generation. Stem Cells Dev. 2014;23:1285–300. doi: 10.1089/scd.2013.0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Balmer NV, Leist M. Epigenetics and transcriptomics to detect adverse drug effects in model systems of human development. Basic Clin. Pharmacol. Toxicol. 2014;115:59–68. doi: 10.1111/bcpt.12203. [DOI] [PubMed] [Google Scholar]
- (26).Khan SR, Baghdasarian A, Fahlman RP, Michail K, Siraki AG. Current status and future prospects of toxicogenomics in drug discovery. Drug Discovery Today. 2014;19:562–578. doi: 10.1016/j.drudis.2013.11.001. [DOI] [PubMed] [Google Scholar]
- (27).Lötsch J, Schneider G, Reker D, Parnham MJ, Schneider P, Geisslinger G, Doehring A. Common non-epigenetic drugs as epigenetic modulators. Trends Mol. Med. 2013;19:742–753. doi: 10.1016/j.molmed.2013.08.006. [DOI] [PubMed] [Google Scholar]
- (28).Nordmann P, Poirel L, Bonnin RA. Screening and deciphering antibiotic resistance in Acinetobacter baumannii: A state of the art. Expert Rev. Anti-Infect. Ther. 2013;11:571–583. doi: 10.1586/eri.13.38. [DOI] [PubMed] [Google Scholar]
- (29).Claes B, Buysschaert I, Lambrechts D. Pharmaco-epigenomics: Discovering therapeutic approaches and biomarkers for cancer therapy. Heredity. 2010;105:152–160. doi: 10.1038/hdy.2010.42. [DOI] [PubMed] [Google Scholar]
- (30).Schamberger AC, Mise N, Meiners S, Eickelberg O. Epigenetic mechanisms in COPD: Implications for pathogenesis and drug discovery. Expert Opin. Drug Discovery. 2014;9:609–628. doi: 10.1517/17460441.2014.913020. [DOI] [PubMed] [Google Scholar]
- (31).Helton SG, Lohoff FW. Serotonin pathway polymorphisms and the treatment of major depressive disorder and anxiety disorders. Pharmacogenomics. 2015;16:541–553. doi: 10.2217/pgs.15.15. [DOI] [PubMed] [Google Scholar]
- (32).McGrath J, Trojer P. Targeting histone lysine methylation in cancer. Pharmacol. Ther. 2015;150:1–22. doi: 10.1016/j.pharmthera.2015.01.002. [DOI] [PubMed] [Google Scholar]
- (33).Hamm CA, Costa FF. Epigenomes as therapeutic targets. Pharmacol. Ther. 2015;151:72–86. doi: 10.1016/j.pharmthera.2015.03.003. [DOI] [PubMed] [Google Scholar]
- (34).Müller T. Catechol-O-methyltransferase inhibitors in Parkinson’s disease. Drugs. 2015;75:157–174. doi: 10.1007/s40265-014-0343-0. [DOI] [PubMed] [Google Scholar]
- (35).Dhanak D, Jackson P. Development and classes of epigenetic drugs for cancer. Biochem. Biophys. Res. Commun. 2014;455:58–69. doi: 10.1016/j.bbrc.2014.07.006. [DOI] [PubMed] [Google Scholar]
- (36).Hronová K, Šíma M, Světlík S, Matoušková O, Slanař O. Pharmacogenetics and immunosuppressive drugs. Expert Rev. Clin. Pharmacol. 2014;7:821–835. doi: 10.1586/17512433.2014.966811. [DOI] [PubMed] [Google Scholar]
- (37).Dalhoff C, Lukinavičius G, Klimašauskas S, Weinhold E. Direct transfer of extended groups from synthetic cofactors by DNA methyltransferases. Nat. Chem. Biol. 2006;2:31–32. doi: 10.1038/nchembio754. [DOI] [PubMed] [Google Scholar]
- (38).Zhang C, Weller RL, Thorson JS, Rajski SR. Natural product diversification using a non-natural cofactor analogue of S-adenosyl-l-methionine. J. Am. Chem. Soc. 2006;128:2760–2761. doi: 10.1021/ja056231t. [DOI] [PubMed] [Google Scholar]
- (39).Law BJC, Struck A-W, Bennett MR, Wilkinson B, Micklefield J. Site-specific bioalkylation of rapamycin by the RapM 16-O-methyltransferase. Chem. Sci. 2015;6:2885–2892. doi: 10.1039/c5sc00164a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Stecher H, Tengg M, Ueberbacher BJJ, Remler P, Schwab H, Griengl H, Gruber-Khadjawi MM. Biocatalytic Friedel-Crafts alkylation using non-natural cofactors. Angew. Chem. 2009;121:9710–9712. doi: 10.1002/anie.200905095. [DOI] [PubMed] [Google Scholar]
- (41).Winter JM, Chiou G, Bothwell IR, Xu W, Garg NK, Luo M, Tang Y. Expanding the structural diversity of polyketides by exploring the cofactor tolerance of an inline methyltransferase domain. Org. Lett. 2013;15:2011–2014. doi: 10.1021/ol401723h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Singh S, Zhang J, Huber TD, Sunkara M, Hurley K, Goff RD, Wang G, Zhang W, Liu C, Rohr J, Van Lanen SG, Morris AJ, Thorson JS. Facile chemoenzymatic strategies for the synthesis and utilization of S-adenosyl-l-methionine analogues. Angew. Chem., Int. Ed. 2014;53:3965–3969. doi: 10.1002/anie.201308272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Lee BWKK, Sun HG, Zang T, Kim BJ, Alfaro JF, Zhou ZS, Ju-Kim B, Alfaro JF, Zhou ZS, Kim BJ, Alfaro JF, Zhou ZS, Ju-Kim B, Alfaro JF, Zhou ZS, Kim BJ, Alfaro JF, Zhou ZS. Enzyme-catalyzed transfer of a ketone group from an S-adenosylmethionine analogue: A tool for the functional analysis of methyltransferases. J. Am. Chem. Soc. 2010;132:3642–3643. doi: 10.1021/ja908995p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Islam K, Zheng W, Yu H, Deng H, Luo M. Expanding cofactor repertoire of protein lysine methyltransferase for substrate labeling. ACS Chem. Biol. 2011;6:679–684. doi: 10.1021/cb2000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Wang R, Zheng W, Yu H, Deng H, Luo M. Labeling substrates of protein arginine methyltransferase with engineered enzymes and matched S-Adenosyl-l-methionine analogues. J. Am. Chem. Soc. 2011;133:7648–7651. doi: 10.1021/ja2006719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Binda O, Boyce M, Rush JS, Palaniappan KK, Bertozzi CR, Gozani O. A chemical method for labeling lysine methyltransferase substrates. ChemBioChem. 2011;12:330–334. doi: 10.1002/cbic.201000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Islam K, Bothwell I, Chen Y, Sengelaub C, Wang R, Deng H, Luo M. Bioorthogonal profiling of protein methylation using azido derivative of S-adenosyl-l-methionine. J. Am. Chem. Soc. 2012;134:5909–5915. doi: 10.1021/ja2118333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Willnow S, Martin M, Lüscher B, Weinhold E. A selenium-based click AdoMet analogue for versatile substrate labeling with wild-type protein methyltransferases. ChemBioChem. 2012;13:1167–1173. doi: 10.1002/cbic.201100781. [DOI] [PubMed] [Google Scholar]
- (49).Wang R, Islam K, Liu Y, Zheng W, Tang H, Lailler N, Blum G, Deng H, Luo M. Profiling genome-wide chromatin methylation with engineered posttranslation apparatus within living cells. J. Am. Chem. Soc. 2013;135:1048–1056. doi: 10.1021/ja309412s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Thomsen M, Vogensen SB, Buchardt J, Burkart MD, Clausen RP. Chemoenzymatic synthesis and in situ application of S-adenosyl-l-methionine analogs. Org. Biomol. Chem. 2013;11:7606–7610. doi: 10.1039/c3ob41702f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Wang R, Luo M. A journey toward bioorthogonal profiling of protein methylation inside living cells. Curr. Opin. Chem. Biol. 2013;17:729–37. doi: 10.1016/j.cbpa.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Dalhoff C, Lukinavičius G, Klimašauskas S, Weinhold E. Synthesis of S-adenosyl-l-methionine analogs and their use for sequence-specific transalkylation of DNA by methyltransferases. Nat. Protoc. 2006;1:1879–1886. doi: 10.1038/nprot.2006.253. [DOI] [PubMed] [Google Scholar]
- (53).Lukinavičius G, Lapienė V, Staševskij Z, Dalhoff C, Weinhold E, Klimašauskas S. Targeted labeling of DNA by methyltransferase-directed transfer of activated groups (mTAG) J. Am. Chem. Soc. 2007;129:2758–2759. doi: 10.1021/ja0691876. [DOI] [PubMed] [Google Scholar]
- (54).Dalhoff C, Hüben M, Lenz T, Poot P, Nordhoff E, Köster H, Weinhold E. Synthesis of S-adenosyl-l-homocysteine capture compounds for selective photoinduced isolation of methyl-transferases. ChemBioChem. 2010;11:256–265. doi: 10.1002/cbic.200900349. [DOI] [PubMed] [Google Scholar]
- (55).Motorin Y, Burhenne J, Teimer R, Koynov K, Willnow S, Weinhold E, Helm M. Expanding the chemical scope of RNA:methyltransferases to site-specific alkynylation of RNA for click labeling. Nucleic Acids Res. 2011;39:1943–1952. doi: 10.1093/nar/gkq825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Lukinavičius G, Tomkuvienė M, Masevičius V, Klimašauskas S. Enhanced chemical stability of AdoMet analogues for improved methyltransferase-directed labeling of DNA. ACS Chem. Biol. 2013;8:1134–1139. doi: 10.1021/cb300669x. [DOI] [PubMed] [Google Scholar]
- (57).Wijayasinghe YS, Blumenthal RM, Viola RE. Producing proficient methyl donors from alternative substrates of S-adenosylmethionine synthetase. Biochemistry. 2014;53:1521–1526. doi: 10.1021/bi401556p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Zhang J, Zheng YG. SAM/SAH analogues as versatile tools for SAM-dependent methyltransferases. ACS Chem. Biol. 2015;11:583–597. doi: 10.1021/acschembio.5b00812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Wang F, Singh S, Zhang J, Huber TD, Helmich KE, Sunkara M, Hurley KA, Goff RD, Bingman CA, Morris AJ, Thorson JS, Phillips GN. Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus. FEBS J. 2014;281:4224–4239. doi: 10.1111/febs.12784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Iwig DF, Booker SJ. Insight into the polar reactivity of the onium chalcogen analogues of S-adenosyl-l-methionine. Biochemistry. 2004;43:13496–13509. doi: 10.1021/bi048693+. [DOI] [PubMed] [Google Scholar]
- (61).Hoffman JL. Chromatographic analysis of the chiral and covalent instability of S-adenosyl-l-methionine. Biochemistry. 1986;25:4444–4449. doi: 10.1021/bi00363a041. [DOI] [PubMed] [Google Scholar]
- (62).Bentley R. Role of sulfur chirality in the chemical processes of biology. Chem. Soc. Rev. 2005;34:609–624. doi: 10.1039/b418284g. [DOI] [PubMed] [Google Scholar]
- (63).Coward JK, Motola NC, Moyer JD. Polyamine biosynthesis in rat prostate. substrate and inhibitor properties of 7-deaza analogues of decarboxylated S-adenosylmethionine and 5′-methylthioadenosine. J. Med. Chem. 1977;20:500–505. doi: 10.1021/jm00214a008. [DOI] [PubMed] [Google Scholar]
- (64).Berti PJ, McCann JAB. Toward a detailed understanding of base excision repair enzymes: Transition state and mechanistic analyses of N-glycoside hydrolysis and N-glycoside transfer. Chem. Rev. 2006;106:506–555. doi: 10.1021/cr040461t. [DOI] [PubMed] [Google Scholar]
- (65).McCann JAB, Berti PJ. Transition state analysis of acid-catalyzed dAMP hydrolysis. J. Am. Chem. Soc. 2007;129:7055–7064. doi: 10.1021/ja067371l. [DOI] [PubMed] [Google Scholar]
- (66).Herr RJ. 5-Substituted-1H-tetrazoles as carboxylic acid isosteres: Medicinal chemistry and synthetic methods. Bioorg. Med. Chem. 2002;10:3379–3393. doi: 10.1016/s0968-0896(02)00239-0. [DOI] [PubMed] [Google Scholar]
- (67).Malik MA, Wani MY, Al-Thabaiti SA, Shiekh RA. Tetrazoles as carboxylic acid isosteres: Chemistry and biology. J. Inclusion Phenom. Macrocyclic Chem. 2014;78:15–37. [Google Scholar]
- (68).Hansch C, Leo A. Exploring QSAR. American Chemical Society; Washington, DC: 1995. [Google Scholar]
- (69).Matta CF, Arabi AA, Weaver DF. The bioisosteric similarity of the tetrazole and carboxylate anions: Clues from the topologies of the electrostatic potential and of the electron density. Eur. J. Med. Chem. 2010;45:1868–1872. doi: 10.1016/j.ejmech.2010.01.025. [DOI] [PubMed] [Google Scholar]
- (70).Liljebris C, Larsen SD, Ogg D, Palazuk BJ, Bleasdale JE. Investigation of potential bioisosteric replacements for the carboxyl groups of peptidomimetic inhibitors of protein tyrosine phosphatase 1B: Identification of a tetrazole-containing inhibitor with cellular activity. J. Med. Chem. 2002;45:1785–1798. doi: 10.1021/jm011100y. [DOI] [PubMed] [Google Scholar]
- (71).Sureshbabu VV, Venkataramanarao R, Naik SA, Chennakrishnareddy G. Synthesis of tetrazole analogues of amino acids using Fmoc chemistry: Isolation of amino free tetrazoles and their incorporation into peptides. Tetrahedron Lett. 2007;48:7038–7041. [Google Scholar]
- (72).Itoh F, Yukishige K, Wajima M, Ootsu K, Akimoto H. Non-glutamate type pyrrolo[2,3-d]pyrimidine antifolates. I: Synthesis and biological properties of pyrrolo[2,3-d]pyrimidine antifolates containing tetrazole congener of glutamic acid. Chem. Pharm. Bull. 1995;43:230–235. doi: 10.1248/cpb.43.230. [DOI] [PubMed] [Google Scholar]
- (73).Pozdnev VF. Activation of carboxylic acids by pyrocarbonates. Application of di-tert-butyl pyrocarbonate as condensing reagent in the synthesis of amides of protected amino acids and peptides. Tetrahedron Lett. 1995;36:7115–7118. [Google Scholar]
- (74).Campagna F, Carotti A, Casini G. A convenient synthesis of nitriles from primary amides under mild conditions. Tetrahedron Lett. 1977;18:1813–1815. [Google Scholar]
- (75).Aureggi V, Sedelmeier G. 1,3-Dipolar cyclo-addition: Click chemistry for the synthesis of 5-substituted tetrazoles from organoaluminum azides and nitriles. Angew. Chem., Int. Ed. 2007;46:8440–8444. doi: 10.1002/anie.200701045. [DOI] [PubMed] [Google Scholar]
- (76).Demko ZP, Sharpless KB. An expedient route to the tetrazole analogues of α-amino acids. Org. Lett. 2002;4:2525–2527. doi: 10.1021/ol020096x. [DOI] [PubMed] [Google Scholar]
- (77).Spero DM, Kapadia SR. Enantioselective synthesis of α,α-disubstituted amino acid derivatives via enzymatic resolution: Preparation of a thiazolyl-substituted α-methyl α-benzyl amine. J. Org. Chem. 1996;61:7398–7401. doi: 10.1021/jo9610671. [DOI] [PubMed] [Google Scholar]
- (78).Demko ZP, Sharpless KB. Preparation of 5-substituted 1H-tetrazoles from nitriles in water. J. Org. Chem. 2001;66:7945–7950. doi: 10.1021/jo010635w. [DOI] [PubMed] [Google Scholar]
- (79).Madduri K, Torti F, Colombo AL, Hutchinson CR. Cloning and sequencing of a gene encoding carminomycin 4-O-methyltransferase from Streptomyces peucetius and its expression in Escherichia coli. J. Bacteriol. 1993;175:3900–3904. doi: 10.1128/jb.175.12.3900-3904.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Di Marco A, Gaetanzi M, Orezzi P, Scarpinato BM, Silvestrini R, Soldati M, Dasdia T, Valentini L. Daunomycin”, a new antibiotic of the rhodomycin group. Nature. 1964;201:706–707. doi: 10.1038/201706a0. [DOI] [PubMed] [Google Scholar]
- (81).Grocholski T, Dinis P, Niiranen L, Niemi J, Metsä-Ketelä M. Divergent evolution of an atypical S-adenosyl-l-methionine–dependent monooxygenase involved in anthracycline biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 2015;112:9866–9871. doi: 10.1073/pnas.1501765112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Dickens ML, Priestley ND, Strohl WR. In vivo and in vitro bioconversion of ε-rhodomycinone glycoside to doxorubicin: Functions of DauP, DauK, and DoxA. J. Bacteriol. 1997;179:2641–2650. doi: 10.1128/jb.179.8.2641-2650.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Kim NY, Kim JH, Lee YH, Lee EJ, Kim J, Lim J, Chong Y, Ahn JH. O-Methylation of flavonoids using DnrK based on molecular docking. J. Microbiol. Biotechnol. 2007;17:1991–1995. [PubMed] [Google Scholar]
- (84).Jansson A, Koskiniemi H, Mäntsälä P, Niemi J, Schneider G. Crystal structure of a ternary complex of DnrK, a methyltransferase in daunorubicin biosynthesis, with bound products. J. Biol. Chem. 2004;279:41149–41156. doi: 10.1074/jbc.M407081200. [DOI] [PubMed] [Google Scholar]
- (85).Gantt RW, Peltier-Pain P, Cournoyer WJ, Thorson JS. Using simple donors to drive the equilibria of glycosyltransferase-catalyzed reactions. Nat. Chem. Biol. 2011;7:685–691. doi: 10.1038/nchembio.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Zhang J, Kulik HJ, Martinez TJ, Klinman JP. Mediation of donor–acceptor distance in an enzymatic methyl transfer reaction. Proc. Natl. Acad. Sci. U. S. A. 2015;112:7954–7959. doi: 10.1073/pnas.1506792112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Pegg AE. S-Adenosylmethionine decarboxylase. Essays Biochem. 2009;46:25–45. doi: 10.1042/bse0460003. [DOI] [PubMed] [Google Scholar]
- (88).Seela F, Mersmann K, Grasby JA, Gait MJ. 7-Deazaadenosine: Oligoribonucleotide building block synthesis and autocatalytic hydrolysis of base-modified hammerhead ribozymes. Helv. Chim. Acta. 1993;76:1809–1820. [Google Scholar]
- (89).Lipson JM, Thomsen M, Moore BS, Clausen RP, La Clair JJ, Burkart MD. A tandem chemoenzymatic methylation by S-adenosyl-l-methionine. ChemBioChem. 2013;14:950–3. doi: 10.1002/cbic.201300221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.