ABSTRACT
21A is an Alu non-coding (nc) RNA transcribed by RNA polymerase (pol) III. While investigating the biological role of 21A ncRNA we documented an inverse correlation between its expression level and the rate of cell proliferation. The downregulation of this ncRNA not only caused a boost in cell proliferation, but was also associated to a transient cell dedifferentiation, suggesting a possible involvement of this RNA in cell dedifferentiation/reprogramming. In this study, we explored the possibility to enhance proliferation and dedifferentiation of cells of interest, by 21A down-regulation, using a mixture of chemically modified Anti-21A RNAs. Our results confirmed the validity of this approach that allows the amplification of specific cell populations, in a controlled manner and without inducing permanent effects. In addition to induce cell proliferation, the procedure did not decrease the tissue regeneration potential of progenitor cells in two different cell systems.
KEYWORDS: Alu, antisense RNAs, cell proliferation, differentiation, mesenchimal stem cells, non-coding RNAs
Introduction
Recent advances in mammalian genome studies highlight the occurrence of a widespread transcription of non-coding (nc) RNAs, mostly devoted to the regulation of the expression of protein coding genes.1-3 In this scenario, a significant fraction of RNA molecules is ascribable to the Alu class of repetitive sequences that represents about one-tenth of the whole human genome.4 Up to date, a long series of biological roles have been attributed to Alu short transcripts5 including RNA editing,6 alternative splicing regulation,7 chromosomal recombination,8 gene-expression regulation,9 cell stress response,10,11 putative miRNA targets.12 Notably, the observation that the transcription of Alu elements is dependent on RNA polymerase (pol) III highlights a previously unexpected role in gene-expression regulation of this machinery that deserves further investigations.2
In a previous study, a computer search for upstream pol III promoter elements of small nuclear RNA transcriptional units allowed us to identify a novel set of approximately 30 ncRNA elements mostly mapping within protein-coding regions and/or sharing a high sequence homology (either in sense or antisense configuration) with pol II–transcribed genomic regions. On these basis, we proposed a synergic mechanism of gene expression control, in which pol III elements might act as trans-locus regulators of their correspondent pol II target genes by interfering with their mRNA maturation or translation.2 In the recent years we defined the transcriptional role of several of these ncRNA.13–19 While investigating the biological role of one of these transcription units (named 21A), we documented an inverse correlation between the expression level of 21A ncRNA and the rate of cell proliferation.2 Indeed, the transfection of cells with a construct expressing 21A ncRNA in antisense configuration led to a marked increase of cell proliferation. This finding suggested the possibility that, using a controlled interference with 21A RNA expression, one could promote, in a controlled manner, the proliferation of poorly proliferating cells or of cells existing in a limited number for different biotechnological applications including therapeutical purposes. Since the transfection of plasmid DNA in cells intended for cell therapy procedures is per se unsafe, a possible reduction of 21A expression might be obtained transfecting transiently chemically-modified anti-21A dsRNAs whose half-life in the cell is less than 48 hours. However, since proliferation and differentiation are known to be inversely correlated in the cell, it was reasonably expected that the increased proliferation driven by anti-21A RNA transfection, might be also accompanied by effects on the cell differentiation, thus suggesting a further investigation of this aspect.
Here, we report that a specific mixture of chemically modified, 32 to 38 nt-long, anti-21A RNAs induced a transient/recoverable increase of cell proliferation in vitro. This treatment, tested in neuron-like differentiated neuroblastoma cells, reproducibly led to a boost of cell proliferation, while maintaining their susceptibility to differentiation stimuli. We also showed that the cell treatment with chemically modified anti-21A RNAs induced the proliferation of human bone-marrow derived mesenchymal stem cells (MSC) without reducing their osteogenic potential and avoiding the permanent activation of undesired pathways that might lead to adverse effects including tumorigenesis.
Results
Definition of a safe protocol to down-regulate 21A RNAs in cells
In a previous work we documented a remarkable increase of HeLa cell proliferation as a consequence of 21A ncRNA downregulation and/or silencing.2 This finding suggested that the induction of a transient and controlled “boost” of cell proliferation and the associated partial de-commitment, might be of relevant interest for the in vitro expansion of poorly proliferating cells or of cells limited in number and intended for biotechnological applications including cell therapy. In order to define a safe protocol for the down-regulation of 21A RNAs, we excluded the use of a plasmid DNA harboring 21A transcriptional region in antisense (AS) configuration as it might be detrimentally integrated in the genome of the host cell. In addition, in our experimental plan we also considered that RNA:RNA pairing of long antisense RNA molecules (more than 40 nts) most likely would induce interferon responses.20 Therefore, to overcome the above issues we synthesized three chemically-stabilized (orthomethylated) short ssRNAs (38nt-, 38nt- and 32nt-long, hereafter referred to as α, β and γ respectively), each of them matching a specific region of 21A transcript sequence and possibly folded in different secondary structures with peculiar susceptibility to degradation (see supplementary material online 1).
To investigate the molecular details of the anti-21A RNAs-dependent cell proliferation increase, we transfected a genetically modified neuroblastoma cell line SKNBE2-S113 characterized by a reduced cell proliferation rate and a partially differentiated phenotype. This cell line tool was appropriate not only to highlight the effects of anti-21A RNAs transfection on the proliferation of more mature cells, but also to identify possible effects on the cell differentiation status.13,21
First, we tested the kinetics of decay of an equimolar mixture (2 × 10−3 picomols/cell) of α, β and γ anti-21A RNAs (hereafter referred to as anti-21A RNAs mix) in the cells. Given their increased stability, orthomethylated RNA molecules were chosen to exert prolonged effects, but not long enough to lead to a permanent perturbation of the gene expression profile of the transfected cells. In order to determine the degradation rate of the orthomethylated RNAs, we transfected SKNBE2-S1 cells with anti-21A RNAs mix marked with Platinum Bright 570 Red/Orange and monitored the fluorescence of the cells at different times. The red fluorescence signal was clearly detected on the green fluorescence background of SKNBE2-S1 cells 24 and 48 hours after transfection, but it became undetectable after 72 hours. Since, because of their short half-life in the cells, the RNA molecules were not likely to induce permanent perturbations of the gene expression, the Anti-21A RNAs mix was considered a good candidate also for transfection of cells intended for clinical applications (Fig. 1A).
Figure 1.
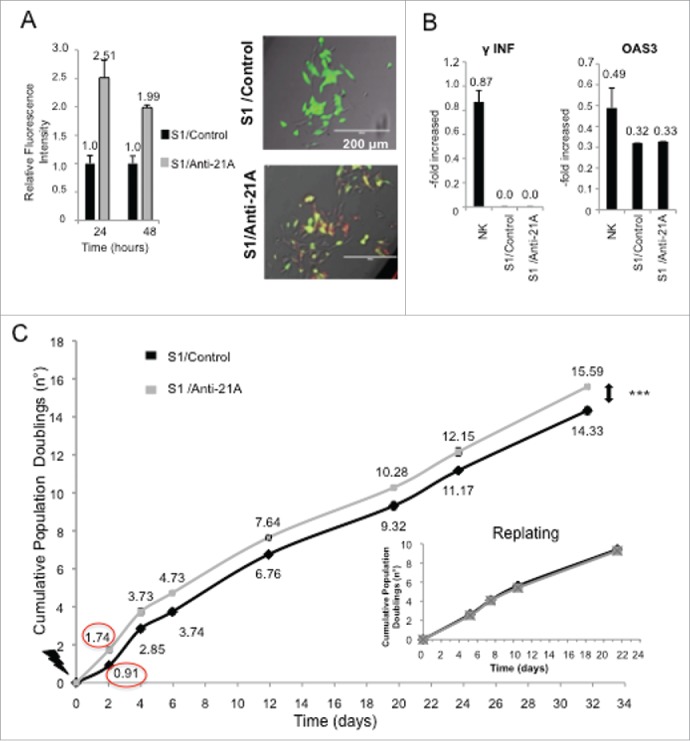
Anti-21A RNAs transfection leads to a transient cell proliferation boost. (A) Quantification of red-labeled Anti-21A RNAs signal in S1 neuroblastoma cells at different time points (seven microscope fields were averaged for each time point). Microscope fields here reported represent S1 cells 24 hours after Anti-21A RNAs transfection. Scale bar: 200 μm. (B) Real time RT-PCR analysis of interferon response biomarkers in S1 cells transfected with Anti-21A RNAs or with water as control; gene expression levels are presented as normalized to GAPDH. Bulk Natural Killer population (NK) was used as positive control. (C) Population doubling of S1 cells transfected with Anti-21A RNAs or water as control, calculated in cell counting experiment. The inset represents the Population Doubling of S1cells replated 20 d after transfection with Anti-21A RNAs or water as control. (***p < 0.001).
To assure that the administration of nucleic acids did not exert toxic effects, we measured the induction of interferon-gamma (IFN γ and of the interferon-induced antiviral biomarker oligoadenylate synthetase 3 (OAS3), 24 hours after transfection. The transfection of anti-21A RNAs in SKNBE2-S1 cells did not induce the expression of these proteins, confirming the safety of the chemically modified 32 to 38 nt-long RNAs mix (Fig. 1B).
The transient transfection of the anti-21A RNAs mix led to an increased proliferation rate of the neuroblastoma cell line.
Next, we determined the extent of the proliferation boost induced by anti-21A RNAs in transfected cells by cell counting and/or the xCELLigence RTCA DP technology. Twenty-four hours after transfection, the number of cells was approximately doubled as compared to the control cells. A preliminary analysis of the xCELLigence growth curves showed that the increase of the proliferation rate occurred within the first 24 hours, whereas, over a longer time, cells recovered the physiological population doubling time (data not shown). Indeed, by cell counting we showed that, after the initial proliferation boost of the 21A-AS RNAs-treated cell, the doubling trend of transfected and un-transfected cells proceeded in parallel during the 32 day-long period of analysis (Fig. 1C). In agreement with this finding, 20 d after the treatment (and after several in vitro passages) cells transfected with anti-21A RNAs and mock transfected control cells, collected by trypsin digestion and replated in new culture dishes at the same cell concentration, presented superimposable population doubling time (inset in Fig. 1C). Altogether these results evidenced that the transfection with anti-21A RNAs mix promoted a transient increase in cell proliferation in vitro and that the gain of an additional cell duplication was promptly followed by the resumption of the physiological proliferation rate.
The proliferation increase driven by anti-21A RNAs in vitro is coincident with neuroblastoma cell dedifferentiation.
We investigated if the increase in cell proliferation induced in neuroblastoma cells by anti-21A RNAs was accompanied by a transient reversion of the neuron-like differentiated phenotype inducing a de-committed stage toward a de-differentiation process. To assess the possible alterations in the expression of genes that play critical roles in the cell cycle and/or differentiation/stemness regulation, we measured, by real time RT-PCR, the expression level of a panel of onco-suppressors, oncogenes and genes related to pluripotency 24, 48, and 168 hours after transfection. We observed a moderate increase of expression of the cell cycle-related/cancer markers c-Myc (after 24 and 48 hours from the transfection) and c-Fos (after 48 hours) with a return to control levels after 7 d. This profile of overexpression suggested that the transience of the anti-21A effects did not promote any permanent cell transformation. In agreement with this finding, we observed a concurrent slight increase in the expression of two onco-suppressors (TP53 and Rb) after 24–48 hours, followed by a return to levels close to the ones of control cells after 1week (Fig. 2A).
Figure 2.
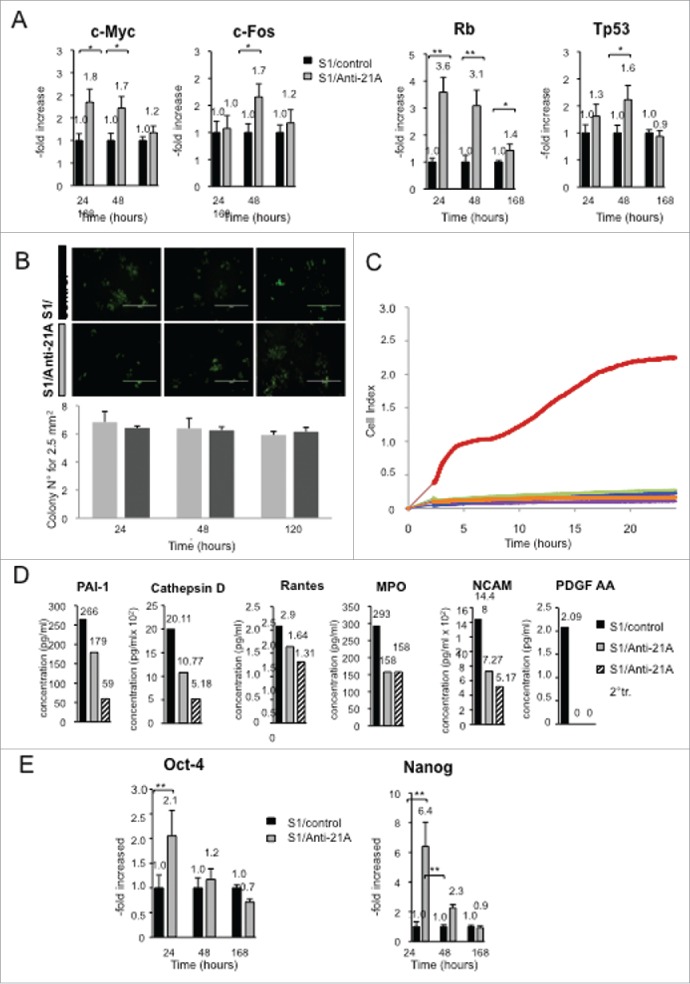
Anti-21A RNAs-dependent proliferation boost does not lead to the acquisition of tumorigenic potential. (A) Real time RT-PCR analysis of oncogenes and oncosuppressors in S1 cells transfected with Anti-21A RNAs; results (gray columns) are normalized to the water-transfected samples (black columns). (B) Colony-forming potential in Methocult of S1 cells transfected with Anti-21A RNAs and/or water (control). Scale bar: 1000 μm. (C) Real-time monitoring migration and invasion of S1 cells transfected with Anti-21A RNAs using the xCELLigence Migration and invasion assay. Blue, Huvec FBS- cells; green, S1 FBS-, Azure, S1 Anti-21A RNAs-transfected FBS-; magenta, HUVEC FBS+; violet, S1 FBS+; orange, S1 Anti-21A RNAs-transfected FBS+. Cells were seeded and their migration was monitored for 24 hours. Cells maintained in serum-free media served as a control. (D) Analysis of secreted cytokines in S1 cells transfected with Anti-21A RNAs (empty bars) or water control (full bars) by MILLIPLEX technology; the amount of cytokines secreted after a 2° treatment of Anti-21A are reported (striped bars). (E) Real time RT-PCR expression analysis of stemness markers; results are reported as Anti-21A/water control ratio. (*p < 0.05; **p < 0.01).
We investigated the possible acquisition of a tumorigenic potential by SKNBE2-S1 cells treated with anti-21A RNAs analyzing their capacity to form colonies on methylcellulose. We found that anti-21A RNAs did not alter the clonogenic potential of these cells that was retained in both anti-21A-transfected and control cells (Fig. 2B).
Next, we investigated possible variations of cell migration capacity induced by anti-21A-treatment by means of the xCELLigence migration assay.22 As shown in figure 2C, in line with the above described data, no differences were observed between transfected and control cells.
We also searched for possible changes in protein secretion driven by anti-21A RNA transfection. Supernatants of 21A RNA-treated and control cells were analyzed using a Human Cytokine detection kit (Luminex technology). We measured the amount of released cytokines that play a relevant role in the nervous system and are characteristically produced by SKNBE2-S1 cells.21 We found, after anti-21A RNAs transfection, a 30 to 70% of observed inhibition of protein release, including PAI-1 (plasminogen activator inhibitor-1),23 the protease cathepsin D,24 the chemokine RANTES (CCL5),25 the MPO (myeloperoxidase),26 and the N-CAM (neural cell adhesion molecule, CD56), (Fig. 2D). In addition, in agreement with the postulated dedifferentiation induced by anti-21A RNAs mix, we found that the secretion of N-CAM (neural cell adhesion molecule, CD56), a hemophilic binding glycoprotein involved in cell-cell adhesion, neurite outgrowth and synaptic plasticity,27 was decreased in anti-21A RNA-transfected cells, thus providing a possible molecular basis for the observed decrease in cell adhesion capacity. In agreement with the loss of neuron-like traits, PDGF-AA (platelet-derived growth factor AA), a neuron-secreted factor28 playing fundamental role in the development of the central nervous system,29 while effectively released in control cells, was not detected in the conditioned medium of SKNBE2-S1 cells after anti-21A transfection.
However, since one could have expected that the induction of the SKNBE2-S1 cell proliferation was sustained by the loss of the differentiated neuron-like phenotype and the acquisition of stemness traits, we measured, by real time RT-PCR, the expression level of two stemness-specific markers (Nanog and Oct4) in both the cells treated with anti-21A RNAs and the control cells. We observed an increased expression of these two genes 24 hours post-transfection, followed by a prompt return to levels closed to the basal, after 48 hours (Fig. 2E).
Anti-21A AS/RNAs drive transient proliferation boost in cultures of mesenchymal stem cell (MSC), as paradigm of primary cells expanded in vitro and intended for cell therapy
As human MSCs are the cells currently most frequently used for cell therapy applications30 these cells were chosen to test the efficiency of the proliferation boost of primary cells induced by the transfection of different combinations of the AS RNAs. Cell proliferation rate was measured by [3H]-thymidine incorporation assay. In the MSCs transfected with an equimolar amount of the three Anti-21A RNAs, after 3 d in culture, DNA synthesis, was increased up to 1.8-folds, without side effects on cell viability (Fig. 3A, B).
Figure 3.
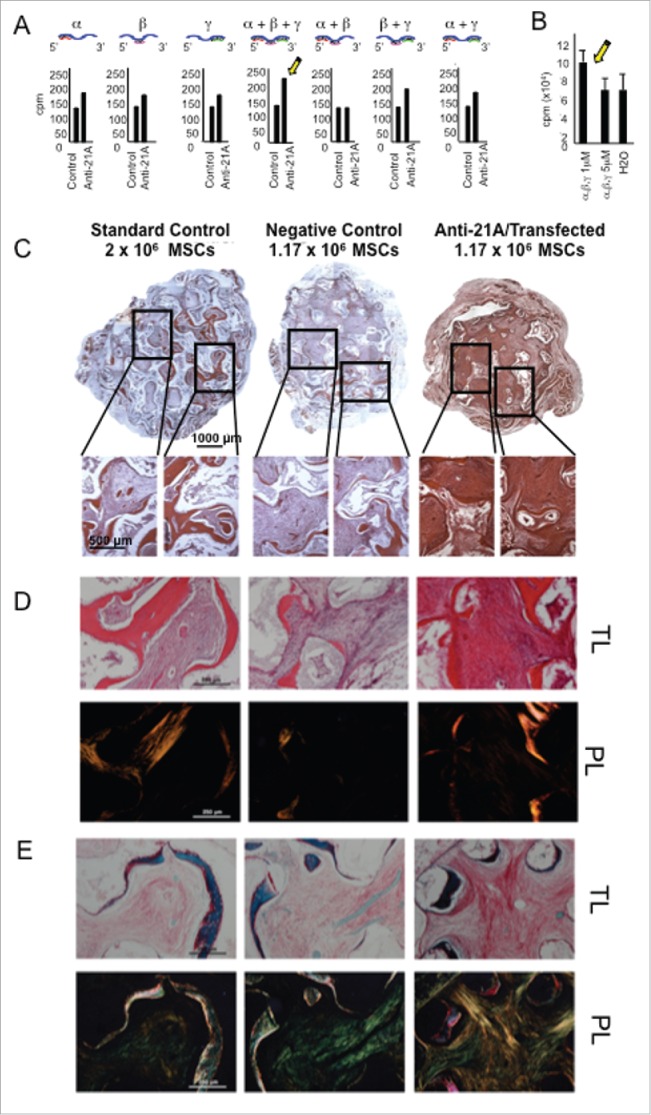
Anti-21A RNAs-transfected MSCs implanted in vivo drive the differentiation of an increased portion of compact fibrous tissue. A) [3H]-thymidine incorporation assay of hMSCs transfected with different combination of α, β and γ Anti-21A RNAs and B) effects on cell proliferation of different concentrations of the equimolar mix (α +β +γ) of Anti-21A RNAs. C) Reconstituted Hematoxilin/Eosin (H/E)-stained skelite sections of different implant conditions and their 10x respective magnifications. D) 10x magnification of Hematoxilin/Eosin-stained sections analyzed under transmitted light (T/L) and under polarized light (P/L). Scale bar: 250 μm. E) Picrosirius red/Fast green-stained skelite sections analyzed under transmitted light (T/L) microscopy and polarized light (P/L) microscopy. Mature collagen type I is stained in red, collagen type I in the maturation process in yellow, and collagen type III in light green in polarized light. All samples were obtained 45 d post-implantation. Scale bar: 250 μm.
The pre-treatment with Anti-21A RNAs did not decrease the osteogenic potential of MSC in a mouse model of ectopic bone formation
Given that the increased expression of Nanog and Oct-4 RNAs, here observed in anti-21A-transfected cells, is usually associated to a cell reprogramming, we hypothesized that the anti-21A RNAs might exert a possible transient dedifferentiating effect on MSCs. We wanted to test if anti/21A-transfected MSCs exhibited a modified susceptibility to microenvironmental stimuli, thus being altered in their tissue regeneration potential. We selected the ectopic mouse model of bone regeneration, in which human bone marrow-derived MSCs seeded on a suitable ceramic scaffold are subcutaneously implanted in the back immunocompromised mice.31 In particular, we investigated whether the treatment with anti-21A RNAs induced differences in bone formation in terms of quantity and/or quality of the engineered tissue. Considering that the treatment with anti-21A RNAs resulted in a 1.8-fold increase in the number of MSCs, we seeded the scaffolds with two different amounts of cells: i) 2 × 106 non-transfected MSCs (standard cell concentration in this type of assay); ii) 1.17 × 106 for anti-21A transfected and non-transfected control MSCs (resulting from the ratio between the standard cell concentration and the population doubling increase induced by anti-21A RNAs). The histological analysis of implanted scaffolds extracted after 45 d showed that transfected MSCs did not induce an increase in bone matrix deposition in comparison to the negative control (Fig. 3C and supplementary material online 2). Interestingly, by histomorphometric analysis of the hematoxylin/eosin stained sections and by the analysis of organized collagen fibers observed under polarized light (Fig. 3C, D), we found an increase in the content of compact fibrous tissue, potentially prone to differentiate toward bone.
To better characterize the phenotype of the observed compact tissue, sections derived from the three experimental groups were stained with picrosirius red/fast green dye, in order to define the thickness of fibers and to discriminate the collagen fibers at different stages of maturation.32,33 The compact tissue present within the pores of scaffolds seeded with transfected MSCs was characterized by a predominant yellow-staining of fibers as expected for collagen type I in the maturation process. On the contrary, the green-stained collagen fibers (characteristic of collagen type III) were more abundant in standard and negative controls, indicating that the transfection of the implanted cells led to increased quantity of compact tissue in the maturation stage (Fig. 3E).
Next, in light of the above results, we investigated the effects of the transfection of anti-21A RNAs during a prolonged implantation time, to allow a more complete osteogenic differentiation of the compact tissues within the scaffolds. In order to avoid individual variability in our experimental setting, each mouse was implanted with 3 scaffolds loaded with i) the standard control (2 × 106 cells untreated), ii) the negative control (1.17 × 106 cells untreated) and iii) the transfected cells (1.17 × 106 cells) respectively. Mice were sacrificed 60 d after implantation and serial sections of the scaffolds were examined. Although the histological analysis showed mineralized tissue in all the experimental groups, the bone thickness distribution analysis showed a statistically significant increase of bone thickness in implants seeded with transfected cells (Fig. 4A and Supplementary material online 3A). In particular, scaffolds seeded with anti/21A-transfected MSCs exhibited a remarkable increase of mineralized areas with the highest thickness distribution in the range between 100 and 200 µm and over (Fig. 4B and Supplementary Material online 3B). On the whole, these results show that the MSC treatment with anti-21A RNAs favors the deposition of mineralized bone after their implantation in vivo.
Figure 4.
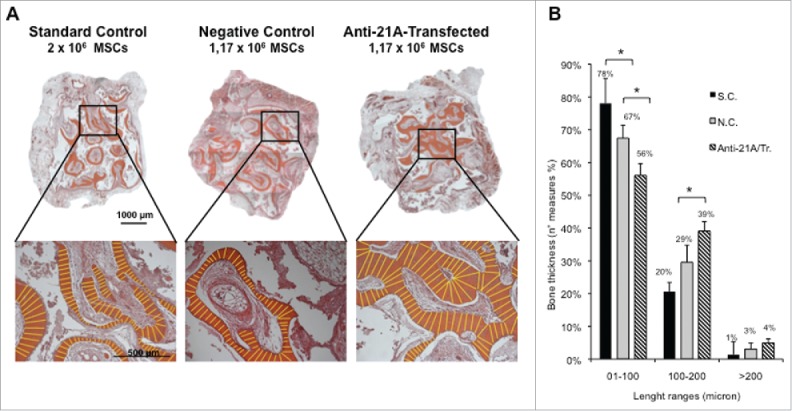
An increased bone mineralized area thickness is detected in implants from Anti-21A RNAs-transfected MSCs. (A) Reconstituted Hematoxilin/Eosin-stained skelite sections of different implant conditions and their respective magnifications. (B) Thickness distribution of mineralized tissue bone matrix deposition area. (SC: standard control group; NC: negative control group; Anti-21A/Tr: Anti-21A RNAs-transfected group).
Anti-21A RNAs enhance the efficiency of tube formation in a model of angiogenesis in vitro
To additionally validate the use of anti-21A RNAs as a tool for regenerative medicine, we investigated its capacity to boost a different tissue regeneration system, such as the formation of three-dimensional tubular structures by endothelial cells in vitro.
To this aim we plated primary cultures of human umbilical vein endothelial cells (HUVECs) onto a layer of Matrigel (composed by laminin and specific growth factors) that promotes cell adhesion, migration and drives cells to differentiate to tube-shaped structures, a process that recapitulates blood vessel formation. Cells plated on Matrigel, treated with anti-21A RNAs or vehicle (control cells), were incubated for 20 hours to allow tubule formation. The determination of the average tube length in treated and control cells showed a remarkable increase (+14%) in anti-21A RNAs-treated cultures with respect to the controls (Fig. 5A, B). This result strengthen the possible application of Anti-21A RNAs as enhancer of the tissue regeneration potential in cells intended for biomedical practices.
Figure 5.
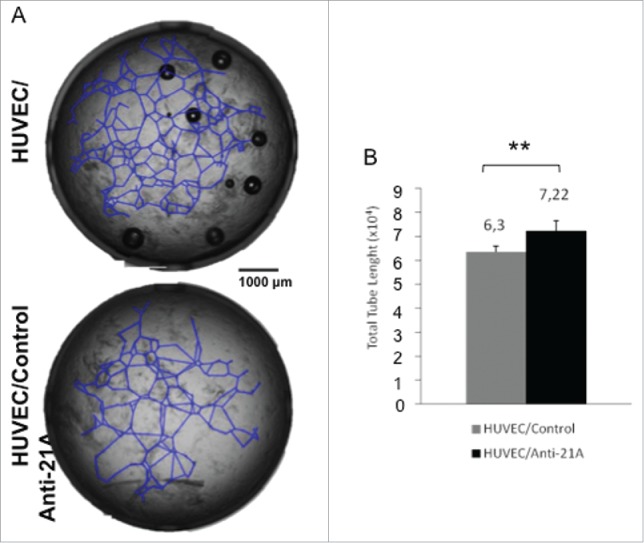
Effect of Anti-21A RNAs treatment on HUVEC's tubule formation potential in vitro. (A) Tubule forming HUVECs plated onto Matrigel, 20 hours after treatment with Anti-21A RNAs (or water as control). (B) Quantitative analysis of tube length.
The proliferation increase driven by anti-21A RNAs in vitro is not associated to the acquisition of a tumorigenic potential
We ruled out the acquisition of a tumor promoting phenotype inoculating subcutaneously 3 × 106 anti-21A RNA-treated or untreated MSCs in nude mice. After 6 months we did not observe tumor formation in both mice groups, confirming that the treatment did not lead to the acquisition of anti-21A RNA-dependent tumorigenic properties (data not shown).
Discussion
In a recent work we documented an inverse correlation between the expression of 21A ncRNA and cell proliferation rate. Indeed, whereas the overexpression of 21A ncRNA led to a reduction of cell proliferation, its decrease (obtained by a siRNA-mediated silencing or by antisense technology) resulted in a dramatic increase of cell proliferation. These findings suggested the possibility to use anti-21A RNAs to increase cell proliferation rate in culture of poorly proliferating cells or when the number of available cells become limiting such as in different biotechnological applications including cell therapy.
To this aim we used equimolar mixture of three chemically-modified anti-21A RNAs to promote a controlled and transient increase of cell proliferation. We demonstrated that for every cycle of transfection of anti-21A RNAs the cells gained an additional doubling in 24/48 hours. Notably, despite a transient increase of oncogenes and oncosuppressors observed in the first few hours after transfection, the physiological control of the cell cycle in anti/21A-transfected cells was promptly recovered in the next few hours and the cells do not acquire tumorigenic potential. Interestingly, the transient upregulation of c-myc is in agreement with the contribute of this gene to the process of somatic cells reprogramming to Induced Pluripotent Stem Cells, and is also corroborated by the enhanced expression of Oct4 and Nanog, two key components of cell stemness and reprogramming. This result is also strengthen by the analysis of proteins secreted after anti-21A treatment, that does not support the activation of tumorigenic pathways and by the transience of the changes of expression of stemness genes observed after cell booster transfection. Therefore, these results suggest the possible use of this procedure for biomedical purposes.
Next, taking advantage of a model of bone regeneration we have shown that MSCs transfected with anti-21A RNAs increase their osteogenic potential in vivo, leading to an increased osteoid thickness. Next, in order to extend the use of Anti-21A RNAs in different tissue engineering contexts, we have demonstrated an increased angiogenic potential of Anti-21A RNAs-treated endothelial cells suggesting a potential widespread application of anti-21A RNAs as tool to increase regenerative capacity of different cell types.
In summary, the results presented here point out the possible use of this procedure to expand and amplify in vitro cell populations of interest and/or to increase the regenerative potential of progenitor cells in tissue engineering procedures.
Materials and methods
Anti-21A RNAs mixture
Anti-21A RNAs equimolar mixture was composed by the following Ortho-methylated synthetic antisense RNAs (GE Healthcare Dharmacon Inc., Lafayette, CO, USA):
α, 5′-ugggcucaagugauccuccugccucagccucccaaagu-3′;
β, 5′-AGAUGAGGUCUCACUAUGUUGCCCAAGCUGGUCUCAAA-3′;
γ,5′-GACCACAGGUGUGCACUACCACACCCAGCUAA-3′.
Cell cultures and transfection
SKNBE2 human neuroblastoma cells were grown in RPMI 1640 (Sigma-Aldrich, St. Louis, MO, USA), supplemented with 10% FBS (fetal bovine serum; Gibco® Life Technologies, Carlsbad, CA, USA), L-glutamine (2mM; Euroclone, Milano, Italy) penicillin/streptomycin (100 U/ml/ 100 µg/ml; Euroclone, Milano, Italy) (SKNBE2-complete medium). Cells were transfected using polyethylenimine (PEI, P3143; Sigma-Aldrich, St. Louis, MO, USA) with p_EGFPN1-NDM29 (hereafter referred to as NDM29). G418 (Geneticin; Invitrogen™ Life Technologies, Carlsbad, CA, USA) was used in culture medium as mean of selection up to 1000 µg/ml, until resistant clones were identified. After selection, the clones were preserved in 200 µg/ml G418 in standard culture conditions. One or two d prior microporation, SKNBE2-NDM29 cells were transferred into flask with fresh medium in order to obtain 70–90% confluency on the day of the experiment. Cells were detached with 0.05% trypsin in 0.01 % EDTA (Sigma-Aldrich, St. Louis, MO, USA), washed with PBS (phosphate-buffered saline, pH 7.2) and resuspended in Buffer R at a final density of 3.5 × 106 cells/ml for microporation with 10 µl Tip (NeonTM Transfection System, 10 µl Kit; Invitrogen™ Life Technologies). Microporation was performed using a Microporator MP-100 (Invitrogen™ Life Technologies, Carlsbad, CA, USA). Anti-21A (AS/RNAs equimolar mixture or water as control) (2µM) were added prior microporation. The optimal pulse condition were previously set for SKNBE-NDM29 cell line following procedures for the fast optimization of electric parameters. After the pulse, cells were transferred immediately into pre-warmed SKNBE2-complete medium deprived of penicillin/streptomycin.
Bone marrow derived mesenchymal stem cells (MSCs) were obtained from iliac crest marrow aspirates of 3 healthy donors (age range: 18–38 years). All the procedures were approved by the Institutional Ethical Review Committee. Bone marrow samples were washed twice with PBS (phosphate-buffered saline, pH 7.2) and mononucleated cells were counted using nuclear staining (0.1% methyl violet in 0.1 M citric acid). Bone marrow cells were then cultured in Coon's modified Ham's F12 medium (Biochrom, Berlin, Germany) supplemented with 10% FCS (fetal calf serum; Invitrogen™ Life Technologies, Carlsbad, CA, USA) L-glutamine (2mM; Euroclone, Milano, Italy), penicillin/streptomycin (100 U/ml/ 100 µg/ml; Euroclone, Milano, Italy) and 1 ng/ml FGF-2 (Peprotech EC, London, UK) (MSCs-complete medium) and plated at 2–5 × 106 cells/100-mm dish. Medium was changed 3 d after plating and twice a week thereafter. Cells from confluent cultures (20–25 d in culture) were detached with 0.05% trypsin in 0.01% EDTA (Sigma-Aldrich, St. Louis, MO, USA), counted and frozen. For microporation cells were thawed and plated at 5 × 105 cells/100-mm dish. Medium was changed twice a week and cells from confluent cultures were detached with 0.05% trypsin in 0.01 % EDTA (Sigma-Aldrich, St. Louis, MO, USA), washed with PBS (phosphate-buffered saline, pH 7.2) and resuspended in Buffer R at a final density of 5 × 106 cells/ml for microporation with 100 µl Tip (NeonTM Transfection System, 100 µl Kit; Invitrogen™ Life Technologies, Carlsbad, CA, USA). Anti-21A RNAs cocktail or water as control were added to cell suspension prior microporation. The pulse condition were set as suggested in microporator cell database. After the pulse, cells were transferred immediately into pre-warmed MSC-complete medium deprived of penicillin/streptomycin. This transfection technique does not affect the proliferative, differentiative or migratory activity of the cells.34
Optical microscope imaging
Transmission images of control and transfected cells were captured using EVOS fl microscope (Advanced Microscopy Group, Bothell, WA, USA), at 20x magnification.
RNA synthesis labeling and transfection optimization
Anti-21A RNAs were synthetized by Thermo Scientific. Different Anti-21A RNAs conditions were tested on MSCs (α; β; γ; α + γ; α + β; γ + β; γ + β + α) (see Fig. 3A). Different α + β + γ anti-21A RNAs concentration conditions were tested (1 µM and 5 µM) (see Fig. 3B). To test the decay kinetics of the anti-21A RNA cocktail, SKNBE-NDM29 cells were microporated with Red labeled anti-21A RNAs (Platinum Bright 570 Red/Orange nucleic acid labeling kit; Leica Biosystems Kreatech™ Inc., Nussloch, Germany) or water as control and seeded in 96-well culture plate black (Thermo Fischer Scientific, Waltham, MA, USA). After 24 and 48 hours the plate fluorescence was measured using a fluorimeter (Genios Pro reader, Tecan Group AG, Männedorf, Switzerland).
Proliferation assays
A) For cell counting assay, control or anti-21A RNA transfected SKNBE2-S1 cells were seeded in 6-well culture plates at the concentration of 10.5 × 104 cells/well, incubated in SKNBE2-standard medium, counted with a hemocytometer after 24 hours and expanded. Cell were counted and expanded in triplicate at different time points.
B) For [3H]-thymidine incorporation assay, control or anti-21A RNA transfected MSCs were plated in 24-well plates; cells were incubated in standard medium for 24 hours and pulsed with [3H]-thymidine (0.3 μ Ci/500 μl/well) (GE Healthcare, New York, NY, USA) for the last 14 hours. We calculated the average proliferation rate based on the thymidine uptake.
C) Cell proliferation was also assessed by xCELLigence RTCA MP System (Roche, Penzberg, Germany) that monitors cellular events in real time by measuring electrical impedance across interdigitated gold micro-electrodes integrated on the bottom of tissue culture plates. The impedance measurement provides quantitative information about the biological status of the cells, including cell number, viability, and morphology. Cell-sensor impedance is expressed as an arbitrary unit called Cell Index (CI). In order to determine the Cell Index, 104 cells were seeded into 100 μL of standard medium in 96×microtiter plates (E-Plate; Roche, Penzberg, Germany). Background impedance was determined using 50 μl of standard medium. Cell attachment, spreading and proliferation were monitored every 15 min using the xCELLigence system. Experimental results were performed using RTCA Software 1.2 that calculated the population doubling by fitting the curve to an exponential equation.35
Migration assay
Cell migration was analyzed on control or anti-21A RNA transfected SKNBE2-S1 cells by the new technique of real-time migration monitoring, using the CIM-Plate 16 and xCELLigence System RTCA DP Instrument (E-Plate; Roche, Penzberg, Germany). HUVEC cells were used as positive control for the cell migration assay. The day before the assay, cells were plated in starvation medium (standard medium with a reduced concentration of FBS to 0.5%). After 24 hours, 4 × 104 cells, suspended in 100 µl of serum-free medium, were seeded in the upper chamber of a CIM-Plate 16. The upper chamber was then placed on the lower chamber of the CIM-Plate 16 containing growth medium supplemented with 20% FBS as an attractant, or without FBS (negative control). Time-dependent cell migration was monitored up to 18 hours, and experimental results analyzed using RTCA Software 1.2, considering the interval of 0–6 hours. Traces show the average of triplicate wells.
Methylcellulose colony formation assay
The clonogenic assay for SKNBE2-S1 cells was performed using a methylcellulose medium consisting of RPMI 1640 with 0.9% methylcellulose (Methocult H4100; StemCell Technologies, Vancouver, BC, Canada), 10% FBS, 100 U/ml penicillin/streptomycin and 2 mM L-glutamine. Cells were plated at a density of 400 cells in 2 ml volume of methylcellulose medium in humidified 6-well plates. For each assay, suspension cells were plated in triplicate. Colonies were counted at 12 d for 14 d after plating.
Cytokine/Chemokine immunoassay
To quantify cytokine and chemokine production, SKNBE2-S1 cells (106 cells) transfected with anti-21A RNAs or water as control were cultured in SKNBE2-complete medium. After 48 hours of incubation, culture supernatants were collected and analyzed using MILLIPLEX Human Neurodegenerative Panel 3 kit (Millipore, St Charles, MO, USA), that allows the simultaneous quantification of 10 human cytokines: brain-derived neurotrophic factor (BDNF); soluble vascular cell adhesion molecule 1 (sVCAM-1); soluble inter-cellular adhesion molecule 1 (sICAM-1); neural cell adhesion molecule (NCAM); the chemokine RANTES (CCL5); myeloperoxidase (MPO); plasminogen activator inhibitor-1 (PAI-1; total); Cathepsin D; platelet-derived growth factor (PDGF)-AA; PDGF-AB/BB. The analysis was performed according to manufacturer's recommended protocols. Data were normalized to cell number and expressed in pg/mL.
Real time quantitative RT-PCR analysis
Total RNAs from samples were extracted using TRIzol reagent (Invitrogen™ Life Technologies, Carlsbad, CA, USA) according to the manufacturer's protocol, DNAse I-digested and subjected to reverse transcription by Transcriptor High Fidelity cDNA Synthesis Kit (Roche, Penzberg, Germany) following manufacturer's instructions. Total RNA from samples was measured by real-time quantitative RT-PCR using the 7500 Fast Real-Time PCR System (Applied Biosystems® Life Technologies, Carlsbad, CA, USA) and SybrGreen method as described (Castelnuovo et al., 2010). The sequences of forward and reverse primers were: γINF, forward 5′-CCAGGACCCATATGTAAAAG-3′ and reverse 5′-TGGCTCTGCATTATTTTTC-3′ (Cupedo et al., 2009); OAS3, forward 5′-AGAGACGGGACATCTGTGCAG-3′ and reverse 5′-GGTTGGGCTGGAGAAATTCA-3′; c-Myc, forward 5′-CGACCACAAGGCCCTCAGTA-3′ and reverse 5′-TTGGAGGAGGAACGCCGCTT-3′; Rb, forward 5′-GCGCTCTTGAGGTTGTAATGG-3′ and reverse 5′-CCATGGGAAAGACAAATCTGTTC-3′; p53, forward 5′-GCTGCCCCCACCATGAG-3′ and reverse 5′-CCTTCCACTCGGATAAGATGCT-3′; c-Fos, forward 5′-CGAGCGCAGAGCATTGG-3′ and reverse 5′-CCTTCGGATTCTCCTTTTCTCTT-3′; Nanog, forward 5′-CCAGCAGATGCAAGAACTCTCC-3′ and reverse 5′-GGCCAGTTGTTTTTCTGCCA-3′; Oct4, forward 5′-ACTGCAGCAGATCAGCCACA-3′ and reverse 5′-CTGGCGCCGGTTACAGAAC-3′. For endogenous control, the expression of Glyceraldehyde 3 phosphate dehydrogenase (GAPDH) was examined. The sequences for human GAPDH primers were 5′-GAAGGTGAAGGTCGGAGTC-3′ and 5′-GAAGATGGTGATGGGATTTC-3′. Relative transcript levels were determined from the relative standard curve constructed from stock cDNA dilutions, and divided by the target quantity of the calibrator following manufacturer's instructions.
Tube formation assay
Matrigel was used to coat 96-wells plates (50 μl/well) and allowed to solidify at 37°C for 30 minutes. HUVECs were microporated with Anti-21A RNAs (at the final concentration of 2 μM) or water (control) as described above, and seeded onto the Matrigel-coated wells at the concentration of 20 × 104 cells/well. HUVECs were incubated with M199 media (Euroclone, Milano, Italy) supplemented with 10% FBS (fetal bovine serum; Gibco® Life Technologies, Carlsbad, CA, USA), L-glutamine (2mM; Euroclone, Milano, Italy), and with growth factors according to manufacturer's instructions (S.S. Banca Biologica e Cell Factory, IRCCS AOU San Martino – IST, Genova, Italy), for up to 20 hours. In order to quantify tube-shaped structures formation we captured images at low magnification (4x) and total wells were reconstructed with photomerge function of Adobe Photoshop CS 8.0 software (http://www.adobe.com). The total length of tube- shaped structures, the averages and the standard deviations observed were worked out for each well using NIH ImageJ software (http://rsb.info.nih.gov/ij/).
Ectopic bone formation assay
Bioceramic scaffolds (SkeliteTM) were 100% synthetic calcium phosphate multiphase biomaterials containing 67% silicon stabilized tricalcium phosphate (TCP). These scaffolds had 60–65% porosity and were produced by Octane Medical Group (Kingston, Ontario, Canada). All scaffolds were cubes of about 4 × 4 × 4 mm dimensions. After microporation, MSCs were washed in serum-free medium, and suspended in aliquots at different cell number (2 × 106 cells, standard cell concentration; 1.17 × 106 cells, anti-21A transfected and not transfected control cells) per 20 µl fibrinogen (2.5 mg/mL; Baxter, Deerfield, IL, USA). Each aliquot was seeded onto a scaffold to which 20 µl of murine thrombin (500 IU/mL; Baxter, Deerfield, IL, USA) were then applied. Recipient Nude/Nude mice were subcutaneously implanted with the MSCs/bioscaffold constructs. In the first preliminary experiment, nine mice received one ectopic implant (three mouse for condition) and were sacrificed after 45 d post-implantation. In the second experiment, six mice were used. Each mouse was implanted with 3 scaffolds and each scaffold was loaded with cells from one of the three experimental conditions above described. Location of the implants of each experimental group was rotated in the different mice and the mice were sacrificed after 60 d.
Histological analysis of the ectopic implants and bone quantification
Formalin-fixed scaffolds were processed as previously reported.36 Briefly, samples were decalcified with Osteodec (BIO OPTICA, Milano, Italy) and embedded in paraffin using standard histological techniques. Four-micrometer serial sections were cut. Sections were stained with hematoxylin and eosin (H/E) to reveal bone tissue and with Picrosirius Red/Fast Green (SR/FG) and counterstained with hematoxylin of Weigher to distinguish between collagen and non-collagenous proteins and to reveal collagen distribution under polarized light microscopy. Images were captured by transmitted light microscopy with a Zeiss Axiovert 200M inverted microscope (Zeiss, Jena, Germany) and processed by using Axio Vision 4.8 software (Zeiss, Jena, Germany). Stained sections were analyzed under polarized light microscopy with a microscope Eclipse 80i (Nikon, Tokyo, Japan).
For H/E staining, images were captured at 10x magnification and total implant sections were reconstructed with photomerge function of Adobe Photoshop CS 8.0 software (www.adobe.com). Bone matrix deposition areas and bone thickness were quantified using NIH ImageJ software (http://rsb.info.nih.gov/ij/). In the second experiment, from the Picrosiriusred/Fast Green stained sections, 7 images for section were captured at 20x magnification and the proportion of different colored collagen fibers was determined using ImageJ software.32 Images were converted in HSB (Hue Saturation Brightness) and threshold manually adjusted using the following hue definition: orange 10–38, yellow 39–51, green 52–128. The number of pixels within each hue range was determined and expressed as a percentage of the total number of collagen pixels.
In vivo tumorigenicity assay
A cell suspension of MSCs (3 × 106 cells in 200 µl di PBS) was subcutaneously injected into 5 NOD/SCID mice for each experimental group. Mice were observed weekly for the development of tumors at injection sites. After six months mice were sacrificed and autopsies performed.
Statistical analysis
Data are reported as mean values ± standard error (SE). Statistical significance of observed differences among different experimental groups was examined using the unpaired Student's t-test, as reported.37
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
A.P. was supported by the IRCCS-AOU San Martino-IST, Genoa-Italy (Progetto 5 × 1000), by the University of Genoa, Genoa-Italy (Progetti di Ricerca di Ateneo 2013) and by the Associazione Italiana per la Lotta al Neuroblastoma/Fondazione Neuroblastoma (Genoa, Italy). T.F. is supported by a grant from the Italian Association for Cancer Research (AIRC, IG13563) and Ricerca Corrente IZS-PLV (2008–2009).
References
- [1].Mattick JS. RNA regulation: a new genetics? Nat Rev Genet 2004; 5:316-23; PMID:15131654; http://dx.doi.org/ 10.1038/nrg1321 [DOI] [PubMed] [Google Scholar]
- [2].Pagano A, Castelnuovo M, Tortelli F, Ferrari R, Dieci G, Cancedda R. New small nuclear RNA gene-like transcriptional units as sources of regulatory transcripts. PLoS Genet 2007; 3:0174-84; http://dx.doi.org/ 10.1371/journal.pgen.0030001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Reis EM, Nakaya HI, Louro R, Canavez FC, Flatschart AVF, Almeida GT, Egidio CM, Paquola AC, Machado A, Festa F, et al.. Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer. Oncogene [Internet] 2004; 23:6684-92. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15221013; PMID:15221013; http://dx.doi.org/ 10.1038/sj.onc.1207880 [DOI] [PubMed] [Google Scholar]
- [4].Schmid CW, Jelinek WR. The Alu family of dispersed repetitive sequences. Science 1982; 216:1065-70; PMID:6281889; http://dx.doi.org/ 10.1126/science.6281889 [DOI] [PubMed] [Google Scholar]
- [5].Berger A, Strub K. Multiple Roles of Alu-Related Noncoding RNAs. Prog Mol Subcell Biol [Internet] 2011; 51:119-46. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21287136; PMID:21287136; http://dx.doi.org/ 10.1007/978-3-642-16502-3_6 [DOI] [PubMed] [Google Scholar]
- [6].Levanon K, Eisenberg E, Rechavi G, Levanon EY. Letter from the editor: Adenosine-to-inosine RNA editing in Alu repeats in the human genome. EMBO Rep 2005; 6:831-5; PMID:16138094; http://dx.doi.org/ 10.1038/sj.embor.7400507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced. Genome Res 2002; 12:1060-7; PMID:12097342; http://dx.doi.org/ 10.1101/gr.229302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sen SK, Han K, Wang J, Lee J, Wang H, Callinan PA, Dyer M, Cordaux R, Liang P, Batzer MA. Human genomic deletions mediated by recombination between Alu elements. Am J Hum Genet 2006; 79:41-53; PMID:16773564; http://dx.doi.org/ 10.1086/504600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Stuart JJ, Egry LA, Wong GH, Kaspar RL. The 3′ UTR of human MnSOD mRNA hybridizes to a small cytoplasmic RNA and inhibits gene expression. Biochem Biophys Res Commun 2000; 274:641-8; PMID:10924331; http://dx.doi.org/ 10.1006/bbrc.2000.3189 [DOI] [PubMed] [Google Scholar]
- [10].Berger A, Ivanova E, Gareau C, Scherrer A, Mazroui R, Strub K. Direct binding of the Alu binding protein dimer SRP9/14 to 40S ribosomal subunits promotes stress granule formation and is regulated by Alu RNA. Nucleic Acids Res [Internet] 2014; 42:11203-17. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4176187&tool=pmcentrez&rendertype=abstract; PMID:25200073; http://dx.doi.org/ 10.1093/nar/gku822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Liu WM, Chu WM, Choudary PV, Schmid CW. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res 1995; 23:1758-65; PMID:7784180; http://dx.doi.org/ 10.1093/nar/23.10.1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Smalheiser NR, Torvik VI. Alu elements within human mRNAs are probable microRNA targets. Trends Genet 2006; 22:532-6; PMID:16914224; http://dx.doi.org/ 10.1016/j.tig.2006.08.007 [DOI] [PubMed] [Google Scholar]
- [13].Castelnuovo M, Massone S, Tasso R, Fiorino G, Gatti M, Robello M, Gatta E, Berger A, Strub K, Florio T, et al.. An Alu-like RNA promotes cell differentiation and reduces malignancy of human neuroblastoma cells. FASEB J 2010; 24:4033-46; PMID:20581224; http://dx.doi.org/ 10.1096/fj.10-157032 [DOI] [PubMed] [Google Scholar]
- [14].Ciarlo E, Massone S, Penna I, Nizzari M, Gigoni A, Dieci G, Russo C, Florio T, Cancedda R, Pagano A. An intronic ncRNA-dependent regulation of SORL1 expression affecting A formation is upregulated in post-mortem Alzheimer disease brain samples. Dis Model Mech 2012; 433:424-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Garritano S, Gigoni A, Costa D, Malatesta P, Florio T, Cancedda R, Pagano A. A novel collection of snRNA-like promoters with tissue-specific transcription properties. Int J Mol Sci 2012; 13:11323-32; PMID:23109855; http://dx.doi.org/ 10.3390/ijms130911323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Massone S, Ciarlo E, Vella S, Nizzari M, Florio T, Russo C, Cancedda R, Pagano A. NDM29, a RNA polymerase III-dependent non coding RNA, promotes amyloidogenic processing of APP and amyloid β secretion. Biochim Biophys Acta - Mol Cell Res [Internet] 2012; 1823:1170-7. Available from: http://dx.doi.org/ 10.1016/j.bbamcr.2012.05.001; http://dx.doi.org/ 10.1016/j.bbamcr.2012.05.001 [DOI] [PubMed] [Google Scholar]
- [17].Massone S, Vassallo I, Fiorino G, Castelnuovo M, Barbieri F, Borghi R, Tabaton M, Robello M, Gatta E, Russo C, et al.. 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol Dis 2011; 41:308-17; PMID:20888417; http://dx.doi.org/ 10.1016/j.nbd.2010.09.019 [DOI] [PubMed] [Google Scholar]
- [18].Penna I, Vassallo I, Nizzari M, Russo D, Costa D, Menichini P, Poggi A, Russo C, Dieci G, Florio T, et al.. A novel snRNA-like transcript affects amyloidogenesis and cell cycle progression through perturbation of Fe65L1 (APBB2) alternative splicing. Biochim Biophys Acta - Mol Cell Res 2013; 1833:1511-26; http://dx.doi.org/ 10.1016/j.bbamcr.2013.02.020 [DOI] [PubMed] [Google Scholar]
- [19].Massone S, Vassallo I, Castelnuovo M, Fiorino G, Gatta E, Robello M, Borghi R, Tabaton M, Russo C, Dieci G, et al.. RNA polymerase III drives alternative splicing of the potassium channel-interacting protein contributing to brain complexity and neurodegeneration. J Cell Biol 2011; 193:851-66; PMID:21624954; http://dx.doi.org/ 10.1083/jcb.201011053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, Khosravi R, et al.. Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol [Internet] 2003; 21:379-86. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12640466; PMID:12640466; http://dx.doi.org/ 10.1038/nbt808 [DOI] [PubMed] [Google Scholar]
- [21].Gavazzo P, Vella S, Marchetti C, Nizzari M, Cancedda R, Pagano A. Acquisition of neuron-like electrophysiological properties in neuroblastoma cells by controlled expression of NDM29 ncRNA. J Neurochem 2011; 119:989-1001; PMID:21933186; http://dx.doi.org/ 10.1111/j.1471-4159.2011.07492.x [DOI] [PubMed] [Google Scholar]
- [22].Rahim S, Üren A. A real-time electrical impedance based technique to measure invasion of endothelial cell monolayer by cancer cells. J Vis Exp 2011; 10–3:2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Harbeck N, Kates RE, Gauger K, Willems A, Kiechle M, Magdolen V, Schmitt M. Urokinase-type plasminogen activator (uPA) and its inhibitor PAI-I: Novel tumor-derived factors with a high prognostic and predictive impact in breast cancer. Thromb Haemost 2004; 92:47-53. [DOI] [PubMed] [Google Scholar]
- [24].Dian D, Heublein S, Wiest I, Barthell L, Friese K, Jeschke U. Significance of the tumor protease cathepsin D for the biology of breast cancer. Histol. Histopathol 2014; 29:433-8. [DOI] [PubMed] [Google Scholar]
- [25].Levy JA. The unexpected pleiotropic activities of RANTES. J Immunol 2009; 182:3945-6; PMID:19299688; http://dx.doi.org/ 10.4049/jimmunol.0990015 [DOI] [PubMed] [Google Scholar]
- [26].Van Der Veen BS, Petersen AH, Belperio JA, Satchell SC, Mathieson PW, Molema G, Heeringa P. Spatiotemporal expression of chemokines and chemokine receptors in experimental anti-myeloperoxidase antibody-mediated glomerulonephritis. Clin Exp Immunol 2009; 158:143-53; PMID:19737241; http://dx.doi.org/ 10.1111/j.1365-2249.2009.03993.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dodla MC, Mumaw J, Stice SL. Role of astrocytes, soluble factors, cells adhesion molecules and neurotrophins in functional synapse formation: implications for human embryonic stem cell derived neurons. Curr Stem Cell Res Ther [Internet] 2010; 5:251-60. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20214556; PMID:20214556; http://dx.doi.org/ 10.2174/157488810791824520 [DOI] [PubMed] [Google Scholar]
- [28].Richardson WD, Pringle N, Mosley MJ, Westermark B, Dubois-Dalcq M. A role for platelet-derived growth factor in normal gliogenesis in the central nervous system. Cell 1988; 53:309-19; PMID:2834067; http://dx.doi.org/ 10.1016/0092-8674(88)90392-3 [DOI] [PubMed] [Google Scholar]
- [29].Fruttiger M, Karlsson L, Hall AC, Abramsson A, Calver AR, Boström H, Willetts K, Bertold CH, Heath JK, Betsholtz C, et al.. Defective oligodendrocyte development and severe hypomyelination in PDGF-A knockout mice. Development 1999; 126:457-67; PMID:9876175 [DOI] [PubMed] [Google Scholar]
- [30].Fossett E, Khan WS. Optismising human mesenchymal stem cell numbers for clinical application: A literature review. Stem Cells Int 2012; 2012:465259; PMID:22448172; http://dx.doi.org/ 10.1155/2012/465259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Muraglia A, Martin I, Cancedda R, Quarto R. A nude mouse model for human bone formation in unloaded conditions. Bone 1998; 22(5 Suppl):131S-134S. [DOI] [PubMed] [Google Scholar]
- [32].Rich L, Whittaker P. Collagen and picrosirius red staining: a polarized light assessment of fibrillar hue and spatial distribution. Braz J Morphol Sci [Internet] 2005; 22:97-104. Available from: http://jms.org.br/PDF/v22n2a06.pdf [Google Scholar]
- [33].Allegrini S, Rumpel E, Kauschke E, Fanghänel J, König B. Hydroxyapatite grafting promotes new bone formation and osseointegration of smooth titanium implants. Ann Anat 2006; 188:143-51; PMID:16551011; http://dx.doi.org/ 10.1016/j.aanat.2005.08.019 [DOI] [PubMed] [Google Scholar]
- [34].Madeira C, Ribeiro SC, Pinheiro ISM, Martins SAM, Andrade PZ, da Silva CL, Cabral JMS. Gene delivery to human bone marrow mesenchymal stem cells by microporation. J Biotechnol 2011; 151:130-6; PMID:21087644; http://dx.doi.org/ 10.1016/j.jbiotec.2010.11.002 [DOI] [PubMed] [Google Scholar]
- [35].Galante D, Corsaro A, Florio T, Vella S, Pagano A, Sbrana F, Vassalli M, Perico A, D'Arrigo C. Differential toxicity, conformation and morphology of typical initial aggregation states of Aβ1-42 and Aβpy3-42 β-amyloids. Int J Biochem Cell Biol 2012; 44:2085-93; PMID:22903022; http://dx.doi.org/ 10.1016/j.biocel.2012.08.010 [DOI] [PubMed] [Google Scholar]
- [36].Tasso R, Augello A, Boccardo S, Salvi S, Caridà M, Postiglione F, Fais F, Truini M, Cancedda R, Pennesi G. Recruitment of a host's osteoprogenitor cells using exogenous mesenchymal stem cells seeded on porous ceramic. Tissue Eng Part A 2009; 15:2203-12; PMID:19265473; http://dx.doi.org/ 10.1089/ten.tea.2008.0269 [DOI] [PubMed] [Google Scholar]
- [37].Costa D, Gigoni A, Würth R, Cancedda R, Florio T, Pagano A. Metformin inhibition of neuroblastoma cell proliferation is differently modulated by cell differentiation induced by retinoic acid or overexpression of NDM29 non-coding RNA. [Internet]. Cancer Cell Int 2014; 14:59. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4128937&tool=pmcentrez&rendertype=abstract; PMID:25120382; http://dx.doi.org/ 10.1186/1475-2867-14-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


