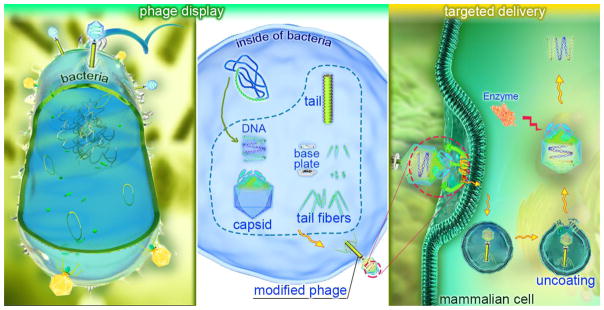
Abstract
The main goal of drug delivery systems is to target therapeutic cargoes to desired cells and to ensure their efficient uptake. Recently a number of studies have focused on designing bio-inspired nanocarriers, such as bacteriophages, and synthetic carriers based on the bacteriophage structure Bacteriophages are viruses that specifically recognize their bacterial hosts. They can replicate only inside their host cell and can act as natural gene carriers. Each type of phage has a particular shape, a different capacity for loading cargo, a specific production time, and their own mechanisms of supramolecular assembly, that have enabled them to act as tunable carriers. New phage-based technologies have led to the construction of different peptide libraries, and recognition abilities provided by novel targeting ligands. Phage hybridization with non-organic compounds introduces new properties to phages and could be a suitable strategy for construction of bio-inorganic carriers. In this review we try to cover the major phage species that have been used in drug and gene delivery systems, and the biological application of phages as novel targeting ligands and targeted therapeutics.
Graphical abstract
1. Introduction
In nature, viruses are natural gene carriers that transfer biological information between different species, which can range from prokaryotic cells such as bacteria to more complicated biological systems like mammalian cells. A wide variety of viruses have been used as drug delivery systems (DDSs) with varying properties. The most commonly-employed eukaryotic viruses such as adenoviruses, retroviruses and lentiviruses, although efficient in infecting target cells, might stimulate immune responses and could cause cancer by disrupting tumor suppressor genes. These possible drawbacks have encouraged researchers to consider bacteriophages and other virus-like particles that are considered safer. The basic structure of viruses has inspired the construction of newly engineered viral particles by a process called pseudotyping. This involves changing their surface proteins and glycoproteins that can lead to generation of de novo particles such as virosomes, that consist of a virion-like phospholipid bilayer that can be coated with specifically chosen surface glycoproteins.
Among all viruses, bacteriophages have a wide range of potential applications in both clinical and non-clinical fields. Phages have been used for food preservation in the food industry[1], as an eradicator of bacterial biofilms[2], and in bio-sensing in wastewater treatment[3]. The fundamental benefits of bacteriophage systems include their ease of use and development, a great capacity for packaging cargos, an ability to be loaded with non-DNA cargos, and their relative safety in humans. These favorable properties suggest that phages can be considered as nanocarriers[1, 3, 4]. Moreover bacteriophages do not undergo alterations in their natural tropism or mutation which is an advantage compared to eukaryotic viruses. This property makes them safer to be used for treatment and prevention of infections in human and animal populations. Bacteriophages have two main stages in their life cycle, which are the lytic cycle and the lysogenic cycle, although few phages are capable of carrying out both[4]. Since DNA and RNA molecules are unstable in their native form in body fluids [5–60 minutes for RNA and 10 minutes for DNA] because they are destroyed by nucleases, phages are ideal vehicles for transferring nucleic acids. Their simple production and purification methods as well as their large capacity for containing genetic material are additional advantages.
The therapeutic application of phages began in the former Soviet Union and Eastern Europe and their unique properties suggested they could be promising agents in clinical applications as anti-infectives. Phages are highly specific for their target and their self-regulating systems, such as a limitation of propagation to specific cells, make them versatile and modifiable carriers in targeted delivery systems[5]. Rather than using intact phages, some phage-derived molecular products can also be used as therapeutic agents. For instance phage endolysins specifically hydrolyze the peptidoglycan component of different bacterial species and are therefore bactericidal[6].In recent years, more researchers have studied new application of phages in the pharmaceutical and biotechnology industries. In this review we are will cover recent studies that have focused on phages as delivery vehicles.
2. Basic properties of bacteriophages
Bacteriophages are viruses with a highly efficient ability for compressing and wrapping DNA to form a compact particle. The phage genome encodes only a few to hundreds of genes and is considered a useful model to study DNA replication[7]. The discovery of bacteriophages first took place in 1915 by Fredrick Twort[8]. In 1917 D’Herelle discovered the potential ability of phages to kill the bacteria they infected via the lytic phase. He suggested that phages could be used as antimicrobial agents in a therapeutic application if they were targeted to bacteria that caused human diseases [8]. After the discovery of penicillin in the 1940s, the application of phages in therapy for human infectious disease was abandoned. Concurrent studies categorized phages as obligate intracellular parasites, and research on-phages provided fundamental discoveries of biological systems such as the identification of nucleic acids as the genetic material. During these studies the nature, chemical composition and life cycles of phages were identified. It became clear that the phage was composed of a protein envelope encapsulating nucleic acids. Thanks to the efforts of Martha Chase and Al Hershey who discovered that the nucleic acids entered the bacterial cells during phage infection while the proteins did not, this led to the realization that hereditary information was solely transmitted by DNA molecules[9]. The properties of phages vary depending on the regulatory mechanism of host cells, their genetic content and physiological variants[10]. Based on their properties, phages have been classified into different groups of which the double stranded (ds) DNA tailed phages are the most common type. It has been calculated that there are no less than 1031 different phages that exist on the planet Earth, making them the most diverse grouping of biological entities in existence. The new classification system called the “Phage Proteomic Tree”, has classified phages in a sequence-based taxonomic system according to their genome in which average distance between pairs of phages is determined and phages are grouped in the context of all other phages[11]. Improvements in electron microscopy have provided more information about the phage morphology, and their interactions with bacterial cells[12]. Studies in this area have found different morphologies for the phage structure. Each type of phage has a particular shape and shows different mechanisms of supramolecular assembly. Some basic morphological and other properties of selected phages are shown in Table 1.
Table 1.
Some basic morphological and other properties of selected phages
| Selected phage | Morphology | Type of genetic material | Size | Host | M.W of intact phage |
|---|---|---|---|---|---|
| T4 |

|
ds DNA | 200 nm | E. coli | 110 kD |
| T5 |

|
ds DNA | 160 nm | E. coli | 75 kD |
| T7 |

|
ds DNA | 46 nm | E. coli | 38 kD |
| ØX174 |
|
Circular ssDNA | 31 nm | E. coli | 1.7 kD |
| F2 |
|
ssRNA | E. coli | 1.2 kD | |
| M13 |

|
Circular/ss DNA | 890 nm | E. coli | 2.4 kD |
| MS2 |
|
ssRNA | 27 nm | E. coli | 3.7 kD |
A variety of phages exist with a different range of hosts and mechanisms of production. Phages are divided into three main classes based on their production and generation: lytic phages such as T4; temperate phages like Lambda; and lysogenic phages such as M13. M13 is a filamentous phage that converts the host cell into a generation factory without lytic disruption. This phage can cross the bacterial membrane to escape by leaking out without disruption of the cell wall and membrane. By contrast in lytic phage infections, bacterial cells are lysed and disrupted by replication of the virions inside the bacterial cell and the infection is rapidly transferred to new hosts[13]. The third type, temperate phages can choose between a lytic and lysogenic pathway of development. In temperate phages, the phage genes become integrated into the bacterial genome. The phages replicate through the natural bacterial cell cycles and are transmitted into the next generation of bacteria, although in stressful conditions such as UV irradiation, the phages become activated and the lytic phase begins.
The genome sequence of phages that have been integrated into bacteria is called the “prophage” and 2/3rd of the genomes of “low CG” bacteria contain these sequences. This phenomenon can alter the properties of the host bacterial cells and can alter their pathogenicity and virulence by changing their physiology and pathology[14]. The actual mechanism of lysogeny is due to the presence of repressor genes that are encoded in temperate phages. These genes generate repressor proteins that repress various bacterial operons. They bind to the phage genomes and interrupt the formation of intact phage particles, but the prophage sequence is translated in bacteria and leads to expression of new properties in the host cells. This phenomenon, which is called “lysogenic conversion”, is important in medicine. Some bacteria such as Corynebacterium diphtheriae, streptococci and Clostridium botulinum are not able to produce their toxins without prophage sequences.
Cell penetration of the phage into the bacterial cell occurs through the recognition and attachment of phages to specific receptors that are expressed on the surface of the host cell, and the surface properties of the host membrane also affect this phenomenon[15]. The tolerance of the phage surface to be chemically altered provides opportunity for insertion of foreign peptides such as RGD (Arg-Gly-Asp) or DGEA (Asp-Gly-Glu-Ala). These moieties can have therapeutic effects in a targeted delivery approach. Studies of Young’s group showed that not only could the surface of phages be modified, but also their genetic material can be manipulated to enhance their ability to be used in drug and gene delivery applications[16]. Genes and drugs that need to be delivered by phages are usually bound to coat proteins of engineered bacteriophage species without significant effects on the phage structure. These bound moieties could involve targeting ligands such as antibodies against specific receptors or targeting peptides, or could include therapeutic moieties such as different types of growth factors, drugs or metabolites[17]. Another strategy to enhance the targeting of bacteriophages to mammalian cells is their hybridization with synthetic polymers (e.g. cationic polymers). In such a strategy, cytotoxic genes could be transferred into different cancer cells in a safe and controlled manner, and the major limitation of phages for mammalian cell targeting could be overcome[18]. In several in vivo studies, it has been shown that phages could be applied for specific targeted drug delivery (that is more effective than un-targeted drug) in both the killing of pathogenic bacteria and in the destruction of cultured tumor cells[1, 3, 9].
The genetic material of phages can also play a carrier role in the delivery of small interfering RNAs and other nucleic acid-based therapeutics. The DNA packaging systems of bacteriophages have been identified as a special type of nano-vehicle for gene delivery. Previously it was shown that DNA packaging motor of Phi29 could be an efficient machine for packaging viral ds-DNA which is used in DNA sequencing technology. Phi29 RNA has high a tendency to form dimers, trimers and hexamers[19]. By using this property of Phi 29 bacteriophage RNA, Hox at al. used molecular engineering to produce a nanocage. This biological RNA used for packaging consisted of three main branches of helical domains and a single stranded internal loop that could efficiently interact with other small RNA molecules[20]. Later Lee et al. conjugated folic acid to a pRNA-3-way junction derived from Phi29 to generate targeted RNA nanoparticles for gene silencing in glioblastoma cancer cells (that have high expression of folate receptors) in the brain[21]. Additionally, it has been reported that the packaging RNA (pRNA) of bacteriophage phi29 could self-assemble into multivalent nanoparticles to deliver siRNA to silence metallothionein-IIa (MT-IIA) and survin mRNA, and could trigger the expression of interferon alpha and beta and thereby enhance inhibition of ovarian cancer cells[22].
3. Phage display
Phage display techniques have been widely used as an efficient tool for target discovery in pharmacology. This technique, used for the first time in 1985[23], is based on the insertion of DNA coding for heterologous peptide sequences into the phage genome leading to expression of exogenous proteins joined to coat proteins on the surface of the phage (Figure 1) . Indeed, phage display is used to produce a physical association between phenotype and genotype. The first step in phage display technology is the construction of a phage display library. This library is obtained by generating random oligonucleotide sequences through recombinant DNA technology. Then the randomized DNA sequences are introduced into specific sites of the phage DNA, which are responsible for encoding the surface proteins and the process leads to expression of varying fusion peptides on the surface of the phage nanoparticles. Although the entire library contains a huge number of peptides, each phage expresses only one specific peptide according to the incorporated sequence. Next, the relevant surface ligands [expressed peptides] are selected by affinity-based screening of the phage display libraries. Those phages that exhibit tight binding with the target cells are isolated in cell culture after a suitable period of time, and the rest of the phages are washed out. The collected tight-binding phages are then further amplified in host bacteria, and the associated DNA sequences are determined by gene sequencing techniques. The result of this process is a newly discovered high-affinity ligand that binds to the desired cells and can be used to improve targeted delivery systems[24, 25].
Figure 1.
Phage display and phage-based targeted delivery technology; A: Insertion of ligand-encoding hybrid oligonucleotides into a gene encoding a capsid (e.g. gene III) and consequent display of the ligand on the surface of phage and B: targeted delivery of the loaded genes or therapeutic compounds into the desired cells.
The peptides selected by phage display technology usually require further modification. A common way to modify the peptide is by extending the length by adding specific amino acids such as GGGSAK-amide to the C-terminal end of hydrophobic peptides to increase the solubility, ease the purification by adding biotin, and allow the detection of the peptides by adding a fluorescent dye. Moreover peptides with the ability to bind to more than one target domain can be linked together to enhance the specificity and binding affinity of the obtain molecules[26]. Modifications to the peptides also can be carried out by using enzymes, when the displayed peptide contains a domain subject to enzymatic conversion[27]. Among different types of phage, the filamentous phages particularly M13, are the main platform used in display libraries[28]. T4, T7 and lambda phages have also been used to a lesser extent as alternative vectors to construct display libraries. Lambda phage with its high capacity for gene insertion is able to express more than one peptide located in the head and the tail, either individually or simultaneously[29]. Filamentous phages possess critical advantages compared to other viruses. Their length is variable according to the size of the packaged DNA, because the vPIII subunit can be altered to accommodate different DNA sequences, while their diameter remains constant leading to a more flexible structure. The special anisotropic shape of M13, and its long rod-like structure shows liquid-crystal like properties, and due to their high length to diameter (aspect) ratio, their penetration into the targeted cells is improved due to a higher number of ligand-receptor interactions. In contrast to spherical-shaped phages, filamentous phages have a tendency to migrate toward the blood-vessel walls when injected in vivo, and therefore have a better chance to interact with their receptors[30].
Although the basic principles of phage display technology were introduced around 30 years ago, new applications and developments of this technology are constantly emerging. This technology can not only facilitate the recognition of de novo ligands, but can enable the production of recombinant peptide libraries, that are unobtainable by traditional techniques such as using the heavy chain binding protein of antibodies[31]. Furthermore, phage display technology enhances in vivo selection of different peptide targets as well allows their real time detection[32]. For instance, Chen et al.[33] discovered that peptide-431 was a specific ligand for the insulin-like growth factor 2 receptor in hepatic stellate cells to allow targeted delivery of antifibrotic agents into the liver. The combination of phage display with other techniques has enabled the production of difficult hydrophobic proteins such as HIV-1 nef (negative factor). The integration of sequences of unusual amino acids into the phage genome, combined with covalent binding of synthetic molecules to the wild-type phage coat produced a new hybrid system for the identification of ligands with unique properties[34].
On the other hand, in vivo applications of phage display have led to enhanced targeted delivery of therapeutic and diagnostic nanocarriers to target tissues. Chen’s group[35] used the synthetic peptide ACSSSPSKHCG to improve the transdermal delivery of proteins such as human growth hormone and insulin. Hofmeister et al.[36]found that the peptide GSPREYTSYMPH (PREY) could guide the nanocarriers to bind to atherosclerotic vasculature.
In phage display technology, ligands such as antibodies, specific peptides and receptor-selective proteins can be loaded onto the surface of the engineered phages[37]. Phage antibody display can be used to manipulate the affinity of antibodies in vivo. Enzyme display, improvements in vaccines, and production of fusion phages for gene therapy are other applications of phage display technology[26].
4. Bacteriophages as nanocarriers
Phages can be used as designed nanocarriers for the targeted delivery of both therapeutic agents and diagnostic reporter molecules, and represents a new aspect of nanotechnology in drug delivery systems. Targeted ligand displays on the surface of the phage provide versatility and high specificity in this regard. Large amounts of drugs can be conjugated into the phage either by genetic manipulation or by chemical interaction. Specific recognition of targeted cells can then lead to controlled release of the payloads. Phage-based nanocarriers can also be efficiently used for eradication of both pathogenic microorganisms and also tumor cells, when compared to unbound free drug, particularly in the case of poorly water-soluble drugs. In this section we cover the major reports concerning phages that have been used as nanocarriers in the delivery of drugs and genes.
4.1 Filamentous phage based nanocarriers
Filamentous phages are defined by their special structure in which a long rod-shaped protein coat encircles single-stranded DNA. These phages are able to infect their host cells without killing them, and become integrated into their hosts that are mostly Gram-negative bacteria such as species of Escherichia and Pseudomonas. The filamentous phages that infect Escherichiacoli are the most fertile bacteriophages in the nature and are called “Ff phages” (this group is sub-divided into f1, M13 and fd phages)[38]. The major coat protein in these phages is called pVIII that consists of just 50 amino acids. pVIII is common among all types of filamentous phages, although its configuration might be slightly different. Furthermore, additional proteins such as pIII, pVI, pVII and pIX play co-constructive roles in the construction of the phage coat. The size of these phages is determined by the length of the DNA core, and can be altered by insertion and deletion of nucleic acids. Filamentous phages follow two main life cycles; some of them produce very large numbers of individual phages, while others integrate into the chromosome and only produce 1 phage per 10–100 cells, and are able to switch between one cycle to another under poorly understood conditions. Insertion of the phages into the bacterial host takes place via interaction with specific receptors. The primary receptors are structures called pili on the surface of the bacterial cell wall that help the phage to come very close to the bacterial surface, while the secondary receptors are located in the inner membrane of Gram-negative bacteria. As a result of this interaction the phages gain entry into the bacterial cytoplasm. By contrast, the entry of the phage nanocarrier into experimental eukaryotic cells takes place by the process of endocytosis[39]. After entry of the phage genome into the host cells, bacterial enzymes convert the single stranded DNA genome to a double stranded replicative form that serves as a template for expression of the phage genes. The genome of the episomal filamentous phages is significantly shorter than that of chromosomally integrated phages; they lack required sequences for host genome integration and expression of regulatory genes. The genome of episomal phages replicates in the bacterial cytoplasm, whereas the genome of chromosomally integrated lysogenic phages is transferred into the host chromosome. The assembly process of the newly produced phages is performed by coating the phage DNA with the pVIII and pIII proteins[40].
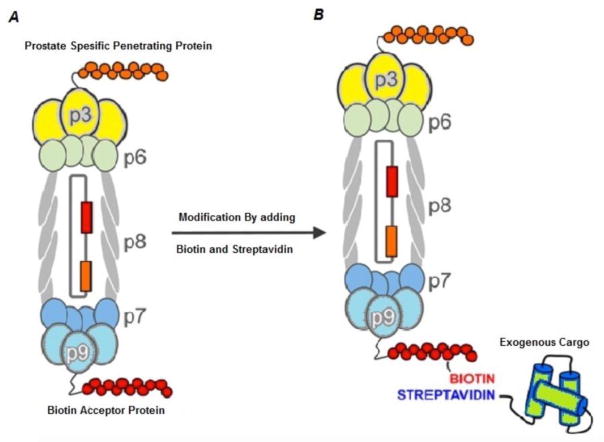
Filamentous phages are highly immunogenic; and this is a property that is taken advantage of in applications in vaccination. This immunogenic property is probably due to the repeated structure of the coat proteins, and the relatively large size of filamentous phages[41]. Fusion peptides with pVIII have higher immunogenicity compared to fusion proteins with pIII, which could be due to the higher number of pVII protein molecules in the surface coat of the phage[42]. Experiments have shown that displaying the targeted epitope fused to pVIII can trigger high titers of antibody[43]. Pioneering studies in this regard have reported the production of vaccines against HIV-1 and malaria parasites, in which epitopes of gp120 and gp 41 of HIV, and epitopes of circumsporozoite proteins of malaria were coated onto the surface of phages[44, 45]. Therapeutic antibodies against other disorders such as cancer and Alzheimer’s disease have also been generated by these phages[46–48]. By utilizing filamentous phages, an anti-IgE antibody was generated that could reduce the allergic immune-response by reducing IgE, and could be a promising method to control allergic reactions[49]. In addition, filamentous phages can be used to present antigenic epitopes to T-cells to produce specific cytotoxic T-cells. Phages displaying an HIV epitope[50], and an epitope derived from the hepatitis B virus[51] could protect experimental animals against these infectious diseases. Sartorius et al.[52]reported that if two different epitopes were simultaneously displayed on both the pVIII and pIII proteins, the immune response by cytotoxic T-cells would be significantly stronger. In their study the ovalbumin CTL epitope was displayed fused to pVIII protein and a dendritic cell marker (DEC-205) was expressed on the pIII protein. This T-cell dual-stimulating phage was used against B16-OVA expressing tumors in mice. Paul and Marks in 1999, first showed targeted delivery of the GFP gene-expression cassette into breast cancer cells, when Fd phages were selected to recognize HER2-expressing cancer cells, and the GFP gene was engineered into the phage[53]. Ghosh’s group[54]displayed prostate cancer cell targeting ligands on the pIII protein of M13 phage, and then loaded doxorubicin onto the pVIII protein to generate a targeted treatment for prostate cancer in vitro. Furthermore, Deporter at al.[55], used genetic manipulation of M13 phage to express a cell-penetrating peptide on the pIII protein and a biotin acceptor peptide (BAP) on the pIX protein. The BAP provided a biotin binding site in which a biotinylated exogenous therapeutic cargo could be attached, and taken up into prostate cancer cells (Figure 2). Jin et al.[56]displayed a collagen mimetic peptide and a streptavidin binding peptide (which was used to attach a streptavidin-conjugated fluorescent dye) to produce a phage-based nanoconstruct that could lead to degradation of collagen in A549 human lung adenocarcinoma cells with an additional diagnostic capability. Li’s group[57] showed that genetic manipulation was not compulsory for tumor targeting of the phage nanoparticles. They suggested that chemical modification of tyrosine residues of pVIII would create a chemical binding site on the surface of the phage, and subsequently two different moieties could be attached to the phage surface; one (such as folic acid) for tumor targeting, and another as a diagnostic agent such as a fluorescent dye. Since previous studies have shown the role of opsonization in the control of tumor growth by stimulating the immune response[58], phage display constructs using tumor specific antigens or Fab antibody fragments would be a potential method to disrupt tumor growth mainly by stimulation and recruitment of immune cells to the tumor.
Figure 2.
Genetic manipulation of M13 bacteriophage for prostate tumor targeting; A) the phage is genetically manipulated to express prostate-specific penetrating peptide on PIII protein and biotin acceptor protein (BAP) on PIX protein. B) Further modification of BAP by biotin-streptavidin provides a binding site for conjugation of exogenous cargo to be transferred into the targeted cells. Reprinted with permission from[55]. Copyright (2016) American Chemical Society.
One application of phages is using phage-based nanocariers for the delivery of antibiotics. In this regard, Vaks’ group[59] displayed an antibody on the pIII minor coating protein of f1 phage and chemically conjugated that with chloramphenicol as a selected antibiotic. The conjugation took place between the amine group provided by neomycin-chloramphenicol and the carboxyl group of the phage surface. This type of conjugation process could change the properties of the phage; the immunogenicity of the phages diminished and their infectivity and toxicity was significantly reduced, whereas the half-life of the conjugated phage increased in the bloodstream. On the other hand, this experiment confirmed that the toxicity and side effects of hazardous drugs could be controlled by conjugation to the filamentous phages. After injection of 1011 antibiotic-loaded phage particles per dose, no toxic side effects of either the drug or the phage were observed. Displaying tissue targeting peptides on the phage coat surface, is another strategy for delivery of therapeutic compounds into a specific cell type. Zahid at al[60] showed that modifying the M13 surface proteins with the APWHLSSQYSRT peptide sequence led to attachment of the phages cardiac myoblasts. More interestingly filamentous phages also possess the ability to penetrate the blood-brain barrier and this property could make them suitable to be used in treatment of Alzheimer’s disease[61]. By using this property, Rakover et al[62] applied the myelin glycoprotein as an immune-recognized epitope for the treatment of multiple sclerosis by inactivation of demyelinating antibodies in the central nervous system. These studies show promise for the ability of the phages to carry large amounts of therapeutic agents as well as their capability to access otherwise unavailable tissue. Filamentous phage nanocarriers can also be used in bio-detection of a particular antigen in solution. This property is possible due to the separation between the location of the pIII protein at one pole of the phage structure, and a large number (thousands) of pVIII proteins in the lateral parts of the phage surface. The pIII protein can be used to display a targeting sequence, whereas the pVIII protein can be modified with an external diagnostic reporter moiety. Lee et al[63] showed that pVIII modification with plasmon-resonant gold nanoparticles could generate a rapid and sensitive sensor for the targeted antigen that was recognized by peptides displayed on the pIII protein.
Despite the large capacity of the filamentous phages to display different peptides, the lack of a wide range of chemically distinct amino acids on their surface has limited their modification. To solve this problem, Carrico et al[64] suggested a new method to convert the pVIII N-terminal amino group into a ketone group which is then converted to alkoxyamine groups. They showed that fluorophores and polyethyleneglycol could be attached to the phage coat, while the pIII displayed peptides could direct the phage to recognize breast cancer cells, while the properties of phages remained unchanged. Although application of filamentous phages in delivery of the therapeutic materials is interesting, they may still produce severe adverse reactions in clinical treatment due to two main limitations; firstly, since they are assembled in a bacterial host, they may contain variable levels of lipopolysaccharide (LPS) that must be reduced. To this end, a more effective purification system is required, and the amount of LPS could be diminished in the final product by using size exclusion columns combined with polymyxin B binding chromatography, and finally treating with a detergent like Triton. Secondly, the use of phages might be restricted in high-risk patients owing to their immune status and the heterogeneity of the particles[28].
4.2 MS2 phages as nanocarriers
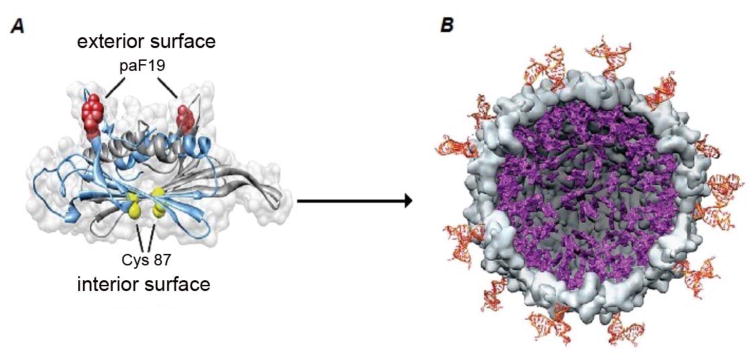
MS2 bacteriophage was discovered in 1961 by Alvin John Clark[65]. This phage contains a single-stranded RNA in its genome, and a symmetric icosahedral capsid structure formed from 180 protein-subunits that surrounds its genome[66]. The MS2 genome, with 3569 nucleic acids, encodes only four proteins and is known as one of the shortest genome sequences in nature[67]. The average size of this phage is around 27 nm, and the DNA-free protein envelope can be generated by recombinant technology. The presence of 32 pores each with a radius of 1.8 nm on its surface, provide a good platform for modification[66]. Some interesting properties of MS2 phages suggest they could be good candidates in drug delivery systems. They can be easily produced, their capsid is able to be genetically and chemically modified, and after removing their genome they are able to encapsulate other modified cargos such as RNA[68]. Since the purification of phage capsid is relatively easy though an E. coli expression system, researchers have used its functionalized capsid as virus-like particles (VLP). The empty envelope of this phage is resistant to pH fluctuations in the range of 3–10, and to severe environmental conditions including temperature. To enhance the modification of this phage coat-protein, a cysteine residue(C87) is added to the sequence of each monomer in the interior portion of the coat[69]. The exterior surface of the phage can be used to display an unnatural amino acid, p-aminophenylalanine(paF). This amino acid provides an aniline group to be conjugated with an N,N-dialkyl-phenylenediamine derivative[70]. This genetic manipulation, chemical conjugation, and the removal of DNA creates an empty nano-vehicle to be loaded with different drugs and fluorescent dyes.
Ashley et al.[71]introduced these phages as an efficient cargo vehicle for delivery of diagnostic agents, therapeutic compounds and RNA molecules. Stephanopoulos et al.[72]by dual modification of the MS2 coat protein, generated a therapeutic nanocarrier targeted against Jurkat leukemic T-cells. They conjugated a cell specific DNA sequence on the exterior surface of the phage envelope and attached porphyrins on the inner surface. In the targeted cells, after cargo delivery, illumination of the porphyrins generated a large volume of reactive oxygen radicals by the photodynamic effect, leading to destruction of the cells only 20 min after the treatment. The basic structure of this carrier is shown in Figure 3. In another study, Glasgow et al.[73]proposed two methods for encapsulation of a protein inside the MS2 capsid in the presence of osmolyte molecules (charged polymers or peptides). In one application, any protein larger than 1.8 nm could be modified with negatively-charged polymers and the phage capsid could be reassembled around it, while in the second method, negatively-charged tagging linkers could be genetically encoded in the phage capsid, and the protein encapsulated into the phage.
Figure 3.
Basic structure of MS2 nanocarriers: A) the external surface of the MS2 coat displays the modified aminoacid p-aminophenyllalamine(paF19) and the protein on the internal phage surface displays cysteine residues (Cys 87). B) After removing the phage genome, the Cys 87 is modified with a maleimide-conjugated porphyrin, and a Jurkat leukemia T cell-specific DNA aptamer is attached to the paF19. After the carrier enters into the targeted cell and is illuminated, reactive oxygen species are produced and the cancerous cells are disrupted. (Reprinted from ref.[72] Copyright (2016) American Chemical Society).
Owing to high instability of free RNA, its sensitivity to RNase enzymes, and the difficulty of using dendritic cell in RNA transduction, MS2 VLP could be a promising nanocarrier for delivery of the RNA. Wei’s group[74] used MS2 VLP for transferring antisense RNA into Huh-7 cells containing HCV reporter systems. They applied a heterobiofunctional cross-linker consisting of sulfosuccinidyl-4-(p-maleidophenyl)-butyrate (sulfo-SMPB) for the attachment of tat peptide as a specific targeting moiety. Pan et al[75] found that the attachment of the RNA to the MS2 VLP, did not change the RNA topological structure. They expressed a micro-RNA146 on the surface of VLP and allowed it to interact with the anti-nuclear antibodies (ANA) that cause the lupus autoimmune disorder. The results showed that this carrier efficiently neutralized the antibody and could be a promising therapeutic vehicle to control this disease, and could also be used against allergies and other antibody-mediated disorders. Gene therapy with a reduced risk of permanent integration into the nucleus, could be achieved by direct transfer of mRNA into the cytoplasm of desired cells. Production of MS2-coat-retrovirus chimeras is a recent strategy for direct transfer of mRNA inducing protein expression. Prel et al[76] recombined the MS2-coat protein with lentiviral RNA derived from HIV-1. This vehicle contained more than two different RNA sequences with the ability of gene transduction. They transferred mRNA coding an osteogenic transcription factor into bone marrow-derived mesenchymal stem cells and shifted their transcription program.
The capsid of this phage also has been used for tissue-specific delivery of loaded cargo by adding targeting peptides such as VOWMEPAYQRFLGGG that recognizes breast cancer and neuroblastoma cells[77]. Further recent studies have suggested MS2 VLP as an efficient carrier for delivery of mRNA as a vaccine carrier into prostate cancer cells[78]. MS2 empty capsids have some limitation of use in vivo conditions due to their high number of surface modification sites. These vacant sites might conjugate with other biologically active molecules to form a corona (especially a protein corona) when applied in vivo. To prevent this, Farkas’ group[79] applied dual surface-modification of the capsid; they first modified the phage surface with desired signaling peptides, and then chemically capped all the remaining free sites with polyethylene glycol and Cu. By this dual modification the MS2 capsid nanocarrier could more efficiently deliver the cargo to the targeted cells, take up by the spleen was reduced, and no immune response was observed. Although we mentioned that formation of a protein corona could change the fate of phage-based nanocarriers to produce deleterious effects, however it is possible that in the future a controlled formation of a corona with desired biological activity could be a new strategy. A surface-engineered phage-based nanocarrier possessing a peptide with a high affinity to apolipoproteins was developed by a phage display strategy, that could recruit a lipoprotein-based protein corona as an agent for crossing the blood-brain barrier(BBB). It was shown that nanoparticles possessing apolipoproteins on their surface survived in the blood circulation and could cross the BBB[80].
4.3 Lambda bacteriophage-based nanocarriers
Lambda phage is a member of coliphage family that was discovered in 1950 by Esther Lederberg, and possesses the ability to undergo both lysogenic and lytic life cycles[81]. The structure of lambda phage consists of three main parts; the head or capsid, the tail and tail nanofibers, and a dsDNA sequence as the genome encapsulated by the capsid. The lambda genome contains 48490 bp ds-DNA and two 12 bp sticky ends at both 5′ terminals[82]. The host infection of the phage takes place by the specific interaction of the tail J protein with the LamB protein on the E. coli bacteria, and is followed by genome injection from the tail end into the host cytoplasm[83]. The lambda phage has been widely used in molecular genetics either as a “shuffling cloning vector” or as an efficient vehicle for specific cargo delivery. This phage also is ideal for phage display application due to ability for vaccine presentation and its capacity for gene delivery. Two main structural proteins of this phage are called gpD in the capsid and gpV in the tail region. These proteins are produced in the cytoplasm and form the procapsid, then the phage genome is encapsulated into them, and finally the completed is released into the environment after host lysis[84–86]. The capacity of the lambda phage to carry large nucleic acid sequences, and its unique formation of the gpD capsid protein that can be modified with a variety of ligands, offers several applications for this phage. The gpD protein of the lambda phage is incorporated into the icosahedral phage head with 405–420 copies on a mature phage head as strongly protruding thimble-shaped trivalent spikes. These are promising for multiple tag expression by being attached to different chemical or biological ligands to improve the specificity of cellular uptake, reduce the unwanted immune responses and improve the pharmacokinetics of lambda nanoparticles[87]. Chang at al.[88]introduced the lambda capsid as a designed nanoparticle, and incorporated a variety of synthetic moieties and genetically integrated peptides, that were simultaneously displayed on the phage surface.
The first application of lambda phage as a cloning vector took place in the 1980s when Okayama transduced lambda packaged with cDNA into mammalian cells[89]. Gene therapy by lambda phage began with the work of Merril et al[90] in 1971 in which the gene for galactose transferase was delivered into human fibroblast cells isolated from a child with a deficiency in the galactose transferase enzyme. Thereafter, Lankes et al.[91]displayed an integrin-binding peptide on the lambda phage and transferred genes into dendritic cells in vivo to express luciferase. This experiment expanded the application of lambda phage by providing real time visualization of gene delivery. Additionally, using the integrin-binding peptide led to internalization of the phage through pinocytosis, and prevented fusion of the phage with the mammalian cell membrane[92]. Sapinoro et al. [93] suggested an antibody-dependent enhancement (ADE) method to increase the transformation of phage-encapsulated genetic material into the targeted cells that were recognized by the antibody. Although this method increased gene expression, it would require pre-immunization and also needs the presence of the specific receptor on the targeted cells.
Further studies that led to the development of Cre-loxP a method for site-specific recombination, that could be used to facilitate peptide expression on the surface of gpD[94]. The high potential of lambda phage to allow multiple numbers of the same antigen on the surface makes it a more efficient candidate for vaccine production compared to low antigen-displaying phage[94]. Clark and Marck[95] demonstrated the immune stimulation ability of lambda phage delivered via the oral administration route. They cloned the V-antigen of Yersinia pestis into the phage genome, and reported a higher immune response after absorption in the oral mucosa. Ghaemi et al[96] encapsulated the gene for GFP and the E7 gene of human papilloma virus (HPV) into the lambda-ZAP-CMV vector to enhance anti-tumor immune response against HPV-expressing cancers. Dual modification of lambda phage for targeted delivery of genes of interest to the immune system is a promising method for combating various diseases. The surface of the lambda phage displays specific ligands and the capsid encapsulates the selected gene sequence that is protected from degradation by enzymes[97]. Thoma et al[98] also confirmed that this strategy could produce higher immune response in the absence of any adjuvant and at physiological pH. Recently, Pavoni et al[29]introduced a new strategy for modification of lambda phage. They modified gpD with a targeting moiety and gpV with GFP, while the capsid could still carry a therapeutic agent or a gene such as alkaline phosphatase.
4.4 T bacteriophage-based nano carrier
The T family of the E. coli infecting bacteriophages has been divided into seven types based on their morphological and genetic properties. Both T-even and T-odd phages possess lytic life cycles and incorporate ds-DNA. These members are Type 1 (T1), Type 2 (T2), Type 3 (T3) and so on, in which T2, T4 and T6 have similar structures and T3 and T7 are shorter in size. T-even phages have been used in studies in fundamental biology and (as mentioned above) T2 phage was used by Hershey and Chase in 1952, to show that genetic materials were the only requirement for phage replication[99]. Among all types of T phages, T4 and T4 like phages are more useful as a cargo delivery vehicle due to their comparatively larger size with a high capacity for gene integration, the presence of 105 functional groups in capsid proteins on the surface, that can be used for modification, or for efficient peptide display[100]. The most useful member of the T-odd phages for delivery systems, is T7 that was discovered in 1945. T7 possesses an icosahedral head with 55 nm diameter and a tail 19 nm long, with two main capsid proteins including 10A and 10B that encapsulate a 40 kbp genome sequence coding for 55 proteins. In optimal conditions, each T7 phage reproduces within 11 minutes and produces about 1013 progenitors in 1 hour of replication cycle[101]. The T7 promoter sequence has been widely used in molecular biology studies owing to its high affinity for the T7 RNA polymerase and resulting good expression levels[102].
The high replication rate of T7 means it is an efficient agent to control bacterial infections. Moreover, the titer of phage increases only in the presence of its host bacterial cells, and therefore provides a self-limiting system to control infections. Lu’s group[103] modified the T7 genome with a biofilm-degrading enzyme to digest the exopolysaccharide components of bacterial biofilm. The lysozyme enzyme of T4 phage and the lytic transglycosylase enzymes of T7 also can destroy the peptidoglycan in bacterial cell walls, and have the potential to be used as antimicrobial agents. Despite the above-mentioned advantages, phages might also have some limitation when applied in clinical situations. Phage attack may change the normal balance of the population of commensal bacteria, and in the case of the high degrees of bacterial disruption, toxic compounds derived from bacteria (such as LPS and enterotoxin) can be released into the body[104]. Vascular endothelial growth factor (VEGF) is one of the basic compounds implicated in cancer progression, and its expression on the surface of T7 phage could break immunologic tolerance to this protein, and thereby reduce the invasion of Lewis lung cancer cells[105]. One of the critical challenges in drug delivery systems is targeting of therapeutic cargos into the liver. Although these therapeutics can be administered by injection, their recognition by Kupffer cells (that are also found in the liver) might stimulate an undesirable immune response. The labeling of the tail fiber protein (P17) of T7 phage with the asialoglycoprotein receptor (ASGPr) could guarantee the delivery of the cargo into hepatocytes. Wong et al[106]. reported that adding a targeting ligand composed of 33 amino acids to P17 could transfer cargos including siRNA, liposomes, and DNA polyplexes into hepatocytes.
T4 phage is a comparatively large particle, 90 nm wide and 200 nm long, with an icosahedral head that encapsulates the genome, a hollow structural tail to transfer the genome, and tail nanofibers for specific recognition of receptors on the host surface. The genome of T4 phage encodes 289 proteins coded by 169 kbp ds DNA[107, 108]. The T4 capsid proteins are divided into two main categories, including major essential capsid proteins, gp23 which forms the hexagonal capsid lattice gp24 which forms pentamers at 11 of the 12 vertices, and gp20, which forms the unique dodecameric portal vertex through which DNA enters during packaging. There is also a non-essential capsid protein, called highly immunogenic outer capsid protein (Hoc) and a small outer capsid protein (Soc)[109, 110]. The interaction of T4 phage with the mammalian cell surface takes place as a result of interaction between the integrin-binding site (KGD) of the gp 24 and integrins on the cell surface; therefore the tail of the phage plays no role and can be removed. Moreover deletion of Hoc improves gp24 exposure, and increases uptake by mammalian cells[107]. Furthermore, Wu et al[111]proposed that displaying two antigenic peptides on SOC and HOC could elevate the immune responses against a particular pathogen. T4 phages do not contain normal cytosine, but replace it with hydroxymethyl-cytosine to protect the phage genome from degradation by endonucleases, and further modification by attachment of glycosides occurs[112].
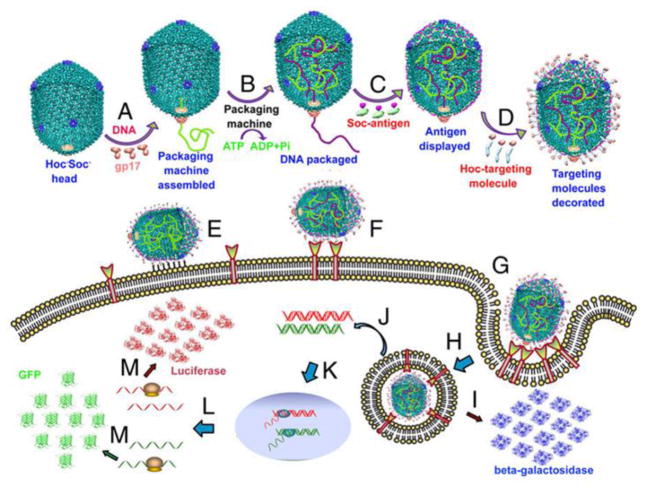
The DNA packaging mechanism of the T4 phage is very rapid and powerful (up to ~2000 bp/Sec) and can display more than 1000 molecules on the capsid surface. Tao et al. suggested the DNA packaging mechanism of T4 phage would be a good strategy for gene delivery for combating cancer and infections such as HIV and TB. The existence of two envelope proteins on the surface of the T4 head also allows its utilization in vaccine applications[113]. The basic principles of this application are depicted in Figure 4. The enzymes of T4 phage such as polynucleotide kinase and ligase are used in the assembly of other phage carriers such as those based on lambda phage[114, 115].
Figure 4.
Mechanism of gene delivery by T4 capsid-based nanoparticles. A) Initiation of DNA packaging by aggregation of gp17; B) DNA encapsulation into the head by ATP utilization; C) Adding the Soc proteins; D) Fusing the Hoc- targeting peptide; E) Nonspecific binding of the nanoparticle to the cell surface; F) Receptor mediated binding to the cell surface; G) As a result of cell binding internalization takes place; (H and I) Either the transferred gene produces the displayed peptide such as beta-galactosidase; (K, L,M) Or DNA is transformed in the nucleus and encodes the desired protein such as luciferase (Reprinted from ref.[113] with permission from PNAS).
Ren et al.[116] found that displaying the appropriate peptide on the T4 capsid could stimulate an immune response. They, previously had shown that the separate respective genetic modification of SOC and HOC with the polyprotein and proteinase of foot-and-mouth disease virus (FMDV) produced 100% immunogenicity compared to the wild type of T4[117]. Next, they recombined the Fms-like tyrosine kinase 4 gene into the T4 phage and stimulated an immune response against Lewis lung carcinoma, although this intervention was not actually able to inhibit the tumor growth, but it could prevent its metastasis. They went on to display murine VEGF receptor 2 (mVEGFR2) and generated T4-mVEGFR2 as a new vaccine vector, and reported that this specific immunotherapy completely inhibited tumor growth and increased the survival time of the experimental animals[118]. Further studies have shown that application of T4 phage could trigger an immune response against infectious bacteria. Tao et al[113]. genetically modified the T4 capsid with the F1-V protein of Y. pestis and targeted it to dendritic cells to induce antibody-dependent immune protection. Production of synthetic proteins with desired motifs could be a promising strategy for drug delivery systems, but the presence of the β-helical configuration has limited this strategy. Yokoi et al. suggested[119] that the baseplate protein (gp5) assembly system of T4 could be a suitable method to solve this obstacle. They determined that the C-terminus of the triple β-helix gp5 ((gp5C) 3) was the most rigid area of the baseplate and generated a nanotube-shaped structural template in which different classes of expressed protein could be aligned.
5. Hybrid bacteriophage based nanomaterials
Several organic and inorganic materials have been tested for construction of nanocarriers and many ligands have been introduced to target them towards particular cells and tissues[120–123]. A combination of organic structures with inorganic compounds can generate a hybrid vehicle which is another approach in drug delivery systems. Bacteriophages are a multivalent versatile platform that can be modified by combination with different moieties to form synthetic hybrid bacteriophage-based nanocarriers.
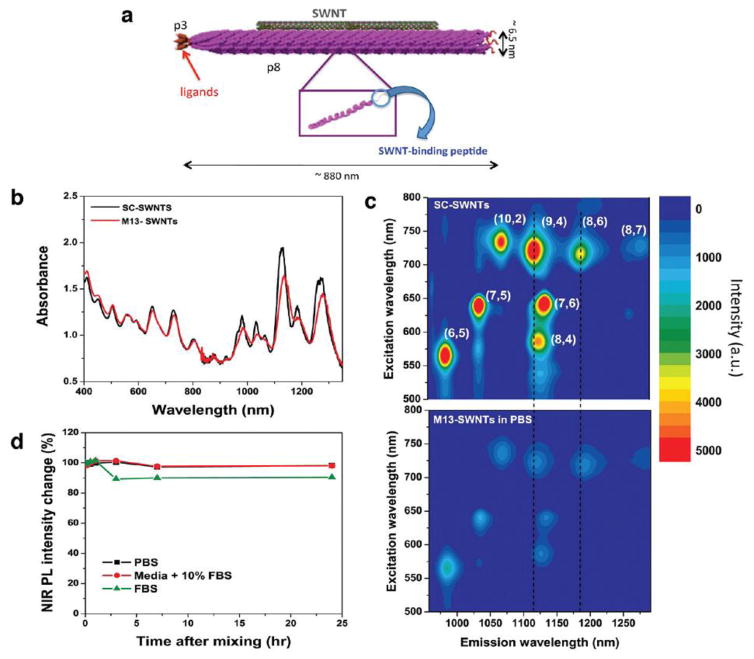
Although phages have a large capacity for integration of external genetic material, they are not able to broadly compete with mammalian viruses, due to the lack of inherent mechanisms for recognition and cargo delivery into mammalian cells[124]. Genes coding for the capsid proteins can be modified to ultimately lead to generation of hybrid peptide libraries. These hybrid phages have been widely used as an adjuvant-free vaccine to stimulate both cellular and humoral immunity[125, 126]. Recently, Wang et al[127] displayed the SLAQVKYTSASSI peptide as a targeting agent for Th cells on the surface of hybrid phages containing the recombinant protein called “secreted aspartic proteins “(Saps) to trigger efficient immune-protection against Candida albicans. The monodispersity of the phage coat protein would be an ideal advantage for hybrid preparation via the phage self-assembly process. Filamentous phages with their rod-shaped structure have been extensively studied as a liquid crystal model system; factors such as the ionic strength of the solution, the concentration of phages, and application of external force have encouraged the formation the liquid crystal phase[28, 128, 129].The introduction of inorganic materials onto the surface of phages displaying ligands, imparts novel properties to the phage carriers. Previous studies have shown that phages displaying peptides are able to recognize the specific topography of semiconductor substrates, and can also distinguish differences between amine groups and hydroxyl groups of a surface. Moreover, some particular amino acids can influence the interaction between the phage surface and inorganic molecules. For example ala-nine may play an important role in this reaction, and the aromatic ring of tryptophan residues can bind to graphite, and help the attachment of carbon nanotubes[130]. Ghosh et al.[131] conjugated fluorescent-labeled single wall carbon nanotubes (SWCNT) to the surface of engineered M13 phage that displayed peptides specific for prostate cancer to generate a deep-tissue diagnostic imaging agent. This M13-SWCNT phage had four times higher accumulations in the target tumor and provided clearer fluorescent images of the tumor site. This carrier is depicted in Figure 5.
Figure 5.
Basic properties of M13-SWCNT fluorescent-labeled nanocarrier; A) basic structure of the imaging vector: SWCNT is embedded in the pVIII structure and pIII is engineered to bind to prostate cancer; B) UV-absorption spectra at different wavelengths; C) comparison of PL excitation of M13-SWNTs in sodium cholate (SC) and phosphate-buffer saline (PBS); D) stability test of SWNTs in serum(Reprinted from ref.[54] Copyright (2016) American Chemical Society).
Conventional methods for constructing hybrid bio-inorganic nanostructures include directional freezing[132], polymer-templating[133], bio-templating[134], and electrospinning. He’s group[135] proposed a new strategy based on self-assembly of M13 phage by using hydroxyapatite (HAP) as an inorganic precursor and the phage as a model bio-fiber. First, they displayed negatively charged E8 peptide on the pVIII protein to produce an anionic nanofiber, and then allowed cationic ions, such as Ca2+, to electrostatically interact with the nanofiber, and finally added anionic compounds such as PO43− to produce nanofiber bundles that aggregated to form HAP-coated fibers. Gold nanoparticles (GNP) have been applied as a component in a phage assembly due to its unique plasmon resonance properties. A combination of positively charged GNPs with the highly negatively charged phage surface through electrostatic interaction generated a highly ordered scaffold for biomedical application[136].
Hajitou et al.[137] combined an adeno-associated virus (AAV) with M13 phage to generate a new hybrid virus that had a mammalian virus packaging system and a phage envelope to prevent tropism in resulting vector. This virus was genetically modified by displaying CDCRGDCFC (called RGD-4C) peptide to target cancer cells. Recently, Yata’s group[18] modified the M13 phage with the RGD4C-ligand (an integrin receptor) and encapsulated the luciferase gene. The whole vector was then covered by two cationic polymers, poly-D-lysine and DEAE-DEX. They reported that the positive charge the polymers produced on the phage surface, increased the particle size, and thus enhanced the phage interaction with the target surface, and subsequently enhanced endosomal escape of the particle. The same hybridization strategy was also used by Dasa’s group[138], where they genetically modified T7 phage with the RGD-4C peptide at the C-terminus of the phage capsid and a polyhistidine (6His) motif at the extra GGGS arm for binding copper ions. The chemical attachment of copper-64 PET isotope to the resulting phage enhanced the specific binding of this nanocarrier to MCF-7 cancer cells as well as allowed PET imaging of the carrier site. Then they converted copper ions into copper metal on the phage surface to enhance the stability and specificity of the carrier. Phage-based hybrid carriers have other advantages. They can increase the solubility of poorly-water soluble drugs by attachment of appropriate moieties to the phage surface that can dissolve and sequester drug molecules. These hybrids can be designed to have better stability under environmental conditions such as pH and temperature. Hybrid phages can be rapidly produced both in vitro and in vivo conditions[71] and they are able to transfer multiple cargoes into either multiple separate targets, or alternatively to different regions on the same target[139]. On the other hand, hybrid phages might have hazardous properties due to their high versatility and multivalent reactivity[77]. These phages could stimulate both cellular and humeral immune responses which might remain active for a longer time[113].
6. Targeting using phage-based nanoparticles
Targeting modalities provide several advantages including: (1) reduced side-effects increasing specificity for the target; 2) need for decreased amounts of therapeutic agents; 3) can be used for a range of various drugs with different pharmaceutical properties[77]. Targeting of nanocarriers is influenced by their specific properties such as size, shape, composition, surface charge, structure, and physiological characteristics. Viruses have a significant ability to recognize their cognate cell receptors, and deliver their payload of genes or drugs into the target cells. In addition to their inexpensive production, they offer versatile surface functionality through genetic engineering and tailoring their coat proteins. Their specific tropism for their host cells, the combination of genetic and proteomic information in one structure, and their potential utilization in recombinant technology makes them an ideal candidate for targeted delivery systems[129]
The tunable surface functionality of phages and their comparatively good safety profile have led to phages being more often considered as nanocarriers compared to other viral and polymeric carriers[129, 140]. Phage targeting methods can be divided into two main categories, passive targeting, and active targeting. In passive targeting, nanoparticles avoid the immune systems by their nano-size, and accumulate in tissues due to the leaky vasculature found in sites of inflammation and tumors. Some phage based delivery systems have intrinsic passive targeting ability due to prolonged circulation times and ability to escape clearance by the reticuloendothelial system (RES)[140]. In addition, the enhanced permeability and retention effect (EPR) allows them to preferentially accumulate in tumors[141–143]. Two strains of long-circulating lambda phage, Argo 1 and Argo 2, showed a high capacity to escape the RES and a long circulation half-life[144], resulting in higher accumulation at pathological sites. The long circulation time was provided by the amino acid residues displayed on the major head protein E exposed on the phage surface. These results highlighted the importance of the capsid protein; alteration of the capsid reduces the rapid clearance of phage particles by the host defense. Similarly, mammalian viruses and dsDNA phages have demonstrated similar results with various levels of DNA encapsulation, different structural capsid proteins and packaging proteins[145, 146]. All drug delivery systems that have been marketed up to the present date, are based on passive targeting; however this approaches suffers from limitations such as poor specificty and potential induction of multiple-drug resistance (MDR)[143, 147]. On the other hand, active targeting uses surface ligands to mediate specific molecular recognition with motifs expressed on the target cells or tissues. Specific binding may improve the accumulation of drugs and the cell uptake, thereby, reducing side-effects[142, 143]. Different surface ligands can be used for active targeting such as folate (vitamin), transferrin (protein), aptamers (RNA), antibodies and peptides. Moreover, insertion of DNA sequences by recombinant DNA technology producing peptide libraries is now used for enhanced active targeting of phage nanocarriers[129, 140].
Several extrinsic and intrinsic mechanisms have been used for active targeting of phages. In extrinsic mechanisms, gene insertions and additional modification with chemical moieties are used for enhancement of the targeting properties of the carriers. By contrast, the intrinsic mechanism uses the inherent recognition ability properties of phages for specific proteins on their host cell. Different chemical compounds can be used for surface modification, and amino acids with reactive side chains (such as glutamate, aspartate, cysteine and lysine) are used to attach surface modifications, for conjugation of drugs, peptides, antibodies, carbohydrates and oligonucleotides, with the help of different chemical linkers[148]. Chemical modification methods have been used variously for phage-based delivery systems, as it is considered a straightforward strategy, with potential application using a wide range of different chemical moieties[149]. Table 2 summarizes the applications of chemical modification of phage-based delivery systems in recent years.
Table 2.
Chemical modification of phage-based nanocarriers.
| Cargo | Nanocarrier | Chemical agent | Target | Ref |
|---|---|---|---|---|
| Green fluorescent protein [GFP] encoding gene | Phage lambda | Human holotransferrin | Human 293T cell line | [149] |
| Fluorescence dye | HK97 phages | Transferrin | HeLa and HT-29 tumor cells | [150] |
| Nanoparticles, chemotherapeutic drugs, siRNA cocktails, and protein | MS2 | SP94 peptide | Human hepatocellular carcinoma | [71] |
| Doxorubicin [DOX] | M13-polymer hybrid | Folate | KB cells | [151] |
| Fluorophore | Phage lambda | Transferrin | HeLa cells | [152] |
| GFP expression cassette | M13 | Transferrin | Human AGS cell line | [153] |
| Fluorescent dye | MS2 | Aptamer | Jurkat T cells | [69] |
| Photodynamic agents | MS2 | Aptamer | Jurkat T cells | [72] |
Depending on the target cell/tissue, suitable ligand can be utilized. For example, different cancer tissues have over-expressed markers and antigens that can be targeted by using appropriate antibodies[143]. Application of peptides as a targeting agent is another efficient way, especially in intracellular targeting strategies[147]. Inexpensive chemical synthesis of short peptides, their possibility for large scale production, small size and non-immunogenicity make them advantageous compared to other ligands such as antibodies and proteins[122]. For example conjugation of human immunodeficiency virus-1 (HIV-1) derived Tat peptide to MS2 phage by using sulfosuccinimidyl-4-(p-maleimidophenyl)-butyrate (sulfo-SMPB) cross-linker allowed RNA delivery into Huh-7 cells that effectively inhibited HCV translation[74].
As noted earlier, phage-based nanocarriers have genetic flexibility that can be used for different surface modification. The genomic flexibility of phages makes them appropriate choices for genetic modification to display targeting ligands in the proteins that compose the phage surface, and these ligands can direct the carrier to the desired cells via binding to specific receptors. Genetic engineering and the resulting modified proteins can be used for different applications including peptide-based affinity tags, reactive handles for bioconjugation, as targeting peptides, and as stimulators of immune response[148]. For example a study used recombinant DNA technology to engineer the F88.4 phage to express cyclic-RGD (cRGD) peptides[154]. Enhanced internalization into HeLa cells was reported using this structure, in comparison with the linear RGD-modified M13 phages. In addition, treatment of autoimmune encephalomyelitis using the immunodominant epitope of myelin oligodendrocyte glycoprotein expressed in a filamentous phage[62], and targeting of A549 cells through deletion of the Hoc and Soc capsid proteins in T4 phage[54] have been reported using this method. Table 3 summarizes some peptides that have been obtained from phage-display libraries that recognize various targets.
Table 3.
Some examples of targeting peptides obtained from phage libraries and used to target various cargos.
| Cargo | Carrier | Peptide | Target | Ref |
|---|---|---|---|---|
| DOX | Liposomes | HSYWLRS ALAAHKL |
Neuroblastoma | [155] |
| Coumarin 6 | Poly [ethyleneglycol]-poly [lactic-coglycolic acid] [PEG-PLGA] nanoparticles | TGNYKALHPHNG | Brain | [118] |
| Anticancer drugs | M13 | ETAPLSTMLSPY | Gastric cancer multidrug resistant cells | [156] |
| - | Phage Clone | CTSTSAPYC | Brain | [157] |
| siRNA | Liposome-phage hybrid | DMPGTVLP | Breast cancer | [158] |
| miR-145 | Polyethylenimine [PEI] polymer | CRIMRILRILKLAR RIMRILRILKLARC |
Breast cancer | [159] |
| miR-145 | Polyethylenimine [PEI] polymer | CRIMRILRILKLAR RIMRILRILKLARC |
Breast cancer | [159] |
| GAPDH siRNA | Phage-like nanoparticles/Nanophage | VEEGGYIAA DWRGDSMDS |
Breast cancer | [160] |
| VEGF siR-NA and SHP1 and Fas siRNA | Poly [cystaminebisacrylamide-diaminohexane, CBA-DAH] polymer | WLSEAGPVVTVR ALRGTGSW |
Cardiomyocyte | [161] |
| BH4 | Liposome | GSPREYTSYMPH | Atheroprone vasculature | [36] |
| - | M13 | -CVNHPAFAC-HTMYYHHYQHHL-NH2 CVNHPAFAC-GYGHT-MYYHHYQHHL-NH2 |
Melanoma | [162] |
| miR-92a | Polyelectrolyte complex micelles | VHPKQHR | Endothelial cells | [163] |
| Docetaxel | Docetaxel nanoparticles | LPLTPLP | Non-small cell lung cancer | [164] |
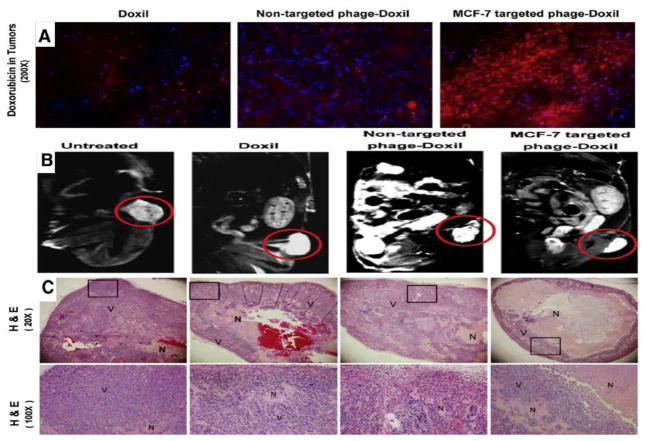
Many studies have investigated liposomal nanocarriers with phage display techniques for enhanced targeting[165–167]. A study was conducted for in vivo assessment of the anti-tumor activity of MCF-7-specific phage fusion protein-modified liposomal doxorubicin (Doxil)[168]. Enhanced tumor accumulation of “Phage-Doxil” and high anticancer activity with low side-effects was reported in tumor-bearing treated mice (Figure 6). Some other hybrid nanocarriers and their applications for targeting of different cells/organs, especially cancer tissues are summarized in Table 4.
Figure 6.
Efficient activity of MCF-7-targeted “Phage-Doxil”. A) Fluorescence images of tumor accumulation of DOX (Red fluorescence). (Blue fluorescence: DAPI-stained nuclei). B) Antitumor activity of Doxil with MR images of tumors in different groups (Doxil, non-targeted Phage-Doxil and MCF-7 targeted Phage-Doxil). C) Antitumor activity images of tumor sections with H & E staining. Necrotic cells (N) and viable cells (V) showing eosinophilic cytosol (pink) accompanied by hematoxylin stained nuclei (blue) (Reproduced from ref. [168] with permission from ELSEVIER).
Table 4.
Phage-based targeting using hybrid nanocarriers.
| Nanoparticle | Cargo | Phage/peptide | Target | Ref |
|---|---|---|---|---|
| Poly[ε-caprolactone] | 5-fluorouracil | Gene E from the phage | Colon cancer | [150] |
| Liposomes | - | Filamentous phages | Breast cancer | [148] |
| Liposomes | - | DMPGTVLP | MCF-7 cancer cell | [151] |
| Poly[caprolactone-b-2-vinylpyridine] | DOX | M13 | KB cancer cells | [126] |
| Liposome | siRNA | DMPGTVLP | MCF-7 cancer cells | [139] |
| Liposomes | DOX | VPEGAFSS | PC3 prostate cancer cells | [147] |
| PEG-PLGA nanoparticles | Coumarin 6 | TGNYKALHPHNG | Brain | [148] |
7. Applications of targeted phage-based nanocarriers
Phage therapy has been used as a treatment for various bacterial infections since the 1930s. However the discovery of antibiotics in the 1940s was a major setback for phage therapy. However since the emergence of multi-antibiotic resistance as a world-wide problem, phage therapy may finally be starting to stage a comeback. Three important issues regarding the use of phages as an antimicrobial therapy (phage therapy) need to be commented upon. The first issue is the emergence of drug-resistant bacterial clones. This could be restricted using antibiotiaprobiotic bacteria categorized as safe and beneficial in humans. The safety of pharmaceutical preparations of bacteriophages is also an important issue. Purification techniques have greatly improved in recent years, and standard operating procedures may be developed, improving the homogeneity and quality control of these preparations[5, 171–174]. Some pharmaceutical companies have tried to screen small-molecule chemical libraries against new protein targets that are broadly conserved in clinically relevant bacterial species. Large amounts of evidence show that this high-throughput screening has not been able to make such a big effect in the anti-bacterial drug discovery pipeline as was first expected. By contrast high-throughput screening (HTS) efforts in other therapeutic areas, usually yield numerous leads for each screen carried out[175, 176]. There are two major problems with target selection using HTS antibacterial screens: the limitation of chemical variety in currently utilized compound libraries; and targets that are susceptible to fast development of resistance when new drugs are isolated. Anti-bacterial drugs existing in the pharmaceutical market seemingly do not follow Lipinski’s rule of five, which explains drug-like structure of chemical compounds through solubility, hydrophobicity, and permeability features[177, 178]. Moreover the restricted return on investment and the long timelines required for drug development have rendered antibacterial HTS efforts less tempting for pharmaceutical companies,. However progress in small-molecule libraries could make HTS more productive and economic[179–182]. The development of techniques based on phages may have an impact on antibacterial drug discovery[183, 184]. The major problem of antibiotic resistance has been aggravated by constantly using new drugs that are simply variants of older, timeworn wide-spectrum antibiotics. Antimicrobial agents which are commercially available target only a few families of bacterial proteins, while genome-wide studies have found that bacteria need in fact encode ten times more essential proteins, especially for in vivo growth, and to maximize pathogenesis and virulence[185–189]. Consequently, it is highly likely that there are a lot of unexploited targets for antibacterial drug discovery. A favorable outcome depends on choosing targets with the highest potential for commercial success[190]. Historically, many researchers have studied the applications of bacteriophage therapy for the treatment of resistant bacterial infections. In this context bacteriophages have numerous advantages over the traditional antibiotic treatments, such as reduced side-effects and enhanced bacterial targeting[191]. Traditional phage therapy is based on lytic bacteriophages that lead to rupture of the the bacterial cell membrane, and as a result cause cell death. This strategy may cause side-effects through the release of endotoxins into the circulation[103]. Phagemids are a recent concept that employs bacteriophage proteins and selective packaging of a totally synthetic plasmid. This approach offers advantages over standard bacteriophage therapies with less risk of side-effects and high throughput delivery and expression[191]. For example Krom et al. designed a M13 bacteriophage-based modular bacterial phagemid system that expressed various nonlytic antimicrobial peptides (AMPs) and also toxic proteins[191]. The engineered system had two plasmids, one for desired antibacterial genes and the phagemid helper system. Rapid, non-lytic bacterial cell death was found both in vitro and in vivo, and high efficacy was reported using this system against E. coli.
Broxmeyer and coworkers investigated the efficacy of phage treatment against an intracellular human pathogen using a mouse model[192]. The work verified the ability of a myco-bacteriophage developed with the aid of a nonvirulent mycobacterium species, to kill Mycobacterium tuberculosis and M. avium(both common diseases in sufferers from AIDS). Vacuoles were developed inside macrophages to contain the phage, and they found that treatment of M. avium-infected (and to an even greater extent) M. tuberculosis-infected cells with the phage led to decreased bacterial numbers. They concluded that infecting macrophages with both M. aviumand M. smegmatis resulted in fusion of the vacuoles containing the two bacteria, which in turn led to delivery of the phage to the target bacteria.
Phages can act as gene delivery carriers in gene therapy applications[87, 193–196]. The motivation of utilizing phages for targeted gene delivery is similar to that of applying phages for delivery of DNA vaccines in which the capsid coat preserves the inner DNA from degradation. Phages are able to display foreign proteins on their surfaces to enable them to target specific cell types which is needed for successful gene therapy[197, 198]. Artificial covalent conjugation combined with phage display are used to display targeting molecules on the surfaces of phages[199–202]. The use of phages, displaying sequences like fibroblast growth factor (FGF)-derived peptides have been proposed to the target cells having the suitable receptors[87, 122, 203, 204]. Increasing the uptake and endosomal release of phages can be accomplished with protein sequences like the “penton base of adenovirus” which facilitates entry and endosomal release[122, 140, 160, 205].Larocca and coworkers were pioneer to demonstrate that FGF2-targeted filamentous phages were able to mediate transient and stable transduction of FGF receptor-positive mammalian cells[199]. They used phagemids with a cytomegalovirus driven reporter gene that were packaged with the helper phage M13KO7, and were targeted to cell surface-bound receptors making use of avidin-biotin-linked FGF2. This approach for targeted and phage-mediated transduction required FGF2 targeting and was phage concentration-dependent; however the overall transduction efficiency was low.
Researchers have proposed that the number of targeting ligands displayed on the phage particles could dictate the efficiency of cell-surface binding and of internalization[201, 206–208]. Multivalent display of targeting-molecules is said to facilitate both high-avidity binding and receptor-dimerization, by making available a multi-valent interaction between the phages and the cells, and in turn triggering internalization. Transduction efficiencies on the scale of 8% were found for human prostate carcinoma cells that were targeted by a display of EGF[208]. Because most cells seem to internalize phages fairly well, some groups have proposed that the transduction frequency remains limited by post-uptake events. Recently another approach has been tested. It was known that genotoxic interventions (camptothecin or UV irradiation) to cancer cells can increase the transduction efficiency of single-stranded adeno-associated virus. This approach was also investigated to increase phagemid-mediated transduction of mammalian cells[201, 209]. Inhibition of topoisomerase I increased EGF-targeted phage transduction in each of the cell lines examined and in prostate tumor xenografts.
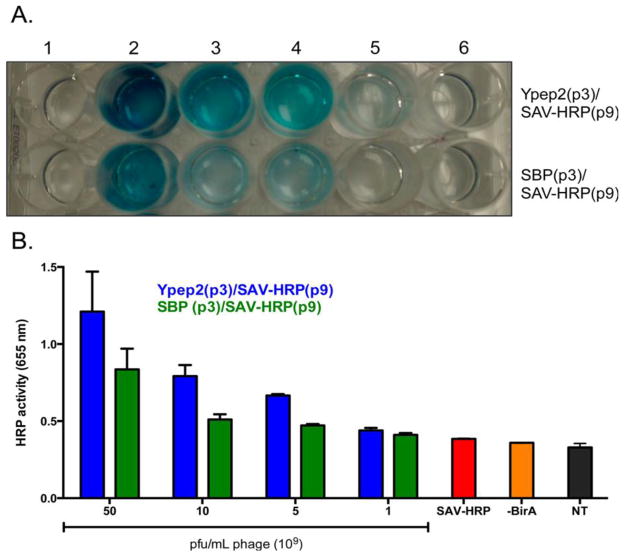
The use of polymeric nanoparticles to deliver various therapeutic cargos has limitations with regard to the need for multiple synthetic and purification steps, and difficulties in designing the precise size, shape, composition, and rigidity of the carrier[55] Genetically-engineered nanoparticles may be solutions for these limitations with a well-defined set of external ligands for better targeting. Furthermore, delivery of multiple protein cargos can be achieved by these phage-based nanocarriers [55]. For example a study engineered M13 phage to display a prostate cancer cell-penetrating peptide (Ypep2) in pIII and a biotin-acceptor peptide on pIX and horseradish peroxidase [HRP] as a reported molecule. By using E. coli biotin ligase, conjugation of the protein cargo with the phage was obtained. Enhanced delivery of the protein payload to PC-3 cells and significant cell death was reported using this nanocarrier (Figure 7).Debadyuti Ghosh et al. redesigned the M13 genome with overlapping of the end of gene VII and the beginning of gene IX [54]. They attached DOX to pVII. pIII displayed the peptides and pIX was biotinylated and loaded with a fluorophore for simultaneous in vitro imaging and killing of prostate cancer cells.
Figure 7.
A) Delivery of horseradish peroxidase (HRP) to prostate cancer cells. 1) no treatment; 2) 50 × 109; 3) 10 × 109; 4) 5 × 109; 5) 1 × 109 Ypep2(p3)/SAVHRP(p9) or SPARC binding peptide (SBP)(p3)/SAV-HRP(p9) pfu/mL phage; 6) ~1.6nM recombinant SAV-HRP. (B) Quantitative demonstration of image (A) results (SAV-HRP: streptavidin- horseradish peroxidase) (Reprinted from ref. [55] with permission from American Chemical Society).
Some clinical studies on phage therapy in humans have been reported with the most coming from centers in Eastern Europe and the former Soviet Union[4]. A serious study evaluating the use of therapeutic phages for prophylaxis against infectious diseases was carried out in Tbilisi, Georgia (1963–1964) using phages against bacterial dysentery[210]. Phages have been claimed to be successful in treating varying bacterial infections like cerebrospinal meningitis in an infant[211], skin infections caused by Staphylococcus, Proteus, Pseudomonas, Klebsiella, E. coli, subhepatic abscesses, recurrent subphrenic abscesses, Staphylococcal lung infections, P. aeruginosa lung infections in cystic fibrosis patients, neonatal sepsis, eye infections, urinary tract infections, and cancer[212–215]. Abdul-Hassan and coworkers described the therapy of 30 cases of burn-wound-related antibiotic-resistant P. aeruginosa infections[216]. Three times daily bandages that had been soaked with 10(10) phages/mL were applied to the burns. About half of the cases were observed to make a full recovery. Markoishvili and coworkers, described the use of “PhagoBioDerm”, which is a biodegradable polymer loaded with bacteriophages, as a wound-healing and antibacterial treatment for infected venous-stasis skin ulcers[217]. PhagoBioDerm was applied to ulcers-that had not responded to many other treatment strategies. Complete healing of ulcers-was reported in most of the patients. In one study, PhagoBioDerm combined with ciprofloxacin was used in three Georgian laborers who were exposed to a strontium-90 source. Besides systemic radiation effects, two of them developed acute local radiation burns which became infected with S. aureus. About 30 days after the incident, therapy with PhagoBioDerm was begun. Purulent drainage ceased in less than 7 days. The clinical improvement was accompanied with rapid and complete removal of the S. aureus, including strains that were resistant to ciprofloxacin [218].
7. Conclusions
Bacteriophages have led to many discoveries, and to some extent could be considered to be one of the cornerstones of modern molecular biology. In addition to the discovery that DNA was the sole source of heritable information, phages were used to confirm the theory of natural selection and have provided most of the enzymes that are ubiquitously used in laboratory protocols. Phage display technology has been used in many combinatorial and high throughput screening approaches to drug discovery and target discovery. Phage therapy was discovered before the advent of antibiotics, but was laid aside in the face of the biggest advance in pharmaceutical drugs ever made, the antibacterial antibiotics, that save countless millions of lives. With the resurgence of untreatable bacterial infections, many authorities have suggested that phage therapy could be revisited, if certain well-known problems could be overcome. These problems include the risk of large-scale lytic disruption of bacteria releasing LPS and other toxins, the possibility of contamination by the bacterial hosts used in production, and ensuring that have the correct tropism for the infecting bacteria.
However it is the use of phages as drug and gene delivery vehicles that concerns us here. The wide adoption of the principles and techniques of nanotechnology has led to an astonishing array of different nanocarriers that have been introduced for the controlled, targeted, and smart-release delivery of therapeutic cargos that include drugs, imaging reporters and genes. Phages have many advantages amongst this panoply of different kinds of nanocarriers. The most impressive is their enormous versatility due to their ability to express a wide range of peptides and proteins attached to their coat proteins using genetic engineering techniques. This phage display approach can be used to attach targeting peptides, to provide handles for chemical modification, and to allow attachment of moieties for loading cargos. Various hybrid phages have been developed including inorganic hybrid phages. The large capacity for introducing foreign genetic material in the phage genome has suggested that phages could be used in gene therapy. The advantage of phages over more conventional viral vectors would then be a greater level of safety, as phages could not reproduce naturally in mammalian hosts. The disadvantages of phages are as follows. They have a high level of immunogenicity (which makes them good in vaccine applications) but would be problematic in repeated application of a therapeutic cargo. Phages tend to have rapid clearance by the host RES, which necessitates special measures to avoid recognition and uptake.
Acknowledgments
Michael R Hamblin was supported by US NIH grant R01AI050875.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Greer GG. Bacteriophage control of foodborne bacteria. Journal of Food Protection®. 2005;68:1102–1111. doi: 10.4315/0362-028x-68.5.1102. [DOI] [PubMed] [Google Scholar]
- 2.Sutherland IW, Hughes KA, Skillman LC, Tait K. The interaction of phage and biofilms. FEMS Microbiology Letters. 2004;232:1–6. doi: 10.1016/S0378-1097(04)00041-2. [DOI] [PubMed] [Google Scholar]
- 3.Withey S, Cartmell E, Avery L, Stephenson T. Bacteriophages—potential for application in wastewater treatment processes. Science of the total environment. 2005;339:1–18. doi: 10.1016/j.scitotenv.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 4.Sulakvelidze A, Alavidze Z, Morris JG. Bacteriophage therapy. Antimicrobial agents and chemotherapy. 2001;45:649–659. doi: 10.1128/AAC.45.3.649-659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu TK, Koeris MS. The next generation of bacteriophage therapy. Current opinion in microbiology. 2011;14:524–531. doi: 10.1016/j.mib.2011.07.028. [DOI] [PubMed] [Google Scholar]
- 6.Entenza J, Loeffler J, Grandgirard D, Fischetti V, Moreillon P. Therapeutic effects of bacteriophage Cpl-1 lysin against Streptococcus pneumoniae endocarditis in rats. Antimicrobial agents and chemotherapy. 2005;49:4789–4792. doi: 10.1128/AAC.49.11.4789-4792.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang I, Jang H, Cho JC. Complete genome sequences of two Persicivirga bacteriophages, P12024S and P12024L. Journal of virology. 2012;86:8907–8908. doi: 10.1128/JVI.01327-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Twort F. THE TWORT-D’HERELLE PHENOMENON. The Lancet. 1926;207:416. [Google Scholar]
- 9.Hershey AD, Chase M. Independent functions of viral protein and nucleic acid in growth of bacteriophage. The Journal of general physiology. 1952;36:39–56. doi: 10.1085/jgp.36.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hendrix RW. Bacteriophages: evolution of the majority. Theoretical population biology. 2002;61:471–480. doi: 10.1006/tpbi.2002.1590. [DOI] [PubMed] [Google Scholar]
- 11.Edwards RA, Rohwer F. Viral metagenomics. Nature Reviews Microbiology. 2005;3:504–510. doi: 10.1038/nrmicro1163. [DOI] [PubMed] [Google Scholar]
- 12.Mullins NC. The development of a scientific specialty: The phage group and the origins of molecular biology. Minerva. 1972;10:51–82. [Google Scholar]
- 13.Hashida Y, Tanaka H, Zhou S, Kawakami S, Yamashita F, Murakami T, Umeyama T, Imahori H, Hashida M. Photothermal ablation of tumor cells using a single-walled carbon nanotube peptide composite. Journal of controlled release. 2014;173:59–66. doi: 10.1016/j.jconrel.2013.10.039. [DOI] [PubMed] [Google Scholar]
- 14.Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brüssow H. Phage as agents of lateral gene transfer. Current opinion in microbiology. 2003;6:417–424. doi: 10.1016/s1369-5274(03)00086-9. [DOI] [PubMed] [Google Scholar]
- 15.Carrasco B, Escobedo S, Alonso JC, Suárez JE. Modulation of Lactobacillus casei bacteriophage A2 lytic/lysogenic cycles by binding of Gp25 to the early lytic mRNA. Molecular microbiology. 2015 doi: 10.1111/mmi.13234. [DOI] [PubMed] [Google Scholar]
- 16.Yoo SY, Jin HE, Choi DS, Kobayashi M, Farouz Y, Wang S, Lee SW. M13 Bacteriophage and Adeno-Associated Virus Hybrid for Novel Tissue Engineering Material with Gene Delivery Functions. Advanced healthcare materials. 2015 doi: 10.1002/adhm.201500179. [DOI] [PubMed] [Google Scholar]
- 17.Jones CH, Chen M, Gollakota A, Ravikrishnan A, Zhang G, Lin S, Tan M, Cheng C, Lin H, Pfeifer BA. Structure Function Assessment of Mannosylated Poly (β-amino esters) upon Targeted Antigen Presenting Cell Gene Delivery. Biomacromolecules. 2015;16:1534–1541. doi: 10.1021/acs.biomac.5b00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yata T, Lee KY, Dharakul T, Songsivilai S, Bismarck A, Mintz PJ, Hajitou A. Hybrid Nanomaterial Complexes for Advanced Phage-guided Gene Delivery. Molecular Therapy—Nucleic Acids. 2014;3:e185. doi: 10.1038/mtna.2014.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee TJ, Schwartz C, Guo P. Construction of bacteriophage phi29 DNA packaging motor and its applications in nanotechnology and therapy. Annals of biomedical engineering. 2009;37:2064–2081. doi: 10.1007/s10439-009-9723-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hao C, Li X, Tian C, Jiang W, Wang G, Mao C. Construction of RNA nanocages by re-engineering the packaging RNA of Phi29 bacteriophage. Nature communications. 2014;5 doi: 10.1038/ncomms4890. [DOI] [PubMed] [Google Scholar]
- 21.Lee T, Haque F, Shu D, Yoo J, Li H, Yokel R, Horbinski C, Kim T, Kim S, Kwon C. RNA nanoparticle as a vector for targeted siRNA delivery into glioblastoma mouse model. Oncotarget. 2015 doi: 10.18632/oncotarget.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tarapore P, Shu Y, Guo P, Ho SM. Application of phi29 motor pRNA for targeted therapeutic delivery of siRNA silencing metallothionein-IIA and survivin in ovarian cancers. Molecular Therapy. 2011;19:386–394. doi: 10.1038/mt.2010.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 24.Phage FF. Novel Expression Vectors That Display Cloned Antigens On The Virion Surface, G. Smith. Science. 1985;228:1315–1316. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 25.Bakhshinejad B. Phage display and targeting peptides: surface functionalization of nanocarriers for delivery of small non-coding RNAs. Frontiers in Genetics. 2015;6 doi: 10.3389/fgene.2015.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pande J, Szewczyk MM, Grover AK. Phage display: concept, innovations, applications and future. Biotechnology advances. 2010;28:849–858. doi: 10.1016/j.biotechadv.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Park JP, Do M, Jin HE, Lee SW, Lee H. M13 Bacteriophage Displaying DOPA on Surfaces: Fabrication of Various Nanostructured Inorganic Materials without Time-Consuming Screening Processes. ACS applied materials & interfaces. 2014;6:18653–18660. doi: 10.1021/am506873g. [DOI] [PubMed] [Google Scholar]
- 28.Henry KA, Arbabi-Ghahroudi M, Scott JK. Beyond phage display: non-traditional applications of the filamentous bacteriophage as a vaccine carrier, therapeutic biologic, and bioconjugation scaffold. Frontiers in microbiology. 2015;6 doi: 10.3389/fmicb.2015.00755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pavoni E, Vaccaro P, D’Alessio V, De Santis R, Minenkova O. Simultaneous display of two large proteins on the head and tail of bacteriophage lambda. BMC biotechnology. 2013;13:79. doi: 10.1186/1472-6750-13-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bakhshinejad B, Karimi M, Khalaj-Kondori M. Phage display: development of nanocarriers for targeted drug delivery to the brain. Neural regeneration research. 2015;10:862. doi: 10.4103/1673-5374.158330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azzazy HM, Highsmith WE. Phage display technology: clinical applications and recent innovations. Clinical biochemistry. 2002;35:425–445. doi: 10.1016/s0009-9120(02)00343-0. [DOI] [PubMed] [Google Scholar]
- 32.Liu GW, Livesay BR, Kacherovsky NA, Cieslewicz M, Lutz E, Waalkes A, Jensen MC, Salipante SJ, Pun SH. Efficient identification of murine M2 macrophage peptide targeting ligands by phage display and next-generation sequencing. Bioconjugate chemistry. 2015;26:1811–1817. doi: 10.1021/acs.bioconjchem.5b00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Z, Jin W, Liu H, Zhao Z, Cheng K. Discovery of Peptide Ligands for Hepatic Stellate Cells Using Phage Display. Molecular pharmaceutics. 2015 doi: 10.1021/acs.molpharmaceut.5b00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bratkovič T. Progress in phage display: evolution of the technique and its applications. Cellular and molecular life sciences. 2010;67:749–767. doi: 10.1007/s00018-009-0192-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y, Shen Y, Guo X, Zhang C, Yang W, Ma M, Liu S, Zhang M, Wen LP. Transdermal protein delivery by a coadministered peptide identified via phage display. Nature biotechnology. 2006;24:455–460. doi: 10.1038/nbt1193. [DOI] [PubMed] [Google Scholar]
- 36.Hofmeister LH, Lee SH, Norlander AE, Montaniel KRC, Chen W, Harrison DG, Sung HJ. Phage-Display-Guided Nanocarrier Targeting to Atheroprone Vasculature. ACS nano. 2015;9:4435–4446. doi: 10.1021/acsnano.5b01048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willats WG. Phage display: practicalities and prospects. Plant molecular biology. 2002;50:837–854. doi: 10.1023/a:1021215516430. [DOI] [PubMed] [Google Scholar]
- 38.Tzagoloff H, Pratt D. The initial steps in infection with coliphage M13. Virology. 1964;24:372–380. doi: 10.1016/0042-6822(64)90174-6. [DOI] [PubMed] [Google Scholar]
- 39.Tian Y, Wu M, Liu X, Liu Z, Zhou Q, Niu Z, Huang Y. Probing the Endocytic Pathways of the Filamentous Bacteriophage in Live Cells Using Ratiometric pH Fluorescent Indicator. Advanced healthcare materials. 2015;4:413–419. doi: 10.1002/adhm.201400508. [DOI] [PubMed] [Google Scholar]
- 40.Rakonjac J. Filamentous Bacteriophages: Biology and Applications. eLS. 2011 doi: 10.1101/pdb.over107754. [DOI] [PubMed] [Google Scholar]
- 41.van Houten NE, Henry KA, Smith GP, Scott JK. Engineering filamentous phage carriers to improve focusing of antibody responses against peptides. Vaccine. 2010;28:2174–2185. doi: 10.1016/j.vaccine.2009.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greenwood J, Willis AE, Perham RN. Multiple display of foreign peptides on a filamentous bacteriophage: peptides from Plasmodium falciparum circumsporozoite protein as antigens. Journal of molecular biology. 1991;220:821–827. doi: 10.1016/0022-2836(91)90354-9. [DOI] [PubMed] [Google Scholar]
- 43.Grabowska A, Jennings R, Laing P, Darsley M, Jameson C, Swift L, Irving W. Immunisation with phage displaying peptides representing single epitopes of the glycoprotein G can give rise to partial protective immunity to HSV-2. Virology. 2000;269:47–53. doi: 10.1006/viro.2000.0185. [DOI] [PubMed] [Google Scholar]
- 44.Demangel C, Lafaye P, Mazie J. Reproducing the immune response against the Plasmodium vivax merozoite surface protein 1 with mimotopes selected from a phage-displayed peptide library. Molecular immunology. 1996;33:909–916. doi: 10.1016/s0161-5890(96)00058-2. [DOI] [PubMed] [Google Scholar]
- 45.Scala G, Chen X, Liu W, Telles JN, Cohen OJ, Vaccarezza M, Igarashi T, Fauci AS. Selection of HIV-specific immunogenic epitopes by screening random peptide libraries with HIV-1-positive sera. The Journal of Immunology. 1999;162:6155–6161. [PubMed] [Google Scholar]
- 46.Frenkel D, Dewachter I, Van Leuven F, Solomon B. Reduction of β-amyloid plaques in brain of transgenic mouse model of Alzheimer’s disease by EFRH-phage immunization. Vaccine. 2003;21:1060–1065. doi: 10.1016/s0264-410x(02)00609-6. [DOI] [PubMed] [Google Scholar]
- 47.Lavie V, Becker M, Cohen-Kupiec R, Yacoby I, Koppel R, Wedenig M, Hutter-Paier B, Solomon B. EFRH-phage immunization of Alzheimer’s disease animal model improves behavioral performance in Morris water maze trials. Journal of Molecular Neuroscience. 2004;24:105–113. doi: 10.1385/JMN:24:1:105. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka K, Nishimura M, Yamaguchi Y, Hashiguchi S, Takiguchi S, Yamaguchi M, Tahara H, Gotanda T, Abe R, Ito Y. A mimotope peptide of Aβ42 fibril-specific antibodies with Aβ42 fibrillation inhibitory activity induces anti-Aβ42 conformer antibody response by a displayed form on an M13 phage in mice. Journal of neuroimmunology. 2011;236:27–38. doi: 10.1016/j.jneuroim.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 49.Rudolf MP, Vogel M, Kricek F, Ruf C, Zürcher AW, Reuschel R, Auer M, Miescher S, Stadler BM. Epitope-specific antibody response to IgE by mimotope immunization. The Journal of Immunology. 1998;160:3315–3321. [PubMed] [Google Scholar]
- 50.Mascolo D, Barba P, De Berardinis P, Di Rosa F, Del Pozzo G. Phage display of a CTL epitope elicits a long-term in vivo cytotoxic response. FEMS Immunology & Medical Microbiology. 2007;50:59–66. doi: 10.1111/j.1574-695X.2007.00229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wan Y, Wu Y, Bian J, Wang X, Zhou W, Jia Z, Tan Y, Zhou L. Induction of hepatitis B virus-specific cytotoxic T lymphocytes response in vivo by filamentous phage display vaccine. Vaccine. 2001;19:2918–2923. doi: 10.1016/s0264-410x(00)00561-2. [DOI] [PubMed] [Google Scholar]
- 52.Sartorius R, Bettua C, D’Apice L, Caivano A, Trovato M, Russo D, Zanoni I, Granucci F, Mascolo D, Barba P. Vaccination with filamentous bacteriophages targeting DEC-205 induces DC maturation and potent anti-tumor T-cell responses in the absence of adjuvants. European journal of immunology. 2011;41:2573–2584. doi: 10.1002/eji.201141526. [DOI] [PubMed] [Google Scholar]
- 53.Poul MA, Marks JD. Targeted gene delivery to mammalian cells by filamentous bacteriophage. Journal of molecular biology. 1999;288:203–211. doi: 10.1006/jmbi.1999.2678. [DOI] [PubMed] [Google Scholar]
- 54.Ghosh D, Kohli AG, Moser F, Endy D, Belcher AM. Refactored M13 bacteriophage as a platform for tumor cell imaging and drug delivery. ACS synthetic biology. 2012;1:576–582. doi: 10.1021/sb300052u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DePorter SM, McNaughton BR. Engineered M13 Bacteriophage Nanocarriers for Intracellular Delivery of Exogenous Proteins to Human Prostate Cancer Cells. Bioconjugate chemistry. 2014;25:1620–1625. doi: 10.1021/bc500339k. [DOI] [PubMed] [Google Scholar]
- 56.Jin HE, Farr R, Lee SW. Collagen mimetic peptide engineered M13 bacteriophage for collagen targeting and imaging in cancer. Biomaterials. 2014;35:9236–9245. doi: 10.1016/j.biomaterials.2014.07.044. [DOI] [PubMed] [Google Scholar]
- 57.Li K, Chen Y, Li S, Nguyen HG, Niu Z, You S, Mello CM, Lu X, Wang Q. Chemical modification of M13 bacteriophage and its application in cancer cell imaging. Bioconjugate chemistry. 2010;21:1369–1377. doi: 10.1021/bc900405q. [DOI] [PubMed] [Google Scholar]
- 58.Battella S, Cox MC, Santoni A, Palmieri G. Natural killer (NK) cells and anti-tumor therapeutic mAb: unexplored interactions. Journal of leukocyte biology. 2016;99:87–96. doi: 10.1189/jlb.5VMR0415-141R. [DOI] [PubMed] [Google Scholar]
- 59.Vaks L, Benhar I. In vivo characteristics of targeted drug-carrying filamentous bacteriophage nanomedicines. Journal of nanobiotechnology. 2011;9:1–10. doi: 10.1186/1477-3155-9-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zahid M, Phillips BE, Albers SM, Giannoukakis N, Watkins SC, Robbins PD. Identification of a cardiac specific protein transduction domain by in vivo biopanning using a M13 phage peptide display library in mice. PloS one. 2010;5:e12252. doi: 10.1371/journal.pone.0012252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ksendzovsky A, Walbridge S, Saunders RC, Asthagiri AR, Heiss JD, Lonser RR. Convection-enhanced delivery of M13 bacteriophage to the brain. Journal of neurosurgery. 2012;117:197. doi: 10.3171/2012.4.JNS111528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rakover IS, Zabavnik N, Kopel R, Paz-Rozner M, Solomon B. Antigen-specific therapy of EAE via intranasal delivery of filamentous phage displaying a myelin immunodominant epitope. Journal of neuroimmunology. 2010;225:68–76. doi: 10.1016/j.jneuroim.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 63.Lee JH, Domaille DW, Cha JN. Amplified protein detection and identification through DNA-conjugated M13 bacteriophage. ACS nano. 2012;6:5621–5626. doi: 10.1021/nn301565e. [DOI] [PubMed] [Google Scholar]
- 64.Carrico ZM, Farkas ME, Zhou Y, Hsiao SC, Marks JD, Chokhawala H, Clark DS, Francis MB. N-terminal labeling of filamentous phage to create cancer marker imaging agents. ACS nano. 2012;6:6675–6680. doi: 10.1021/nn301134z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Valegård K, Liljas L, Fridborg K, Unge T. Structure determination of the bacteriophage MS2. Acta Crystallographica Section B: Structural Science. 1991;47:949–960. doi: 10.1107/s0108768191006821. [DOI] [PubMed] [Google Scholar]
- 66.Laguna, Bacteriophage MS2 capsid protein. Sci Photo Libr, Sci Photo Libr. 2013 Available at: http://www.sciencephoto.com/media/471076/enlarge.
- 67.Fiers W, Contreras R, Duerinck F, Haegmean G, Merregaert J, Jou WM, Raeymakers A, Volckaert G, Ysebaert M. A-protein gene of bacteriophage MS2. Nature. 1975;256:273–278. doi: 10.1038/256273a0. [DOI] [PubMed] [Google Scholar]
- 68.Sun S, Li W, Sun Y, Pan Y, Li J. A new RNA vaccine platform based on MS2 virus-like particles produced in Saccharomyces cerevisiae. Biochemical and biophysical research communications. 2011;407:124–128. doi: 10.1016/j.bbrc.2011.02.122. [DOI] [PubMed] [Google Scholar]
- 69.Tong GJ, Hsiao SC, Carrico ZM, Francis MB. Viral capsid DNA aptamer conjugates as multivalent cell-targeting vehicles. Journal of the American Chemical Society. 2009;131:11174–11178. doi: 10.1021/ja903857f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carrico ZM, Romanini DW, Mehl RA, Francis MB. Oxidative coupling of peptides to a virus capsid containing unnatural amino acids. Chemical Communications. 2008:1205–1207. doi: 10.1039/b717826c. [DOI] [PubMed] [Google Scholar]
- 71.Ashley CE, Carnes EC, Phillips GK, Durfee PN, Buley MD, Lino CA, Padilla DP, Phillips B, Carter MB, Willman CL. Cell-specific delivery of diverse cargos by bacteriophage MS2 virus-like particles. ACS nano. 2011;5:5729–5745. doi: 10.1021/nn201397z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stephanopoulos N, Tong GJ, Hsiao SC, Francis MB. Dual-surface modified virus capsids for targeted delivery of photodynamic agents to cancer cells. ACS nano. 2010;4:6014–6020. doi: 10.1021/nn1014769. [DOI] [PubMed] [Google Scholar]
- 73.Glasgow JE, Capehart SL, Francis MB, Tullman-Ercek D. Osmolyte-mediated encapsulation of proteins inside MS2 viral capsids. ACS nano. 2012;6:8658–8664. doi: 10.1021/nn302183h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wei B, Wei Y, Zhang K, Wang J, Xu R, Zhan S, Lin G, Wang W, Liu M, Wang L. Development of an antisense RNA delivery system using conjugates of the MS2 bacteriophage capsids and HIV-1 TAT cell penetrating peptide. Biomedicine & Pharmacotherapy. 2009;63:313–318. doi: 10.1016/j.biopha.2008.07.086. [DOI] [PubMed] [Google Scholar]
- 75.Pan Y, Jia T, Zhang Y, Zhang K, Zhang R, Li J, Wang L. MS2 VLP-based delivery of microRNA-146a inhibits autoantibody production in lupus-prone mice. International journal of nanomedicine. 2012;7:5957. doi: 10.2147/IJN.S37990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Prel A, Caval V, Gayon R, Ravassard P, Duthoit C, Payen E, Maouche-Chretien L, Creneguy A, Nguyen TH, Martin N. Highly efficient in vitro and in vivo delivery of functional RNAs using new versatile MS2-chimeric retrovirus-like particles. Molecular therapy Methods & clinical development. 2015;2:15039. doi: 10.1038/mtm.2015.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ma Y, Nolte RJ, Cornelissen JJ. Virus-based nanocarriers for drug delivery. Advanced drug delivery reviews. 2012;64:811–825. doi: 10.1016/j.addr.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 78.Li J, Sun Y, Jia T, Zhang R, Zhang K, Wang L. Messenger RNA vaccine based on recombinant MS2 virus-like particles against prostate cancer. International Journal of Cancer. 2014;134:1683–1694. doi: 10.1002/ijc.28482. [DOI] [PubMed] [Google Scholar]
- 79.Farkas ME, Aanei IL, Behrens CR, Tong GJ, Murphy ST, O’Neil JP, Francis MB. PET imaging and biodistribution of chemically modified bacteriophage MS2. Molecular pharmaceutics. 2012;10:69–76. doi: 10.1021/mp3003754. [DOI] [PubMed] [Google Scholar]
- 80.Wagner S, Zensi A, Wien SL, Tschickardt SE, Maier W, Vogel T, Worek F, Pietrzik CU, Kreuter J, Von Briesen H. Uptake mechanism of ApoE-modified nanoparticles on brain capillary endothelial cells as a blood-brain barrier model. PloS one. 2012;7:e32568. doi: 10.1371/journal.pone.0032568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lech K, Reddy K, Sherman L. Preparing lambda DNA from phage lysates. Current Protocols in Molecular Biology. 1990:1.13. 11–11.13. 10. doi: 10.1002/0471142727.mb0113s10. [DOI] [PubMed] [Google Scholar]
- 82.Galikowska E, Kunikowska D, Tokarska-Pietrzak E, Dziadziuszko H, Łoś JM, Golec P, Węgrzyn G, Łoś M. Specific detection of Salmonella enterica and Escherichia coli strains by using ELISA with bacteriophages as recognition agents. European journal of clinical microbiology & infectious diseases. 2011;30:1067–1073. doi: 10.1007/s10096-011-1193-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Erni B, Zanolari B, Kocher H. The mannose permease of Escherichia coli consists of three different proteins. Amino acid sequence and function in sugar transport, sugar phosphorylation, and penetration of phage lambda DNA. Journal of Biological Chemistry. 1987;262:5238–5247. [PubMed] [Google Scholar]
- 84.Mikawa YG, Maruyama IN, Brenner S. Surface display of proteins on bacteriophage λ heads. Journal of molecular biology. 1996;262:21–30. doi: 10.1006/jmbi.1996.0495. [DOI] [PubMed] [Google Scholar]
- 85.Christensen AC. Bacteriophage lambda-based expression vectors. Molecular biotechnology. 2001;17:219–224. doi: 10.1385/MB:17:3:219. [DOI] [PubMed] [Google Scholar]
- 86.Beghetto E, Gargano N. Lambda-display: a powerful tool for antigen discovery. Molecules. 2011;16:3089–3105. doi: 10.3390/molecules16043089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nicastro J, Sheldon K, Slavcev RA. Bacteriophage lambda display systems: developments and applications. Applied microbiology and biotechnology. 2014;98:2853–2866. doi: 10.1007/s00253-014-5521-1. [DOI] [PubMed] [Google Scholar]
- 88.Chang JR, Song EH, Nakatani-Webster E, Monkkonen L, Ratner DM, Catalano CE. Phage Lambda Capsids as Tunable Display Nanoparticles. Biomacromolecules. 2014;15:4410–4419. doi: 10.1021/bm5011646. [DOI] [PubMed] [Google Scholar]
- 89.Okayama H, Berg P. Bacteriophage lambda vector for transducing a cDNA clone library into mammalian cells. Molecular and cellular biology. 1985;5:1136–1142. doi: 10.1128/mcb.5.5.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Merril CR, Geier MR. Bacterial virus gene expression in human cells. Nature. 1971;233:398–400. doi: 10.1038/233398a0. [DOI] [PubMed] [Google Scholar]
- 91.Lankes H, Zanghi C, Santos K, Capella C, Duke C, Dewhurst S. In vivo gene delivery and expression by bacteriophage lambda vectors. Journal of applied microbiology. 2007;102:1337–1349. doi: 10.1111/j.1365-2672.2006.03182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vaccaro P, Pavoni E, Monteriù G, Andrea P, Felici F, Minenkova O. Efficient display of scFv antibodies on bacteriophage lambda. Journal of immunological methods. 2006;310:149–158. doi: 10.1016/j.jim.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 93.Sapinoro R, Volcy K, Rodrigo WSI, Schlesinger JJ, Dewhurst S. Fc receptor-mediated, antibody-dependent enhancement of bacteriophage lambda-mediated gene transfer in mammalian cells. Virology. 2008;373:274–286. doi: 10.1016/j.virol.2007.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Adhya S, Merril CR, Biswas B. Therapeutic and prophylactic applications of bacteriophage components in modern medicine. Cold Spring Harbor perspectives in medicine. 2014;4:a012518. doi: 10.1101/cshperspect.a012518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Clark JR, March JB. Bacterial viruses as human vaccines? 2004 doi: 10.1586/14760584.3.4.463. [DOI] [PubMed] [Google Scholar]
- 96.Ghaemi A, Soleimanjahi H, Gill P, Hassan Z, Jahromi SRM, Roohvand F. Research Recombinant λ-phage nanobioparticles for tumor therapy in mice models. 2010 doi: 10.1186/1479-0556-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Min JJ, Nguyen VH, Gambhir SS. Molecular imaging of biological gene delivery vehicles for targeted cancer therapy: beyond viral vectors. Nuclear medicine and molecular imaging. 2010;44:15–24. doi: 10.1007/s13139-009-0006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Thomas BS, Nishikawa S, Ito K, Chopra P, Sharma N, Evans DH, Tyrrell DLJ, Bathe OF, Rancourt DE. Peptide vaccination is superior to genetic vaccination using a recombineered bacteriophage λ subunit vaccine. Vaccine. 2012;30:998–1008. doi: 10.1016/j.vaccine.2011.12.070. [DOI] [PubMed] [Google Scholar]
- 99.O’Connor C. Isolating hereditary material: Frederick Griffith, Oswald Avery, Alfred Hershey, and Martha Chase. Nature Education. 2008;1:105. [Google Scholar]
- 100.Mullaney JM, Black LW. Activity of foreign proteins targeted within the bacteriophage T4 head and prohead: implications for packaged DNA structure. Journal of molecular biology. 1998;283:913–929. doi: 10.1006/jmbi.1998.2126. [DOI] [PubMed] [Google Scholar]
- 101.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. Journal of molecular biology. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 102.Lindner T, Kolmar H, Haberkorn U, Mier W. DNA libraries for the construction of phage libraries: Statistical and structural requirements and synthetic methods. Molecules. 2011;16:1625–1641. doi: 10.3390/molecules16021625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lu TK, Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. Proceedings of the National Academy of Sciences. 2007;104:11197–11202. doi: 10.1073/pnas.0704624104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Drulis-Kawa Z, Majkowska-Skrobek G, Maciejewska B. Bacteriophages and Phage-Derived Proteins Application Approaches. Current medicinal chemistry. 2015;22:1757–1773. doi: 10.2174/0929867322666150209152851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li XH, Tang L, Liu D, Sun HM, Zhou CC, Tan LS, Wang LP, Zhang PD, Zhang SQ. Antitumor effect of recombinant T7 phage vaccine expressing xenogenic vascular endothelial growth factor on Lewis lung cancer in mice. Ai zheng= Aizheng= Chinese journal of cancer. 2006;25:1221–1226. [PubMed] [Google Scholar]
- 106.Wong SC, Wakefield D, Klein J, Monahan SD, Rozema DB, Lewis DL, Higgs L, Ludtke J, Sokoloff AV, Wolff JA. Hepatocyte targeting of nucleic acid complexes and liposomes by a T7 phage p17 peptide. Molecular pharmaceutics. 2006;3:386–397. doi: 10.1021/mp050108r. [DOI] [PubMed] [Google Scholar]
- 107.Robertson KL, Soto CM, Archer MJ, Odoemene O, Liu JL. Engineered T4 viral nanoparticles for cellular imaging and flow cytometry. Bioconjugate chemistry. 2011;22:595–604. doi: 10.1021/bc100365j. [DOI] [PubMed] [Google Scholar]
- 108.Yaung SJ, Esvelt KM, Church GM. Complete genome sequences of T4-like bacteriophages RB3, RB5, RB6, RB7, RB9, RB10, RB27, RB33, RB55, RB59, and RB68. Genome announcements. 2015;3:e01122–01114. doi: 10.1128/genomeA.01122-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fokine A, Battisti AJ, Bowman VD, Efimov AV, Kurochkina LP, Chipman PR, Mesyanzhinov VV, Rossmann MG. Cryo-EM study of the Pseudomonas bacteriophage φKZ. Structure. 2007;15:1099–1104. doi: 10.1016/j.str.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 110.Fokine A, Islam MZ, Zhang Z, Bowman VD, Rao VB, Rossmann MG. Structure of the three N-terminal immunoglobulin domains of the highly immunogenic outer capsid protein from a T4-like bacteriophage. Journal of virology. 2011;85:8141–8148. doi: 10.1128/JVI.00847-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu J, Tu C, Yu X, Zhang M, Zhang N, Zhao M, Nie W, Ren Z. Bacteriophage T4 nanoparticle capsid surface SOC and HOC bipartite display with enhanced classical swine fever virus immunogenicity: a powerful immunological approach. Journal of virological methods. 2007;139:50–60. doi: 10.1016/j.jviromet.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 112.Black LW. Old, new, and widely true: The bacteriophage T4 DNA packaging mechanism. Virology. 2015;479:650–656. doi: 10.1016/j.virol.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tao P, Mahalingam M, Marasa BS, Zhang Z, Chopra AK, Rao VB. In vitro and in vivo delivery of genes and proteins using the bacteriophage T4 DNA packaging machine. Proceedings of the National Academy of Sciences. 2013;110:5846–5851. doi: 10.1073/pnas.1300867110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wu M, Sherwin T, Brown WL, Stockley PG. Delivery of antisense oligonucleotides to leukemia cells by RNA bacteriophage capsids, Nanomedicine: Nanotechnology. Biology and Medicine. 2005;1:67–76. doi: 10.1016/j.nano.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 115.Shoae-Hassani A, Mortazavi-Tabatabaei SA, Sharif S, Madadi S, Rezaei-Khaligh H, Verdi J. Recombinant λ bacteriophage displaying nanobody towards third domain of HER-2 epitope inhibits proliferation of breast carcinoma SKBR-3 cell line. Archivum immunologiae et therapiae experimentalis. 2013;61:75–83. doi: 10.1007/s00005-012-0206-x. [DOI] [PubMed] [Google Scholar]
- 116.Ren S-x, Ren Z-j, Zhao M-y, Wang X-b, Zuo S-g, Yu F. Antitumor activity of endogenous mFlt4 displayed on a T4 phage nanoparticle surface. Acta Pharmacologica Sinica. 2009;30:637–645. doi: 10.1038/aps.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ren ZJ, Tian CJ, Zhu QS, Zhao MY, Xin AG, Nie WX, Ling SR, Zhu MW, Wu JY, Lan HY. Orally delivered foot-and-mouth disease virus capsid protomer vaccine displayed on T4 bacteriophage surface. 100% protection from potency challenge in mice. Vaccine. 2008;26:1471–1481. doi: 10.1016/j.vaccine.2007.12.053. [DOI] [PubMed] [Google Scholar]
- 118.Li J, Feng L, Fan L, Zha Y, Guo L, Zhang Q, Chen J, Pang Z, Wang Y, Jiang X. Targeting the brain with PEG PLGA nanoparticles modified with phage-displayed peptides. Biomaterials. 2011;32:4943–4950. doi: 10.1016/j.biomaterials.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yokoi N, Inaba H, Terauchi M, Stieg AZ, Sanghamitra NJM, Koshiyama T, Yutani K, Kanamaru S, Arisaka F, Hikage T. Construction of Robust Bio-nanotubes using the Controlled Self-Assembly of Component Proteins of Bacteriophage T4. small. 2010;6:1873–1879. doi: 10.1002/smll.201000772. [DOI] [PubMed] [Google Scholar]
- 120.Choy JH, Oh JM, Choi SJ. Bio-Inorganic Conjugates for Drug and Gene Delivery. Bio-inorganic Hybrid Nanomaterials: Strategies, Syntheses, Characterization and Applications. 2008:401–418. [Google Scholar]
- 121.Bronstein LM. Virus-Based Nanoparticles with Inorganic Cargo: What Does the Future Hold? Small. 2011;7:1609–1618. doi: 10.1002/smll.201001992. [DOI] [PubMed] [Google Scholar]
- 122.Bakhshinejad B, Karimi M, Sadeghizadeh M. Bacteriophages and medical oncology: targeted gene therapy of cancer. Medical Oncology. 2014;31:1–11. doi: 10.1007/s12032-014-0110-9. [DOI] [PubMed] [Google Scholar]
- 123.Karimi M, Solati N, Amiri M, Mirshekari H, Mohamed E, Taheri M, Hashemkhani M, Saeidi A, Estiar MA, Kiani P. Carbon nanotubes part I: preparation of a novel and versatile drug-delivery vehicle. Expert opinion on drug delivery. 2015:1–17. doi: 10.1517/17425247.2015.1003806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Stoneham CA, Hollinshead M, Hajitou A. Clathrin-mediated endocytosis and subsequent endolysosomal trafficking of adeno-associated virus/phage. Journal of Biological Chemistry. 2012;287:35849–35859. doi: 10.1074/jbc.M112.369389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Soto CM, Ratna BR. Virus hybrids as nanomaterials for biotechnology. Current opinion in biotechnology. 2010;21:426–438. doi: 10.1016/j.copbio.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 126.Marek M, van Oers MM, Devaraj FF, Vlak JM, Merten OW. Engineering of baculovirus vectors for the manufacture of virion-free biopharmaceuticals. Biotechnology and bioengineering. 2011;108:1056–1067. doi: 10.1002/bit.23028. [DOI] [PubMed] [Google Scholar]
- 127.Wang Y, Su Q, Dong S, Shi H, Gao X, Wang L. Hybrid phage displaying SLAQVKYTSASSI induces protection against Candida albicans challenge in BALB/c mice. Human vaccines & immunotherapeutics. 2014;10:1057–1063. doi: 10.4161/hv.27714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Merzlyak A, Indrakanti S, Lee SW. Genetically engineered nanofiber-like viruses for tissue regenerating materials. Nano letters. 2009;9:846–852. doi: 10.1021/nl8036728. [DOI] [PubMed] [Google Scholar]
- 129.Farr R, Choi DS, Lee SW. Phage-based nanomaterials for biomedical applications. Acta biomaterialia. 2014;10:1741–1750. doi: 10.1016/j.actbio.2013.06.037. [DOI] [PubMed] [Google Scholar]
- 130.Merzlyak A, Lee SW. Phage as templates for hybrid materials and mediators for nanomaterial synthesis. Current opinion in chemical biology. 2006;10:246–252. doi: 10.1016/j.cbpa.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 131.Yi H, Ghosh D, Ham MH, Qi J, Barone PW, Strano MS, Belcher AM. M13 phage-functionalized single-walled carbon nanotubes as nanoprobes for second near-infrared window fluorescence imaging of targeted tumors. Nano letters. 2012;12:1176–1183. doi: 10.1021/nl2031663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang H, Hussain I, Brust M, Butler MF, Rannard SP, Cooper AI. Aligned two-and three-dimensional structures by directional freezing of polymers and nanoparticles. Nature materials. 2005;4:787–793. doi: 10.1038/nmat1487. [DOI] [PubMed] [Google Scholar]
- 133.Hedden RC, Bauer BJ, Smith AP, Gröhn F, Amis E. Templating of inorganic nanoparticles by PAMAM/PEG dendrimer star polymers. Polymer. 2002;43:5473–5481. [Google Scholar]
- 134.Hall SR, Shenton W, Engelhardt H, Mann S. Site-Specific Organization of Gold Nanoparticles by Biomolecular Templating. ChemPhysChem. 2001;2:184–186. doi: 10.1002/1439-7641(20010316)2:3<184::AID-CPHC184>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 135.He T, Abbineni G, Cao B, Mao C. Nanofibrous Bio-inorganic Hybrid Structures Formed Through Self-Assembly and Oriented Mineralization of Genetically Engineered Phage Nanofibers. Small. 2010;6:2230–2235. doi: 10.1002/smll.201001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sawada T, Kang S, Watanabe J, Mihara H, Serizawa T. Hybrid hydrogels composed of regularly assembled filamentous viruses and gold nanoparticles. ACS Macro Letters. 2014;3:341–345. doi: 10.1021/mz500073t. [DOI] [PubMed] [Google Scholar]
- 137.Hajitou A, Trepel M, Lilley CE, Soghomonyan S, Alauddin MM, Marini FC, Restel BH, Ozawa MG, Moya CA, Rangel R. A hybrid vector for ligand-directed tumor targeting and molecular imaging. Cell. 2006;125:385–398. doi: 10.1016/j.cell.2006.02.042. [DOI] [PubMed] [Google Scholar]
- 138.Dasa SSK, Jin Q, Chen CT, Chen L. Target-Specific Copper Hybrid T7 Phage Particles. Langmuir. 2012;28:17372–17380. doi: 10.1021/la3024919. [DOI] [PubMed] [Google Scholar]
- 139.Shu Y, Cinier M, Shu D, Guo P. Assembly of multifunctional phi29 pRNA nanoparticles for specific delivery of siRNA and other therapeutics to targeted cells. Methods. 2011;54:204–214. doi: 10.1016/j.ymeth.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bakhshinejad B, Sadeghizadeh M. Bacteriophages as vehicles for gene delivery into mammalian cells: prospects and problems. Expert opinion on drug delivery. 2014;11:1561–1574. doi: 10.1517/17425247.2014.927437. [DOI] [PubMed] [Google Scholar]
- 141.Panyam J, Labhasetwar V. Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Advanced drug delivery reviews. 2003;55:329–347. doi: 10.1016/s0169-409x(02)00228-4. [DOI] [PubMed] [Google Scholar]
- 142.Farokhzad OC, Langer R. Impact of nanotechnology on drug delivery. ACS nano. 2009;3:16–20. doi: 10.1021/nn900002m. [DOI] [PubMed] [Google Scholar]
- 143.Zhang XQ, Xu X, Bertrand N, Pridgen E, Swami A, Farokhzad OC. Interactions of nanomaterials and biological systems: Implications to personalized nanomedicine. Advanced drug delivery reviews. 2012;64:1363–1384. doi: 10.1016/j.addr.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Merril CR, Biswas B, Carlton R, Jensen NC, Creed GJ, Zullo S, Adhya S. Long-circulating bacteriophage as antibacterial agents. Proceedings of the National Academy of Sciences. 1996;93:3188–3192. doi: 10.1073/pnas.93.8.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Benson SD, Bamford JK, Bamford DH, Burnett RM. Does common architecture reveal a viral lineage spanning all three domains of life? Molecular cell. 2004;16:673–685. doi: 10.1016/j.molcel.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 146.Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. Journal of virology. 2005;79:14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nanocarriers as an emerging platform for cancer therapy. Nature nanotechnology. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 148.Yildiz I, Shukla S, Steinmetz NF. Applications of viral nanoparticles in medicine. Current opinion in biotechnology. 2011;22:901–908. doi: 10.1016/j.copbio.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Khalaj-Kondori M, Sadeghizadeh M, Behmanesh M, Saggio I, Monaci P. Chemical coupling as a potent strategy for preparation of targeted bacteriophage-derived gene nanocarriers into eukaryotic cells. The journal of gene medicine. 2011;13:622–631. doi: 10.1002/jgm.1617. [DOI] [PubMed] [Google Scholar]
- 150.Huan R, Steinmetz N, Fu C, Manchester M, Johnson J. Transferrin-mediated targeting of bacteriophage HK97 nanoparticles into tumour cells. Nanomedicine (Lond) 2011;6:55–68. doi: 10.2217/nnm.10.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Suthiwangcharoen N, Li T, Li K, Thompson P, You S, Wang Q. M13 bacteriophage-polymer nanoassemblies as drug delivery vehicles. Nano Research. 2011;4:483–493. [Google Scholar]
- 152.Koudelka KJ, Ippoliti S, Medina E, Shriver LP, Trauger SA, Catalano CE, Manchester M. Lysine Addressability and Mammalian Cell Interactions of Bacteriophage λ Procapsids. Biomacromolecules. 2013;14:4169–4176. doi: 10.1021/bm401577f. [DOI] [PubMed] [Google Scholar]
- 153.KHALAJ-KONDORI M, KAVOOSI M, RAHMATI-YAMCHI M, KADIVAR M. Preparation of a transferrin-targeted M13-based gene nanocarrier and evaluation of its efficacy for gene delivery and expression in eukaryote cells. Turkish Journal of Biology. 2016:40. [Google Scholar]
- 154.Choi DS, Jin HE, Yoo SY, Lee SW. Cyclic RGD peptide incorporation on phage major coat proteins for improved internalization by HeLa Cells. Bioconjugate chemistry. 2014;25:216–223. doi: 10.1021/bc4003234. [DOI] [PubMed] [Google Scholar]
- 155.Loi M, Di Paolo D, Soster M, Brignole C, Bartolini A, Emionite L, Sun J, Becherini P, Curnis F, Petretto A. Novel phage display-derived neuroblastoma-targeting peptides potentiate the effect of drug nanocarriers in preclinical settings. Journal of Controlled Release. 2013;170:233–241. doi: 10.1016/j.jconrel.2013.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kang J, Zhao G, Lin T, Tang S, Xu G, Hu S, Bi Q, Guo C, Sun L, Han S. A peptide derived from phage display library exhibits anti-tumor activity by targeting GRP78 in gastric cancer multidrug resistance cells. Cancer letters. 2013;339:247–259. doi: 10.1016/j.canlet.2013.06.016. [DOI] [PubMed] [Google Scholar]
- 157.Li J, Zhang Q, Pang Z, Wang Y, Liu Q, Guo L, Jiang X. Identification of peptide sequences that target to the brain using in vivo phage display. Amino acids. 2012;42:2373–2381. doi: 10.1007/s00726-011-0979-y. [DOI] [PubMed] [Google Scholar]
- 158.Bedi D, Musacchio T, Fagbohun OA, Gillespie JW, Deinnocentes P, Bird RC, Bookbinder L, Torchilin VP, Petrenko VA. Delivery of siRNA into breast cancer cells via phage fusion protein-targeted liposomes, Nanomedicine: Nanotechnology. Biology and Medicine. 2011;7:315–323. doi: 10.1016/j.nano.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lee HJ, Namgung R, Kim WJ, Kim JI, Park IK. Targeted Delivery of MicroRNA-145 to Metastatic breast cancer by peptide conjugated branched PEI gene carrier. Macromolecular Research. 2013;21:1201–1209. [Google Scholar]
- 160.Bedi D, Gillespie JW, Petrenko VA, Jr, Ebner A, Leitner M, Hinterdorfer P, Petrenko VA. Targeted delivery of siRNA into breast cancer cells via phage fusion proteins. Molecular pharmaceutics. 2013;10:551–559. doi: 10.1021/mp3006006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nam HY, Kim J, Kim SW, Bull DA. Cell targeting peptide conjugation to siRNA polyplexes for effective gene silencing in cardiomyocytes. Molecular pharmaceutics. 2012;9:1302–1309. doi: 10.1021/mp200589z. [DOI] [PubMed] [Google Scholar]
- 162.Matsuo AL, Juliano MA, Figueiredo CR, Batista WL, Tanaka AS, Travassos LR. A new phage-display tumor-homing peptide fused to antiangiogenic peptide generates a novel bioactive molecule with antimelanoma activity. Molecular Cancer Research. 2011;9:1471–1478. doi: 10.1158/1541-7786.MCR-10-0501. [DOI] [PubMed] [Google Scholar]
- 163.Kuo C-H, Leon L, Chung EJ, Huang R-T, Sontag TJ, Reardon CA, Getz GS, Tirrell M, Fang Y. Inhibition of atherosclerosis-promoting microRNAs via targeted polyelectrolyte complex micelles. Journal of Materials Chemistry B. 2014 doi: 10.1039/C4TB00977K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Jiang Y, Yang N, Zhang H, Sun B, Hou C, Ji C, Zheng J, Liu Y, Zuo P. Enhanced in vivo antitumor efficacy of dual-functional peptide-modified docetaxel nanoparticles through tumor targeting and Hsp90 inhibition. Journal of Controlled Release. 2016;221:26–36. doi: 10.1016/j.jconrel.2015.11.029. [DOI] [PubMed] [Google Scholar]
- 165.Jayanna PK, Bedi D, Gillespie JW, DeInnocentes P, Wang T, Torchilin VP, Bird RC, Petrenko VA. Landscape phage fusion protein-mediated targeting of nanomedicines enhances their prostate tumor cell association and cytotoxic efficiency. Nanomedicine: Nanotechnology, Biology and Medicine. 2010;6:538–546. doi: 10.1016/j.nano.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wang T, D’Souza GG, Bedi D, Fagbohun OA, Potturi LP, Papahadjopoulos-Sternberg B, Petrenko VA, Torchilin VP. Enhanced binding and killing of target tumor cells by drug-loaded liposomes modified with tumor-specific phage fusion coat protein. Nanomedicine. 2010;5:563–574. doi: 10.2217/nnm.10.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang T, Kulkarni N, Bedi D, D’Souza GG, Papahadjopoulos-Sternberg B, Petrenko VA, Torchilin VP. In vitro optimization of liposomal nanocarriers prepared from breast tumor cell specific phage fusion protein. Journal of drug targeting. 2011;19:597–605. doi: 10.3109/1061186X.2010.550920. [DOI] [PubMed] [Google Scholar]
- 168.Wang T, Hartner WC, Gillespie JW, Praveen KP, Yang S, Mei LA, Petrenko VA, Torchilin VP. Enhanced tumor delivery and antitumor activity in vivo of liposomal doxorubicin modified with MCF-7-specific phage fusion protein. Nanomedicine: Nanotechnology Biology and Medicine. 2014;10:421–430. doi: 10.1016/j.nano.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ortiz R, Prados J, Melguizo C, Arias JL, Ruiz MA, Alvarez PJ, Caba O, Luque R, Segura A, Aranega A. 5-Fluorouracil-loaded poly (ε-caprolactone) nanoparticles combined with phage E gene therapy as a new strategy against colon cancer. International journal of nanomedicine. 2012;7:95. doi: 10.2147/IJN.S26401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wang T, Kulkarni N, D’Souza GG, Petrenko VA, Torchilin VP. On the mechanism of targeting of phage fusion protein-modified nanocarriers: only the binding peptide sequence matters. Molecular pharmaceutics. 2011;8:1720–1728. doi: 10.1021/mp200080h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Dorval Courchesne NM, Parisien A, Lan CQ. Production and application of bacteriophage and bacteriophage-encoded lysins. Recent patents on biotechnology. 2009;3:37–45. doi: 10.2174/187220809787172678. [DOI] [PubMed] [Google Scholar]
- 172.Merabishvili M, Pirnay JP, Verbeken G, Chanishvili N, Tediashvili M, Lashkhi N, Glonti T, Krylov V, Mast J, Van Parys L. Quality-controlled small-scale production of a well-defined bacteriophage cocktail for use in human clinical trials. PloS one. 2009;4:e4944. doi: 10.1371/journal.pone.0004944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Fenton M, McAuliffe O, O’Mahony J, Coffey A. Recombinant bacteriophage lysins as antibacterials. Bioengineered bugs. 2010;1:9–16. doi: 10.4161/bbug.1.1.9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Gill JJ, Hyman P. Phage choice, isolation, and preparation for phage therapy. Current pharmaceutical biotechnology. 2010;11:2–14. doi: 10.2174/138920110790725311. [DOI] [PubMed] [Google Scholar]
- 175.Freire-Moran L, Aronsson B, Manz C, Gyssens IC, So AD, Monnet DL, Cars O, Group EEW. Critical shortage of new antibiotics in development against multidrug-resistant bacteria—time to react is now. Drug resistance updates. 2011;14:118–124. doi: 10.1016/j.drup.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 176.Laxminarayan R, Duse A, Wattal C, Zaidi AKM, Wertheim HFL, Sumpradit N, Vlieghe E, Hara GL, Gould IM, Goossens H. Antibiotic resistance—the need for global solutions. The Lancet infectious diseases. 2013;13:1057–1098. doi: 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- 177.Lipinski CA. Drug-like properties and the causes of poor solubility and poor permeability. Journal of pharmacological and toxicological methods. 2000;44:235–249. doi: 10.1016/s1056-8719(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 178.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced drug delivery reviews. 2012;64:4–17. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 179.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nature Reviews Drug Discovery. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 180.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiology and Molecular Biology Reviews. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gwynn MN, Portnoy A, Rittenhouse SF, Payne DJ. Challenges of antibacterial discovery revisited. Annals of the New York Academy of Sciences. 2010;1213:5–19. doi: 10.1111/j.1749-6632.2010.05828.x. [DOI] [PubMed] [Google Scholar]
- 182.Cushnie TPT, Lamb AJ. Recent advances in understanding the antibacterial properties of flavonoids. International journal of antimicrobial agents. 2011;38:99–107. doi: 10.1016/j.ijantimicag.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 183.Miller ES, Kutter E, Mosig G, Arisaka F, Kunisawa T, Rüger W. Bacteriophage T4 genome. Microbiology and molecular biology reviews. 2003;67:86–156. doi: 10.1128/MMBR.67.1.86-156.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Roucourt B, Lavigne R. The role of interactions between phage and bacterial proteins within the infected cell: a diverse and puzzling interactome. Environmental microbiology. 2009;11:2789–2805. doi: 10.1111/j.1462-2920.2009.02029.x. [DOI] [PubMed] [Google Scholar]
- 185.Forsyth R, Haselbeck RJ, Ohlsen KL, Yamamoto RT, Xu H, Trawick JD, Wall D, Wang L, Brown-Driver V, Froelich JM. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Molecular microbiology. 2002;43:1387–1400. doi: 10.1046/j.1365-2958.2002.02832.x. [DOI] [PubMed] [Google Scholar]
- 186.Knuth K, Niesalla H, Hueck CJ, Fuchs TM. Large-scale identification of essential Salmonella genes by trapping lethal insertions. Molecular microbiology. 2004;51:1729–1744. doi: 10.1046/j.1365-2958.2003.03944.x. [DOI] [PubMed] [Google Scholar]
- 187.Lange RP, Locher HH, Wyss PC, Then RL. The targets of currently used antibacterial agents: lessons for drug discovery. Current pharmaceutical design. 2007;13:3140–3154. doi: 10.2174/138161207782110408. [DOI] [PubMed] [Google Scholar]
- 188.Baba T, Huan H-C, Datsenko K, Wanner BL, Mori H. Microbial Gene Essentiality: Protocols and Bioinformatics. Springer; 2008. The applications of systematic in-frame, single-gene knockout mutant collection of Escherichia coli K-12; pp. 183–194. [DOI] [PubMed] [Google Scholar]
- 189.Silver LL. Challenges of antibacterial discovery. Clinical microbiology reviews. 2011;24:71–109. doi: 10.1128/CMR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Brown ED. Drugs against superbugs: private lessons from bacteriophages. Trends in biotechnology. 2004;22:434–436. doi: 10.1016/j.tibtech.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 191.Krom RJ, Bhargava P, Lobritz MA, Collins JJ. Engineered Phagemids for Nonlytic, Targeted Antibacterial Therapies. Nano letters. 2015;15:4808–4813. doi: 10.1021/acs.nanolett.5b01943. [DOI] [PubMed] [Google Scholar]
- 192.Broxmeyer L, Sosnowska D, Miltner E, Chacón O, Wagner D, McGarvey J, Barletta RG, Bermudez LE. Killing of Mycobacterium avium and Mycobacterium tuberculosis by a mycobacteriophage delivered by a nonvirulent mycobacterium: a model for phage therapy of intracellular bacterial pathogens. Journal of Infectious Diseases. 2002;186:1155–1160. doi: 10.1086/343812. [DOI] [PubMed] [Google Scholar]
- 193.Barry MA, Dower WJ, Johnston SA. Toward cell targeting gene therapy vectors: Selection of cell binding peptides from random peptide presenting phage libraries. Nature medicine. 1996;2:299–305. doi: 10.1038/nm0396-299. [DOI] [PubMed] [Google Scholar]
- 194.Dunn IS. Mammalian cell binding and transfection mediated by surface-modified bacteriophage lambda. Biochimie. 1996;78:856–861. doi: 10.1016/s0300-9084(97)84338-6. [DOI] [PubMed] [Google Scholar]
- 195.Podgornik A, Janez N, Smrekar F, Peterka M. Continuous Production of Bacteriophages. Continuous Processing in Pharmaceutical Manufacturing. 2014:297–338. [Google Scholar]
- 196.Li J, Feng L, Jiang X. In vivo phage display screen for peptide sequences that cross the blood cerebrospinal-fluid barrier. Amino acids. 2015;47:401–405. doi: 10.1007/s00726-014-1874-0. [DOI] [PubMed] [Google Scholar]
- 197.Clark JR, March JB. Bacteriophages and biotechnology: vaccines, gene therapy and antibacterials. Trends in biotechnology. 2006;24:212–218. doi: 10.1016/j.tibtech.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 198.Clark JR. Bacteriophage therapy: history and future prospects. Future Virology. 2015;10:449–461. [Google Scholar]
- 199.Larocca D, Witte A, Johnson W, Pierce GF, Baird A. Targeting bacteriophage to mammalian cell surface receptors for gene delivery. Human gene therapy. 1998;9:2393–2399. doi: 10.1089/hum.1998.9.16-2393. [DOI] [PubMed] [Google Scholar]
- 200.Larocca D, Kassner PD, Witte A, Ladner RC, Pierce GF, Baird A. Gene transfer to mammalian cells using genetically targeted filamentous bacteriophage. The FASEB journal. 1999;13:727–734. doi: 10.1096/fasebj.13.6.727. [DOI] [PubMed] [Google Scholar]
- 201.Seow Y, Wood MJ. Biological gene delivery vehicles: beyond viral vectors. Molecular Therapy. 2009;17:767–777. doi: 10.1038/mt.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Tan WS, Ho KL. Phage display creates innovative applications to combat hepatitis B virus. World journal of gastroenterology: WJG. 2014;20:11650. doi: 10.3748/wjg.v20.i33.11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Hart SL, Knight AM, Harbottle RP, Mistry A, Hunger HD, Cutler DF, Williamson R, Coutelle C. Cell binding and internalization by filamentous phage displaying a cyclic Arg-Gly-Asp-containing peptide. Journal of Biological Chemistry. 1994;269:12468–12474. [PubMed] [Google Scholar]
- 204.Sperinde JJ, Choi SJ, Szoka FC. Phage display selection of a peptide DNase II inhibitor that enhances gene delivery. The journal of gene medicine. 2001;3:101–108. doi: 10.1002/jgm.165. [DOI] [PubMed] [Google Scholar]
- 205.Piersanti S, Cherubini G, Martina Y, Salone B, Avitabile D, Grosso F, Cundari E, Di Zenzo G, Saggio I. Mammalian cell transduction and internalization properties of λ phages displaying the full-length adenoviral penton base or its central domain. Journal of molecular medicine. 2004;82:467–476. doi: 10.1007/s00109-004-0543-2. [DOI] [PubMed] [Google Scholar]
- 206.Becerril B, Poul MA, Marks JD. Toward selection of internalizing antibodies from phage libraries. Biochemical and biophysical research communications. 1999;255:386–393. doi: 10.1006/bbrc.1999.0177. [DOI] [PubMed] [Google Scholar]
- 207.Ivanenkov VV, Felici F, Menon AG. Uptake and intracellular fate of phage display vectors in mammalian cells. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research. 1999;1448:450–462. doi: 10.1016/s0167-4889(98)00162-1. [DOI] [PubMed] [Google Scholar]
- 208.Larocca D, Baird A. Receptor-mediated gene transfer by phage-display vectors: applications in functional genomics and gene therapy. Drug discovery today. 2001;6:793–801. doi: 10.1016/s1359-6446(01)01837-2. [DOI] [PubMed] [Google Scholar]
- 209.Burg MA, Jensen-Pergakes K, Gonzalez AM, Ravey P, Baird A, Larocca D. Enhanced phagemid particle gene transfer in camptothecin-treated carcinoma cells. Cancer research. 2002;62:977–981. [PubMed] [Google Scholar]
- 210.Babalova EG, Katsitadze KT, Sakvarelidze LA, Imnaishvili N, Sharashidze TG, Badashvili VA, Kiknadze GP, Me pariani AN, Gendzekhadze ND, Machavariani EV. Preventive value of dried dysentery bacteriophage. Zhurnal mikrobiologii, epidemiologiii immunobiologii. 1968;45:143–145. [PubMed] [Google Scholar]
- 211.Stroj L, Weber-Dabrowska B, Partyka K, Mulczyk M, Wojcik M. Successful treatment with bacteriophage in purulent cerebrospinal meningitis in a newborn. Neurologia I neurochirurgia polska. 1998;33:693–698. [PubMed] [Google Scholar]
- 212.Cisło M, Dabrowski M, Weber-Dabrowska B, Woytoń A. Bacteriophage treatment of suppurative skin infections. Archivum immunologiae et therapiae experimentalis. 1986;35:175–183. [PubMed] [Google Scholar]
- 213.Kaczkowski H, Weber-Dabrowska B, Dabrowski M, Zdrojewicz Z, Cwioro F. Use of bacteriophages in the treatment of chronic bacterial diseases. Wiadomości lekarskie (Warsaw, Poland: 1960) 1990;43:136. [PubMed] [Google Scholar]
- 214.Pavlenishvili I, Tsertsvadze T. Bacteriophagotherapy and enterosorbtion in treatment of sepsis of newborns caused by gram-negative bacteria. Pren Neon Infect. 1993;11:104. [Google Scholar]
- 215.Weber-Dabrowska B, Mulczyk M, Górski A. Bacteriophage therapy for infections in cancer patients. Clinical and Applied Immunology Reviews. 2001;1:131–134. [Google Scholar]
- 216.Abdul-Hassan HS, El-Tahan E, Massoud B, Gomaa R. Bacteriophage therapy of Pseudomonas burn wound sepsis. Ann Medit Burn Club. 1990;3:262–264. [Google Scholar]
- 217.Markoishvili K, Tsitlanadze G, Katsarava R, Glenn J, Sulakvelidze A. A novel sustained-release matrix based on biodegradable poly (ester amide) s and impregnated with bacteriophages and an antibiotic shows promise in management of infected venous stasis ulcers and other poorly healing wounds. International journal of dermatology. 2002;41:453–458. doi: 10.1046/j.1365-4362.2002.01451.x. [DOI] [PubMed] [Google Scholar]
- 218.Jikia D, Chkhaidze N, Imedashvili E, Mgaloblishvili I, Tsitlanadze G, Katsarava R, Glenn Morris J, Sulakvelidze A. The use of a novel biodegradable preparation capable of the sustained release of bacteriophages and ciprofloxacin, in the complex treatment of multi-drug-resistant Staphylococcus aureus-infected local radiation injuries caused by exposure to Sr90. Clinical and experimental dermatology. 2005;30:23–26. doi: 10.1111/j.1365-2230.2004.01600.x. [DOI] [PubMed] [Google Scholar]