Abstract
Monkeypox virus (MPXV) infection fails to activate the host anti-viral protein, PKR, despite lacking a full-length homologue of the vaccinia virus (VACV) PKR inhibitor, E3. Since PKR can be activated by dsRNA produced during a viral infection, we have analyzed the accumulation of dsRNA in MPXV-infected cells. MPXV infection led to less accumulation of dsRNA than VACV infection. Because in VACV infections accumulation of abnormally low amounts of dsRNA is associated with mutations that lead to resistance to the anti-poxvirus drug isatin beta-thiosemicarbazone (IBT), we investigated the effects of treatment of MPXV-infected cells with IBT. MPXV infection was eight-fold more resistant to IBT than wild-type vaccinia virus (wtVACV). These results demonstrate that MPXV infection leads to the accumulation of less dsRNA than wtVACV, which in turn likely leads to a decreased capacity for activation of the dsRNA-dependent host enzyme, PKR.
Keywords: Monkeypox virus, Poxvirus pathogenesis, dsRNA, E3 gene, F3 gene, Innate immune evasion, IBT resistance, Poxvirus transcription, Virulence, Vaccinia virus
1. Introduction
The VACV E3L gene is highly conserved amongst the chordopoxvirinae. The gene is only absent in the avipoxviruses and in molluscum contagiosum virus (Senkevich et al., 1997, Tulman et al., 2004). Two proteins are expressed from the E3L gene (Chang et al., 1992, Yuwen et al., 1993), a full length 190-amino-acid protein, p25, and a protein truncated of 37 N-terminal amino acids, p20, that is the product of leaky scanning past the first AUG codon. In the majority of chordopoxviruses, the protein consists of two conserved domains. The C-terminal domain is both necessary and sufficient for binding to dsRNA (Chang and Jacobs, 1993, Chang et al., 1995), and contains a consensus dsRNA binding motif. This domain is necessary for pathogenesis in the mouse model (Brandt et al., 2005, Brandt and Jacobs, 2001), and is necessary for replication in most cells in culture (Chang and Jacobs, 1993, Chang et al., 1995), with the exception of RK13, BHK, and CEF cells. This domain is also necessary for interferon resistance (IFNR) of VACV in RK13 cells (Shors et al., 1997). Our data is consistent with this domain acting possibly as a dsRNA “sponge”, sequestering dsRNA to prevent its recognition by the host anti-viral machinery.
The majority of poxvirus E3-like proteins also contain a conserved N-terminal domain. The N-terminal domain is necessary for pathogenesis in the mouse model (Brandt et al., 2005, Brandt and Jacobs, 2001), but until recently had been dispensable for replication and IFNR in all cells in culture we had tested. We have recently shown that a complete N-terminal domain is necessary for IFNR in 129 MEFs (White and Jacobs, 2012) and for replication in mouse JC cells (Arndt et al., 2015). In 129 MEFs, HeLa cells, and infected mice, the N-terminus is required to fully inhibit PKR (White and Jacobs, 2012, Langland and Jacobs, 2004). Since PKR is activated by dsRNA, these results suggest that the N-terminus of E3 is in some way counteracting the effects of dsRNA.
dsRNA is formed in copious amounts during intermediate and late gene expression in VACV-infected cells (Jacobs and Langland, 1996). Despite the accumulation of dsRNA, VACV does not lead to activation of PKR or the 2′5′ OAS/RNase L pathway, and VACV is a poor inducer of type I IFN (Langland et al., 2006a, Langland et al., 2006b, Xiang et al., 2002). E3 protein is necessary for inhibition of PKR and the 2′5′ OAS/RNase L pathway and for inhibition of type I IFN gene expression. Both conserved domains are necessary for full inhibition of PKR and 2′5′ OAS (Langland and Jacobs, 2004). The mechanism by which the N-terminus of E3 inhibits PKR is at present unclear. Nonetheless, infection with VACV containing an N-terminally truncated E3 leads to activation of PKR and 2′5′ OAS at very late times post-infection (Langland and Jacobs, 2004), to eIF2α phosphorylation in infected mice (Langland and Jacobs, 2004), and activation of PKR seems to be responsible for the IFN-sensitivity of VACVE3LΔ83N in strain 129 MEFs (White and Jacobs, 2012).
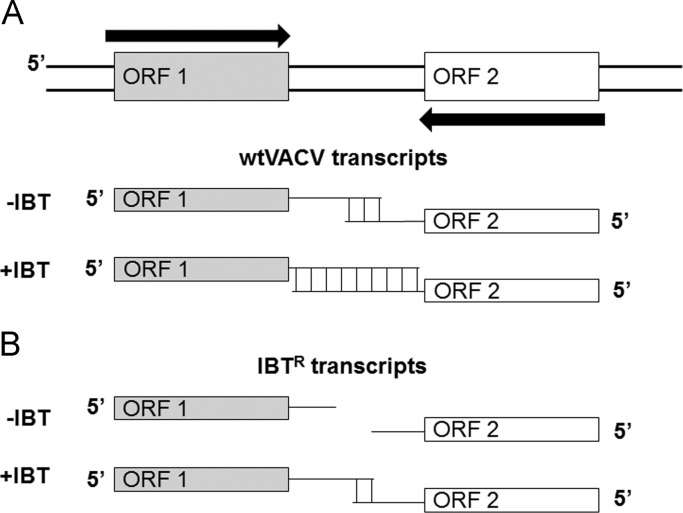
The majority of dsRNA produced during infection with VACV is a product of intermediate and late gene expression (Colby and Duesberg, 1969, Duesberg and Colby, 1969). There is no precise termination of intermediate and late transcription during VACV infection, and the result is that long run-on transcripts are formed (see Fig. 1). If these transcripts contain sequences derived from genes coded on the opposite strand of DNA, these complementary transcripts can potentially hybridize to form dsRNA. Processivity of the VACV polymerase, and thus the length of intermediate and late transcripts which can form dsRNA during VACV infection, is regulated by both positive and negative factors (Bayliss and Condit, 1993, Cresawn and Condit, 2007, Cresawn et al., 2007, Prins et al., 2004). The genetics of dsRNA formation during VACV infection has been primarily brought to light through use of the anti-poxvirus drug, IBT. IBT increases processivity of the VACV intermediate/late RNA polymerase and leads to synthesis of very long mRNAs that can form excess dsRNA (Cresawn and Condit, 2007). This excess dsRNA can lead to activation of PKR and the 2′5′ OAS/RNase L pathway despite the presence of E3 (Xiang et al., 1998). Thus, wtVACV is IBT sensitive (IBTS). Numerous IBTR and IBT-dependent (IBTD) mutants of VACV have been identified and characterized. IBTR and IBTD mutants of VACV still react to IBT by making increased amounts of longer transcripts (Cresawn and Condit, 2007). However, in the absence of IBT these mutants express a polymerase with decreased processivity, resulting in shorter intermediate/late mRNAs (Cresawn and Condit, 2007). IBT increases the length of these mRNAs, but since the polymerase has decreased processivity to start with, the resulting transcripts are not long enough to make excess dsRNA and overcome the effects of E3 in these IBT mutants. IBT-resistance maps to two subunits of the VACV polymerase, A24R and J6R, and to three positive late transcription factors, G2R, J3R and H5R (Cresawn and Condit, 2007, Cresawn et al., 2007). At restrictive temperatures, a temperature sensitive (ts) mutant of A18R, ts23, is phenotypically similar to treatment of wtVACV with IBT, suggesting that A18R is a negative regulator of elongation, which when inactivated leads to accumulation of increased amounts of long post-replicative transcripts and increased amounts of dsRNA (Pacha and Condit, 1985, Pacha et al., 1990).
Fig. 1.
dsRNA accumulation in orthopoxvirus-infected cells. (A) dsRNA is thought to accumulate from convergent transcripts. At intermediate and late times post-infection, termination of transcription is imprecise, leading to long run-on transcripts. In wild-type VACV, these transcripts can base-pair with transcripts read from the opposite strand of the genome, forming dsRNA. The anti-poxvirus drug IBT increases processivity of the viral polymerase, leading to longer transcripts and accumulation of more dsRNA. (B) IBTR mutants of VACV generate shorter transcripts with less overlap, and thus accumulate less dsRNA. IBT treatment of cells infected with IBTR mutants of VACV still increases the length of viral transcripts, but the length of these transcripts is thought to be less than the length of transcripts in IBT-treated cells infected with wtVACV.
Among the chordopoxviruses only MPXV and the leporipoxviruses do not contain full N-terminal domains in their E3 homologues. The sequence of the MPXV genome suggests that its E3L homologue, F3L, will not express the 37 N-terminal amino acids (Shchelkunov et al., 2001, Shchelkunov et al., 2002), and thus is predicted to encode a protein similar to the VACV E3L-encoded protein, p20. We have recently shown that MPXV only expresses a p20-equivalent protein (Arndt et al., 2015). The leporipox viruses, myxoma and Shope fibroma virus, are missing 89 N-terminal amino acids (Cameron et al., 1999, Willer et al., 1999). The 37 N-terminal amino acids of E3 contain hydrophobic residues involved in folding of the N-terminal domain, and a VACV peptide fragment truncated of 37 N-terminal amino acids fails to fold properly when expressed in Escherichia coli (A. Rich, personal communication). Despite containing an N-terminally truncated E3 homologue, MPXV is fully IFNR, does not lead to activation of PKR, and replicates efficiently in JC cells (Arndt et al., 2015).
Since the N-terminal domain of the VACV E3 protein has been implicated in counteracting the host response to dsRNA (White and Jacobs, 2012, Langland and Jacobs, 2004), we were interested in asking if MPXV, which codes for a partially truncated N-terminal domain, might differ from VACV, which codes for a full-length protein, in terms of dsRNA metabolism. In this manuscript, we demonstrate that MPXV infection leads to the accumulation of less dsRNA than VACV. Since accumulation of decreased amounts of dsRNA during VACV infection has been associated with resistance to the anti-poxvirus drug, IBT, we have analyzed sensitivity of MPXV to IBT. MPXV is in fact more resistant to IBT than VACV.
2. Materials and methods
2.1. Cells and viruses
HeLa (kind gift of George Pavlakis, NCI) and BSC-40 cells (ATCC) were maintained in Dulbecco's Modified-Minimal Essential Medium (DMEM; Cellgro) supplemented with 5% Fetal Bovine Serum (FBS; HyClone). JC (murine adenocarcinoma cells, ATCC) were maintained in RPMI (ATCC) supplemented with 10% FBS. RK-E3L cells [RK13 cells (ATCC) stably transfected with a plasmid expressing the E3L gene, using the Tet-Off system from Clontech] were maintained in Eagle's minimal essential medium (MEM; Cellgro) supplemented with 5% FBS. All cells were incubated at 37 °C in the presence of 5% CO2. VACV Copenhagen (VC-2) and Western Reserve (WR) strains, both designated as VACV, were used as the parental viruses for all the recombinant viruses used throughout this study. VACV containing a 37-amino-acid N-terminal truncation of E3 (VACV-E3L∆37N) was generated as previously described (Chang et al., 1995, Shors et al., 1997). Monkeypox virus strains (MPXV) 7–61 (WRAIR), US2003, and Zaire 79 (V79-I-005), were used in this study. All MPXV-Zaire experiments as well as experiments with recombinant non-Select Agent MPXV strains were performed in a biosafety level 3 laboratory (BSL-3) in accordance with protocols approved by Arizona State University, by The National Institutes of Health, and by the Centers for Disease Control and Prevention (CDC). Wild type non-Select Agent monkeypox strains (7-61 and US2003) were used in a controlled BSL-2 dedicated virus lab.
2.2. Immunofluorescent microscopy
HeLa cells were seeded on poly l-lysine-treated coverslips in 6-well dishes. HeLa cells were infected at a multiplicity of infection (MOI) of 5 with VACV, VACV-E3L∆37N, and MPXV. At 3, 6, 9, and 12 h post infection (hpi) the cells were rinsed twice with phosphate buffered saline (PBS). Subsequently, ice cold methanol was added and the cells were placed at −20 °C for 20 min. The cells were then washed twice with ice cold PBS and blocked with blocking buffer (0.3% gelatin in PBS, 0.1% triton X-100) at room temperature for 30 min. Primary antibody [mouse monoclonal, J2 anti-dsRNA (Schonborn et al., 1991)] was diluted in 0.3% gelatin, 0.1% triton x-100 in PBS (GTPBS) and incubated overnight at 4 °C. The cells were then washed five times with GTPBS for 10 min/wash. The second primary antibody (rabbit polyclonal anti-E3) was diluted in GTPBS and the procedure was repeated. Secondary antibodies [Alexa Fluor 488 and 594 (Invitrogen)] diluted in GTPBS were added and incubated for 1 h at room temperature in the dark. After incubation, the cells were washed 3 times with GTPBS for 10 min/wash, then twice with PBS. 4′6-diamino-2-phenylindole (DAPI, Invitrogen) was added at a concentration of 5 µg/mL for 15 min at room temperature in the dark. The cells were rinsed with H2O and then mounted onto slides with ProLong Gold anti-fade mounting reagent (Invitrogen). The samples were allowed to cure overnight. Samples were analyzed using the Zeiss Duo confocal microscope.
2.3. Real-time PCR
HeLa cells were infected at an MOI of 5 with VACV, VACV-E3L∆37N, and MPXV. Total RNA was extracted 1, 2, 4, 6, 8, 10, and 12 hpi using the RNeasy Mini Kit (Qiagen) according to manufacturer's instructions. cDNA was generated with 500 ng of total RNA diluted in 17.5 µL of H2O (RNase, DNase free) followed by addition of 1 µL Oligo dT (500 µg/mL, Promega) and incubation at 70 °C for 5 min. Samples were then chilled on ice and the Reverse Transcription mix [10 µL of 5X M-MLV Real Time PCR Buffer (Promega), 20 µL of 1 mM dNTPs, 1 µL of M-MLV Reverse Transcriptase (200 U/mL, Promega), 0.5 µL of RNasin® Plus RNase Inhibitor (40 U/µLPromega)] was added. The samples were incubated at 37 °C for 1 h and 95 °C for 10 min, and then placed on ice. Real Time PCR was performed with MJ Mini (BioRad) thermocycler under the following condition: 95 °C for 10 min, followed by 35 cycles of 95 °C for 15 s, 55 °C for 30 s, and 72 °C for 30 s. Two hundred ng of cDNA was added to Real Time PCR mix [12.5 µL of 2X iQ SYBR Green Supermix (BioRad) and 2.5 µL of 10 mM Forward and Reverse Real-Time primers, 5.5 µL H2O for a total volume of 25 µL]. VACV and MPXV M1L transcripts were detected with primer pairs M1L-F (5′–AAC GGA CCA CAT CCT TCT TC–3′) and M1L-R (5′–ATC CAA ACG CGT GTG ATA AA–3′). VACV and MPXV G8R transcripts were detected with primer pairs G8R-F (5′–GCG GAT CTG TAA ACA TTT GG–3′) and G8R-R (5′–CCT TGG ACA CAG GAA GAT TAA A–3′) and the MPXV specific primer, MPXV-G8R-R (5′–CCT TGG ACA CTG GAA GGT TAA A–3′). VACV A5L transcripts were detected with primer pairs A5L-F (5′–TTT CCA TCC GAT TGT TGT GT–3′) and A5L-R (5′–AGT TCA CTC CTT CCA GCG TT–3′) and MPXV A5L transcripts were detected with MPXV specific primer pairs, MPXV-A5L-F (5′–CTT CCA TCC GAT TGT TGT GT–3′) and MPXV-A5L-R (5′–AGT ACA CTC CTT CCA GCG TT–3′). For VACV and MPXV genome Real-Time PCR, 500 ng of DNA was added to the Real Time PCR mix and the same thermocycling conditions and G8R primer sets were used as described above. GAPDH was used as an internal loading control and standardization among samples using the primer pairs GAPDH-F (5′–CCT GTT CGA CAG TCA GCC G–3′) and GAPDH-R (5′–CGA CCA AAT CCG TTG ACT CC–3′).
2.4. Slot Blot for dsRNA
HeLa cells were mock-infected or infected with VACV, MPXV 7-61, or MPXV Zaire. Total RNA was extracted with the RNeasy Mini Kit, using QiaShredder homogenization and in-column DNase treatment as described by the manufacturer (Qiagen). An equal volume of total RNA (2 µL) was diluted in ddH2O (198 µL). Diluted RNA was applied through a VacuSlot VS manifold and transferred onto BrightStar-Plus Positively Charged Nylon Membrane (Ambion). The membrane was UV crosslinked with a Stratagene Stratalinker 1800 (Stratagene). Western blot analysis was performed with the J2 monoclonal anti-dsRNA antibody (Scicons). Goat anti-mouse HRP conjugate was used as the secondary antibody. Probe specificity was verified using dsRNA and ssRNA ladders (New England Biolabs) as positive and negative controls respectively. As additional controls, RNaseIII (NEB) and RnaseA (ThermoFisher) digestions were performed as described by the manufacturer; RNaseA digestion was performed with 0.5 µg/mL enzyme in 2x SSC buffer (300 mM NaCl and 30 mM Sodium Citrate).
2.5. ELISA for Detection of dsRNA
Microtiter plates were coated overnight at 4 °C with 0.4 µg Protein A per well. Free binding sites on the plates were blocked with 2% bovine serum albumen (BSA) in PBS. The plate was washed with 1X PBS and stored at 4 °C. Plates were incubated with J2 mouse monoclonal dsRNA capture antibody (1 µg/mL, IgG2a, English Scientific Consulting) in ELISA binding buffer (0.1 M Na HPO, pH 9.0) overnight at 4 °C. Plates were then blocked for 2 h at room temperature with ELISA blocking buffer (PBS, 1% BSA), followed by washing three times with ELISA wash buffer (PBS, 0.05% Tween-20). Five hundred ng of total RNA was diluted in ELISA dilution buffer (blocking buffer +0.05% Tween-20) to a final volume of 100 µL. The diluted RNA was added to the wells and allowed to incubate overnight at 4 °C. Plates were then washed 4 times with wash buffer followed by incubation for 2 h at room temperature with the K2 mouse monoclonal dsRNA detection antibody (IgM, hybridoma supernatant diluted 1:4 in blocking buffer +0.05% Tween-20, English Scientific Consulting). The plate was then washed 4 times with wash buffer, followed by incubation with anti-mouse IgM antibody (1:1000 in blocking buffer +0.05% Tween-20, Santa Cruz Biotechnology) conjugated to HRP for 30 min. The plates were subsequently washed 5 times with wash buffer followed by addition of 100 µL/well of SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific). dsRNA signal was quantified by a luminescent plate reader.
2.6. In vitro PKR kinase assay
Semi-confluent monolayers of HeLa cells were infected with VACV, or MPXV at an MOI of 5. Total RNA was isolated using the RNeasy Mini Kit (Qiagen) at 1, 6, 12, and 24 hpi, according to manufacturer's instructions. For cytoplasmic extracts of PKR, HeLa cell monolayers were pretreated with rhIFN A-D for 18 h prior to extraction and subsequently washed and scraped into ice cold PBS. The cells were pelleted by centrifugation at 1000×g for 10 min at 4 °C and resuspended in Nonidet P-40 lysis buffer [20 mM HEPES, pH 7.5, 120 mM KCl, 5 mM MgCl, 1 mM dithiothreitol (DTT), 10% (vol/vol) glycerol, 0.5% Nonidet P-40] at a volume of 100 µL per 100 mm dish of cells at 80% confluency. Nuclei and cell debris were removed by centrifugation at 10,000×g for 10 min at 4 °C. Cytoplasmic extracts were stored on ice, until needed. 500 ng of total RNA was added to in vitro kinase buffer (5 mM MgOAc, 5 mM MnCl, 20 mM HEPES, pH 7.5, 10% Glycerol, 100 mM KCl, 1 mM DTT, 1 mM Benzamidine, 100 µM ATP). A 30 µL reaction containing 10 µL of NP-40 extract and the 500 ng of total RNA was added to the in vitro kinase buffer and incubated at 30 °C for 10 min, after which 30 µL of 2X SDS-PAGE loading buffer was added. Samples were boiled and analyzed on 12% polyacrylamide gels by SDS-PAGE and Western blot for phosphorylated PKR as described above.
2.7. Western blotting
Infected or mock-infected cells were lysed with an SDS solution containing a protease/phosphatase inhibitor and β-mercaptoethanol, and proteins were harvested from a QiaShredder column (Qiagen). Harvested samples were heated at 95 °C for five minutes, then run on 10% polyacrylamide gels, followed by transfer to nitrocellulose membranes. Western blots were probed with primary antibodies overnight at 4 °C, rinsed with tris-buffered saline-Tween 20 (TBST), then probed with appropriate secondary antibodies, and washed three times with TBST. Visualization was done with Dura Western Blotting Kit (Thermo Scientific) according to the manufacturer’s instructions.
2.8. Plaque reduction assay
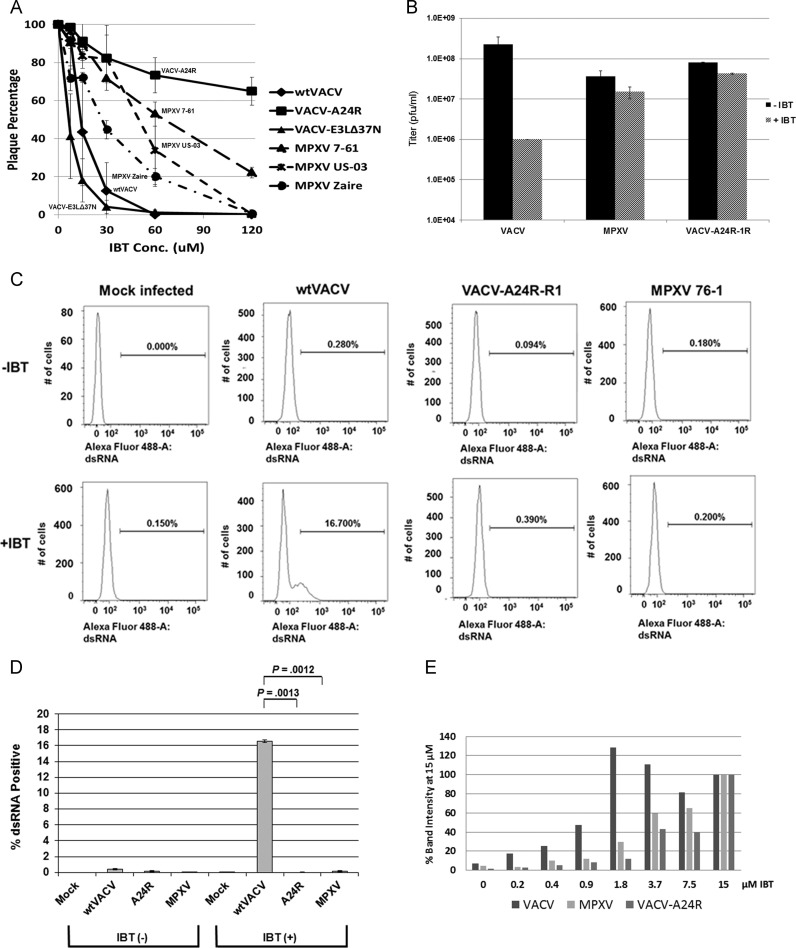
BSC-40 cells in 6-well plates were infected with ~100 pfu of virus and incubated in media containing IBT at 2-fold increasing concentrations as indicated in Fig. 8A. When plaques appeared, two to three days after infection, cells were stained with crystal violet and counted.
Fig. 8.
MPXV sensitivity to IBT. (A) MPXV sensitivity to IBT: Plaque Reduction. BSC-40 cells were infected with ~100 pfu of virus and treated with two-fold increasing concentrations of IBT. Viruses used were VACV, VACV-A24R-R1, VACV-E3LΔ37N, two West African strains of MPXV (7-61 and US-2003), and MPXV Zaire (Central African strain). Plaques were stained with crystal violet and counted. Results appear as the percentage of the number of plaques present in the untreated cells from three replicates. (B) MPXV sensitivity to IBT: Multi-cycle Growth. Multi-cycle growth kinetics assays were performed in BSC-40 cells. The cells were infected with VACV, MPXV, and VACV-A24R-R1 at an MOI of 0.01 in the presence of absence of 60 µM IBT. Viruses were harvested at 0 and 72 hpi and titered by plaque assay in RK-E3L cells. Data presented are means with standard error of multiple experiments. (C) MPXV sensitivity to IBT: Accumulation of Free dsRNA. HeLa cells were mock infected or infected with either wt VACV, VACV A24R-R1 or MPXV. Infected cells were either untreated or treated with 45 µM IBT. After 6 hpi, cells were trypsinized and fixed. Cells were stained with antibodies against total VACV and dsRNA and were subjected to flow cytometry the next day. Total cells were gated to exclude non-viable cells. (D) Quantitation of replicate results from Panel C. Statistical difference was determined by t test using Holm-Sidak method with alpha=5%. (E) MPXV sensitivity to IBT: PKR Phosphorylation. Hela cells were infected with VACV, MPXV, or VACV-A24R-R1 at an MOI of 5 and treated one hour post infection with two-fold increasing concentrations of IBT. Western blot analyses were performed with antibodies directed against the phosphorylated form of PKR and with antibodies to the viral protein E3/F3. Western blot band intensities were measured with Image Quant, normalized to GAPDH levels, and shown as a percentage of the band intensity at the highest (15 µM) level of IBT treatment.
2.9. IBT sensitivity assay
HeLa cells in 6-well plates were infected at an MOI of 5 with VACV, VACV-A24R-R1, MPXV, VACV-∆E3L, or were mock-infected. One hpi, media containing IBT was added at final concentrations of 1.8–120 µM IBT. At 9 hpi, media was removed and cells were washed with PBS and prepared as described above for Western blotting. Blots were probed with 1:1000 anti-phospho PKR (Millipore) or 1:5000 anti-E3/F3, followed by a horseradish peroxidase-conjugated secondary.
2.10. Flow cytometry analysis
HeLa cells were mock-infected or infected with either wtVACV, VACV-A24R-R1, or MPXV. Infected cells were either untreated or treated with 45 µM IBT. At 6 hpi, cells were trypsinized and fixed with CytoPerm/Fix kit (BD). Cells were stained with rabbit anti-VACV and mouse anti-dsRNA (Scicons) overnight at 4 °C. Cells were washed and stained with anti-rabbit-IgG Pacific Blue (BD) and anti-mouse-IgG 488 (Abcam) for 4 °C. Cells were washed, resuspended in FACS buffer (1% FBS, 0.9% sodium azide in PBS) and were analyzed by flow cytometry using a BD LSRFortessa Cell Analyzer. Up to 50,000 total cells were collected and then gated to exclude non-viable cells.
2.11. Bioinformatics
The VACV genes A24R, G2R, H5R, J3R, and J6R have been shown to be mutable to IBTR (Cresawn and Condit, 2007, Cresawn et al., 2007, Latner et al., 2000). Mutations in A18R can phenotypically copy IBT treatment (Bayliss and Condit, 1993). BLASTp (Altschul et al., 1997) was used to compare the wild type products of these genes to their homologues in MPXV. Table 1 lists the sequences used in this analysis. MPXV homologues were also searched for mutations known to confer IBT resistance in VACV. Strains used for analysis were VACV Western Reserve and MPXV WRAIR7-61.
Table 1.
VACV IBTR genes and MPXV homologues compared with BLASTp.
| Protein name | VACV gene | MPXV gene | VACV protein ref sequence | MPXV protein ref sequence |
|---|---|---|---|---|
| DNA helicase | A18R | WRAIR124 | YP_233020.1 | AAU01334.1 |
| DNA-dependent RNA polymerase subunit rpo132 | A24R | WRAIR170 | YP_233026.1 | AAU01340.1 |
| Late transcription elongation | G2R | WRAIR065 | YP_232962.1 | AAU01276.1 |
| VLTF-4 | H5R | WRAIR089 | YP_232985.1 | AAU01299.1 |
| Multifunctional poly-A polymerase subunit | J3R | WRAIR081 | YP_232977.1 | AAU01291.1 |
| DNA-dependent RNA polymerase subunit rpo147 | J6R | WRAIR084 | YP_232980.1 | AAU01294.1 |
VLTF-4, the product of H5R, was found to differ the most between MPXV and VACV, including a lack of several amino acids from VACV that were found in MPXV. To determine how well conserved H5R is across the orthopoxviruses, ClustalW2 (Goujon et al., 2010, Larkin et al., 2007) was used to align and compare the protein product of H5R and its homologues in several orthopoxviruses. Table 2 lists the sequences used in this analysis.
Table 2.
VLTF-4 homologues compared with ClustalW2.
| Species | Gene | Protein ref sequence |
|---|---|---|
| Vaccinia virus | H5R | YP_232985.1 |
| Monkeypox virus | WARAIR089 | AAU01299.1 |
| Variola virus | I5R | NP_042132.1 |
| Camelpox virus | CamMLVgp101 | NP_570491.1 |
| Myxoma virus | m073R | NP_571787.1 |
| Tetrapox virus | TATV_DAH68_106 | YP_717412.1 |
| Ectromelia virus | EVM087 | NP_671605.1 |
| Horsepox virus | HSPV104 | ABH08210.1 |
3. Results
3.1. MPXV produces less dsRNA than VACV in HeLa cells
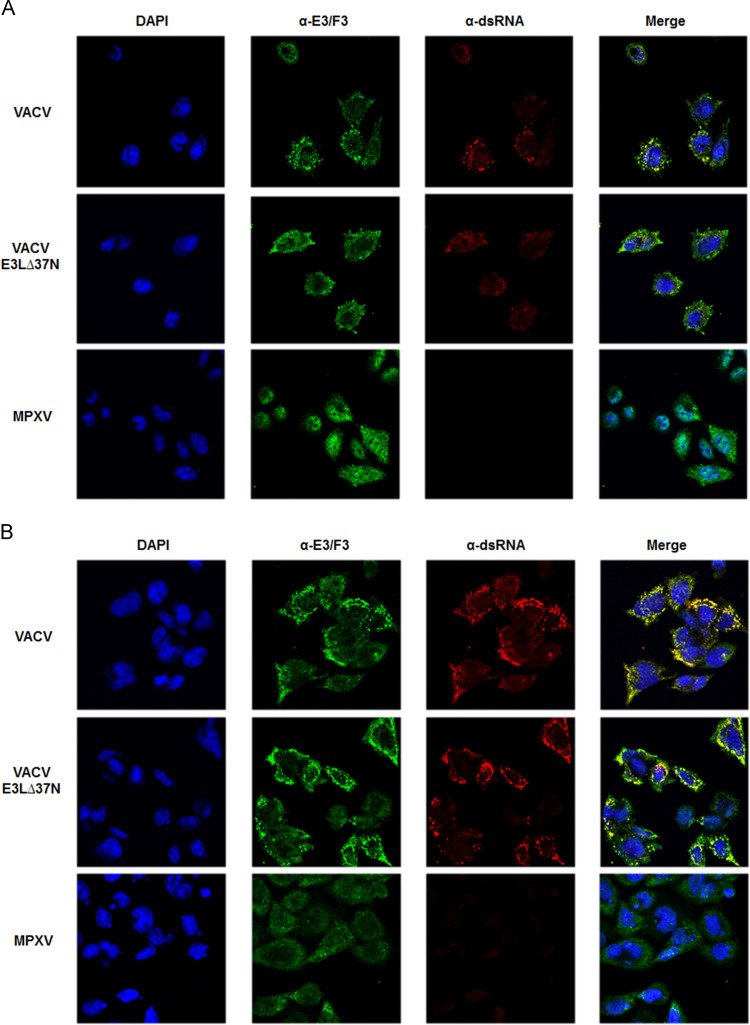
We hypothesized that there is a second viral mechanism compensating for the lack of a full length N-terminus of F3 in MPXV (Arndt et al., 2015). Since the N-terminus of E3 has been shown to be necessary for inhibition of dsRNA-dependent enzymes (Langland and Jacobs, 2004), we further hypothesized that MPXV could compensate for the lack of a full-length N-terminal domain on its E3 homologue by producing less dsRNA. Therefore, we decided to compare the accumulation of dsRNA during a MPXV and VACV infection. dsRNA accumulation in methanol-fixed HeLa cells was visualized by immunofluorescence with antibodies that specifically recognize dsRNA (Schonborn et al., 1991). We co-stained cells for E3 protein to ensure that all the cells were infected. At 3 hpi, E3 and F3 proteins were detected in VACV-, VACV-E3L∆37N-, and MPXV-cells, but no dsRNA was detected at this time (data not shown). At 6 hpi, both E3 protein and dsRNA were detected in VACV- and VACV-E3L∆37N-infected cells ( Fig. 2A). Punctate staining was observed for both E3 protein and dsRNA and the merge showed that E3 protein overlays with a significant portion of the dsRNA. This suggests that E3 protein and dsRNA may be interacting in the infected cells, most likely through E3 protein's C-terminal dsRNA binding domain (BD). Remarkably, no dsRNA staining was observed in MPXV-infected cells. The lack of dsRNA accumulation in these cells was not due to a low level of MPXV infection since F3 protein was detected in all the cells (Fig. 2A). Infected HeLa cells stained at 9 hpi showed high levels of dsRNA for both the VACV and VACV-E3L∆37N with E3 protein co-localizing with the dsRNA (Fig. 2B). Again, no accumulation of dsRNA was observed in the MPXV-infected cells at this time, despite positive staining for F3 protein (Fig. 2B).
Fig. 2.
VACV and MPXV dsRNA levels in HeLa cells at 6 hpi and 9 hpi. HeLa cells were infected at an MOI of 5 with VACV, VACV-E3L∆37N and MPXV. At 6 hpi (A) or 9 hpi (B) the cells were fixed with methanol and stained for the presence of E3 or F3 (green) and dsRNA (Red). Nuclei (blue) were stained with DAPI. Merge panels represent an overlay of all three images. Yellow indicates the co-localization of E3 or F3 and dsRNA.
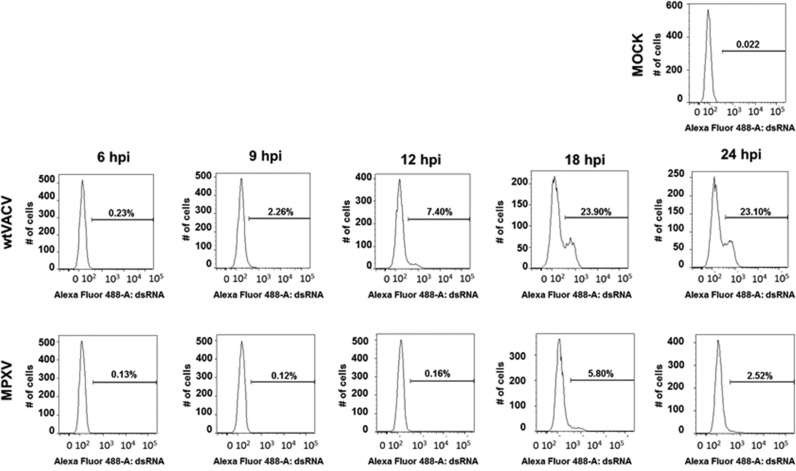
We also tested for dsRNA detectable by flow cytometry. HeLa cells were either mock-infected or infected with VACV or MPXV ( Fig. 3). At 6, 9, 12, 18, or 24 hpi cells were fixed with formaldehyde, permeabilized, and stained for dsRNA. They were then analyzed in a flow cytometer. At 9 hpi, 2% of VACV-infected cells stained positive for dsRNA. This level of MPXV- infected cells positive for dsRNA was not observed until 12 hpi. By this time, more than 20% of VACV-infected cells contain detectable dsRNA. The maximum % of MPXV-infected cells staining positive for dsRNA was 5.8%, four-fold less than the maximum for VACV-infected cells. At every time point throughout the infection, VACV had a greater percentage of cells with detectable dsRNA than MPXV.
Fig. 3.
Flow cytometry assay of dsRNA in infected cells. HeLa cells were mock infected or infected with either wtVACV or MPXV. At 6, 9, 12, 18 or 24 hpi, cells were trypsinized and fixed with CytoPerm/Fix kit (BD). Cells were stained with antibodies against dsRNA and analyzed by flow cytometry the next day. Total cells were gated to exclude non-viable cells.
While cell-based methods for detecting dsRNA likely detect pre-existing RNA, they may be confounded by the presence of dsRNA-binding proteins, which may partially mask dsRNA and interfere with detection by dsRNA-binding antibodies. In fact, quantitative differences seen between immunohistochemistry and flow cytometry detection of dsRNA-staining cells may be due to differences in the proteins bound to dsRNA following differing fixation methods (Huynh, Cotsmire, and Jacobs, unpublished observations). In order to eliminate problems associated with the presence of dsRNA-binding proteins, we used several techniques to detect the presence of dsRNA in de-proteinized RNA extracts from infected cells.
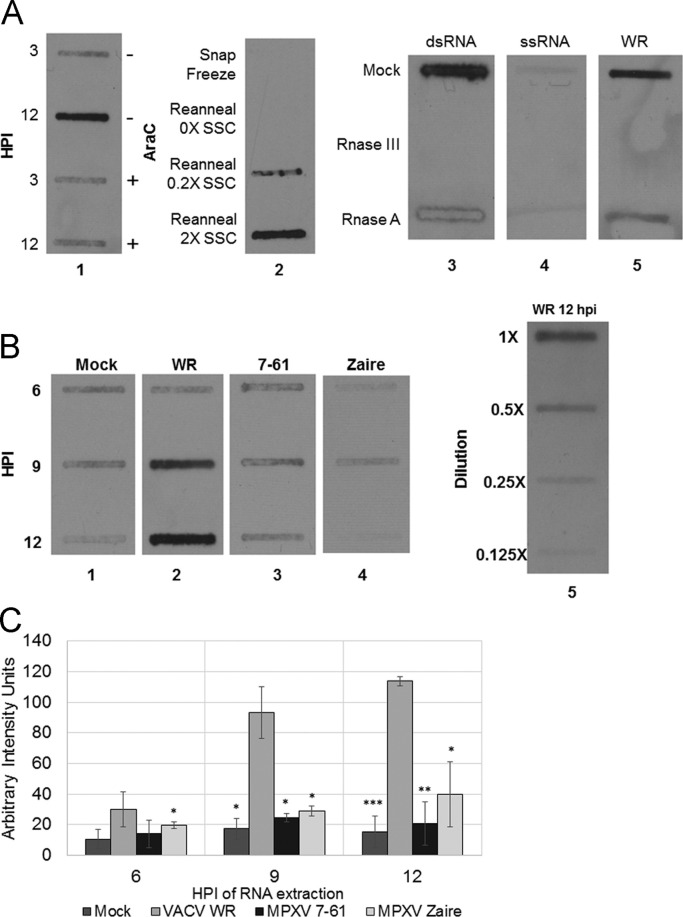
First we analyzed potential dsRNA levels with a modified slot blot using antibodies to detect dsRNA in RNA that had been isolated from infected cells. We initially performed several control assays to confirm the specificity of the procedure. Increased staining was detected at 12 hpi with VACV ( Fig. 4A, lane 1): this staining was sensitive to araC treatment, which blocks intermediate and late gene expression and blocks accumulation of viral dsRNA (Kibler et al., 1997). Denatured and snap-frozen RNA from infected cells did not give a detectable signal, but renaturation restored antibody detection (Fig. 4A, lane 2). Commercially available dsRNA gave a signal, while commercially available ssRNA did not (Fig. 4A, lanes 3 and 4). The signal from either commercially available dsRNA (Fig. 4A, lane 3) or RNA from infected cells (lane 5) was sensitive to treatment with RNase III, but relatively insensitive to treatment with RNase A. These controls demonstrate that this assay specifically recognizes dsRNA.
Fig. 4.
Slot blot detection of dsRNA in extracts from virus-infected cells. Cells were infected with virus and then total RNA was extracted at multiple time points post-infection using the RNeasy Kit (Qiagen). Equal volumes of extract were then transferred onto BrightStar®-Plus Positively Charged Nylon Membrane (Ambion) using a vacuum slot apparatus. dsRNA was visualized on the blot by probing with J2 anti-dsRNA antibodies and Goat anti-mouse HRP conjugate secondary antibody. (A) To verify the specificity of the J2 antibody in a slot blot, the following controls were performed: Lane 1: HeLa cells were mock pretreated or pretreated with 40 µg/mL AraC 1 h prior to infection with VACV WR. Total RNA was extracted at 3 and 12 hpi. Lane 2: BSC-40 cells were infected with WR and total RNA was extracted at 8 hpi. RNA was denatured by boiling at 95 °C for 5 min; samples where then either snap frozen or left at room temperature to reanneal in either no, low, or high salt conditions. Lanes 3–5: dsRNA ladder, ssRNA ladder, and WR 12 hpi-extract were either mock treated or treated with RNAse III (dsRNA specific) or RnaseA (ssRNA specific) RnaseA digestion was done in 2XSSC (300 mM NaCl and 30 mM Sodium Citrate). (B) Lanes 1–4: HeLa cells were mock infected or infected with VACV WR, MPXV 7-61, or MPXV Zaire at an MOI of 5. Total RNA was collected at 6, 9, and 12 hpi. RNA extractions were performed in triplicate; figure shows representative results. Lane 5: Serial 2-fold dilutions of the VACV WR 12 hpi-extract were included to verify that the exposure was in the linear range. (C) Band intensity of slot blots was analyzed with TotalLab Quant. The intensity of the WR 12 hpi-extraction was calibrated to 100 arbitrary units. The triplicate extraction intensities were averaged; error bars show standard deviation. Significance was calculated with unpaired 2-tailed t-test. Asterisks indicate significance of difference from corresponding VACV WR time point. *p<0.05, **p<0.01, and ***p<0.005.
HeLa cells were infected using VACV, VACV-E3L∆37N, and MPXV at an MOI of 5 and total RNA was isolated at 3, 6, 9, and 12 hpi. Isolated RNA was slot-blotted onto a charged nylon membrane and probed with antibodies to dsRNA. VACV and VACV-E3L∆37N generated detectable amounts of dsRNA at 6 hpi and dsRNA levels continued to increase through 12 hpi (Fig. 4B and C). In contrast, MPXV dsRNA was not detected until 9 hpi and increased slightly through 12 hpi; however, the total amount of dsRNA generated by MPXV appeared to be less than the levels observed for VACV and VACV-E3L∆37N (Fig. 4B and C).
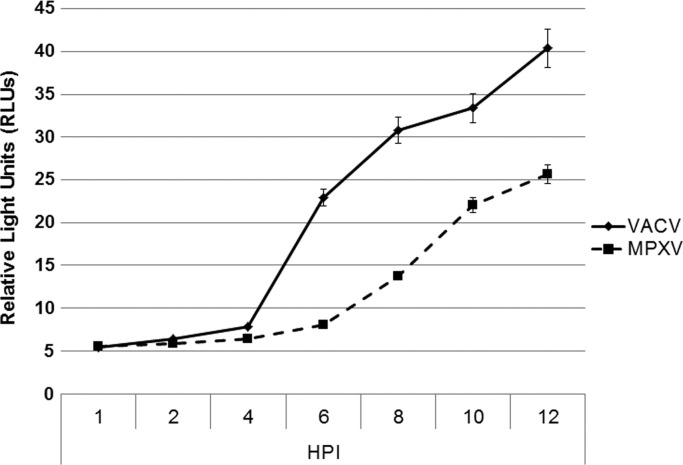
An ELISA-based assay with the J2 (IgG2a) anti-dsRNA antibody as the capture antibody and the K2 (IgM) anti-dsRNA antibody as the detection antibody was also used to determine quantitatively the amount of potential dsRNA produced during virus infection. Total RNA was extracted from HeLa cells infected with VACV and MPXV and added to an ELISA plate containing the capture antibody. MPXV dsRNA levels were an average of 40–50% lower when compared to VACV between 6 and 12 hpi ( Fig. 5).
Fig. 5.
ELISA for VACV and MPXV dsRNA levels. HeLa cells were infected with VACV and MPXV at an MOI 5. At 1, 2, 4, 6, 8, 10, and 12 hpi cells were harvested, total RNA was extracted, and 500 ng of total RNA was added to ELISA plate. Signal was quantified by Luminescent plate reader. Data presented are means with standard error of multiple experiments.
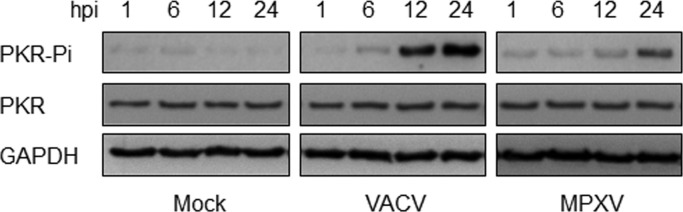
Finally, we performed an in vitro PKR assay to compare the amounts of potential dsRNA capable of activating PKR that is generated during a VACV or MPXV infection ( Fig. 6). For this assay, RNA was isolated at 1, 6, 12 and 24 hpi from mock-, VACV-, and MPXV-infected cells. Isolated RNA was incubated with cytoplasmic extracts containing unphosphorylated PKR, and ATP was added as the substrate for PKR. The amount of phosphorylated PKR was determined by Western blot with antibodies specific for the phosphorylated form of PKR. At 6 hpi, RNA from VACV-infected cells induced PKR phosphorylation. The ability of RNA isolated from VACV-infected cells to activate PKR increased through 24 hpi. At 6 hpi the levels of phosphorylated PKR observed for RNA isolated from MPXV-infected cells were similar to levels seen for mock-infected cells. MPXV RNA isolated at 12 hpi induced PKR phosphorylation, and the ability to activate PKR increased through 24 hpi. However, RNA isolated from MPXV-infected cells as late as 24 hpi was less efficient at activating PKR than RNA isolated from VACV-infected cells at 12 or 24 hpi.
Fig. 6.
Use of PKR activation to detect dsRNA in extracts from the infected cells. HeLa cells were either mock infected or infected with VACV or MPXV. RNA was extracted at 1, 6, 12, and 24 hpi. RNA was added to an in vitro PKR activation assay with IFN-treated HeLa lysate. The lysates were than analyzed by Western blot, using antiserum that recognizes PKR phosphorylation at Threonine 446.
3.2. Accumulation of viral RNA and DNA from MPXV-infected cells
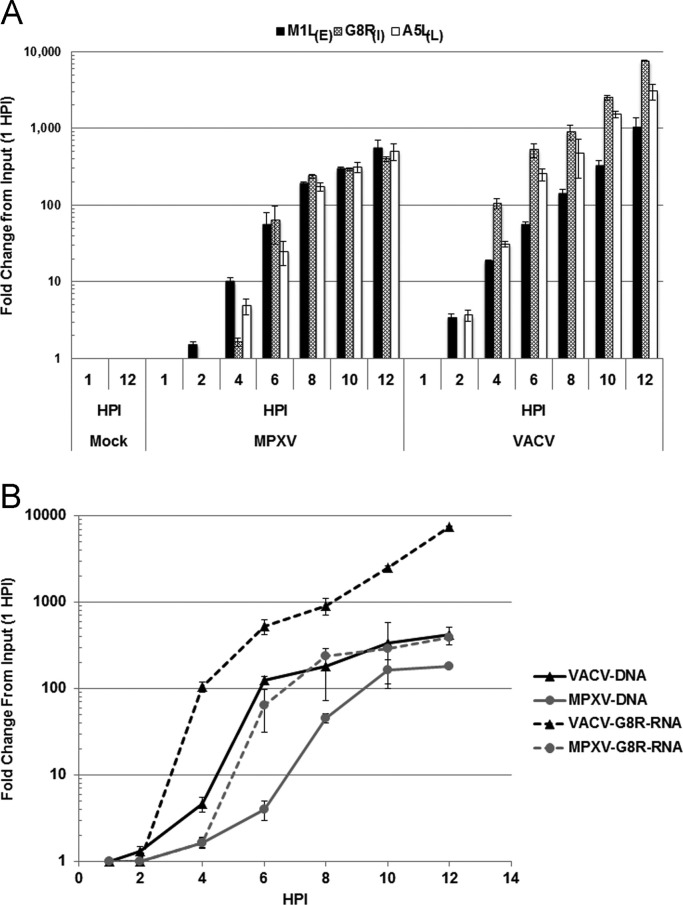
To determine if the decreased accumulation of dsRNA during MPXV infection was due to a decrease in accumulation of mRNA, we examined the production of early, intermediate, and late viral transcripts using real-time quantitative PCR with primers specific for early (M1L), intermediate (G8R), and late (A5L) viral transcripts ( Fig. 7A). The majority of dsRNA produced during a poxvirus infection is due to accumulation of intermediate and late transcripts (Duesberg and Colby, 1969). In VACV-infected cells, a considerable increase of intermediate and late viral transcripts was detected by 4 hpi and their levels continued to increase throughout the infection (Fig. 7A). Prior to 4 hpi, the levels of intermediate and late viral transcripts were relatively low, consistent with the immunofluorescence and slot blot data at 3 hpi, which did not show accumulation of detectable dsRNA. Considerable increases in MPXV intermediate and late viral transcripts were detected by 6 hpi and continued to increase throughout the infection (Fig. 7A). These data suggest that MPXV viral transcription may be delayed by two hours as compared to VACV transcription. However, this does not explain why dsRNA was not observed by immunofluorescence in the MPXV-infected cells at both 6 and 9 hpi (Fig. 2A and B), as the levels of MPXV viral transcripts at 8 hpi were similar to those levels observed for VACV at 6 hpi. At later times, VACV infection leads to higher levels of intermediate and late mRNA synthesis, which may account for the difference in accumulation of dsRNA later than 9 hpi. Since intermediate and late viral transcription occurs immediately following the initiation of genomic replication (Keck et al., 1990, Wright et al., 1991, Wright and Moss, 1989), we have also analyzed accumulation of viral genomic DNA using real-time quantitative PCR (Fig. 7B). qPCR for viral genomic DNA was performed with the G8R gene specific primer set on DNA extracted from HeLa cells infected with VACV and MPXV. VACV genomic DNA was first detectable at approximately 2 hpi and increased considerably from 4 to 6 hpi (Fig. 7B). MPXV genomic DNA was first detectable at approximately 4 hpi and increased significantly from 6 to 10 hpi. Ultimately, VACV and MPXV generated similar levels of viral genome by 10 hpi. These data suggest that the MPXV life cycle may be delayed by two hours in HeLa cells as compared to VACV. Again, this is unlikely to be the cause of the decreased accumulation of dsRNA in a MPXV-infected cell.
Fig. 7.
Accumulation of viral RNA and DNA in HeLa cells. HeLa cells were either mock infected or infected with VACV and MPXV at an MOI of 5. RNA (A) or DNA (B) was extracted at 1, 2, 4, 6, 8, 10, and 12 hpi. (A) Real-Time quantitative reverse-transcriptase PCR was performed with gene-specific primers for M1L (early), G8R (intermediate) and A5L (late) viral transcripts. (B) Real-Time quantitative PCR for viral DNA was performed with G8R-specific primers on 500 ng of total DNA.
3.3. MPXV is resistant to the anti-poxvirus drug IBT
The anti-poxvirus drug IBT has been shown to increase the amount of dsRNA present during an infection by inducing longer than normal intermediate and late viral transcripts (Prins et al., 2004). This increase in transcript length results in an increase in dsRNA accumulation, presumably from transcripts being read from opposing strands of the genome, in a convergent manner (see Fig. 1). IBTR mutants of VACV accumulate less dsRNA than wtVACV and it is suspected this is due to the generation of shorter intermediate and late transcripts in the presence or absence of IBT (Bayliss and Condit, 1993) (see Fig. 1). Since MPXV appears to be producing less dsRNA than VACV, we hypothesized that MPXV may be naturally resistant to IBT, leading to the reduced accumulation of dsRNA observed in MPXV-infected cells.
Fig. 8A shows results of plaque reduction assays with multiple viruses as a function of IBT concentration. Plaque formation with both wtVACV and VACV-E3LΔ37N were highly sensitive to IBT treatment, with an IC50 of less than 15 µM. Plaque formation by VACV-A24R-R1, a known IBTR virus (kindly provided by Dr. Rich Condit), was highly resistant to IBT treatment with an IC50 greater than 120 µM. Three different strains of MPXV all showed intermediate sensitivity to IBT, with IC50s of 20–60 µM.
We also performed multi-step growth kinetics with VACV, MPXV, and VACV-A24R-R1 in BSC-40 cells in the presence of 60 µM IBT (Fig. 8B). In the absence of IBT, VACV, MPXV, and VACV-A24R-R1 replicated to similar titers; however, in the presence of IBT, VACV replication was reduced by two logs whereas replication of MPXV and VACV-A24R-R1 was relatively unaffected (Fig. 8B). Together, these results demonstrate that MPXV has an IBTR phenotype.
Accumulation of detectable dsRNA after IBT treatment was determined by flow cytometry analysis (Fig. 8C and D). At 6 hpi, few of the infected cells had accumulated dsRNA that was detectable by flow cytometry. Treatment with 45 µM IBT led to the accumulation of detectable dsRNA in 10–20% of cells infected with wtVACV. Less than 1% dsRNA+ cells were detected after IBT treatment of VACV-A24R-R1- or MPXV-infected cells.
To determine if treatment with IBT of cells infected with these viruses led to the dsRNA-dependent activation of anti-viral pathways we monitored phosphorylation of PKR. Western blot analyses on protein lysates isolated at 9 hpi from HeLa cells infected with VACV, MPXV, and VACV-A24R-R1, treated with increasing amounts of IBT show an increase in phosphorylated PKR for both VACV and MPXV (Fig. 8E). Phosphorylated PKR was detected at low levels of IBT for VACV in treated cells (0.2 µM), and peaked at 1.8 µM IBT. Phosphorylated PKR was not detected in VACV-A24R-R1-infected cells until much higher levels (3.7 µM) and peaked at 15 µM IBT. Phosphorylated PKR was first detected at intermediate levels of IBT in MPXV-infected cells (1.8 µM) and peaked at 15 µM IBT. This is consistent with the relative sensitivities of these viruses to IBT.
4. Discussion
In this manuscript we demonstrate that less dsRNA accumulates during MPXV infection compared to infection with VACV. While the MPXV life cycle appeared to be delayed by 2 h compared to the VACV life cycle, MPXV produced less dsRNA at relative time points. Additionally, the decreased accumulation of dsRNA did not correlate with a decreased accumulation of late transcripts. However, the accumulation of less dsRNA was associated with partial resistance to the anti-poxviral drug IBT. Thus, MPXV appears to be naturally resistant to IBT. It is possible that the decreased accumulation of dsRNA is compensating for the lack of a full length E3 homologue in MPXV.
Poxviruses, despite containing a dsDNA genome, are notorious for inducing the synthesis of dsRNA (Colby and Duesberg, 1969, Duesberg and Colby, 1969). The majority of the dsRNA induced during VACV infection is thought to be the product of symmetrical transcription from intermediate and late genes read from opposite strands of the viral genome (Langland et al., 2006a), although early dsRNA has been detected in cells infected with virus deleted of the host range gene, K1L (Willis et al., 2011). The accumulation of post-replicative dsRNA is not due to overlapping genes, but to imprecise termination of transcription for at least some transcripts (Colby and Duesberg, 1969, Duesberg and Colby, 1969).
Regulation of the size of post-replicative transcripts, and thus of accumulation of dsRNA, has been studied extensively during infection with VACV. The anti-poxvirus drug, IBT, has been shown to increase the length of post-replicative transcripts (Cresawn and Condit, 2007, Cresawn et al., 2007), leading to accumulation of excess dsRNA, and the activation of PKR and the 2′5′ OAS/RNase L pathway (Xiang et al., 1998). IBTR and IBTD mutants of VACV have been isolated and have been shown to lead to the accumulation of smaller post-replicative transcripts in the absence of IBT treatment (Cresawn and Condit, 2007, Cresawn et al., 2007). IBT treatment increases the size of transcripts generated from IBTR and IBTD mutants of VACV (Cresawn and Condit, 2007, Cresawn et al., 2007), but presumably not to the point of leading to the accumulation of excess dsRNA. IBTR and IBTD mutants map to three positive regulators of elongation, G2R, J3R, and H5R, and to two subunits of the viral DNA-dependent RNA polymerase, J6R and A24R (Cresawn and Condit, 2007, Cresawn et al., 2007). A temperature sensitive mutant of A18R is a phenocopy of IBT treatment, suggesting that A18R is a negative regulator of elongation, and that loss of function of A18R leads to longer post-replicative transcripts and accumulation of excess dsRNA (Pacha and Condit, 1985, Pacha et al., 1990).
For four of the five genes that are known to harbor IBT mutations in VACV, G2R, J3R, J6R and A24R, the MPXV homologues are 98–99% identical at the amino acid level, and do not contain polymorphisms at the known sites leading to IBTR. A18R is also very similar between VACV and MPXV, with 97% amino acid identity. Only the product of H5R, VLTF-4, differs appreciably between VACV and MPXV, with an amino acid identity of only 89%. VACV VLTF-4 is smaller than MPXV VLTF-4. Alignment of VLTF-4 homologues of multiple orthopoxviruses, including VACV, MPXV, variola virus, and camelpox virus indicates that VACV has accumulated two unique deletions relative to the other orthopoxviruses. It is at present unclear if the differences in sensitivity to IBT between VACV and MPXV are due to changes in H5R, as yet uncharacterized polymorphisms in the other candidate IBT genes, or to polymorphisms in other as yet uncharacterized genes. Mapping of the IBT genes in MPXV will be needed to answer these questions.
It is tempting to speculate that the accumulation of decreased amounts of dsRNA is compensating for the lack of a full-length N-terminal domain on the E3 homologue of MPXV (F3). The N-terminal domain of VACV E3 is required for full inhibition of PKR activation in both infected cells in culture and in mice (Langland and Jacobs, 2004). The N-terminal domain is also required to inhibit the 2′5′ OAS/RNase L pathway in some infected cells (Langland and Jacobs, 2004). Since both pathways are activated by dsRNA, it is likely that the N-terminal domain is required to fully counteract the effects of dsRNA in infected cells. In HeLa cells, the N-terminus is only needed to inhibit PKR activation at very late times post-infection (Langland and Jacobs, 2004), times at which high levels of dsRNA accumulate in infected cells. Thus, it is possible that accumulation of decreased amounts of dsRNA could abrogate the need for a full length N-terminus at very late times post-infection. Determining if the IBT alleles of MPXV rescue the phenotypes associated with lack of a full length N-terminal domain on E3 will provide support for this hypothesis.
In conclusion, in this manuscript we demonstrate that MPXV leads to accumulation of less dsRNA than VACV and is more resistant to the anti-poxvirus drug IBT than VACV. This, along with the partial truncation of the dsRNA-binding F3 protein in MPXV, suggests that the paradigm for synthesis and modulation of the effects of dsRNA that has arisen from studies of VACV is likely not universal for all orthopoxviruses.
Acknowledgments
The authors like to thank Connie Chamberlain and Nobuko Fukushima for excellent technical assistance, and Dr. Richard Condit for provision of IBT viruses and for helpful discussions.
The work described in this manuscript was supported by NIH grant R01 AI095394.
References
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt W.D., Cotsmire S., Trainor K., Harrington H., Hauns K., Kibler K.V., Huynh T.P., Jacobs B.L. Evasion of the innate immune type I interferon system by monkeypox virus. J. Virol. 2015;89:10489–10499. doi: 10.1128/JVI.00304-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss C.D., Condit R.C. Temperature-sensitive mutants in the vaccinia virus A18R gene increase double-stranded RNA synthesis as a result of aberrant viral transcription. Virology. 1993;194:254–262. doi: 10.1006/viro.1993.1256. [DOI] [PubMed] [Google Scholar]
- Brandt T., Heck M.C., Vijaysri S., Jentarra G.M., Cameron J.M., Jacobs B.L. The N-terminal domain of the vaccinia virus E3L-protein is required for neurovirulence, but not induction of a protective immune response. Virology. 2005;333:263–270. doi: 10.1016/j.virol.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Brandt T.A., Jacobs B.L. Both carboxy- and amino-terminal domains of the vaccinia virus interferon resistance gene, E3L, are required for pathogenesis in a mouse model. J. Virol. 2001;75:850–856. doi: 10.1128/JVI.75.2.850-856.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron C., Hota-Mitchell S., Chen L., Barrett J., Cao J.X., Macaulay C., Willer D., Evans D., McFadden G. The complete DNA sequence of myxoma virus. Virology. 1999;264:298–318. doi: 10.1006/viro.1999.0001. [DOI] [PubMed] [Google Scholar]
- Chang H.W., Jacobs B.L. Identification of a conserved motif that is necessary for binding of the vaccinia virus E3L gene products to double-stranded RNA. Virology. 1993;194:537–547. doi: 10.1006/viro.1993.1292. [DOI] [PubMed] [Google Scholar]
- Chang H.W., Uribe L.H., Jacobs B.L. Rescue of vaccinia virus lacking the E3L gene by mutants of E3L. J. Virol. 1995;69:6605–6608. doi: 10.1128/jvi.69.10.6605-6608.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.W., Watson J.C., Jacobs B.L. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc. Natl. Acad. Sci. USA. 1992;89:4825–4829. doi: 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby C., Duesberg P.H. Double-stranded RNA in vaccinia virus infected cells. Nature. 1969;222:940–944. doi: 10.1038/222940a0. [DOI] [PubMed] [Google Scholar]
- Cresawn S.G., Condit R.C. A targeted approach to identification of vaccinia virus postreplicative transcription elongation factors: genetic evidence for a role of the H5R gene in vaccinia transcription. Virology. 2007;363:333–341. doi: 10.1016/j.virol.2007.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresawn S.G., Prins C., Latner D.R., Condit R.C. Mapping and phenotypic analysis of spontaneous isatin-beta-thiosemicarbazone resistant mutants of vaccinia virus. Virology. 2007;363:319–332. doi: 10.1016/j.virol.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesberg P.H., Colby C. On the biosynthesis and structure of double-stranded RNA in vaccinia virus-infected cells. Proc. Natl. Acad. Sci. USA. 1969;64:396–403. doi: 10.1073/pnas.64.1.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon M., McWilliam H., Li W., Valentin F., Squizzato S., Paern J., Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs B.L., Langland J.O. When two strands are better than one: the mediators and modulators of the cellular responses to double-stranded RNA. Virology. 1996;219:339–349. doi: 10.1006/viro.1996.0259. [DOI] [PubMed] [Google Scholar]
- Keck J.G., Baldick C.J., Jr., Moss B. Role of DNA replication in vaccinia virus gene expression: a naked template is required for transcription of three late trans-activator genes. Cell. 1990;61:801–809. doi: 10.1016/0092-8674(90)90190-p. [DOI] [PubMed] [Google Scholar]
- Kibler K.V., Shors T., Perkins K.B., Zeman C.C., Banaszak M.P., Biesterfeldt J., Langland J.O., Jacobs B.L. Double-stranded RNA is a trigger for apoptosis in vaccinia virus-infected cells. J. Virol. 1997;71:1992–2003. doi: 10.1128/jvi.71.3.1992-2003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langland J.O., Jacobs B.L. Inhibition of PKR by vaccinia virus: role of the N- and C-terminal domains of E3L. Virology. 2004;324:419–429. doi: 10.1016/j.virol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Langland J.O., Cameron J.M., Heck M.C., Jancovich J.K., Jacobs B.L. Inhibition of PKR by RNA and DNA viruses. Virus Res. 2006;119:100–110. doi: 10.1016/j.virusres.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Langland J.O., Kash J.C., Carter V., Thomas M.J., Katze M.G., Jacobs B.L. Suppression of proinflammatory signal transduction and gene expression by the dual nucleic acid binding domains of the vaccinia virus E3L proteins. J. Virol. 2006;80:10083–10095. doi: 10.1128/JVI.00607-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Latner D.R., Xiang Y., Lewis J.I., Condit J., Condit R.C. The vaccinia virus bifunctional gene J3 (nucleoside-2′-O-)-methyltransferase and poly(A) polymerase stimulatory factor is implicated as a positive transcription elongation factor by two genetic approaches. Virology. 2000;269:345–355. doi: 10.1006/viro.2000.0243. [DOI] [PubMed] [Google Scholar]
- Pacha R.F., Condit R.C. Characterization of a temperature-sensitive mutant of vaccinia virus reveals a novel function that prevents virus-induced breakdown of RNA. J. Virol. 1985;56:395–403. doi: 10.1128/jvi.56.2.395-403.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacha R.F., Meis R.J., Condit R.C. Structure and expression of the vaccinia virus gene which prevents virus-induced breakdown of RNA. J. Virol. 1990;64:3853–3863. doi: 10.1128/jvi.64.8.3853-3863.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins C., Cresawn S.G., Condit R.C. An isatin-beta-thiosemicarbazone-resistant vaccinia virus containing a mutation in the second largest subunit of the viral RNA polymerase is defective in transcription elongation. J. Biol. Chem. 2004;279:44858–44871. doi: 10.1074/jbc.M408167200. [DOI] [PubMed] [Google Scholar]
- Schonborn J., Oberstrass J., Breyel E., Tittgen J., Schumacher J., Lukacs N. Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res. 1991;19:2993–3000. doi: 10.1093/nar/19.11.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkevich T.G., Koonin E.V., Bugert J.J., Darai G., Moss B. The genome of molluscum contagiosum virus: analysis and comparison with other poxviruses. Virology. 1997;233:19–42. doi: 10.1006/viro.1997.8607. [DOI] [PubMed] [Google Scholar]
- Shchelkunov S.N., Totmenin A.V., Babkin I.V., Safronov P.F., Ryazankina O.I., Petrov N.A., Gutorov V.V., Uvarova E.A., Mikheev M.V., Sisler J.R., Esposito J.J., Jahrling P.B., Moss B., Sandakhchiev L.S. Human monkeypox and smallpox viruses: genomic comparison. FEBS Lett. 2001;509:66–70. doi: 10.1016/S0014-5793(01)03144-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shchelkunov S.N., Totmenin A.V., Safronov P.F., Mikheev M.V., Gutorov V.V., Ryazankina O.I., Petrov N.A., Babkin I.V., Uvarova E.A., Sandakhchiev L.S., Sisler J.R., Esposito J.J., Damon I.K., Jahrling P.B., Moss B. Analysis of the monkeypox virus genome. Virology. 2002;297(2):172–194. doi: 10.1006/viro.2002.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shors T., Kibler K.V., Perkins K.B., Seidler-Wulff R., Banaszak M.P., Jacobs B.L. Complementation of vaccinia virus deleted of the E3L gene by mutants of E3L. Virology. 1997;239:269–276. doi: 10.1006/viro.1997.8881. [DOI] [PubMed] [Google Scholar]
- Tulman E.R., Afonso C.L., Lu Z., Zsak L., Kutish G.F., Rock D.L. The genome of canarypox virus. J. Virol. 2004;78:353–366. doi: 10.1128/JVI.78.1.353-366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S.D., Jacobs B.L. The amino terminus of the vaccinia virus E3 protein is necessary to inhibit the interferon response. J. Virol. 2012;86:5895–5904. doi: 10.1128/JVI.06889-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer D.O., McFadden G., Evans D.H. The complete genome sequence of shope (rabbit) fibroma virus. Virology. 1999;264:319–343. doi: 10.1006/viro.1999.0002. [DOI] [PubMed] [Google Scholar]
- Willis K.L., Langland J.O., Shisler J.L. Viral double-stranded RNAs from vaccinia virus early or intermediate gene transcripts possess PKR activating function, resulting in NF-kappaB activation, when the K1 protein is absent or mutated. J. Biol. Chem. 2011;286:7765–7778. doi: 10.1074/jbc.M110.194704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright C.F., Keck J.G., Tsai M.M., Moss B. A transcription factor for expression of vaccinia virus late genes is encoded by an intermediate gene. J. Virol. 1991;65:3715–3720. doi: 10.1128/jvi.65.7.3715-3720.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright C.F., Moss B. Identification of factors specific for transcription of the late class of vaccinia virus genes. J. Virol. 1989;63:4224–4233. doi: 10.1128/jvi.63.10.4224-4233.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Condit R.C., Vijaysri S., Jacobs B., Williams B.R., Silverman R.H. Blockade of interferon induction and action by the E3L double-stranded RNA binding proteins of vaccinia virus. J. Virol. 2002;76:5251–5259. doi: 10.1128/JVI.76.10.5251-5259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Simpson D.A., Spiegel J., Zhou A., Silverman R.H., Condit R.C. The vaccinia virus A18R DNA helicase is a postreplicative negative transcription elongation factor. J. Virol. 1998;72:7012–7023. doi: 10.1128/jvi.72.9.7012-7023.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuwen H., Cox J.H., Yewdell J.W., Bennink J.R., Moss B. Nuclear localization of a double-stranded RNA-binding protein encoded by the vaccinia virus E3L gene. Virology. 1993;195:732–744. doi: 10.1006/viro.1993.1424. [DOI] [PubMed] [Google Scholar]