Abstract
Organogenesis requires the coordination of many highly-regulated developmental processes, including cell fate determination, cell division and growth, and cell-cell communication. For tissue- and organ-scale coordination, a network of regulators enables molecular events in individual cells to translate into multicellular changes in structure and functional capacity. One recurrent theme in plant developmental networks is a central role for plant hormones, especially auxin. Here, we focus first on describing recent advances in understanding lateral root development, one of the best-studied examples of auxin-mediated organogenesis. We then use this framework to examine the parallel process of emergence of lateral organs in the shoot—a process called phyllotaxy. This comparison reveals a high degree of conservation, highlighting auxin's pivotal role determining overall plant architecture.
Keywords: root branching, shoot architecture, hormone signaling
INTRODUCTION
Plants retain the capacity to generate new organs throughout their adult life, and developmental responses are essential for surviving environmental stresses. Plant form is principally determined by the outgrowth of lateral organs along the major axis running from stem cells at the top of the primary shoot (shoot apical meristem, SAM) to the stem cell niche at the tip of the primary root. The SAM produces leaves, inflorescence stems and floral meristems that become flowers. Lateral root fate is specified in the root stem cell niche, but there is a delay in time and space before organogenesis is initiated. Specification of new organs at both root and shoot occurs at regular intervals and, once initiated, follows a stereotypical progression of cellular events. Yet the diversity in plant architecture is breathtaking, suggesting these programs are readily rewired throughout evolution. Understanding the molecular details underlying each of these core developmental programs has already yielded valuable insights into how molecular events are coordinated across temporal and spatial scales, and there is still much to be discovered.
AUXIN SIGNALING, BIOSYNTHESIS, AND TRANSPORT
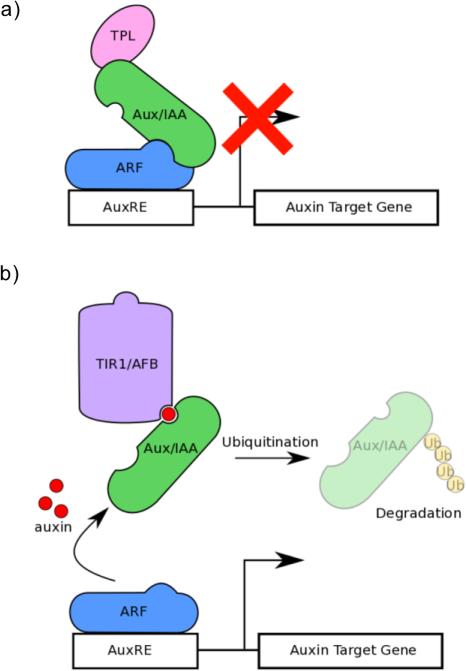
Plant hormones have long been implicated in the coordination of development (Vanstraelen and Benkova, 2012), and no hormone is more ubiquitous a developmental regulator than auxin. There are multiple hypotheses for how auxin can regulate such diverse, context-specific developmental processes, including fate specification (Smit and Weijers, 2015), anisotropic cell expansion (Kutschera et al., 1987), and even senescence (Lim et al., 2010). Nuclear perception of auxin occurs by a classic “repressor of a repressor” genetic module (reviewed here: Pierre-Jerome et al., 2013). In low auxin conditions, the activity of AUXIN RESPONSE FACTOR (ARF) transcription factors is inhibited by interactions with Auxin/INDOLE-3-ACETIC ACID (Aux/IAAs) repressor proteins and the co-repressor TOPLESS (TPL) (Fig 1A). Members of the TRANSPORT INHIBITOR RESPONSE1/ AUXIN SIGNALING F-BOX PROTEINS (TIR1/AFBs) bind auxin in a complex with the Aux/IAAs, which serve as auxin co-receptors. This interaction leads to ubiquitination and proteasome-mediated degradation of the Aux/IAAs. This relieves repression on the ARFs, allowing auxin-induced transcription to proceed (Fig 1B) (Pierre-Jerome et al., 2013). ARFs, Aux/IAAs, and AFBs have diversified gene families, and different combinations or “modules” of these auxin signaling proteins are required for different developmental programs (Goh et al., 2012). However, to date there has been only limited biochemical evidence for the specificity of different protein interactions in the auxin response (Hardtke et al., 2004; Vernoux et al., 2011; Guilfoyle, 2015; Boer et al., 2014). Co-expression of many members of each family within the same tissues and even the same cells adds to the mystery of how auxin can promote such varied responses (Piya et al., 2014).
Figure 1.
The auxin signaling pathway. (a) In low auxin conditions, the activity of AUXIN RESPONSE FACTOR (ARF) transcription factors is inhibited by interactions with Auxin/INDOLE-3-ACETIC ACID (Aux/IAAs) repressor proteins and the co-repressor TOPLESS (TPL). (b) Auxin acts a molecular glue between TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX PROTEINS (TIR1/AFBs) and Aux/IAAs, leading to ubiquitination and proteasome-mediated degradation of the Aux/IAAs. This relieves repression on the ARFs, allowing auxin-induced transcription to proceed. This simplified schematic incorporates the essential players and logic of auxin response, but does not include the higher order complexes resulting from homo- and heterotypic interactions among many of these components.
Auxin biosynthesis and transport are additional mechanisms for regulating auxin-responsive signaling during development (reviewed in Rosquete et al., 2012 and Grones and Friml, 2015, respectively). Plants produce multiple auxinic compounds, with indole-3-acetic acid (IAA) being the most abundant. The main auxin biosynthesis pathway in Arabidopsis requires the sequential action of the TAA1 and YUCCA (YUC) biosynthetic enzymes (Won et al., 2011). Local production of auxin is important for normal development. Mutants with reduced capacity to synthesize auxin have developmental defects that cannot be rescued by exogenous auxin application but can be rescued through cell-type-specific expression of a bacterial gene that enables auxin biosynthesis (Cheng et al., 2006). Auxin transport augments local biosynthesis as a means of changing auxin levels in different tissues. Influx of auxin into cells is mediated by AUXIN RESISTANT1 and LIKE AUXIN RESISTANT (AUX1/LAX) proteins. Auxin efflux is facilitated by P-glycoproteins of the ATP-binding cassette transporter family or by PIN-FORMED (PIN) proteins. The PINs are of particular interest in developmental biology as they are frequently localized in a polar fashion, allowing directional auxin transport and the establishment of local auxin maxima and minima. Whether auxin acts as a morphogen or simply a threshold catalyst remains unresolved. If auxin is a morphogen, cells that receive more auxin should have different fates than those that receive less, such that auxin concentration itself is a fate regulator (Bhalerao and Bennett, 2003). Such auxin gradients do seem to govern the differentiation of xylem in secondary cell wall synthesis (Uggla et al., 1996). In contrast, several studies have established that a very low concentration of auxin is sufficient to induce an auxin response, suggesting that response specificity comes from factors other than concentration (Lau et al., 2011; Finet and Jaillais, 2012). New technologies to examine how auxin response correlates to auxin concentration will help answer this question. For instance, an input biosensor of auxin concentrations, DII-Venus, fuses a fast-maturing fluorescent reporter to the degron region of the Aux/IAA protein. The inverse of DII-Venus fluorescence consequently shows auxin activity at the cellular level, and can track auxin dynamic responses (Brunoud et al., 2012).
AUXIN AND ROOT ARCHITECTURE
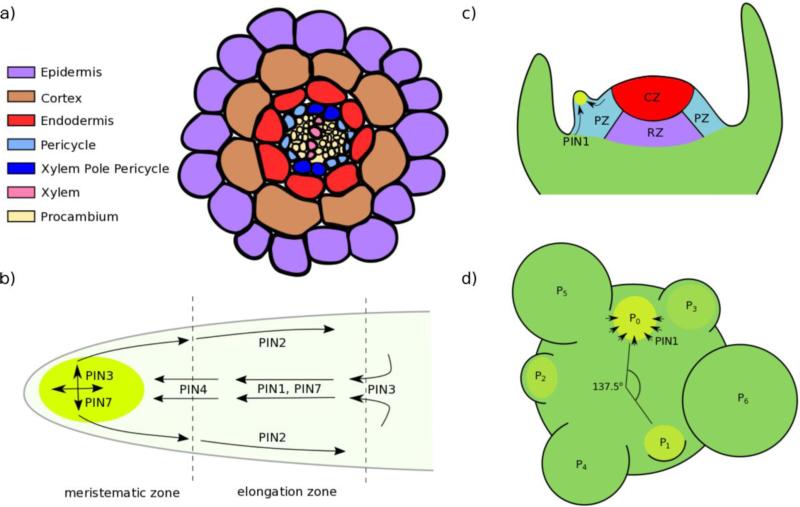
The establishment of the root-shoot juncture is one of the earliest steps in plant embryogenesis. During embryo-suspensor differentiation, the root stem cell niche and the SAM are generated. The stem cell niche produces the different tissue types of the root, which are arranged in concentric circles (Fig 2A) (reviewed in Overvoorde et al., 2010). The innermost cylinder of tissue is the vascular tissue, which is surrounded sequentially by the pericycle, the endodermis, the cortex, and the epidermis. Stem cell initials for each of these types of tissue reside in the stem cell niche and their division and the subsequent asymmetric fate of their progeny generates each cell file. Cell division occurs at the meristem, and newly divided cells then quickly expand longitudinally in the elongation zone, finally reaching their mature cell fate in the differentiation zone. Lateral roots emerge only from fully differentiated sections of the primary root.
Figure 2.
Auxin flow in the root and shoot. (a-d) Auxin maxima are shown in bright green. (a) Transverse section of the Arabidopsis root, with different cell types indicated. (b) Diagram indicating the flow of auxin in the primary root and the PINs primarily responsible for this movement. The meristematic zone and elongation zone of the root are divided by dashed lines. (c) The shoot apical meristem. Auxin transported by PIN1 generates maxima triggering the formation of primordia. CZ: central zone, RM: rib meristem, PZ: peripheral zone. (d) Top-down view of an Arabidopsis inflorescence meristem, showing the sites of developing primordia (P). PIN1 is localized towards the site of incipient primordia (P0), leading to high local concentrations of auxin.
The flow of auxin in the primary root follows a regular pattern, termed the “inverted fountain” model (Fig 2B) (Benkova et al., 2003). The majority of auxin in the root is first synthesized in aerial tissues and transported to the root via the phloem via specific PIN efflux carriers (Blilou et al., 2005). Auxin accumulates in the root cap and is then transported back up the outer cell files of the root via transporter proteins (Overvoorde et al., 2010). This predictable movement of auxin through the primary roots makes possible the auxin pulsatile signals and sustained maxima that are hallmarks of lateral root specification. One mechanism for coordinating metabolism across the plant body is by changing the levels of auxin made in the shoot and transported rootward. Sucrose-mediated upregulation of auxin biosynthesis in photosynthetic tissue causes an increase in rootward auxin transport, signaling an increased demand for root-acquired nutrients like nitrogen (Lilley et al., 2012). High auxin concentrations in the root tip are required for the maintenance of the meristem, as high expression of the auxin-responsive PLETHORA (PLT) genes maintains meristematic activity at the root tip. Auxin indirectly helps to form a gradient of PLT concentration by controlling cell division and promoting cell expansion rates. High auxin levels at the stem cell niche promote PLT transcription, and as cells move further away from this local auxin maximum they expand, decreasing PLT abundance by growth dilution (Mahonen et al., 2014). Loss of PLT activity promotes cellular differentiation (Galinha et al., 2007).
Lateral root organogenesis passes through three general stages: specification, initiation, and emergence (reviewed in Peret et al., 2009). Lateral roots are specified by a transient auxin maximum in the basal meristem of the pericycle cell file, which borders the vasculature. In Arabidopsis, lateral roots arise only from xylem pole pericycle (XPP) cells (Dubrovsky et al., 2000), while in maize lateral roots only form from the phloem pole pericycle cells (Jansen et al., 2012). The next step, initiation, sets the pattern of lateral roots on the primary root axis, as nearly all initiated lateral roots will eventually emerge. Initiation starts when two longitudinally-neighboring pericycle cells in a single cell file nuclei round and migrate towards their shared cell border (De Rybel et al., 2010). This migration occurs in two radially neighboring pairs of pericycle cells and is immediately followed by an anticlinal cell division, the first in the new lateral root. A series of periclinal and anticlinal cell divisions follow in a stereotypical pattern. When the lateral root has passed through several stages of development, it will emerge perpendicularly to the primary root axis, requiring the cellular remodeling of the endodermal cell layers that encircle the pericycle to allow the root through (Vermeer et al., 2014). The lateral root then establishes a similar meristematic zone and cell files as occurs in the primary root (Laskowski et al., 1995). The lateral root also establishes an auxin gradient with a maximum at the emerging root tip. This gradient is promoted by relocalization of PIN1 polarly to the membranes of cells within the primordium facing the primordium tip (Benkova et al., 2003). The patterning of lateral roots along the primary root forms the root system architecture and is essential for nutrient and water uptake. This patterning can be regulated at every step of lateral root development.
Regulation of auxin levels and distribution is integral to maintaining auxin maxima with distinct dynamics at each stage of organogenesis. During lateral root specification, pericycle cells progress through multiple tiers of competency. A pulsatile auxin response signal in the basal meristem (the region of the root directly shootward of the stem cell niche) oscillates in phase with a large number of genes (Moreno-Risueno et al., 2010). In a process known as lateral root priming, pericycle cells that have the potential to form lateral roots are resident in the basal meristem during the auxin maximum; such cells have been called “prebranch sites.” Very recently, the cause of the oscillatory auxin response maxima has been found—regular periodic programmed cell death (PCD) in lateral root cap cells releases auxin into surrounding tissue (Xuan et al., 2016). This auxin is transported to the basal meristem via AUX1. Genetic interference with the timing of root cap PCD or with AUX1 function causes less frequent oscillations in the basal meristem and fewer prebranch sites, resulting in fewer lateral roots (Xuan et al., 2016).
Although imaging suggests that all pericycle cells in the basal meristem receive the initial auxin signal (De Smet et al., 2007), typically only two xylem pole pericycle cells within a cell file retain a robust maximum of auxin response beyond the basal meristem. These two longitudinally neighboring cells are called lateral root founder cells and have the ability to initiate lateral root organogenesis by the migration of their nuclei towards their bordering membranes. Lateral roots never emerge from both poles of the xylem in the same transverse section, a phenomenon explored by a recent paper that modeled the regulation of transport by hormone levels (el-Showk et al., 2015). The model demonstrates that upregulation of AUX1 in pericycle cells is sufficient to promote the influx of auxin into these cells and maintain the transient auxin maximum from the prebranch site. Polar localization of PIN1 and PIN7 in the pericycle to the xylem pole could then generate a stable auxin maximum from this transient AUX1 signal at only one pole of the xylem, causing founder cell identity in only that cell. Interestingly, auxin transport inhibitors or high exogenous auxin treatments allow lateral root primordia to form opposite each other on the same transverse portion of the root (Kircher and Schopfer, 2015), supporting this model. Additionally, mutations in the GDP/GTP exchange factor GNOM, which regulates the localization of PIN proteins, causes a loss of the sustained auxin maxima and decreased lateral root initiation (Okumura et al., 2013).
The transition from founder cell to lateral root initiation is regulated by multiple inputs. Not all founder cells that have sustained auxin maxima will initiate the first division of lateral root development and produce a lateral root. Auxin transport in the endodermis plays a key role in determining where and whether initiation will occur (Marhavy et al., 2013). PIN3 expression is upregulated not only in pericycle founder cells and lateral root primordia, but also in the overlaying endodermal tissue. This endodermal upregulation happens only after the auxin response maxima in the pericycle, so it is not required for founder cell specification. In most endodermal cells, PIN3 is non-polarly localized; however, in endodermal cells that surround lateral root founder cells, PIN3 localizes to the inner lateral membrane that borders the pericycle. Gravity-induced root bending, which strongly induces initiation of a new root at the outer bend, relocalizes endodermal PIN3 to the inner lateral membrane. This relocalization of PIN3 suggests a mechanism by which auxin reflux from the endodermis to the pericycle enforces the sustained auxin maximum needed to trigger initiation. Null pin3 mutants have fewer lateral root primordia (Benkova et al., 2003; Laskowski et al., 2008) but have more lateral root founder cells (Marhavy et al., 2013), suggesting that PIN3 controls this switch from founder cell identity to lateral root initiation. Consistent with this model, endodermal-specific expression of PIN3 rescues founder cell number and lateral root density.
A recent paper further elaborates the central role of PIN3 in lateral root initiation (Chen et al., 2015). Combining genetics and mathematical modeling the authors describe a coherent feed-forward loop that drives PIN3 expression at lateral root founder cells to sustain auxin levels and provide a cellular memory for meristematic exposure to auxin. PIN3 transcription is regulated both by ARF7 directly and by a second ARF7-induced transcription factor called FOURLIPS (FLP). ARF7-dependent upregulation of FLP transcription is required for normal lateral root density. As in pin3 mutants, loss of FLP leads to a decrease of initiated lateral roots but an increase in lateral root founder cells. Although binding of either transcription factor to the PIN3 promoter is sufficient to upregulate PIN3 expression, both FLP and ARF7 activity are required for wild type lateral root densities. The proposed coherent feed forward architecture could allow for a longer response to auxin, connecting the initial auxin pulse to lateral root initiation. If this feed-forward loop occurs in specific cell files surrounding the founder cell, such as the endodermis, this motif would further support the transport of auxin into the pericycle by increasing the amount of PIN3 protein at the border between these cell files.
Following the first initiation division, subsequent divisions and cell expansion allow the new root to grow out, establish its own meristem and reiterate the cellular organization of the primary root—all processes subject to auxin regulation (reviewed in Peret et al., 2009). As lateral roots develop from the internal pericycle cell file, cell wall remodeling of external cell files is required for cell separation and emergence of the new organ. The auxin influx carrier LAX3 is expressed in cortical and epidermal cells directly overlaying new primordia (Swarup et al., 2008), and this upregulation is auxin-dependent. Intriguingly, lax3 mutant plants, which have fewer emerged lateral roots, have three times more lateral root primordia than wild type. This result indicates that auxin transport by LAX3 regulates lateral root organogenesis post-initiation but pre-emergence. The expression pattern of LAX3 is regulated in part by the localization of the auxin source, and LAX3 transcription is promoted by PIN3 upregulation in the cortex (Peret et al., 2013). LAX3 activity then induces the expression of a number of cell-wall remodeling enzymes like PECTIN METHYLESTERASES (PMEs), which prime pectin for cleavage. IAA14-dependent auxin pathways then upregulate additional pectin cleavage enzymes such as pectin lysases in the primary root cell files surrounding lateral root primordia, thereby facilitating cell separation and lateral root emergence (Laskowski et al., 2006).
Further evidence for auxin regulation of the cell wall comes from a transcriptomic analysis of lateral root development (Lewis et al., 2013). Of a set of 72 auxin-induced cell wall remodeling genes, the authors found eight mutants with a lateral root phenotype. One of these genes, CELLULASE 3 (CEL3), encodes an enzyme that belongs to a family that catalyzes the breakdown of cellulose, although it is a member of a subclass that lacks a cellulose binding domain (Urbanowicz et al., 2007). cel3 plants have fewer late stage primordia. The authors did not examine whether early stage primordia or founder cell number increase in these mutants, which would suggest a defect in emergence as observed for lax3. Another gene, LEUCINE-RICH REPEAT/EXTENSIN 2 (LRX2), is an extensin protein important for cell wall development (Draeger et al., 2015). lrx2 mutants have fewer emerged lateral roots and increased earlier stage primordia (Lewis et al., 2013).
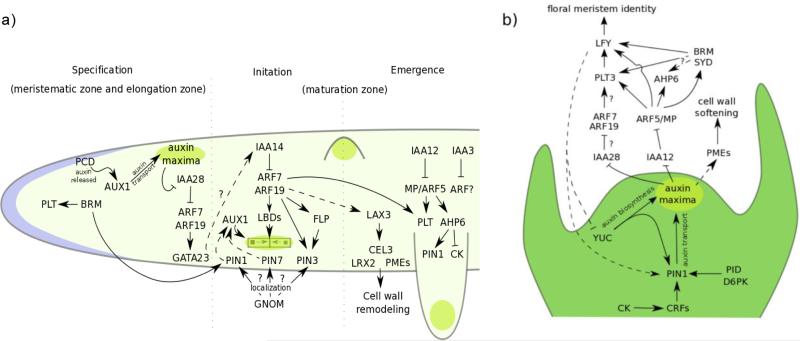
Auxin is sensed by different components of the auxin nuclear signaling pathway at each stage of lateral root organogenesis (Fig 3A). How these molecular modules interact and what determines the timing of action is still an open question. Specification is linked to degradation of the repressor IAA28 and subsequent activation of its targets ARF7 and ARF19 (Rogg et al., 2001; De Rybel et al., 2010). IAA28 degradation triggers induction of GATA23 in both the basal meristem and lateral root founder cells, sites of auxin maxima that are competent to form lateral roots. Reduction in the expression levels of GATA23 or stabilization of IAA28 decreases the number of founder cells with stable auxin maxima, suggesting that auxin-induction of GATA23 is necessary for the specification stage of lateral root organogenesis. Initiation is regulated by IAA14 degradation and the further activation of ARF7 and ARF19. Stabilized iaa14 mutants produce no lateral roots, and few if any cases of successful initiation (Fukaki et al., 2002). If the auxin-induced degradation rate of IAA14 is slowed, lateral root initiation is also delayed. (Guseman et al., 2015). IAA14 degradation leads to induction of a number of LOB DOMAIN CONTAINING (LBD) genes, which are required for the polar nuclear migration at the start of initiation (Goh et al., 2012). The degradation of IAA12 and activation of ARF5/MP (De Smet et al., 2010), as well as the degradation of IAA3 (Goh et al., 2012), further promote lateral initiation and emergence. Stabilization of either IAA12 or IAA3 lowers the number of lateral root primordia.
Figure 3.
Network of auxin-regulated genes involved in lateral organ development. (a,b) Auxin maxima are shown in bright green. (a) Network of auxin-regulated genes in lateral root development. The root is divided into three developmental stages going from left (tipwards) to right (shootwards). The corresponding root developmental zones are indicated in parentheses. Pictographic representations of each stage are included: an oscillatory auxin pulse during specification, two neighboring pericycle cell nuclei migrating towards each other at initiation, and an emerging and emerged lateral root. The position of the root cap is shown in blue. (b) Network of auxin-regulated genes in phyllotaxy. The shoot apical meristem is shown in two-dimensions; the auxin maxima is on the meristem periphery, extending outwards towards the viewer. Barred lines indicate repressive interactions, while arrows represent positive or activating interactions. Indirect regulation is shown with dashed lines, and questions marks are included where interactions have not been validated experimentally. CK: cytokinin.
AUXIN AND INFLORESCENCE ARCHITECTURE
In contrast to lateral root development, specification and initiation of new aerial lateral organs such as flowers and leaves happen in rapid succession without significant intervening cell elongation. The shoot apical meristem can be divided into three zones: (1) the central zone, involved in meristem maintenance and stem cell generation, (2) the rib zone, whose cells ultimately become inner stem tissue, and (3) the peripheral zone, in which cells differentiate to form new organs such as leaves and flowers (Fig 2C) (Tucker and Laux, 2007). The patterning of organ emergence around the shoot axis is termed phyllotaxy. Arabidopsis inflorescence meristems exhibit spiral phyllotaxy, where floral meristem primordia are initiated one at a time with an average angle of 137.5° between each primordium. A variety of factors can alter the timing of primordia initiation or the angle of divergence between primordia, including meristem size and meristem determinacy (Landrein et al., 2015; Bartlett and Thompson, 2014).
As with lateral root formation, auxin transport is required for production of lateral primordia at the SAM. The auxin efflux carrier PIN1 was originally identified through its striking mutant phenotype: a naked or pin-like inflorescence stem with few if any lateral organs (Okada et al., 1991). PIN1 proteins direct auxin flow towards the site of incipient primordia, leading to high local concentrations of auxin (Fig 2D). These auxin maxima function as auxin sinks, depleting the surrounding cells of auxin. This is thought to create an inhibitory field that sets the spacing between successive primordia (Reinhardt et al., 2003). Once a primordium has been established, PIN1 polarity within this domain reverses to allow auxin to flow away from the distal tip and allow further progression through development (Heisler et al., 2005). The precise mechanism that allows this stage-dependent PIN1 localization is an area of active investigation. AGCVIII protein kinases D6 PROTEIN KINASE (D6PK) and PINOID (PID) are thought to play a role in this process by phosphorylating PINs, localizing them to apical membranes and activating PIN-dependent auxin efflux (Barbosa and Schwechheimer, 2014). PIN polarity also appears to rely on phosphorylation-independent mechanisms that are not yet well understood (Ganguly et al., 2014). Along with transport, auxin biosynthesis is important for post-embryonic organ formation. For example, knocking down auxin synthesis and transport, as seen in the yuc1yuc4pin1 and yuc1yuc2yuc4yuc6aux1 mutants, prevents leaf and flower formation (Cheng et al., 2007).
Similarly to lateral root development, organogenesis in the shoot ultimately requires changes in the cell wall of differentiating tissues. As in the root, in the stem, auxin-induced cell wall softening via PMEs is necessary for organ outgrowths from the inflorescence meristem (Braybrook and Peaucelle, 2013). Blocking pectin de-methyl-esterification prevents the formation of lateral organs; plants overexpressing an inducible form of PECTIN METHYLESTERASE INHIBITOR 3 have a pin1-like meristem after induction (Peaucelle et al., 2008). In addition to localized cell wall softening, auxin is also thought to induce cortical microtubule disorganization, leading to isotropic cell walls (Sassi et al., 2014). This combination of cell wall remodeling and induced isotropy is predicted to be sufficient for organ outgrowth.
Unlike in the root, however, the specific players in nuclear auxin response regulating specification, initiation, and outgrowth of SAM primordia have been difficult to identify with forward genetics. This may reflect increased redundancy within the network, less quantifiable phenotypes, or the potential requirement for the same players at earlier stages of development where viability depends on a functional SAM. Despite this limitation, there are strong indications that auxin triggers critical transcriptional events in shoot organ formation just as it does in lateral roots. Indeed, a number of the same auxin-regulated genes that control lateral root development are important in the SAM (Fig 3B). For example, plants that express slower-degrading variants of IAA28 have altered phyllotaxy (Moss et al., 2015). ARF7 and ARF19 positively regulate the expression of PLT3, PLT5, and PLT7, genes also important for lateral root formation. In plt3plt5plt7 triple mutants, inflorescence phyllotaxy is significantly altered, with flowers initiating at a 180° or 90° angle of divergence (Prasad et al., 2011). This change in phyllotactic patterning is thought to reflect decreased auxin biosynthesis via YUC1 and YUC4 in the shoot meristem (Pinon et al., 2013). plt3plt5plt7 mutant plants also exhibit lateral root defects, including aberrant spacing and few fully emerged primordia (Hofhuis et al., 2013). It is not yet known whether these phenotypes can be attributed to altered PIN expression.
A number of genes with specific roles in SAM fate specification have also been connected to auxin. For example, SHOOT MERISTEMLESS (STM) is required for maintenance of stem cell function (Scofield et al., 2014), and is downregulated at the same time that PIN1 expression increases in incipient primordia (Heisler et al., 2005). Interestingly, STM expression in the meristem is restricted by the LBD proteins JAGGED LATERAL ORGANS (JLO) and ASYMMETRIC LEAVES2 (AS2). LBD proteins also play an important role in lateral root initiation (Rast and Simon, 2012). Expression of LEAFY (LFY), the master regulator of the vegetative-to-floral transition, rises dramatically following the formation of the auxin maximum (Heisler et al., 2005; Li et al., 2013). The LFY promoter contains several auxin regulatory elements (AuxREs) that are bound by ARF5/MP and IAA12 (Yamaguchi et al., 2013). ARF5/MP and IAA12 are known to have key roles in inflorescence development; plants carrying weak arf5/mp alleles forms pin-like inflorescence stems lacking flowers (Przemeck et al., 1996) that cannot be rescued by application of exogenous auxin. In addition to ARF5/MP, PLT3 has also been recently reported to upregulate LFY in response to auxin (Yamaguchi et al., 2016). As with PIN3 expression in the root, LFY may be regulated by an auxin-driven feed-forward loop in which ARF5/MP activates expression of LFY and PLT3, and PLT3 further promotes LFY expression (Yamaguchi et al., 2016). LFY itself may also regulate auxin signaling, as overexpression of LFY increases expression of the auxin reporter DR5 while decreasing expression of genes required for auxin biosynthesis (Li et al., 2013).
Despite numerous parallels between shoot and root branching, there are many auxin-regulated pathways involved in lateral root development that do not seem to play a role in the shoot. The IAA14-mediated module of lateral root initiation, for example, has no known shoot equivalent.
FINE-TUNING THROUGH AUXIN AND CYTOKININ CROSS-REGULATION
Auxin interacts with many signaling pathways, including those of many other hormones. In particular, cytokinin is well-known for acting in concert with auxin, and plays a key role in root and shoot meristem maintenance (Gordon et al., 2009; Dello Ioio et al., 2007). The cytokinin response inhibitor ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEIN 6 is a well-studied case of such integration. AHP6 regulates organogenesis in roots (Moreira et al., 2013) and shoots (Besnard et al., 2014) by fine-tuning the balance between auxin and cytokinin activity. AHP6 is activated by auxin, and is directly downstream of ARF5/MP. In roots, the inhibition of cytokinin activity by AHP6 is necessary for the proper orientation of cell divisions during early stages of lateral root initiation. The auxin maxima in lateral root founder cells promote the expression of AHP6 via ARF5/MP, which in turn inhibits cytokinin signaling in these cells. Like many determinants of early stages of lateral root development, AHP6 is expressed both at the basal meristem and in lateral root founder cells in the pericycle file of the primary root, as well as in all cells of early stage primordia. PIN1 is mislocalized in ahp6 mutant lateral root primordia, suggesting that AHP6 may regulate the orientation of cell divisions in early lateral root stages by promoting proper PIN1 localization (Moreira et al., 2013).
In addition to their lateral root defects, ahp6-1 mutants exhibit abnormal phyllotaxy due to a change in the time delay between each floral organ initiation (the plastochron) (Besnard et al., 2014). Contrary to what was observed in the roots, ahp6-1 was not found to affect PIN1 expression or localization in the inflorescence meristem. Instead, AHP6 is thought to regulate the spatiotemporal pattern of cytokinin signaling by negatively regulating several cytokinin response genes in the shoot. This suggests that while auxin is necessary to set up phyllotactic patterning, inhibitory fields set up by cytokinin signaling are required to fine-tune the plastochron itself.
Cytokinins can also more directly regulate PIN gene expression (Simaskova et al., 2015). Both PIN1 and PIN7 have PIN CYTOKININ RESPONSE ELEMENTs (PCREs) in their promoters. Yeast-one-hybrid and chromatin immunoprecipitation experiments found that these PIN1 and PIN7 PCREs can be bound by CYTOKININ RESPONSE FACTORS (CRFs). Furthermore, crf2, crf3, and crf3crf6 loss-of-function mutants have significantly reduced PIN1 and PIN7 expression and display reduced numbers of lateral roots.
NEW APPROACHES
The large gene families and complex feedback dynamics involved in auxin response complicate traditional genetic analysis. New methods in building and analyzing models of gene regulatory networks (GRNs) offer a complementary approach. By using a transcriptomic time course to reconstruct the GRN of lateral root initiation, Lavenus et al. were able to link early genetic players in initiation to later cell fate markers through a heavily feedback-regulated genetic cascade (Lavenus et al., 2015). The authors formulated a new algorithm to infer GRNs from time courses called time-delay correlation (TDCor), and extensively validated their system by identifying known ARF7 targets and their relationships in an ARF7-inducible transcriptomic dataset. In their lateral root organogenesis time course, the authors found that their GRN was divided into two antagonistic modules, one promoted by ARF7 and ARF19 activity and the other by ARF5/MP activity. They further found that these modules could be arranged chronologically, with ARF7 and ARF19 promoting early-acting fate-specification programs and ARF5/MP promoting later cell cycle and division regulators. The linking players between these two modules are the PLT proteins, which, as discussed above, are transcription factors that also play a pivotal role in shoot organogenesis (Prasad et al., 2011).
This transcriptomic approach provides support for the proposal that different stages of lateral root development are regulated by different auxin response modules—auxin-induced turnover of IAA28 and IAA14 leads to an early spike in ARF7 and ARF19 activity that is followed by later turnover of IAA12 leading to sustained ARF5/MP activation. But by mapping out the GRN the authors showed how these modules were interconnected and cross-regulated. This sequential action by the ARFs suggest a mechanism by which transient, quick pulses of early LR fate specifiers, such as GATA23, can be turned on and then quickly repressed when initiation begins. In contrast, auxin response targets essential for primordium outgrowth, such as the LBDs and cell expansion regulators, are more slowly repressed. Notably, this study found many examples of feed-forward motif in their lateral root organogenesis gene network that may, similarly to the ARF7-FLP regulation of PIN3 (Chen et al., 2015), allow for sustained action of auxin-regulated targets that regulate later stages of organogenesis. Feed-forward loops are also overrepresented in the GRN surrounding xylem development, suggesting that such gene regulatory motifs may play an important role in precisely controlling shifts in cell fate (Taylor-Teeples et al., 2015).
Complementary to GRN analysis, synthetic reconstruction of auxin signaling allows for direct functional analysis of isolated auxin response circuits implicated in specific developmental responses. Time-lapse flow cytometry of engineered yeast strains expressing specific auxin signaling components allows high throughput, quantitative measures of auxin-induced degradation of Aux/IAAs and activation of downstream transcription (Havens et al., 2012; Pierre-Jerome et al., 2014). This data facilitates the building of mathematical models to describe and analyze circuit dynamics. Functional parameters specific to different AFB/IAA and ARF/IAA pairs have been examined systematically and modeled using this approach and have successfully predicted in planta behavior. For example, the rate of auxin-induced degradation was the most sensitive parameter for tuning activation of synthetic auxin circuits. Transgenic plants revealed that the timing of lateral root initiation was exquisitely sensitive to the turnover rate of IAA14 (Guseman et al., 2015). Many open questions in auxin signaling, especially how the different auxin response modules are enacted over time, may be amenable to more complex synthetic reconstructions.
Epigenetics likely also contribute to differential auxin responses. A recent screen of chromatin modifiers identified two SWI/SNF subgroup ATPases, BRAHMA (BRM) and SPLAYED (SYD), as essential for floral meristem initiation (Wu et al., 2015). Co-immunoprecipitations and yeast-two-hybrid experiments revealed that ARF5/MP interacts physically with BRM and SYD, but that this interaction is blocked in the presence of Aux/IAA proteins known to associate with ARF5/MP. Based on ChIP assays, BRM and SYD were also found to bind the same regulatory regions as ARF5/MP and increase accessibility to these targets. The authors conclude that ARF5/MP recruits these SWI/SNF factors to increase accessibility of the DNA for induction of key regulators of flower primordia initiation. Notably, brm loss-of-function mutants have more lateral roots, but reduced expression of PIN and PLT genes (Yang et al., 2015). It will be very exciting to see if these epigenetic mechanisms apply more broadly to other ARFs and other developmental contexts.
CONCLUSIONS
One of the most fascinating and frustrating aspect of auxin signaling is how it can direct so many developmental processes and yet show striking specificity. The large protein families involved in the auxin nuclear response were initially thought to accommodate this paradox—specific modules of regulators could code for different developmental programs. Unfortunately, biochemical studies do not support this model in many cases (Hardtke et al., 2004; Vernoux et al., 2011). Instead, a more nuanced hypothesis has emerged with recent structural papers (Korasick et al., 2014; Nanao et al., 2014; Boer et al., 2014). ARFs, which dimerize at both their DNA binding and distal PB1 domains, have the potential to form heterodimers, and possibly higher-order structures, with other ARFs and Aux/IAAs. ARF interactions with other ARFs and other transcription factors could provide a combinatorial code for specific action in different contexts. The regulation of PIN3 by both ARF7 and the ARF7-responsive transcription factor FLP in lateral root initiation could be an example of such context-specific encoding.
The GRNs shaping root and shoot architecture show remarkable similarities, perhaps reflecting a common evolutionary origin. More work in plants from more basal lineages will be helpful in determining if this is indeed the case. The liverwort Marchantia polymorpha has a simple auxin transcriptional response including one IAA, one AFB, and three ARFs (Flores-Sandoval et al., 2015; Kato et al., 2015). This pathway is necessary for the transition from gametophyte to sporophyte, as well as for patterning of gemma cups, gemmae, and rhizoids. The authors posit that auxin acts as a facilitator rather than a determiner of cell fate, a signal that can be co-opted by various developmental pathways to regulate their progression and patterning. It is possible that auxin-directed mechanisms that pattern aerial tissues could have later been reused for patterning the root as well. As plants invaded new niches during evolution, the modularity of the core auxin circuit and the regulation of auxin by transport, feedback, and other developmental pathways may have provided a means to rapidly elaborate more complex organs with greater environmental adaptability. Comparative approaches, across organs and organisms, will help elucidate the key themes that allow auxin signaling to function in natural contexts and point to attractive targets for plant engineering in the future.
Highlights.
Auxin plays a critical role in shaping plant architecture.
Similar auxin-driven gene modules impact development in shoots and roots.
New approaches elucidate how auxin GRNs translate into developmental outcomes.
ACKNOWLEDGEMENTS
We thank Jessica Guseman, Orlando de Lange, and Stacey Lowman for careful reading of our manuscript, and members of the Nemhauser and Imaizumi Labs for helpful discussions. Work on auxin in the Nemhauser Lab is supported by the National Science Foundation (MCB-1411949 and IOS-1539834) and National Institute of Health (R01-GM107084). A.L. is supported by a National Science Foundation Graduate Research Fellowship (DGE-1256082).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Barbosa IC, Schwechheimer C. Dynamic control of auxin transport-dependent growth by AGCVIII protein kinases. Curr Opin Plant Biol. 2014;22:108–115. doi: 10.1016/j.pbi.2014.09.010. [DOI] [PubMed] [Google Scholar]
- Bartlett ME, Thompson B. Meristem identity and phyllotaxis in inflorescence development. Front Plant Sci. 2014;5:508. doi: 10.3389/fpls.2014.00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- Besnard F, Refahi Y, Morin V, Marteaux B, Brunoud G, Chambrier P, Rozier F, Mirabet V, Legrand J, Laine S, Thevenon E, Farcot E, Cellier C, Das P, Bishopp A, Dumas R, Parcy F, Helariutta Y, Boudaoud A, Godin C, Traas J, Guedon Y, Vernoux T. Cytokinin signalling inhibitory fields provide robustness to phyllotaxis. Nature. 2014;505:417–421. doi: 10.1038/nature12791. [DOI] [PubMed] [Google Scholar]
- Bhalerao RP, Bennett MJ. The case for morphogens in plants. Nat Cell Biol. 2003;5:939–943. doi: 10.1038/ncb1103-939. [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;433:39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- Boer DR, Freire-Rios A, van den Berg WA, Saaki T, Manfield IW, Kepinski S, Lopez-Vidrieo I, Franco-Zorrilla JM, de Vries SC, Solano R, Weijers D, Coll M. Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell. 2014;156:577–589. doi: 10.1016/j.cell.2013.12.027. [DOI] [PubMed] [Google Scholar]
- Braybrook SA, Peaucelle A. Mechano-chemical aspects of organ formation in Arabidopsis thaliana: the relationship between auxin and pectin. PLoS One. 2013;8:e57813. doi: 10.1371/journal.pone.0057813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, Beeckman T, Kepinski S, Traas J, Bennett MJ, Vernoux T. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature. 2012;482:103–106. doi: 10.1038/nature10791. [DOI] [PubMed] [Google Scholar]
- Chen Q, Liu Y, Maere S, Lee E, Van Isterdael G, Xie Z, Xuan W, Lucas J, Vassileva V, Kitakura S, Marhavy P, Wabnik K, Geldner N, Benkova E, Le J, Fukaki H, Grotewold E, Li C, Friml J, Sack F, Beeckman T, Vanneste S. A coherent transcriptional feed-forward motif model for mediating auxin-sensitive PIN3 expression during lateral root development. Nat Commun. 2015;6:8821. doi: 10.1038/ncomms9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006;20:1790–1799. doi: 10.1101/gad.1415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell. 2007;19:2430–2439. doi: 10.1105/tpc.107.053009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B, Vassileva V, Parizot B, Demeulenaere M, Grunewald W, Audenaert D, Van Campenhout J, Overvoorde P, Jansen L, Vanneste S, Moller B, Wilson M, Holman T, Van Isterdael G, Brunoud G, Vuylsteke M, Vernoux T, De Veylder L, Inze D, Weijers D, Bennett MJ, Beeckman T. A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr Biol. 2010;20:1697–1706. doi: 10.1016/j.cub.2010.09.007. [DOI] [PubMed] [Google Scholar]
- De Smet I, Lau S, Voss U, Vanneste S, Benjamins R, Rademacher EH, Schlereth A, De Rybel B, Vassileva V, Grunewald W, Naudts M, Levesque MP, Ehrismann JS, Inze D, Luschnig C, Benfey PN, Weijers D, Van Montagu MC, Bennett MJ, Jurgens G, Beeckman T. Bimodular auxin response controls organogenesis in Arabidopsis. Proc Natl Acad Sci U S A. 2010;107:2705–2710. doi: 10.1073/pnas.0915001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, Frei dit Frey N, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audenaert D, Inze D, Bennett MJ, Beeckman T. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development. 2007;134:681–690. doi: 10.1242/dev.02753. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol. 2007;17:678–682. doi: 10.1016/j.cub.2007.02.047. [DOI] [PubMed] [Google Scholar]
- Draeger C, Ndinyanka Fabrice T, Gineau E, Mouille G, Kuhn BM, Moller I, Abdou MT, Frey B, Pauly M, Bacic A, Ringli C. Arabidopsis leucine-rich repeat extensin (LRX) proteins modify cell wall composition and influence plant growth. BMC Plant Biol. 2015;15:155. doi: 10.1186/s12870-015-0548-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Doerner PW, Colon-Carmona A, Rost TL. Pericycle cell proliferation and lateral root initiation in Arabidopsis. Plant Physiol. 2000;124:1648–1657. doi: 10.1104/pp.124.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Showk S, Help-Rinta-Rahko H, Blomster T, Siligato R, Maree AF, Mahonen AP, Grieneisen VA. Parsimonious Model of Vascular Patterning Links Transverse Hormone Fluxes to Lateral Root Initiation: Auxin Leads the Way, while Cytokinin Levels Out. PLoS Comput Biol. 2015;11:e1004450. doi: 10.1371/journal.pcbi.1004450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finet C, Jaillais Y. Auxology: when auxin meets plant evo-devo. Dev Biol. 2012;369:19–31. doi: 10.1016/j.ydbio.2012.05.039. [DOI] [PubMed] [Google Scholar]
- Flores-Sandoval E, Eklund DM, Bowman JL. A Simple Auxin Transcriptional Response System Regulates Multiple Morphogenetic Processes in the Liverwort Marchantia polymorpha. PLoS Genet. 2015;11:e1005207. doi: 10.1371/journal.pgen.1005207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 2002;29:153–168. doi: 10.1046/j.0960-7412.2001.01201.x. [DOI] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature. 2007;449:1053–1057. doi: 10.1038/nature06206. [DOI] [PubMed] [Google Scholar]
- Ganguly A, Park M, Kesawat MS, Cho HT. Functional Analysis of the Hydrophilic Loop in Intracellular Trafficking of Arabidopsis PIN-FORMED Proteins. Plant Cell. 2014;26:1570–1585. doi: 10.1105/tpc.113.118422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh T, Joi S, Mimura T, Fukaki H. The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development. 2012;139:883–893. doi: 10.1242/dev.071928. [DOI] [PubMed] [Google Scholar]
- Goh T, Kasahara H, Mimura T, Kamiya Y, Fukaki H. Multiple AUX/IAA-ARF modules regulate lateral root formation: the role of Arabidopsis SHY2/IAA3-mediated auxin signalling. Philos Trans R Soc Lond B Biol Sci. 2012;367:1461–1468. doi: 10.1098/rstb.2011.0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM. Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc Natl Acad Sci U S A. 2009;106:16529–16534. doi: 10.1073/pnas.0908122106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grones P, Friml J. Auxin transporters and binding proteins at a glance. J Cell Sci. 2015;128:1–7. doi: 10.1242/jcs.159418. [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ. The PB1 domain in auxin response factor and Aux/IAA proteins: a versatile protein interaction module in the auxin response. Plant Cell. 2015;27:33–43. doi: 10.1105/tpc.114.132753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guseman JM, Hellmuth A, Lanctot A, Feldman TP, Moss BL, Klavins E, Calderón Villalobos LI, Nemhauser JL. Auxin-induced degradation dynamics set the pace for lateral root development. Development. 2015;142:905–909. doi: 10.1242/dev.117234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Ckurshumova W, Vidaurre DP, Singh SA, Stamatiou G, Tiwari SB, Hagen G, Guilfoyle TJ, Berleth T. Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development. 2004;131:1089–1100. doi: 10.1242/dev.00925. [DOI] [PubMed] [Google Scholar]
- Havens KA, Guseman JM, Jang SS, Pierre-Jerome E, Bolten N, Klavins E, Nemhauser JL. A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiol. 2012;160:135–142. doi: 10.1104/pp.112.202184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol. 2005;15:1899–1911. doi: 10.1016/j.cub.2005.09.052. [DOI] [PubMed] [Google Scholar]
- Hofhuis H, Laskowski M, Du Y, Prasad K, Grigg S, Pinon V, Scheres B. Phyllotaxis and rhizotaxis in Arabidopsis are modified by three PLETHORA transcription factors. Curr Biol. 2013;23:956–962. doi: 10.1016/j.cub.2013.04.048. [DOI] [PubMed] [Google Scholar]
- Jansen L, Roberts I, De Rycke R, Beeckman T. Phloem-associated auxin response maxima determine radial positioning of lateral roots in maize. Philos Trans R Soc Lond B Biol Sci. 2012;367:1525–1533. doi: 10.1098/rstb.2011.0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Ishizaki K, Kouno M, Shirakawa M, Bowman JL, Nishihama R, Kohchi T. Auxin-Mediated Transcriptional System with a Minimal Set of Components Is Critical for Morphogenesis through the Life Cycle in Marchantia polymorpha. PLoS Genet. 2015;11:e1005084. doi: 10.1371/journal.pgen.1005084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Schopfer P. Priming and positioning of lateral roots in Arabidopsis. An approach for an integrating concept. J Exp Bot. 2015 doi: 10.1093/jxb/erv541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick DA, Westfall CS, Lee SG, Nanao MH, Dumas R, Hagen G, Guilfoyle TJ, Jez JM, Strader LC. Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc Natl Acad Sci U S A. 2014;111:5427–5432. doi: 10.1073/pnas.1400074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutschera U, Bergfeld R, Schopfer P. Cooperation of epidermis and inner tissues in auxin-mediated growth of maize coleoptiles. Planta. 1987;170:168–180. doi: 10.1007/BF00397885. [DOI] [PubMed] [Google Scholar]
- Landrein B, Refahi Y, Besnard F, Hervieux N, Mirabet V, Boudaoud A, Vernoux T, Hamant O. Meristem size contributes to the robustness of phyllotaxis in Arabidopsis. J Exp Bot. 2015;66:1317–1324. doi: 10.1093/jxb/eru482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski M, Biller S, Stanley K, Kajstura T, Prusty R. Expression profiling of auxin-treated Arabidopsis roots: toward a molecular analysis of lateral root emergence. Plant Cell Physiol. 2006;47:788–792. doi: 10.1093/pcp/pcj043. [DOI] [PubMed] [Google Scholar]
- Laskowski M, Grieneisen VA, Hofhuis H, Hove CA, Hogeweg P, Maree AF, Scheres B. Root system architecture from coupling cell shape to auxin transport. PLoS Biol. 2008;6:e307. doi: 10.1371/journal.pbio.0060307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski MJ, Williams ME, Nusbaum HC, Sussex IM. Formation of lateral root meristems is a two-stage process. Development. 1995;121:3303–3310. doi: 10.1242/dev.121.10.3303. [DOI] [PubMed] [Google Scholar]
- Lau S, De Smet I, Kolb M, Meinhardt H, Jurgens G. Auxin triggers a genetic switch. Nat Cell Biol. 2011;13:611–615. doi: 10.1038/ncb2212. [DOI] [PubMed] [Google Scholar]
- Lavenus J, Goh T, Guyomarc'h S, Hill K, Lucas M, Voss U, Kenobi K, Wilson MH, Farcot E, Hagen G, Guilfoyle TJ, Fukaki H, Laplaze L, Bennett MJ. Inference of the Arabidopsis lateral root gene regulatory network suggests a bifurcation mechanism that defines primordia flanking and central zones. Plant Cell. 2015;27:1368–1388. doi: 10.1105/tpc.114.132993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DR, Olex AL, Lundy SR, Turkett WH, Fetrow JS, Muday GK. A kinetic analysis of the auxin transcriptome reveals cell wall remodeling proteins that modulate lateral root development in Arabidopsis. Plant Cell. 2013;25:3329–3346. doi: 10.1105/tpc.113.114868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhou Y, Liu X, Yu P, Cohen JD, Meyerowitz EM. LEAFY controls auxin response pathways in floral primordium formation. Sci Signal. 2013;6:ra23. doi: 10.1126/scisignal.2003937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley JL, Gee CW, Sairanen I, Ljung K, Nemhauser JL. An endogenous carbon-sensing pathway triggers increased auxin flux and hypocotyl elongation. Plant Physiol. 2012;160:2261–2270. doi: 10.1104/pp.112.205575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim PO, Lee IC, Kim J, Kim HJ, Ryu JS, Woo HR, Nam HG. Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. J Exp Bot. 2010;61:1419–1430. doi: 10.1093/jxb/erq010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahonen AP, ten Tusscher K, Siligato R, Smetana O, Diaz-Trivino S, Salojarvi J, Wachsman G, Prasad K, Heidstra R, Scheres B. PLETHORA gradient formation mechanism separates auxin responses. Nature. 2014;515:125–129. doi: 10.1038/nature13663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhavy P, Vanstraelen M, De Rybel B, Zhaojun D, Bennett MJ, Beeckman T, Benkova E. Auxin reflux between the endodermis and pericycle promotes lateral root initiation. EMBO J. 2013;32:149–158. doi: 10.1038/emboj.2012.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira S, Bishopp A, Carvalho H, Campilho A. AHP6 inhibits cytokinin signaling to regulate the orientation of pericycle cell division during lateral root initiation. PLoS One. 2013;8:e56370. doi: 10.1371/journal.pone.0056370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science. 2010;329:1306–1311. doi: 10.1126/science.1191937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss BL, Mao H, Guseman JM, Hinds TR, Hellmuth A, Kovenock M, Noorassa A, Lanctot A, Villalobos LI, Zheng N, Nemhauser JL. Rate Motifs Tune Auxin/Indole-3-Acetic Acid Degradation Dynamics. Plant Physiol. 2015;169:803–813. doi: 10.1104/pp.15.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanao MH, Vinos-Poyo T, Brunoud G, Thevenon E, Mazzoleni M, Mast D, Laine S, Wang S, Hagen G, Li H, Guilfoyle TJ, Parcy F, Vernoux T, Dumas R. Structural basis for oligomerization of auxin transcriptional regulators. Nat Commun. 2014;5:3617. doi: 10.1038/ncomms4617. [DOI] [PubMed] [Google Scholar]
- Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y. Requirement of the Auxin Polar Transport System in Early Stages of Arabidopsis Floral Bud Formation. Plant Cell. 1991;3:677–684. doi: 10.1105/tpc.3.7.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura K, Goh T, Toyokura K, Kasahara H, Takebayashi Y, Mimura T, Kamiya Y, Fukaki H. GNOM/FEWER ROOTS is required for the establishment of an auxin response maximum for arabidopsis lateral root initiation. Plant Cell Physiol. 2013;54:406–417. doi: 10.1093/pcp/pct018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde P, Fukaki H, Beeckman T. Auxin control of root development. Cold Spring Harb Perspect Biol. 2010;2:a001537. doi: 10.1101/cshperspect.a001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaucelle A, Louvet R, Johansen JN, Hofte H, Laufs P, Pelloux J, Mouille G. Arabidopsis phyllotaxis is controlled by the methyl-esterification status of cell-wall pectins. Curr Biol. 2008;18:1943–1948. doi: 10.1016/j.cub.2008.10.065. [DOI] [PubMed] [Google Scholar]
- Peret B, De Rybel B, Casimiro I, Benkova E, Swarup R, Laplaze L, Beeckman T, Bennett MJ. Arabidopsis lateral root development: an emerging story. Trends Plant Sci. 2009;14:399–408. doi: 10.1016/j.tplants.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Peret B, Larrieu A, Bennett MJ. Lateral root emergence: a difficult birth. J Exp Bot. 2009;60:3637–3643. doi: 10.1093/jxb/erp232. [DOI] [PubMed] [Google Scholar]
- Peret B, Middleton AM, French AP, Larrieu A, Bishopp A, Njo M, Wells DM, Porco S, Mellor N, Band LR, Casimiro I, Kleine-Vehn J, Vanneste S, Sairanen I, Mallet R, Sandberg G, Ljung K, Beeckman T, Benkova E, Friml J, Kramer E, King JR, De Smet I, Pridmore T, Owen M, Bennett MJ. Sequential induction of auxin efflux and influx carriers regulates lateral root emergence. Mol Syst Biol. 2013;9:699. doi: 10.1038/msb.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre-Jerome E, Jang SS, Havens KA, Nemhauser JL, Klavins E. Recapitulation of the forward nuclear auxin response pathway in yeast. Proc Natl Acad Sci U S A. 2014;111:9407–9412. doi: 10.1073/pnas.1324147111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre-Jerome E, Moss BL, Nemhauser JL. Tuning the auxin transcriptional response. J Exp Bot. 2013;64:2557–2563. doi: 10.1093/jxb/ert100. [DOI] [PubMed] [Google Scholar]
- Pinon V, Prasad K, Grigg SP, Sanchez-Perez GF, Scheres B. Local auxin biosynthesis regulation by PLETHORA transcription factors controls phyllotaxis in Arabidopsis. Proc Natl Acad Sci U S A. 2013;110:1107–1112. doi: 10.1073/pnas.1213497110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piya S, Shrestha SK, Binder B, Stewart CN, Jr., Hewezi T. Protein-protein interaction and gene co-expression maps of ARFs and Aux/IAAs in Arabidopsis. Front Plant Sci. 2014;5:744. doi: 10.3389/fpls.2014.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad K, Grigg SP, Barkoulas M, Yadav RK, Sanchez-Perez GF, Pinon V, Blilou I, Hofhuis H, Dhonukshe P, Galinha C, Mahonen AP, Muller WH, Raman S, Verkleij AJ, Snel B, Reddy GV, Tsiantis M, Scheres B. Arabidopsis PLETHORA transcription factors control phyllotaxis. Curr Biol. 2011;21:1123–1128. doi: 10.1016/j.cub.2011.05.009. [DOI] [PubMed] [Google Scholar]
- Przemeck GK, Mattsson J, Hardtke CS, Sung ZR, Berleth T. Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta. 1996;200:229–237. doi: 10.1007/BF00208313. [DOI] [PubMed] [Google Scholar]
- Rast MI, Simon R. Arabidopsis JAGGED LATERAL ORGANS acts with ASYMMETRIC LEAVES2 to coordinate KNOX and PIN expression in shoot and root meristems. Plant Cell. 2012;24:2917–2933. doi: 10.1105/tpc.112.099978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C. Regulation of phyllotaxis by polar auxin transport. Nature. 2003;426:255–260. doi: 10.1038/nature02081. [DOI] [PubMed] [Google Scholar]
- Rogg LE, Lasswell J, Bartel B. A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell. 2001;13:465–480. doi: 10.1105/tpc.13.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosquete MR, Barbez E, Kleine-Vehn J. Cellular auxin homeostasis: gatekeeping is housekeeping. Mol Plant. 2012;5:772–786. doi: 10.1093/mp/ssr109. [DOI] [PubMed] [Google Scholar]
- Sassi M, Ali O, Boudon F, Cloarec G, Abad U, Cellier C, Chen X, Gilles B, Milani P, Friml J, Vernoux T, Godin C, Hamant O, Traas J. An auxin-mediated shift toward growth isotropy promotes organ formation at the shoot meristem in Arabidopsis. Curr Biol. 2014;24:2335–2342. doi: 10.1016/j.cub.2014.08.036. [DOI] [PubMed] [Google Scholar]
- Scofield S, Dewitte W, Murray JA. STM sustains stem cell function in the Arabidopsis shoot apical meristem and controls KNOX gene expression independently of the transcriptional repressor AS1. Plant Signal Behav. 2014;9 doi: 10.4161/psb.28934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simaskova M, O'Brien JA, Khan M, Van Noorden G, Otvos K, Vieten A, De Clercq I, Van Haperen JM, Cuesta C, Hoyerova K, Vanneste S, Marhavy P, Wabnik K, Van Breusegem F, Nowack M, Murphy A, Friml J, Weijers D, Beeckman T, Benkova E. Cytokinin response factors regulate PIN-FORMED auxin transporters. Nat Commun. 2015;6:8717. doi: 10.1038/ncomms9717. [DOI] [PubMed] [Google Scholar]
- Smit ME, Weijers D. The role of auxin signaling in early embryo pattern formation. Curr Opin Plant Biol. 2015;28:99–105. doi: 10.1016/j.pbi.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Swarup K, Benkova E, Swarup R, Casimiro I, Peret B, Yang Y, Parry G, Nielsen E, De Smet I, Vanneste S, Levesque MP, Carrier D, James N, Calvo V, Ljung K, Kramer E, Roberts R, Graham N, Marillonnet S, Patel K, Jones JD, Taylor CG, Schachtman DP, May S, Sandberg G, Benfey P, Friml J, Kerr I, Beeckman T, Laplaze L, Bennett MJ. The auxin influx carrier LAX3 promotes lateral root emergence. Nat Cell Biol. 2008;10:946–954. doi: 10.1038/ncb1754. [DOI] [PubMed] [Google Scholar]
- Taylor-Teeples M, Lin L, de Lucas M, Turco G, Toal TW, Gaudinier A, Young NF, Trabucco GM, Veling MT, Lamothe R, Handakumbura PP, Xiong G, Wang C, Corwin J, Tsoukalas A, Zhang L, Ware D, Pauly M, Kliebenstein DJ, Dehesh K, Tagkopoulos I, Breton G, Pruneda-Paz JL, Ahnert SE, Kay SA, Hazen SP, Brady SM. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature. 2015;517:571–575. doi: 10.1038/nature14099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker MR, Laux T. Connecting the paths in plant stem cell regulation. Trends Cell Biol. 2007;17:403–410. doi: 10.1016/j.tcb.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Uggla C, Moritz T, Sandberg G, Sundberg B. Auxin as a positional signal in pattern formation in plants. Proc Natl Acad Sci U S A. 1996;93:9282–9286. doi: 10.1073/pnas.93.17.9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowicz BR, Bennett AB, Del Campillo E, Catala C, Hayashi T, Henrissat B, Hofte H, McQueen-Mason SJ, Patterson SE, Shoseyov O, Teeri TT, Rose JK. Structural organization and a standardized nomenclature for plant endo-1,4-beta-glucanases (cellulases) of glycosyl hydrolase family 9. Plant Physiol. 2007;144:1693–1696. doi: 10.1104/pp.107.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanstraelen M, Benkova E. Hormonal interactions in the regulation of plant development. Annu Rev Cell Dev Biol. 2012;28:463–487. doi: 10.1146/annurev-cellbio-101011-155741. [DOI] [PubMed] [Google Scholar]
- Vermeer JE, von Wangenheim D, Barberon M, Lee Y, Stelzer EH, Maizel A, Geldner N. A spatial accommodation by neighboring cells is required for organ initiation in Arabidopsis. Science. 2014;343:178–183. doi: 10.1126/science.1245871. [DOI] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, Legrand J, Oliva M, Das P, Larrieu A, Wells D, Guedon Y, Armitage L, Picard F, Guyomarc'h S, Cellier C, Parry G, Koumproglou R, Doonan JH, Estelle M, Godin C, Kepinski S, Bennett M, De Veylder L, Traas J. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol. 2011;7:508. doi: 10.1038/msb.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, Kasahara H, Kamiya Y, Chory J, Zhao Y. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci U S A. 2011;108:18518–18523. doi: 10.1073/pnas.1108436108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MF, Yamaguchi N, Xiao J, Bargmann B, Estelle M, Sang Y, Wagner D. Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. Elife. 2015;4:e09269. doi: 10.7554/eLife.09269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan W, Band LR, Kumpf RP, Van Damme D, Parizot B, De Rop G, Opdenacker D, Moller BK, Skorzinski N, Njo MF, De Rybel B, Audenaert D, Nowack MK, Vanneste S, Beeckman T. Cyclic programmed cell death stimulates hormone signaling and root development in Arabidopsis. Science. 2016;351:384–387. doi: 10.1126/science.aad2776. [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Jeong CW, Nole-Wilson S, Krizek BA, Wagner D. AINTEGUMENTA and AINTEGUMENTA-LIKE6/PLETHORA3 Induce LEAFY Expression in Response to Auxin to Promote the Onset of Flower Formation in Arabidopsis. Plant Physiol. 2016;170:283–293. doi: 10.1104/pp.15.00969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Wu MF, Winter CM, Berns MC, Nole-Wilson S, Yamaguchi A, Coupland G, Krizek BA, Wagner D. A molecular framework for auxin-mediated initiation of flower primordia. Dev Cell. 2013;24:271–282. doi: 10.1016/j.devcel.2012.12.017. [DOI] [PubMed] [Google Scholar]
- Yang S, Li C, Zhao L, Gao S, Lu J, Zhao M, Chen CY, Liu X, Luo M, Cui Y, Yang C, Wu K. The Arabidopsis SWI2/SNF2 Chromatin Remodeling ATPase BRAHMA Targets Directly to PINs and Is Required for Root Stem Cell Niche Maintenance. Plant Cell. 2015;27:1670–1680. doi: 10.1105/tpc.15.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]