Abstract
Periodontitis is a biofilm-induced inflammatory disease characterized by dysbiosis of the commensal periodontal microbiota. It is unclear how natural regulation of inflammation affects the periodontal biofilm. Promoters of active resolution of inflammation including Resolvin E1 (RvE1) effectively treat inflammatory periodontitis in animal models. The goals of this study were 1) to compare periodontal tissue gene expression in different clinical conditions, 2) to determine the impact of local inflammation on the composition of subgingival bacteria, and 3) to understand how inflammation impacts these changes. Two clinically-relevant experiments were performed in rats: prevention and treatment of ligature-induced periodontitis with RvE1 topical treatment. The gingival transcriptome was evaluated by RNA-seq sequencing of mRNA. The composition of the subgingival microbiota was characterized by 16S rDNA sequencing. Periodontitis was assessed by bone morphometric measurements and histomorphometry of block sections. H&E and, tartrate resistant acid phosphatase staining were used to characterize and quantify inflammatory changes. RvE1 treatment prevented bone loss in ligature induced periodontitis. Osteoclast density and inflammatory cell infiltration in the RvE1 groups were lower than those in the placebo group. RvE1 treatment reduced expression of inflammation-related genes returning the expression profile to one more similar to health. Treatment of established periodontitis with RvE1 reversed bone loss, reversed inflammatory gene expression and reduced osteoclast density. Assessment of the rat subgingival microbiota after RvE1 treatment revealed marked changes in both prevention and treatment experiments. The data suggest that modulation of local inflammation has a major role in shaping the composition of the subgingival microbiota.
Introduction
Chronic periodontitis is a multifactorial inflammatory disease with high prevalence in many populations (1). Periodontitis is characterized by dysbiosis of the commensal microbiota associated with inflammation; however, the temporal sequence and interplay between specific bacteria and inflammation has not been described (2). Mechanical debridement has long been the standard treatment for periodontitis (3). It is believed that removal of subgingival plaque is sufficient to reduce the inflammatory response stimulated by bacteria and stop tissue destruction. However, subgingival debridement has limitations and cleansing of diseased areas is often incomplete (4). Regardless of completeness of debridement, bacteria grow back gradually after mechanical therapy in the absence of further treatment or maintenance and the disease associated hyper-inflammation rapidly returns (5). Adjunctive use of antibiotics improves clinical outcomes, but antibiotic resistance and other side effects are always a concern (6).
In recent years, it has become apparent that susceptibility to and pathogenesis of periodontal disease are mediated by the host (7–10). Subjects with severe periodontitis exhibit measurable excess inflammation that includes cytokine production (11) and oxidative stress (12, 13), and the presence of inflammation predicts diseases progression (14, 15). Early studies attempting to control the host inflammatory response have shown that COX inhibitors slow disease progression without modifying bacterial composition in animal studies (16–18) and clinical studies (19–21), but the long term use of cyclooxygenase inhibitors has serious side effects, such as increased risks of gastrointestinal tract bleeding and cardiovascular diseases (22), that preclude their prolonged clinical use for periodontitis. There is a critical need to develop rational and safe host modulation therapies to prevent and treat periodontitis.
Specialized pro-resolving mediators (SPMs), including lipoxins, resolvins, protectins, and maresins, play an important role in the resolution phase of acute inflammation (23). In the resolution phase, SPMs actively reduce neutrophil infiltration, promote neutrophil apoptosis (24), and recruit non-phlogistic macrophages to clear apoptotic neutrophils and remaining bacteria signaling through specific G protein-coupled receptors. To complete the resolution phase, macrophages are themselves cleared by efferocytosis (25, 26). SPMs, which are inflammation regulators rather than inhibitors, can replace NSAIDs in treating inflammation by actively inducing and enhancing resolution of inflammation. Resolvin E1 (RvE1), one of the resolvins derived from the omega-3 polyunsaturated fatty acid, eicosapentaenoic acid (EPA), has been shown to be effective in treating periodontitis in animals (27). RvE1 promotes inflammation resolution (28) and regulates bone remodeling (29, 30). In rabbits, local application of RvE1 prevents the onset of periodontitis (31) and promotes regeneration of the periodontium (27). An interesting observation in these studies was the apparently spontaneous disappearance of the periodontal pathogen P. gingivalis without the use of mechanical or antimicrobial therapy (27). The dynamics of microbial shifts resulting from resolution of inflammation that permit tissue regeneration following RvE1 application are not clear.
The goal of this study was to begin to understand the temporal dynamics of inflammation induced dysbiosis of the periodontal microbiota. To accomplish this goal, periodontitis was induced in the rat using ligatures to observe changes in the microbiota and the host without the addition of exogenous pathogens. Global differential gene expression in periodontal tissues was evaluated in health, disease and after treatment with RvE1 to provide an unbiased assessment of inflammatory changes. In parallel, 16S rDNA sequencing was employed in the same experiments to determine the impact of local inflammation on the composition of subgingival bacteria. The data suggest a dynamic, temporal interaction between local inflammation and the composition of the periodontal microbiota.
Materials and Methods
Experimental Periodontitis
The impact of inflammation on the subgingival microbiota in ligature induced periodontitis in the rat was assessed in two separate experiments: prevention of ligature induced periodontitis (prevention experiment) and treatment of established periodontitis (treatment experiment). The protocols were approved by Institutional Animal Care and Use Committee (IACUC) of the Forsyth Institute. Six-week old male Wistar rats (weight 180–200 g, Charles River Laboratories, Wilmington, MA) were maintained in a controlled temperature (22°C to 25° C) and dark/light cycle (12/12 hours) facility. Rats received standard laboratory chow diet and water ad libitum. Rats were sedated with 2–3% isoflurane and anesthetized with ketamine (80mg/kg, intraperitoneally) and xylazine (16 mg/kg, intraperitoneally) for all procedures. In the prevention experiment, 16 rats were divided into four groups (no ligature (health), ligature + vehicle (disease), ligature + RvE1 (0.1μg/μl), ligature + RvE1 (0.5μg/μl)). Two μl vehicle alone, RvE1 at 0.1μg/μl (0.28mM), or 0.5μg/μl (1.4mM) was topically applied with a Hamilton syringe (25G needle, 5μl syringe) three times a week (M, W, F). 5–0 silk ligatures were placed subgingivally on the maxillary right and left second molars of all rats. The experimental period was 4 weeks. Ligatures were checked at every application. RvE1 (FW: 350.5; purity > 97%, λmax: 272 nm) was obtained from Cayman Biochemicals. Untreated, unligatured animals were sacrificed to obtain healthy tissue samples. Subgingival plaque samples were collected on day 0, 7, 10, 14, 17, 21, 24, and 28. Rats were euthanized by CO2 inhalation; gingival tissue and maxillae were harvested.
In the treatment experiment, 18 rats were divided into four groups (no ligature (health): n=6, ligature alone: n=6, ligature + vehicle: n=3, ligature + RvE1: n=3). 3–0 silk ligatures were placed subgingivally on the maxillary right and left second molars at baseline. The entire experimental period lasted six weeks beginning with a three-week disease induction phase followed by the subsequent three-week treatment phase. Six rats in the no ligature group provided healthy tissue. At the end of the disease induction phase (21st day), the six rats in the ligature alone group were sacrificed to quantify the amount of disease induced. During the treatment phase (weeks four to six), 4 μl of a 0.25 μg/μl solution of RvE1 (0.7mM) was topically applied to ligated teeth (RvE1 group) every other day. 4 μl application of vehicle alone following the same regimen served as the treatment control. The dose was based on the estimated optimal dose determined in the prevention experiment. At the end of week six, remaining rats were sacrificed. Subgingival plaque samples were collected on day 0, 8, 12, 16, 20, 24, 28, 32, 36, and 40.
Morphometric analysis
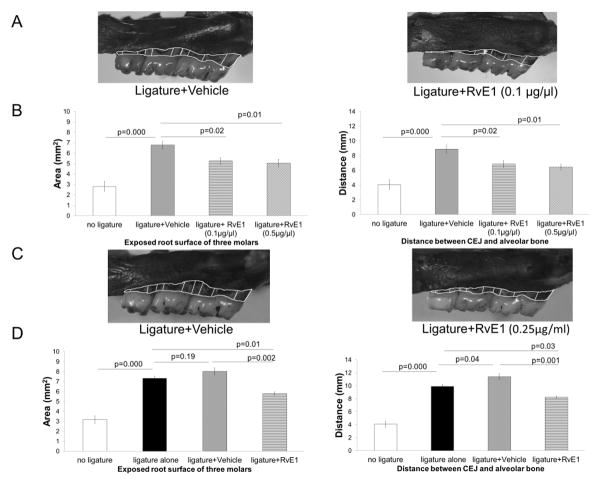
The dissected maxillae were defleshed, stored in 0.3% hydrogen peroxide for 24 hours and dried completely before methylene blue staining. Images (0.63X10) were taken under dissecting microscope (Axio observer A1, ZEISS) using AxioVision 4.8 software. The area of exposed root surface of three maxillary molars was measured buccally and palatally (Fig. 1). The distance between the alveolar bone and cementoenamel junction (CEJ) was measured at nine sites as indicated of three maxillary molars at the buccal site and the palatal site along the axis of the tooth root (Fig. 1) using computer software (ImageJ).
Figure 1. RvE1 treatment prevents and reverses alveolar bone loss.
(A) Prevention of periodontitis: alveolar bone loss in the disease (vehicle + ligature) group prevented by RvE1 (0.1μg/μl). The area of exposed root surface and the distance between CEJ and alveolar bone level are outlined. (B) The area of bone loss in mm2 and linear bone loss in distance from the CEJ to bone in mm are significantly reduced (~30–40%) in both the RvE1 groups (0.1μg/μl) and (0.5μg/μl). (n=4 in the no ligature group, the ligature + vehicle group, and the ligature+ RvE1 (0.1μg/μl) group; n=3 in the ligature+RvE1 (0.5μg/μl) group). (C) Regeneration of alveolar bone: established periodontitis was treated by topical application of RvE1 as a monotherapy. ~30–40% of lost bone was restored with RvE1 therapy. (D) The area of bone loss in mm2 and linear bone loss in distance from the CEJ to bone in mm are significantly reduced (~30–40%) with RvE1 treatment, but alveolar bone loss in the vehicle group worsened. (n=6 in the no ligature group and the ligature alone group; n=3 in the ligature + vehicle group and the ligature + RvE1 group).
Histomorphometry
Half of the maxilla was decalcified in 20 volumes of 10% EDTA and the decalcification was confirmed by calcium oxalate precipitation. Specimens were dehydrated in increasing concentrations of ethanol sequentially, and embedded in paraffin. Serial mesiodistal sections (6 mm) parallel to the long axis of the teeth were cut and stained with H&E and identification of the cellular composition, or tartrate-resistant acid phosphatase to quantify osteoclastic activity with light microscopy.
Similar anatomic positions were selected for quantitative measurements and identification of samples was masked to avoid bias. The number of neutrophils, lymphocytes and plasma cells was counted at 400X. The total number of multinucleated osteoclasts in the interproximal bone was counted at 200X and osteoclast density expressed as number of osteoclasts per μm2 using computer software (ImageJ).
RNA-Seq and differential expression analysis
Gingival samples were collected from two rats in each group from the prevention experiment and processed for RNA sequencing (RNA-Seq) on the Illumina platform. Total RNA was extracted from homogenized gingival tissue with an RNeasy® Mini kit (QIAGEN Inc., Valencia, CA). RNA quality was assessed with the Agilent RNA 6000 NanoLabChip Kit of the Bioanalyzer system (Santa Clara, CA, USA). RNA-Seq cDNA libraries were made using the Illumina Truseq stranded mRNA Sample Prep kit and were sequenced using the NextSeq v3, 2 x 75 cycle cartridge on a NextSeq sequencer. The RNA-Seq reads were uploaded to the Illumina BaseSpace cloud platform (http://basespace.illumina.com) and the RNAExpress application was used to align reads to the rat genome (Rattus norvegicus UCSC version rn5), calculate read count per gene using the STAR aligner (32), and perform differential gene expression analysis with DESeq2 (33). The raw RNA sequencing reads were also submitted to the National Center for Biotechnology Information Sequence Read Archive database (http://www.ncbi.nlm.nih.gov/Traces/sra/, BioProject number PRJNA321482).
The iPathwayGuide was used to score the biological pathways using impact analysis (34, 35). Impact analysis uses two types of evidence: i) the overrepresentation of differentially expressed (DE) genes in a given pathway and ii) the perturbation of that pathway computed by propagating the measured expression changes across the pathway topology. These aspects are captured by two independent probability values, pORA representing the probability of obtaining a number of DE genes on the given pathway greater or equal to chance observations (36), and pAcc calculated based on the amount of perturbation measured in each pathway. pORA and pAcc are then combined in a unique global p-value using Fisher's method. This p-value is then corrected for multiple comparisons using the False Discovery Rate (FDR) method.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
qRT-PCR was utilized to confirm the results of RNA-seq. A total of 1 μg RNA from each sample assessed in RNA-seq was converted to cDNA using a high-capacity cDNA reverse transcriptase kit (Applied Biosystems, Grand Island, NY). qRT-PCR was performed using primers and TaqMan probes for ten differentially expressed genes (Acp5, Angptl4, Ccl9, Cxcl1, Cxcl2, Ide, Il1b, Mmp13, Nfkibia, Rac2) in RNA-seq (Applied Bio-systems, TaqMan gene expression assays). β-actin (Actb) was the internal standard amplified using preformulated TaqMan probes (Applied Biosystems, Endogenous Control). Quantification was performed in an automated thermal cycler (StepOnePlus™ System, Applied Biosystems); 50°C for 2 min (one cycle), 95°C for 20 sec (one cycle), 95°C for 1 sec, and 60°C for 20 sec (40 cycles) and relative expression (2−Δ ΔCT) calculated.
RNA-seq was not performed on samples from the treatment experiment, but qRT-PCR was used to assess expression of candidate genes. Total RNA was extracted from homogenized gingival tissue with Trizol reagent (Invitrogen, Grand Island, NY). The concentration and purity of RNA was estimated spectrophotometrically (ratio of absorbance at 260 and 280 nm: NanoDrop 2000c, Thermo Fisher Scientific, Waltham, MA). Total RNA was reversely transcribed to cDNA and cDNA was quantified following the same protocol utilized in the prevention experiment. Three inflammation related genes, Cxcl1 (IL-8 equivalent), Nos2 (inducible nitric oxide synthase), and Ptgs2 (COX2) were assessed and β-actin (Actb) was the internal standard (Applied Bio-systems, TaqMan gene expression assays).
Bacterial DNA extraction and Amplification
Subgingival plaque samples were collected using the tip of a 2-Whiteside scaler (Hu-Friedy, Chicago, IL) from the palatal side of the maxillary second molars. DNA was extracted using MolYsis Basic (CaerusBio Inc., Downingtown, PA) and QIAamp® mini kits (QIAGEN Inc., Valencia, CA). The MolYsis Basic kit excludes DNA of mammalian cells (37, 38). The extracted bacterial DNA was amplified with a multiple displacement amplification (MDA) kit (GenomiPhi™V3 DNA amplification kit, GE Healthcare Bio-Sciences, Pittsburgh, PA) enabling whole genomic amplification of DNA targets (39, 40). The concentration of amplified DNA samples was measured with fluorescent nucleic acid stain (Quant-iT™ PicoGreen® dsDNA Assay, Life Technologies, Grand Island, NY).
16S rDNA sequencing
The microbial composition of plaque samples (50ng DNA/10μl) was characterized by sequencing the hyper-variable V3 and V4 regions of the 16S rDNA gene using the Illumina MiSeq® platform. The 2 x 250-bp paired-end sequencing allows the capture of the entire 440-bp V3-V4 region by merging the read pairs based on the overlap. Successfully merged reads were processed through the QIIME pipeline (41) using a 97% identity threshold and the SILVA reference database (SILVA/Greengenes) (42, 43). Taxonomic assignments to operational taxonomic units (OTUs) from phylum to genus level were completed through QIIME’s use of the given reference database (SILVA/Greengenes) and the UCLUST algorithm (44). To understand the diversity and composition of microbial community, further analyses included alpha diversity (e.g. richness), beta diversity (weighted UniFrac) (45), principal coordinate analysis (PCoA) and relative abundance profiles of microbial community in the samples. The relative abundance of taxa was determined by dividing the hits of a specific taxon by the total hits of all taxa in the sample.
Statistical Analysis
In the prevention experiment, sample size was calculated based on published data (27) and preliminary results. We hypothesized that the change in exposed root surface in disease can be prevented 50% by treatment and the clinical effect is the same in the two treatment groups. At least three animals per group were needed in order to detect the difference in exposed root surface between four groups with 80% power. The sample size in the treatment experiment was also calculated based on the preliminary results and the assumption that the treatment can regenerate 50% of lost bone surface. At least two animals per group were needed to detect the difference in exposed surface between four groups with 80% power. Multiple comparisons of bone morphometry, histomorphometry, and relative gene expression were analyzed by one-way analysis of variance (ANOVA) and post hoc analysis (pairwise t-test). All values are expressed as mean ± standard error of the mean. P-value <0.05 was considered statistically significant
For 16S rDNA sequencing experiments, descriptive statistics are presented due to the exceptionally large number of potential comparisons. Values are expressed as mean ± standard error of the mean.
For RNA-seq, significance of differentially expressed genes was obtained using a threshold of 0.6 for absolute log2 expression change and 0.05 (unless otherwise stated) false discovery rate, or family-wise error rate (Benjamini–Hochberg procedure). Wald test on logarithmic transformed data was used to compare the gene expression level between two groups in each comparison. These data were analyzed in the context of pathways obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (release 73.0+/0316, March 15) (46, 47), the gene ontology from the Gene Ontology Consortium database (September 19, 2014) (48, 49), miRNAs from the miRBase (release 21) and MicroCosm Targets version 5 (updated March 2015) databases (50–53), and diseases from the KEGG database (release 73.0+/0316, March 15) (46, 47). Correlation between mean fold changes of genes in RNA-seq and mean fold changes of genes in qRT-PCR for each sample was assessed by a Spearman rank correlation coefficient.
Results
RvE1 treatment prevents and reveres alveolar bone loss
Ligature placement induced significant bone loss in both experiments (Fig. 1A, 1B, 1C, 1D). In the prevention experiment, prophylactic RvE1 treatment at low and high doses reduced alveolar bone loss by 1.51 and 1.73 mm2, respectively (95% confidence interval [CI]= (0.33, 2.70) and (0.40, 3.06); p=0.02 and 0.01, respectively compared with vehicle control) expressed as area of exposed root surface. The distance between the alveolar bone and CEJ was reduced by 2.03 and 2.45 mm, respectively (95% CI= (0.27, 3.80) and (0.59, 4.30); p=0.02 and 0.01, respectively compared with vehicle control) (Fig. 1A, 1B). In the treatment experiment, RvE1 treatment of established periodontitis reversed alveolar bone loss induced by ligature placement. Specifically, bone loss expressed as area of exposed root surface was reduced by 2.24 mm2 (95% CI= (1.11, 3.37), p=0.002 and by 3.17 mm in distance between the alveolar bone and CEJ compared with vehicle (95% CI= (1.69, 4.65), p=0.001, Fig. 1C, 1D).
RvE1 treatment inhibits inflammation and osteoclast activity
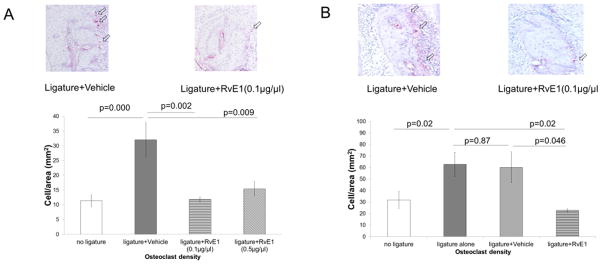
Ligature placement significantly increased inflammatory cell infiltration and osteoclast density (Fig. 2A, 2B; Supplemental Fig. 1B, 1D). Prophylactic RvE1 treatment significantly reduced inflammatory cell infiltration (Supplemental Fig. 1B) and osteoclast density (Fig. 2A) compared to vehicle at both doses. There was no significant difference between the two RvE1 doses (ANOVA; Fig. 2A).
Figure 2. RvE1 treatment controls osteoclast activity.
(A) Osteoclast density was defined as active osteoclast count (TRAP positive multinucleated osteoclasts) around the interproximal bone divided by the total area of the interproximal bone. Prevention of periodontitis: the osteoclast density in the RvE1 groups (0.1μg/μl) and (0.5μg/μl) is significantly lower than the ligature + vehicle group. (n=4 in the no ligature group, the ligature + vehicle group, and the ligature+ RvE1 (0.1μg/μl) group; n=3 in the ligature+RvE1 (0.5μg/μl) group). (B) Periodontal bone regeneration: RvE1 treatment reduced the osteoclast density by 60% compared to the vehicle group and the ligature alone group. (n=6 in the no ligature group and the ligature alone group; n=3 in the ligature + Vehicle group and the ligature + RvE1 group).
In the treatment experiment, the mean count of inflammatory cells in the RvE1 group trended lower (p=0.57) (Supplemental Fig.1D). RvE1 treatment significantly reduced osteoclast density compared to vehicle (Fig. 2B).
Impact of RvE1 treatment on periodontal tissue gene expression
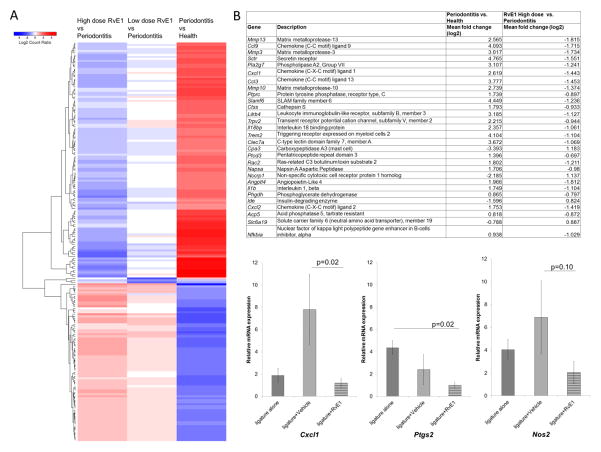
Fig. 3A, which illustrates the top 200 differentially expressed genes in the heat map comparing periodontitis versus health, and RvE1 versus periodontitis treatment at low and high doses in the prevention experiment, reveals that prophylactic RvE1 treatment appeared to prevent the gene expression pattern of the diseased state, being very similar to the expression pattern of health. A clear dose response was observed. Further analyses were done with the high RvE1 dose (0.5 μg/μl).
Figure 3. Impact of RvE1 on Periodontal Gene Expression.
(A) A heat map of top 200 differentially expressed genes (absolute log2 fold change) compares periodontitis vs. health, Low dose RvE1 (0.1μg/μl) vs. periodontitis, and High dose RvE1 (0.5μg/μl) vs. periodontitis. The color intensity reflects the log 2 fold change levels for each comparison with red for up- and blue for down-regulation. Genes (in rows) were ordered based on the dendrogram derived from hierarchical clustering of log2 values of all rows, therefore, genes with similar differential expression patterns were grouped together. (B) Genes differentially expressed (mean fold change ≥ 2^0.6, adjusted p-value <0.1) in the comparison between disease (ligature + vehicle) and health (no ligature) and between RvE1 (0.5μg/μl) and disease (ligature+ vehicle). The genes are listed by the order of adjusted p-value in the comparison between disease and health. The p-values are adjusted using the Benjamini–Hochberg procedure. (C) Relative mRNA expression (2−ΔΔCT) was used to analyze the gene expression levels of inflammation related genes in different groups. In the treatment experiment, gene expression of Cxcl1 in the RvE1 group is significantly lower than that in the vehicle group. Gene expression of Ptgs2 in the RvE1 group is significantly lower than that in the ligature alone group.
Twenty nine differentially expressed genes exhibited an expression fold change ≥ 2^0.6 (adjusted p-value <0.1). All genes exhibited opposite expression patterns with RvE1 treatment (Fig. 3B). Most genes were identified as inflammatory response genes (Ccl3, CxCl1, Cxcl2, Il1b, Il18bp, Mmp3, Mmp10, Mmp13, Nfkbia) or bone resorption associated genes (Acp5, Ccl9).
There were six significant biological pathways identified as associated with the observed changes (Table I). These KEGG pathways were related to the infection response (chemokine signaling pathway, Chagas disease), bone resorption (rheumatoid arthritis, osteoclast differentiation) and metabolic activity (pancreatic secretion, bile secretion). Five genes (Ccl3, Ccl9, Cxcl1, Cxcl2, Rac2) belonging to chemokine signaling pathway were upregulated in the diseases state, but were downregulated following RvE1 treatment. RvE1 treatment downregulated bone resorption pathway gene expression (Acp5, Ccl3, Mmp3, Trem2) and pancreatic secretion pathway gene expression (Sctr).
Table 1.
Biological pathways in the two comparisons (Disease vs. Health; RvE1 (0.5μg/μl) vs. Disease)
| Disease vs. Health | RvE1(0.5μg/ml) vs. Disease | ||
|---|---|---|---|
| Pathways | False Discovery Rate (FDR) | Pathways | False Discovery Rate (FDR) |
| Phagosome | 2.35E-09 | *Chemokine signaling pathway | 6.98E-05 |
| Lysosome | 4.01E-09 | Salmonella infection | 2.18E-03 |
| Protein digestion and absorption | 1.18E-07 | TNF signaling pathway | 1.03E-02 |
| Hematopoietic cell lineage | 3.10E-06 | *Chagas disease (American trypanosomiasis) | 1.03E-02 |
| Tuberculosis | 3.88E-05 | NOD-like receptor signaling pathway | 1.03E-02 |
| *Rheumatoid arthritis | 8.20E-05 | Legionellosis | 1.03E-02 |
| *Chemokine signaling pathway | 8.41E-05 | PPAR signaling pathway | 2.07E-02 |
| Renin-angiotensin system | 8.41E-05 | *Rheumatoid arthritis | 2.07E-02 |
| Leishmaniasis Antigen processing and presentation | 8.41E-05 | *Bile secretion | 3.38E-02 |
| *Osteoclast differentiation | 9.69E-05 | *Osteoclast differentiation | 3.75E-02 |
| ECM-receptor interaction | 1.53E-04 | ||
| Systemic lupus erythematosus | 1.54E-04 | ||
| Staphylococcus aureus infection | 2.13E-04 | ||
| Asthma | 2.13E-04 | ||
| Cytokine-cytokine receptor interaction | 3.93E-04 | ||
| Inflammatory bowel disease (IBD) | 7.15E-04 | ||
| Intestinal immune network for IgA production | 1.04E-03 | ||
| Amoebiasis | 1.17E-03 | ||
| Graft-versus-host disease | 1.46E-03 | ||
| Focal adhesion | 1.97E-03 | ||
| Type I diabetes mellitus | 2.39E-03 | ||
| *Pancreatic secretion | 5.28E-03 | ||
| Leukocyte transendothelial migration | 5.82E-03 | ||
| Autoimmune thyroid disease | 6.05E-03 | ||
| Allograft rejection | 6.05E-03 | ||
| Natural killer cell mediated cytotoxicity | 7.68E-03 | ||
| Herpes simplex infection | 8.45E-03 | ||
| Malaria | 9.14E-03 | ||
| Primary immunodeficiency | 1.17E-02 | ||
| Platelet activation | 1.27E-02 | ||
| PI3K-Akt signaling pathway | 1.45E-02 | ||
| *Chagas disease (American trypanosomiasis) | 2.43E-02 | ||
| *Bile secretion | 2.43E-02 | ||
| Complement and coagulation cascades | 4.41E-02 | ||
| Platelet activation | 1.27E-02 | ||
The biological pathways found by using impact analysis are listed by the order of false discovery rate (<0.05).
The common pathways are in boldface.
Comparing mean fold changes (log2) of 10 selected genes in RNA-seq and qRT-PCR, 10 genes have similar expression patterns (Table II). Gene expression assessed by RNA-seq and qRT-PCR are comparable (Spearman's rho = 0.88, P = 0.0000).
Table II.
Verification of RNA-seq with qRT-PCR
| Gene | Comparison | Mean fold change (Log2) | Comparison | Mean fold change (Log2) | ||
|---|---|---|---|---|---|---|
| RNA-seq | qRT-PCR | RNA-seq | qRT-PCR | |||
| Acp5 | Disease vs. Health | 0.818 | 1.301 | RvE1 (0.5μg/μl) treatment vs Disease | −0.872 | −0.905 |
| Angptl4 | 1.966 | 2.051 | −1.812 | −2.612 | ||
| Ccl9 | 4.093 | 5.295 | −1.175 | −2.398 | ||
| Cxcl1 | 2.619 | 3.851 | −1.443 | −2.474 | ||
| Cxcl2 | 1.753 | 3.712 | −1.419 | −2.823 | ||
| Ide | −1.596 | −0.117 | 0.824 | 0.241 | ||
| Il1b | 1.749 | 3.143 | −1.104 | −2.913 | ||
| Mmp13 | 2.565 | 2.882 | −1.815 | −2.123 | ||
| Nfkbia | 0.938 | 1.338 | −1.029 | −1.126 | ||
| Rac2 | 1.802 | 1.724 | −1.211 | −1.499 | ||
Two samples in each group (Health, Disease, and RvE1 (0.5μg/μl) treatment groups) were used in the two analyses. The genes are listed in the alphabetic order.
In the treatment experiment, three candidate genes were assessed by qRT-PCR. Cxcl1, Nos2, and Ptgs2 in the RvE1 group were lower than controls, and RvE1 treatment significantly reduced gene expression level of Cxcl1 compared to vehicle application (p=0.02) (Fig. 3C).
RvE1 treatment and the subgingival microbiota
Considering the variation in relative abundance and limited sample size, a threshold p-value of 0.3 (Student’s t-test) was chosen to screen the potential genera representing subgingival microbiota in different groups in an attempt to eliminate background noise. Results are presented at the genus level, since the threshold of 97% sequence similarity was used to assign OTU.
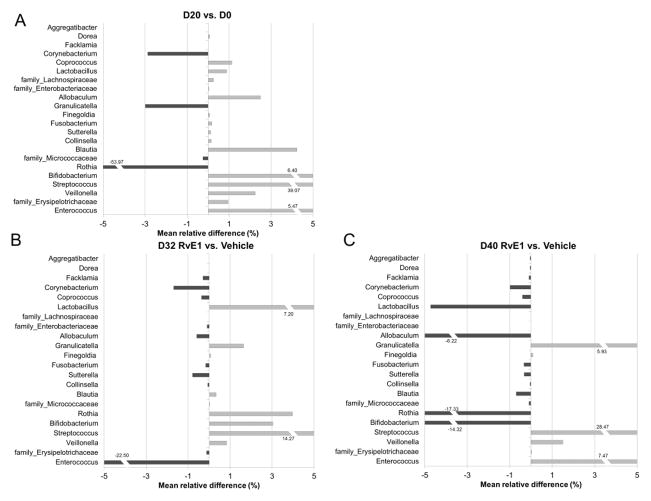
The composition of subgingival microbiota at baseline was markedly different from disease (Fig. 4A). One genus, Rothia, was dominant with consistent relative abundance in all the samples at baseline (59.32 ±19.20 %). After disease induction, increased diversity was observed with a loss of Rothia (53.97%) and a loss of Granulicatella (3.0%) and Corynebacterium (2.9%) and overgrowth and emergence of Streptococcus (39.07%), Enterococcus (5.47%), Bifidobacterium (6.4%) and Blautia (4.23 %), among others including Lactobacillus (0.88%).
Figure 4. Shifts in subgingival microbiota associated with periodontal regeneration following RvE1 treatment.
(A) Mean relative abundance difference between day 0 and day 20 is calculated by subtracting mean relative abundance at baseline (day 0) from mean relative abundance at 20 days post ligature. Positive values indicate the mean relative abundance on day 20 is larger than the mean relative abundance on day 0. OTUs that do not match any known genera in the reference data base are named at the family level. The most notable changes are the loss of Rothia spp. And the overgrowth of Streptococcus spp. as disease develops. (B)(C) Microbial shifts associated with the periodontal bone regeneration induced by RvE1 (0.25 μg/ml) treatment. Day 32-Panel B and day 40-Panel C are shown. Positive values indicate the mean relative abundance of the RvE1 group is larger than the mean relative abundance of the Vehicle group. Notably, most of the genera with increased relative abundance after disease induction exhibit a trend to return to level more associate with a healthy periodontium. (n=6 in the no ligature group and the ligature alone group; n=3 in the ligature + Vehicle group and the ligature + RvE1 group)”.
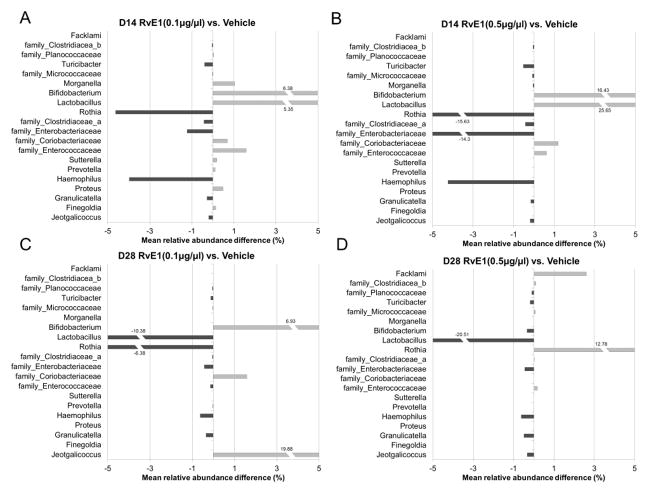
In the prevention experiment, RvE1 treatment (0.1μg/μl and 0.5μg/μl) altered the mean relative abundance of each genus measured compared to vehicle on day 14 (Fig. 5A, 5B) and day 28 (Fig. 5C, 5D). By day 28, 0.1μg/ml RvE1 reduced overgrowth of Lactobacillus, but not of Bifidobacterium; it also did not reverse the loss of the dominant species in health, Rothia. At day 28 with 0.5μg/ml RvE1, the dominance of Rothia had re-emerged, Lactobacillus was further reduced, Bifidobacterium overgrowth was reduced and the complexity of the flora was simpler (Supplemental Fig. 2). The compositional shifts in subgingival alpha and beta diversity were associated with different degrees of inflammation in the local periodontal environment (Supplemental Fig. 2). In general, the relative abundance of most of these genera in the RvE1 groups was lower than the disease group. Data are expressed as relative abundance; direct comparisons of bacterial numbers were not made.
Figure 5. Shifts in subgingival microbiota composition during periodontal disease initiation and development.
Prevention of periodontitis: shifts in the composition of the microbiota assessed by 16S rDNA sequencing were assessed during development of ligature induced periodontitis and compared to the low and high doses treatment with RvE1 during disease induction. (A)(B) Mean relative abundance difference between the 0.1μg/μl- Panel A, or 0.5μg/μl- Panel B. RvE1 treatment and vehicle treated control is calculated by subtracting mean relative abundance of the vehicle treatment from mean relative abundance of the RvE1treated doses at 14 days post ligature. Positive values indicate the mean relative abundance of the RvE1 group is larger than the mean relative abundance of the vehicle group. The family name of the bacterium is listed for the OTUs that are not assigned to any known genus. (C)(D) Mean relative abundance differences on day 28 for the two doses of RvE1 compared to vehicle. RvE1 treatment induces major shifts in the subgingival microbiota. Over time, RvE1 treatment tends to shift the microbiota toward a composition more associated with periodontal health. (n=4 in the no ligature group, the ligature + vehicle group, and the ligature+ RvE1 (0.1μg/μl) group; n=3 in the ligature+RvE1 (0.5μg/μl) group).
In the treatment experiment, there was a clear shift in the subgingival microbiota following ligature placement (Fig. 4A). Two genera, Streptococcus (overgrowth) and Rothia (loss), exhibited the largest changes in mean relative abundance during the disease induction phase. In the treatment phase, the subgingival microbiota shift was compared between the ligature + RvE1 group and the ligature + vehicle group. Four genera (Enterococcus, Veillonella, Allobaculum, Lactobacillus) demonstrated the greatest shifts on day 32. On day 40, eleven genera (Coprococcus, Lactobacillus, Blautia, Sutterella, Veillonella, Streptococcus, Corynebacterium, Collinsella, Facklamia, Dorea, Aggregatibacter) exhibited the largest shifts (Fig. 4B, 4C). Although there was a trend for returning the microbiota to one more associated with health that was consistent with the clinical outcome (reversal of bone loss and reduced inflammation), the shift was incomplete in the timeframe of the experiment.
Discussion
It is well established that RvE1 inhibits neutrophil infiltration, induces neutrophil apoptosis, attracts non-phlogistic macrophages that phagocytize apoptotic neutrophils and bacteria and clear the chronic inflammatory lesions through efferocytosis (23). Release of pro-inflammatory cytokines and chemokines is reduced during the resolution of inflammation (23, 28). Resolvins provide a feed forward, receptor agonist driven enhancement of resolution of inflammation, not inhibition of inflammation as with NSAID inhibition of COX or receptor antagonists. The present study demonstrates, in separate experiments, that RvE1 application prevents and successfully treats ligature induced periodontitis in the rat. Treatment was associated with regeneration of alveolar bone lost to periodontitis, inhibition of inflammation and osteoclast activity, and downregulation of inflammation associated genes in gingival tissues. We also demonstrate, to our knowledge for the first time, that shifts in the subgingival microbiota induced with ligature are markedly changed by control of local inflammation. The experiments tested the hypothesis that inflammation plays a major role in the induction and maintenance of dysbiosis. We demonstrate that regulation of inflammation with resolvin E1 has a significant impact on gingival gene expression and subgingival bacterial composition that is coincident with prevention and treatment of disease.
Periodontitis is a disease of microbial dysbiosis (54, 55). Shifts in the composition and bacterial counts within the subgingival microbiota have been associated with periodontal disease in myriad studies (56–58). From an ecological point of view, in active periodontitis many disease-associated species overgrow by obtaining required nutrients from the inflamed environment. Therefore, the diseased microbial community is more diverse at any one site (alpha diversity) than is the healthy microbial community. The microbial community of samples from different disease sites becomes more similar (beta diversity) compared to the microbial community of different healthy samples (59–63). The ecological plaque hypothesis proposed by Marsh (64, 65) emphasizes a direct link between local environmental conditions and the activity and composition of the microbial community. The changes in local environment trigger the changes in microbiota, and vice versa.
Inflammation and change of local environment appear to drive the shifts in subgingival microbiota (Figs. 4, 5). However, this does not minimize the role of overgrowth of specific bacteria and expression of virulence factors that coincide with inflammation. It has been recently demonstrated in a bacterial transcriptomic analyses that putative virulence factors are upregulated by bacteria (both putative pathogens and health associated commensals) in progressing periodontitis sites (66, 67). Bacteria are necessary to induce periodontitis in the ligature model (68), but the change of microbiota in different clinical conditions has never been explored. Previously reported studies in rabbits show that microbial shifts are prevented and reversed by RvE1, but these studies are indirect using probes to human microorganisms (27). The results of the unbiased characterization of the microbiota of the rat performed here emphasize the differences between human and rat microbiota associated with health and disease. This study did not aim to investigate the roles of these genera in pathogenesis of experimental periodontitis in rats.
RvE1 treatment induced marked changes in the microbiota. The compositional shifts in subgingival alpha and beta diversity were associated with different degrees of inflammation in the local periodontal environment (Supplemental Fig. 2). The data suggest that most of these shifts are driven by the changes in inflammation, since RvE1 was the only treatment variable and it is known to have no direct antimicrobial activity (31, 69). Since experimental periodontitis was induced consistently in these animals, the shifts in subgingival microbiota and the emergence of new bacteria can be attributed to changes in inflammation. There are at least two possible mechanisms: (i) RvE1 regulates killing activity of immune cells; and/or (ii) resolution of inflammation alters the local environment removing the source of nutrients for certain bacteria, particularly those associated with periodontitis. Asaccharolytic bacteria that comprise periodontitis associated bacteria depend on peptides and other metabolites derived from host-tissue degradation as nutrients to survive (7, 10). The composition of metabolites in periodontal tissue fluid is associated with periodontal inflammation (7, 71, 72). The impact of RvE1 on phagocytosis and production of reactive oxygen species (ROS) by immune cells has been demonstrated (24, 69, 70). Taken together, the data suggest that inflammation regulated by RvE1 leads to the observed shifts in microbial composition.
The pattern of differential gene expression in rat periodontal tissue between periodontitis and health is similar to that between the periodontitis group and the healthy group in human gingiva (73). In addition to the down-regulation of immune response stimulation related genes (Ccl3, CxCl1, Cxcl2, Il1b), immune response inhibition genes (Il18bp, Lilrb4, Nfkbia) were also down-regulated following RvE1 treatment. These genes may be expressed in the disease state to compensate excessive inflammation and down-regulated when the inflammation was controlled by RvE1 application (Figs. 3A, 3B). Several genes encoding proteins, such as MMPs-3, 10, 13, which are biomarkers for periodontitis in humans, were also highly expressed in the rat gingiva with periodontal disease (74–76). The expression of these genes was down-regulated by RvE1 treatment. The expression of the genes associated with vascularization as well as angiogenesis (Angptl4) (77) and superoxide production as well as neutrophil migration (Rac2) (78) were increased in disease and decreased upon resolution This was also observed in a human experimental gingivitis study (ANGPTL1) (79) and human periodontitis-affected gingiva (RAC2) (80). The gene, Cxcl1, was significantly downregulated following RvE1 application in both prevention and treatment studies (Figs. 3B, 3C). Cxcl1 is the major chemoattractant of neutrophils in rodents (81, 82) and neutrophils are the major cells initiating periodontitis in humans. These results demonstrate RvE1 application actions in experimental periodontitis at the molecular level. In gene ontology analysis, the genes in the top three significant biological processes (response to lipopolysaccharide, response to external stimulus, response to molecules of bacterial origin) were downregulated following RvE1 treatment (Supplemental Table I).
The results also demonstrate that RvE1 regulates the NOD-like receptor signaling pathway and the peroxisome proliferator-activated receptor (PPAR) signaling pathway. These two pathways have been implicated as important for the inflammatory response in periodontitis (83, 84). From the available evidence, RvE1 and other SPMs do not directly regulate NOD-like receptors or PPARs (85), but indirectly regulate PPAR activity via upstream regulation of inflammation (86). Further studies are needed to clarity the role of resolvins in the regulation of these two pathways.
Generally, gene expression analyzed by RNA-seq or qRT-PCR is highly correlated, but the strength of gene expression fold change, specific genes selected for the analysis, the design of primers, and the characteristics of samples may result in discrepancies (87–89). In the present study, results of gene expression in RNA-seq and qRT-PCR were well correlated (Table II).
The dose-response to RvE1 was most apparent in RNA sequencing experiments. Low dose RvE1 might be able to control early-stage inflammation reducing bone resorption (90) and inflammatory cell infiltration. Gene expression in gingiva appears to be more sensitive than morphometric changes. High dose RvE1 tended to inhibit the growth of bacteria more than low dose (Fig. 5). The shifts in subgingival microbiota reflect the change of microenvironment. Taken together, there appears to be an optimal dose of RvE1 for treating periodontitis.
The data suggest that reduced osteoclast activity following RvE1 application plays an important role in controlling alveolar bone loss in this model. It was previously shown that RvE1 inhibits osteoclast differentiation in vitro (30) and promotes osteoblast formation in vivo (29). In addition, the regeneration of bone following RvE1 treatment likely involves enhanced recruitment of non-phlogistic macrophages in the resolution phase given that macrophages of the proper phenotype are necessary for tissue regeneration (91, 92). RNA-seq data demonstrate that bone resorption related pathways are regulated by RvE1, but using the KEGG analysis, none of the regeneration related biological pathways or processes were significant. Further studies are required to clarify the mechanism of tissue regeneration induced by RvE1.
In conclusion, prophylactic RvE1 treatment significantly prevents alveolar bone loss in the ligature induced periodontitis model in the rat. The disease associated changes in gene expression profile of periodontal tissues is reversed by RvE1 treatment. RvE1 treatment of existing ligature induced periodontitis significantly regenerates lost alveolar bone. The shifts in the rat subgingival microbiota in experimental periodontitis following RvE1 application suggest that inflammation has a significant impact on the local environment and bacterial growth conditions. The results suggest that changes in the composition of subgingival microbiota emerge as a result of inflammation and fill an important gap in knowledge of the actions of RvE1 regulation of the host inflammatory response at the transcriptome level.
Supplementary Material
Acknowledgments
This work was supported in part by USPHS grant DE25020 (TVD) from the National Institute of Dental and Craniofacial Research.
We would like to thank Dr. Keerthana Krishnan for processing the samples in the sequencing analysis. Dr. Van Dyke holds unlicensed patents relating to RvE1 in periodontal disease. Other authors have no other potential conflicts to report.
Abbreviations used in this article
- RvE1
resolvin E1
- DE
differentially expressed
- 95% CI
95% confidence interval
Footnotes
The raw RNASeq sequence reads used in this study have been deposited to the NCBI Sequence Read Archive database (http://www.ncbi.nlm.nih.gov/Traces/sra/) and are accessible under the umbrella BioProject ID PRJNA321482.
References
- 1.Eke PI, Dye BA, Wei L, Slade GD, Thornton-Evans GO, Borgnakke WS, Taylor GW, Page RC, Beck JD, Genco RJ. Update on Prevalence of Periodontitis in Adults in the United States: NHANES 2009 to 2012. Journal of periodontology. 2015;86:611–622. doi: 10.1902/jop.2015.140520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nature reviews Microbiology. 2012;10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heitz-Mayfield LJ, Lang NP. Surgical and nonsurgical periodontal therapy. Learned and unlearned concepts. Periodontology 2000. 2013;62:218–231. doi: 10.1111/prd.12008. [DOI] [PubMed] [Google Scholar]
- 4.Slots J. Selection of antimicrobial agents in periodontal therapy. Journal of periodontal research. 2002;37:389–398. doi: 10.1034/j.1600-0765.2002.00004.x. [DOI] [PubMed] [Google Scholar]
- 5.Haffajee AD, Socransky SS. Introduction to microbial aspects of periodontal biofilm communities, development and treatment. Periodontology 2000. 2006;42:7–12. doi: 10.1111/j.1600-0757.2006.00190.x. [DOI] [PubMed] [Google Scholar]
- 6.Feres M, Figueiredo LC, Soares GM, Faveri M. Systemic antibiotics in the treatment of periodontitis. Periodontology 2000. 2015;67:131–186. doi: 10.1111/prd.12075. [DOI] [PubMed] [Google Scholar]
- 7.Van Dyke TE. Proresolving lipid mediators: potential for prevention and treatment of periodontitis. Journal of clinical periodontology. 2011;38(Suppl 11):119–125. doi: 10.1111/j.1600-051X.2010.01662.x. [DOI] [PubMed] [Google Scholar]
- 8.Van Dyke TE. Commentary: periodontitis is characterized by an immuno-inflammatory host-mediated destruction of bone and connective tissues that support the teeth. Journal of periodontology. 2014;85:509–511. doi: 10.1902/jop.2014.130701. [DOI] [PubMed] [Google Scholar]
- 9.Moutsopoulos NM, Konkel J, Sarmadi M, Eskan MA, Wild T, Dutzan N, Abusleme L, Zenobia C, Hosur KB, Abe T, Uzel G, Chen W, Chavakis T, Holland SM, Hajishengallis G. Defective neutrophil recruitment in leukocyte adhesion deficiency type I disease causes local IL–17-driven inflammatory bone loss. Science translational medicine. 2014;6:229ra240. doi: 10.1126/scitranslmed.3007696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moutsopoulos NM, Chalmers NI, Barb JJ, Abusleme L, Greenwell-Wild T, Dutzan N, Paster BJ, Munson PJ, Fine DH, Uzel G, Holland SM. Subgingival microbial communities in Leukocyte Adhesion Deficiency and their relationship with local immunopathology. PLoS pathogens. 2015;11:e1004698. doi: 10.1371/journal.ppat.1004698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graves D. Cytokines that promote periodontal tissue destruction. Journal of periodontology. 2008;79:1585–1591. doi: 10.1902/jop.2008.080183. [DOI] [PubMed] [Google Scholar]
- 12.Kantarci A, Oyaizu K, Van Dyke TE. Neutrophil–mediated tissue injury in periodontal disease pathogenesis: findings from localized aggressive periodontitis. Journal of periodontology. 2003;74:66–75. doi: 10.1902/jop.2003.74.1.66. [DOI] [PubMed] [Google Scholar]
- 13.Chapple IL, Matthews JB. The role of reactive oxygen and antioxidant species in periodontal tissue destruction. Periodontology 2000. 2007;43:160–232. doi: 10.1111/j.1600-0757.2006.00178.x. [DOI] [PubMed] [Google Scholar]
- 14.Tanner AC, Kent R, Jr, Kanasi E, Lu SC, Paster BJ, Sonis ST, Murray LA, Van Dyke TE. Clinical characteristics and microbiota of progressing slight chronic periodontitis in adults. Journal of clinical periodontology. 2007;34:917–930. doi: 10.1111/j.1600-051X.2007.01126.x. [DOI] [PubMed] [Google Scholar]
- 15.Kinney JS, Morelli T, Oh M, Braun TM, Ramseier CA, Sugai JV, Giannobile WV. Crevicular fluid biomarkers and periodontal disease progression. Journal of clinical periodontology. 2014;41:113–120. doi: 10.1111/jcpe.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams RC, Jeffcoat MK, Kaplan ML, Goldhaber P, Johnson HG, Wechter WJ. Flurbiprofen: a potent inhibitor of alveolar bone resorption in beagles. Science. 1985;227:640–642. doi: 10.1126/science.3969553. [DOI] [PubMed] [Google Scholar]
- 17.Offenbacher S, Braswell LD, Loos AS, Johnson HG, Hall CM, McClure H, Orkin JL, Strobert EA, Green MD, Odle BM. Effects of flurbiprofen on the progression of periodontitis in Macaca mulatta. Journal of periodontal research. 1987;22:473–481. doi: 10.1111/j.1600-0765.1987.tb02058.x. [DOI] [PubMed] [Google Scholar]
- 18.Williams RC, Jeffcoat MK, Howell TH, Reddy MS, Johnson HG, Hall CM, Goldhaber P. Ibuprofen: an inhibitor of alveolar bone resorption in beagles. Journal of periodontal research. 1988;23:225–229. doi: 10.1111/j.1600-0765.1988.tb01363.x. [DOI] [PubMed] [Google Scholar]
- 19.Williams RC, Jeffcoat MK, Howell TH, Rolla A, Stubbs D, Teoh KW, Reddy MS, Goldhaber P. Altering the progression of human alveolar bone loss with the non-steroidal anti-inflammatory drug flurbiprofen. Journal of periodontology. 1989;60:485–490. doi: 10.1902/jop.1989.60.9.485. [DOI] [PubMed] [Google Scholar]
- 20.Howell TH. Blocking periodontal disease progression with anti-inflammatory agents. Journal of periodontology. 1993;64:828–833. doi: 10.1902/jop.1993.64.8s.828. [DOI] [PubMed] [Google Scholar]
- 21.Jeffcoat MK, Reddy MS, Haigh S, Buchanan W, Doyle MJ, Meredith MP, Nelson SL, Goodale MB, Wehmeyer KR. A comparison of topical ketorolac, systemic flurbiprofen, and placebo for the inhibition of bone loss in adult periodontitis. Journal of periodontology. 1995;66:329–338. doi: 10.1902/jop.1995.66.5.329. [DOI] [PubMed] [Google Scholar]
- 22.Yu Y, Ricciotti E, Scalia R, Tang SY, Grant G, Yu Z, Landesberg G, Crichton I, Wu W, Pure E, Funk CD, FitzGerald GA. Vascular COX-2 modulates blood pressure and thrombosis in mice. Science translational medicine. 2012;4:132ra154. doi: 10.1126/scitranslmed.3003787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serhan CN. Pro-resolving lipid mediators are leads for resolution physiology. Nature. 2014;510:92–101. doi: 10.1038/nature13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El Kebir D, Gjorstrup P, Filep JG. Resolvin E1 promotes phagocytosis-induced neutrophil apoptosis and accelerates resolution of pulmonary inflammation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:14983–14988. doi: 10.1073/pnas.1206641109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stables MJ, Shah S, Camon EB, Lovering RC, Newson J, Bystrom J, Farrow S, Gilroy DW. Transcriptomic analyses of murine resolution-phase macrophages. Blood. 2011;118:e192–208. doi: 10.1182/blood-2011-04-345330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oh SF, Pillai PS, Recchiuti A, Yang R, Serhan CN. Pro-resolving actions and stereoselective biosynthesis of 18S E-series resolvins in human leukocytes and murine inflammation. The Journal of clinical investigation. 2011;121:569–581. doi: 10.1172/JCI42545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hasturk H, Kantarci A, Goguet-Surmenian E, Blackwood A, Andry C, Serhan CN, Van Dyke TE. Resolvin E1 regulates inflammation at the cellular and tissue level and restores tissue homeostasis in vivo. Journal of immunology. 2007;179:7021–7029. doi: 10.4049/jimmunol.179.10.7021. [DOI] [PubMed] [Google Scholar]
- 28.Serhan CN, Petasis NA. Resolvins and protectins in inflammation resolution. Chemical reviews. 2011;111:5922–5943. doi: 10.1021/cr100396c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao L, Faibish D, Fredman G, Herrera BS, Chiang N, Serhan CN, Van Dyke TE, Gyurko R. Resolvin E1 and chemokine-like receptor 1 mediate bone preservation. Journal of immunology. 2013;190:689–694. doi: 10.4049/jimmunol.1103688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu M, Van Dyke TE, Gyurko R. Resolvin E1 regulates osteoclast fusion via DC-STAMP and NFATc1. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2013;27:3344–3353. doi: 10.1096/fj.12-220228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hasturk H, Kantarci A, Ohira T, Arita M, Ebrahimi N, Chiang N, Petasis NA, Levy BD, Serhan CN, Van Dyke TE. RvE1 protects from local inflammation and osteoclast- mediated bone destruction in periodontitis. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2006;20:401–403. doi: 10.1096/fj.05-4724fje. [DOI] [PubMed] [Google Scholar]
- 32.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R. A systems biology approach for pathway level analysis. Genome research. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tarca AL, Draghici S, Khatri P, Hassan SS, Mittal P, Kim JS, Kim CJ, Kusanovic JP, Romero R. A novel signaling pathway impact analysis. Bioinformatics. 2009;25:75–82. doi: 10.1093/bioinformatics/btn577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Draghici S, Khatri P, Bhavsar P, Shah A, Krawetz SA, Tainsky MA. Onto-Tools, the toolkit of the modern biologist: Onto-Express, Onto-Compare, Onto-Design and Onto-Translate. Nucleic acids research. 2003;31:3775–3781. doi: 10.1093/nar/gkg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horz HP, Scheer S, Huenger F, Vianna ME, Conrads G. Selective isolation of bacterial DNA from human clinical specimens. Journal of microbiological methods. 2008;72:98–102. doi: 10.1016/j.mimet.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 38.Horz HP, Scheer S, Vianna ME, Conrads G. New methods for selective isolation of bacterial DNA from human clinical specimens. Anaerobe. 2010;16:47–53. doi: 10.1016/j.anaerobe.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 39.Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, Sun Z, Zong Q, Du Y, Du J, Driscoll M, Song W, Kingsmore SF, Egholm M, Lasken RS. Comprehensive human genome amplification using multiple displacement amplification. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brito LC, Teles FR, Teles RP, Franca EC, Ribeiro-Sobrinho AP, Haffajee AD, Socransky SS. Use of multiple-displacement amplification and checkerboard DNA-DNA hybridization to examine the microbiota of endodontic infections. Journal of clinical microbiology. 2007;45:3039–3049. doi: 10.1128/JCM.02618-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and environmental microbiology. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic acids research. 2013;41:D590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 45.Lozupone C, Hamady M, Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at GenomeNet. Nucleic acids research. 2002;30:42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gene Ontology, C. Creating the gene ontology resource: design and implementation. Genome research. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Molecular cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic acids research. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome research. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic acids research. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nature reviews Microbiology. 2010;8:481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- 55.Jiao Y, Hasegawa M, Inohara N. The Role of Oral Pathobionts in Dysbiosis during Periodontitis Development. Journal of dental research. 2014;93:539–546. doi: 10.1177/0022034514528212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. Journal of clinical periodontology. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 57.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. Bacterial diversity in human subgingival plaque. Journal of bacteriology. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Teles R, Teles F, Frias-Lopez J, Paster B, Haffajee A. Lessons learned and unlearned in periodontal microbiology. Periodontology 2000. 2013;62:95–162. doi: 10.1111/prd.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, Leys EJ. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. The ISME journal. 2012;6:1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu B, Faller LL, Klitgord N, Mazumdar V, Ghodsi M, Sommer DD, Gibbons TR, Treangen TJ, Chang YC, Li S, Stine OC, Hasturk H, Kasif S, Segre D, Pop M, Amar S. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PloS one. 2012;7:e37919. doi: 10.1371/journal.pone.0037919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, Gamonal J, Diaz PI. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. The ISME journal. 2013;7:1016–1025. doi: 10.1038/ismej.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Y, He J, He Z, Zhou Y, Yuan M, Xu X, Sun F, Liu C, Li J, Xie W, Deng Y, Qin Y, VanNostrand JD, Xiao L, Wu L, Zhou J, Shi W, Zhou X. Phylogenetic and functional gene structure shifts of the oral microbiomes in periodontitis patients. The ISME journal. 2014;8:1879–1891. doi: 10.1038/ismej.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi B, Chang M, Martin J, Mitreva M, Lux R, Klokkevold P, Sodergren E, Weinstock GM, Haake SK, Li H. Dynamic changes in the subgingival microbiome and their potential for diagnosis and prognosis of periodontitis. mBio. 2015;6:e01926–01914. doi: 10.1128/mBio.01926-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marsh PD. Microbial ecology of dental plaque and its significance in health and disease. Advances in dental research. 1994;8:263–271. doi: 10.1177/08959374940080022001. [DOI] [PubMed] [Google Scholar]
- 65.Marsh PD. Are dental diseases examples of ecological catastrophes? Microbiology. 2003;149:279–294. doi: 10.1099/mic.0.26082-0. [DOI] [PubMed] [Google Scholar]
- 66.Duran-Pinedo AE, Chen T, Teles R, Starr JR, Wang X, Krishnan K, Frias-Lopez J. Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. The ISME journal. 2014;8:1659–1672. doi: 10.1038/ismej.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yost S, Duran-Pinedo AE, Teles R, Krishnan K, Frias-Lopez J. Functional signatures of oral dysbiosis during periodontitis progression revealed by microbial metatranscriptome analysis. Genome medicine. 2015;7:27. doi: 10.1186/s13073-015-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rovin S, Costich ER, Gordon HA. The influence of bacteria and irritation in the initiation of periodontal disease in germfree and conventional rats. Journal of periodontal research. 1966;1:193–204. doi: 10.1111/j.1600-0765.1966.tb01860.x. [DOI] [PubMed] [Google Scholar]
- 69.Chiang N, Fredman G, Backhed F, Oh SF, Vickery T, Schmidt BA, Serhan CN. Infection regulates pro-resolving mediators that lower antibiotic requirements. Nature. 2012;484:524–528. doi: 10.1038/nature11042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Herrera BS, Hasturk H, Kantarci A, Freire MO, Nguyen O, Kansal S, Van Dyke TE. Impact of resolvin E1 on murine neutrophil phagocytosis in type 2 diabetes. Infection and immunity. 2015;83:792–801. doi: 10.1128/IAI.02444-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barnes VM, Ciancio SG, Shibly O, Xu T, Devizio W, Trivedi HM, Guo L, Jonsson TJ. Metabolomics reveals elevated macromolecular degradation in periodontal disease. Journal of dental research. 2011;90:1293–1297. doi: 10.1177/0022034511416240. [DOI] [PubMed] [Google Scholar]
- 72.Barnes VM, Kennedy AD, Panagakos F, Devizio W, Trivedi HM, Jonsson T, Guo L, Cervi S, Scannapieco FA. Global metabolomic analysis of human saliva and plasma from healthy and diabetic subjects, with and without periodontal disease. PloS one. 2014;9:e105181. doi: 10.1371/journal.pone.0105181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Davanian H, Stranneheim H, Bage T, Lagervall M, Jansson L, Lundeberg J, Yucel-Lindberg T. Gene expression profiles in paired gingival biopsies from periodontitis-affected and healthy tissues revealed by massively parallel sequencing. PloS one. 2012;7:e46440. doi: 10.1371/journal.pone.0046440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kiili M, Cox SW, Chen HY, Wahlgren J, Maisi P, Eley BM, Salo T, Sorsa T. Collagenase-2 (MMP-8) and collagenase-3 (MMP-13) in adult periodontitis: molecular forms and levels in gingival crevicular fluid and immunolocalisation in gingival tissue. Journal of clinical periodontology. 2002;29:224–232. doi: 10.1034/j.1600-051x.2002.290308.x. [DOI] [PubMed] [Google Scholar]
- 75.Letra A, Silva RM, Rylands RJ, Silveira EM, de Souza AP, Wendell SK, Garlet GP, Vieira AR. MMP3 and TIMP1 variants contribute to chronic periodontitis and may be implicated in disease progression. Journal of clinical periodontology. 2012;39:707–716. doi: 10.1111/j.1600-051X.2012.01902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ahn SJ, Rhim EM, Kim JY, Kim KH, Lee HW, Kim EC, Park SH. Tumor necrosis factor-alpha induces matrix metalloproteinases-3, -10, and -13 in human periodontal ligament cells. Journal of periodontology. 2014;85:490–497. doi: 10.1902/jop.2013.130063. [DOI] [PubMed] [Google Scholar]
- 77.Chong HC, Chan JS, Goh CQ, Gounko NV, Luo B, Wang X, Foo S, Wong MT, Choong C, Kersten S, Tan NS. Angiopoietin-like 4 stimulates STAT3-mediated iNOS expression and enhances angiogenesis to accelerate wound healing in diabetic mice. Molecular therapy : the journal of the American Society of Gene Therapy. 2014;22:1593–1604. doi: 10.1038/mt.2014.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gu Y, Filippi MD, Cancelas JA, Siefring JE, Williams EP, Jasti AC, Harris CE, Lee AW, Prabhakar R, Atkinson SJ, Kwiatkowski DJ, Williams DA. Hematopoietic cell regulation by Rac1 and Rac2 guanosine triphosphatases. Science. 2003;302:445–449. doi: 10.1126/science.1088485. [DOI] [PubMed] [Google Scholar]
- 79.Offenbacher S, Barros SP, Paquette DW, Winston JL, Biesbrock AR, Thomason RG, Gibb RD, Fulmer AW, Tiesman JP, Juhlin KD, Wang SL, Reichling TD, Chen KS, Ho B. Gingival transcriptome patterns during induction and resolution of experimental gingivitis in humans. Journal of periodontology. 2009;80:1963–1982. doi: 10.1902/jop.2009.080645. [DOI] [PubMed] [Google Scholar]
- 80.Abe D, Kubota T, Morozumi T, Shimizu T, Nakasone N, Itagaki M, Yoshie H. Altered gene expression in leukocyte transendothelial migration and cell communication pathways in periodontitis-affected gingival tissues. Journal of periodontal research. 2011;46:345–353. doi: 10.1111/j.1600-0765.2011.01349.x. [DOI] [PubMed] [Google Scholar]
- 81.Bozic CR, Kolakowski LF, Jr, Gerard NP, Garcia-Rodriguez C, von Uexkull-Guldenband C, Conklyn MJ, Breslow R, Showell HJ, Gerard C. Expression and biologic characterization of the murine chemokine KC. Journal of immunology. 1995;154:6048–6057. [PubMed] [Google Scholar]
- 82.Chintakuntlawar AV, Chodosh J. Chemokine CXCL1/KC and its receptor CXCR2 are responsible for neutrophil chemotaxis in adenoviral keratitis. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2009;29:657–666. doi: 10.1089/jir.2009.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jiao Y, Darzi Y, Tawaratsumida K, Marchesan JT, Hasegawa M, Moon H, Chen GY, Nunez G, Giannobile WV, Raes J, Inohara N. Induction of bone loss by pathobiont-mediated Nod1 signaling in the oral cavity. Cell host & microbe. 2013;13:595–601. doi: 10.1016/j.chom.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Andriankaja OM, Galicia J, Dong G, Xiao W, Alawi F, Graves DT. Gene expression dynamics during diabetic periodontitis. Journal of dental research. 2012;91:1160–1165. doi: 10.1177/0022034512465292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Krishnamoorthy S, Recchiuti A, Chiang N, Yacoubian S, Lee CH, Yang R, Petasis NA, Serhan CN. Resolvin D1 binds human phagocytes with evidence for proresolving receptors. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1660–1665. doi: 10.1073/pnas.0907342107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liao Z, Dong J, Wu W, Yang T, Wang T, Guo L, Chen L, Xu D, Wen F. Resolvin D1 attenuates inflammation in lipopolysaccharide-induced acute lung injury through a process involving the PPARgamma/NF-kappaB pathway. Respiratory research. 2012;13:110. doi: 10.1186/1465-9921-13-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shi Y, He M. Differential gene expression identified by RNA-Seq and qPCR in two sizes of pearl oyster (Pinctada fucata) Gene. 2014;538:313–322. doi: 10.1016/j.gene.2014.01.031. [DOI] [PubMed] [Google Scholar]
- 88.Wang C, Gong B, Bushel PR, Thierry-Mieg J, Thierry-Mieg D, Xu J, Fang H, Hong H, Shen J, Su Z, Meehan J, Li X, Yang L, Li H, Labaj PP, Kreil DP, Megherbi D, Gaj S, Caiment F, van Delft J, Kleinjans J, Scherer A, Devanarayan V, Wang J, Yang Y, Qian HR, Lancashire LJ, Bessarabova M, Nikolsky Y, Furlanello C, Chierici M, Albanese D, Jurman G, Riccadonna S, Filosi M, Visintainer R, Zhang KK, Li J, Hsieh JH, Svoboda DL, Fuscoe JC, Deng Y, Shi L, Paules RS, Auerbach SS, Tong W. The concordance between RNA-seq and microarray data depends on chemical treatment and transcript abundance. Nature biotechnology. 2014;32:926–932. doi: 10.1038/nbt.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu AR, Neff NF, Kalisky T, Dalerba P, Treutlein B, Rothenberg ME, Mburu FM, Mantalas GL, Sim S, Clarke MF, Quake SR. Quantitative assessment of single-cell RNA-sequencing methods. Nature methods. 2014;11:41–46. doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.de Lima V, Bezerra MM, de Menezes Alencar VB, Vidal FD, da Rocha FA, de Castro Brito GA, de Albuquerque Ribeiro R. Effects of chlorpromazine on alveolar bone loss in experimental periodontal disease in rats. European journal of oral sciences. 2000;108:123–129. doi: 10.1034/j.1600-0722.2000.00766.x. [DOI] [PubMed] [Google Scholar]
- 91.Yin Y, Henzl MT, Lorber B, Nakazawa T, Thomas TT, Jiang F, Langer R, Benowitz LI. Oncomodulin is a macrophage-derived signal for axon regeneration in retinal ganglion cells. Nature neuroscience. 2006;9:843–852. doi: 10.1038/nn1701. [DOI] [PubMed] [Google Scholar]
- 92.Aurora AB, Porrello ER, Tan W, Mahmoud AI, Hill JA, Bassel-Duby R, Sadek HA, Olson EN. Macrophages are required for neonatal heart regeneration. The Journal of clinical investigation. 2014;124:1382–1392. doi: 10.1172/JCI72181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.