Abstract
Large variance in reproductive success is the primary factor that reduces effective population size (Ne) in natural populations. In sequentially hermaphroditic (sex-changing) fish, the sex ratio is typically skewed and biased towards the ‘first' sex, while reproductive success increases considerably after sex change. Therefore, sex-changing fish populations are theoretically expected to have lower Ne than gonochorists (separate sexes), assuming all other parameters are essentially equal. In this study, we estimate Ne from genetic data collected from two ecologically similar species living along the eastern coast of South Africa: one gonochoristic, the ‘santer' sea bream Cheimerius nufar, and one protogynous (female-first) sex changer, the ‘slinger' sea bream Chrysoblephus puniceus. For both species, no evidence of genetic structuring, nor significant variation in genetic diversity, was found in the study area. Estimates of contemporary Ne were significantly lower in the protogynous species, but the same pattern was not apparent over historical timescales. Overall, our results show that sequential hermaphroditism may affect Ne differently over varying time frames, and that demographic signatures inferred from genetic markers with different inheritance modes also need to be interpreted cautiously, in relation to sex-changing life histories.
Introduction
The amount of genetic diversity in a species reflects the effective population size (Ne), which in practical terms informs on the number of breeders that contribute to the offspring, generation after generation; this number, especially in some marine animals, has been estimated to be several orders of magnitude lower than census size (Nc) (Hauser and Carvalho, 2008). Census size fluctuations and ecological perturbations are known to reduce Ne, especially in fragmented populations. However, life history traits also have a fundamental role in determining the effective size of a population (reviewed in Caballero (1994)). Lee et al. (2011) suggest that delayed age at maturity and lowered juvenile survival reduce Ne/Nc. Recently, age at maturity and adult lifespan were shown to explain half of the variance in Ne/Nc among 63 animal and plant species (Waples et al., 2013). Thus, variation in key life history traits related to mating success and survival rates, through their effect on individual lifetime reproductive success, appear to shape Ne differences among populations.
In teleosts, older and larger females generally produce more and larger eggs (Chambers and Leggett, 1996; Palumbi, 2004). Larger size also improves the mating capacity of males through behaviours such as dominance and protection of territories (Warner, 1988). Hence, growth rate is also likely a key factor determining reproductive success. However, additional reproductive traits in marine fish might account for further components of lifetime variance in reproductive success. Sequential hermaphrodites first mature as one sex and, after changing, reproduce as the opposite sex. Since younger and smaller individuals of the first sex are generally more abundant than older and larger individuals of the second sex, sequential hermaphrodite species typically present skewed sex ratios compared with gonochoristic (separate sexes) species (Allsop and West, 2004). According to the size advantage model (Ghiselin, 1969; Warner, 1975), reproductive success in sequential hermaphrodites tends to increase considerably after sex change, with individuals of the ‘second sex' expected to have a greater contribution to the next generation. Therefore, the age at which individuals change sex—which has been shown to fluctuate in response to environmental factors (Hamilton et al., 2007; Mariani et al., 2013)—will have a significant impact on the lifetime reproductive success in sequential hermaphrodites. As a result, intrinsically biased sex ratios (Wright, 1931) and variance in reproductive success are theoretically expected to result in lower Ne in sequentially hermaphroditic species, compared with gonochoristic ones, assuming that other characteristics are somewhat equal. This is inevitably complicated by the flexible nature of age-/size-at-sex change in natural populations (see Ross, 1990, Avise and Mank, 2009; Mariani et al., 2013, for discussion).
In the present study, by directly examining empirical data, we investigate whether sex-changing life history may indeed determine a reduction in Ne as a result of increased lifetime variance of reproductive success and skewed sex ratio. We compared genetic data in two closely related and sympatric species (family: Sparidae) with largely comparable habitat, ecology, abundance, behaviour and life-history traits, with the exception of their reproductive modes: one species being protogynous (the slinger sea bream Chrysoblephus puniceus, which first matures as female, later turning to male) and the other being gonochoristic (santer sea bream Cheimerius nufar maturing either as male or female; Garratt, 1985a, b; 1986, 1993). C. puniceus is endemic and restricted to the south-east coast of southern Africa, while C. nufar is distributed over a wider area of the Western Indian Ocean. Both species are targeted by the same local commercial and recreational line fisheries, and together represent a large proportion of catches from this region (Mann and Fennessy, 2013a, b). Both species are opportunistic predators found in shoals around coastal reefs feeding on crustaceans, mollusks and small fish (Garratt, 1986). Sex ratios were found to fluctuate greatly in C. puniceus, both spatially, between southern Mozambique and the KwaZulu-Natal region (Garratt, 1985a), and temporally (Mariani et al., 2013), with changes to some extent influenced by the degree of fishing pressure. In contrast, even sex ratios were found in C. nufar (Garratt, 1985a). Results obtained using nuclear and mitochondrial molecular markers are interpreted as a function of inheritance mode and reproductive strategy. The findings enhance our understanding of the role of life history in population genetics and may have implications for the management of exploited populations.
Materials and methods
Sampling
C. puniceus and C. nufar specimens were collected from commercial ski-boat line fishermen between May and July 2007 at three locations along the KwaZulu-Natal coast: Port Edward, Park Rynie and Richards Bay (RB) (Figure 1). A total of 138 C. puniceus (122 females and 16 males) and 139 C. nufar (69 females and 67 males) were collected.
Figure 1.
Map: sampling locations off the KwaZulu-Natal Coast of South Africa.
Fork length and weight were measured and sex was assessed by macroscopic gonad identification. We used previously published length–age relationships to derive age from fork length for both C. puniceus (Garratt et al., 1993) and C. nufar (Druzhinin, 1975; Coetzee and Baird, 1981). Fin clips were taken from the pectoral fins and stored in absolute ethanol for later DNA extraction.
Molecular analyses
DNA was extracted using a modified phenol–chloroform protocol (Sambrook and Russell, 2001). DNA concentration and quality were estimated on a NanoDropTM ND-1000 (Thermo Fisher Scientific Inc., Wilmington, DE, USA) spectrophotometer. Samples of both species were screened at 11 microsatellite loci, some specifically developed for this study (Chopelet et al., 2009a). Of the 11 microsatellites designed for C. puniceus, 5 cross-amplified in C. nufar. Six supplementary microsatellites were specifically developed for C. nufar, using the same protocol as in Chopelet et al. (2009a) (see Table 1 for details). Microsatellites were amplified using fluorescence-labelled forward primers (Applied Biosystems, Waltham, MA, USA) and 2X Multiplex PCR Master Mix (Qiagen, Hilden, Germany) in a final volume of 10 μl. Depending on size and dye, fragments were amplified into two multiplexed reactions for C. puniceus (Chopelet et al., 2009a), while another microsatellite (SL3) was amplified separately. For C. nufar, one reaction contained SL25 and SA2, and the other included the nine remaining loci (Table 1). All amplifications were carried out using the same conditions. An initial step of 15 min at 95 °C was followed by 30 cycles of 45 s at 94 °C, 45 s at 60 °C and 45 s at 72 °C, and a final extension step at 72 °C for 45 min. PCR products were sized on an ABI 3130xl alongside a GS600 ladder. Genemapper v 4 (Applied Biosystems) was used for allele scoring.
Table 1. Variability of 10 polymorphic microsatellite loci in santer Cheimerius nufar from South Africa (n=131) grouped into two multiplex reactions (I and II).
| Locus name | GenBank accession number | Dye label | Primer sequences (5′–3′) | Repeat motif | Ta | Size range (bp) | NA | He | Ho |
|---|---|---|---|---|---|---|---|---|---|
| SA1 (I) | 6FAM | F:CAGCGATGCACAGTAAAGTACC R:AGCATACAGAGGCCTTCAGC | (TG)33 | 58 | 253–293 | 19 | 0.93 | 0.94 | |
| SA2 (II) | NED | F:GAGCCAGACTCCAGACATCC R:CCGGACAGGAGTATTGAAGC | (GTCT)11 | 58 | 190–234 | 8 | 0.75 | 0.73 | |
| SA3 (I) | PET | F:CCAGAGTCTGTGCTGTGAGTGC R:TCCTTGTGGTCCACTTTACG | (CA)17 | 58 | 386–420 | 13 | 0.89 | 0.77 | |
| SA6 (I) | 6FAM | F:AGCTGCTGCTCATCTCACG R:GCAGTGTTAACATCTTCGAATGC | (TG)12 | 58 | 187–209 | 9 | 0.75 | 0.79 | |
| SA10 (I) | 6FAM | F:GAAGCCAAACGAGGACAGC R:GTGAGGAGCATGCTAATACCG | (GT)15GA(GT)26 GAGTGAGTGA(GT)18 | 58 | 428–532 | 42 | 0.96 | 0.92 | |
| SA25 (I) | VIC | F:GGAGGAAATGAACCGATGG R:GCAGCTGGTCAATAGTGTGG | (TG)7CA(TG)6 | 58 | 152–220 | 19 | 0.61 | 0.70 | |
| SL25 (II) | FJ526983 | NED | F:GGTACTGTTTGGCCCTTGC R:GCCTGGTAATATGCCTGAGC | (GA)11TG(GA)3TA(GA)4GTCA (GA)2AA(GA)9CA(GA)2AA(GA)12 | 58 | 207–271 | 7 | 0.61 | 0.66 |
| SL26 (I) | FJ526984 | PET | F:TGAAGGTGCTGATGACTTTCC R:CAGTCCTGCCTCTGACTGG | (TC)2TT(TC)9GC(TC)4TGCCTT (TC)5GC(TC)7 | 58 | 239–247 | 5 | 0.38 | 0.32 |
| SL27 (I) | FJ526985 | VIC | F:CAGCCTCAGCTCATTTCTCC R:CCTGCCCTCCTGTAGATGC | (TG)55 | 58 | 183–201 | 6 | 0.35 | 0.32 |
| SL34 (I) | FJ526988 | VIC | F:GCGTGCACACTCTTACAGTACC R:TCGGATGTGCATCTCATAGG | (CA)17 | 58 | 321–361 | 19 | 0.84 | 0.85 |
Abbreviations: He, expected heterozygosity; Ho, observed heterozygosity; NA, number of alleles observed; Ta, annealing temperature (°C).
For slinger Chrysoblephus puniceus, see Chopelet et al. (2009a).
Universal primers Hsp1 and Lsp1 were also used to amplify the first hypervariable region of the mitochondrial DNA (mtDNA) control region (Ostellari et al., 1996). Each reaction was carried out using 300 ng of genomic DNA in Ready Mix (Applied Biosystems) in a final volume of 25 μl. PCR cycles were as follows: (a) 95 °C (5 min); (b) 30 cycles at 95 °C (50 s), 56 °C 1 min and 72 °C (2 min); and (c) a final 10 min elongation at 72 °C (10 min). Amplified products were purified with exonuclease I and shrimp alkaline phosphatase (Hanke & Wink 1994) and sequenced at GATC-Biotech (Konstanz, Germany).
Statistical analyses
Genetic diversity
Genetic analyses were first performed to detect patterns of spatial structure and estimate diversity within and between locations. For microsatellite data, frequencies of null alleles were estimated using FREENA (Chapuis and Estoup, 2007). For each location sample, number of alleles (Na), observed (Ho) and expected (He) heterozygozity and inbreeding coefficient (FIS) were assessed using GENEPOP ON THE WEB (http://genepop.curtin.edu.au/; Raymond and Rousset, 1995; Rousset, 2008). Marker neutrality was tested in LOSITAN (Antao et al., 2008). To correct for variance in the sample size among populations, we further estimated the allelic richness (AR) based on the minimum sample size (Table 1) using FSTAT 2.9.3 (Goudet, 1995) and the number of private alleles using the rarefaction method (Kalinowski, 2005) implemented in ADZE 1.0 (Szpiech et al., 2008). Population differentiation was estimated using the θ estimator of FST (Weir and Cockerham, 1984) and relative confidence intervals using 10 000 permutations on the individuals in GENETIX v.4.05.2 (Belkhir et al., 1996–2004). Although the mutation rate was in our instance likely to be orders of magnitude smaller than migration rates, the corrected F′ST (Hedrick, 2005) was calculated, and Jost's Dest (Jost, 2008) estimated using SMOGD (Crawford, 2010), and reported as additional information on genetic substructure. POWSIM was used to evaluate the power of the data set to detect genetic differentiation (Ryman and Palm, 2006). Five hundred replications and different Ne/t ratios (500/0; 2000/2; 1000/5; 1000/10; and 500/10) were used to obtain the expected FST according to this equation: FST=1−(1−1/2Ne)t, with t being the number of generations of isolation (Ryman and Palm, 2006). Bayesian assignment was performed in STRUCTURE 2.3 (Pritchard et al., 2000; Falush et al., 2003, 2007) to infer the most likely number of genetic clusters (K) present in the data sets using the admixture model, and 500 000 iterations, after 50 000 burn-in. The number of clusters was calculated by averaging the mean posterior probability of the data L(K) over 10 independent runs.
Nucleotide and haplotype diversities were estimated from mtDNA sequences using DNASP v4.5 (Rozas et al., 2003). Median-Joining networks were constructed for both species using POPART (http://popart.otago.ac.nz).
Estimating effective population size
We used the linkage disequilibrium (LD) method implemented in LDNe (Waples and Do, 2008) to estimate contemporary Ne for each location from the microsatellite data, both including and excluding the markers that were not under Hardy-Weinberg equilibrium according to the exact test performed in Genepop. The LD method is based on the theoretical relationship (Hill, 1981) between a measure of LD (r2=squared correlation of alleles at pairs of unlinked gene loci), sample size (N) and Ne. LDNe implements a modification of Hill's method that accounts for bias from ignoring second order terms in N and Ne. LDNe allows one to screen out rare alleles, which tend to upwardly bias Ne estimates, by selecting a minimum allowable allele frequency (PCrit). We focused especially on PCrit=0.02, which (given the minimum sample sizes of N=39–45; Table 2) ensured that any alleles that occurred in a single copy were not used (Waples and Do, 2010). Another estimate of effective population size was obtained using the Approximate Bayesian Computation method implemented in DIYABC 2.01 (Cornuet et al., 2014). Calculations were performed for each species, pooling samples from all locations in order to reflect the lack of genetic substructure detected in our data. Three simple scenarios were simulated. Each one represented one single population whose Ne remained constant (scenario 1), one where Ne increased after a time t1 (scenario 2) and the third where Ne decreased after t1. Priors were as follows: effective population size was between 10 and 106; and t1 between 10 and 104 generations.
Table 2. Genetic diversity of slinger Chrysoblephus puniceus and santer Cheimerius nufar estimated with microsatellites and mitochondrial DNA.
|
Microsatellites |
Mitochondrial DNA |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Location | N | NA | AR | He | Ho | FIS | Ne | Ne(HWE)LD | Ne(HWE)ABC | N | Nhap | Hd | π | |
| C. puniceus (slinger) | PE | 43 | 18.5 | 18 | 0.83 | 0.78 | 0.05 | 835 (287 to ∞) | 707 (259 to ∞) | 24 | 23 | 0.996 | 0.03 | |
| PR | 41 | 18.7 | 18.2 | 0.83 | 0.81 | 0.02 | 128 (93 to 187) | 535 (228 to ∞) | 25 | 24 | 0.997 | 0.03 | ||
| RB | 38 | 18 | 18 | 0.83 | 0.79 | 0.04 | 166 (114 to 292) | 462 (198 to ∞) | 26 | 26 | 1 | 0.03 | ||
| All | 125 | 18.4 | 18.1 | 0.83 | 0.79 | 0.04 | 371 (289 to 511) | 601 (404 to 1124) | 560 000 (52 000 to 980 000) | 75 | 65 | 0.996 | 0.03 | |
| C. nufar (santer) | PE | 43 | 11.6 | 11.8 | 0.72 | 0.74 | 0.06 | ∞ (−261 to ∞) | ∞ (327 to ∞) | 25 | 22 | 0.99 | 0.04 | |
| PR | 45 | 12.4 | 12.4 | 0.74 | 0.76 | −0.04 | ∞ (−340 to ∞) | ∞ (733 to ∞) | 26 | 20 | 0.969 | 0.04 | ||
| RB | 42 | 12.1 | 12.1 | 0.72 | 0.70 | 0.02 | ∞ (405 to ∞) | ∞ (353-∞) | 26 | 24 | 0.994 | 0.04 | ||
| All | 131 | 12 | 12.1 | 0.73 | 0.73 | 0.01 | 2236 (654 to ∞) | 3233 (743 to ∞) | 370 000 (23 000 to 960 000) | 77 | 55 | 0.984 | 0.04 | |
Abbreviations: AR, allelic richness; FIS, inbreeding coefficient; Hd, haplotype diversity; He, expected heterozygosity; Ho, observed heterozygosity; HWE, hardy-weinberg equilibrium; N, number of samples analysed; NA, number of alleles; Nhap, number of haplotypes; π, nucleotide diversity; PE, Port Edward; PR, Park Rynie; RB, Richards Bay.
The three Ne estimates refer to, in order: (i) estimates based on LDNe using all loci; (ii) estimates based on LDNe excluding loci departing from HW equilibrium; (iii) estimates based on DIYABC.
A longer-term view of effective population size was also obtained through estimates of historical female Ne from mtDNA data. We first used the Watterson estimator of the mutation parameter theta (θ) obtained from the number of polymorphic sites (S) (Watterson, 1975). In DNASP v4.5, θ is defined as 2Neμ for mtDNA, where Ne is the effective population size and μ is the mutation rate per DNA sequence per generation (Tajima 1996). We estimated the female effective population size (Nef) from the haplotype mutation rate and generation time (T) according to this equation:
We assumed a widely accepted rate μ=11% per site per million year for the Sparid mtDNA control region (Bargelloni et al., 2003; Sala-Bozano et al., 2009; Coscia et al., 2012), equal to 0.055 substitutions per site per million years. The age at maximum egg production was estimated with Linf=47 cm for C. puniceus and Linf=75 cm for C. nufar (where Linf, a parameter of the von Bertalanffy growth equation, is defined as the length that an individual would reach if it grew indefinitely). This, according to Beverton (1992), can be used as an approximation of generation time (T=5 for C. puniceus and 7 for C. nufar). Therefore, to account for life-history plasticity, we estimated Ne in both species with generation time encompassing these values: T=3, 5 and 8.
Furthermore, we applied the Bayesian Skyline Plot approach implemented in BEAST v 1.7 (Drummond et al., 2012) to estimate trends in past effective population size. Firstly, jModelTest 0.1.1 (Posada, 2008; Guindon and Gascuel, 2003) was used to select the best model of substitution for each data set via the AIC (Akaike Information Criterion): GTR (Generalised Time Reversible described in Tavaré (1986)) was selected for C. nufar and HKY (Hasegawa 1985) for C. puniceus. To avoid convergence issues, several independent runs (each 106 generations and 10% burn-in) were used for each species until each effective sample size value reached ~200 as per the user's manual.
Results
Species and population characteristics
Males (375–1404 g) were larger than females (257–952 g) in C. puniceus, while male (420–1436 g) and female sizes (440–2820 g) overlapped in C. nufar (Supplementary Figures S1 and S2). Individuals from the most northern location, Richards Bay, had slightly larger sizes in both species (Supplementary Figure S2). Fifteen male C. puniceus were found in Richards Bay and one male in Park Rynie, while only females were collected in Port Edward (southernmost location). A recent study has shown that the likely age-at-sex-change for C. puniceus is around 301 mm, significantly lower than three decades ago (Mariani et al., 2013), and potentially decreased as a result of increased fishing pressure.
Genetic variation
No evidence of null alleles was detected within the data set. At least one marker in each of the two species deviated from equilibrium expectations across all locations with strongly positive associated FIS values (SA3 in C. nufar and SL35 in C. puniceus, Supplementary Table S2). As LOSITAN did not detect any signature of selection (data not shown) and no significant FIS was recorded for any species at each location (Table 2), all markers were retained in the subsequent analyses.
Expected and observed heterozygosities were 0.83 in all locations for C. puniceus, and varied between 0.72 (Port Edward and Richards Bay) and 0.74 (Park Rynie) for C. nufar. Allelic richness and the number of alleles were higher in C. puniceus (overall NA=18.4 and AR=18.1) than C. nufar (overall NA=12 and AR=12.1; Table 2). No significant FST values were found within either species (Supplementary Table S1; C. puniceus overall FST=0.0011, P=0.19; C. nufar overall FST=0.0004, P=0.37). No significant genetic differentiation was found between the three samples, for each species, irrespective of the method used (overall Dest was 0.0036 for C. puniceus and 0.0010 for C. nufar) or corrections used (Hedrick's corrected G′ST was 0.0047 for C. puniceus and 0.0019 for C. nufar). Hence, as expected, STRUCTURE detected one genetic cluster in each species (K=1, Supplementary Figure S3). According to the power estimations implemented in POWSIM, the probability that our data sets can detect low genetic differentiations up to an FST of 0.005 is 100% (Supplementary Figure S4), with high probabilities (70–80%) also for values around 0.0025. This suggests a lack of genetic structuring among the three locations sampled at microsatellites (Supplementary Table S1).
A total of 75 individual C. puniceus and 77 C. nufar were sequenced (179 and 223 bp long fragments of the control region, respectively; Genbank Accession Nos: KX358082–KX358233). Overall, nucleotide diversity π was 0.03 for C. puniceus and 0.04 for C. nufar, while haplotype diversity Hd was 0.996 for C. puniceus, with 65 haplotypes, and 0.984 for C. nufar, with 55 haplotypes. Within species, both π and Hd did not vary, with the former being 0.03 and 0.04 for C. puniceus and C. nufar, respectively, across all sampling locations (Table 2). Hd ranged between 1 (Richards Bay) and 0.996 (Port Edward) for C. puniceus, and between 0.994 (Richards Bay) and 0.969 (Park Rynie) for C. nufar. No significant ΦST was detected for any of the two species between any location (Supplementary Table S1), and visual inspection of median joining networks also showed a lack of geographical structure (Supplementary Figures S5 and S6).
Effective population size
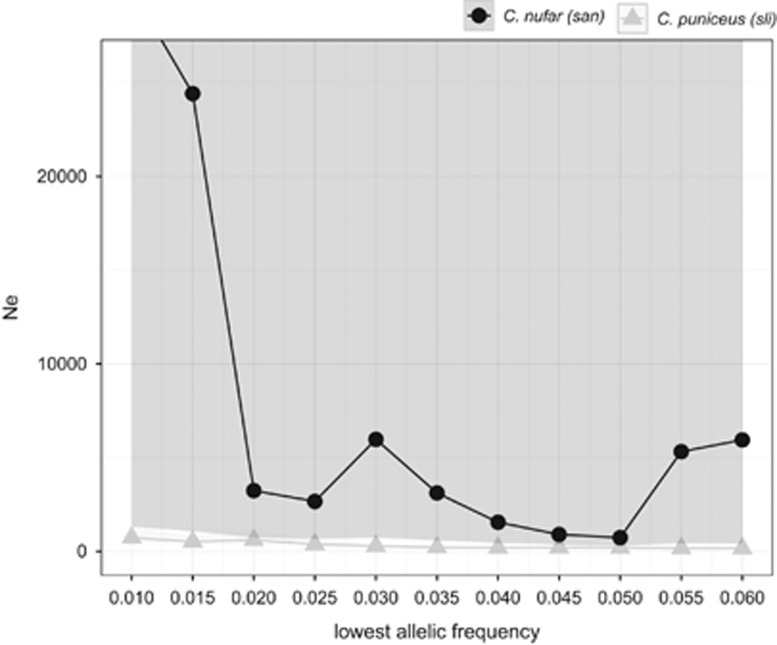
Ne estimates using LDNe tended to increase with extreme values of allelic frequencies (Pcrit) (Figure 2). However, C. puniceus consistently had much smaller Ne and smaller variances than C. nufar, irrespective of the allelic frequencies included (Figure 2). Estimates of effective population size (Ne) at Pcrit=0.02 are reported in Table 2. In C. nufar, negative Ne were interpreted as infinite (Waples and Do, 2010); the lower boundaries of the confidence intervals for C. nufar did not overlap with the highest boundaries of the C. puniceus estimates, using all loci, and only marginally overlapped when we excluded the locus not in hardy-weinberg equilibrium (Table 2). The pattern was found to be robust and consistent, whether samples were pooled or treated as separate geographical collections. Historical Ne was estimated from microsatellites using DIYABC. Of the three scenarios simulated, the second one was by far the most likely for C. puniceus (Ne increased in time), while scenarios 1 and 2 were equally likely for C. nufar. For consistency with C. puniceus data—and in line with population expansion results from mtDNA data (see below)—we used scenario 2 for C. nufar too. Estimates of effective population size (t0) were slightly higher for C. puniceus than C. nufar, but the confidence limits largely overlapped (Table 2).
Figure 2.
Effective population size estimates: effective population size of Cheimerius nufar (santer, in black) and Chrysoblephus puniceus (slinger, in grey) estimated with different allelic frequencies (or Pcrit). The three sampled populations were pooled.
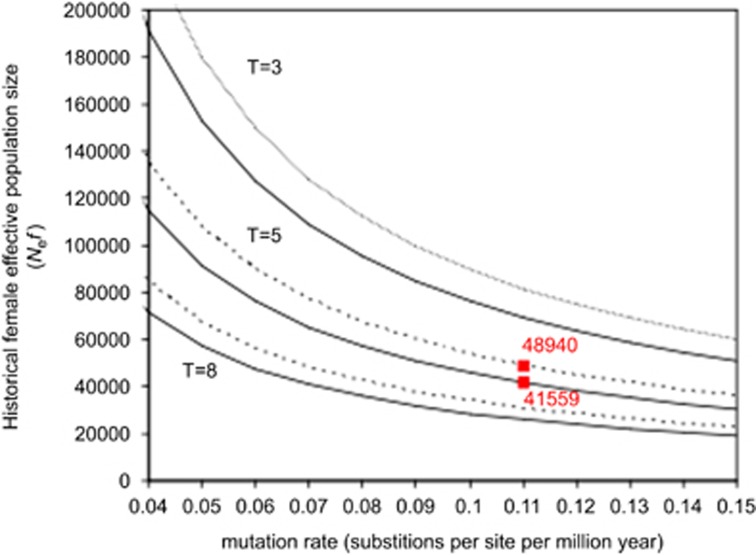
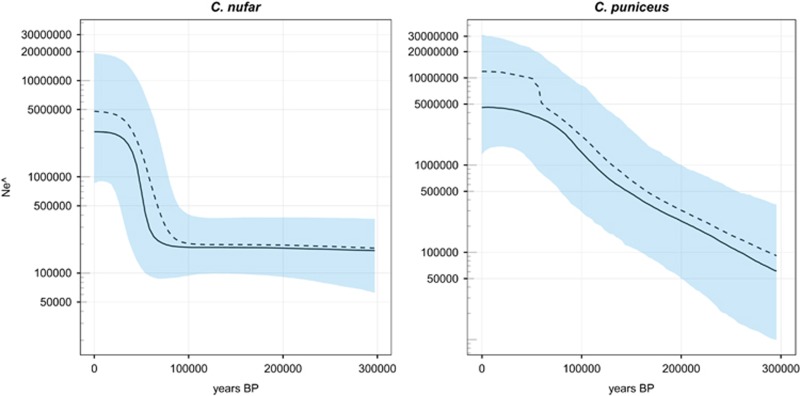
Historical female population size inferred from mtDNA offered further insight into demographic aspects, but the obtained estimates proved to be of different orders of magnitude depending on the method used. The Watterson method returned estimates in the order of the tens of thousands (Figure 3), slightly larger in C. nufar than in C. puniceus, all else being equal; however, considering the longer generation time in the latter, estimates for the protogynous C. puniceus are slightly greater than for C. nufar (Figure 3). The Bayesian Skyline method (Figure 4) also indicates larger effective size in C. puniceus, although the confidence limits largely overlap.
Figure 3.
Historical female effective population size: historical female effective population size (Nef) of slinger Chrysoblephus puniceus (continuous line) and santer Cheimerius nufar (dotted line) based on θ for different mutation rates (μ) and three generation lengths (T=3, 5 and 8 years). Red values correspond to Nef estimations for μ=11% and T=5. A full color version of this figure is available at the Heredity journal online.
Figure 4.
Bayesian Skyline plots: Bayesian Skyline plots for santer Cheimerius nufar and slinger Chrysoblephus puniceus. The continuous and dashed lines represent the median and mean values, respectively.
Discussion
No significant genetic divergence was detected among populations of either species, using either microsatellites or the variable mitochondrial control region, suggesting that the three sampled populations for each species do not belong to independent evolutionarily significant units. This lack of genetic differentiation is also mirrored by similar levels of genetic diversity among locations. Similar findings for C. puniceus were recently reported by Duncan et al. (2015) who reported a single panmictic stock. The life histories and behaviour of both species are consistent with the lack of genetic differentiation within this area: they live in large shoals in the vicinity of rocky and coral reefs and are capable of migration across tens of kilometres (Garratt, 1984; Griffiths and Wilke, 2002). On the basis of studies of histological changes in the ovaries, it appears that spawning in C. puniceus occurs in large aggregations off the coasts of southern Mozambique and Northern KwaZulu-Natal (mostly to the north of Durban) and decreases to the south of Park Rynie (Garratt, 1985a, b). Most C. puniceus males sampled in this study were from Richards Bay, in accordance with this pattern. The majority of eggs are fertilized and scattered above coral and rocky reefs from northern locations, and the developing larvae drift southward influenced by the strong Agulhas Current in this region (Garratt, 1985a, b). Juveniles settling out to the south of Park Rynie, where no spawning takes place, are believed to move northward later in life to reproduce (Maggs et al., 2013). Accordingly, average sizes were found to be higher in the northernmost location of Richards Bay. Such southward larval dispersal followed by northward adult migration could result in a constant redistribution of alleles across the area, ultimately ensuring population cohesion.
C. nufar has an extended distribution along the African east coast. The spawning area stretches from the Eastern Cape to the Gulf of Aden and individuals in spawning condition are found regularly in both Eastern Cape and KwaZulu-Natal waters (Coetzee, 1983; Garratt, 1985a, b). Patterns of adult migration and settlement of juveniles are less well known for C. nufar (Griffiths and Wilke, 2002), but it is likely that other separate spawning aggregations exist in more northern, unsampled areas of the Western Indian Ocean.
This study examined population genetic inference of Ne in the context of sex change in marine fish. Other sources of Ne variation were minimized by comparing two species with very similar taxonomical and biological characteristics, and by obtaining replicate samples from the same biological units and dwelling in the same locations. No estimates of census size exist for these species, but these are also likely to be similar in the study area: C. puniceus and C. nufar are known to be the two most abundant commercial line-fishery targets in the South-African east coast, with annual landings around 300 000 and 100 000 individuals, respectively, but with the fishery that targets a habitats slightly more suited to the former (Winker et al., 2012).
All methods to calculate effective population size will assume discrete generations; therefore, underestimation of Ne may occur here, but will likely affect both species similarly (Waples et al., 2014). Predictions from population genetics theory led to the expectation that a sex changing species would present smaller Ne than a gonochoristic one for the following reasons: (a) the sex ratio of the sex-changing species is biased towards the ‘first sex', which is known to reduce Ne (Wright, 1931; Chopelet et al., 2009b); and (b) the dynamic balance of age-at-sex change, which affects individual variance in reproductive success (Vk). Using the LD method, we find agreement with these expectations, with substantially lower effective sizes in the protogynous C. puniceus; while Ne estimates in C. nufar were one order or magnitude larger, with infinite upper confidence boundaries, indicating that more precise estimates in this species would ideally require greater sample sizes (Palstra and Ruzzante, 2008). Obviously, the Ne/Nc ratios of these species may be different, but unpublished surveys indicate that local abundances are in the same order of magnitude (Winker et al., 2012). Protogynous populations can still therefore sustain large numbers through a relatively small number of breeders; over short timescales, a population may be dominated by a relatively small number of strong, large breeding males, belonging to 2- or 3-year classes, and able to pass their gene combinations to most of the new cohorts. On the other hand, ABC computation provides no evidence that Ne differs significantly between breeding strategies. Point estimates and confidence limits are very similar in the two species, suggesting that over evolutionary timescales, some factors are at play in reducing the expected high variance in individual reproductive success of sex changers. One such scenario could be the inherently ‘bet-hedging' nature of a sex-changing life history, whereby successful genotypes with optimal timing of first maturation and sex change are able to maximize reproductive output as both females and males, hence reducing long-term stochasticity.
Effective population size estimates based on mtDNA do not reveal substantial differences between C. nufar and C. puniceus, except for the magnitude of the point estimates (that is, millions with BEAST and tens of thousands with the Watterson method). Although the Bayesian Skyline Plot shows overlapping confidence limits, Ne appears larger in C. puniceus; with the Watterson method—once taken into account that the generation time for C. puniceus is lower (Garratt, 1985a, b; Mann et al., 2014)—there also seems to be a trend towards greater Ne in C. puniceus. Interestingly, in a protogynous species like C. puniceus, virtually every individual has the potential to reproduce as a female, with only a fraction becoming males. This is likely to maintain a large mtDNA diversity over evolutionary timescales. In gonochoristic species like C. nufar, only about half of individuals (the females) from every new cohort will transmit their mtDNA, which may explain the patterns detected here using maternally inherited loci. On the basis of this, protogynous species can be expected to have mtDNA diversity similar or higher, while protandrous species would likely be lower, than that estimated for gonochoristic species.
This is potentially an important fact to be considered in conservation biology, and presumably similar (yet opposite) considerations should be applied to protandrous species (where only a smaller proportion of individuals will reproduce as females). This hypothesis can potentially be tested already using the available literature. However, expectations will always require adjustment according to the degree of plasticity of sex-changing patterns; for instance, a recent stock assessment of the C. puniceus population in South Africa has revealed strong evidence of recovery since the introduction of a cut in commercial fishing effort in 2003–2006 (Winker et al., 2012). This may have been facilitated by earlier age-at-sex-change inferred by Mariani et al. (2013).
Much remains to be understood about the interaction among life history traits in determining lifetime variance in reproductive success and, by reflection, the effective size of a population, especially when the added complexities of sex change, its timing and its extent are added to the picture. Beyond fecundity, longevity and age at maturation, sex change is a particularly labile trait: individuals of the first sex might sometimes be larger than individuals of the second sex, it is not always the largest individuals of a group that change sex (Munoz and Warner, 2003), and some individuals in the population might not change sex at all (Mann and Buxton, 1998). In the case of C. puniceus it is likely that the large females that do not change sex make a significant contribution to future generations (Berkeley et al., 2004; Palumbi, 2004). Furthermore, the social status of individuals within the group will affect the timing of sex change, as will behavioural and/or biochemical (pheromones) interactions (Munday et al., 2006). Our initial empirical analyses show that theoretical predictions may be supported over contemporary time scales, but other mechanisms may play bigger roles over evolutionary periods.
In a population, it is generally possible to distinguish different groups such as individuals of the same age or with the same sex. The reproductive success (the average number of gametes transmitted to the next generations) of these groups can differ significantly (males can produce and transmit more gametes than females, or vice versa, depending on operational sex ratios). In sex-changing species the partitioning of reproductive success depends on the age-at-sex change. Some individuals change sex earlier and might present higher overall lifetime reproductive success than those changing sex at older stages, although shifts in age-at-sex-change will likely be linked with trade-offs with growth, maturity and longevity (Allsop and West, 2003). The next step towards offering generality to this indication will require a detailed understanding of how age-at-sex-change and other life-history traits shape the reproductive success and Ne. Furthermore, the remarkably diverse magnitude range of Ne estimates obtained through different methods, using the same data sets, suggests that, while these can be useful in a within-method comparative approach, much remains to be done to match method estimation with the appropriate timescales (Waples, 2005). We expect that the analysis of genetic data in larger sets of sex-changing and gonochoristic species pairs will be required to shed more light on the significance of sex change in affecting Ne and the evolutionary trajectories of populations and species.
Data archiving
Microsatellite data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.g36ch. Mitochondrial data available from GenBank: accession numbers KX358082—KX358233.
Acknowledgments
This study is part of a project funded by Science Foundation Ireland. We are grateful to all the fishermen of the KwaZulu-Natal coast and the staff at the Oceanographic Research Institute, who helped with sampling. The authors are grateful to Allan McDevitt for help with data analysis and for his insightful comments on the manuscript. The paper was improved through the suggestions of the associate editor and three reviewers.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Allsop D, West S. (2003). Changing sex at the same relative body size—similar forces may select for gender switching across taxa in all animals with this facility. Nature 425: 783–784. [DOI] [PubMed] [Google Scholar]
- Allsop D, West S. (2004). Sex-ratio evolution in sex changing animals. Evolution 58: 1019–1027. [DOI] [PubMed] [Google Scholar]
- Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G. (2008). LOSITAN: a workbench to detect molecular adaptation based on a FST-outlier method. BMC Bioinformatics 9: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise JC, Mank J. (2009). Evolutionary perspectives on hermaphroditism in fishes. Sex Dev 3: 152–163. [DOI] [PubMed] [Google Scholar]
- Bargelloni L, Alarcón JA, Alvarez MC, Penzo E, Magoulas A, Reis C et al. (2003). Discord in the family Sparidae (Teleostei): divergent phylogeographical patterns across the Atlantic-Mediterranean divide. J Evol Biol 16: 1149–1158. [DOI] [PubMed] [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. (1996–2004) Laboratoire Génome, Populations, Interactions. CNRS UMR 5171, Université de Montpellier II: Montpellier, France. [Google Scholar]
- Berkley S, Chapman C, Sogard S. (2004). Maternal age as a determinant of larval growth and survival in a marine fish, Sebastes melanops. Ecology 85: 1258–1264. [Google Scholar]
- Beverton RJH. (1992). Patterns of reproductive strategy parameters in some marine teleost fishes. J Fish Biol 41: 137–160. [Google Scholar]
- Caballero A. (1994). Developments in the prediction of effective population size. Heredity 73: 657–679. [DOI] [PubMed] [Google Scholar]
- Chambers RC, Leggett WC. (1996). Maternal influences on variation in egg sizes in temperate marine fishes. Am Zool 36: 180–196. [Google Scholar]
- Chapuis MP, Estoup A. (2007). Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol 24: 621–631. [DOI] [PubMed] [Google Scholar]
- Chopelet J, Helyar S, Mann B, Mariani S. (2009. a). Novel polymorphic microsatellite loci for the protogynous hermaphrodite slinger sea bream (Chrysoblephus puniceus, Sparidae). Mol Ecol Resour 9: 1223–1226. [DOI] [PubMed] [Google Scholar]
- Chopelet J, Waples RS, Mariani S. (2009. b). Sex change and the genetic structure of marine fish populations. Fish Fish 10: 329–343. [Google Scholar]
- Coetzee P. (1983). Seasonal histological and microscopical changes in the gonads of Cheimerius nufar (Ehrenberg, 1820) (Sparidae: Pisces). S Afr J Zool 18: 76–88. [Google Scholar]
- Coetzee P, Baird D. (1981). Age, growth and food of Cheimerius nufar (Ehrenberg, 1820) (Sparidae), collected off St Croix Island, Algoa Bay. S Afr J Zool 18: 76–88. [Google Scholar]
- Cornuet J-M, Pudlo P, Veyssier J, Dehne-Garcia A, Gautier M, Leblois R et al. (2014). DIYABC v2.0: a software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 30: 1187–1189. [DOI] [PubMed] [Google Scholar]
- Coscia I, Vogiatzi E, Kotoulas G, Tsigenopoulos C, Mariani S. (2012). Exploring neutral and adaptive processes in expanding populations of gilthead sea bream, Sparus aurata L., in the North-East Atlantic. Heredity 108: 537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NG. (2010). SMOGD: software for the measurement of genetic diversity. Mol Ecol Resour 10: 556–557. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druzhinin A. (1975). Some data on sparid fishes (Fam. Sparidae) of the Gulf of Aden region. J Ichthyol 15: 531–541. [Google Scholar]
- Duncan M, James N, Fennessy S, Mutombene R, Mwale M. (2015). Genetic structure and consequences of stock exploitation of Chrysoblephus puniceus, a commercially important sparid in the South West Indian Ocean. Fish Res 164: 64–72. [Google Scholar]
- Falush D, Stephens M, Pritchard JK. (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. (2007). Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7: 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garratt PA. (1984). The biology and fishery of Chrysoblephus puniceus (Gilchrist and Thompson, 1971) and Cheimerus nufar (Ehrenberg, 1830), two offshore sparids in Natal waters. University of Natal Unpublished M.Sc. thesis.
- Garratt P. (1985. a). The offshore linefishery of Natal: I. Exploited population structures of the sparids Chrysoblephus puniceus and Cheimerius nufar. Oceanographic Research Institure: Durban, South Africa. Investigational Report No. 62, pp 1–18.
- Garratt P. (1985. b). The offshore linefishery of Natal: II. Reproductive biology of the sparids Chrysoblephus puniceus and Cheimerius nufar. Oceanographic Research Institute: Durban, South Africa, Investigational Report No. 62, pp 1–21.
- Garratt P. (1986). Protogynous hermaphroditism in the slinger Chrysoblephus puniceus (Gilchrist & Thompson 1908) (Teleostei: Sparidae). J Fish Biol 28: 297–306.
- Garratt P, Comparative aspects of the reproductive biology of seabreams (Pisces: Sparidae). PhD thesis, Rhodes University, Grahamstown, South Africa, 1993.
- Garratt P, Govender A, Punt A. (1993). Growth acceleration at sex change in the protogynous hermaphrodite Chrysoblephus puniceus (Pisces: Sparidae). S Afr J Mar Sci 13: 187–193. [Google Scholar]
- Ghiselin MT. (1969). Evolution of hermaphroditims among animals. Q Rev Biol 44: 189–208. [DOI] [PubMed] [Google Scholar]
- Goudet J. (1995). FSTAT (Version 1.2): A computer program to calculate F-statistics. J Hered 86: 485–486. [Google Scholar]
- Griffiths M, Wilke C. (2002). Long-term movement patterns of five temperate reef-fishes (Pisces: Sparidae): implications for marine reserves. Mar Freshw Res 53: 233–244. [Google Scholar]
- Guindon S, Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704. [DOI] [PubMed] [Google Scholar]
- Hamilton S, Caselle J, Standish J, Schroeder D, Love M, Rosales-Casian J et al. (2007). Size-selective harvesting alters life histories of a temperate sex-changing fish. Ecol Appl 17: 2268–2280. [DOI] [PubMed] [Google Scholar]
- Hanke M, Wink M. (1994). Direct DNA sequencing of PCR-amplified vector inserts following enzymatic degradation of primers and dNTPs. BioTechniques 17: 858–860. [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano T. (1985). Dating of human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22: 160–174. [DOI] [PubMed] [Google Scholar]
- Hauser L, Carvalho GR. (2008). Paradigm shifts in marine fisheries genetics: ugly hypotheses slain by beautiful facts. Fish Fish 9: 333–362. [Google Scholar]
- Hedrick PW. (2005). A standardized genetic differentiation measure. Evolution 59: 1633–1638. [PubMed] [Google Scholar]
- Hill W. (1981). Estimation of effective population size from data on linkage disequilibrium. Genet Res 38: 209–216. [Google Scholar]
- Jost L. (2008). G(ST) and its relatives do not measure differentiation. Mol Ecol 17: 4015–4026. [DOI] [PubMed] [Google Scholar]
- Kalinowski ST. (2005). HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5: 187–189. [Google Scholar]
- Lee AM, Engen S, Saether B-E. (2011). The influence of persistent individual differences and age at maturity on effective population size. Proc Biol Sci 278: 3303–3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggs J, Mann B, Cowley P. (2013). Reef fish display station-keeping and ranging behaviour in the Pondoland Marine Protected Area on the east coast of South Africa. Afr J Mar Sci 35: 183–193. [Google Scholar]
- Mann B, Buxton C, Russell B, Pollard D, Carpenter KE. (2014) Chrysoblephus puniceus. The IUCN Red List of Threatened Species 2014: e.T170259A1303743. Available at http://dx.doi.org/10.2305/IUCN.UK.2014-3.RLTS.T170259A1303743.en.
- Mann B, Buxton CD. (1998). The reproductive biology of Diplodus sargus capensis and D. cervinus hottentotus (Sparidae) off the south-east Cape coast, South Africa. Cybium 22: 31–47. [Google Scholar]
- Mann B, Fennessy S. (2013. a) Chrysoblephus puniceus. In: Mann B (ed), Southern African Marine Linefish Species Profiles vol. 9, Oceanographic Research Institute: Durban, South Africa, pp 212–214. [Google Scholar]
- Mann B, Fennessy S. (2013. b) Cheimerius nufar. In: Mann B (ed), Southern African Marine Linefish Species Profiles Vol. 9, Oceanographic Research Institute: Durban, South Africa, pp 225–227. [Google Scholar]
- Mariani S, Sala-Bozano M, Chopelet J, Benvenuto C. (2013). Spatial and temporal patterns of size-at-sex-change in two exploited coastal fish. Environ Biol Fishes 96: 535–541. [Google Scholar]
- Munday PL, White JW, Warner RR. (2006). A social basis for the development of primary males in a sex-changing fish. Proc Biol Sci 273: 2845–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz RC, Warner RR. (2003). A new version of the size-advantage hypothesis for sex change: incorporating sperm competition and size-fecundity skew. Am Nat 161: 749–761. [DOI] [PubMed] [Google Scholar]
- Ostellari L, Bargelloni L, Penzo E, Patarnello P, Patarnello T. (1996). Optimization of single-strand conformation polymorphism and sequence analysis of the mitochondrial control region in Pagellus bogaraveo (Sparidae, Teleostei): rationalized tools in fish population biology. Anim Genet 27: 423–427. [DOI] [PubMed] [Google Scholar]
- Palstra FP, Ruzzante DE. (2008). Genetic estimates of contemporary effective population size: what can they tell us about the importance of genetic stochasticity for wild population persistence? Mol Ecol 17: 3428–3447. [DOI] [PubMed] [Google Scholar]
- Palumbi S. (2004). Why mothers matter. Nature 430: 621–622. [DOI] [PubMed] [Google Scholar]
- Posada D. (2008). jModelTest: phylogenetic model averaging. Mol Biol Evol 25: 1253–1256. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. (1995). GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86: 248–249. [Google Scholar]
- Ross R. (1990). The evolution of sex-change mechanisms in fishes. Environ Biol Fishes 29: 81–93. [Google Scholar]
- Rousset F. (2008). GENEPOP' 007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour 8: 103–106. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. (2003). DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- Ryman N, Palm S. (2006). POWSIM: a computer program for assessing statistical power when testing for genetic differentiation. Mol Ecol Notes 6: 600–602. [DOI] [PubMed] [Google Scholar]
- Sala-Bozano M, Ketmaier V, Mariani S. (2009). Contrasting signals for multiple markers illuminate population connectivity in a marine fish. Mol Ecol 18: 4811–4826. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell D. (2001) Molecular Cloning, 3rd edn. Cold Spring Harbor Laboratory Press: New York, NY, USA. [Google Scholar]
- Szpiech ZA, Jakobsson M, Rosenberg NA. (2008). ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics 24: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. (1996). The amount of DNA polymorphism maintained in a finite population when the neutral mutation rate varies among sites. Genetics 143: 1457–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavaré S. (1986). Some probabilistic and statistical problems in the analysis of dna sequences. In American Mathematical Society: Lectures on Mathematics in the Life Sciences, vol 17. American Mathematical Society: Providence, RI, USA, pp 57–86.
- Waples RS. (2005). Genetic estimates of contemporary effective population size: to what time periods do the estimates apply? Mol Ecol 14: 3335–3352. [DOI] [PubMed] [Google Scholar]
- Waples RS, Antao T, Luikart G. (2014). Effects of overlapping generations on linkage disequilibrium estimates of effective population size. Genetics 197: 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples RS, Do C. (2008). LDNE: a program for estimating effective population size from data on linkage disequilibrium. Mol Ecol Resour 8: 753–756. [DOI] [PubMed] [Google Scholar]
- Waples RS, Do C. (2010). Linkage disequilibrium estimates of contemporary N-e using highly variable genetic markers: a largely untapped resource for applied conservation and evolution. Evol Appl 3: 244–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples RS, Luikart G, Faulkner JR, Tallmon DA. (2013). Simple life-history traits explain key effective population size ratios across diverse taxa. Proc Biol Sci 280: 20131339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner RR. (1975). Adaptive significance of sequential hermaphroditism in animals. Am Nat 109: 61–82. [Google Scholar]
- Warner RR. (1988). Sex change and the size-advantage model. Trends Ecol Evol 3: 133–136. [DOI] [PubMed] [Google Scholar]
- Watterson G. (1975). On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Weir B, Cockerham C. (1984). Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- Winker H, Kerwath S, Attwood C. (2012). South African Department of Agriculture, Forestry and Fisheries, Linefish Scientific Working Group Report No. 3, pp 65.
- Wright S. (1931). Evolution in Mendelian populations. Genetics 16: 97–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.