Abstract
Stromal fibrosis is a prominent histological characteristic of pancreatic ductal adenocarcinoma (PDAC), but how stromal fibroblasts are regulated in the tumor microenvironment (TME) to support tumor growth is largely unknown. Here we show that PDAC cells can induce DNA methylation in cancer associated fibroblasts (CAF). Upon direct contact with PDAC cells, DNA methylation of SOCS1 and other genes is induced in mesenchymal stem cells (MSC) or in CAF that lack SOCS1 methylation at baseline. Silencing or decitabine treatment to block the DNA methylation enzyme DNMT1 inhibited methylation of SOCS1. In contrast, SOCS1 gene methylation and downregulation in CAF activated STAT3 and induced IGF-1 expression to support PDAC cell growth. Moreover, CAF facilitated methylation-dependent growth of PDAC tumor xenografts in mice. The ability of patient-derived CAF with SOCS1 methylation to promote PDAC growth was more robust than CAF without SOCS1 methylation. Overall, our results reveal how PDAC cells can reprogram CAF to modify tumor-stromal interactions in the tumor microenvironment which promote malignant growth and progression.
Keywords: Pancreatic ductal adenocarcinoma, Tumor microenvironment, Cancer associated fibroblasts, DNA Methylation, SOCS1
INTRODUCTION
The tumor microenvironment(TME) plays a complex role in supporting cancer initiation, progression, and metastasis(1–3). Pancreatic ductal Adenocarcinoma(PDAC) with its characteristic fibrotic stroma compartment that occupies the vast majority of the tumor mass (4, 5) is an ideal disease model for the study of tumor-stroma interaction.
Accumulated evidence has suggested that the malignant tumor stroma is a major obstacle to effective treatments of pancreatic cancer(6–9). However, experiments targeting stromal fibroblasts in mouse models of PDAC showed controversial results(10, 11), and highlighted the importance of investigating the specific molecular mechanisms on how stromal fibroblasts are regulated in the TME as a result of interaction with tumor cells. Particularly when tumor formation is initiated or a metastatic tumor cell is homing to a distant organ, tumor cells need to change their surrounding environment, which could otherwise be naturally hostile to them. Although genetic alterations have been reported in CAFs from breast and ovarian cancers(12, 13), such genetic mutations are extremely rare. It is more likely that the phenotypes of CAFs are regulated at the epigenetic level, as previously suggested by a genome-wide analysis of breast cancer stromal cells(14). By contrast, epigenetic regulation such as DNA methylation can be dynamically modulated through DNA methylation enzymes. Epigenetic regulation has been conceived to provide a dynamic and reversible modulation of stromal cells(15, 16) such as CAFs within the tumor microenvironment. This epigenetic footprint, particularly in the form of DNA methylation, can be stably passed through cell generations in high fidelity. Thus, through epigenetic regulation, the tumor cell, depending on its need, can create either a stable or dynamic TME that expresses growth factors to support the tumor growth.
In this study, we show that PDAC tumor cells are able to induce DNA methylation in CAFs. Tumor-induced DNA methylation in CAFs subsequently promotes the growth of PDAC xenografts in mice. This study thus reveals a novel mechanism that mediates the reprogramming of TME for promoting tumor growth.
Materials and Methods
Tissue specimens and cultures
Primary cultures of PDAC tumor cells, including 3.27T, 1.30T, 3.30T, 9.05T, 3.29T and 7.07T, and CAFs, including 2.15F, 10.29F, 5.101F, 2.01F, 7.02F, 10.09F, 9.07F, 9.28F, 5.10F, 3.16F, 7.09F, 3.05F, 3.27F, 1.30F, 3.30F, 9.05F, 2.29F, 7.12F, 1.23F and 7.21F, were established from banked, surgically resected PDAC specimens between 2008 and 2014 and the Panc10.05 cell line was established in 1998 in accordance with the Johns Hopkins Medical Institution Institutional Review Board(JHMI IRB)-approved protocols and authenticated by DNA and gene expression profiling as previously described(17,18). These cell cultures were maintained in RPMI1640(Life Technology) containing 10% fetal bovine serum(FBS) as described previously(19). Human mesenchymal stem cells(MSCs) obtained from Texas A&M Health Science Center (http://medicine.tamhsc.edu/irm/msc-distribution.html) were cultured in Alpha Modified Eagle Medium media containing 10% premium selected FBS(Atlanta Biologicals) and passaged for fewer than 6 months after resuscitation in 2013 and authenticated by DNA and gene expression profiling. Human colon adenocarcinoma cell lines including SW620 obtained from the American Type Culture Collection were cultured in Leibovitz's L-15 media containing 10% FBS and passaged for fewer than 6 months after resuscitation in 2014 and authenticated by DNA profiling. Archived, formalin fixed paraffin embedded, surgically resected pancreatic specimens from patients with PDAC were obtained in accordance with the JHMI IRB approved protocols and were used for microdissection and immunohistochemistry.
5-Aza-2’-deoxycytidine(Decitabine; DAC) treatment
Cells were plated at 2×105 cells per T75 flask and treated with 1 µmol/L DAC(Sigma Aldrich) on the following day for 4 days, with changes of media and drug every 24 hours(20). Cells were harvested at the end of the 4-day treatment course for co-culture or xenograft experiment.
Bisulfite conversion, methylation specific PCR and MethySYBR PCR
Genomic DNA extraction from microdissected tissues or Dynabeads bound cells was performed by using QIAamp DNA micro kit(Qiagen). DNA from tumor or CAF cultures was extracted by Blood and Tissue DNeasy kit(Qiagen). Extracted DNA was bisulfite-modified with EZ DNA methylation Kit(Zymo Research) which converted all unmethylated cytosines to uracils while leaving methylcytosines unaltered. Bisulfite-converted DNA was amplified by methylation specific PCR(MSP) as previous described(21). The quantity of target methylation was determined by MethySYBR, a SYBR green fluorescence based real quantitative PCR as described previously(22,23). DNA from DNA methyltransferases DNMT1(−/−) and DNMT3b(−/−) double knock out cell line(DKO) was used as negative control(24). In MethySYBR, the Alu-based control PCR reaction was used to normalize the methylated products(25). The methylation ratio was calculated as (SOCS1 MSP in tested sample)/ (ALU PCR in tested sample) / (SOCS1 MSP in IVD)/(ALU PCR in IVD).
Tumor conditioned medium(TCM)
TCM was obtained by passing PDAC culture supernatant through a Corning sterile 50 mL filtration system with a 0.22 µm polyethersulfone membrane. Ten times concentrated TCM was obtained by centrifuging TCM at 3,500g in Centricon® Plus-70 Centrifugal Filter Units (EMD) for 30 min.
Fibroblast Cell Isolation
Fibroblast cells (CAFs or MSCs) and tumor cells were plated at a ratio of 1:3. After co-culturing for 24h, fibroblast cell isolation was performed using the Cellection Biotin Binder Kit (Life Technology, Carlsbad, CA, USA) and the sheep anti-human FAP biotinylated affinity purified antibody (R&D Systems Inc. Minneapolis, MN, USA) according to the Direct Technique isolation instructions.
Quantitative real time RT-PCR
Total RNA was extracted from cells by RNeasy Micro Kit(Qiagen). cDNA was synthesized by Superscript III First Strand Synthesis Supermix Kit(Life Technology). Quantitative real-time RT-PCR(qPCR) was performed on the StepOnePlus Real Time PCR System(Life Technology) and analyzed by the Stepone software V2.1. The expression of SOCS1 was quantified by the Taqman probe system(Life Technology). The expression of DNMT1, DNMT3a, DNMT3b and IGF-1 was measured by SYBR Green-based qPCR. Information on primers and probes is provided in Table S1.
RNA interference
shRNA experiments were performed with the lentivirus encoding validated short hairpin RNA directed against SOCS1(Thermo Scientific), DNMT1(OriGene Technologies, Rockville, MD, USA) or corresponding controls, as previously described(19). Transfection of MSCs with miR-29b mimic (hsa-miR-29b-3p; Ambion) or the control microRNA (negative control #1; Ambion) was performed according to the manufacturer’s instructions.
In vivo tumorigenesis assays
NOD-SCID mouse strains were maintained and subjected to the experiments at 8 weeks old in accordance with the protocols approved by the Johns Hopkins Animal Care and Use Committee. For DAC treatment, mice were treated by intraperitoneal injection of 1 mg/g DAC of body weight for 5 days. Panc10.05Tumor cells(2.5×105) were injected subcutaneously, alone or mixed with 5×105 MSCs. Tumor growth was monitored daily until tumor was palpable. Tumor free survival was measured from the day of tumor inoculation to the day when tumor long axes was less than 2mm. Then, tumor diameter was measured with calipers twice a week. Mice were euthanized at week 7 following tumor inoculation. The long(L) and short(S) axes of each tumor were measured on harvested tumors with calipers. Tumor volume(V) was calculated as V = (L × S2)/2.
DNA methylation and gene expression microarray analysis
Merged analysis of DNA methylation array and gene expression array was performed on the Illumina Infinium Methylation450K Beadarray (including CpG islands in both promoter regions and gene bodies) and the Illumina H-12 Expression Beadarray, respectively. Genes identified to be both more methylated and downregulated in MSCs or 09.05CAFs were subjected to the gene clustering analysis with a web based tool, CROC (http://metagenomics.uv.es/CROC/)(26).
Statistical analysis
Statistical analyses as indicated were performed using SPSS and GraphPad Prism software. The quantitative data were presented as the means ± standard error of the mean(SEM). A p values <0.05 was considered to be statistical significance.
RESULTS
SOCS1 is frequently methylated in pancreatic CAFs
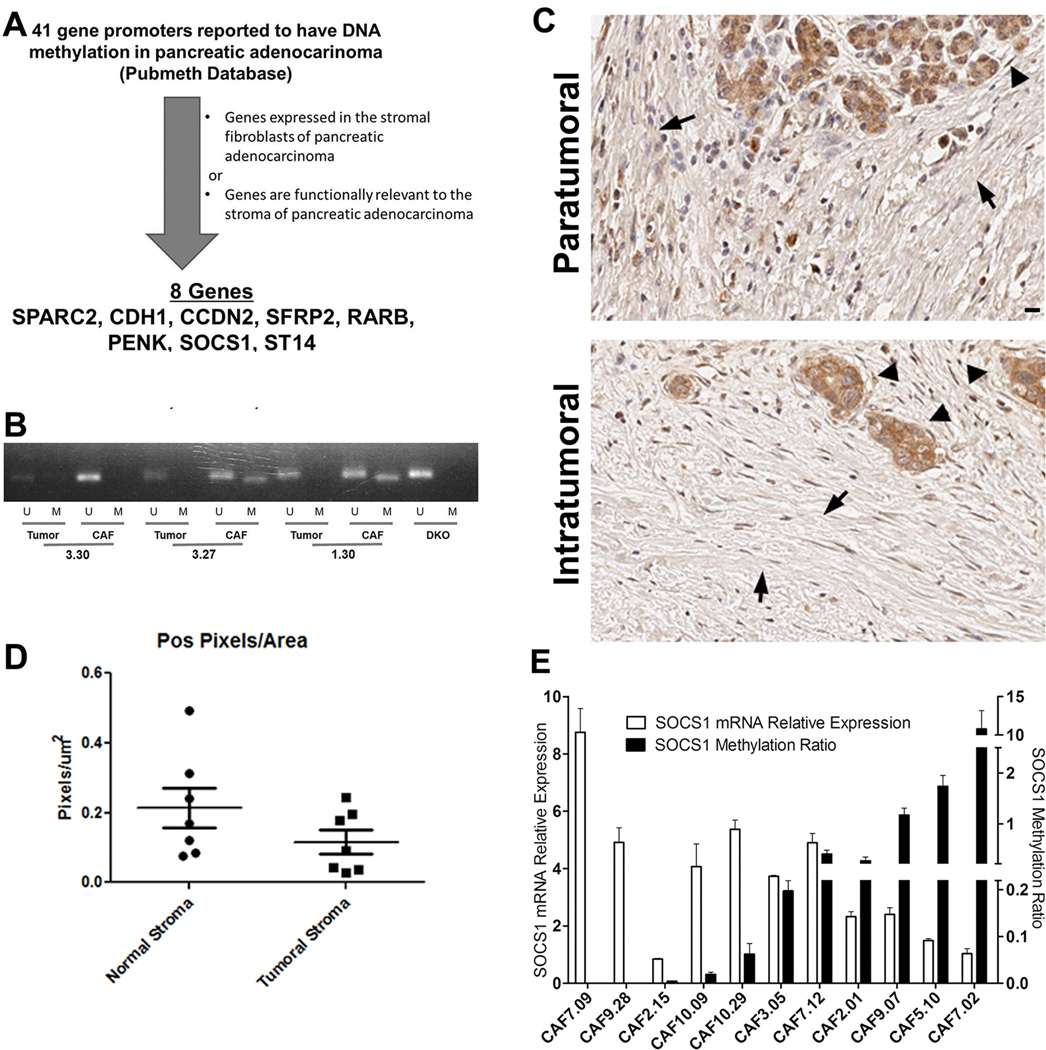
To understand how CAFs in PDACs may be regulated through DNA methylation, we first sought to identify genes methylated in CAFs. We found 41 gene promoters that were reported to have DNA methylation in pancreatic adenocarcinoma by searching the Pubmeth Database that includes methylated genes validated by methylation-specific sequencing or PCR analyses in the literature. We then selected 7 genes known to be expressed in or functionally relevant to the stromal fibroblasts of PDAC, including SPARC2, CCND2, SFRP2, ARB, PENK, SOCS1 and ST14(Fig. 1A) as well as CDH1 as an epithelial gene control. With methylation specific PCR(MSP), we checked the promoter methylation status of these eight genes in 3 matched primary PDAC neoplasm cultures and their associated fibroblasts, which were expanded in short-term cultures just to achieve adequate yield and purity. All genes except SOCS1 were either methylated in both PDACs and matched CAFs, or in PDACs but not in matched CAFs(Supplementary Table S2). Interestingly, none of the 3 PDACs were methylated in the SOCS1 promoter whereas 2 of 3 CAFs showed SOCS1 promoter methylation(Fig. 1B). The SOCS1 gene encodes a member of the suppressor of cytokine signaling(SOCS) family(27–29). Subsequently, with quantitative MSP, we found that none of 8 primary PDAC neoplasm cultures at early passages(< 4 passages) had detectable SOCS1 promoter methylation, whereas 16 out of 20 CAFs demonstrated various levels of SOCS1 methylation. Consistently, both MSP and methylation pyrosequencing(Supplementary Table S3) showed SOCS1 methylation in the stroma microdissected from 15 out of 16 human PDAC specimens(Table 1). By contrast, none of the paratumoral normal tissues including both acinar cells and stroma showed SOCS1 methylation on the nucleotide sites tested with pyrosequencing. To compare the intratumoral stroma specially to the stroma in the paratumoral normal tissues, we examined the expression of SOCS1 in human PDAC specimens by the immunohistochemistry(IHC) staining(Fig. 1C) and found that SOCS1 expression is significantly decreased in the intratumoral stroma compared to the paratumoral stroma(Fig. 1D). In primary CAF cultures, the levels of SOCS1 mRNA expression appeared to be negatively related to the levels of promoter methylation in a statistically significant manner(Fig. 1E).
Figure 1. SOCS1 is frequently methylated in pancreatic cancer associated fibroblasts.
A, The schema of candidate gene selection process. B, Promoter methylation status of 8 genes as indicated was checked in 3 paired primary PDAC cells and CAFs by Methylation Specific PCR (MSP)(Supplemental Table 2). C, IHC of SOCS1 was performed on paraffin-embedded slides of the seven human PDAC specimens as described (41). Representative IHC images of paratumoral areas(upper panel) and intratumoral areas(lower panel) from the same slide were shown. In upper panel, arrowhead indicates the normal acinar cells; in lower panel, arrowhead indicates the PDAC tumor cells. In both panels, arrows indicate stromal fibroblasts. Scale bar, 20 µm. D, protein expression was quantified by the Image Analysis Software(Aperio) as the total pixel number of positive staining signals in stroma divided by the total area size of the stroma, as previously described (42). The comparison between intratumoral stroma(Tumoral Stroma) and paratumoral normal stroma (Normal Stroma) was conducted by a paired t-Test(p=0.0487). E, SOCS1 methylation was quantified by MethySYBR real-time PCR. SOCS1 mRNA expression was quantified by real-time RT-PCR(qPCR); and GAPDH was used for normalization. SOCS1 mRNA expression were significantly correlated with SOCS1 promoter methylation in the simple linear regression analysis(p=0.021).
Table 1.
Summary of the results of SOCS1 methylation in multiple primary PDAC tumor cell cultures and primary CAFs analyzed by MSP and those in stroma dissected from PDAC tissues analyzed by pryosequencing
| Unmethylated | Methylated | |
|---|---|---|
| Primary PDAC tumor cells | 8 | 0 |
| Primary PDAC CAFs | 4 | 16 |
| Stroma dissected from PDAC tumors |
1 | 15 |
| Paratumoral normal tissues dissected from PDAC |
6 | 0 |
Induction of SOCS1 methylation in CAFs by interacting with tumor cells of PDAC
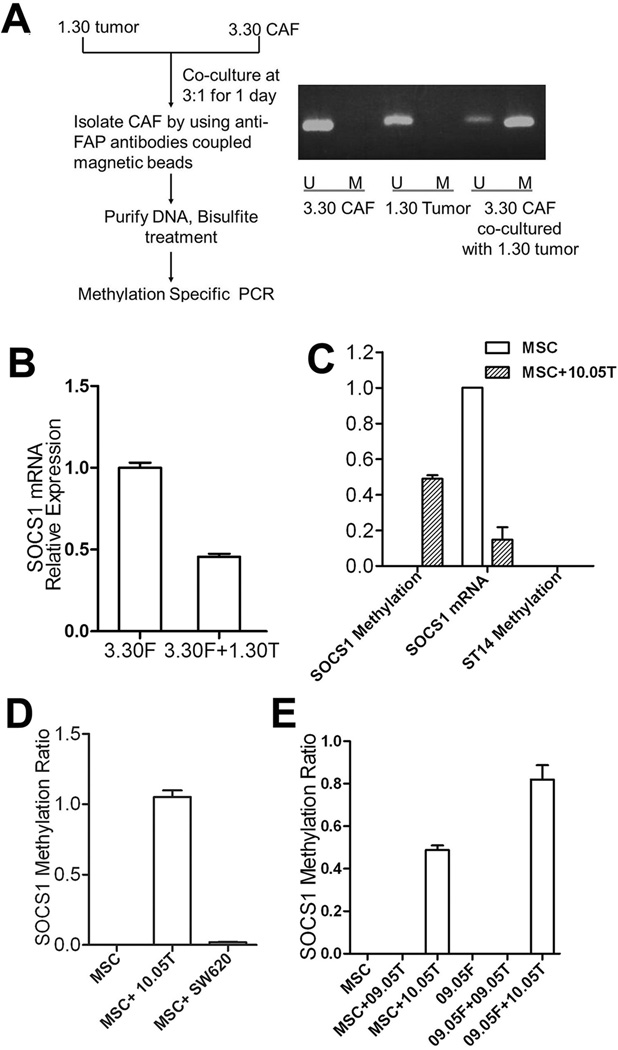
Next, we examined whether SOCS1 methylation in CAFs is pre-established or is induced by tumor-stroma interactions. We noticed that CAFs from some PDACs, such as 3.30CAFs from the Panc3.30 PDAC, did not have SOCS1 methylation. If SOCS1 methylation in CAFs is not pre-established, but is induced by tumor cells, we would anticipate the induction of SOCS1 methylation in 3.30CAFs co-cultured with Panc1.30 tumor cells. Twenty-four hours later, 3.30CAFs were separated from the co-culture by anti-FAP antibodies coupled with magnetic beads. The result did show that SOCS1 in 3.30CAFs was methylated and its mRNA expression was decreased after co-culturing with 1.30Tumor(Fig. 2A,B).
Figure 2. SOCS1 methylation in CAFs is induced by co-culture with pancreatic tumor cells.
A, Experimental schema for CAF/tumor co-culture and CAF purification. SOCS1 methylation in primary fibroblasts 3.30CAFs, 1.30Tumor, and CAFs purified from the co-culture of 3.30CAFs and 1.30Tumor were examined. SOCS1 methylation was measured by regular MSP in this panel and quantified by MethySYBR in the remaining panels. B, SOCS1 mRNA expression was quantified in 3.30CAFs and CAFs purified from the co-culture of 3.30CAFs and 1.30 Tumor by qPCR; and GAPDH was used for normalization. C, SOCS1 and ST14 methylation was examined in MSCs and fibroblasts purified from Panc10.05Tumor and MSC co-culture, respectively. SOCS1 mRNA expression was also examined by qPCR. D, SOCS1 methylation was examined in fibroblasts purified from the Panc10.05Tumor-MSC co-culture and the SW620Tumor-MSC co-culture, respectively. E, SOCS1 methylation was not detectable in fibroblasts isolated from the Panc09.05Tumor-MSC co-culture(MSC+09.05T) or the Panc09.05Tumor-09.05CAF co-culture(09.05F+09.05T), but was detectable in those from the Panc10.05Tumor-MSC co-culture(MSC+10.05T) and the Panc10.05Tumor-09.05CAF co-culture(09.05F+10.05T).
As CAFs have already been exposed to tumor cells in vivo, we sought to examine whether SOCS1 methylation can be induced in human mesenchymal stem cells(MSCs), which are thought to be the bone marrow derived cell origin of CAFs(30). As shown in Fig. 2C, SOCS1 methylation was induced and SOCS1 mRNA expression was decreased in MSCs after co-culture with Panc10.05 tumor cells for 24 hours. As a control, the ST14 promoter remained unmethylated. By contrast, methylated SOCS1 could not be induced in MSCs by colon cancer cell lines such as SW620(Fig. 2D). To confirm the initial MSP results of SOCS1 methylation in Fig. 1B, using another pair of matched PDAC tumor(Panc09.05Tumor) and CAF(09.05CAF) cells that do not have baseline SOCS1 methylation, we found that SOCS1 methylation in 09.05CAF can be induced by Panc10.05Tumor, but not its matched Panc09.05Tumor cells(Fig. 2E). The same results were observed when MSC, 3.30CAF or 09.05CAF was co-cultured with other human PDAC cell lines that are available to us and whose matched CAFs were found to have SOCS1 methylation(Fig. 2E). We also confirmed the MSP results (Fig. 1B&2C) by more quantitative, pyrosequencing assays (Supplementary Fig. S1;Table S4).
Induction of SOCS1 methylation in CAFs requires direct tumor-CAF contact
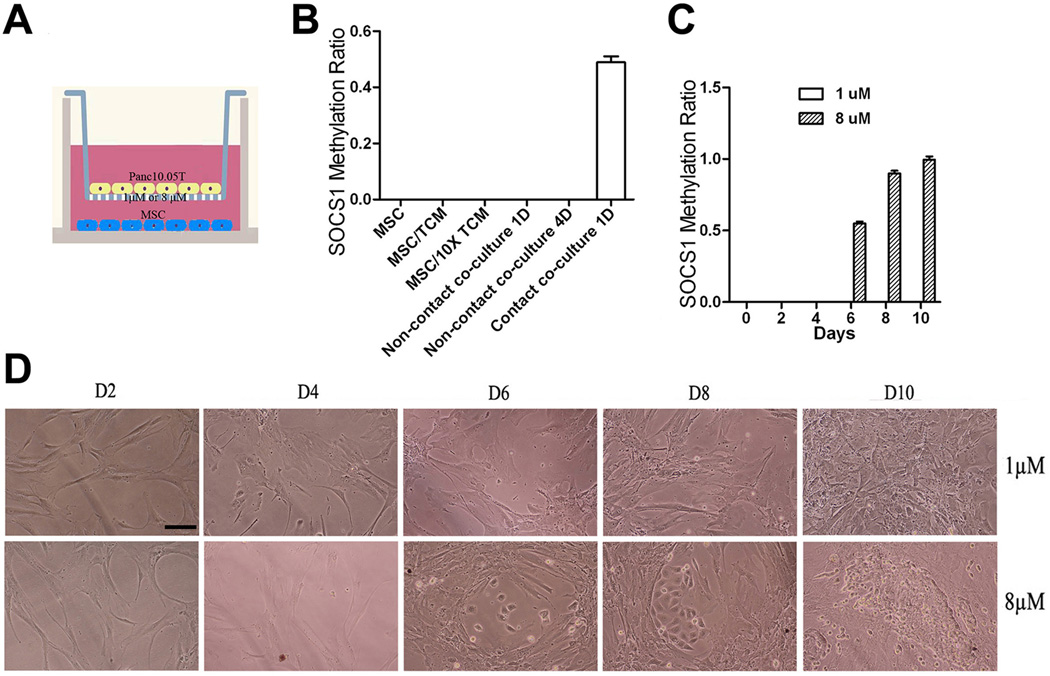
Next we sought to examine whether the tumor-stroma interaction that leads to the induction of SOCS1 methylation in CAFs or MSCs is mediated by paracrine signals or by direct cell-cell contact(Fig. 3). As shown in Fig. 3B, no SOCS1 methylation was induced in MSCs by tumor conditioned medium(TCM) or 10 times concentrated TCM. When tumor cells and MSCs were separated by two chambers in a trans-well apparatus (Fig. 3A), SOCS1 methylation was not induced by tumor cells (Fig. 3B & C). However, if the pore size of the trans-well filter was increased from 1µM to 8µM allowing tumor cells to migrate to the MSCs seeded in lower chamber (Fig. 3D), increasing levels of SOCS1 methylation was observed when increasing numbers of tumor cells migrated to the lower chamber (Fig. 3C).
Figure 3. Induction of SOCS1 methylation in MSCs requires direct cell-cell contact between tumor cells and MSCs.
A, Schematic illustration of the trans-well system. B, No induction of SOCS1 methylation in MSCs cultured with fresh tumor conditioned medium(TCM), 10 times concentrated TCM, or in no contact co-culture, where MSC and Panc10.05Tumor cells were separated by a 1 µM semitransparent membrane that tumor cells are not able to migrate through. C, MSCs and Panc10.05Tumor cells were separated by either 1 µM or 8µM semitransparent membrane. Panc10.05Tumor cells seeded above the 1µM membrane could not induce SOCS1 methylation in MSCs seeded in the bottom chamber. Panc10.05Tumor cells that migrated through the 8 µM membrane could induce the SOCS1 methylation in MSCs on and after Day 6. D, Panc10.05T cells(lower panel) on the 8 µM-pore membrane migrated to the bottom chamber as seen on and after Day 6. Panc10.05T cells(upper panel) were not able to migrate cross the 1 µM-pore membrane. Scale bar, 20 µm.
SOCS1 methylation was up-regulated by DNA methyltransferase 1 overexpression in fibroblast
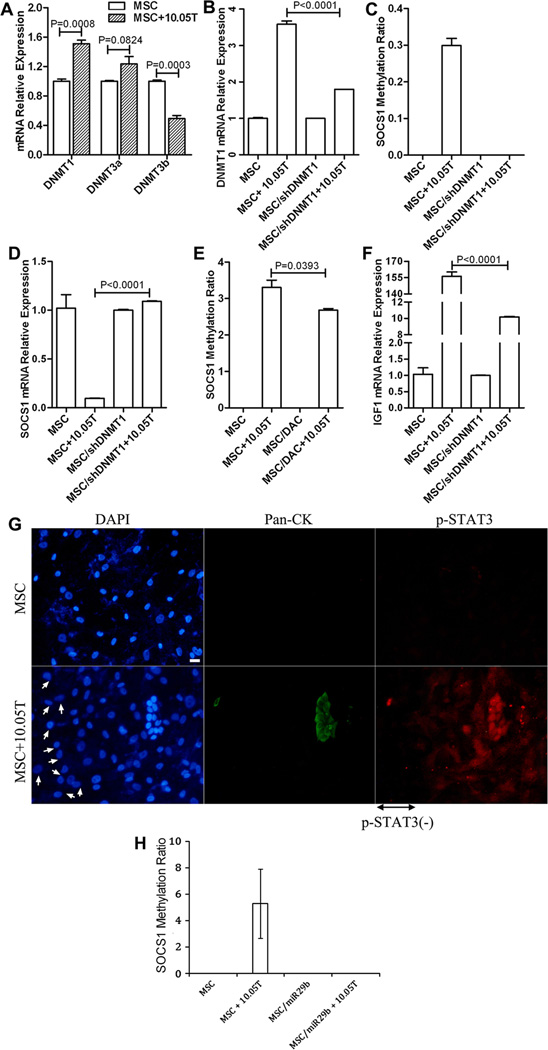
To examine whether DNA methyltransferases(DNMTs) account for the induction of SOCS1 methylation, we examined the mRNA expression of DNMT1, 3a, and 3b, the three major enzymes responsible for DNA methylation in mammals(31, 32), in MSCs in comparison to MSCs co-cultured with PDAC tumor cells. As shown in Fig. 4A, DNMT1 mRNA expression showed a significant increase in MSCs after co-culture with tumor cells. By contrast, when DNMT1 in MSCs was knocked down by the shRNA of DNMT1 carried by lentivirus(Supplementary Fig. S2) inhibiting DNMT1 expression in MSCs (Fig. 4B), the methylation of SOCS1 in MSC could not be induced(Fig. 4C). Downregulation of SOCS1 expression in MSCs after co-culture with tumor cells was also reversed by DNMT1 knock-down (Fig. 4D). Consistently, methylation of SOCS1 in MSCs was inhibited by pretreating MSCs with a DNMTs inhibitor, decitabine(DAC)(Fig. 4E). These results suggest that the induction of SOCS1 methylation is not a spontaneous process, but a highly regulated, DNMT-dependent process.
Figure 4. DNMT1 regulated SOCS1 methylation leads to the down-regulation of SOCS1 expression and subsequent activation of STAT3 and induction of IGF-1 expression.
Lentivirus-carried shRNA targeting DNMT1(shDNMT1) was used to knock down the expression of DNMT1(Supplementary Fig. S2). GAPDH expression was used for normalization of all qPCR results. A, Expression of DNMT1, DNMT3a and DNMT3b measured by qPCR in MSCs before and after co-culture with Panc10.05Tumor(10.05T) for 24 hours was compared. B, DNMT1 expression in MSCs before and after co-culture with 10.05T was compared with that in MSCs with knockdown of DNMT1. C, SOCS1 methylation examined by MethySYBR in MSCs before and after co-culture with 10.05T was compared with that in MSCs with knockdown of DNMT1. D, SOCS1 expression in MSCs before and after co-culture with 10.05T was compared with that in MSCs with knockdown of DNMT1. E, SOCS1 methylation in MSCs before and after co-culture with 10.05T was compared with that in MSCs pretreated with DAC. F, IGF-1 expression measured by qPCR in MSCs before and after co-culture with 10.05T was compared with that in MSCs with knockdown of DNMT1. G, MSCs were co-cultured with Panc10.05Tumor cells in an 8 µM trans-well system for 5 days as described in Fig. 3. Cells in the bottom chamber were fixed and subjected to a dual immunofluorescence staining with FITC-conjugated antibodies recognizing Pan-CK to mark epithelial tumor cells(Green) and PE-conjugated antibodies either for STAT3 or for p-STAT3 (Tyr705)(Red), and counter-stained with DAPI for nuclei(Blue). Scale bar, 20 µm. H, SOCS1 methylation examined by MethySYBR in MSCs transfected with the control microRNA before and after co-culture with 10.05T was compared with that in MSCs transfected with miR-29b. In A–F&H, comparisons were analyzed by a two-tail unpaired Student’s t-Test with p values shown.
STAT3 phosphorylation and IGF-1 expression were induced in CAFs after co-culture with PDACs in a DNMT1 dependent manner
SOCS1 is a known suppressor of many pro-cancerous cytokines and growth factors(29). Therefore, we attempted to understand whether the induction of SOCS1 methylation in CAFs through tumor-stroma interaction also regulates this biological function of SOCS1. To this end, we examined the expression of the insulin like growth factor 1 (IGF-1), a representative SOCS1-downstream pro-cancerous growth factor that is primarily secreted by fibroblasts(3, 33, 34). IGF-1 is chosen also because as its associated signaling pathway is among the pathways that are the most significantly upregulated in CAF or MSC upon co-culture with PDAC tumor cells in the Ingenuity pathway analysis of the mRNA expression microarray data described below(Supplementary Fig. S3). As shown in Fig. 4F, IGF-1 mRNA expression in MSCs co-cultured with tumor cells was increased by more than 100 times comparing to the single cultured MSCs. By contrast, this increase was diminished when DNMT1 was knocked down from MSCs.
It is known that the activation of the signal transducer and activator of transcription(STAT) family, particularly STAT3 and STAT5, mediates the suppressive role of SOCS1 in regulating the transcription of many pro-cancerous cytokines and growth factors including IGF-1(35, 36). In the absence of SOCS1, STAT3 is phosphorylated and assumes an activated form (p-STAT3) to promote gene transcription. Thus, we examined STAT3 staining pattern in MSCs surrounding tumor cells. As shown in Fig. 4G, p-STAT3 nuclear staining was more obvious in MSCs immediately adjacent to tumor cells whereas pSTAT3 is not detectable in MSCs further away from tumor cells. It was previously reported that miR-29b induces SOCS1 expression by promoter demethylation, leading to the reduced STAT3 phosphorylation(35). Thus, we hypothesized that miR-29b serves as a negative regulator of tumor-induced SOCS1 methylation in MSCs. Consistent with this notion, as shown in Fig. 4H, SOCS1 methylation was not induced in the PDAC co-cultured MSCs transfected with miR-29b. Taken together, our results suggest that PDAC tumor cells induce DNA methylation of SOCS1 genes in CAFs leading to the downregulation of SOCS1 expression and subsequently the activation of STAT3 and high expression of pro-cancerous growth factors such as IGF-1.
PDAC tumor cells induced DNA methylation in a global panel of clustered genes and caused their down-regulation in CAFs
SOCS1 is unlikely the only gene that is methylated in CAFs as a result of tumor-stroma interaction. More likely, a global panel of genes, which are programmed to be unmethylated in fibroblasts or their origins such as MSCs under normal conditions, are reprogrammed to be methylated in CAFs upon interacting with tumor cells. A combined array analysis of DNA methylation and gene expression (GEO Accession Number GSE80369) showed that a panel of approximately 1585 genes, including SOCS1, is both methylated and downregulated in MSCs upon interacting with Panc10.05Tumor(Table 2;Supplementary Table S5). A near complete overlapping panel of genes are both methylated and downregulated in 09.05CAF upon co-culture with PDAC cells. Many of these genes do not appear to be distributed randomly, but are clustered, in a number of chromosomal regions(Table 2;Supplementary Table S5), suggesting that, in addition to DMNT1, other chromatin remodeling mechanisms may also be involved in determining which clusters of genes in specific chromatin regions are simultaneously methylated and co-regulated in CAFs upon interacting with PDAC cells. As shown in Supplementary Table S6, gene expression alterations in CAFs upon interacting with PDAC cells may also be mediated by other epigenetic mechanisms.
Table 2.
The CROC analysis of genomic clustering of the genes both methylated and downregulated in CAFs upon interacting with PDAC tumor cells
| Total numbers of genes with induced methylation and expression downregulation in CAFs* |
1585 |
|---|---|
| Number of clusters with a minimum of 4 genes | 71 |
| Number of genes involved in 4-gene clusters | 421 |
| Number of clusters with a minimum of 5 genes | 64 |
| Number of genes involved in 5-gene clusters | 397 |
| Number of clusters with a minimum of 6 genes | 22 |
| Number of genes involved in 6-gene clusters | 149 |
Data from two paired samples (MSC before and after co-culture with PDAC tumor cells and 09.05CAFs before and after co-culture with PDAC tumors) were included in the analysis.
Tumor-induced DNA methylation and SOCS1 downregulation are important for stromal fibroblasts in supporting PDAC growth on mice
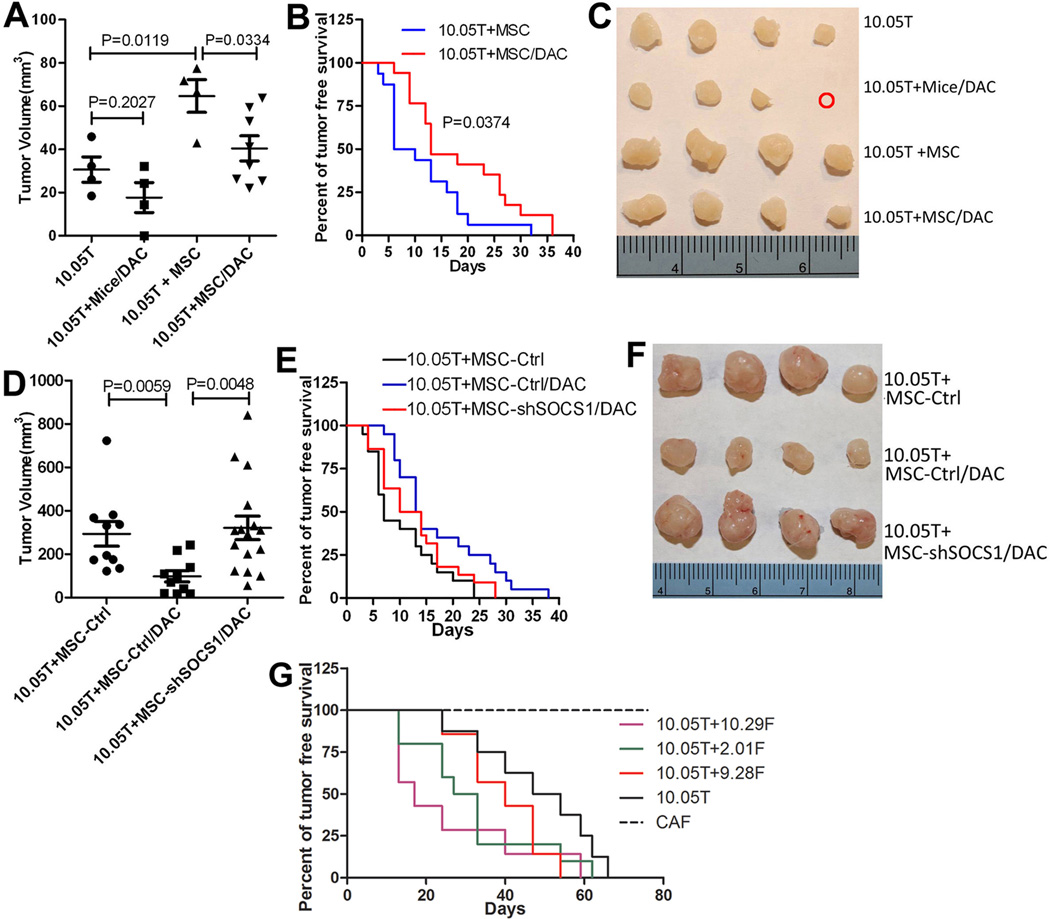
Fibroblasts were previously shown to promote tumor growth in a subcutaneous tumor model when co-injected with tumor cells(36, 37). We therefore investigated whether this pro-cancerous activity would be inhibited when methylation was blocked in CAFs. Panc10.05Tumor cells were inoculated into non-obese diabetic-severe combined immunodeficiency(NOD-SCID) mice subcutaneously alone or with MSCs. Seven weeks later, xenografts formed by Panc10.05Tumor plus MSCs were significantly bigger than those formed by Panc10.05Tumor(Fig. 5A). However, when MSCs were pre-treated with DAC for 4 days during the cell culture, they lost their capacity to promote xenograft growth. Similarly, if mice were pretreated with DAC five days before Panc10.05 inoculation, the xenografts grew slower. This suggests that the stromal fibroblasts from the host mice lost the capacity to support xenograft growth due to the transient inhibition of DNA methylation(Fig. 5A,B). It is likely that the xenografts eventually grew in DAC-treated groups because the effect of transient DAC treatment stopped. However, the tumor free survival of NOD-SCID mice with Panc10.05Tumor co-inoculated with DAC-pretreated MSCs was significantly longer than that of mice with Panc10.05Tumor co-inoculated with untreated MSCs(Fig. 5C). The pro-cancerous effect of DAC-pretreated MSCs, measured by tumor volume(Fig. 5D,F), was restored when SOCS1 was knocked down by shRNA from MSCs(Supplementary Fig. S4). Even if the MSCs were pretreated with DAC, they were still able to promote the growth of Panc10.05Tumor xenografts when SOCS1 was knocked down (Fig. 5D,E). These results suggest that DNA methylation in stromal fibroblasts is important for supporting PDAC tumor growth and also suggest that SOCS1 downregulation in stromal fibroblasts mediates the role of DNA methylation in supporting PDAC growth. Interestingly, the pro-cancerous effect of DAC-pretreated MSCs was not completely restored when SOCS1 was knocked down by shRNA from MSCs, as measured by tumor free survival and judged by the insignificant p value(p=0.1204) comparing 10.05T+MSC-ctrl/DAC vs. 10.05T+MSC-shSOCS1/DAC(Fig. 5E). Thus, other genes(Table 2) identified to be methylated in CAFs upon interacting with PDAC tumor cells may also be important for supporting PDAC growth.
Figure 5. DNA methylation and SOCS1 downregulation in CAFs are critical for supporting PDAC growth on mice.
A, Volume measurements of xenograft tumors harvested at week 5 following inoculation of Panc10.05Tumor in one representative experiment. 10.05T, mice inoculated with Panc10.05Tumor alone(n=4); 10.05T+Mice/DAC, DAC-pretreated mice inoculated with 10.05T(n=4); 10.05T+MSC, 10.05T mixed with MSCs inoculated on untreated mice(n=4); 10.05T+MSC/DAC, 10.05T mixed with MSCs pretreated with DAC(n=8). B, Tumor free survival was compared between the 10.05T+MSC group(n=16) and the 10.05T+MSC/DAC group(n=17). C, Tumors harvested from the experiment in A are shown. The red cycle indicates no tumor. Four biggest tumors were shown as representatives for the 10.05T+MSC/DAC group. D, Volume measurements of xenograft tumors harvested at week 7 following inoculation of Panc10.05Tumor. 10.05T+MSC-ctrl, 10.05T mixed with MSCs controlled for shRNA transduction(n=10); 10.05T+MSC-ctrl/DAC, 10.05T mixed with MSCs controlled for shRNA transduction and pretreated with DAC(n=10); 10.05T+MSC-shSOCS1/DAC, 10.05T mixed with MSCs transduced with shRNA targeting SOCS1 and pretreated with DAC(n=16). E, Tumor free survival was compared between groups shown in D. 10.05T+MSC-ctrl(n=20) vs. 10.05T+MSC-ctrl/DAC(n=20), p=0.009; 10.05T+MSC-shSOCS1/DAC(n=21) vs. 10.05T+MSC-ctrl/DAC(n=20), p=0.1204. F, Four largest tumors as representatives from each group in D are shown. G, Tumor free survival was compared between the groups of NOD-SCID mice inoculated with 10.05T alone or in combination with different patient derived CAFs. 10.05T, mice inoculated with 10.05T alone; 10.05T+9.28F, mice inoculated with 10.05T and CAF 9.28F; 10.05T+10.29F, mice inoculated with 10.05T and CAF 10.29F; 10.05T+2.01F, mice inoculated with 10.05T and CAF 2.01F; CAF, a representative CAF sample alone. N=10 per group. 10.05T+10.29F vs. 10.05T, p=0.017; 10.05T+2.01F vs. 10.05T, p=0.044; 10.05T+9.28F vs. 10.05T, p=0.217. All experiments were repeated twice. In A&D, comparisons were analyzed by a two-tail unpaired Student’s t-Test; and in B,E,G, comparisons were analyzed by the Log-rank(Mantel-Cox) test.
Finally, we attempted to determine whether patients derived CAFs may promote PDAC xenograft growth on mice. Panc10.05Tumor were inoculated into NOD-SCID mice subcutaneously alone or with the patient derived CAFs that were in sufficient quantity for this experiments. Among these CAFs, 9.28F showed no methylation in SOCS whereas 10.29F and 2.01F showed methylation (Fig. 1). The tumor free survival of NOD-SCID mice co-inoculated with Panc10.05Tumor and CAF 10.29F or 2.01F was significantly shorter than that of mice inoculated with Panc10.05Tumor alone whereas the tumor free survival of NOD-SCID mice co-inoculated with Panc10.05Tumor and CAF 9.28F was minimally shorter than that of mice with Panc10.05Tumor inoculated alone (Fig. 5G). CAFs themselves did not form any tumor. This result suggested that the ability of patient derived CAFs with SOCS1 methylation in promoting PDAC growth is stronger than that of CAFs without SOCS1 methylation.
DISUSSION
This is the first report demonstrating that CAFs within the TME are modulated by PDAC tumor cells through epigenetic regulation in the form of DNA methylation. By inducing methylation in CAFs, SOCS1 expression is suppressed, which leads to the activation of its downstream signaling pathways, such as the phosphorylation of STAT3. As a result, cytokines and growth factors such as IGF-1 are induced in the TME. Thus, cancer cells create a TME that supports their growth. This may be a novel and critical mechanism for tumor initiation, progression, and/or metastasis formation. Our findings also implicate that stromal fibroblasts at their baseline may be hostile to the tumorigenesis of PDACs if they are not modulated by tumor cells at the epigenetic level. Stromal fibroblasts modulated by tumor-induced methylation may subsequently reprogram the TME from an anti-cancerous one to a pro-cancerous one. Hence, our findings are consistent with the recently published studies showing that general depletion of stromal fibroblasts, presumably prior to being modulated by tumor-induced methylation, facilitate PDAC development and metastasis in the mouse model(10, 11).
SOCS1 promotor hypermethylation and decreased expression of SOCS1 have been reported in pancreatic epithelial tumor cell lines and minimally dissected PDAC tissues(38). We did not observe SOCS1 hypermethylation in low-passage primary cultures of PDAC cells. It is possible that SOCS1 promoter hypermethylation had occurred spontaneously during the continuous culture of established PDAC cell lines.
DNA methylation is reversible, thus targeting methylation susceptible genes provide promising targets for cancer treatment and prevention. This study suggests that cell surface receptors mediate direct contact between tumor cells and CAFs. Such cell surface receptors, once identified, are ideal drug targets for modifying DNA methylation mechanisms specifically in CAFs. Both integrin and adheren pathways are significantly upregulated at the mRNA level in CAFs and MSCs upon co-culture with PDAC tumor cells(Supplementary Fig. S3), supporting the investigation of both integrin and adheren families of cell surface molecules as candidates that may mediate direct contact between tumor cells and CAFs. Another future step is to determine whether methylation in stromal SOCS1 influences the clinical outcomes of patients with PDACs. Interestingly, the patients (3.30, 7.09, 9.28) whose CAFs did not show SOCS1 methylation all have an overall survival more than 3 years. This is consistent with our result with a limited sample number of patient derived CAFs showing that CAFs without SOCS1 methylation did not appear to have the ability in promoting PDAC growth in mice in contrast to those with SOCS1 methylation. Therefore, the prognostic value of SOCS1 methylation in the stroma will need to be investigated in a larger number of stroma specimens dissected from PDACs.
We observed a global effect on gene methylation in CAFs upon interacting with PDAC tumor cells whereas a previous study found very few methylated genes that responded to the 5-aza-2'-deoxycytidine treatment in CAF cell lines(20). This discrepancy is likely attributed to the difference between primary CAF cultures in the current study and immortalized CAF lines used in the previous study(20). Consistently, Shakya et al. reported expression changes of a large panel of genes that are reversible with DAC treatment in CAFs from a transgenic mouse model of pancreatic cancer(39). It was also recently reported that breast carcinoma cells altered the expression of genes, particularly ADAMTS1, in breast fibroblasts, not via promoter methylation, but via epigenetic alterations involving EZH2/H3K27(40), suggesting that other epigenetic regulating mechanisms may be involved in reprograming CAFs. In addition, although we show that colon cancer lines tested did not induce SOCS1 methylation in MSCs, we found that a similar tumor-induced methylation mechanism involving either the same or different genes expressed in stromal fibroblasts may exist for other types of malignancies. Moreover, a similar tumor-induced DNA methylation process may occur universally in other stromal cells including tumor infiltrating immune cells, thus highlighting the significance of our findings for further exploration.
Supplementary Material
Acknowledgments
This work was conducted at the Johns Hopkins University School of Medicine. We thank Dr. Ben Park for his comments on the manuscript, Dr. Michael Goggins for his insightful discussion, and Dr. Wayne Yu at the Microarray Core for his help with microarray analyses. The gene expression and methylation microarray data have been uploaded in GEO (Accession Number GSE80369).
Grant Support
This work was supported in part by National Institutes of Health (R01CA169702 and K23 CA148964-01 to L.Zheng.), Viragh Foundation and the Skip Viragh Pancreatic Cancer Center at Johns Hopkins (E.M.Jaffee, L.Zheng.), Lefkofsky Family Foundation (L.Zheng), the National Cancer Institute Specialized Programs of Research Excellence in Gastrointestinal Cancers P50 CA062924 (E.M.Jaffee, L.Zheng), Lustgarten Foundation (L.Zheng), the Sol Goldman Pancreatic Cancer Center grants (L.Zheng). Q.Xiao. is supported by the Chinese Scholarship Council Foundation 201206320056; and D.Zhou. is supported by the Chinese Scholarship Council Foundation 2011632121.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed by the authors.
REFERENCES
- 1.Tlsty TD, Coussens LM. Tumor stroma and regulation of cancer development. Annu Rev Pathol. 2006;1:119–150. doi: 10.1146/annurev.pathol.1.110304.100224. [DOI] [PubMed] [Google Scholar]
- 2.Orimo A, Weinberg RA. Stromal fibroblasts in cancer: a novel tumor-promoting cell type. Cell Cycle. 2006;5:1597–1601. doi: 10.4161/cc.5.15.3112. [DOI] [PubMed] [Google Scholar]
- 3.Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432:332–337. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolfgang CL, Herman JM, Laheru DA, Klein AP, Erdek MA, Fishman EK, et al. Recent progress in pancreatic cancer. CA Cancer J Clin. 2013;63:318–348. doi: 10.3322/caac.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rucki AA, Zheng L. Pancreatic cancer stroma: understanding biology leads to new therapeutic strategies. World J Gastroenterol. 2014;20:2237–2246. doi: 10.3748/wjg.v20.i9.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Apte MV, Wilson JS, Lugea A, Pandol SJ. A starring role for stellate cells in the pancreatic cancer microenvironment. Gastroenterology. 2013;144:1210–1219. doi: 10.1053/j.gastro.2012.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feig C, Gopinathan A, Neesse A, Chan DS, Cook N, Tuveson DA. The pancreas cancer microenvironment. Clin Cancer Res. 2012;18:4266–4276. doi: 10.1158/1078-0432.CCR-11-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stromnes IM, DelGiorno KE, Greenberg PD, Hingorani SR. Stromal reengineering to treat pancreas cancer. Carcinogenesis. 2014;35:1451–1460. doi: 10.1093/carcin/bgu115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waghray M, Yalamanchili M, di Magliano MP, Simeone DM. Deciphering the role of stroma in pancreatic cancer. Curr Opin Gastroenterol. 2013;29:537–543. doi: 10.1097/MOG.0b013e328363affe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ozdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu CC, Simpson TR, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25:719–734. doi: 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25:735–747. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patocs A, Zhang L, Xu Y, Weber F, Caldes T, Mutter GL, et al. Breast-cancer stromal cells with TP53 mutations and nodal metastases. N Engl J Med. 2007;357:2543–2551. doi: 10.1056/NEJMoa071825. [DOI] [PubMed] [Google Scholar]
- 13.Tuhkanen H, Anttila M, Kosma VM, Heinonen S, Juhola M, Helisalmi S, et al. Frequent gene dosage alterations in stromal cells of epithelial ovarian carcinomas. Int J Cancer. 2006;119:1345–1353. doi: 10.1002/ijc.21785. [DOI] [PubMed] [Google Scholar]
- 14.Qiu W, Hu M, Sridhar A, Opeskin K, Fox S, Shipitsin M, et al. No evidence of clonal somatic genetic alterations in cancer-associated fibroblasts from human breast and ovarian carcinomas. Nat Genet. 2008;40:650–655. doi: 10.1038/ng.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu M, Yao J, Cai L, Bachman KE, van den Brule F, Velculescu V, et al. Distinct epigenetic changes in the stromal cells of breast cancers. Nat Genet. 2005;37:899–905. doi: 10.1038/ng1596. [DOI] [PubMed] [Google Scholar]
- 16.Mitra AK, Zillhardt M, Hua Y, Tiwari P, Murmann AE, Peter ME, et al. MicroRNAs reprogram normal fibroblasts into cancer-associated fibroblasts in ovarian cancer. Cancer Discov. 2012;2:1100–1108. doi: 10.1158/2159-8290.CD-12-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walter K, Omura N, Hong SM, Griffith M, Goggins M. Pancreatic cancer associated fibroblasts display normal allelotypes. Cancer Biol Ther. 2008;7:882–888. doi: 10.4161/cbt.7.6.5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaffee EM, Schutte M, Gossett J, Morsberger LA, Adler AJ, Thomas M, et al. Development and characterization of a cytokine-secreting pancreatic adenocarcinoma vaccine from primary tumors for use in clinical trials. Cancer J Sci Am. 1998;4:194–203. [PubMed] [Google Scholar]
- 19.Zheng L, Foley K, Huang L, Leubner A, Mo G, Olino K, et al. Tyrosine 23 phosphorylation-dependent cell-surface localization of annexin A2 is required for invasion and metastases of pancreatic cancer. PLoS One. 2011;6:e19390. doi: 10.1371/journal.pone.0019390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu J, Walter K, Omura N, Hong SM, Young A, Li A, et al. Unlike pancreatic cancer cells pancreatic cancer associated fibroblasts display minimal gene induction after 5-aza-2'-deoxycytidine. PLoS One. 2012;7:e43456. doi: 10.1371/journal.pone.0043456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.House MG, Guo M, Iacobuzio-Donahue C, Herman JG. Molecular progression of promoter methylation in intraductal papillary mucinous neoplasms (IPMN) of the pancreas. Carcinogenesis. 2003;24:193–198. doi: 10.1093/carcin/24.2.193. [DOI] [PubMed] [Google Scholar]
- 22.Chan MW, Chu ES, To KF, Leung WK. Quantitative detection of methylated SOCS-1, a tumor suppressor gene, by a modified protocol of quantitative real time methylation-specific PCR using SYBR green and its use in early gastric cancer detection. Biotechnol Lett. 2004;26:1289–1293. doi: 10.1023/B:BILE.0000044922.43572.2d. [DOI] [PubMed] [Google Scholar]
- 23.Lo PK, Watanabe H, Cheng PC, Teo WW, Liang X, Argani P, et al. MethySYBR, a novel quantitative PCR assay for the dual analysis of DNA methylation and CpG methylation density. J Mol Diagn. 2009;11:400–414. doi: 10.2353/jmoldx.2009.080126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ting AH, Jair KW, Schuebel KE, Baylin SB. Differential requirement for DNA methyltransferase 1 in maintaining human cancer cell gene promoter hypermethylation. Cancer Res. 2006;66:729–735. doi: 10.1158/0008-5472.CAN-05-1537. [DOI] [PubMed] [Google Scholar]
- 25.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 26.Pignatelli M, Serras F, Moya A, Guigo R, Corominas M. CROC: finding chromosomal clusters in eukaryotic genomes. Bioinformatics. 2009;25:1552–1553. doi: 10.1093/bioinformatics/btp248. [DOI] [PubMed] [Google Scholar]
- 27.Heinrich PC, Behrmann I, Muller-Newen G, Schaper F, Graeve L. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem J. 1998;334(Pt 2):297–314. doi: 10.1042/bj3340297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alexander WS, Starr R, Metcalf D, Nicholson SE, Farley A, Elefanty AG, et al. Suppressors of cytokine signaling (SOCS): negative regulators of signal transduction. J Leukoc Biol. 1999;66:588–592. doi: 10.1002/jlb.66.4.588. [DOI] [PubMed] [Google Scholar]
- 29.Sasi W, Sharma AK, Mokbel K. The role of suppressors of cytokine signalling in human neoplasms. Mol Biol Int. 2014;2014:630797. doi: 10.1155/2014/630797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scarlett CJ. Contribution of bone marrow derived cells to the pancreatic tumor microenvironment. Front Physiol. 2013;4:56. doi: 10.3389/fphys.2013.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramaniam D, Thombre R, Dhar A, Anant S. DNA methyltransferases: a novel target for prevention and therapy. Front Oncol. 2014;4:80. doi: 10.3389/fonc.2014.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin B, Li Y, Robertson KD. DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer. 2011;2:607–617. doi: 10.1177/1947601910393957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahmed SF, Farquharson C. The effect of GH and IGF1 on linear growth and skeletal development and their modulation by SOCS proteins. J Endocrinol. 2010;206:249–259. doi: 10.1677/JOE-10-0045. [DOI] [PubMed] [Google Scholar]
- 34.Mueller MM, Fusenig NE. Friends or foes - bipolar effects of the tumour stroma in cancer. Nat Rev Cancer. 2004;4:839–849. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- 35.Amodio N, Bellizzi D, Leotta M, Raimondi L, Biamonte L, D'Aquila P, et al. miR-29b induces SOCS-1 expression by promoter demethylation and negatively regulates migration of multiple myeloma and endothelial cells. Cell Cycle. 2013;12:3650–3662. doi: 10.4161/cc.26585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan XL, Jia YL, Chen L, Zeng Q, Zhou JN, Fu CJ, et al. Hepatocellular carcinoma-associated mesenchymal stem cells promote hepatocarcinoma progression: role of the S100A4-miR155-SOCS1-MMP9 axis. Hepatology. 2013;57:2274–2286. doi: 10.1002/hep.26257. [DOI] [PubMed] [Google Scholar]
- 37.Mallette FA, Calabrese V, Ilangumaran S, Ferbeyre G. SOCS1, a novel interaction partner of p53 controlling oncogene-induced senescence. Aging (Albany NY) 2010;2:445–452. doi: 10.18632/aging.100163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fukushima N, Sato N, Sahin F, Su GH, Hruban RH, Goggins M. Aberrant methylation of suppressor of cytokine signalling-1 (SOCS-1) gene in pancreatic ductal neoplasms. Br J Cancer. 2003;89:338–343. doi: 10.1038/sj.bjc.6601039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shakya R, Gonda T, Quante M, Salas M, Kim S, Brooks J, et al. Hypomethylating therapy in an aggressive stroma-rich model of pancreatic carcinoma. Cancer Res. 2013;73:885–896. doi: 10.1158/0008-5472.CAN-12-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tyan SW, Hsu CH, Peng KL, Chen CC, Kuo WH, Lee EY, et al. Breast cancer cells induce stromal fibroblasts to secrete ADAMTS1 for cancer invasion through an epigenetic change. PLoS One. 2012;7:e35128. doi: 10.1371/journal.pone.0035128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komazaki T1, Nagai H, Emi M, Terada Y, Yabe A, Jin E, et al. Hypermethylation-associated inactivation of the SOCS-1 gene, a JAK/STAT inhibitor, in human pancreatic cancers. Jpn J Clin Oncol. 2004;34:191–194. doi: 10.1093/jjco/hyh035. [DOI] [PubMed] [Google Scholar]
- 42.Foley K, Rucki AA, Xiao Q, Zhou D, Leubner A, Mo G, et al. Semaphorin 3D autocrine signaling mediates the metastatic role of annexin A2 in pancreatic cancer. Sci Signal. 2015;8:ra77. doi: 10.1126/scisignal.aaa5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.