Abstract
Hepatitis B virus X protein (HBx) plays an important role in the development of hepatocellular carcinoma (HCC). In addition, hepatoma upregulated protein (HURP) is a cellular oncogene that is upregulated in a majority of HCC cases. We highlight here recent findings demonstrating a link between HBx, HURP and anti-apoptosis effects observed in cisplatin-treated HCC cells. We observed that Hep3B cells overexpressing HBx display increased HURP mRNA and protein levels, and show resistance to cisplatin-induced apoptosis. Knockdown of HURP in HBx-expressing cells reverses this effect, and sensitizes cells to cisplatin. The anti-apoptotic effect of HBx requires activation of the p38/MAPK pathway as well as expression of SATB1, survivin and HURP. Furthermore, silencing of HURP using short-hairpin RNA promotes accumulation of p53 and reduces cell proliferation in SK-Hep-1 cells (p53+/–), whereas these effects are not observed in p53-mutant Mahlavu cells. Similarly, HURP silencing does not affect the proliferation of H1299 lung carcinoma cells or Hep3B HCC cells which lack p53. Silencing of HURP sensitizes SK-Hep-1 cells to cisplatin. While HURP overexpression promotes p53 ubiquitination and degradation by the proteasome, HURP silencing reverses these effects. Inoculation of SK-Hep-1 cancer cells in which HURP has been silenced produces smaller tumors than control in nude mice. Besides, gankyrin, a positive regulator of the E3 ubiquitin ligase MDM2, is upregulated following HURP expression, and silencing of gankyrin reduces HURP-mediated downregulation of p53. In addition, we observed a positive correlation between HURP and gankyrin protein levels in HCC patients (r2 = 0.778; n = 9). These findings suggest a role for the viral protein HBx and the host protein HURP in preventing p53-mediated apoptosis during cancer progression and establishment of chemoresistance.
Keywords: Hepatitis B virus X protein, Hepatocellular carcinoma, Hepatitis B virus, Hepatoma upregulated protein, p53, Gankyrin, SATB1
Core tip: Hepatitis B virus X protein (HBx) plays a critical role in the development of hepatocellular carcinoma (HCC). Hepatoma upregulated protein (HURP) is an oncogene that is upregulated in a majority of HCC cases. However, the role of these proteins in the response of HCC cells to chemotherapeutic drugs remains unclear. We show here that the HBx/SATB1/HURP axis plays a critical role in down-regulating p53 and upregulating anti-apoptotic proteins in vitro and in vivo. We discuss the regulation of this novel pathway and its implications in resistance of HCC cells to chemotherapy.
INTRODUCTION
Chronic hepatitis B virus (HBV) infection represents an important risk factor for the development of hepatocellular carcinoma (HCC)[1-3]. The hepatitis B virus X protein (HBx) is produced by HBV and is required for viral replication in host cells[4,5]. HBx interferes with a variety of cellular functions in host cells. It forms a heterodimeric complex with the host protein HBX interacting protein, and this interaction dysregulates centrosome dynamics and chromosomal stability[6]. HBx also interacts with the tumor suppressor p53 and modulates cellular apoptosis in the presence of various stimuli[7-9]. A recent study indicates that HBx binds to the DNA-repair protein damaged DNA binding protein 1 (DDB1), and redirects the CUL4-DDB1 E3 complex, a protein complex with ubiquitin ligase activity that is involved in regulating DNA replication and repair, transcription and signal transduction in host cells[10].
Recent studies suggest that HBx plays a role in HCC pathogenesis[11-14]. However, the effect of HBx on apoptosis remains incompletely understood as some authors have reported pro-apoptotic[15-19] as well as anti-apoptotic effects[8,20-23]. Importantly, experiments performed in laboratory animals indicate that the HBx protein may induce resistance to the anti-cancer drug cisplatin in hepatoma cells[16]. Here, I present recent experimental evidence highlighting a prominent pathway used by HBx to upregulate hepatoma upregulated protein (HURP) and avoid apoptosis in HCC cells.
HURP AS A MARKER IN HCC
HURP was initially shown to be overexpressed in HCC based on a bioinformatics analysis of sequence tags expressed in the human liver[24]. Also known as discs large homolog 7 or disks large-associated protein 5[25,26], HURP was previously thought to represent a stem cell marker as this protein is not detected in fully differentiated cells[27]. Previous reports indicate that HURP overexpression in differentiated cells blocks apoptosis and increases cell growth in response to serum starvation[24,28]. HURP also appears to regulate the cell cycle and act specifically during mitosis. More specifically, HURP represents a kinetochore protein that stabilizes microtubules in the vicinity of chromosomes[29-31]. That is, HURP is associated with the mitotic spindle where it helps to determine spindle bipolarity and participates in the growth of microtubules toward chromosomes during mitosis. Furthermore, HURP forms a Ran-dependent complex[29], and is a target of the serine/threonine kinase aurora-A, which possesses oncogenic properties[28]. Aurora-A thus phosphorylates HURP and this process may represent a cellular mechanism that controls mitotic spindle assembly and functions[32]. Therefore, HURP is implicated in stem cell functions[25-27] and carcinogenesis in cancer cells of human origins[24,28]. Analysis of gene expression showed that HURP represents a marker of cancer prognosis that can be used to distinguish between benign and malignant adrenocortical tumors[33,34]. In addition, HURP undergoes proteolysis following phosphorylation by Cdk1-cyclin B and recognition by the Fbx7-associated SCF complex that functions as an E3 ubiquitin ligase[35]. These results indicate that HURP is involved in control of the cell cycle, specifically during mitosis, suggesting that this protein may regulate apoptosis and be involved in tumor development. However, the role of HURP in HCC and apoptosis, and how this protein is regulated is incompletely understood.
HBx UPREGULATES HURP EXPRESSION IN HCC CELLS
Given that the viral protein HBx plays a critical role in the development of HCC and HURP is upregulated in a majority of HCC cases, we examined the possible link between HBx, HURP, and cisplatin resistance in HCC cells. Hep3B cells expressing HBx showed not only elevated HURP mRNA and protein levels but also resistance to apoptosis induced by cisplatin. HURP silencing in cells expressing HBx reversed this process and enhanced sensitization of Hep3B cells to apoptosis. Notably, HBx overexpression induced SATB1, a global gene regulator that is upregulated in breast cancer. However, the role of SATB1 in the regulation of cell survival is unclear. We found that the anti-apoptotic effect of HBx requires p38/MAPK pathway activation in Hep3B cells. HBx also induced the expression of the anti-apoptotic protein survivin in an HURP-dependent manner[36]. We observed that the HBx produces anti-apoptotic effects in HCC cells, a process that may lead to chemoresistance. Enhanced chemoresistance of HCC cells that express HBx was associated with increased activity of several proteins, including STAB1, HURP, and survivin. Previous reports indicate that PKC negatively regulates SATB1 transactivation activity[37]. Our group showed that SATB1 and the p38/MAPK pathway mediates the anti-apoptotic activity of HBx. Therefore, it appears that HBx upregulates SATB1 and MAPK or HURP transcription. HURP induced survivin expression in HBx-expressing cells. ERK inhibition also inhibited surviving activity[36,38]; however, HURP protein levels remained constant in the presence of ERKi, an observation which suggested that HBx may induce survivin via another pathway that requiresthe ERK kinase (Figure 1). Our results may explain, at least in part, the cellular mechanism underlying the anti-apoptotic effect of HBx during the development of HBV-associated HCC. In agreement with our results, previous studies have shown that stable expression of HBx can stimulate PI3-kinase activity and suppress TGF-beta-induced apoptosis in Hep3B cells[8,22].
Figure 1.
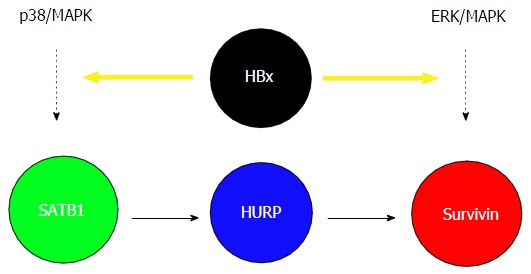
Proposed model to explain the link between hepatitis B virus X protein, p38/MAPK, SATB1, hepatoma upregulated protein, and survivin in mediating anti-apoptotic effects during cisplatin treatment. HBx upregulates the anti-apoptotic protein survivin through induction of p38/MAPK and ERK/MAPK pathways. Another less defined ERK/MAPK pathway which may regulate survivin independently of HURP is also shown. HBx: Hepatitis B virus X protein; HURP: Hepatoma upregulated protein.
SATB1, a chromatin organizer, is involved in gene regulation and the formation of chromosome loops, in addition to its role in the organization of transcriptionally poised chromatin[39]. SATB1 was initially described as a protein mediator of apoptosis[40]. We have shown the role of SATB1 in upregulating surviving and preventing apoptosis during cancer progression and establishment of chemoresistance[36]. SATB1 phosphorylation also appears to control interleukin-2 transcription as shown based on results obtained in a T-cell activation model; a similar mechanism may potentially be associated with SATB1 and its gene regulation activity[37]. In addition, SATB1 cleavage via sumoylation-directed caspase activity appears to regulate gene expression or may lead to clearance of immune cells[41]. Furthermore, SATB1 promotes cancer cell metastasis and overexpression of this protein increases resistance to chemotherapeutic drugs in breast cancer cells[42,43]. These observations suggest that HBx induces HURP expression by activating the p38/MAPK pathway and SATB1, leading to accumulation of survivin. We conclude that activation of the HBx/SATB1/HURP axis may increase chemoresistance in hepatic cancer cells.
HURP/GANKYRIN/p53 AXIS IN REGULATING HCC APOPTOSIS
The tumor suppressor p53 inhibits cancer development by inducing cell cycle arrest and apoptosis[44,45]. Some human tumors (10%) are characterized by overexpression of MDM2, an E3 ubiquitin ligase known for its role in the ubiquitination of p53 and its subsequent degradation by the proteasome[46]; this phenomenon may account for the development of many cancers, even those in which the p53 gene is no longer functional[47]. We found that overexpressing HURP in HEK293 cells induces p53 ubiquitination and degradation of the protein by the proteasome[48]. Conversely, HURP silencing with short-hairpin RNA reverses these processes. Knockdown of HURP promotes p53 accumulation and reduces cell proliferation in SK-Hep-1 cells (p53+/–), while Mahlavu cells (p53-mutant) are not affected. HURP silencing showed no effect on cellular proliferation in Hep3B and H1299 cells (lung carcinoma) (both lack p53 activity). In comparison, HURP silencing sensitized SK-Hep-1 cells to cisplatin. HURP overexpression not only reduced exogenous p53 expression in H1299 and Hep3B cells but also reduced sensitivity of these cells to cisplatin. Notably, HURP expression induced HEK293 cell proliferation in an anchorage-independent manner; moreover, injection of SK-Hep-1 cancer cells in which HURP had been silenced produced tumors of smaller size in immunocompromised mice compared to control[48].
The ankyrin-repeat oncoprotein gankyrin[49] has also been shown to be upregulated in HCC. Previous work indicated that this protein interacts with the product of retinoblastoma (Rb) gene as well as a subunit of the 26S proteasome subunit (S6 ATPase), a process that increases degradation of Rb[50,51]. Gankyrin is part of the 19S cap of the proteasome. This protein has an ankyrin repeat that forms alpha helices[51]. Gankyrin can increase the E3 ubiquitin ligase activity of MDM2, which regulates the degradation of the tumor suppressors p53 and Rb, which are both often mutated in human tumors[52,53]. Gankyrin regulates the cell cycle by mediating protein-protein interactions involving CDK4 (a cyclin-dependent kinase). Rb may inhibit the activity of gankyrin and lead to inhibition of MDM2-mediated p53 ubiquitination in HCC cells[54]. In our study[48], we observed that HURP represents an oncogene that reduces the level of p53 in normal cells and cancerous cells. Gankyrin was upregulated following HURP overexpression, and silencing of gankyrin reduced downregulation of p53 mediated by HURP. Importantly, high HURP levels positively correlated with gankyrin protein levels in HCC patients (n = 9; r2 = 0.778).
We propose a mechanism to explain the activity of HURP and its action on gankyrin accumulation in cancer cells (Figure 2). In this model, HURP prevents the ubiquitination and degradation of gankyrin but in a process that does not involve the disruption of the interaction between MDM2 and gankyrin. Alternatively, HURP may regulate the activity of other deubiquitination enzymes by inducing binding to the gankyrin/MDM2 protein complex (not illustrated in the model shown in Figure 2), which may subsequently inhibit MDM2’s effects on gankyrin degradation. More experimental data are needed to determine if HURP affects deubiquitination enzymes which interact with the MDM2/gankyrin protein complex. The degradation of p53 mediated by HURP may therefore be relevant to the development of HCC. Our findings identify a novel pathway for the malignant transformation induced by HURP and involving degradation of p53 and accumulation of gankyrin.
Figure 2.
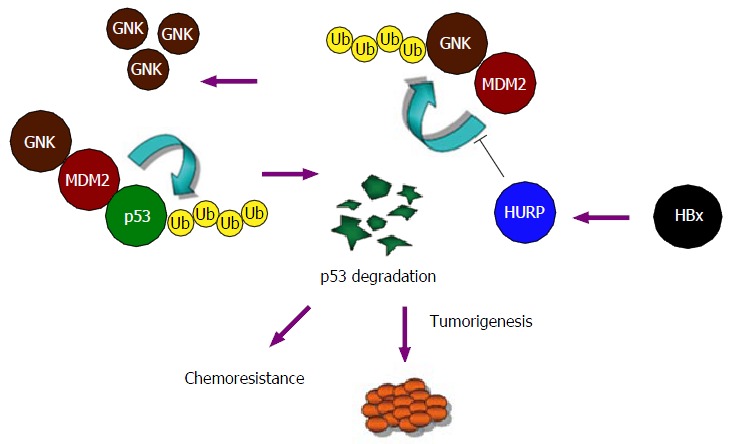
Simplified model illustrating the oncogenic properties of hepatitis B virus X protein and hepatoma upregulated protein in human liver cancer. In this cycle of gankyrin/MDM2-enhanced p53 degradation, HURP reduces MDM2-mediated ubiquitination of gankyrin, leading to accumulation of gankyrin in both normal and tumorigenic cells. Downstream effects of HURP appear to include malignant cell transformation and prevention of apoptosis induced by chemotherapeutic drug, processes which may in turn lead to the development of a chemoresistant cellular phenotype. HBx: Hepatitis B virus X protein; HURP: Hepatoma upregulated protein.
CONCLUSION
Our observations suggest that HBx induces HURP expression via the p38/MAPK pathway and SATB1 activity. This process leads to accumulation survivin, which possesses anti-apoptotic properties. Our results also identify a novel cellular pathway in which the oncogenic protein HURP induces cancer transformation by inducing p53 degradation and gankyrin accumulation. The processes of cell survival and apoptosis have been shown to be regulated by differential activation of p53 target genes[55]. For instance, CAS may bind to p53 on chromatin and this process may induce expression of a set of genes that facilitate apoptosis[56]. In contrast, interaction between the zinc-finger protein Hzf and p53 activates expression of growth-arrest genes and promotes cell survival[56,57]. HURP-mediated p53 degradation thus appears to be relevant for the development of HCC. In conclusion, recent advances regarding the oncogenic proteins HBx and HURP as described here offer new strategies to defeat human liver cancer.
ACKNOWLEDGMENTS
The author would like to thank Ms. Chiaying Yang for technical assistance as well Dr. Kuo TC (Kuo JY) for helpful discussions.
Footnotes
Conflict-of-interest statement: No conflict of interest.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Taiwan
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
Peer-review started: April 22, 2016
First decision: June 6, 2016
Article in press: July 22, 2016
P- Reviewer: Piiper A, Tomizawa M S- Editor: Gong ZM L- Editor: A E- Editor: Li D
References
- 1.Bouchard MJ, Schneider RJ. The enigmatic X gene of hepatitis B virus. J Virol. 2004;78:12725–12734. doi: 10.1128/JVI.78.23.12725-12734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang H, Oishi N, Kaneko S, Murakami S. Molecular functions and biological roles of hepatitis B virus x protein. Cancer Sci. 2006;97:977–983. doi: 10.1111/j.1349-7006.2006.00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yen TS. Hepadnaviral X Protein: Review of Recent Progress. J Biomed Sci. 1996;3:20–30. doi: 10.1007/BF02253575. [DOI] [PubMed] [Google Scholar]
- 4.McClain SL, Clippinger AJ, Lizzano R, Bouchard MJ. Hepatitis B virus replication is associated with an HBx-dependent mitochondrion-regulated increase in cytosolic calcium levels. J Virol. 2007;81:12061–12065. doi: 10.1128/JVI.00740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouchard MJ, Puro RJ, Wang L, Schneider RJ. Activation and inhibition of cellular calcium and tyrosine kinase signaling pathways identify targets of the HBx protein involved in hepatitis B virus replication. J Virol. 2003;77:7713–7719. doi: 10.1128/JVI.77.14.7713-7719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wen Y, Golubkov VS, Strongin AY, Jiang W, Reed JC. Interaction of hepatitis B viral oncoprotein with cellular target HBXIP dysregulates centrosome dynamics and mitotic spindle formation. J Biol Chem. 2008;283:2793–2803. doi: 10.1074/jbc.M708419200. [DOI] [PubMed] [Google Scholar]
- 7.Kim H, Lee H, Yun Y. X-gene product of hepatitis B virus induces apoptosis in liver cells. J Biol Chem. 1998;273:381–385. doi: 10.1074/jbc.273.1.381. [DOI] [PubMed] [Google Scholar]
- 8.Shih WL, Kuo ML, Chuang SE, Cheng AL, Doong SL. Hepatitis B virus X protein inhibits transforming growth factor-beta -induced apoptosis through the activation of phosphatidylinositol 3-kinase pathway. J Biol Chem. 2000;275:25858–25864. doi: 10.1074/jbc.M003578200. [DOI] [PubMed] [Google Scholar]
- 9.Su F, Schneider RJ. Hepatitis B virus HBx protein sensitizes cells to apoptotic killing by tumor necrosis factor alpha. Proc Natl Acad Sci USA. 1997;94:8744–8749. doi: 10.1073/pnas.94.16.8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li T, Robert EI, van Breugel PC, Strubin M, Zheng N. A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery. Nat Struct Mol Biol. 2010;17:105–111. doi: 10.1038/nsmb.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feitelson M. Hepatitis B virus infection and primary hepatocellular carcinoma. Clin Microbiol Rev. 1992;5:275–301. doi: 10.1128/cmr.5.3.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murakami S. Hepatitis B virus X protein: structure, function and biology. Intervirology. 1999;42:81–99. doi: 10.1159/000024969. [DOI] [PubMed] [Google Scholar]
- 13.Robinson WS. Molecular events in the pathogenesis of hepadnavirus-associated hepatocellular carcinoma. Annu Rev Med. 1994;45:297–323. doi: 10.1146/annurev.med.45.1.297. [DOI] [PubMed] [Google Scholar]
- 14.Kew MC. Hepatitis B virus x protein in the pathogenesis of hepatitis B virus-induced hepatocellular carcinoma. J Gastroenterol Hepatol. 2011;26 Suppl 1:144–152. doi: 10.1111/j.1440-1746.2010.06546.x. [DOI] [PubMed] [Google Scholar]
- 15.Kim SY, Kim JK, Kim HJ, Ahn JK. Hepatitis B virus X protein sensitizes UV-induced apoptosis by transcriptional transactivation of Fas ligand gene expression. IUBMB Life. 2005;57:651–658. doi: 10.1080/15216540500239697. [DOI] [PubMed] [Google Scholar]
- 16.Koike K, Moriya K, Yotsuyanagi H, Iino S, Kurokawa K. Induction of cell cycle progression by hepatitis B virus HBx gene expression in quiescent mouse fibroblasts. J Clin Invest. 1994;94:44–49. doi: 10.1172/JCI117343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin N, Chen HY, Li D, Zhang SJ, Cheng ZX, Wang XZ. Apoptosis and its pathway in X gene-transfected HepG2 cells. World J Gastroenterol. 2005;11:4326–4331. doi: 10.3748/wjg.v11.i28.4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miao J, Chen GG, Chun SY, Lai PP. Hepatitis B virus X protein induces apoptosis in hepatoma cells through inhibiting Bcl-xL expression. Cancer Lett. 2006;236:115–124. doi: 10.1016/j.canlet.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 19.Su F, Theodosis CN, Schneider RJ. Role of NF-kappaB and myc proteins in apoptosis induced by hepatitis B virus HBx protein. J Virol. 2001;75:215–225. doi: 10.1128/JVI.75.1.215-225.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng AS, Wong N, Tse AM, Chan KY, Chan KK, Sung JJ, Chan HL. RNA interference targeting HBx suppresses tumor growth and enhances cisplatin chemosensitivity in human hepatocellular carcinoma. Cancer Lett. 2007;253:43–52. doi: 10.1016/j.canlet.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Murata M, Matsuzaki K, Yoshida K, Sekimoto G, Tahashi Y, Mori S, Uemura Y, Sakaida N, Fujisawa J, Seki T, et al. Hepatitis B virus X protein shifts human hepatic transforming growth factor (TGF)-beta signaling from tumor suppression to oncogenesis in early chronic hepatitis B. Hepatology. 2009;49:1203–1217. doi: 10.1002/hep.22765. [DOI] [PubMed] [Google Scholar]
- 22.Shih WL, Kuo ML, Chuang SE, Cheng AL, Doong SL. Hepatitis B virus X protein activates a survival signaling by linking SRC to phosphatidylinositol 3-kinase. J Biol Chem. 2003;278:31807–31813. doi: 10.1074/jbc.M302580200. [DOI] [PubMed] [Google Scholar]
- 23.Wu BK, Li CC, Chen HJ, Chang JL, Jeng KS, Chou CK, Hsu MT, Tsai TF. Blocking of G1/S transition and cell death in the regenerating liver of Hepatitis B virus X protein transgenic mice. Biochem Biophys Res Commun. 2006;340:916–928. doi: 10.1016/j.bbrc.2005.12.089. [DOI] [PubMed] [Google Scholar]
- 24.Tsou AP, Yang CW, Huang CY, Yu RC, Lee YC, Chang CW, Chen BR, Chung YF, Fann MJ, Chi CW, et al. Identification of a novel cell cycle regulated gene, HURP, overexpressed in human hepatocellular carcinoma. Oncogene. 2003;22:298–307. doi: 10.1038/sj.onc.1206129. [DOI] [PubMed] [Google Scholar]
- 25.Nomura N, Miyajima N, Sazuka T, Tanaka A, Kawarabayasi Y, Sato S, Nagase T, Seki N, Ishikawa K, Tabata S. Prediction of the coding sequences of unidentified human genes. I. The coding sequences of 40 new genes (KIAA0001-KIAA0040) deduced by analysis of randomly sampled cDNA clones from human immature myeloid cell line KG-1. DNA Res. 1994;1:27–35. doi: 10.1093/dnares/1.1.27. [DOI] [PubMed] [Google Scholar]
- 26.Bassal S, Nomura N, Venter D, Brand K, McKay MJ, van der Spek PJ. Characterization of a novel human cell-cycle-regulated homologue of Drosophila dlg1. Genomics. 2001;77:5–7. doi: 10.1006/geno.2001.6570. [DOI] [PubMed] [Google Scholar]
- 27.Gudmundsson KO, Thorsteinsson L, Sigurjonsson OE, Keller JR, Olafsson K, Egeland T, Gudmundsson S, Rafnar T. Gene expression analysis of hematopoietic progenitor cells identifies Dlg7 as a potential stem cell gene. Stem Cells. 2007;25:1498–1506. doi: 10.1634/stemcells.2005-0479. [DOI] [PubMed] [Google Scholar]
- 28.Yu CT, Hsu JM, Lee YC, Tsou AP, Chou CK, Huang CY. Phosphorylation and stabilization of HURP by Aurora-A: implication of HURP as a transforming target of Aurora-A. Mol Cell Biol. 2005;25:5789–5800. doi: 10.1128/MCB.25.14.5789-5800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koffa MD, Casanova CM, Santarella R, Köcher T, Wilm M, Mattaj IW. HURP is part of a Ran-dependent complex involved in spindle formation. Curr Biol. 2006;16:743–754. doi: 10.1016/j.cub.2006.03.056. [DOI] [PubMed] [Google Scholar]
- 30.Silljé HH, Nagel S, Körner R, Nigg EA. HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr Biol. 2006;16:731–742. doi: 10.1016/j.cub.2006.02.070. [DOI] [PubMed] [Google Scholar]
- 31.Wong J, Fang G. HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J Cell Biol. 2006;173:879–891. doi: 10.1083/jcb.200511132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong J, Lerrigo R, Jang CY, Fang G. Aurora A regulates the activity of HURP by controlling the accessibility of its microtubule-binding domain. Mol Biol Cell. 2008;19:2083–2091. doi: 10.1091/mbc.E07-10-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Betz MJ, Beuschlein F. Diagnosis: Novel molecular signatures for adrenocortical carcinoma. Nat Rev Endocrinol. 2009;5:297–299. doi: 10.1038/nrendo.2009.93. [DOI] [PubMed] [Google Scholar]
- 34.de Reyniès A, Assié G, Rickman DS, Tissier F, Groussin L, René-Corail F, Dousset B, Bertagna X, Clauser E, Bertherat J. Gene expression profiling reveals a new classification of adrenocortical tumors and identifies molecular predictors of malignancy and survival. J Clin Oncol. 2009;27:1108–1115. doi: 10.1200/JCO.2008.18.5678. [DOI] [PubMed] [Google Scholar]
- 35.Hsu JM, Lee YC, Yu CT, Huang CY. Fbx7 functions in the SCF complex regulating Cdk1-cyclin B-phosphorylated hepatoma up-regulated protein (HURP) proteolysis by a proline-rich region. J Biol Chem. 2004;279:32592–32602. doi: 10.1074/jbc.M404950200. [DOI] [PubMed] [Google Scholar]
- 36.Kuo TC, Chao CC. Hepatitis B virus X protein prevents apoptosis of hepatocellular carcinoma cells by upregulating SATB1 and HURP expression. Biochem Pharmacol. 2010;80:1093–1102. doi: 10.1016/j.bcp.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 37.Pavan Kumar P, Purbey PK, Sinha CK, Notani D, Limaye A, Jayani RS, Galande S. Phosphorylation of SATB1, a global gene regulator, acts as a molecular switch regulating its transcriptional activity in vivo. Mol Cell. 2006;22:231–243. doi: 10.1016/j.molcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 38.Wiesenauer CA, Yip-Schneider MT, Wang Y, Schmidt CM. Multiple anticancer effects of blocking MEK-ERK signaling in hepatocellular carcinoma. J Am Coll Surg. 2004;198:410–421. doi: 10.1016/j.jamcollsurg.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 39.Kumar PP, Bischof O, Purbey PK, Notani D, Urlaub H, Dejean A, Galande S. Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat Cell Biol. 2007;9:45–56. doi: 10.1038/ncb1516. [DOI] [PubMed] [Google Scholar]
- 40.Zweyer M, Riederer BM, Ochs RL, Fackelmayer FO, Kohwi-Shigematsu T, Bareggi R, Narducci P, Martelli AM. Association of nuclear matrix proteins with granular and threaded nuclear bodies in cell lines undergoing apoptosis. Exp Cell Res. 1997;230:325–336. doi: 10.1006/excr.1996.3415. [DOI] [PubMed] [Google Scholar]
- 41.Tan JA, Sun Y, Song J, Chen Y, Krontiris TG, Durrin LK. SUMO conjugation to the matrix attachment region-binding protein, special AT-rich sequence-binding protein-1 (SATB1), targets SATB1 to promyelocytic nuclear bodies where it undergoes caspase cleavage. J Biol Chem. 2008;283:18124–18134. doi: 10.1074/jbc.M800512200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 2008;452:187–193. doi: 10.1038/nature06781. [DOI] [PubMed] [Google Scholar]
- 43.Li QQ, Chen ZQ, Xu JD, Cao XX, Chen Q, Liu XP, Xu ZD. Overexpression and involvement of special AT-rich sequence binding protein 1 in multidrug resistance in human breast carcinoma cells. Cancer Sci. 2010;101:80–86. doi: 10.1111/j.1349-7006.2009.01372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 45.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 46.Yang Y, Li CC, Weissman AM. Regulating the p53 system through ubiquitination. Oncogene. 2004;23:2096–2106. doi: 10.1038/sj.onc.1207411. [DOI] [PubMed] [Google Scholar]
- 47.Michael D, Oren M. The p53-Mdm2 module and the ubiquitin system. Semin Cancer Biol. 2003;13:49–58. doi: 10.1016/s1044-579x(02)00099-8. [DOI] [PubMed] [Google Scholar]
- 48.Kuo TC, Chang PY, Huang SF, Chou CK, Chao CC. Knockdown of HURP inhibits the proliferation of hepacellular carcinoma cells via downregulation of gankyrin and accumulation of p53. BiochemPharmacol. 2012;83:758–768. doi: 10.1016/j.bcp.2011.12.034. [DOI] [PubMed] [Google Scholar]
- 49.Higashitsuji H, Itoh K, Nagao T, Dawson S, Nonoguchi K, Kido T, Mayer RJ, Arii S, Fujita J. Reduced stability of retinoblastoma protein by gankyrin, an oncogenic ankyrin-repeat protein overexpressed in hepatomas. Nat Med. 2000;6:96–99. doi: 10.1038/71600. [DOI] [PubMed] [Google Scholar]
- 50.Dawson S, Apcher S, Mee M, Higashitsuji H, Baker R, Uhle S, Dubiel W, Fujita J, Mayer RJ. Gankyrin is an ankyrin-repeat oncoprotein that interacts with CDK4 kinase and the S6 ATPase of the 26 S proteasome. J Biol Chem. 2002;277:10893–10902. doi: 10.1074/jbc.M107313200. [DOI] [PubMed] [Google Scholar]
- 51.Krzywda S, Brzozowski AM, Higashitsuji H, Fujita J, Welchman R, Dawson S, Mayer RJ, Wilkinson AJ. The crystal structure of gankyrin, an oncoprotein found in complexes with cyclin-dependent kinase 4, a 19 S proteasomal ATPase regulator, and the tumor suppressors Rb and p53. J Biol Chem. 2004;279:1541–1545. doi: 10.1074/jbc.M310265200. [DOI] [PubMed] [Google Scholar]
- 52.Higashitsuji H, Liu Y, Mayer RJ, Fujita J. The oncoproteingankyrin negatively regulates both p53 and RB by enhancing proteasomal degradation. Cell Cycle. 2005;4:1335–1337. doi: 10.4161/cc.4.10.2107. [DOI] [PubMed] [Google Scholar]
- 53.Higashitsuji H, Higashitsuji H, Itoh K, Sakurai T, Nagao T, Sumitomo Y, Masuda T, Dawson S, Shimada Y, Mayer RJ, et al. The oncoproteingankyrin binds to MDM2/HDM2, enhancing ubiquitylation and degradation of p53. Cancer Cell. 2005;8:75–87. doi: 10.1016/j.ccr.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 54.Qiu W, Wu J, Walsh EM, Zhang Y, Chen CY, Fujita J, Xiao ZX. Retinoblastoma protein modulates gankyrin-MDM2 in regulation of p53 stability and chemosensitivity in cancer cells. Oncogene. 2008;27:4034–4043. doi: 10.1038/onc.2008.43. [DOI] [PubMed] [Google Scholar]
- 55.Aylon Y, Oren M. Living with p53, dying of p53. Cell. 2007;130:597–600. doi: 10.1016/j.cell.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 56.Tanaka T, Ohkubo S, Tatsuno I, Prives C. hCAS/CSE1L associates with chromatin and regulates expression of select p53 target genes. Cell. 2007;130:638–650. doi: 10.1016/j.cell.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 57.Das S, Raj L, Zhao B, Kimura Y, Bernstein A, Aaronson SA, Lee SW. Hzf Determines cell survival upon genotoxic stress by modulating p53 transactivation. Cell. 2007;130:624–637. doi: 10.1016/j.cell.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]


