Abstract
RNA transcripts are bound and regulated by RNA-binding proteins (RBPs). Current methods for identifying in vivo targets of a RBP are imperfect and not amenable to examining small numbers of cells. To address these issues, we developed TRIBE (Targets of RNA-binding proteins Identified By Editing), a technique that couples an RBP to the catalytic domain of the Drosophila RNA editing enzyme ADAR and expresses the fusion protein in vivo. RBP targets are marked with novel RNA editing events and identified by sequencing RNA. We have used TRIBE to identify the targets of three RBPs (Hrp48, dFMR1 and NonA). TRIBE compares favorably to other methods, including CLIP, and we have identified RBP targets from as little as 150 specific fly neurons. TRIBE can be performed without an antibody and in small numbers of specific cells.
Introduction
Post-transcriptional regulation of gene expression is mediated by a host of proteins which bind to pre-mRNA and mRNA. Their activity is essential for the correct splicing, localization and translation of cellular components, and their dysregulation is implicated in numerous human diseases (Lukong et al., 2008, Kang et al., 2013, Nussbacher et al., 2015). A complete functional understanding of any RNA-binding protein (RBP) requires the identification of its RNA targets. However, identifying biologically relevant RBP targets is challenging. Although there are very good approaches to define in vitro targets, in vivo target identification is a more complicated exercise. There is growing appreciation that even seemingly homogenous tissues are composed of different cell types that can exhibit striking differences in gene expression, proteome and phenotypic output (Jaitin et al., 2014). Cell types can be broken down even further into subpopulations, and single-cell transcriptional studies have revealed substantial gene expression differences even between individual cells of the same apparent type (Shalek et al., 2013). Therefore, because the targets of many RBPs are also likely to be different between tissue and cell types, it is crucial to identify cell-specific targets.
Traditional methods of RBP target identification, typically immunoprecipitation of RBP-bound target RNAs, are generally performed on mixed tissues and as such are plagued by issues such as post-lysis in vitro association of RBPs with spurious targets (Mili and Steitz, 2004). Furthermore, results can change dramatically with seemingly subtle differences in experimental conditions. The long list of candidate targets for FMRP (an RBP associated with Fragile X Syndrome) with little overlap between labs, is testament to this fact (Darnell et al., 2005). Similarly, multiple studies performed on the ALS-associated RBP TDP-43 have also yielded strikingly non-overlapping sets of targets (Buratti et al., 2013).
More sophisticated methods include the current gold standard for the identification of RNA-binding protein (RBP) targets in vivo, CLIP (Cross Linking and Immunoprecipitation) and variants thereof (here we use the term CLIP to include all variants) (Ule et al., 2003, Ule et al., 2005, Hafner et al., 2010, Huppertz et al., 2014, Moore et al., 2014). These methods are based on immunoprecipitation and involve creating covalent interactions between the RBP and its targets within cells and tissues, digesting unprotected RNA and sequencing the remaining ’bound’ RNA. Despite its myriad advantages, CLIP has several disadvantages (Darnell, 2010, Moore et al., 2014). Prominent among them is the requirement for a high affinity, specific antibody and the inefficiency of cross-linking (generally 1-5%) (Darnell, 2010). CLIP therefore requires rather large amounts of material (currently millions of cells), and as such is best suited to the examination of targets in whole tissue rather than specific cells.
It should be possible to identify cell-specific RBP targets using CLIP (using an epitope-tagged RBP expressed in a cell-specific manner), but this is still technically challenging. Crosslinking is compromised by limited UV penetration in some tissues and more importantly by the low amounts of material in restricted cell populations. Although CLIP was first described in 2005 (Ule et al., 2005), to our knowledge no such cell-specific experiments have yet been published.
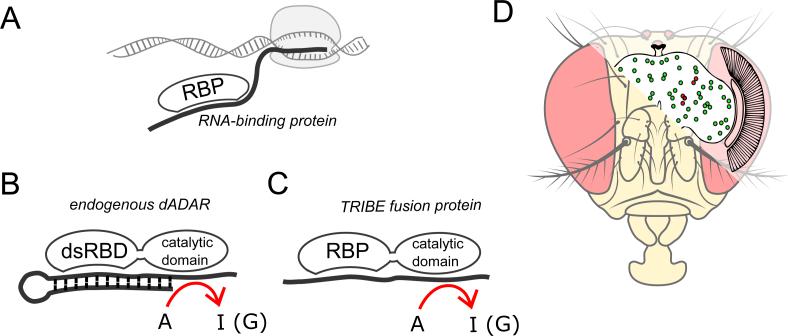
To circumvent these issues, we developed an orthogonal technique to identify RNA-binding protein (RBP) substrates. It is called TRIBE, Targets of RNA-binding proteins Identified By Editing, and is particularly well-suited to identify target RNAs within small numbers of cells. TRIBE is conceptually similar to the DNA-oriented ‘targeted DamID’ (van Steensel and Henikoff, 2000, Southall et al., 2013) and entails in vivo expression of a fusion protein between an RBP and the catalytic domain of the RNA-editing enzyme ADAR. The transcriptome is then sequenced to identify novel editing events introduced by the RPB-ADARcd.
Endogenous Drosophila ADAR is a modular enzyme consisting of two double-stranded RNA-binding domains (dsRBDs) as well as a catalytic domain, which deaminates adenosine to inosine (Bass and Weintraub, 1988, Keegan et al., 2004). Inosine is recognized as guanosine both in vitro and in vivo. Because the TRIBE fusion protein only contains the catalytic domain of ADAR (ADARcd) and is lacking the RNA recognition features of ADAR, the specificity of the RBP should dictate its target transcripts (analogous to other methods of directing ADARcd-meditated editing) (Montiel-Gonzalez et al., 2013, Vogel et al., 2014). Importantly, the editing event is irreversible, marking the target RNA permanently after the interaction. No biochemistry is required, RNA is simply extracted from cells of interest and sequenced. Novel editing events define RNA targets, namely, transcripts that interacted with the fusion protein. Comparison with CLIP data indicates that TRIBE works well for multiple RBPs and is capable of identifying cell-specific targets.
Results
TRIBE involves the fusion of an RBP to the catalytic domain of an RNA-editing enzyme (ADARcd). Because the double stranded RNA-binding (dsRBD) regions of ADAR are missing from the fusion protein, its editing specificity is determined by the RNA recognition features of the RBP; target transcripts are edited in vivo and then identified by RNA sequencing (Figure 1). TRIBE is compatible with cell culture but also applicable to the identification of cell-specific RBP targets. For example, purification and sequencing of RNA from specific cells, achieved by co-expression of fluorescent protein and FACS or manual cell-sorting, allows for the identification of targets in tiny numbers of neurons (see below).
Figure 1. Targets of RNA-binding proteins Identified By Editing (TRIBE): A fusion protein of an RNA-binding protein and the catalytic domain of ADAR will edit the target transcripts of the RNA-binding protein.
A) The aim of this technique is to identify the binding target transcripts of a specific RNA-binding protein (RBP). B) Native Drosophila ADAR is composed of two double stranded RNA-binding domains (dsRBDs) that mediates its target specificity, and a deaminase domain that catalyzes adenosine to inosine conversion. C) The dsRBDs of ADAR are replaced with the RBP of interest. The editing specificity of the fusion protein is determined by the RNA recognition features of the RBP and the target transcript is permanently marked by a novel editing event. D) Cell-specific expression of the fusion protein will allow identification of targets in discrete populations of cells in vivo. Co-expression of a fluorescent protein allows for enrichment of RNA from the cells of interest. Examples of Drosophila neuronal subsets examined here are the core circadian pacemaker neurons (pdf expressing, ~ 16 cell/brain (red)) and dopaminergic neurons (tyrosine hydroxylase expressing, ~1000 cells/brain (green)).
The Hrp48-TRIBE fusion protein has editing activity and Hrp48-determined specificity
Hrp48 (also called Hrb27C) is a homolog of the mammalian hnRNP A/B family and implicated in splicing regulation, mRNA localization and translation (Huynh et al., 2004, Blanchette et al., 2005, Nelson et al., 2007, Blanchette et al., 2009). We chose it as an initial RBP because it has well-characterized targets, there is an excellent antibody (Hammond et al., 1997), and we had preliminary data that it participates in circadian regulation, a major interest of our lab (data not shown).
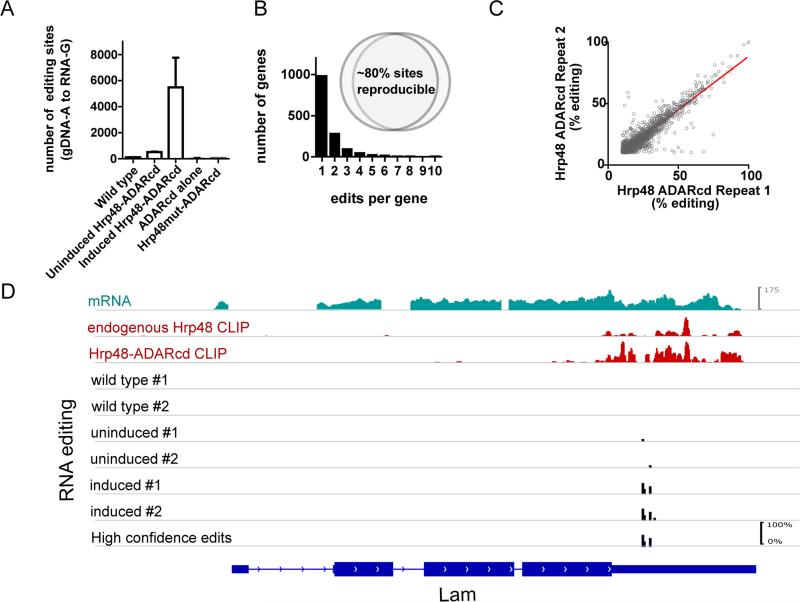
Initial experiments were performed by creating stable Drosophila S2 cell lines that express the Hrp48-ADARcd fusion protein (henceforth referred to as Hrp48-TRIBE) under inducible control. Expression of this protein in S2 cells, which have extremely low levels of endogenous editing, lead to a dramatic increase in the number of detected editing events (approximately 20 fold, Fig. 2A). They are confined to the correct base conversion, A to G (data not shown), indicating that the fusion protein is catalyzing the appropriate deamination reaction. Induction of the ADAR catalytic domain alone results in no increase in editing sites, despite stable protein expression (Fig. 2A). Similarly, Hrp48-TRIBE with mutated Hrp48 RNA-binding domains is stably expressed (not shown) and also causes no increase in editing sites, indicating a requirement for the RNA-binding ability of Hrp48 (Fig. 2A).
Figure 2. The TRIBE fusion protein reproducibly edits certain sites.
(A) An increase in A to G editing events is observed upon induction of the fusion protein in S2 cells. No increase in editing sites are observed when an ADAR catalytic domain alone is expressed, or when Hrp48mut-ADARcd (Hrp48 with mutated RNA-binding domains). The same genes and the same sites are reproducibly edited across biological replicates at similar efficiencies (B inset, C). A frequency histogram of number of edits per target genes show that most genes have only one editing site but the TRIBE protein has strong specificity for certain sites (B), and that those sites are edited to a similar degree between biological repeats (R2 = 0.859). D) Endogenous and fusion protein have similar binding patterns and TRIBE editing reflects the pattern of the CLIP signal. An example gene, Lam, showing mRNA expression and CLIP signal (top three panels), and editing tracks for wild type cells, stable cells lines (Hrp48-TRIBE) without and with induction of expression of the fusion protein. Editing events are indicated by black bars, the height of the bar indicates the percentage editing at that site.
The majority of target genes are marked by a single editing site and these events are reproducible both in their position and frequency (% editing), indicating that Hrp48-TRIBE exhibits specificity (Fig. 2B,C). Genes that are marked by editing sites in both biological replicas are defined as high confidence TRIBE targets (Fig. 2B,D, Table S1). For Hrp48, these 1401 targets constitute approximately 20% of all expressed genes (Fig. 2B), and have a wide range of expression levels (data not shown), indicating that TRIBE is selective.
CLIP and TRIBE agree that Hrp48 preferentially binds to the 3'UTR of transcripts
To assess if editing sites reflect true Hrp48 target genes, we performed a series of CLIP-seq experiments to address target preference by this more traditional method. Firstly, CLIP of endogenous Hrp48 shows the same binding pattern as CLIP of the Hrp48-TRIBE fusion protein. Both proteins are strongly enriched in the 3’ UTR, as is seen in an example gene Lam (Fig. 2D.) In a well-characterized target of Hrp48, Hsp83 both Hrp48 and Hrp48-TRIBE CLIP show binding throughout the 3’ UTR surrounding a specific element previously shown to be bound by Hrp48 (Fig. S1) (Bashirullah et al., 1999, Nelson et al., 2007). This 3’ UTR binding pattern holds transcriptome-wide (Fig. 3A) and suggests that fusion to the ADARcd does not markedly interfere with the ability of the RBP to recognize and bind to its normal targets. Similarly, fusion of the ADARcd to Hrp48 did not alter its largely cytoplasmic localization pattern (as assayed by immunocytochemistry and subcellular fractionation, data not shown).
Figure 3. TRIBE demonstrates Hrp48 preferentially binds the 3’ UTR of transcripts.
A) Both CLIP signal and editing sites are enriched in the 3'UTR. Metagene quantification of the location of either CLIP peaks or TRIBE edits. Background indicates the proportion of the fly transcriptome composed of the indicated regions. The majority of TRIBE editing sites are near CLIP peaks (B). The fraction of editing sites within a certain distance of a CLIP peak was quantified for both endogenous Hrp48 and Hrp48-TRIBE. 0 indicates that the editing site was within the bounds of a CLIP peak. C) TRIBE targets are a subset of CLIP targets. Venn diagram overlap of genes between all expressed genes, all TRIBE target genes and all genes with at least one statistically significant CLIP peak. D) TRIBE targets are more CLIP enriched. Frequency distribution of per gene CLIP enrichments of all CLIP target genes and TRIBE genes that have CLIP signal. The overlap between the top 25% ranked CLIP targets (highlighted in pink) and TRIBE targets is inset.
The 3’ UTR binding preference of Hrp48 identified by CLIP is closely mirrored by the pattern of Hrp48-TRIBE editing sites. Transcriptome-wide, approximately 50% of Hrp48-TRIBE editing sites and 50% of Hrp48 CLIP peaks, are found in the 3’ UTR (Fig. 3A). This suggests that TRIBE editing marks endogenous targets quite close to the region of the RBP binding site.
Indeed, Hrp48-TRIBE edits within the previously characterized binding element of Hsp83 (Fig. S1), showing that TRIBE can mark an RBP binding site. Overall, ~10% of TRIBE editing sites are located within a CLIP peak (as in the Hsp83 example), and ~80% are less than 500bp from a CLIP peak (Fig. 3B). Motif analysis of the Hrp48 CLIP data identify TA-rich binding motifs similar to that previously published using in vitro selection (Blanchette et al., 2009), but TA-rich motifs were not found surrounding editing sites (Fig. S2). This suggests that TRIBE usually edits near but not at the binding site, consistent with fusion of the editing moiety to the C-terminus of Hrp48. However, some more distant editing might be due to RNA flexibility and/or the dsRNA preference of the ADARcd (see Discussion).
Hrp48-TRIBE defines many fewer targets than Hrp48 CLIP. Most expressed genes have at least one statistically significant CLIP peak, with similar results from endogenous Hrp48 and the Hrp48-TRIBE fusion protein (Fig. 3C). This is in contrast to TRIBE targets, which are more restricted and comprise only ~20% of expressed genes (Fig. 3C). These results are from very deep sequencing (~200 M reads), suggesting that the more restricted target set is not merely due to lack of sensitivity. Because a single, statistically significant CLIP peak is not a stringent threshold for defining a target, total CLIP enrichment (relative to expression) was calculated per gene. Compared to the whole population of genes identified by CLIP, Hrp48-TRIBE target genes are biased toward higher CLIP enrichment (right shifted distribution, Fig 3D); indeed, 45% of the top quarter ranked CLIP targets are also TRIBE targets, suggesting that Hrp48-TRIBE preferentially detects stronger Hrp48 CLIP targets (Fig 3D, inset). Structure prediction modeling of the regions surrounding TRIBE editing events suggest that editing occurs preferentially at a bulge embedded within dsRNA (Fig. S3A), similar to an RNA structure edited by the human ADAR2cd when expressed in S. cerevisae (Fig. S3B) (Yi-Brunozzi et al., 2001, Gupta et al., 2012, Eifler et al., 2013). This structure is absent from CLIP binding regions that lack an editing site (Fig. S3A). Taken together, the cell culture data for Hrp48-TRIBE indicate that TRIBE labels many endogenous Hrp48 targets and in the correct metagene location.
dFMR1-TRIBE preferentially edits coding sequence, reflecting prior CLIP data
A second TRIBE protein was created by fusing dFMR1, the Drosophila ortholog of FMRP (Fragile X Mental Retardation Protein) to the ADARcd. dFMR1 has roles in mRNA localization and translational regulation in neurons (Dictenberg et al., 2008, Darnell et al., 2011).
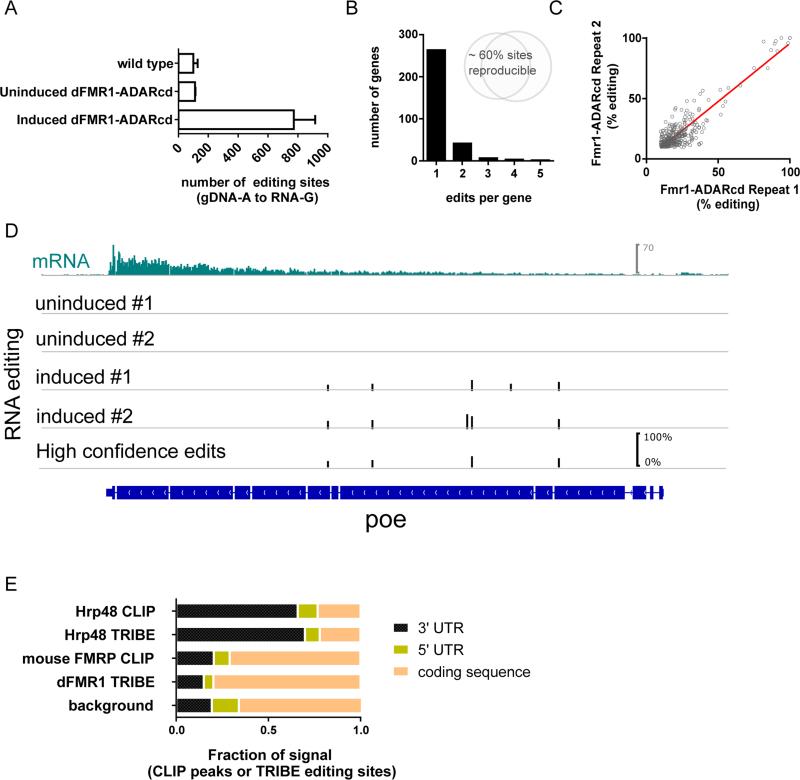
As observed for Hrp48-TRIBE, induction of dFMR1-TRIBE expression in Drosophila S2 cells led to a robust increase in editing events (Fig. 4A). The majority of targets are marked by only one editing site, and these individual editing events are reproducible both in their position and frequency (% editing), indicating that dFMR1-TRIBE manifests specificity (Fig.3b,C). Genes that are marked by editing sites in two biological repeats are classified as high confidence TRIBE targets (Fig. 4B; Table S1). In the case of dFMR1, these 315 targets constitute approximately 5% of all expressed genes.
Figure 4. TRIBE demonstrates dFMR1 preferentially binds the coding sequence of transcripts.
A) The dFMR1-TRIBE protein retains deaminase activity, an increase in A to G editing events is observed upon induction of the fusion protein in S2 cells. The same genes and the same sites are reproducibly edited across biological replicates at similar efficiencies (B,C). A frequency histogram of number of edits per target genes show that most genes have only one editing site but the TRIBE protein has strong specificity for certain sites (B), and that those sites are edited to a similar degree between biological repeats (R2 = 0.86) (C).
D) An example gene, poe, showing mRNA expression (top panel), and editing tracks for stable cells lines (dFMR1-TRIBE) without and with induction of expression of the fusion protein. Editing events are indicated by black bars, the height of the bar indicates the percentage editing at that site. E) Metagene quantification of the location of either CLIP peaks or TRIBE edits. Background indicates the proportion of the fly transcriptome composed of the indicated regions. Intronic sites are excluded from the analysis here to allow direct comparison to mouse FMRP CLIP data from Darnell et al. (2011).
In stark contrast to the distribution of Hrp48-TRIBE editing events, dFMR1-TRIBE editing events are distributed throughout the coding region of transcripts (Fig. 4D, E). This same pattern has been observed by CLIP of FMRP in mammalian cells (Darnell et al., 2011, Ascano et al., 2012) and is consistent with the role of dFMR1 as a translation regulator. Further examination of mouse brain FMRP CLIP targets revealed that the mouse homologs of the dFMR1-TRIBE target genes were biased toward higher CLIP rankings, suggesting that dFMR1-TRIBE identifies conserved targets of FMRP (Fig. S4A) (Darnell et al., 2011).
TRIBE shows that NonA preferentially binds introns
A third Drosophila RBP, NonA, is the ortholog of the mammalian protein NonO and was assayed in a similar manner. NonO is a multifunctional protein involved in the function of nuclear paraspeckles as well as other nuclear events like splicing, mRNA export and the regulation of transcription (Kozlova et al., 2006, Kaneko et al., 2007). As for Hrp48, our interest in NonA is due to its role in circadian biology (Brown et al., 2005).
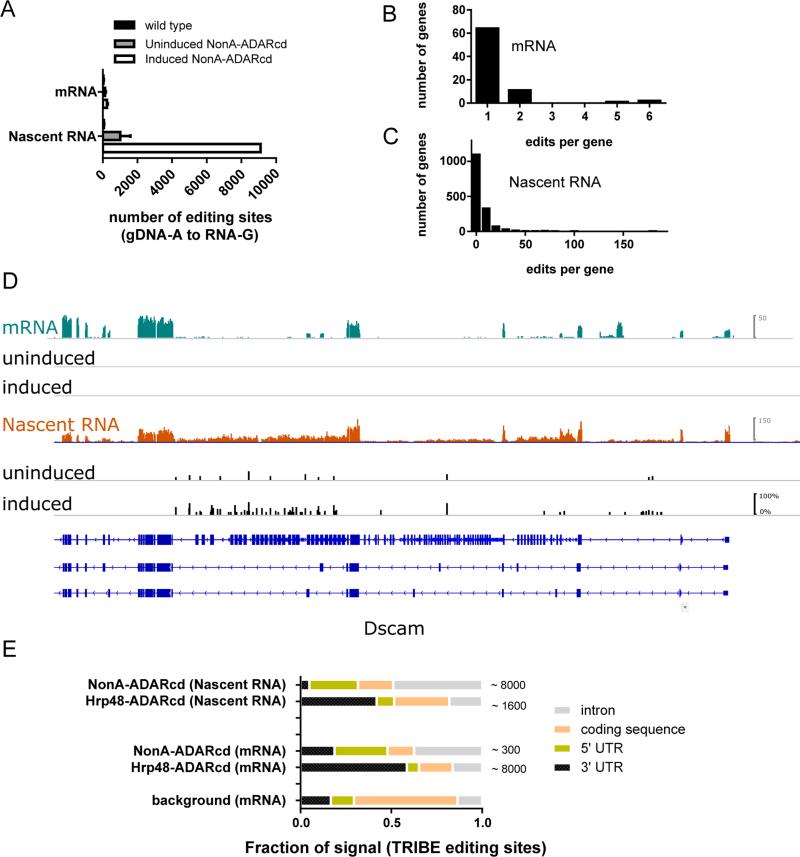
Expression of the NonA-TRIBE fusion protein in S2 cells led to a small increase in mRNA editing (approximately 3-fold; Fig. 5A top), much less than what was observed above with the dFMR1 and Hpr48 fusion proteins. Because of the known nuclear functions of NonA, we considered that NonA might preferentially bind nascently transcribed nuclear RNA. To this end, we isolated and sequenced chromatin-associated RNA from S2 cells (Wuarin and Schibler, 1994, Khodor et al., 2011). Indeed, the nascent RNA from cells expressing NonA-TRIBE had 30-fold more editing sites than mRNA, even at a lower sequencing depth (mRNA, ~300 sites, ~90M mapped reads; Nascent RNA, ~9000 sites, ~60M mapped reads) (Fig. 5A, Fig. S5b).
Figure 5. TRIBE demonstrates NonA preferentially binds the introns of transcripts.
The NonA-TRIBE protein retains deaminase activity, (A) an increase in A to G editing events is observed upon induction of the fusion protein in S2 cells. NonA TRIBE induces very few sites in mRNA but many sites are present in nascently transcribed RNA. B,C) Frequency histograms of number of edits per target genes show that most genes have only one editing site in mRNA, but in nascent RNA many genes have many more edits (median=2, 75th percentile=6, maximum=181 edits per gene). D) An example gene, Dscam, showing mRNA (top) and nascent RNA (middle) expression and editing tracks without and with induction of expression of the fusion protein. Editing events are indicated by black bars, the height of the bar indicates the percentage editing at that site. The full annotation of Dscam is shown above two example splice variants likely expressed in these S2 cells (bottom). E) Metagene quantification of the location of TRIBE edits (NonA-TRIBE and Hrp48-TRIBE) in both mRNA and nascent RNA. Total number of editing sites are marked next to bars. Background indicates the proportion of the fly transcriptome composed of the indicated regions.
The nascent RNA edited by NonA-TRIBE has many genes with multiple editing sites per gene (Fig. 5C), in contrast to the editing events in mRNA due to Hrp48-TRIBE (Fig. 2B), dFMR1-TRIBE (Fig. 4B), or even NonA-TRIBE (Fig. 5B). The median number of sites per gene in nascent RNA is 2, and 25% of genes have greater than 6 sites per gene. These sites are in 1561 target genes, approximately 20% of expressed genes. The maximum number is 181 sites (in Shab; Table S1).
Not surprisingly, NonA-TRIBE editing sites are enriched in intronic regions of the nascent transcripts (Fig. 5D,E). Perhaps owing to retained introns, half of the few (40) NonA-TRIBE editing events in mRNA are also in intronic regions (Fig. S6). Nascent RNA sequenced from cells expressing Hrp48-TRIBE does not show this intronic concentration of editing sites (Fig. 5E). On the contrary, Hrp48-TRIBE maintains its preference for 3’ UTRs even in nascent RNA. This is despite the fact that nascent transcripts contain fewer 3’ UTR reads due to the 5’ enrichment of nascent RNA (Khodor et al., 2011). The NonA moiety therefore dictates the preference of NonA-TRIBE for intronic RNA rather than it being a general RBP property.
Gene ontology (GO) analysis reinforces the different roles of the three RBPs as the targets of each RBP have divergent functions. (Table S2.1). The cell culture experiments taken together indicate that TRIBE can determine the RNA targets of three RBPs, as well as where on a transcript they bind.
Hrp48-TRIBE can identify RBP targets in specific cells
Because the targets of many RBPs are likely to be different between tissue and cell types, we tested whether TRIBE could identify cell-specific RBP targets within the fly brain. To achieve cell-specific expression of the TRIBE proteins, we employed the Drosophila UAS/Gal4 system (Phelps and Brand, 1998). Transgenic fly lines harboring the Hrp48-TRIBE transgene under the control of the UAS promoter were generated. Cell-specific expression was achieved using a range of Gal4 driver lines, which express the UAS transcriptional activator Gal4 only in specific subsets of neurons. The neuronal groups examined were the core circadian PDF neuropeptide expressing cells (pdf-Gal4, ~16 cells/brain), dopaminergic neurons (Tyrosine hydroxylase, TH-Gal4, ~1000 cells/brain) and all neurons (pan-neuronal driver, elav-Gal4, ~100,000 cells/brain). A fluorescent protein (UAS-eGFP) was co-expressed to allow manual cell sorting of TRIBE protein-expressing and control neurons from dissociated Drosophila brains (Nagoshi et al., 2010, Abruzzi et al., 2015).
Similar to the cell culture result, neuronal expression of the Hrp48-TRIBE protein caused a large increase in the number of editing sites, far more than the level of endogenous editing (range of endogenous editing sites, ~300-2000; range of TRIBE editing sites, ~8000-11,000 (Fig. 6A)). Neuronal expression of only the ADARcd gave rise to no additional editing (data not shown). All endogenous editing events detected in any cell type were excluded from downstream analysis of TRIBE expressing neurons. Like in S2 cells, editing events due to Hrp48-TRIBE were enriched in the 3’ UTRs of neuronal transcripts, which had even more edits per gene than was typical in cultured S2 cells (Table S1). This may partly be a result of the extended length of 3’ UTRs often observed in neurons compared to other cell types (Hilgers et al., 2011).
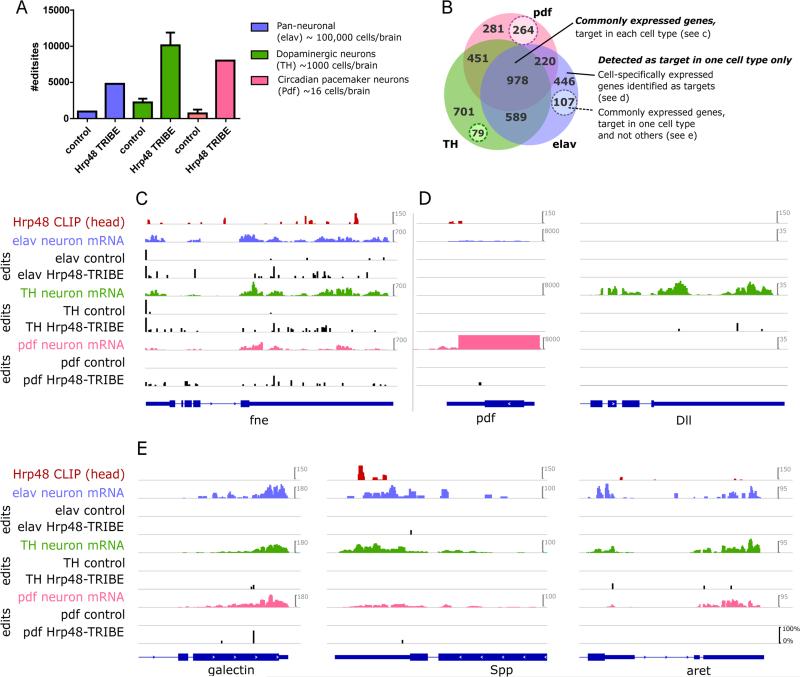
Figure 6. Hrp48-TRIBE can identify RBP targets in specific subsets of cells.
A) The number of editing sites detected in RNA isolated from specific subsets of neurons expressing the Hrp48-TRIBE protein (bars labelled TRIBE) is significantly greater than the background number of endogenous editing sites (bars labelled control). Cell types examined were the core circadian pdf neuropeptide expressing cells (pdf-Gal4), dopaminergic neurons (TH-Gal4) and generic neurons (elav-Gal4). B) Venn diagram of the genes that are identified as Hrp48 targets by TRIBE in different cell types. Example genes from three categories of targets are shown 3 in (C,D,E). C) Many commonly expressed genes are identified as targets in each of the three cell types. D, E) Some genes are identified as targets in only one (or a combination of two) cell types. D) Genes that are expressed only in certain cell types are identified as targets and (E) commonly expressed genes may be identified as a target in one cell type and not the others (dashed insets in B). Note that ‘expression’ here is classified as sufficient sequencing depth at the editing site location, and as such the numbers of cell-specifically expressed genes are likely an overestimation. (C,D,E) Tracks shown are; RNA-seq and Hrp48 CLIP from whole fly heads, RNA-seq and Hrp48-TRIBE editing tracks from indicated isolated neuron subtype, either GFP expressing control (labelled pdf, TH, elav) or Hrp48-TRIBE expressing cells (labelled pdf, TH, elav-TRIBE). Editing events are indicated by black bars, and the height of the bar indicates the percentage editing at that site. The scale for mRNA-seq is constant for each gene, resulting in truncation of signal of pdf in pdf cells.
Similar numbers of Hrp48 target genes were identified in each of the three cell types (~1743-2798); the target gene sets were overlapping but not identical (Fig. 6b, Table S1). Not surprisingly, commonly expressed genes were identified as targets in all three cell types (Fig. 6B,C). For example, the neuronal RBP fne (found in neurons) is identified as a Hrp48 target in all neuronal subsets, other targets in this class include bru-3, mamo, and pum.
However, some genes are identified as a target in only one or two cell types despite being expressed in all three. For example, the genes galectin, aret and Spp are expressed in all three cell types, but aret is identified as a target only in TH cells. Galectin is identified as a target in pdf and TH cells, and Spp is a target in pdf and elav cells (Fig. 6E). These ‘commonly expressed genes’ have sufficient sequencing depth at the given base position (minimum 20 reads) in all three cell types to detect an editing event of roughly comparable frequency, suggesting that editing is indeed cell-type specific.
Another set of targets are identified in only one or two cell types due to bona fide cell-type specific gene expression differences: for example, pdf is only identified as a target in pdf neurons because it is only expressed there; similarly, the transcription factor Dll is only expressed in dopaminergic (TH) neurons where it is also identified as a Hrp48 target (Fig. 6D). Note that there are probably a number of genes inappropriately assigned as cell-specifically expressed (Fig. 6B). This occurs when the specific editing site has insufficient coverage (due to uneven sequencing coverage) despite acceptable overall gene expression.
These Hrp48 target genes were also analyzed by GO terms (Table S2.2). Common target genes are not surprisingly enriched for general neuronal functions, consistent with the newly described role of Hrp48 in axon guidance and branching (Bruckert et al., 2015), whereas cell-specific target gene functions were distinct. This probably reflects cell-specific gene expression patterns as well as cell-specific binding of Hrp48 to commonly expressed genes.
dFMR1-TRIBE can identify different RBP targets in specific cells
dFMR1-TRIBE was also expressed in specific neurons. Based on evidence from mammalian systems that the balance between excitation and inhibition is affected when FMRP is altered (as in the human Fragile X Syndrome)(Contractor et al., 2015), we chose to examine dFMR1 targets in excitatory (cholinergic; Cha-Gal4) and inhibitory (GABAergic; GAD-Gal4) cells. They were purified as described above and the resulting isolated mRNA sequenced.
dFMR1-TRIBE editing sites were found throughout coding regions in neurons, which mirrors the cell culture results described above. Many fewer target genes were identified in GABAergic neurons than in cholinergic neurons (Fig. 7A,B,C; Table S1), possibly due to the lower expression level of the TRIBE protein in GABAergic neurons (data not shown). Due to this difference, Cha targets may not be truly cell-specific, but the smaller number of GAD-Gal4-specific dFMR1 targets are likely to be legitimate cell-specific targets (Fig. 7C,F).
Figure 7. Identification of dFMR1 targets in excitatory and inhibitory neurons.
(A) The number of editing sites detected in RNA isolated from specific subsets of neurons expressing the dFMR1-TRIBE protein (bars labelled TRIBE) is greater than the background number of endogenous editing sites (bars labelled control). Cell types examined were excitatory neurons (cholinergic; Cha-Gal4) and inhibitory neurons (GABA-ergic; GAD-Gal4). (B) Frequency histograms of number of edits per target genes. (C) Venn diagram of the genes that are identified as dFMR1 targets by TRIBE in different cell types. Target editing sites that have sufficient sequencing depth in both cell types are outlined with a dashed line. (D-F) Example genes are shown. (E) Many commonly expressed genes are identified as targets in each of the both cell types, and some genes (D and F) are identified as targets in either one or the other cell type, despite being expressed in both.
Tracks shown are; RNA-seq and dFMR1-ADARcd TRIBE editing tracks from the indicated isolated neuron subtype, either GFP expressing control or dFMR1-ADARcd expressing cells. Editing events are indicated by black bars, and the height of the bar indicates the percentage editing at that site.
GO analysis indicates that common targets are enriched for general neuronal functions, including genes associated with the known role of FMRP as a regulator of the microtubule network (Yao et al., 2011). Overall, 45% of robust mouse brain CLIP targets (Darnell et al., 2011) that have clear fly homologs were also dFMR1 TRIBE targets in excitatory fly neurons (Fig. S4B,C). One of these genes, futsch, the fly homolog of MAP1B, has been identified as a FRMP target by genetic means (Zhang et al., 2001) and was also the #3 ranked CLIP target in Darnell et al. (2011). GAD-specific dFMR1 targets are enriched for nuclear processes including transcription (Fig. 7F, Table S2.3). In contrast, the Cha-specific targets are enriched for cytoplasmic functions, signal transduction and the regulation of GTPase activity (Fig. 7E, Table S2.3). As described above for the Hrp48 cell-specific targets, dFMR1 cell-specific targets may reflect cell-specific gene expression and/or cell-specific binding of dFMR1 to commonly expressed genes.
Discussion
We have developed TRIBE to allow the identification of RBP targets in small numbers of specific cells in vivo. To show that TRIBE is applicable to different types of RBPs we have applied it to three Drosophila RBPs (Hrp48, dFMR1 and NonA). The fusion proteins maintain catalytic activity, and expression of all three TRIBE fusion proteins results in robust and reproducible introduction of new editing sites. The three RBP-fusions have dramatically different editing patterns, indicating that the RBPs play a major role in determining editing specificity. Hrp48-TRIBE editing sites are enriched in the 3'UTR, as is the CLIP signal of both the fusion and endogenous proteins, demonstrating that TRIBE editing correctly reflects the endogenous binding pattern of Hrp48. dFMR1-TRIBE editing sites are dispersed throughout the coding sequence of transcripts, which is the observed binding pattern of mammalian FMRP (Darnell et al., 2011) and is consistent with its role as a translation regulator. The third RPB fusion protein, NonA-TRIBE, edits RNA preferentially in introns, consistent with its published role as a splicing factor (Kozlova et al., 2006, Kaneko et al., 2007).
Negative controls (the truncated ADAR catalytic domain alone, and Hrp48-TRIBE with mutagenized RNA-binding domains) do not result in any additional editing sites, further indicating that editing is specified by the RBP. We are therefore confident that the TRIBE editing sites faithfully mark transcript targets of the RBPs. Additional experiments expressing Hrp48-TRIBE and dFMR1-TRIBE in specific neurons demonstrate the ability to identify cell-specific RBP targets. Comparing these targets between neuronal subtypes illustrates the diversity of cell-type specific RBP targets and therefore the importance of a method for defining RBP-target interactions in individual cell types.
Comparison of TRIBE to CLIP
The current standard for the identification of RBP targets is CLIP and variants thereof. Although most TRIBE sites are within 500bp of a CLIP site, CLIP is essential for determining precise RBP binding position. Nonetheless, the absence of alternative high-resolution methods for measuring in vivo binding has made it difficult to critically assess CLIP data for systematic biases or sources of false positives and false negatives (Lambert et al., 2014).
CLIP false positives are a particular concern as transcripts that are functionally unaffected by knock-down of the RBP are often identified as targets (Lambert et al., 2014). Biases include the preferential crosslinking of uridines; the choice of RNase and fragmentation conditions also have a significant impact on the detected targets (Fecko et al., 2007, Kishore et al., 2011, Sugimoto et al., 2012, Lambert et al., 2014). Most importantly perhaps, the low efficiency steps of the CLIP protocol necessitates large numbers of cells and as such is not amenable to the study of discrete, small numbers of cells.
Many of these drawbacks are avoided by TRIBE. It is not a biochemical technique and requires no antibody. While our manuscript was being revised, a conceptually similar method called RNA tagging was published (Lapointe et al., 2015). In this case, the RBP was fused to a C. elegans poly (U) polymerase, and target RNAs recognized by their U tails. Although it is too soon to compare the two methods, RNA tagging does not identify the region of the mRNA bound by the RBP, and target identification requires a special library protocol. Our method in contrast uses standard library methods and therefore needs very little RNA. Indeed, we have used TRIBE to identify RBP targets in tiny numbers of specific neurons from the fly brain. The smallest group of neurons used in this study are the key circadian pacemaker neurons, the pdf cells, of which there are 16 in a single fly brain. The minimum number of cells from which we generated TRIBE sequencing libraries is ~150 neurons. Given recent advances and the trajectory of developments in RNA-seq, TRIBE could be applicable to individual cells and provide an unprecedented level of resolution on RBP targets.
As TRIBE involves sequencing the transcriptome of specific cells and therefore also captures their specific gene expression features, e.g., alternative splicing and 3’ UTR patterns, it may be possible to correlate transcriptional, post-transcriptional and RBP binding events. (As expression of the TRIBE fusion protein may affect gene expression, transient expression and gene expression analysis of a parallel sample without TRIBE expression are advisable.) In contrast, CLIP is typically done from mixed tissue, making it difficult to correlate the binding data with a specific gene expression state. For example, if CLIP is performed on brain in which the RBP is not ubiquitously expressed (e.g., FMRP is expressed in neurons but not glia or blood vessels), the corresponding transcriptome, expression levels as well as isoform features, are an average of all cell types. This also makes normalization difficult if not impossible, as noted by Darnell et al (2011). As a result, CLIP targets are biased toward highly-expressed and long genes (Ouwenga and Dougherty, 2015). Although TRIBE requires a minimum expression level of target, above that threshold there is no bias toward highly expressed genes.
Although optimization of CLIP can be challenging (e.g., the optimal crosslinking parameters differ between proteins, over-digestion of crosslinked complexes by RNase can affect the results) (Ule et al., 2005, Konig et al., 2011), TRIBE is comparatively simple to perform and should also be amenable to mammalian systems. It only requires cloning and expression, the RNA is then purified and sequenced, and novel editing events detected via a bioinformatics pipeline.
Potential shortcomings of TRIBE
A risk of hijacking the function of ADAR is that some of its own editing selectivity may remain. Obviously the ADARcd-TRIBE protein has an absolute requirement for an editable substrate (an adenosine) proximal to the RBP binding site, the absence of which may preclude the editing of some targets and result in false negatives. In addition, endogenous ADARs have preferences for bases proximal to the editing site and all TRIBE proteins maintain these published preferences (i.e. 5’ U enriched, 3’ G enriched, Fig. S7) (Eggington et al., 2011, Kuttan and Bass, 2012, Porath et al., 2014). This suggests that the editing specificity is at least partially dictated by the deaminase domain.
However the most prominent cause of false negatives is probably the strong preference of the ADAR catalytic domain for double-stranded RNA even without its dsRBDs (Macbeth et al., 2005, Eggington et al., 2011, Montiel-Gonzalez et al., 2013, Vogel et al., 2014, Vogel and Stafforst, 2014, Phelps et al., 2015). Although endogenous ADAR exhibits considerable plasticity, i.e., it edits regions of highly complex structure as well long stretches of duplex RNA, we assume that RBP-ADARcd proteins will not label their bound targets if they are composed exclusively of single stranded RNA. Indeed, the comparison of edited regions with those CLIP targets that have no editing (Fig. S3) indicates that the requirement for a bulged A within a dsRNA region (Eifler et al., 2013) explains why TRIBE has fewer targets than CLIP.
Yet our data suggest that 40-50% of target mRNAs have sufficient double-stranded character near the RBP binding site to be edited by the TRIBE ADARcd (Fig. 3D, Supp. Fig. 3). We therefore suggest that tethering of the ADARcd to its targets by an RBP can take advantage of dynamic structure formation in vivo (Mortimer et al., 2014, Kwok et al., 2015), especially over a time frame of hours, to edit and permanently mark many substrates at a detectable frequency. Indeed, if the TRIBE ADARcd can take advantage of double-stranded RNA features that are dynamic, TRIBE favors the labelling of long-lived interactions between the RBP and its target RNA. CLIP by contrast may take a snapshot that includes many weaker and transient interactions that are false positives. Although this interpretation explains the much larger number of CLIP targets for Hrp48, TRIBE targets have much better overlap with the best CLIP targets, suggesting that a high fraction of them also reflect more stable and therefore meaningful in vivo interactions.
Improvements and extensions
This study is a first demonstration of TRIBE and can be improved and extended. First and as implied above, it would be strengthened by using an additional editing moiety. We attempted to perform TRIBE using cytidine deaminases, which edit single-stranded RNA and catalyze the conversion of Cytidine to Uridine. We tried 7 characterized and putative cytidine deaminases and had modest success only with mouse APOBEC1, but editing was too inefficient to be useful (data not shown). Second, the biases inherent in ADARcd-TRIBE might be ameliorated with a characterized ADAR mutant, which is more catalytically active and has less of a nearest neighbor preference than wild type ADAR (Kuttan and Bass, 2012). Third, the caveats associated with overexpression could be avoided in the future by using endogenous locus knockins to achieve endogenous levels of RBP-TRIBE expression, a strategy that is beyond the scope of this first proof of principle study. Lastly, TRIBE could be extended by fusing the ADARcd to other proteins and expressing them in a cell-type specific manner, e.g., polyA binding protein for transcriptome identification or ribosomes for translational profiling, all done without any biochemistry. One can even imagine adding a light- or chemical-activatable domain to control editing spatially and temporally.
Cell specificity
Experiments expressing Hrp48-TRIBE and dFMR1-TRIBE in specific subsets of neurons identified cell-specific targets. Comparing them between neuronal subtypes illustrates the diversity of cell-type specific RBP targets and thus the importance of TRIBE for defining RBP-target interactions in particular cell types. Although recent advances in sequencing technology have revealed the distinct regulation of individual cell types at the transcriptional and post-transcriptional level, it has been extremely difficult to define the cell-type specific roles and targets of individual RBPs. TRIBE ameliorates this issue and should therefore contribute to addressing the long-standing question of how more broadly expressed proteins have cell-specific effects. This issue is particularly relevant to several RNA-binding proteins associated with human diseases, e.g., FMRP, FUS, and TDP43.
Experimental Procedures
Molecular biology
The RBP of interest was cloned upstream of the Drosophila ADAR catalytic domain (the whole C terminus downstream of the 2nd dsRBD was used, starting Y268 to terminal E669 of AHN59262.1) with minimal linker region into a pMT-A vector (Invitrogen, V4120-20, also harboring a blasticidin resistance gene). Stable S2 cell lines were made by transfecting with the pMT-RBP-ADARcd-V5-Blasticidin plasmid followed by subsequent blasticidin resistance selection. Fusion protein expression was induced by introduction of copper sulphate for 24 hours, prior to harvesting protein and RNA. S2 cell expression of all fusion proteins was assayed by western blot for the V5 tag (Invitrogen, 46-1157). Nascent RNA was extracted from S2 cells, according to Khodor et al. (2011) and depleted of ribosomal RNA according to Pennington et al. (2013) and mRNA (two rounds of pA depletion using Invitrogen Dynabeads Oligo dT, according to manufacturer's protocol).
Fluorescently labelled neurons were isolated from dissected, triturated fly brains by manual sorting using a glass micro-pipette, as described in Abruzzi et al. (2015). RNA sequencing libraries from S2 cells were constructed using the standard Illumina Truseq kit protocol. RNA-sequencing libraries from manually sorted neurons were made as described in Abruzzi et al. (2015).
RNA editing analysis
RNA editing events are defined as loci where there are A > 80% and zero G in genomic DNA and G > 0% in RNA (in the reverse strand, we evaluate the reverse complement). Genomic DNA was also sequenced from either S2 cells or yw flies, to provide a reference which contains SNPs. Analysis of RNA sequencing data was performed as previously published (Rodriguez et al., 2012), with some modifications (see Supplemental Information).
In neurons, removal of endogenous editing events from the analysis was required. A lower threshold (10 reads, 10% editing) was used to define endogenous editing events. All endogenous events identified, including sites from all non-TRIBE expressing negative controls in all lines used in a given experiment, were removed from data from TRIBE expressing neurons. Gene ontology analysis was performed using DAVID (Huang da et al., 2009).
CLIP
CLIP libraries of Hrp48 and Hrp48-ADARcd were constructed as described in Cho et al. (2012), with some modifications as described in Supplemental Information. Significant regions of binding were determined using the CLIPper algorithm (Lovci et al., 2013) and as described in Moore et al. (2014).
Fly lines
TRIBE flies were generated by cloning the RBP-ADARcd-V5 transgene into a modified pJFRC7-20x UAS construct (RBP in locus of removed mCD8-GFP, Addgene #26220), which was injected by BestGene (Chino Hills, CA). UAS-RBP-ADARcd-V5; UAS-eGFP flies (Bloomington stock center #1522) were crossed to a range of driver lines (pdf-Gal4, TH-Gal4, elav-gsg-Gal4, Cha-Gal4, GAD-Gal4) to achieve cell-type specific expression of the fusion protein. Constitutive pan-neuronal expression of Hrp48-TRIBE (elav-Gal4, UAS-Hrp48-ADARcd) was lethal, so adult specific expression was achieved using the gene-switch system (Osterwalder et al., 2001), elav-gsg-Gal4. Young flies (~3 days old) were maintained on food containing RU486 (0.2 μg/ml, Sigma #8046) for 5-8 days prior to dissection and cell sorting.
Raw sequencing data and processed RNA editing tracks are available for download from the NCBI Gene Expression Omnibus database, accession number GSE78065.
See Supplemental Experimental Procedures for additional details.
Supplementary Material
The Hsp83 degradation element (HDE) is known to be bound by Hrp48 (Bashirullah et al., 1999, Nelson et al., 2007), Hrp48-TRIBE causes editing within the consensus Hrp48 binding motif (Blanchette et al., 2009, also see Fig. S2) within the HDE. Data are from S2 cells, editing percentage of total number of reads is shown at the edit sites.
Gene ontology analysis of all TRIBE proteins in S2 cells and neurons.
Reads Sequencing read numbers and mapping percentages.
Editing sites identified in wild type S2 cells and in cells expressing the only the catalytic domain of dADAR.
TRIBE editing sites from all experiments supplied in bedgraph format, with information on chromosomal coordinate, sequencing depth, editing percentages and gene names.
Significant motifs found by MEME analysis are shown. Motifs found by in vitro selection (SELEX, (Blanchette et al., 2009)) or CLIP (endogenous Hrp48 and Hrp48-ADARcd) (A, B, C). Motifs found in regions surrounding Hrp-ADARcd TRIBE editing sites (D, E). For Hrp48-ADARcd TRIBE editing events in S2 cells, an area +/− 20 and 100 bp around the edited base was used for analysis (FDR < 0.001).
Predicted double-strandedness around TRIBE editing sites (orange) or CLIP binding sites that lack TRIBE editing sites (grey) (A). Single nucleotide resolution for Hrp48 binding location was achieved by performing CIMS analysis (Cross Linking induced Mutation Site) on CLIP data. A flanking region of 250nt both 5’ and 3’ of the site (501nt in total) was folded with UNAFold, base pairing was counted in the predicted minimum free energy (MFE) and suboptimal structures (within ΔΔG=5Kcal/mol of the MFE), and the profile is averaged per. All sites were then averaged to yield this plot (mean +/− SEM, n = 17 TRIBE editing sites). B) Schematic modified from Eifler et al, 2013. RNA structure around the sites edited by the catalytic domain of human ADAR2 in yeast resemble the intermediate complex formed when ADAR2 distorts local dsRNA and the sequences flanking the edited adenosine are optimal for deaminase domain binding. Data shown is from Hrp48 TRIBE.
Mouse homologs of dFMR1-TRIBE targets in S2 cells are enriched for higher CLIP ranking targets of FMRP (A). Similarly, the mouse homologs of neuronal dFMR1 TRIBE targets are enriched for higher CLIP rankings. (B). Approximately 50% of the fly homologs of robust mouse brain FMRP CLIP targets are also TRIBE targets in excitatory fly neurons (Cha) (C). Mouse FMRP CLIP data are from Darnell et al, 2011.
The number of editing sites detected in S2 cells expressing Hrp48 TRIBE at different sequencing depths (million mapped reads), employing different coverage thresholds for the identification of an editing sites (A). The more stringent threshold of 20 reads was used throughout this study. The number of editing sites detected in S2 cells expressing different RBP TRIBE constructs (B) The number of sites for a given sequencing depth differs by RBP (data for 20 read threshold editing sites are shown).
A). A modest increase in A to G editing events is observed in mRNA upon induction of the fusion protein in S2 cells (data also shown, along with nascent RNA in Figure 4a). B) Editing percentage is correlated at given sites between biological repeats (R2 = 0.88). C) Classification of editing sites based on refseq annotation. Most ‘intronic’ sites are not also annotated as exonic, i.e., most mRNA NonA intronic sites are not mis-categorization, the sites are actually in introns that are found in the mRNA fraction. D) Example gene, ppn, which has many NonA-TRIBE editing events in an intronic region. The intronic region is clearly expressed in the mRNA, as well as the nascent RNA fraction, and is identified as a binding target in both.
Nearest neighbor preference the three TRIBE proteins (in S2 cells) (A). Nearest neighbor preference for editing sites of different editing percentages for Hrp48-ADARcd (in S2 cells) (B). n indicates the number of sites used.
TRIBE target genes, as well as number of editing sites per gene, are listed for each TRIBE protein in S2 cells and neurons.
Acknowledgements
We thank Jin Billy Li, Donald Rio, Roy Parker, Robert Singer and members of the Rosbash lab, especially Katherine Abruzzi, for helpful comments on the manuscript. We are grateful to Robert Reenan, Tom Jongens, Maureen Hanson, Bora Baysal, Shraddha Sharma and Donald Rio for the gift of plasmids and reagents. This work was initially supported by the Howard Hughes Medical Institute and then by the award of NIH EUREKA R01 grant DA037721.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization, AM, WL and MR; Software, AM and RR; Investigation, AM, RR, HJ, JS and AF; Analysis, AM, RR and HJ; HJ was essential for the CLIP experiments and analysis, RR for bioinformatics and RNA folding, and their contributions were of equal importance; Writing- Original Draft; AM and MR, Writing- Review and Editing; AM, RR, HJ and MR; Visualization, AM; Funding Acquisition, AM and MR;
References
- Abruzzi K, Chen X, Nagoshi E, Zadina A, Rosbash M. RNA-seq profiling of small numbers of Drosophila neurons. Methods in enzymology. 2015;551:369–386. doi: 10.1016/bs.mie.2014.10.025. [DOI] [PubMed] [Google Scholar]
- Ascano M, Jr., Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, Williams Z, Ohler U, Tuschl T. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashirullah A, Halsell SR, Cooperstock RL, Kloc M, Karaiskakis A, Fisher WW, Fu W, Hamilton JK, Etkin LD, Lipshitz HD. Joint action of two RNA degradation pathways controls the timing of maternal transcript elimination at the midblastula transition in Drosophila melanogaster. The EMBO journal. 1999;18:2610–2620. doi: 10.1093/emboj/18.9.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BL, Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988;55:1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- Blanchette M, Green RE, Brenner SE, Rio DC. Global analysis of positive and negative pre-mRNA splicing regulators in Drosophila. Genes & development. 2005;19:1306–1314. doi: 10.1101/gad.1314205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Green RE, MacArthur S, Brooks AN, Brenner SE, Eisen MB, Rio DC. Genome-wide analysis of alternative pre-mRNA splicing and RNA-binding specificities of the Drosophila hnRNP A/B family members. Molecular cell. 2009;33:438–449. doi: 10.1016/j.molcel.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SA, Ripperger J, Kadener S, Fleury-Olela F, Vilbois F, Rosbash M, Schibler U. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science (New York, NY) 2005;308:693–696. doi: 10.1126/science.1107373. [DOI] [PubMed] [Google Scholar]
- Bruckert H, Marchetti G, Ramialison M, Besse F. Drosophila Hrp48 Is Required for Mushroom Body Axon Growth, Branching and Guidance. PloS one. 2015;10:e0136610. doi: 10.1371/journal.pone.0136610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Romano M, Baralle FE. TDP-43 high throughput screening analyses in neurodegeneration: advantages and pitfalls. Molecular and cellular neurosciences. 2013;56:465–474. doi: 10.1016/j.mcn.2013.03.001. [DOI] [PubMed] [Google Scholar]
- Cho J, Chang H, Kwon SC, Kim B, Kim Y, Choe J, Ha M, Kim YK, Kim VN. LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell. 2012;151:765–777. doi: 10.1016/j.cell.2012.10.019. [DOI] [PubMed] [Google Scholar]
- Contractor A, Klyachko VA, Portera-Cailliau C. Altered Neuronal and Circuit Excitability in Fragile X Syndrome. Neuron. 2015;87:699–715. doi: 10.1016/j.neuron.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Fraser CE, Mostovetsky O, Stefani G, Jones TA, Eddy SR, Darnell RB. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes & development. 2005;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, Darnell RB. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell RB. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley interdisciplinary reviews RNA. 2010;1:266–286. doi: 10.1002/wrna.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Developmental cell. 2008;14:926–939. doi: 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggington JM, Greene T, Bass BL. Predicting sites of ADAR editing in double-stranded RNA. Nature communications. 2011;2:319. doi: 10.1038/ncomms1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eifler T, Pokharel S, Beal PA. RNA-Seq analysis identifies a novel set of editing substrates for human ADAR2 present in Saccharomyces cerevisiae. Biochemistry. 2013;52:7857–7869. doi: 10.1021/bi4006539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fecko CJ, Munson KM, Saunders A, Sun G, Begley TP, Lis JT, Webb WW. Comparison of femtosecond laser and continuous wave UV sources for protein-nucleic acid crosslinking. Photochemistry and photobiology. 2007;83:1394–1404. doi: 10.1111/j.1751-1097.2007.00179.x. [DOI] [PubMed] [Google Scholar]
- Gupta A, Rahman R, Li K, Gribskov M. Identifying complete RNA structural ensembles including pseudoknots. RNA biology. 2012;9:187–199. doi: 10.4161/rna.18386. [DOI] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr., Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond LE, Rudner DZ, Kanaar R, Rio DC. Mutations in the hrp48 gene, which encodes a Drosophila heterogeneous nuclear ribonucleoprotein particle protein, cause lethality and developmental defects and affect P-element third-intron splicing in vivo. Molecular and cellular biology. 1997;17:7260–7267. doi: 10.1128/mcb.17.12.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V, Perry MW, Hendrix D, Stark A, Levine M, Haley B. Neural-specific elongation of 3′ UTRs during Drosophila development. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:15864–15869. doi: 10.1073/pnas.1112672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huppertz I, Attig J, D'Ambrogio A, Easton LE, Sibley CR, Sugimoto Y, Tajnik M, Konig J, Ule J. iCLIP: protein-RNA interactions at nucleotide resolution. Methods (San Diego, Calif) 2014;65:274–287. doi: 10.1016/j.ymeth.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh JR, Munro TP, Smith-Litiere K, Lepesant JA, St Johnston D. The Drosophila hnRNPA/B homolog, Hrp48, is specifically required for a distinct step in osk mRNA localization. Developmental cell. 2004;6:625–635. doi: 10.1016/s1534-5807(04)00130-3. [DOI] [PubMed] [Google Scholar]
- Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, Amit I. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science (New York, NY) 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes & development. 2007;21:1779–1789. doi: 10.1101/gad.1565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Kim C, Lee H, Kim W, Lee EK. Post-transcriptional controls by ribonucleoprotein complexes in the acquisition of drug resistance. International journal of molecular sciences. 2013;14:17204–17220. doi: 10.3390/ijms140817204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan LP, Leroy A, Sproul D, O'Connell MA. Adenosine deaminases acting on RNA (ADARs): RNA-editing enzymes. Genome biology. 2004;5:209. doi: 10.1186/gb-2004-5-2-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodor YL, Rodriguez J, Abruzzi KC, Tang CH, Marr MT, 2nd, Rosbash M. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes & development. 2011;25:2502–2512. doi: 10.1101/gad.178962.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore S, Jaskiewicz L, Burger L, Hausser J, Khorshid M, Zavolan M. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nature methods. 2011;8:559–564. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Luscombe NM, Ule J. Protein-RNA interactions: new genomic technologies and perspectives. Nature reviews Genetics. 2011;13:77–83. doi: 10.1038/nrg3141. [DOI] [PubMed] [Google Scholar]
- Kozlova N, Braga J, Lundgren J, Rino J, Young P, Carmo-Fonseca M, Visa N. Studies on the role of NonA in mRNA biogenesis. Exp Cell Res. 2006;312:2619–2630. doi: 10.1016/j.yexcr.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Kuttan A, Bass BL. Mechanistic insights into editing-site specificity of ADARs. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3295–3304. doi: 10.1073/pnas.1212548109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok CK, Tang Y, Assmann SM, Bevilacqua PC. The RNA structurome: transcriptome-wide structure probing with next-generation sequencing. Trends in biochemical sciences. 2015;40:221–232. doi: 10.1016/j.tibs.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Lambert N, Robertson A, Jangi M, McGeary S, Sharp PA, Burge CB. RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Molecular cell. 2014;54:887–900. doi: 10.1016/j.molcel.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe CP, Wilinski D, Saunders HA, Wickens M. Protein-RNA networks revealed through covalent RNA marks. Nature methods. 2015;12:1163–1170. doi: 10.1038/nmeth.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovci MT, Ghanem D, Marr H, Arnold J, Gee S, Parra M, Liang TY, Stark TJ, Gehman LT, Hoon S, Massirer KB, Pratt GA, Black DL, Gray JW, Conboy JG, Yeo GW. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nature structural & molecular biology. 2013;20:1434–1442. doi: 10.1038/nsmb.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends in genetics : TIG. 2008;24:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Macbeth MR, Schubert HL, Vandemark AP, Lingam AT, Hill CP, Bass BL. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science (New York, NY) 2005;309:1534–1539. doi: 10.1126/science.1113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. Rna. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montiel-Gonzalez MF, Vallecillo-Viejo I, Yudowski GA, Rosenthal JJ. Correction of mutations within the cystic fibrosis transmembrane conductance regulator by site-directed RNA editing. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:18285–18290. doi: 10.1073/pnas.1306243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Zhang C, Gantman EC, Mele A, Darnell JC, Darnell RB. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nature protocols. 2014;9:263–293. doi: 10.1038/nprot.2014.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer SA, Kidwell MA, Doudna JA. Insights into RNA structure and function from genome-wide studies. Nature reviews Genetics. 2014;15:469–479. doi: 10.1038/nrg3681. [DOI] [PubMed] [Google Scholar]
- Nagoshi E, Sugino K, Kula E, Okazaki E, Tachibana T, Nelson S, Rosbash M. Dissecting differential gene expression within the circadian neuronal circuit of Drosophila. Nat Neurosci. 2010;13:60–68. doi: 10.1038/nn.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MR, Luo H, Vari HK, Cox BJ, Simmonds AJ, Krause HM, Lipshitz HD, Smibert CA. A multiprotein complex that mediates translational enhancement in Drosophila. The Journal of biological chemistry. 2007;282:34031–34038. doi: 10.1074/jbc.M706363200. [DOI] [PubMed] [Google Scholar]
- Nussbacher JK, Batra R, Lagier-Tourenne C, Yeo GW. RNA-binding proteins in neurodegeneration: Seq and you shall receive. Trends in neurosciences. 2015;38:226–236. doi: 10.1016/j.tins.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouwenga RL, Dougherty J. Fmrp targets or not: long, highly brain-expressed genes tend to be implicated in autism and brain disorders. Molecular autism. 2015;6:16. doi: 10.1186/s13229-015-0008-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington KL, Marr SK, Chirn GW, Marr MT., 2nd Holo-TFIID controls the magnitude of a transcription burst and fine-tuning of transcription. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:7678–7683. doi: 10.1073/pnas.1221712110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps CB, Brand AH. Ectopic gene expression in Drosophila using GAL4 system. Methods (San Diego, Calif) 1998;14:367–379. doi: 10.1006/meth.1998.0592. [DOI] [PubMed] [Google Scholar]
- Phelps KJ, Tran K, Eifler T, Erickson AI, Fisher AJ, Beal PA. Recognition of duplex RNA by the deaminase domain of the RNA editing enzyme ADAR2. Nucleic acids research. 2015;43:1123–1132. doi: 10.1093/nar/gku1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porath HT, Carmi S, Levanon EY. A genome-wide map of hyper-edited RNA reveals numerous new sites. Nature communications. 2014;5:4726. doi: 10.1038/ncomms5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J, Menet JS, Rosbash M. Nascent-seq indicates widespread cotranscriptional RNA editing in Drosophila. Molecular cell. 2012;47:27–37. doi: 10.1016/j.molcel.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Raychowdhury R, Schwartz S, Yosef N, Malboeuf C, Lu D, Trombetta JJ, Gennert D, Gnirke A, Goren A, Hacohen N, Levin JZ, Park H, Regev A. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southall TD, Gold KS, Egger B, Davidson CM, Caygill EE, Marshall OJ, Brand AH. Cell-type-specific profiling of gene expression and chromatin binding without cell isolation: assaying RNA Pol II occupancy in neural stem cells. Developmental cell. 2013;26:101–112. doi: 10.1016/j.devcel.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y, Konig J, Hussain S, Zupan B, Curk T, Frye M, Ule J. Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions. Genome biology. 2012;13:R67. doi: 10.1186/gb-2012-13-8-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods (San Diego, Calif) 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science (New York, NY) 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nature biotechnology. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- Vogel P, Schneider MF, Wettengel J, Stafforst T. Improving site-directed RNA editing in vitro and in cell culture by chemical modification of the guideRNA. Angewandte Chemie (International ed in English) 2014;53:6267–6271. doi: 10.1002/anie.201402634. [DOI] [PubMed] [Google Scholar]
- Vogel P, Stafforst T. Site-directed RNA editing with antagomir deaminases--a tool to study protein and RNA function. ChemMedChem. 2014;9:2021–2025. doi: 10.1002/cmdc.201402139. [DOI] [PubMed] [Google Scholar]
- Wuarin J, Schibler U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Molecular and cellular biology. 1994;14:7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao A, Jin S, Li X, Liu Z, Ma X, Tang J, Zhang YQ. Drosophila FMRP regulates microtubule network formation and axonal transport of mitochondria. Human molecular genetics. 2011;20:51–63. doi: 10.1093/hmg/ddq431. [DOI] [PubMed] [Google Scholar]
- Yi-Brunozzi HY, Stephens OM, Beal PA. Conformational changes that occur during an RNA-editing adenosine deamination reaction. The Journal of biological chemistry. 2001;276:37827–37833. doi: 10.1074/jbc.M106299200. [DOI] [PubMed] [Google Scholar]
- Zhang YQ, Bailey AM, Matthies HJ, Renden RB, Smith MA, Speese SD, Rubin GM, Broadie K. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Hsp83 degradation element (HDE) is known to be bound by Hrp48 (Bashirullah et al., 1999, Nelson et al., 2007), Hrp48-TRIBE causes editing within the consensus Hrp48 binding motif (Blanchette et al., 2009, also see Fig. S2) within the HDE. Data are from S2 cells, editing percentage of total number of reads is shown at the edit sites.
Gene ontology analysis of all TRIBE proteins in S2 cells and neurons.
Reads Sequencing read numbers and mapping percentages.
Editing sites identified in wild type S2 cells and in cells expressing the only the catalytic domain of dADAR.
TRIBE editing sites from all experiments supplied in bedgraph format, with information on chromosomal coordinate, sequencing depth, editing percentages and gene names.
Significant motifs found by MEME analysis are shown. Motifs found by in vitro selection (SELEX, (Blanchette et al., 2009)) or CLIP (endogenous Hrp48 and Hrp48-ADARcd) (A, B, C). Motifs found in regions surrounding Hrp-ADARcd TRIBE editing sites (D, E). For Hrp48-ADARcd TRIBE editing events in S2 cells, an area +/− 20 and 100 bp around the edited base was used for analysis (FDR < 0.001).
Predicted double-strandedness around TRIBE editing sites (orange) or CLIP binding sites that lack TRIBE editing sites (grey) (A). Single nucleotide resolution for Hrp48 binding location was achieved by performing CIMS analysis (Cross Linking induced Mutation Site) on CLIP data. A flanking region of 250nt both 5’ and 3’ of the site (501nt in total) was folded with UNAFold, base pairing was counted in the predicted minimum free energy (MFE) and suboptimal structures (within ΔΔG=5Kcal/mol of the MFE), and the profile is averaged per. All sites were then averaged to yield this plot (mean +/− SEM, n = 17 TRIBE editing sites). B) Schematic modified from Eifler et al, 2013. RNA structure around the sites edited by the catalytic domain of human ADAR2 in yeast resemble the intermediate complex formed when ADAR2 distorts local dsRNA and the sequences flanking the edited adenosine are optimal for deaminase domain binding. Data shown is from Hrp48 TRIBE.
Mouse homologs of dFMR1-TRIBE targets in S2 cells are enriched for higher CLIP ranking targets of FMRP (A). Similarly, the mouse homologs of neuronal dFMR1 TRIBE targets are enriched for higher CLIP rankings. (B). Approximately 50% of the fly homologs of robust mouse brain FMRP CLIP targets are also TRIBE targets in excitatory fly neurons (Cha) (C). Mouse FMRP CLIP data are from Darnell et al, 2011.
The number of editing sites detected in S2 cells expressing Hrp48 TRIBE at different sequencing depths (million mapped reads), employing different coverage thresholds for the identification of an editing sites (A). The more stringent threshold of 20 reads was used throughout this study. The number of editing sites detected in S2 cells expressing different RBP TRIBE constructs (B) The number of sites for a given sequencing depth differs by RBP (data for 20 read threshold editing sites are shown).
A). A modest increase in A to G editing events is observed in mRNA upon induction of the fusion protein in S2 cells (data also shown, along with nascent RNA in Figure 4a). B) Editing percentage is correlated at given sites between biological repeats (R2 = 0.88). C) Classification of editing sites based on refseq annotation. Most ‘intronic’ sites are not also annotated as exonic, i.e., most mRNA NonA intronic sites are not mis-categorization, the sites are actually in introns that are found in the mRNA fraction. D) Example gene, ppn, which has many NonA-TRIBE editing events in an intronic region. The intronic region is clearly expressed in the mRNA, as well as the nascent RNA fraction, and is identified as a binding target in both.
Nearest neighbor preference the three TRIBE proteins (in S2 cells) (A). Nearest neighbor preference for editing sites of different editing percentages for Hrp48-ADARcd (in S2 cells) (B). n indicates the number of sites used.
TRIBE target genes, as well as number of editing sites per gene, are listed for each TRIBE protein in S2 cells and neurons.