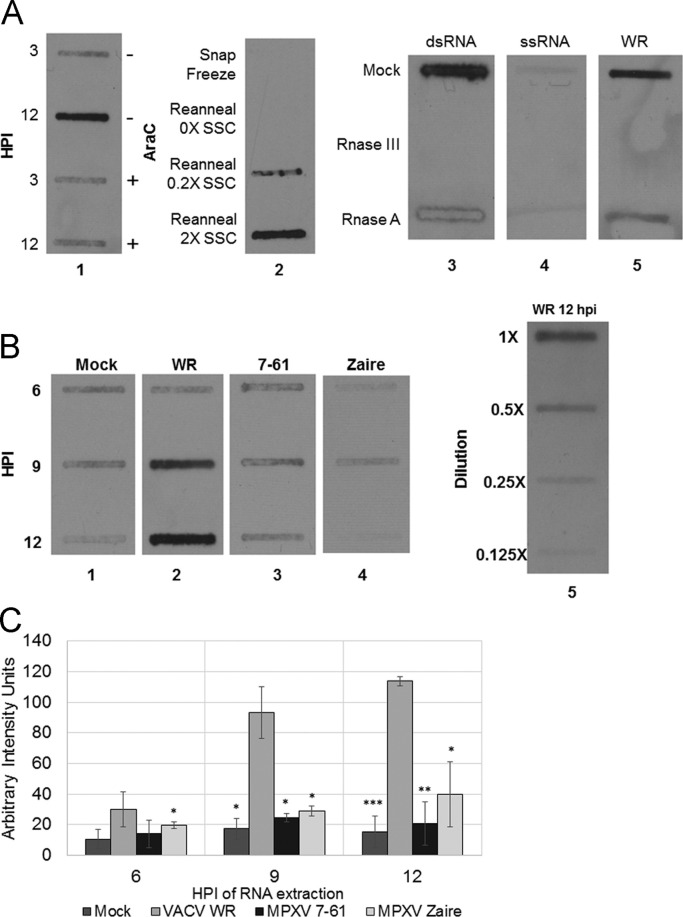
Fig. 4.
Slot blot detection of dsRNA in extracts from virus-infected cells. Cells were infected with virus and then total RNA was extracted at multiple time points post-infection using the RNeasy Kit (Qiagen). Equal volumes of extract were then transferred onto BrightStar®-Plus Positively Charged Nylon Membrane (Ambion) using a vacuum slot apparatus. dsRNA was visualized on the blot by probing with J2 anti-dsRNA antibodies and Goat anti-mouse HRP conjugate secondary antibody. (A) To verify the specificity of the J2 antibody in a slot blot, the following controls were performed: Lane 1: HeLa cells were mock pretreated or pretreated with 40 µg/mL AraC 1 h prior to infection with VACV WR. Total RNA was extracted at 3 and 12 hpi. Lane 2: BSC-40 cells were infected with WR and total RNA was extracted at 8 hpi. RNA was denatured by boiling at 95 °C for 5 min; samples where then either snap frozen or left at room temperature to reanneal in either no, low, or high salt conditions. Lanes 3–5: dsRNA ladder, ssRNA ladder, and WR 12 hpi-extract were either mock treated or treated with RNAse III (dsRNA specific) or RnaseA (ssRNA specific) RnaseA digestion was done in 2XSSC (300 mM NaCl and 30 mM Sodium Citrate). (B) Lanes 1–4: HeLa cells were mock infected or infected with VACV WR, MPXV 7-61, or MPXV Zaire at an MOI of 5. Total RNA was collected at 6, 9, and 12 hpi. RNA extractions were performed in triplicate; figure shows representative results. Lane 5: Serial 2-fold dilutions of the VACV WR 12 hpi-extract were included to verify that the exposure was in the linear range. (C) Band intensity of slot blots was analyzed with TotalLab Quant. The intensity of the WR 12 hpi-extraction was calibrated to 100 arbitrary units. The triplicate extraction intensities were averaged; error bars show standard deviation. Significance was calculated with unpaired 2-tailed t-test. Asterisks indicate significance of difference from corresponding VACV WR time point. *p<0.05, **p<0.01, and ***p<0.005.