Abstract
To identify novel traits associated with alleles known to predispose to alcohol and nicotine use, we conducted a phenome-wide association study (PheWAS) in a large multi-population cohort. We investigated 7688 African-Americans, 1133 Asian-Americans, 14 081 European-Americans, and 3492 Hispanic-Americans from the Women's Health Initiative, analyzing alleles at the CHRNA3–CHRNA5 locus, ADH1B, and ALDH2 with respect to phenotypic traits related to anthropometric characteristics, dietary habits, social status, psychological traits, reproductive history, health conditions, and nicotine/alcohol use. In ADH1B trans-population meta-analysis and population-specific analysis, we replicated prior associations with drinking behaviors and identified multiple novel phenome-wide significant and suggestive findings related to psychological traits, socioeconomic status, vascular/metabolic conditions, and reproductive health. We then applied Bayesian network learning algorithms to provide insight into the causative relationships of the novel ADH1B associations: ADH1B appears to affect phenotypic traits via both alcohol-mediated and alcohol-independent effects. In an independent sample of 2379 subjects, we also replicated the novel ADH1B associations related to socioeconomic status (household gross income and highest grade finished in school). For CHRNA3–CHRNA5 risk alleles, we replicated association with smoking behaviors, lung cancer, and asthma. There were also novel suggestive CHRNA3–CHRNA5 findings with respect to high-cholesterol-medication use and distrustful attitude. In conclusion, the genetics of alcohol and tobacco use potentially has broader implications on physical and mental health than is currently recognized. In particular, ADH1B may be a gene relevant for the human phenome via both alcohol metabolism-related mechanisms and other alcohol metabolism-independent mechanisms.
Introduction
Susceptibility to use and abuse of tobacco and alcohol is genetically influenced (Treur et al, 2014; Wu et al, 2014). Numerous genetic studies, including genome-wide association studies (GWAS), have examined smoking and drinking behaviors, and certain risk/protective alleles have been identified and validated (The Tobacco and Genetics Consortium, 2010; Gelernter et al, 2014; Gelernter et al, 2015; Buhler et al, 2015). The best replicated loci for drinking behaviors map to genes encoding alcohol-metabolizing enzymes. Minor alleles at ADH1B (ie, rs1229984 and rs2066702) are associated with lower levels of drinking (Xu et al, 2015). Consistent with the genetic associations, these variants increase the alcohol oxidization rate, raising acetaldehyde levels and its related aversive symptoms, including facial flushing, nausea, headache, and tachycardia (Edenberg, 2007). Similarly, ALDH rs672 is negatively associated with alcohol use behaviors in Asian populations (this variant is very rare or absent in non-Asian populations) (Quillen et al, 2014) and it causes a complete or near-complete lack of acetaldehyde metabolism activity via this pathway (Peng et al, 2014), producing an accumulation of acetaldehyde and consequently its aversive symptoms. It is also recognized that the ADH1B locus is under strong selection in East Asians and a recent study hypothesized an independent evolution of the same locus also in Europeans (Galinsky et al, 2016). For smoking behaviors, the most recognized risk locus is the CHRNA3–CHRNA5–CHRNB4 gene cluster, and risk alleles in this region (rs8034191, rs1051730, rs12914385, rs2036527, and rs16969968) are associated with smoking behaviors including nicotine dependence and smoking quantity (The Tobacco and Genetics Consortium, 2010; David et al, 2012). On the molecular level, these genetic variants are associated with functional alterations in subunits of nicotinic acetylcholine receptors, reducing their activity and the aversive effects of nicotine (Wen et al, 2016).
As both drinking and smoking behaviors are strongly associated with numerous health consequences, many studies have sought to link genes implicated in risk for smoking and drinking behaviors to the adverse health consequences of these behaviors, obtaining consistent results (Druesne-Pecollo et al, 2009; Li et al, 2011, 2012; Chen et al, 2015; Hallden et al, 2015). However, owing to the potent effects that nicotine, alcohol, and their metabolites have on the human body (Guo and Ren, 2010; Drug Therapeutics Bulletin, 2014), smoking and drinking behaviors and their associated genes (with large effects on nicotine response and acetaldehyde metabolism, respectively) might be involved in a much wider range of health conditions and human behaviors than is currently recognized. Phenome-wide association studies (PheWAS) are a powerful approach to discover novel phenotypic associations of loci previously identified (Denny et al, 2013). Thus, we performed a PheWAS for alleles related to risk for alcohol and nicotine use for a range of physical and mental health traits. We applied this approach to a large multi-ethnic sample of 26 394 subjects from the Women's Health Initiative (WHI) cohorts. The WHI is a major research program designed to address the most common causes of death, disability, and poor quality of life in postmenopausal women (The Women's Health Initiative Study Group, 1998). The WHI subjects were recruited for three clinical trials and one observational study. There are well-known sex differences in smoking and drinking behaviors and their consequences (Pandeya et al, 2013; Cosgrove et al, 2014; White et al, 2015), and the current study provided novel important data regarding the role of alcohol and tobacco use and their genetics in women's health.
Materials and methods
Study Population
The data sets used for the analyses were obtained, after authorized access, from the National Center for Biotechnology Information (NCBI) database of Genotypes and Phenotypes (dbGaP; available at http://www.ncbi.nlm.nih.gov/gap) through dbGaP accession numbers: phs000200.v9.p3 for the WHI Clinical Trial (CT) and Observational Study (OS) (The Women's Health Initiative Study Group, 1998); phs000386.v5.p3 for the WHI SNP Health Association Resource (SHARe) (Hutter et al, 2011); and phs000227.v2.p3 for the Population Architecture using Genomics and Epidemiology (PAGE) WHI (Spencer et al, 2013). Phenotypic data were extracted from the WHI CT and OS (phs000200.v9.p3) that is the top-level WHI study in dbGaP and includes the phenotypic information for all dbGaP WHI cohorts. Genotype data were extracted from WHI SHARe (phs000386.v5.p3) and PAGE WHI (phs000227.v2.p3). The WHI SHARe is part of the SHARe project, funded by the National Heart Lung and Blood Institute (NHLBI). The PAGE WHI is part of the PAGE project, funded by National Human Genome Research Institute (NHGRI). All dbGaP data set versions used in the current study are past their embargo periods.
Phenotypic Data
Phenotypic information available in the WHI CT and OS (phs000200.v9.p3) are available in multiple dbGaP documents. We extracted phenotypic data from the following documents: Breast Cancer Outcome (dbGaP ID: phd004164), Cancer Outcome (dbGaP ID: phd004167), Daily Life (dbGaP ID: phd002002), ECG results (dbGaP ID: phd003370, phd002025, phd003233), Eligibility Screening (dbGaP ID: phd001995), Food Frequency Questionnaire (dbGaP ID: phd003368, phd003177), Medical History (dbGaP ID: phd001996), OS Questionnaire Screening (dbGaP ID: phd002006, phd002009), Personal Habits (dbGaP ID: phd003176, phd002000), Personal Information (dbGaP ID: phd001994, phd002004), Physical Measurements (dbGaP ID: phd002014), Reproductive History (dbGaP ID: phd003174), and Thoughts and Feelings (dbGaP ID: phd002001). These dbGaP documents include 2183 items.
These phenotypic data were reviewed manually, and traits with low variation across individuals were excluded from further analysis. Quantitative traits were normalized using appropriate Box-Cox power transformations before being entered in the analysis. Quantitative traits with non-normal distribution after Box-Cox transformation were excluded. The final list of phenotypes entered in the PheWAS included 360 traits related to 12 main categories: Anthropometric traits, Cancer, Cardiovascular Health, Dietary Habits, Drinking Behaviors, Gastrointestinal Health, General Health, Physical Activity, Psychological Traits, Reproductive Health, Smoking Behaviors, and Socioeconomic Status. Details regarding the demographics and phenotypes extracted from dbGaP are available in Supplementary Tables 1 and 2. An additional sample of 2379 EUR subjects was used to replicate the findings of ADH1B rs1229984 (because this variant generated the most relevant results in the PheWAS conducted). Detailed information about phenotyping and genotyping procedures for the replication cohort are described in our previous GWAS of alcohol- and substance-dependence traits (Gelernter et al, 2014; Gelernter et al, 2015).
Genotype Data
For our PheWAS, we investigated two ADH1B variants (rs1229984 and rs2066702) and ALDH2 (rs671); and variants mapped to the CHRNA3–CHRNA5–CHRNB4 gene cluster locus (rs8034191, rs1051730, rs12914385, rs2036527, and rs16969968). We chose these variants on the basis of numerous GWAS (Gelernter et al, 2014; Xu et al, 2015) and candidate-locus studies (Sherva et al, 2010; Hart et al, 2015; Jensen et al, 2015), and a recent meta-analysis of genetic studies of nicotine and alcohol traits (Buhler et al, 2015) and GWAS performed previously in different population groups (David et al, 2012; Gelernter et al, 2014).
Genotype information was extracted from WHI SHARe (phs000386.v5.p3) and PAGE WHI (phs000227.v2.p3). The WHI SHARe includes mainly African-American and Hispanic women. For this cohort, genetic data were obtained using the Affymetrix 6.0 platform. To characterize genetic ancestry and ethnicity, we conducted a principal component analysis using flashpca software (Abraham and Inouye, 2014) and the WHI SHARe genome-wide data set pruned for linkage disequilibrium (r2>80%). We also imputed the variants that were missing from the Affymetrix array using SHAPEIT(Delaneau et al, 2012) for pre-phasing, IMPUTE2 (Howie et al, 2011) for imputation, and the 1000 Genomes Project Phase 3 (1000 Genomes Project Consortium et al, 2015) as the reference panel. We derived best-guess genotypes for each individual where the genotype probability was greater than 0.8.
The PAGE WHI mainly includes women of European descent. For this cohort, genetic data were generated using Illumina BeadXpress. The custom Illumina BeadXpress arrays covered 685 markers, but with great variation in genotyping completeness across individuals. Accordingly, genotype information was obtained only from the BeadXpress data. As no genome-wide information is available for the PAGE WHI sample, self-reported ancestry and ethnicity were used to stratify the sample. Self-reported information about ancestry and ethnicity (as opposed genetically defined values) is not the best approach to adjust population stratification in genetic studies. However, approximately 30 previous studies successfully conducted genetic investigations on the PAGE WHI cohort using approaches similar to the one applied in the current analysis (Kocarnik et al, 2015). For the African (AFR), Asian (ASN), and European (EUR) ancestry groups, we included those subjects who reported one of these ancestries (Supplementary Table 1) and excluded those with multiple self-reported ancestries. Because the genetic background of the Hispanic population group (HISP) is an admixture primarily of African, European, and Native American ancestries (Gravel et al, 2013), we included subjects who reported Hispanic ethnicity, but used both the AFR and EUR descents as covariates in the genetic analysis. Other population groups were excluded owing to low numbers. Because three variants among those selected for the PheWAS were not genotyped in the PAGE WHI cohort—ADH1B rs2066702, CHRNA5 rs12914385, and CHRNA5 rs2036527—they could not be analyzed in PAGE WHI subjects.
After quality control, the final WHI study population consisted of 26 394 women, including 7688 AFR, 1133 ASN, 14 081 EUR, and 3492 HISP subjects. Details regarding the distribution of ancestry and genotype information in the final sample across WHI cohorts are reported in Supplementary Tables 3 and 4.
Statistical Analysis
Plink 1.07 (Purcell et al, 2007) was used to implement logistic and linear regression analyses to calculate the association between genetic variants and phenotypic traits (binary and quantitative, respectively). The association analysis was stratified by population (AFR, ASN, EUR, and HISP). We considered different covariate sets, depending in part on the available genotype data: age, age-squared, and the first 10 ancestry principal components for the WHI SHARe cohort; age and age-squared for the AFR, ASN, and EUR samples of the PAGE WHI cohort; and age, age-squared, AFR, and EUR for the HISP sample of the PAGE WHI cohort. A trans-population meta-analysis was conducted using the program METAL (Willer et al, 2010). We applied a Bonferroni-based threshold for multiple testing correction based on the 360 traits and 8 genetic markers studied (=2880 tests): p-value <1.74*10−5 (phenome-wide significance, PWS) and p-value<1.74*10−4 (suggestive significance). Bonferroni correction considers the number of independent tests conducted; the correlation among the traits investigated would thus have permitted us to apply a less stringent criterion because the number of independent tests was less, effectively, than the total number conducted. However, we decided to apply the more stringent approach to present only the most reliable results. To verify further the statistical significance of our method empirically, a permutation analysis was also conducted to verify the results of ADH1B rs1229984 in the EUR sample (because this variant generated the PheWAS results with strongest statistical significance). We performed 1000 permutations of ADH1B rs1229984 genotypes in the EUR sample and tested their association with the 360 phenotypic traits (360 000 randomly permutated tests). To help understand the causative relationship of the novel phenotype associations related to ADH1B rs1229984, we performed a causative model analysis based on Bayesian networks. Specifically, we used the R package bnlearn (Scutari, 2010), applying the hill-climbing learning algorithm, and determined the 'best' causative network considering the BIC (Bayesian information criterion) score. For the replication sample, we tested the association of ADH1B rs1229984 with some of the traits that were PWS or suggestive evidences in the WHI sample, adjusting the analysis for age, age-squared, cocaine dependence, cannabis dependence, opioid dependence, and the first 10 ancestry principal components.
Supplementary Figure 1 shows the results of a power analysis to estimate the minimal detectable effects considering the total sample size of WHI data sets (N=26 394) and the phenome-wide significant threshold (p=1.74*10−5) under different scenarios. The calculations were performed using QUANTO software (available at http://biostats.usc.edu/Quanto.html).
Results
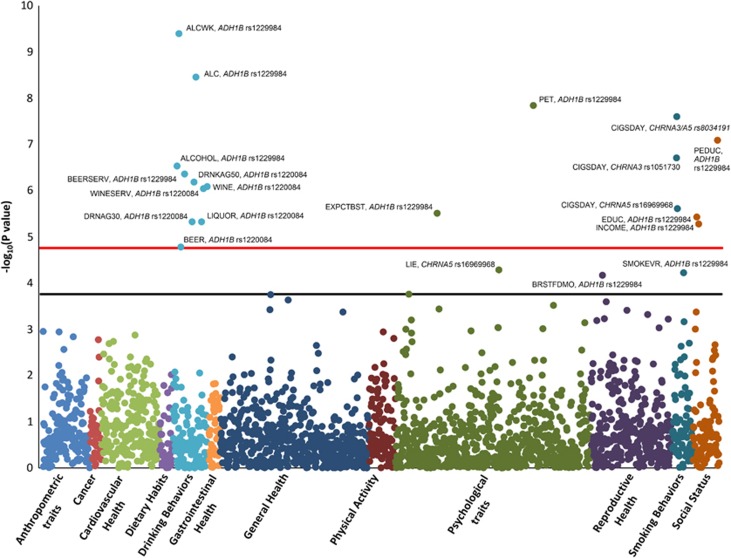
We identified several novel phenotypic associations, and replicated previously known genetic associations in both the population-specific analyses and the trans-population meta-analysis. The PheWAS results of the trans-population meta-analysis are summarized in the Manhattan plot (Figure 1).
Figure 1.
Manhattan plot of the trans-population meta-analysis including both smoking and alcohol-associated alleles. Red and black lines indicate the phenome-wide and the suggestive significance thresholds (p<1.74*10−5 and p<1.74*10−4, respectively). Phenotype abbreviations are defined in Supplementary Table 2. A full color version of this figure is available at the Neuropsychopharmacology journal online.
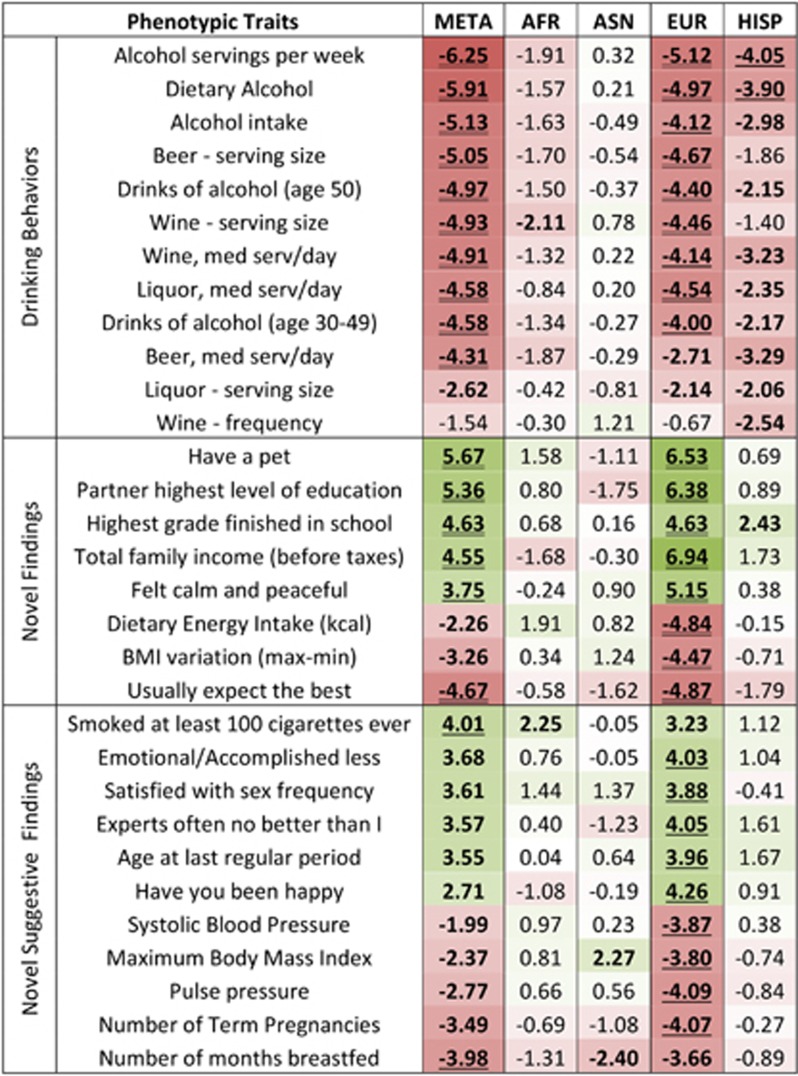
The most interesting findings were for ADH1B rs1229984 (Figure 2; Supplementary Table 5). As expected, the minor allele—known to encode a protein with increased enzymatic activity and to be protective with respect to alcohol use (Edenberg, 2007)—was negatively associated with traits related to drinking behaviors. The replications were at a PWS level (p<1.74*10−5) for trans-population meta-analysis and the EUR sample, at a suggestive significance level (p<1.74*10−4) for the HISP sample, and at nominal level for the AFR sample (p<0.05).
Figure 2.
Phenotypic associations of ADH1B rs1229984 in trans-population meta-analysis (META) and in African (AFR), Asian (ASN), European (EUR), and Hispanic (HISP) samples. In each cell, the correspondent z-score is reported (double underline: phenome-wide significance, p<1.74*10–5 based on Bonferroni correction for number of traits and markers studied; single underline: suggestive significance, p<1.74*10–4; bold: nominal significance, p<0.05). The intensity of the cell shading is proportional to the association strength (negative association: red; positive association: green). A full color version of this figure is available at the Neuropsychopharmacology journal online.
Beyond these previously known associations, novel phenotypic associations were observed for ADH1B rs1229984. At a PWS level, the minor allele was positively associated (ie, opposite direction with respect to that observed for drinking traits) with having a pet (META: N=19 877, z=5.67, p=1.44*10–8; EUR: N=9924, z=6.53, p=6.54*10–11), level of partner education (META: N=11 049, z=5.36, p=8.15*10–8; EUR: N=6351, z=6.38, p=1.78*10–10), level of education (META: N=19 806, z=4.63, p=3.73*10–6; EUR: N=9898, z=4.63, p=3.70*10–6), total family income (META: N=19 254, z=4.55, p=5.27*10–6; EUR: N=9593, z=6.94, p=4.07*10–12), and calm attitude (EUR: N=9956, z=5.15, p=2.59*10–7). Negative associations (ie, same direction as for drinking behavior associations) were observed for dietary energy intake (EUR: N=9958, z=−4.84, p=1.32*10–6), body mass index (BMI) variation (EUR: N=5338, z=−4.47, p=8.02*10–6), and optimistic attitude (META: N=19 797, z=−4.67, p=3.09*10–6; EUR: N=9908, z=−4.87, p=1.13*10–6). Other suggestive findings (p<1.74*10–4) were also observed for ADH1B rs1229984: smoking behaviors (at least 100 cigarettes lifetime); psychological traits (the subject's perception that she accomplished less than planned because of emotional issues, satisfied with sex frequency, negative attitude, happiness); reproductive health (age of onset of menopause, number of term pregnancies, number of months breastfed); and vascular/metabolic status (systolic blood pressure, maximum BMI, pulse pressure).
To confirm the reliability of ADH1B rs1229984 results, we performed 1000 permutations of ADH1B rs1229984 genotypes in the EUR sample (the population group with the strongest results). None of the 360 000 randomly permutated tests reached PWS level (Supplementary Figure 2). In addition, we tested the association of ADH1B rs1229984 with the respect to some of the traits with PWS or suggestive significance evidences (ie, maximum BMI, highest grade finished in school, household gross income, self-reported high blood pressure, number of term pregnancies, and smoked 100 cigarettes lifetime) in an independent cohort of 2379 EUR subjects (Table 1). We obtained replications (ie, concordant findings with WHI results) for household gross income (N=2349, z=1.97, p=0.035) and education (N=2376, z=2.11, p=0.048) at a nominal significance level; and possible replication for maximum BMI (N=2379, z=−1.80, p=0.071) at a trend significance level.
Table 1. Results of ADH1B rs1229984 in the Replication Sample.
| Phenotypic traits | N | Z score | p-value |
|---|---|---|---|
| Highest grade finished in school | 2376 | 1.97 | 0.049 |
| Household gross income | 2349 | 2.11 | 0.035 |
| Self-reported high blood pressure | 2379 | −0.20 | 0.843 |
| Maximum body mass index | 2379 | −1.80 | 0.071 |
| Smoked at least 100 cigarettes ever | 2302 | −0.95 | 0.345 |
| Number of term pregnanciesa | 701 | 0.31 | 0.756 |
Number of term pregnancies was tested in women only.
After quality control, information regarding ALDH2 rs671 genotypes was only available for a sample of ASN women (as expected, there was no variation in the other population groups). We replicated the protective effect of the ALDH2 rs671 allele at a phenome-wide and nominal significance level with respect to drinking behaviors (Supplementary Table 6). No other traits reached PWS or were suggestive. However, the education trait (ie, highest grade finished in school) is positively associated with ALDH2 rs671 at a nominal level (N=280, z=2.08, p=0.039). Considering this result in the context of the ADH1B rs1229984 findings described above, the same trait was PWS in the EUR sample (N=9898, z=4.63, p=3.70*10–6) and nominally significant in the HISP sample (N=2960, z=2.43, p=0.015) with concordant direction. No significant findings were observed for ADH1B rs2066702.
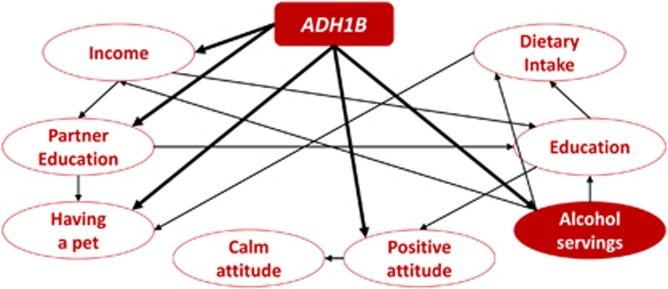
To gain insight into the biological mechanisms underlying the novel ADH1B rs1229984 phenotype associations identified in the WHI cohorts, we conducted an analysis based on Bayesian network learning algorithm. We restricted the analysis to the EUR subjects from the PAGE cohort and considered the top association for drinking behaviors (alcohol servings per week) and the novel PWS findings (having a pet, level of partner education, level of education, total family income, calm attitude, dietary energy intake, BMI variation, and optimistic attitude). The best causative model obtained from the hill-climbing learning algorithm indicated that ADH1B and alcohol servings per week affected the phenotypic traits investigated, but in some cases, the ADH1B associations are best explained as independent from drinking behaviors (Figure 3).
Figure 3.
Best causative model of the novel ADH1B rs1229984 phenotype associations identified in WHI cohorts. Phenotype abbreviations are defined in Supplementary Table 2.
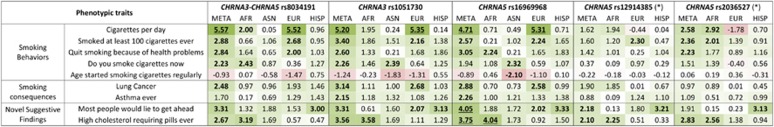
With respect to smoking behaviors, CHRNA3–CHRNA5 rs8034191, rs1051730, and rs16969968, there was replication of the well-known positive association with smoking quantity (ie, cigarettes per day) at a PWS level in trans-population meta-analysis and in the EUR sample (Figure 4; Supplementary Table 7). The CHRNA5 rs2036527 association with smoking quantity (ie, cigarettes per day) was replicated in trans-population meta-analysis and in the AFR sample. Nominally significant replications were observed for the other smoking behaviors across the population groups. We also replicated the association of CHRNA3–CHRNA5 risk alleles (rs8034191, rs1051730, and rs16969968) with lung cancer and asthma, two well-known downstream smoking consequences, in a pattern consistent with the smoking quantity associations. Two novel suggestive phenotypic associations were also observed: CHRNA5 rs16969968 was positively associated with distrustful attitude (N=25 914, z=4.05, p=5.19*10–5) in the trans-population meta-analysis and with the use of medication for high cholesterol (N=6984, z=4.04, p=5.44*10–5) in the AFR sample. Nominal replications of the CHRNA5 rs16969968 suggestive associations were observed for the other CHRNA3–CHRNA5 risk alleles across population groups.
Figure 4.
Phenotypic associations of CHRNA3–CHRNA5 locus (rs8034191, rs1051730, rs12914385, rs2036527, and rs16969968) in trans-population meta-analysis (META) and in African (AFR), Asian (ASN), European (EUR), and Hispanic (HISP) samples. In each cell, the correspondent z-score is reported (double underline: phenome-wide significance, p<1.74*10–5 based on Bonferroni correction for number of traits and markers studied; single underline: suggestive significance, p<1.74*10–4; bold: nominal significance, p<0.05). The intensity of the cell shading is proportional to the association strength (negative association: red; positive association: green). (*) WHI PAGE data set was not available for these markers. A full color version of this figure is available at the Neuropsychopharmacology journal online.
Discussion
Here, we report the first PheWAS for the study of risk alleles identified previously as relevant to drinking and smoking behaviors, in a large multi-ethnic cohort of 26 394 women. PheWAS analyses are considered a useful post-GWAS approach; such analyses can deepen our understanding of the range of phenotypic effects associated with GWAS-identified risk alleles (Bush et al, 2016). The risk alleles we studied are recognized has having large effects on nicotine response and alcohol metabolism (The Tobacco and Genetics Consortium, 2010; Gelernter et al, 2014; Quillen et al, 2014). Our results provide strong support for either pleiotropic or consequent, or both, effects of these risk alleles on other traits—physical and mental. That is, the same risk alleles could be responsible for multiple traits, or could primarily affect one or several traits with the others being a consequence of the primary action.
The strongest results of the present study were observed for ADH1B rs1229984. This functional variant is negatively associated with drinking behaviors, mainly in European populations (Gelernter et al, 2014). We replicated prior findings. As expected, the strongest ADH1B rs1229984 associations with drinking behaviors were observed in EUR women (alcoholic beverage serving per week, p=3.10*10–7). Concordant findings, but with lower statistical significance, were also observed for AFR and HISP samples (wine serving size, AFR p=0.035; alcoholic beverage servings per week, HISP p=5.16*10–5). Although it is well-known that ADH1B rs1229984 has fewer population-level effects in African-ancestry individuals than that those observed in European-ancestry populations because of its allele distribution across populations (Polimanti et al, 2015a), no study with large sample size has previously investigated the role of this variant in Hispanic populations. Our results indicate that ADH1B rs1229984 affects alcohol consumption in Hispanics also; but further studies are needed to understand why ADH1B rs1229984 has less effect on Hispanic subjects than is observed in European ones, despite an allele distribution that is comparable in the two groups. Our previous study on African- and European-Americans indicated that ADH1B population-specific haplotype structure can affect the association of some variants with alcohol use (Polimanti et al, 2015a). In our PheWAS, no relevant results were observed for ADH1B rs1229984 in the ASN sample, which is probably attributable to the power limitations from the small ASN sample available. However, we strongly replicated the protective role of ALDH2 rs671 in drinking behaviors (alcohol intake, p=3.77*10–8) that is associated with the accumulation of blood acetaldehyde and its resultant aversive effects in this ancestry group (Peng et al, 2014; Quillen et al, 2014), despite the very small sample size.
Beyond the replication of known genetic associations, we identified several novel phenotypic associations related to ADH1B rs1229984. A consistent group of psychological traits was identified (at both phenome-wide and suggestive significance levels). We identified having a pet, calm attitude, accomplished less for emotional issues, satisfied with sex frequency, and happiness, which were associated in the same direction as the protective association of ADH1B rs1229984 with drinking behaviors; and positive attitude with opposite association direction. These psychological trait associations seem to indicate that a protective allele for alcohol use behaviors is generally associated with positive psychological circumstances. A consistent epidemiological literature has shown that alcohol use and abuse are associated with psychological distress (Ibanez et al, 2015; Obasi et al, 2015).
We also observed ADH1B association with traits related to education and socioeconomic status, with the same direction as the protective association of ADH1B with drinking behaviors: partner's highest level of education, proband's highest level of education, and total family income. In particular, the association with 'highest level of education' showed PWS in the EUR sample and was replicated at a nominal significance in the HISP sample. A further replication for the same trait was observed in the ASN sample with ALDH2 rs671—that is, the gene encoding the alcohol-metabolizing enzyme that clears acetaldehyde, as distinguished from ADH1B, the protein product of which generates acetaldehyde. In addition, we replicated the education and income findings in an independent sample of 2379 EUR subjects. Our prior polygenic risk score analysis in 9863 subjects from the Generation Scotland: Scottish Family Health Study also indicated that the genetic predisposition to alcohol dependence is negatively associated with cognitive function and socioeconomic deprivation (Clarke et al, 2015). It is recognized that alcohol use and abuse are correlated with both lower income and academic achievement (Latvala et al, 2014; Poonawalla et al, 2014).
Another cluster of novel phenotype associations for ADH1B is related to dietary habits and vascular/metabolic status. Dietary energy intake, BMI variation, pulse pressure, maximum BMI, and systolic blood pressure were associated with ADH1B in a direction opposite that of its protective effect on drinking behaviors. A Mendelian randomization study in 261 991 individuals of European descent indicated that carriers of the ADH1B rs1229984 protective allele have a more favorable cardiovascular profile (Holmes et al, 2014). Furthermore, our recent GWAS of BMI in subjects with alcohol dependence showed that an alcohol-metabolism gene (in that case, ALDH1A1) could be involved in the regulation of body mass in the context of alcohol dependence (Polimanti et al, 2015b). These findings support a relationship between alcohol-metabolism gene variants with cardiovascular and metabolic conditions.
Suggestive findings were also observed in relation to reproductive health: age of onset of menopause concordant in direction with protective effect ADH1B on drinking behaviors; and number of term pregnancies and numbers of months breastfed were opposite in direction to protective effect of ADH1B on drinking behaviors. Alcohol use can have harmful effects on reproductive functions and is associated with significant sex hormone changes in women (Schliep et al, 2015). The association between ADH1B rs1229984 and age of onset of menopause is in agreement with these previous findings: women with a protective allele for drinking behaviors undergo menopause (a physiological process strictly regulated by sex hormones) later than those without the allele.
These novel associations indicate that ADH1B is involved in a wide range of physical and mental conditions. Our analysis based on Bayesian learning algorithms revealed that ADH1B is involved in the traits identified by our PheWAS and drinking behaviors are apparently intermediary for some of them only. Although some alcohol-independent effects of ADH1B on human phenome find support in other functions for the encoded enzyme (Haavik et al, 2008), our analysis should be considered exploratory. Indeed, the WHI assessment for drinking and smoking behaviors is focused primarily on current use rather than lifetime use. This likely caused a systematic underestimation of the effect of alcohol use on the causative relationship analysis of ADH1B associations.
The PheWAS results for CHRNA3–CHRNA5 risk alleles replicated the known associations with smoking behaviors. As previously observed (The Tobacco and Genetics Consortium, 2010), CHRNA3–CHRNA5 rs8034191, rs1051730, and rs16969968 were strongly associated with smoking quantity (ie, cigarettes per day) in European subjects. We also replicated the association of CHRNA5 rs2036527 with the same trait in the AFR sample, consistent with results from a large GWAS of smoking quantity in African-American subjects (David et al, 2012). Unfortunately, we were not able to replicate the associations of CHRNA5 rs12914385, likely because the genotypes for this variant were not available for the PAGE WHI cohort and therefore the data were available in only 36% of the total sample (2% of the total EUR sample). Consistent with the observed smoking quantity associations, we found that CHRNA3–CHRNA5 rs8034191, rs1051730, and rs16969968 were associated with lung cancer and asthma. Although genetic studies of lung cancer identified CHRNA3–CHRNA5 as a risk locus previously (Chen et al, 2015), very few investigations have focused on CHRNA3–CHRNA5 locus and risk of asthma (Torjussen et al, 2012). Although there were no novel PWS results for smoking-related alleles, we observed two suggestive findings. In trans-population meta-analysis, CHRNA5 rs16969968 was associated with distrustful attitude in the same direction as the risk for smoking behaviors. Tobacco use, abuse, and dependence frequently co-occur with several psychiatric traits and disorders (Picciotto and Kenny, 2013) especially with depression and anxiety (Morozova et al, 2015). However, results from a recent Mendelian randomization analysis did not support a causal role of smoking heaviness towards the development of depression and anxiety (Taylor et al, 2014). In our study, individuals with a smoking-related risk allele tended to report more frequently that 'most people lie to get ahead' than those without the risk allele. Further studies are needed to clarify the role of nicotine consumption, and the genetic predisposition to nicotine consumption, in psychological status, considering physiological and pathological conditions. In the AFR sample, a suggestive association was observed between CHRNA5 rs16969968 and cholesterol-lowering medication use: women with a smoking risk allele were more likely to use cholesterol-lowering medication than those without risk allele. Smoking behaviors have previously been associated with several cardiovascular risk factors and strong effects have been observed on cholesterol levels (Forey et al, 2013). Global genetic correlations were found between smoking and lipid levels (cholesterol and triglycerides) based on linkage disequilibrium score regression analysis, considering large GWAS incorporating data relevant to these traits (Bulik-Sullivan et al, 2015). Our data suggest that genetic predisposition to nicotine response may potentially affect lipid levels (either directly or indirectly).
In conclusion, we provided novel information regarding the role of tobacco and alcohol use and some of their best-established associated genetic risk loci on an extensive set of physical and mental traits. Our data indicated that alcohol and nicotine use genetics potentially has much broader implications than is currently recognized. ADH1B appears to be a relevant locus for several phenotypic traits also via mechanisms that may be independent from alcohol use. Although the biological significance for some of the phenotypic associations identified herein is not completely clear, our current data provide new insights regarding the roles of the investigated genes in the human phenome. Further studies are needed to follow up these new possible routes and mechanisms. In particular, Mendelian randomization studies of a larger cohort can facilitate a better understanding of how genetic information can be used to understand the impact of modifiable exposures, such as tobacco and alcohol use (Gage et al, 2016), in relation to the phenotypic traits identified by the current PheWAS.
Funding and disclosure
This study was supported by National Institutes of Health grants RC2 DA028909, R01 DA12690, R01 DA12849, R01 DA18432, R01 AA11330, R01 AA017535, P50 AA012870, the VISN1 and VISN4 MIRECCs, and a NARSAD Young Investigator Award (to RP) from the Brain & Behavior Research Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Dr Kranzler has been an advisory board member, consultant, or CME speaker for Indivior, Lundbeck, and Otsuka. He is also a member of the American Society of Clinical Psychopharmacology's Alcohol Clinical Trials Initiative, which is supported by AbbVie, Alkermes, Ethypharm, Indivior, Lilly, Lundbeck, Pfizer, and XenoPort. The other authors reported no biomedical financial interests or potential conflicts of interest.
Acknowledgments
The publicly available data sets used for the analyses described in this manuscript were obtained from dbGaP through dbGaP accession number phs000200.v9.p3 (WHI CT and OS), phs000386.v5.p3 (WHI SHARe), and phs000227.v2.p3 (PAGE WHI). The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts N01WH22110, 24152, 32100-2, 32105-6, 32108-9, 32111-13, 32115, 32118-32119, 32122, 42107-26, 42129-32, and 44221. This manuscript was not prepared in collaboration with investigators of the WHI, has not been reviewed and/or approved by the WHI, and does not necessarily reflect the opinions of the WHI investigators or the NHLBI. WHI PAGE is funded through the NHGRI PAGE network (Grant Number U01 HG004790). Assistance with phenotype harmonization, SNP selection, data cleaning, meta-analyses, data management and dissemination, and general study coordination was provided by the PAGE Coordinating Center (U01HG004801-01). Funding for WHI SHARe genotyping was provided by NHLBI Contract N02-HL-64278.
Footnotes
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp)
Supplementary Material
References
- 1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM et al (2015). A global reference for human genetic variation. Nature 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham G, Inouye M (2014). Fast principal component analysis of large-scale genome-wide data. PLoS One 9: e93766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler KM, Gine E, Echeverry-Alzate V, Calleja-Conde J, de Fonseca FR, Lopez-Moreno JA (2015). Common single nucleotide variants underlying drug addiction: more than a decade of research. Addict Biol 20: 845–871. [DOI] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR et al (2015). An atlas of genetic correlations across human diseases and traits. Nat Genet 47: 1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush WS, Oetjens MT, Crawford DC (2016). Unravelling the human genome-phenome relationship using phenome-wide association studies. Nat Rev Genet 17: 129–145. [DOI] [PubMed] [Google Scholar]
- Chen LS, Hung RJ, Baker T, Horton A, Culverhouse R, Saccone N et al (2015). CHRNA5 risk variant predicts delayed smoking cessation and earlier lung cancer diagnosis—a meta-analysis. J Natl Cancer Inst 107: pii: djv100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TK, Smith AH, Gelernter J, Kranzler HR, Farrer LA, Hall LS et al (2015). Polygenic risk for alcohol dependence associates with alcohol consumption, cognitive function and social deprivation in a population-based cohort. Addict Biol 21: 469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove KP, Wang S, Kim SJ, McGovern E, Nabulsi N, Gao H et al (2014). Sex differences in the brain's dopamine signature of cigarette smoking. J Neurosci 34: 16851–16855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David SP, Hamidovic A, Chen GK, Bergen AW, Wessel J, Kasberger JL et al (2012). Genome-wide meta-analyses of smoking behaviors in African Americans. Transl Psychiatry 2: e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Marchini J, Zagury JF (2012). A linear complexity phasing method for thousands of genomes. Nat Methods 9: 179–181. [DOI] [PubMed] [Google Scholar]
- Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, Mosley JD et al (2013). Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol 31: 1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druesne-Pecollo N, Tehard B, Mallet Y, Gerber M, Norat T, Hercberg S et al (2009). Alcohol and genetic polymorphisms: effect on risk of alcohol-related cancer. Lancet Oncol 10: 173–180. [DOI] [PubMed] [Google Scholar]
- Drug Therapeutics Bulletin (2014). Republished: Nicotine and health. BMJ 349: 2014 2017 0264rep. [Google Scholar]
- Edenberg HJ (2007). The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health 30: 5–13. [PMC free article] [PubMed] [Google Scholar]
- Forey BA, Fry JS, Lee PN, Thornton AJ, Coombs KJ (2013). The effect of quitting smoking on HDL-cholesterol - a review based on within-subject changes. Biomark Res 1: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage SH, Davey Smith G, Ware JJ, Flint J, Munafo MR (2016). G = E: what GWAS can tell us about the environment. PLoS Genet 12: e1005765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinsky KJ, Bhatia G, Loh PR, Georgiev S, Mukherjee S, Patterson NJ et al (2016). Fast principal-component analysis reveals convergent evolution of ADH1B in Europe and East Asia. Am J Hum Genet 98: 456–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Herman AI, Koesterer R et al (2015). Genome-wide association study of nicotine dependence in American populations: identification of novel risk loci in both African-Americans and European-Americans. Biol Psychiatry 77: 493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH et al (2014). Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry 19: 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Zakharia F, Moreno-Estrada A, Byrnes JK, Muzzio M, Rodriguez-Flores JL et al (2013). Reconstructing Native American migrations from whole-genome and whole-exome data. PLoS Genet 9: e1004023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R, Ren J (2010). Alcohol and acetaldehyde in public health: from marvel to menace. Int J Environ Res Public Health 7: 1285–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haavik J, Blau N, Thony B (2008). Mutations in human monoamine-related neurotransmitter pathway genes. Hum Mutat 29: 891–902. [DOI] [PubMed] [Google Scholar]
- Hallden S, Sjogren M, Hedblad B, Engstrom G, Hamrefors V, Manjer J et al (2015). Gene variance in the nicotinic receptor cluster (CHRNA5-CHRNA3-CHRNB4) predicts death from cardiopulmonary disease and cancer in smokers. J Intern Med 279: 388–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AB, Lynch KG, Farrer L, Gelernter J, Kranzler HR (e-pub ahead pf print 1 April 2015). Which alcohol use disorder criteria contribute to the association of ADH1B with alcohol dependence? Addict Biol. [DOI] [PMC free article] [PubMed]
- Holmes MV, Dale CE, Zuccolo L, Silverwood RJ, Guo Y, Ye Z et al (2014). Association between alcohol and cardiovascular disease: Mendelian randomisation analysis based on individual participant data. BMJ 349: g4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Marchini J, Stephens M (2011). Genotype imputation with thousands of genomes. G3 (Bethesda) 1: 457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter CM, Young AM, Ochs-Balcom HM, Carty CL, Wang T, Chen CT et al (2011). Replication of breast cancer GWAS susceptibility loci in the Women's Health Initiative African American SHARe Study. Cancer Epidemiol Biomarkers Prev 20: 1950–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez MI, Camacho L, Mezquita L, Villa H, Moya-Higueras J, Ortet G (2015). Alcohol expectancies mediate and moderate the associations between big five personality traits and adolescent alcohol consumption and alcohol-related problems. Front Psychol 6: 1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KP, DeVito EE, Herman AI, Valentine GW, Gelernter J, Sofuoglu M (2015). A CHRNA5 smoking risk variant decreases the aversive effects of nicotine in humans. Neuropsychopharmacology 40: 2813–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocarnik JM, Park SL, Han J, Dumitrescu L, Cheng I, Wilkens LR et al (2015). Pleiotropic and sex-specific effects of cancer GWAS SNPs on melanoma risk in the population architecture using genomics and epidemiology (PAGE) study. PLoS One 10: e0120491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latvala A, Rose RJ, Pulkkinen L, Dick DM, Korhonen T, Kaprio J (2014). Drinking, smoking, and educational achievement: cross-lagged associations from adolescence to adulthood. Drug Alcohol Depend 137: 106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhao H, Gelernter J (2011). Strong association of the alcohol dehydrogenase 1B gene (ADH1B) with alcohol dependence and alcohol-induced medical diseases. Biol Psychiatry 70: 504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhao H, Gelernter J (2012). Strong protective effect of the aldehyde dehydrogenase gene (ALDH2) 504lys (*2) allele against alcoholism and alcohol-induced medical diseases in Asians. Hum Genet 131: 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova M, Rabin RA, George TP (2015). Co-morbid tobacco use disorder and depression: A re-evaluation of smoking cessation therapy in depressed smokers. Am J Addict 24: 687–694. [DOI] [PubMed] [Google Scholar]
- Obasi EM, Brooks JJ, Cavanagh L (2015). The relationship between psychological distress, negative cognitions, and expectancies on problem drinking: exploring a growing problem among university students. Behav Modif 40: 51–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandeya N, Olsen CM, Whiteman DC (2013). Sex differences in the proportion of esophageal squamous cell carcinoma cases attributable to tobacco smoking and alcohol consumption. Cancer Epidemiol 37: 579–584. [DOI] [PubMed] [Google Scholar]
- Peng GS, Chen YC, Wang MF, Lai CL, Yin SJ (2014). ALDH2*2 but not ADH1B*2 is a causative variant gene allele for Asian alcohol flushing after a low-dose challenge: correlation of the pharmacokinetic and pharmacodynamic findings. Pharmacogenet Genomics 24: 607–617. [DOI] [PubMed] [Google Scholar]
- Picciotto MR, Kenny PJ (2013). Molecular mechanisms underlying behaviors related to nicotine addiction. Cold Spring Harb Perspect Med 3: a012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polimanti R, Yang C, Zhao H, Gelernter J (2015. a). Dissecting ancestry genomic background in substance dependence genome-wide association studies. Pharmacogenomics 16: 1487–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polimanti R, Zhang H, Smith AH, Zhao H, Farrer LA, Kranzler HR et al (e-pub ahead of print 12 October 2015. b). Genome-wide association study of body mass index in subjects with alcohol dependence. Addict Biol. [DOI] [PMC free article] [PubMed]
- Poonawalla IB, Kendzor DE, Owen MT, Caughy MO (2014). Family income trajectory during childhood is associated with adolescent cigarette smoking and alcohol use. Addict Behav 39: 1383–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillen EE, Chen XD, Almasy L, Yang F, He H, Li X et al (2014). ALDH2 is associated to alcohol dependence and is the major genetic determinant of "daily maximum drinks" in a GWAS study of an isolated rural Chinese sample. Am J Med Genet B Neuropsychiatr Genet 165B: 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliep KC, Zarek SM, Schisterman EF, Wactawski-Wende J, Trevisan M, Sjaarda LA et al (2015). Alcohol intake, reproductive hormones, and menstrual cycle function: a prospective cohort study. Am J Clin Nutr 102: 933–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scutari M (2010). Learning Bayesian Networks with the bnlearn R Package. J Stat Softw 35: 1–22.21603108 [Google Scholar]
- Sherva R, Kranzler HR, Yu Y, Logue MW, Poling J, Arias AJ et al (2010). Variation in nicotinic acetylcholine receptor genes is associated with multiple substance dependence phenotypes. Neuropsychopharmacology 35: 1921–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer KL, Malinowski J, Carty CL, Franceschini N, Fernandez-Rhodes L, Young A et al (2013). Genetic variation and reproductive timing: African American women from the Population Architecture using Genomics and Epidemiology (PAGE) Study. PLoS One 8: e55258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AE, Fluharty ME, Bjorngaard JH, Gabrielsen ME, Skorpen F, Marioni RE et al (2014). Investigating the possible causal association of smoking with depression and anxiety using Mendelian randomisation meta-analysis: the CARTA consortium. BMJ Open 4: e006141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Tobacco and Genetics Consortium (2010). Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet 42: 441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Women's Health Initiative Study Group (1998). Design of the Women's Health Initiative clinical trial and observational study. Control Clin Trials 19: 61–109. [DOI] [PubMed] [Google Scholar]
- Torjussen TM, Lodrup Carlsen KC, Munthe-Kaas MC, Mowinckel P, Carlsen KH, Helms PJ et al (2012). Alpha-nicotinic acetylcholine receptor and tobacco smoke exposure: effects on bronchial hyperresponsiveness in children. Pediatr Allergy Immunol 23: 40–49. [DOI] [PubMed] [Google Scholar]
- Treur JL, Boomsma DI, Lubke GH, Bartels M, Vink JM (2014). The predictive value of smoking expectancy and the heritability of its accuracy. Nicotine Tob Res 16: 359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Jiang K, Yuan W, Cui W, Li MD (2016). Contribution of variants in CHRNA5/A3/B4 gene cluster on chromosome 15 to tobacco smoking: from genetic association to mechanism. Mol Neurobiol 53: 472–484. [DOI] [PubMed] [Google Scholar]
- White A, Castle IJ, Chen CM, Shirley M, Roach D, Hingson R (2015). Converging patterns of alcohol use and related outcomes among females and males in the United States, 2002 to 2012. Alcohol Clin Exp Res 39: 1712–1726. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR (2010). METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SH, Guo Q, Viken RJ, Reed T, Dai J (2014). Heritability of usual alcohol intoxication and hangover in male twins: the NAS-NRC Twin Registry. Alcohol Clin Exp Res 38: 2307–2313. [DOI] [PubMed] [Google Scholar]
- Xu K, Kranzler HR, Sherva R, Sartor CE, Almasy L, Koesterer R et al (2015). Genomewide association study for maximum number of alcoholic drinks in European Americans and African Americans. Alcohol Clin Exp Res 39: 1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.