Abstract
Repetitive stimulation of T cell receptor (TCR) with cognate antigen results in robust proliferation and expansion of the T cells, and also imprints them with replicative senescence signatures. Our previous studies have shown that life-span and anti-tumor function of T cells can be enhanced by inhibiting reactive oxygen species (ROS) or intervening with ROS dependent JNK activation that leads to its activation induced cell death (AICD). Since tumor suppressor protein p53 is also a redox active transcription factor that regulates cellular ROS generation that triggers downstream factor mediating apoptosis, we determined if p53 levels could influence persistence and function of tumor reactive T cells. Using h3T TCR transgenic mice, with human tyrosinase epitope reactive T cells developed on p53 knock-out (KO) background, we determined its role in regulating anti-tumor T cell function. Our data shows that as compared to h3T cells, h3T-p53 KO T cells exhibited enhanced glycolytic commitment that correlated with increased proliferation, IFN-γ secretion, cytolytic capacity, expression of stemness gene signature and decreased TGF-β signaling. This increased effector function correlated to the improved control of subcutaneously established murine melanoma after adoptive transfer of p53-KO T cells. Pharmacological inhibition of human TCR transduced T cells using a combination of p53 inhibitors also potentiated the T cell effector function and improved persistence. Thus, our data highlights the key role of p53 in regulating the tumor reactive T cell response and that targeting this pathway could have potential translational significance in adoptive T cell therapy.
INTRODUCTION
Adoptive transfer of tumor epitope reactive T cell in cancer patients has generated much interest due to promising control of tumor growth (1). However, susceptibility to immunosuppression and reduced survival of effector T cells in an oxidative tumor microenvironment are the key confounding factors in immunotherapy (2,3). We have previously shown that reactive oxygen species (ROS) scavengers can inhibit repetitive TCR stimulation mediated activation induced cell death (AICD) of tumor reactive T cells without interfering with cytokine production (4), a measure of CTL function, placing redox regulation at a central point for therapeutic intervention.
The altered expression of a redox active transcription factor p53 leads to uncontrolled cell proliferation, senescence and cell death (5). However, only a handful of studies have reported the role of p53 in shaping T cell immune response. Grayson et al (6) reported slightly higher memory response in p53-KO mice compared to p53-sufficient mice, and only minor differences in proliferation, apoptosis, or maintenance of ‘non-self’ viral antigen specific T cells. A recent study has shown that in order to mount an effective antigen-specific proliferative responses of CD4+ T cell kinetically down regulate the expression of tumor suppressor p53 until 72-96 hr. (7). Another study showed that p53 inhibits systemic autoimmune diseases by inducing regulatory T cells (Treg’s) (8). Since p53 is also required for TGF-β gene responses by cooperating with Smads (9), we hypothesized that T cells from p53-KO mice will be less prone to TGF-β mediated immunosuppression in a tumor microenvironment, and with less incidence of iTreg generation a durable anti-tumor T cell response could be mounted by targeting p53. Further, p53 negatively regulates glycolysis through activation of TP53-induced glycolysis regulator (TIGAR) (10), and positively regulates oxidative phosphorylation (OXPHOS) through up-regulation of SCO2, a member of the COX-2 assembly involved in the electron-transport chain (11). Since long-term T cell effector and memory response is also metabolically regulated (12), we determined if differences in metabolic signature due to lack of p53 expression co-relates to anti-tumor T cell function.
Our study demonstrates that p53 deficient T cells exhibited enhanced effector function and proliferation while maintaining the CD62LhiCD44hi central memory (Tcm) phenotype. Further, p53-KO T cells are not transformed to iTregs and exhibit elevated cytolytic properties with remarkable tumor control in a mouse melanoma model. Thus, p53 could serve as target for improving ACT.
MATERIALS AND METHODS
Mice
C57BL/6 (Cat # 000664) and p53-KO (Cat # 002101) mice were obtained from Jackson Laboratory (Bar Harbor, ME). Development of h3T TCR transgenic mouse has been described recently (13). Briefly, the class-I restricted human tyrosinase epitope (YMDTMSQV)368-376 reactive TCR isolated from tumor infiltrating lymphocytes of a HLA-A2+ metastatic melanoma patient was used to generate this transgenic mice. Animals were maintained in pathogen-free facilities and procedures approved by IACUC.
Culture conditions
Recombinant cytokines were purchased from BioLegend, San Diego, CA. Complete IMDM (cIMDM) media containing 10% FBS, penicillin, streptomycin was used for T cells differentiation. On day 3 of culture, T cells were harvested and either processed for intracellular cytokine analysis, RNA preparation using Trizol (Invitrogen, CA) or used for adoptive cell therapy.
Adoptive T cell protocol
Mouse melanoma tumor (B16-F10), and human melanoma (624-MEL) were maintained in vitro in cIMDM. B16-F10 (0.25 × 106) and 624-MEL (2.5 × 106) were injected subcutaneously (s.c.) into left flank of C57BL/6 or Rag1−/− C57BL/6 mice or NSG-A2 mice respectively. Twenty-four hour before adoptive transfer of T cells on day tenth, the recipient mice were injected cyclophosphamide (4 mg/mice, i.p).
Activation induced T cell death
Three day post TCR activation transgenic T cells were re-stimulated for 4h with either cognate antigen or non-specific antigen loaded T2-A2 cells at 5:1 ratio. Apoptosis was measured by staining for Annexin V according to the manufacturer’s protocol, followed by flow cytometry. Data were analyzed with FlowJo software (Tree Star, OR).
Glucose consumption, oxygen consumption and glycolytic flux
Cells were stained with fluorescent-labeled deoxy-glucose analog, 2NBDG (Cayman Chemicals, Ann Arbor, MI) according to manufacturer’s protocol. Cells were washed and stained with other fluorochrome-conjugated antibodies and acquired by flow cytometry. All analysis was done on viable cells. Mitochondrial oxygen consumption or glycolytic flux was measured using the XF 24 analyzer (Seahorse Bioscience, MA) as described earlier (14).
Flow cytometry
Detailed protocols for staining the cells for surface markers and intracellular cytokines have been described earlier (15). Data were analyzed with FlowJo software (Tree Star, OR).
Real-time quantitative-PCR
Total RNA was isolated using Trizol reagent (Invitrogen, CA). cDNA was generated from 1 μg total RNA using iScript cDNA Synthesis Kit (SA Biosciences, Frederick, MD). Quantitative real-time PCR was performed using a SYBR Green mix (Biorad, Hercules, CA) in the CFX96 Detection System (BioRad, Hercules, CA). The fold change in expression of molecules in h3T-p53 KO T cells was calculated over h3T cells and expressed as relative fold change. The TGF-β Pathway PCR array (Qiagen) was used to monitor the expression of 84 genes, along with five housekeeping genes and control for genomic DNA contamination, RNA quality, and general PCR performance. Data analysis was performed using Qiagen’s proprietary web-based analysis tool.
Statistical analysis
All data reported are the arithmetic mean from three or five independent experiments performed in triplicate ±SD unless stated otherwise. The unpaired Student’s t-test was used to evaluate the significance of differences observed between groups, accepting p < 0.05 as a threshold of significance. Data analyses were performed using the Prism software (GraphPad, San Diego, CA). In vivo data were analyzed using Kaplan-Meier methods and pairwise comparisons of survival distributions were done via the log-rank test. Mice that did not reach a tumor size of 400 mm3 by the end of the experiment were sacrificed and had survival time censored in the analysis.
RESULTS
p53 knockout (p53-KO) TCR transgenic T cells show increased proliferation, Tcm phenotype and reduced senescence
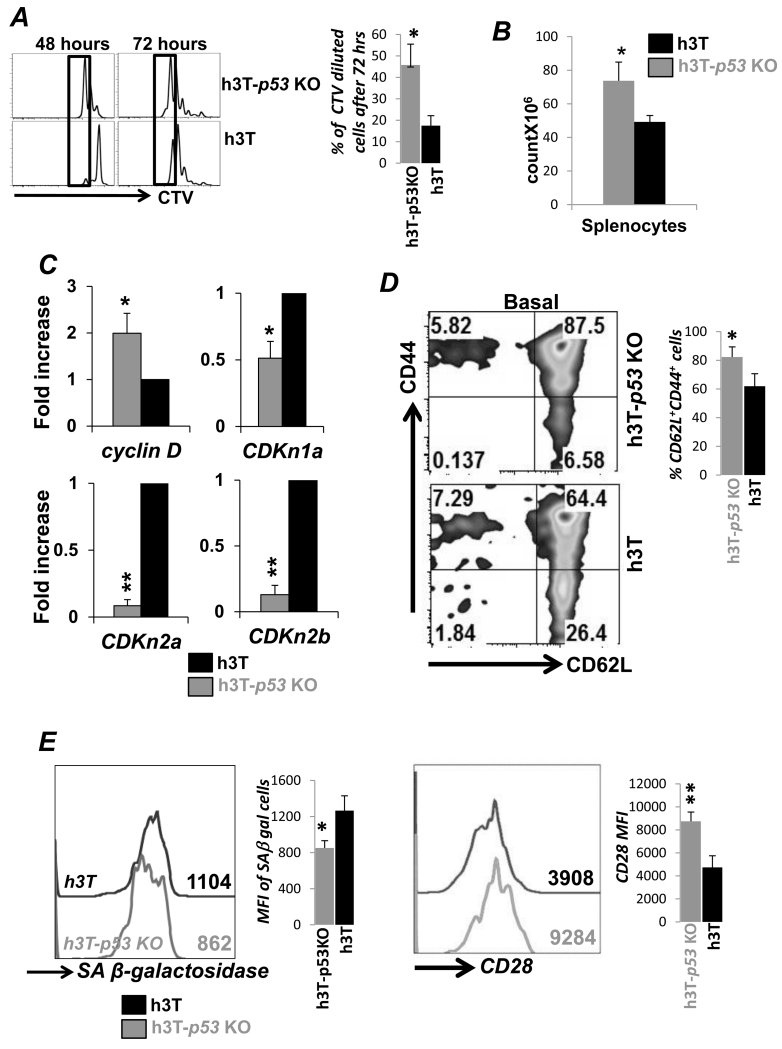
To determine the role of p53 in tumor epitope specific T cells we crossbred p53-KO mice with h3T TCR transgenic mice (13). Figure S1A shows the PCR based genotype screening for the h3T-p53 KO. Using cell trace violet dye we noticed that upon stimulation with cognate antigen the TCR transgenic T cells from h3T-p53 KO proliferated faster until 48 hrs (left panel) as compared to the wild-type (wt) h3T T cells (Figure 1A). The difference persisted even after 72 hours (right panel) of stimulation showing greater cell division in h3T-p53 KO derived T cells. This increased proliferation could be attributed solely to the absence of p53, since the expression of activation induced cell surface molecules as CD69, or CD25 (IL-2Rα) was similar in h3T-p53 KO and h3T derived T cells (Figure S1B). In keeping with the increase in proliferation, higher number of total splenocytes and thymocytes were retrieved from h3T-p53 KO mice (Figure 1B, and Figure S1C). Our data shows that TCR activated h3T-p53 KO derived T cells have higher expression of Cyclin D, a key cyclin protein involved in regulating cell cycle progression and is repressed by p53 (16). The expression of cyclin dependent kinase inhibitors CDKn1a, CDKn2a, and CDKn2b, which are regulated by p53 were also significantly reduced in h3T-p53 KO cells as compared to h3T T cells (Figure 1C). In addition, higher proliferation rate could lead the T cells close to replicative senescence with increased CD62Llo phenotype and susceptibility to cell death (3). A recent study has also shown p53 isoform switching regulates tumor associated replicative senescence T cells (17). However, we observed that h3T-p53 KO T cells not only exhibit higher percentage of CD62L+CD44+ T central memory (Tcm) phenotype as compared to h3T T cells (Figure 1D), but also showed lower expression of senescence associated β-galactosidase and increased CD28 expression (Figure 1E). Thus, reduced expression of p53 modulates cell cycle progression of T cells without inducing replicative senescent phenotype.
Figure 1. p53 KO T cells preserve Tcm phenotype despite increased proliferation.
A) Splenocytes from the h3T and h3T-p53 KO mouse were harvested and stained with cell trace violet (CTV) dye before stimulating with human tyrosinase peptide pulsed irradiated splenic feeder cells from HLA-A2 mouse. The dilution of CTV dye with time was determined using FACS to evaluate antigen specific proliferation. Adjacent bar diagram shows percent increase in proliferating cells from different experiments. B) Bar diagram representing the total viable splenocytes obtained from three individual h3T and h3T-p53 KO mouse as counted using trypan blue dye. C) Real time quantitative PCR analysis for cyclin D and cyclin inhibitors (CDKn1a, CDKn2a, CDKn2b) was done using RNA obtained from h3T and h3T-p53 KO mouse derived splenic T cells. Data from two repeat experiment is shown. D) Basal cell surface expression of CD44 and CD62L was determined using FACS on Vβ12+ gated splenic T cells from h3T and h3T-p53 KO. Adjacent bar diagram shows percent difference in CD62L+CD44+ T cells from repeat experiments. E) TCR activated splenic T cells from h3T and h3T-p53 KO were used to detrmine expression of senescence associated β-galactosidase as per manufacturer’s protocol, and CD28 expression using FACS. Numerical value represents mean fluorescence intensity. Adjacent bar diagram shows cumulative data from different experiments. (N=3, *p < 0.05; **p < 0.01).
Decreased cell death in h3T-p53 KO T cells correlates with higher anti-oxidant capacity
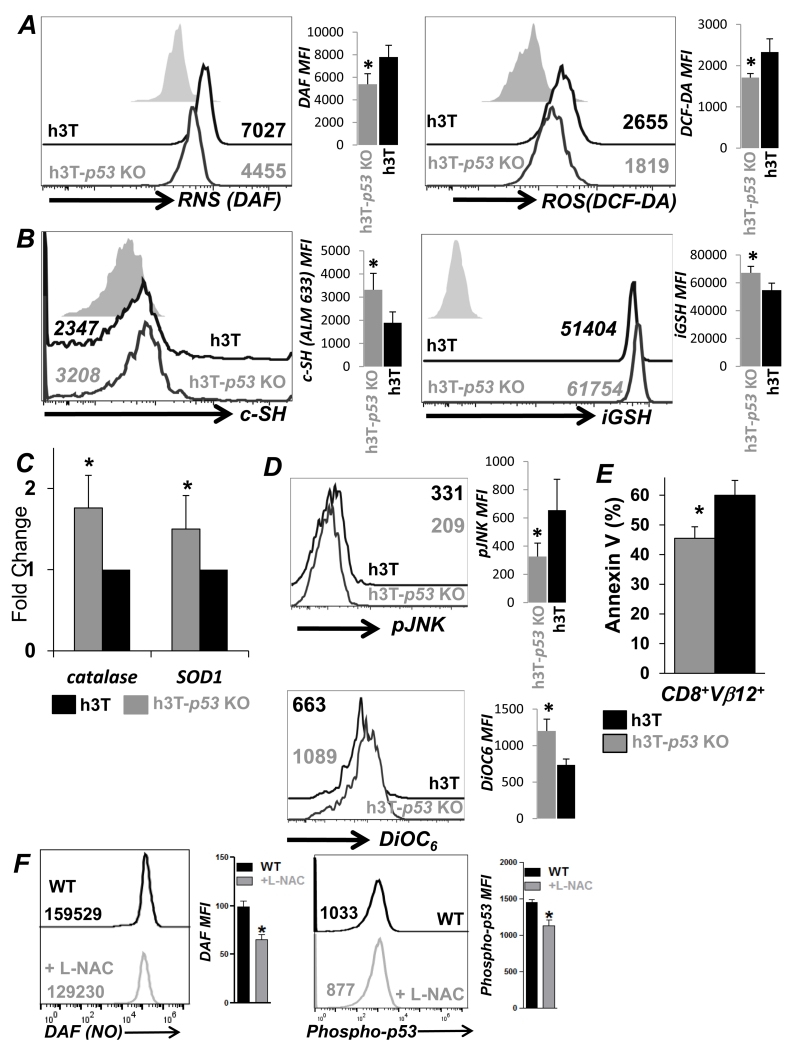
Since Tcm phenotype is associated with higher anti-oxidant capacity and reduced cell death (3), we determined ROS/RNS levels and AICD levels between h3T-p53 KO vs. h3T T cells. Upon TCR restimulation with cognate tyrosinase antigen h3T-p53 KO T cells secreted less ROS (measured using DCFDA dye), and RNS (measured using DAF dye), as compared to h3T T cells (Figure 2A). This also correlated to increased anti-oxidant levels as determined by cell surface thiol (c-SH) (measured using melamide dye) and intracellular glutathione (iGSH) (measured using monocholorobimane dye) (Figure 2B) in p53-KO T cells. Further, a quantitative real time analysis revealed that anti-oxidant enzymes catalase and superoxide dismutase (SOD) levels were also elevated in activated h3T-p53 KO T cells as compared to h3T T cells. While TCR restimulation induced ROS/RNS levels could affect down-stream signaling that involves JNK and leads to T cell death (4), we observed that upon TCR restimulation h3T-p53 KO T cells exhibit reduced JNK phosphorylation (Figure 2D, upper panel), and cell death, as indicated by reduced loss of mitochondrial membrane potential (measured using DiOC6) (Figure 2D, lower panel). Phosphatidyl serine upregulation (using Annexin V) among the Vβ12+CD8+ TCR transgenic T cells was also reduced (Figure 2E). Admittedly, while the difference between the Annexin V+ cells in p53-KO vs. WT was about 10-15 %, the number of cells that were Annexin Vlo (between 0-102 on x-axis) was appreciable (Figure S1D). To further confirm if ROS/RNS levels are important mediators of p53 phosphorylation, the TCR activated wt T cells were either pretreated for 45 min. with anti-oxidant compound L-NAC (10 mM), or left untreated before TCR restimulation. We observed that reduced RNS accumulation (determined by DAF staining) after anti-oxidant L-NAC pretreatment also correlated with reduced p53 phosphorylation (Figure 2F). Thus, the loss of p53 in T cells results in their increased anti-oxidant capacity, which renders them less susceptible to oxidative stress mediated cell death.
Figure 2. Increased anti-oxidant capacity and lower cell death in p53-KO T cells.
TCR activated splenic T cells from h3T and h3T-p53 KO mouse at day three were used: A) after re-stimulation with cognate human tyrosinase antigen pulsed T2-A2 cells for 4 hr and stained using DAF (for NO), or DCF-DA (for H2O2) dyes before analyzing by FACS, B) for staining with alexa-fluor labeled maleamide dye to determine the expression of cell surface thiols (c-SH), and with monochlorobimane dye for determining intracellular glutathione (iGSH) levels using FACS, C) for real time quantitative PCR analysis of anti-oxidant genes catalase and superoxide dismutase using RNA, D) for staining with fluorochrome conjugated phospho-JNK antibody (left panel), and membrane potential dye DiOC6 (right panel) after 4h of re-stimulation, E) for determining AICD 4hr after 4h TCR restimulation by staining with Annexin V and using FACS. Numerical value represents MFI in FACS overlay panels, and adjacent bar diagram represent cumulative data from different experiments. (*p < 0.05). F) Wild type splenic T cells were TCR stimulated cultured for 72h in presence of IL-2 (100 U/ml), after which these were harvested, washed, and then either incubated with NAC (10 mM) for 45 minutes or left untreated. Cells were further TCR re-srimulated and stained with DAF and phopho-p53 antibody following manufacturer’s protocol (BD phosflow), and CD8+ T cells were gated for FACS analysis. N=2.
Loss of p53 enhances glycolysis and pentose phosphate pathway activity in stimulated CD8+T cells
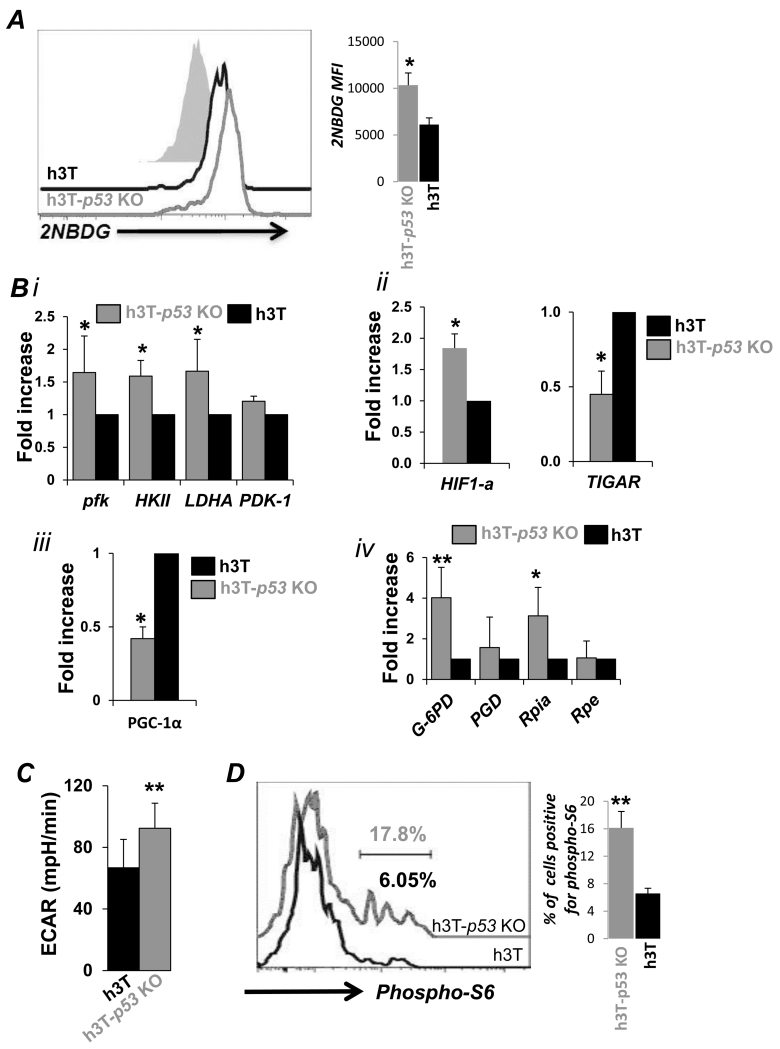
Recent studies have shown that p53 is also involved in regulating various metabolic pathways (18), by balancing glycolysis and oxidative phosphorylation, limiting the production of ROS. While determining how p53 loss regulates T cell metabolism, we observed that uptake of fluorescent glucose 2-NBDG was higher in TCR activated h3T-p53 KO T cells as compared to h3T T cells (Figure 3Ai). Next, we observed significantly higher mRNA levels of glycolytic pathway enzymes in TCR activated p53-KO T cells as compared to the h3T T cells (Figure 3B). Specifically, glycolysis genes, hexokinse (HKII), phosphofructokinase (Pfk), lactate dehydrogenase A (LDHA) (Figure 3Bi), and key glycolysis regulator hypoxia-inducing factor (HIF-1α) (Figure 3Bii, *p ≤ 0.05) (19) were found to be significantly upregulated. Further, the expression of TIGAR (Tp53-induced glycolysis and apoptosis regulator), a known negative regulator of glycolysis that is activated by p53 (10), was also reduced in h3T-p53 KO T cells as compared to h3T T cells (Figure 3Bii). Increased expression of glycolytic genes was also observed when comparing magnetic bead sorted CD8+ T cells from p53-KO T cells to the wt CD8+ T cells (Figure S1E). However, expression level of peroxisome proliferator-activated receptor gamma co-activator 1-alpha (PGC-1α), a key regulator of mitochondrial biogenesis was significantly decreased in h3T-p53 KO T cells as compared to h3T T cells (Figure 3Biii). Further, we also observed increased expression levels of key enzymes involved in regulation of pentose phosphate pathway (PPP) that are required for nucleotide synthesis. In concordance with a recent study that showed p53 inhbits PPP (20), we observed that the mRNA expression of glucose-6 phophate dehydrogenase (G6PD) was 4-fold higher, and that of ribose-5-phophate isomerase (RPIA), was about three-fold higher in activated h3T-p53 KO T as compared to h3T T cells (Figure 3Biv). Since, cells with glycolytic phenotype exhibit significantly higher rates extracellular acidification rate (ECAR) than those dependent upon oxidative phosphorylation which display higher oxygen consumption rate (OCR) (14), we determined the ECAR and OCR levels in real-time using seahorse bioanalyzer. Our data shows that three day activated h3T-p53 KO T exhibit higher ECAR as compared to h3T T cells (Figure 3C). Thus, enhanced glycolysis accompanied by increased commitment to PPP could be contributing to T cell anabolism (21). The higher degree of glycolysis also correlated to the higher usage of mTOR pathway as observed by elevated phosphorylation levels of ribosomal protein S6 (Figure 3D), which is reported to mediate glycolysis (22). Transgenic T cells at the basal level or stimulated with control peptide showed lower pS6 staining, indicating that the increase pS6 in h3T-p53 KO was antigen specific. Thus, this data suggests that increased anti-oxidant capacity and glycolytic commitment of p53-KO T cells could be due to increased expression of PPP molecules - as G6PD reduces nicotinamide adenine dinucleotide phosphate (NADP) to NADPH, and NADPH in turn maintains the level of glutathione to help protect against oxidative damage - a scenario that could be useful in maintaining persistence of tumor reactive T cells in oxidative tumor microenvironment.
Figure 3. Increased glycolytic commitment in p53-KO T cells.
Splenic T cells from h3T and h3T-p53 KO mouse were TCR activated for three days and used: A) for determining the fluoresecent glucose (2NBDG) uptake using FACS as detailed in Material and methods. B) to obtain RNA for analyzing the expression of key glycolytic genes (i), HIF1-α, and TIGAR (ii), mitochondrial biogenesis regulator PGC1-α (iii), and pentose phosphate pathway genes (iv). C) for determining basal extracellular acidification rate (ECAR) using seahorse assay bio-analyzer as per manufacturer’s protocol. D) to determine phosphorylation level of S6 protein after intracellular staining and analusis using FACS. (*p<0.05; **p<0.01). Bar diagram on right of each overlay represent cumulative data from different experiments.
p53 expression inversely correlates to cytokine response and effector function in CD8+ T cells
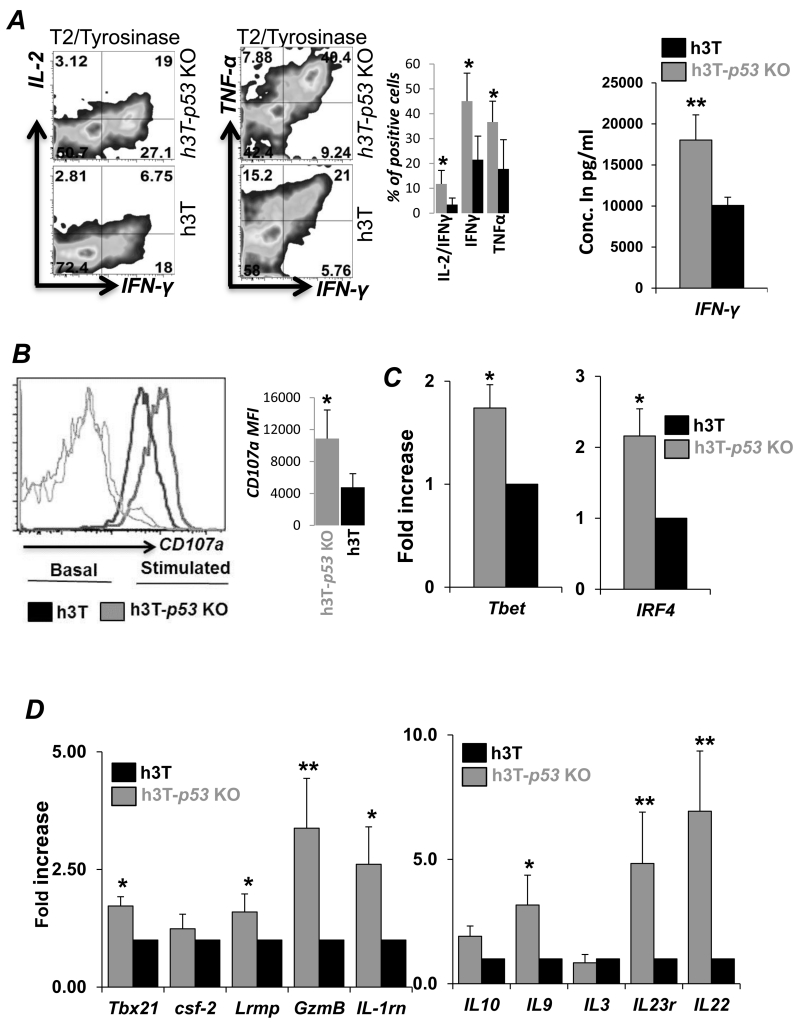
Since loss of p53 results in increased glycolysis, a key metabolic pathway that regulates cytokine IFN-γ (23), we compared expression of effector molecules between h3T-p53 KO vs. h3T T cells. Our data demonstrates that upon TCR stimulation with tyrosinase antigen, the fraction of T cells secreting cytokines IL-2, IFN-γ and TNF-α were about two-fold more as compared to the h3T T cells (Figure 4A). Overnight antigen stimulation also confirmed that h3T-p53 KO T cells secreted twice the amount of IFN-γ as compared to h3T T cells (Figure 4A, right panel). Importantly, h3T-p53 KO T cells also exhibit increased externalization of lysosomal protein CD107a (Figure 4B), an indicator of enhanced cytotoxic granules exocytosis (24), which indicates increased cytolytic ability of h3T-p53 KO T cells over p53 sufficient h3T T cells. We also observed that the expression of signature transcription factors for type-1 cytototoxic (Tc1) cells as T-bet and IRF-4 were higher in p53-KO CD8+ T cells (Figure 4C). In addition, an increased expression of genes related to key effector molecules as GM-CSF, Granzyme B, IL1Rn, IL23R, IL22 was noticed in h3T-p53 KO T cells as compared to h3T T cells (Figure 4D). These data indicate that h3T-p53 KO T cells are highly poly-functional cells and exhibit increased effector function as compared to h3T T cells.
Figure 4. p53-KO T cells exhibit enhanced effector functions.
A) Splenic T cells from h3T and h3T-p53 KO mouse activated for three days were either re-stimulated with cognate human tyrosinase antigen pulsed T2-A2 cells for 6 hr. before performing intracellular staining using flurochrome conjugated anti-cytokine (IL-2, IFN-γ, TNF-α) antibody (left panel), or were re-stimulated overnight to determine the IFN-γ secretion in the supernatant by ELISA (right panel). B) The degree of degranulation was determined in naïve or three day activated h3T and h3T-p53 KO splenic T cells by staining for CD107a expression. RNA isolated from three day activated h3T and h3T-p53 KO mouse were used to determine expression of: C) Transcription factors T-bet and IRF4, and D) Effector molecules and cytokine, cytokine receptors. The fold change in expression of these molecules in h3T-p53 KO T cells was calculated over h3T cells. (*p <0.05; **p <0.01).
Adoptive transfer of p53-KO TCR transgenic CD8+ T cells improves tumour control
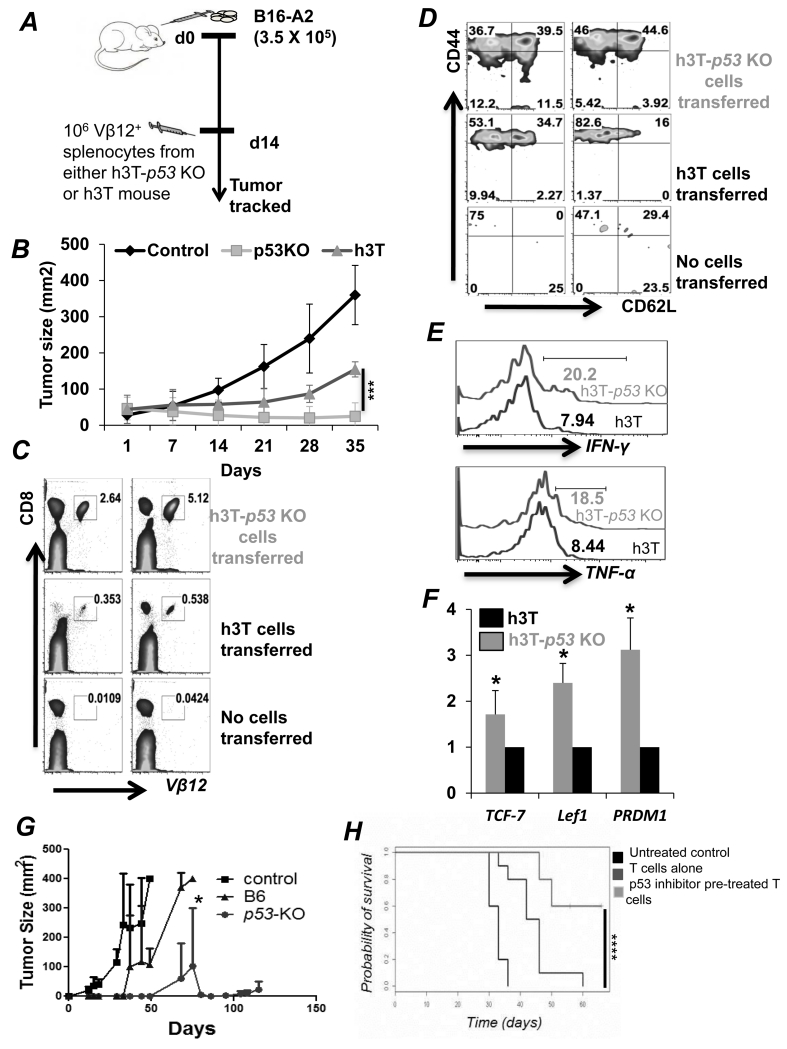
To determine the anti-tumor potential of p53 deficient T cells, B16-A2 tumor melanoma cells were established subcutaneously in HLA-A2 mice for fourteen days before transferring HLA-A2 restricted tyrosinase reactive Vβ12+ TCR transgenic CD8+ splenic T cells from h3T-p53 KO or h3T mouse (schema in Figure 5A). We observed that h3T-p53 KO T cells showed long-term tumor control than those that received h3T T cells (Figure 5B, and Figure S2A). At the experimental end-point ten-fold higher transferred T cells were tracked in the peripheral blood of the recipient group that received h3T-p53 KO splenic T cells (Figure 5C), which exhibited CD62L+CD44+ central memory phenotype (16% in h3T vs. 44% in h3T-p53 KO) (Figure 5D). The retrieved h3T-p53 KO transgenic T cells continued to produce more effector cytokines IFN-γ and TNF-α than retrieved h3T T cells upon re-stimulation (Figure 5E). We also noted that a fraction (4-11%) of h3T-p53 KO transferred T cells exhibited CD44−CD62L+ phenotype that is known to harbor stem-cell memory phenotype cells (25), which correlated with two-to-three fold higher expression of stemness genes Tcf7, Lef1 and PRDM1 as compared h3T T cells (Figure 5F). These data indicate that the quantitative and qualitative differences between the h3T vs. h3T-p53 KO T cells may account for differences in ability to control tumor growth in vivo. Further, to confirm the feasibility of this approach using TCR engineered T cells, we used the splenic T cells from the C57BL/6 wild-type mice and p53-KO mice that were rendered tumor antigen specific by using retroviral transduction of tyrosinase reactive HLA-A2+ TIL1383I TCR (26). Adoptive transfer of the tyrosinase reactive TIL1383I TCR transduced T cells also showed long-term control of subcutaneously established B16-A2 tumors in the HLA-A2 recipient mice (Figure 5G). Next, we tested the efficacy of p53 inhibitors pifithrin-mu (PFT-μ) and pifithrin-alpha (PFT-α) in controlling tumor growth. For this purpose, subcutanelously established B16-F10 murine melanoma in C57BL/6 mice were treated by adoptively transferring 1 × 106 melanoma epitope gp100 reactive T cells that were activated for three days with cognate antigen in presence or absence of inhibitors. We observed that as compared to the activated T cells alone, the p53 inhibitor pre-treated T cells resulted in a significantly improved control of tumor growth and thus survival of the tumor bearing mouse (Figure 5H). This data shows that pharmacological inhibition of p53 could be a clinically translatable ACT approach.
Figure 5. Improved tumor control by adoptively transferred h3T-p53 KO T cells.
A) Schematic representation of the experimental protocol. B) Tumor growth curve (in mm2) obtained after treating subcutaneously established melanoma in C57BL/6 recipient mice by adoptively transferring either 1 × 106 h3T or h3T-p53 KO T cells. Nine mice per group were treated using ACT. The peripheral blood was obtained from the recipient mice and the adoptively transferred Vβ12+ T cells were evaluated for: C) total percent population; D) cell surface expression of CD44 and CD62L; E) cytokine IFN-γ and TNF-α secretion upon restimulation. F) Activated h3T or h3T-p53 KO splenic T cells were used to prepare RNA and determine the expression of genes related to stem cell phenotype using qPCR. G) Splenic CD8+ T cells obtained from C57BL/6 WT or C57BL/6-p53 KO mouse strains were transduced with HLA-A2+ human tyrosinase epitope reactive TIL1383I TCR splenocytes and 107 cells were adoptively transferred to the Rag-A2 recipients with sub-cutaneously established murine melanoma B16-A2. The tumor growth in various groups of recipient mice that were either treated or left un-treated is shown. Seven mice in each group between two experiments were used and showed identical response. H) Melanoma epitope gp100 reactive T cells were obtained from Pmel TCR transgenic mouse, and activated for three days with cognate antigen either alone or in presence of p53 inhibitors (5 μM Pif-α + Pif-μ). The activated T cells were transferred to the B16-F10 murine melanoma bearing C57BL/6 host and tumor growth were measured twice weekly. A total of 12-16 mice in each group were treated in two experiments with similar results. (*p<0.05; **p<0.005, ***p<0.00, ****p = 0.0006).
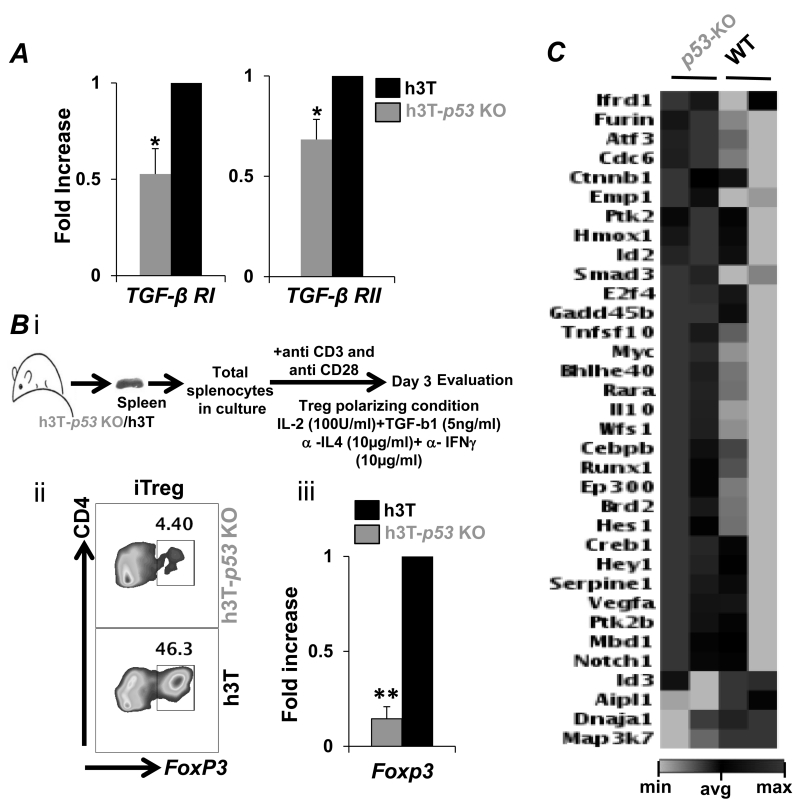
Reduced TGF-β signaling in p53-KO T cells
Next we addressed if the improved tumor control exhibited by h3T-p53 KO T cells is due to reduced susceptibility to immunosuppression or reduced plasticity towards inducible regulatory T cells (iTreg’s) conversion. Importantly, h3T-p53 KO T cells exhibited reduced expression of TGF-βRI and TGF-βRII as compared to the h3T derived splenic T cells (Figure 6A). Further, ex vivo programming conditions that use TGF-β and IL-2 (Figure 6Bi) showed that p53-KO derived splenic CD4+ T cells exhibit less susceptibility to iTreg conversion (Figure 6Bii). The quantitatively reduced iTreg’s also corresponded to the reduced FoxP3 expression in the p53-KO derived splenic CD4+ T cells as compared to wild type T cells (Figure 6Biii). A detailed analysis for differences in TGF-β signaling pathway was performed using the TGF-β Signaling Targets RT2 Profiler PCR Array (Qiagen). Our data in Figure 6C shows that a number of signaling molecules were differentially expressed in T cells obtained from p53-KO mice, which may have contributed to the enhanced anti-tumor phenotype. For example: Furin is a direct target gene of the IL-12/STAT4 pathway, regulates Th1/2 cell balance by limiting conversion to Th2 phenotype, and its expression directly co-relates to the stability and long-term secretion of IL2 by CD4+ T cells (27). Similarly, increased expression of activating transcription factor (ATF) 3 that is a positive regulator of IFN-γ gene expression (28) supports our observation of increased Th1 cytokine response in p53-KO T cells. The cell division cycle 6 (Cdc6), a target of p53 that coordinates S phase and mitosis was also upregulated in p53-KO T cells. (29). Similarly, Ctnnb1, a gene that encodes β-catenin protein is upregulated in p53-KO T cells. Ptk2, protein tyrosine kinase 2, (also known as focal adhesion kinase) that play an important role in T cell–antigen-presenting-cell conjugation, and HMOx1 encoded heme-oxygenase-1 levels, a target of p53 (30), were also increased in p53-KO T cells. The expression of inhibitor of dna binding 2 (Id2), which promotes generation of distinct CD8+ T cell memory was also increased in p53-KO T cells (31). Tumor suppressor p53 is an essential partner of Smads, affecting TGF-β signaling at various points in the pathway (32). Importantly, the expression of mitogen-activated protein kinase kinase kinase 7 (Map3K7) was reduced in p53-KO T cells. This kinase mediates the signal transduction induced by TGF-β and controls a variety of cell functions including transcription regulation and apoptosis (33). Expression of E2F4 – a transcription factor that plays a crucial role in the control of cell cycle and regulating antigen recall response in CD8+ T cells was also elevated in p53-KO T cells (34). The p53-KO T cells expressed higher Gadd45b, which augments anti-tumor immune response by enhancing the expression of IFNγ, granzyme B, CCR5 in T cells (35), and protecting from apoptosis by p38 activation and JNK inhibition (36). WFS1, a gene encoding an endoplasmic reticulum (ER) membrane protein and involved in survival of pancreatic β-cells was also found to be upregulated in p53-KO T cells. As expected, we also found that Myc levels were enhanced in p53-KO T cells, which also showed higher HIF1α and glycolytic commitment. Thus, targeting p53 in T cells result in modulating TGF-β mediated signaling molecules.
Figure 6. Altered TGF-β signaling in p53-KO T cells.
A) Activated h3T or h3T-p53 KO splenic CD8+ T cells obtained after FACS sorting were used to prepare RNA and determine the expression of TGF-β receptors (TGF-βRI and TGF-βRII). B) h3T or h3T-p53 KO splenic T cells were cultured for three days under iTreg polarizing conditions, and FoxP3 expression analysis was done using intracellular staining (left panel), or real-time PCR (right panel). C) RNA from A) was also used to run the TGF-β signaling real-time PCR based 84 gene array. The data obtained is presented in fold change with genes grouped for pathways indicated on left. The data is representative from one of two experiments and genes with similar results in both array experiments are presented.
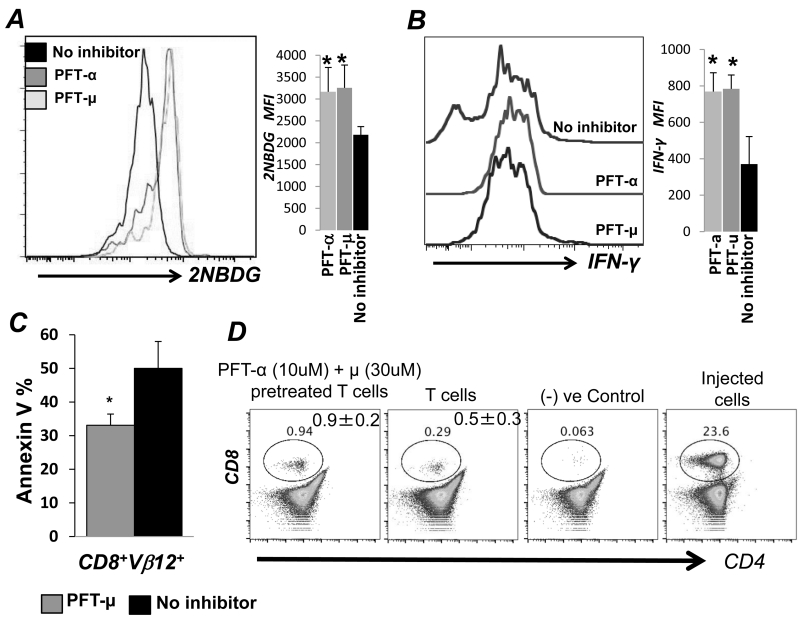
Pharmacological inhibition of p53 inhibitor Pifithrin-μ alters functionality of human TCR transduced CD8+ T cells
To determine the translational potential of inhbiting p53, pharmacological inhibitors PFT-μ and PFT-α pre-treated human tyrosinase TCR TIL1383I transduced T cells were characterized. Upon p53 inhibition, we noticed a significant increase in glucose uptake as measured by 2-NBDG (Figure 7A), which also correlated to an increased fraction of IFNγ secreting cells upon antigen restimulation (Figure 7B). Importantly, AICD was also reduced when PFT-μ pretreated TIL1383I TCR transduced T cells were restimulated with the cognate tyrosinase epitope (Figure 7C). Additionally, p53 inhibtion not only downregulated ROS and RNS, but also reduced the expression of CD95, CD95L, exhaustion molecules Lag3 and PD1 on TCR activated T cells (Figure S2B) Thus, pharmacologically inhibiting p53 in human TCR transduced cells mimicked results of increased metabolic activity in form of glycolysis, with increased effector functions and lower cell death. Further, we observed that pretreatment with either PFT-μ or PFT-α results in fewer human T cells exhibiting FoxP3 expression under iTreg ex vivo programming condition (Figure S2C). Lastly, tracking studies using human T cells transduced with melanoma reactive TIL1383I TCR that were pre-treated with a combination of p53 inhibitors and transferred into NSG-A2 mice showed increased the persistence at 72 hrs. as compared to the untreated counterparts (Figure 7D). Thus, we believe that the strategy to inhibit p53 expression is potentially translatable and could improve the efficacy of ACT.
Figure 7. p53 inhibitor treated TCR transduced T cells exhibit increased effector function and persistence.
Human peripheral blood T cells from normal healthy individuals were retro-virally transduced with melanoma epitope tyrosinase reactive TIL1383I TCR and were either left untreated or were pretreated for an hour with p53 inhibitors Pifithrin-α (30μM) or Pifithrin-μ (10μM) before TCR restimulation with tyrosinase peptide pulsed T2-A2 cells for 4-6 hrs to analyze: A) Glucose uptake using 2NBDG assay, B) Cytokine secretion by intracellular IFN-γ staining, and C) Susceptibility to AICD by Annexin V staining (*p<0.01). Bar diagram on right of each overlay represent cumulative data from different experiments. D) Ten million human T cells engineered with tyrsoinase reactive TIL1383I TCR were either untreated or pretreated with a combination of p53 inhibitors Pifithrin-α and Pifithrin-μ and adoptively transferred to NSG-A2 mice. Peripheral blood and spleens of recipient mice were stained for human Vβ12, human CD8 and CD4 for tracking the persistence of the transferred cells. Data was acquired using FACS. Numerical value is the average from three mice in similar groups.
DISCUSSION
p53 is regarded as the “guardian of genome integrity” due to its complex role in regulating cellular differentiation (37). More than 50% of tumors have a direct mutation of p53, which promotes invasion, metastasis, proliferation and cell survival (38). p53 is also a central regulator of glycolysis and TGF-β signaling pathways (9,19). Therefore, we hypothesized that rendering properties of higher proliferation, lower cell death, increased persistence to CTL’s by lowering its p53 expression could improve adoptive T cell immunotherapy. Our data establishes that p53 deficient tumor specific CD8+ T cells exhibit increased glycolytic commitment that correlates to higher IFN-γ secretion, increased persistence due to high stem-cell related gene expression, reduced susceptibility to immunosuppression and iTreg conversion due to reduced TGF-β signaling. All these features result in an effector phenotype leading to improved tumor control.
A recent study showed that antigen-specific proliferative responses of CD4+ T cells require down-regulation of tumor suppressor p53 (7), and that inhibiting p53-regulating protein Mdm2 resulted in its sustained expression and prevented proliferation. Our data shows that the T cells obtained from the TCR transgenic mice h3T developed on p53-KO background (i.e. h3T-p53 KO) proliferate rapidly, and maintain antigen specificity upon TCR stimulation. Given the role of p53 in negatively regulating cell cycle progression by blocking cyclin D1 (16), we observed that h3T-p53 KO T cells exhibited higher expression of cyclin D and lower levels of cyclin inhibitors that correlate to increased proliferation leading to enlarged spleen and thymus. However, increased proliferation was not associated with shedding of CD62L molecule (3,39), since we observed that h3T-p53 KO T cells exhibit CD62Lhi central memory (Tcm) phenotype. Importantly, h3T-p53 KO T cells also exhibited elevated expression of stem-cell specific transcription factors as Tcf7, Lef-1, and PRDM1. Notably, p53 has been shown to bind at the promoter region of Oct4 and Nanog, which are required for self-renewal and maintenance of embryonic stem cells in an undifferentiated state, and reduce their gene transcription (40). Given a recent report that a subset of memory T cells with stem-cell like phenotype (referred as Tscm) exists within the Tcm fraction (41), it is likely that the Tscm phenotype is increased in the p53-KO T cells.
p53 also regulates aerobic respiration at the glycolytic and OXPHOS steps via transcriptional regulation of its downstream genes TIGAR and SCO2 (10,11). In line with the role of p53 as negative regulator of glycolysis (11), our data shows that p53-KO T cells exhibit higher glycolytic commitment as observed by glucose uptake and increased expression of key glycolytic genes. This increase in glycolysis corresponds to the reduced expression of TIGAR, an inhibitor of the fructose-2, 6-bisphosphate, which is normally activated by p53 to regulate glycolysis (11). Another property of p53 deficient T cells was their ability to persist longer and exhibit lower degree of AICD. It has been shown that p53 leads to up-regulation of a number of pro-oxidant enzymes as quinone oxidoreductase, proline oxidase, BAX, and PUMA leading to oxidative stress and consequently to apoptosis (42-44). Upon its mitochondrial translocation p53 binds to and inhibits MnSOD, playing a direct role in promoting ROS formation and eventually in apoptosis (45). Thus, an inverse correlation between p53 and anti-oxidant capacity may have contributed to the increased persistence and anti-tumor T cell response. Interestingly, a detailed necropsy of the tumor bearing recipient animals showed increased inflammatory reactions, without any evidence to suggest that this was due to the transfer of p53 KO T cells (data not shown).
Another confounding factor that limits long-term tumor control by ACT is suppressive tumor microenvironment, which is abundant with suppressive cytokines as TGF-β. Importantly, key cellular responses to TGF-β signals have been shown to rely on p53 family members (9). This study shows that p53-KO T cells display an impaired response to TGF-β signals. Additionally, Smad and p53 protein complexes converge on separate cis binding elements on a target promoter and synergistically activate TGF-β induced transcription (6). Thus, it is likely that p53-KO T cells displayed diminished transcriptional activation of key TGF-β target genes (32). It has also been shown that p53 enhances the transcription of Treg signature transcription factor Foxp3 by binding to the promoter and the conserved noncoding DNA sequence-2 of the Foxp3 gene (8), and that fewer nTreg’s and iTreg’s are obtained from p53-KO mice. Our data also confirms this observation, and implies that impaired TGF-β signaling molecules may be responsible for reduced plasticity in p53 deficient T cells. It is also likely that increased glycolytic commitment identified in h3T-p53 KO T cells by elevated levels of ECAR values, glycolytic genes and HIF1-α metabolically down regulates Treg differentiation, since HIF-1α has been shown to attenuate Treg development by binding Foxp3 and targeting it for proteasomal degradation (46). Importantly, p53 pharmacological inhibitors also improved T cell mediated tumor control, and pifithrin pretreated murine and human T cells exhibited increased persistence in vivo. Overall, this study shows that p53 is a central regulator of multiple pathways (as glycolysis, ROS, TGF-β signaling), and its inhibition could be important for ACT of tumor.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge Drs. Zihai Li, Radhika Gudi, and Ephraim Ansa-Addo in Department of Microbiology and Immunology at MUSC for their help with this manuscript.
The work was supported in part by NIH grants R21CA137725 and R01CA138930 to SM, and P01CA154778 to MIN.
Footnotes
Conflict of interest: The authors have no conflict of inetest.
REFERENCES
- 1.Feldman SA, Assadipour Y, Kriley I, Goff SL, Rosenberg SA. Adoptive Cell Therapy--Tumor-Infiltrating Lymphocytes, T-Cell Receptors, and Chimeric Antigen Receptors. Seminars in oncology. 2015;42(4):626–39. doi: 10.1053/j.seminoncol.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chatterjee S, Thyagarajan K, Kesarwani P, Song JH, Soloshchenko M, Fu J, et al. Reducing CD73 expression by IL1beta-Programmed Th17 cells improves immunotherapeutic control of tumors. Cancer research. 2014;74(21):6048–59. doi: 10.1158/0008-5472.CAN-14-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kesarwani P, Al-Khami AA, Scurti G, Thyagarajan K, Kaur N, Husain S, et al. Promoting thiol expression increases the durability of antitumor T-cell functions. Cancer research. 2014;74(21):6036–47. doi: 10.1158/0008-5472.CAN-14-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Norell H, Martins da Palma T, Lesher A, Kaur N, Mehrotra M, Naga OS, et al. Inhibition of superoxide generation upon T-cell receptor engagement rescues Mart-1(27-35)-reactive T cells from activation-induced cell death. Cancer research. 2009;69(15):6282–9. doi: 10.1158/0008-5472.CAN-09-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuckerman V, Wolyniec K, Sionov RV, Haupt S, Haupt Y. Tumour suppression by p53: the importance of apoptosis and cellular senescence. The Journal of pathology. 2009;219(1):3–15. doi: 10.1002/path.2584. [DOI] [PubMed] [Google Scholar]
- 6.Grayson JM, Lanier JG, Altman JD, Ahmed R. The role of p53 in regulating antiviral T cell responses. Journal of immunology. 2001;167(3):1333–7. doi: 10.4049/jimmunol.167.3.1333. [DOI] [PubMed] [Google Scholar]
- 7.Watanabe M, Moon KD, Vacchio MS, Hathcock KS, Hodes RJ. Downmodulation of tumor suppressor p53 by T cell receptor signaling is critical for antigen-specific CD4(+) T cell responses. Immunity. 2014;40(5):681–91. doi: 10.1016/j.immuni.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawashima H, Takatori H, Suzuki K, Iwata A, Yokota M, Suto A, et al. Tumor suppressor p53 inhibits systemic autoimmune diseases by inducing regulatory T cells. Journal of immunology. 2013;191(7):3614–23. doi: 10.4049/jimmunol.1300509. [DOI] [PubMed] [Google Scholar]
- 9.Cordenonsi M, Dupont S, Maretto S, Insinga A, Imbriano C, Piccolo S. Links between tumor suppressors: p53 is required for TGF-beta gene responses by cooperating with Smads. Cell. 2003;113(3):301–14. doi: 10.1016/s0092-8674(03)00308-8. [DOI] [PubMed] [Google Scholar]
- 10.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126(1):107–20. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 11.Madan E, Gogna R, Bhatt M, Pati U, Kuppusamy P, Mahdi AA. Regulation of glucose metabolism by p53: emerging new roles for the tumor suppressor. Oncotarget. 2011;2(12):948–57. doi: 10.18632/oncotarget.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: insights into metabolism and lymphocyte function. Science. 2013;342(6155):1242454. doi: 10.1126/science.1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehrotra S, Al-Khami AA, Klarquist J, Husain S, Naga O, Eby JM, et al. A coreceptor-independent transgenic human TCR mediates anti-tumor and anti-self immunity in mice. Journal of immunology. 2012;189(4):1627–38. doi: 10.4049/jimmunol.1103271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrick DA, Neilson A, Beeson C. Advances in measuring cellular bioenergetics using extracellular flux. Drug discovery today. 2008;13(5-6):268–74. doi: 10.1016/j.drudis.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Chatterjee S, Eby JM, Al-Khami AA, Soloshchenko M, Kang HK, Kaur N, et al. A quantitative increase in regulatory T cells controls development of vitiligo. The Journal of investigative dermatology. 2014;134(5):1285–94. doi: 10.1038/jid.2013.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rocha S, Martin AM, Meek DW, Perkins ND. p53 represses cyclin D1 transcription through down regulation of Bcl-3 and inducing increased association of the p52 NF-kappaB subunit with histone deacetylase 1. Molecular and cellular biology. 2003;23(13):4713–27. doi: 10.1128/MCB.23.13.4713-4727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mondal AM, Horikawa I, Pine SR, Fujita K, Morgan KM, Vera E, et al. p53 isoforms regulate aging- and tumor-associated replicative senescence in T lymphocytes. The Journal of clinical investigation. 2013;123(12):5247–57. doi: 10.1172/JCI70355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nature reviews Molecular cell biology. 2015;16(7):393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 19.Yeung SJ, Pan J, Lee MH. Roles of p53, MYC and HIF-1 in regulating glycolysis - the seventh hallmark of cancer. Cellular and molecular life sciences : CMLS. 2008;65(24):3981–99. doi: 10.1007/s00018-008-8224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M, et al. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nature cell biology. 2011;13(3):310–6. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patra KC, Hay N. The pentose phosphate pathway and cancer. Trends in biochemical sciences. 2014;39(8):347–54. doi: 10.1016/j.tibs.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng SC, Quintin J, Cramer RA, Shepardson KM, Saeed S, Kumar V, et al. mTOR- and HIF-1alpha-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345(6204):1250684. doi: 10.1126/science.1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang CH, Curtis JD, Maggi LB, Jr., Faubert B, Villarino AV, O’Sullivan D, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153(6):1239–51. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Betts MR, Brenchley JM, Price DA, De Rosa SC, Douek DC, Roederer M, et al. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. Journal of immunological methods. 2003;281(1-2):65–78. doi: 10.1016/s0022-1759(03)00265-5. [DOI] [PubMed] [Google Scholar]
- 25.Gattinoni L, Klebanoff CA, Restifo NP. Paths to stemness: building the ultimate antitumour T cell. Nature reviews Cancer. 2012;12(10):671–84. doi: 10.1038/nrc3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishimura MI, Avichezer D, Custer MC, Lee CS, Chen C, Parkhurst MR, et al. MHC class I-restricted recognition of a melanoma antigen by a human CD4+ tumor infiltrating lymphocyte. Cancer research. 1999;59(24):6230–8. [PubMed] [Google Scholar]
- 27.Oksanen A, Aittomaki S, Jankovic D, Ortutay Z, Pulkkinen K, Hamalainen S, et al. Proprotein convertase FURIN constrains Th2 differentiation and is critical for host resistance against Toxoplasma gondii. Journal of immunology. 2014;193(11):5470–9. doi: 10.4049/jimmunol.1401629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Filen S, Ylikoski E, Tripathi S, West A, Bjorkman M, Nystrom J, et al. Activating transcription factor 3 is a positive regulator of human IFNG gene expression. Journal of immunology. 2010;184(9):4990–9. doi: 10.4049/jimmunol.0903106. [DOI] [PubMed] [Google Scholar]
- 29.Duursma A, Agami R. p53-Dependent regulation of Cdc6 protein stability controls cellular proliferation. Molecular and cellular biology. 2005;25(16):6937–47. doi: 10.1128/MCB.25.16.6937-6947.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andres NC, Fermento ME, Gandini NA, Romero AL, Ferro A, Donna LG, et al. Heme oxygenase-1 has antitumoral effects in colorectal cancer: involvement of p53. Experimental and molecular pathology. 2014;97(3):321–31. doi: 10.1016/j.yexmp.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 31.Yang CY, Best JA, Knell J, Yang E, Sheridan AD, Jesionek AK, et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nature immunology. 2011;12(12):1221–9. doi: 10.1038/ni.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elston R, Inman GJ. Crosstalk between p53 and TGF-beta Signalling. Journal of signal transduction. 2012;2012:294097. doi: 10.1155/2012/294097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voorneveld PW, Kodach LL, Jacobs RJ, van Noesel CJ, Peppelenbosch MP, Korkmaz KS, et al. The BMP pathway either enhances or inhibits the Wnt pathway depending on the SMAD4 and p53 status in CRC. British journal of cancer. 2015;112(1):122–30. doi: 10.1038/bjc.2014.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bancos S, Cao Q, Bowers WJ, Crispe IN. Dysfunctional memory CD8+ T cells after priming in the absence of the cell cycle regulator E2F4. Cellular immunology. 2009;257(1-2):44–54. doi: 10.1016/j.cellimm.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ju S, Zhu Y, Liu L, Dai S, Li C, Chen E, et al. Gadd45b and Gadd45g are important for anti-tumor immune responses. European journal of immunology. 2009;39(11):3010–8. doi: 10.1002/eji.200839154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta M, Gupta SK, Hoffman B, Liebermann DA. Gadd45a and Gadd45b protect hematopoietic cells from UV-induced apoptosis via distinct signaling pathways, including p38 activation and JNK inhibition. The Journal of biological chemistry. 2006;281(26):17552–8. doi: 10.1074/jbc.M600950200. [DOI] [PubMed] [Google Scholar]
- 37.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358(6381):15–6. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 38.Muller PA, Vousden KH. p53 mutations in cancer. Nature cell biology. 2013;15(1):2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 39.Yang S, Liu F, Wang QJ, Rosenberg SA, Morgan RA. The shedding of CD62L (L-selectin) regulates the acquisition of lytic activity in human tumor reactive T lymphocytes. PloS one. 2011;6(7):e22560. doi: 10.1371/journal.pone.0022560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nature genetics. 2006;38(4):431–40. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 41.Graef P, Buchholz VR, Stemberger C, Flossdorf M, Henkel L, Schiemann M, et al. Serial transfer of single-cell-derived immunocompetence reveals stemness of CD8(+) central memory T cells. Immunity. 2014;41(1):116–26. doi: 10.1016/j.immuni.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 42.Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389(6648):300–5. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 43.Rivera A, Maxwell SA. The p53-induced gene-6 (proline oxidase) mediates apoptosis through a calcineurin-dependent pathway. The Journal of biological chemistry. 2005;280(32):29346–54. doi: 10.1074/jbc.M504852200. [DOI] [PubMed] [Google Scholar]
- 44.Liu B, Chen Y, St Clair DK. ROS and p53: a versatile partnership. Free radical biology & medicine. 2008;44(8):1529–35. doi: 10.1016/j.freeradbiomed.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao Y, Chaiswing L, Velez JM, Batinic-Haberle I, Colburn NH, Oberley TD, et al. p53 translocation to mitochondria precedes its nuclear translocation and targets mitochondrial oxidative defense protein-manganese superoxide dismutase. Cancer research. 2005;65(9):3745–50. doi: 10.1158/0008-5472.CAN-04-3835. [DOI] [PubMed] [Google Scholar]
- 46.Dang EV, Barbi J, Yang HY, Jinasena D, Yu H, Zheng Y, et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146(5):772–84. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.