Abstract
Objectives
To assess the feasibility of quantifying shoulder cartilage morphology and relaxometry in a clinically feasible scan time comparing different pulse sequences and assessing their reproducibility at 3 Tesla.
Methods
Three pulse sequences were compared for morphological assessments of shoulder cartilage thickness and volume (SPGR, MERGE, FIESTA), while a combined T1ρ-T2 sequence was optimized for relaxometry measurements. The shoulders of six healthy subjects were scanned twice with repositioning, and the cartilage was segmented and quantified. The degree of agreement between the three morphological sequences was assessed using Bland-Altman plots, while the morphological and relaxometry reproducibility were assessed with root-mean-square coefficients of variation (RMS-CVs)
Results
Bland-Altman plots indicated good levels of agreement between the morphological assessments of the three sequences. The reproducibility of morphological assessments yielded RMS-CVs between 4.0% and 17.7%. All sequences correlated highly (R>0.9) for morphologic assessments with no statistically significant differences. For relaxometry assessments of humeral cartilage, RMS-CVs of 6.4% and 10.6% were found for T1ρ and T2, respectively.
Conclusions
The assessment of both cartilage morphology and relaxometry is feasible in the shoulder with SPGR, humeral head, and T1ρ being the more reproducible morphological sequence, anatomic region, and quantitative sequence, respectively.
Keywords: Shoulder Cartilage, Cartilage Thickness, MRI, T1ρ mapping, T2 mapping
Introduction
Magnetic resonance imaging (MRI) has been a widely used technique for the evaluation of cartilage with high contrast and spatial resolution. Many studies have demonstrated that the application of three-dimensional post-processing techniques allows for the accurate measurement of cartilage volume and thickness. However, most of these studies have been performed at the knee joint, which is the joint with thickest cartilage in the appendicular skeleton [1–8]. Furthermore, T1ρ and T2 relaxation has been recently used to assess biochemical composition of cartilage in the knee [9; 10] and hip [11; 12] and potentially detect cartilage at risk of developing OA before the irreversible changes of OA occur in the morphological images [13; 14].
MRI-based quantification of shoulder cartilage is challenging for different reasons: the most obvious issue is related to the extremely curved surfaces of the humeral and glenoid cartilage, which are responsible for severe partial volume effects; these artefacts are also due to the non-orthogonal alignment of the images with the surfaces. This is more prominent in the most anterior and most posterior cuts. Another reason is related to the thickness of the shoulder cartilage; the mean cartilage thickness in the shoulder is only approximately 1–1.5 mm vs 2–3 mm in the knee [15–17]. The size of the cartilage requires imaging with high spatial resolution as well as robust postprocessing methods. These issues have contributed to a lack of research in the shoulder cartilage: only a few studies have investigated the feasibility of assessing cartilage morphology in the shoulder [18; 19]. The first study showed the validity of morphological assessment of shoulder cartilage at 1.5 Tesla [18]. The study was performed in human specimens, which enabled high-spatial resolution acquisitions albeit with long acquisition times (~19 min). The second study was performed in-vivo, however it only quantified cartilage of the humeral head. Although the spatial resolution was higher than in the ex-vivo study, the acquisition time was still long to be considered clinically feasible (~12min) [19]. Furthermore, to the best of our knowledge MRI-based biochemical assessments of shoulder cartilage has not been performed yet.
In this study, we have compared three state of the art MR pulse sequences for the morphological assessment of the thin shoulder cartilage within clinical feasible acquisition times: Spoiled Gradient Echo (SPGR), Multi-Echo Recalled Gradient Echo (MERGE), and Fast Imaging Employing Steady-State Acquisition (FIESTA). For biochemical evaluation, we have optimized a combined T1ρ-T2 mapping sequence [10]. Thus, the aim of this study was to show the feasibility of novel MRI pulse sequences for the morphological and biochemical characterization of shoulder cartilage in a clinically feasible time and to assess their precision.
Materials and Methods
The study was approved by the Institutional Review Board and conducted in accordance with the Committee for Human Research at our institution. Informed consent was obtained from each subject before the scans explaining the nature of the study.
Subjects
Six healthy volunteers (age= 22–36 years, mean 29) were recruited for this feasibility and precision study. Subjects had no pain, shoulder range of motion limitations or any other shoulder symptoms. They were scanned twice using a 3T MRI system with an 8-channel shoulder surface coil. The two scans were obtained with repositioning within at least 1 hour apart to allow the subject for stretching out. The combined T1ρ and T2 scan for subject 3 could not be used due to severe motion.
Image Acquisition
Shoulder MR imaging was performed on a 3.0-T scanner (MR750; GE Healthcare, Milwaukee, Wis) using an 8-channel shoulder surface coil (Clinical MR Solutions, Brookfield, Wis). Coronal oblique MR images of the humeral head and the glenoid were acquired using clinical product sequences (SPGR, MERGE and FIESTA). The scan time was kept between 8 and 9 minutes for all sequences. Slice thickness of 2mm and an in-plane resolution of 273µm were applied for all sequences. Echo time TE, repetition time TR, and flip angle were 4.2ms/9.3ms/15° (FIESTA), 15.7ms/34.2ms/20° (MERGE), and 4ms/16ms/20° (SPGR), respectively. For MERGE, 4 echoes were combined to create one image.
Acquisitions for T1ρ and T2 quantification were obtained with a combined research pulse sequence [20]. All images were acquired with a slice thickness of 3mm and 546µm in-plane spatial resolution in 14 minutes scan time for both T1ρ and T2. Acquisition parameters were TR of 5.8ms with times of spin-lock (TSLs) for T1ρ=0ms, 15ms, 30ms, 45ms, and TEs for T2=0ms, 10ms, 20ms, 40ms. A spin-lock frequency of 300 Hz was used for T1ρ. In addition, both B0 magnetic field heterogeneity effects and B1 RF heterogeneity effects were simultaneously corrected by using phase-cycled composite spin lock (PCC-SL) pulses for T1ρ imaging [21].
Segmentation
Segmentation of the glenoid and humeral cartilage was performed by a trained radiologist (5 years of experience in musculoskeletal imaging) using in-house developed software written in MATLAB (The Mathworks, Inc., Natick, MA) [8]. To standardize segmentation the central slice of the oblique coronal plane was identified, where the total volume of the cartilage reaches its maximum and partial volume effects interfered the least with the assessment. This section and the adjacent three more anterior sections as well as the next three consecutive posterior sections were used for cartilage segmentation. The regions of interest (ROIs) for the humeral and glenoid cartilage were anatomically defined as visually inspected on the available sequences. Total time for segmenting the humeral and glenoid cartilage of each subject was 30–35 minutes. For biochemical assessment, cartilage of the humeral head only was segmented from the first TSL/TE acquisitions similar to morphological assessment.
Image Analysis
For the T1ρ and T2 mapping sequence, the first and all subsequent TSL/TE images were carefully checked for motion, and registration was performed if necessary to align all acquisitions in a series to the first TSL/TE. T2 and T1ρ maps were then generated by fitting an exponential decay on a voxel-by-voxel basis according to the following equations respectively:
| [1] |
| [2] |
Cartilage morphology was assessed in the form of 3D cartilage thickness and volume measurements extracted from the 3D GRE sequences based on manual segmentation [8], while cartilage relaxation times were summarized as mean values.
Statistical Analysis
A Bland-Altman plot [22] was used to assess the degree of agreement between the three morphological pulse sequences. In addition, Pearson product-moment correlation coefficients were calculated between the different pulse sequences and repeated measures analysis of variance was used to test for morphological differences between the pulse sequences. The required significance level was set to p<0.05. Precision for cartilage thickness, volume, T1ρ, and T2 measurements was computed based on root-mean-square coefficients of variation (RMS-CVs) according to Glüer et al. [23].
Results
All results of morphological assessments are summarized in Table 1 for the thickness and in Table 2 for the volume. Representative examples of MR images for morphological assessment are shown in Figure 1 for the three pulse sequences. Figure 2 shows the typical range of slices used for segmentation. The Bland-Altman plots for the comparisons of the morphological sequences are shown in Figure 3 for thickness and Figure 4 for volume.
Table 1.
Compiled results of all thickness measurements.
| Cartilage Thickness [mm] |
Baseline | Follow-up | |||||
|---|---|---|---|---|---|---|---|
| SPGR | MERGE | FIESTA | SPGR | MERGE | FIESTA | ||
| Humerus | Subject 1 | 1.12 | 0.98 | 0.96 | 1.04 | 1.05 | 1.35 |
| Subject 2 | 0.77 | 0.7 | 0.7 | 0.74 | 0.7 | 0.88 | |
| Subject 4 | 0.69 | 0.46 | 0.56 | 0.64 | 0.57 | 0.58 | |
| Subject 5 | 0.86 | 0.72 | 0.71 | 0.85 | 0.59 | 0.79 | |
| Subject 6 | 0.67 | 0.56 | 0.56 | 0.71 | 0.57 | 0.42 | |
| Glenoid | Subject 1 | 1.2 | 0.75 | 1.08 | 1.28 | 0.95 | 1.04 |
| Subject 2 | 1.06 | 0.96 | 1.12 | 0.86 | 0.84 | 1.04 | |
| Subject 4 | 0.92 | 0.83 | 0.92 | 0.82 | 0.81 | 0.86 | |
| Subject 5 | 0.79 | 0.68 | 0.65 | 0.99 | 0.81 | 1 | |
| Subject 6 | 0.9 | 0.74 | 0.83 | 0.86 | 0.68 | 0.79 | |
Table 2.
Compiled results of all volume measurements.
| Cartilage Volume [mm3] |
Baseline | Follow-up | |||||
|---|---|---|---|---|---|---|---|
| SPGR | MERGE | FIESTA | SPGR | MERGE | FIESTA | ||
| Humerus | Subject 1 | 1115.23 | 1037.04 | 933.19 | 1052.43 | 1093.77 | 1178.15 |
| Subject 2 | 846.95 | 802.75 | 769.03 | 815.76 | 804.73 | 929.27 | |
| Subject 4 | 676.66 | 556.49 | 623.91 | 671.77 | 645.78 | 599.41 | |
| Subject 5 | 815.85 | 698.26 | 695.66 | 768.37 | 590.4 | 707.04 | |
| Subject 6 | 645.18 | 577.28 | 563.36 | 657.32 | 587.84 | 468.17 | |
| Glenoid | Subject 1 | 413.64 | 327.47 | 345.02 | 387.85 | 280.95 | 312.81 |
| Subject 2 | 350.7 | 322.6 | 361.92 | 307.01 | 263.17 | 328.9 | |
| Subject 4 | 298.43 | 304.68 | 314.76 | 338.24 | 255 | 254.26 | |
| Subject 5 | 213.56 | 245.99 | 212.31 | 333.7 | 237.65 | 281.42 | |
| Subject 6 | 302.21 | 233.17 | 269.92 | 292.7 | 225.84 | 258.29 | |
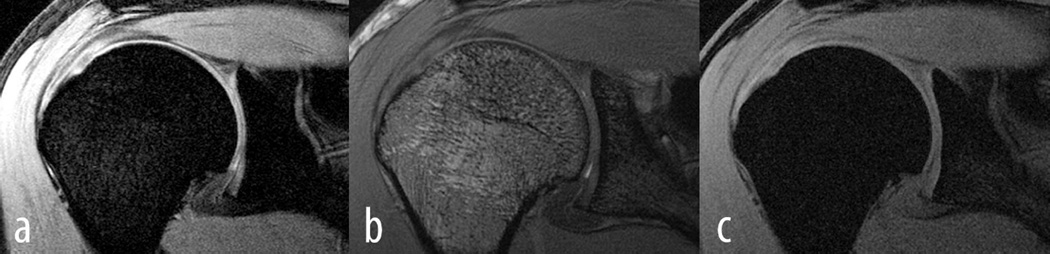
Figure 1.
Shown are a) SPGR, b) FIESTA and c) MERGE from one subject. Both, the humeral cartilage as well as the glenoidal cartilage can be appreciated. It is also clear from these images that SPGR provided the best contrast between cartilage as well as cartilage and surrounding tissues.
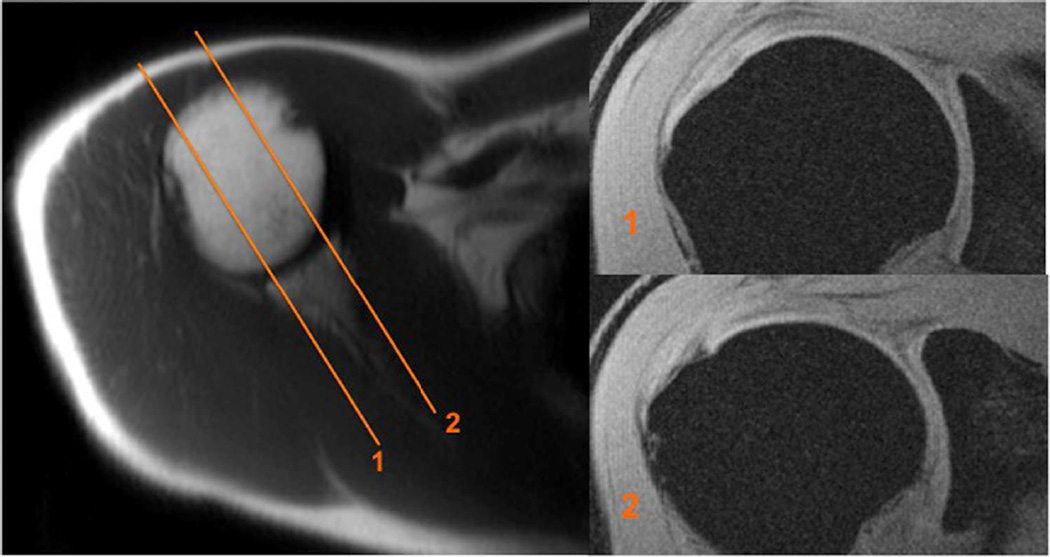
Figure 2.
On the axial image (left) the range of slices used for segmentation is presented. The lateral (1) and medial (2) limits are shown. This range was selected in order to minimize the interference of partial volume effects. The sagittal images on the right depict the first most lateral and last most medial slice accordingly.
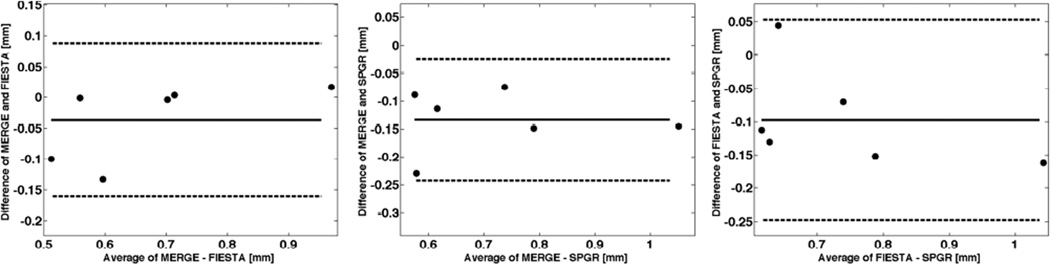
Figure 3.
Bland-Altman plots comparing the cartilage thicknesses for two pairs of morphological sequences at baseline. Shown is the mean difference (solid line) and the limits of agreement (dashed lines) defined as the mean difference plus and minus 1.96 the standard deviation of the differences.
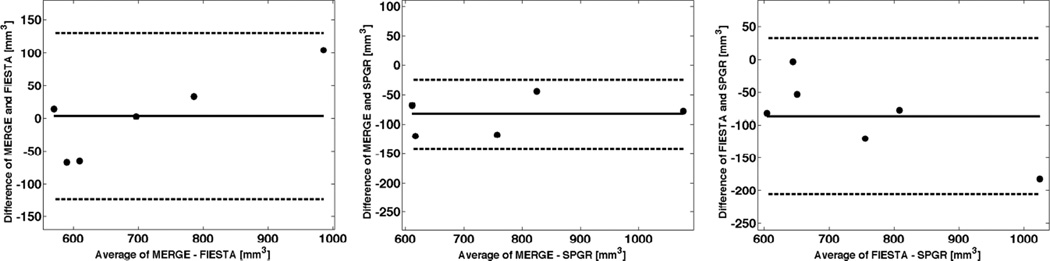
Figure 4.
Bland-Altman plots comparing the cartilage volume for two pairs of morphological sequences at baseline. Shown is the mean difference (solid line) and the limits of agreement (dashed lines) defined as the mean difference plus and minus 1.96 the standard deviation of the differences.
For thickness, the mean difference between MERGE and FIESTA (MERGE-FIESTA) was −0.036mm with a limit of agreement of ±0.12mm (95% of differences lie between these limits). Thus, the values from FIESTA were in general slightly higher than from MERGE. The mean difference between MERGE and SPGR (MERGE-SPGR) was −0.13mm with a limit of agreement of ±0.11mm. Thus, SPGR produced slightly larger thickness values than MERGE. The mean difference between FIESTA and SPGR (FIESTA-SPGR) was −0.10mm with a limit of agreement of ±0.15mm. Hence, mean thickness values calculated from SPGR images were slightly larger than from FIESTA. In summary, SPGR produced the largest mean thickness values followed by FIESTA and MERGE. However, these differences of thickness measurements were not significant in the humeral head. These values were only significantly different in the glenoid for the follow-up scans, which reflects the difficulties of segmenting the thin glenoidal cartilage.
For volume, the mean difference between MERGE and FIESTA (MERGE-FIESTA) was 3.5mm3 with a limit of agreement of ±127mm3. The mean difference between MERGE and SPGR (MERGE-SPGR) was −82.8mm3 with a limit of agreement of ±59.0mm3. Thus, SPGR produced slightly larger volumes than MERGE. The mean difference between FIESTA and SPGR (FIESTA-SPGR) was −86.3mm3 with a limit of agreement of ±118.9mm3. Hence, mean volume values calculated from SPGR images were slightly larger than from FIESTA. In summary, SPGR produced the largest mean volume.
Correlations of R>0.9 were found between all pulse sequences in the humeral head for both thickness and volume.
Table 3 depicts the correlations of the morphological measurements at baseline between the three different pulse sequences indicating lower correlations for the glenoid. Correlations of R>0.9 were found between all pulse sequences in the humeral head for both thickness and volume. Table 3 also presents results from the precision measurements of morphological cartilage assessment as calculated by the RMS-CVs for each pulse sequence in the humeral head and glenoid. As can be seen, SPGR yielded the highest reproducibility followed by MERGE and FIESTA. The precision was much higher for the humeral head than for the glenoid.
Table 3.
Pearson product-moment correlations between morphological sequences at baseline and the RMS-CVs for each sequence.
| R-values | SPGR with MERGE | MERGE with FIESTA | FIESTA with SPGR | |
|---|---|---|---|---|
| Thickness | Head | 0.95 | 0.95 | 0.91 |
| Glenoid | 0.52 | 0.73 | 0.91 | |
| Volume | Head | 0.98 | 0.98 | 0.97 |
| Glenoid | 0.66 | 0.83 | 0.87 | |
| RMS-CVs [%] | SPGR | MERGE | FIESTA | |
| Thickness | Head | 4.89 | 7.98 | 17.73 |
| Glenoid | 9.42 | 12.04 | 12.01 | |
| Volume | Head | 4.01 | 6.30 | 12.39 |
| Glenoid | 14.4 | 11.88 | 10.61 | |
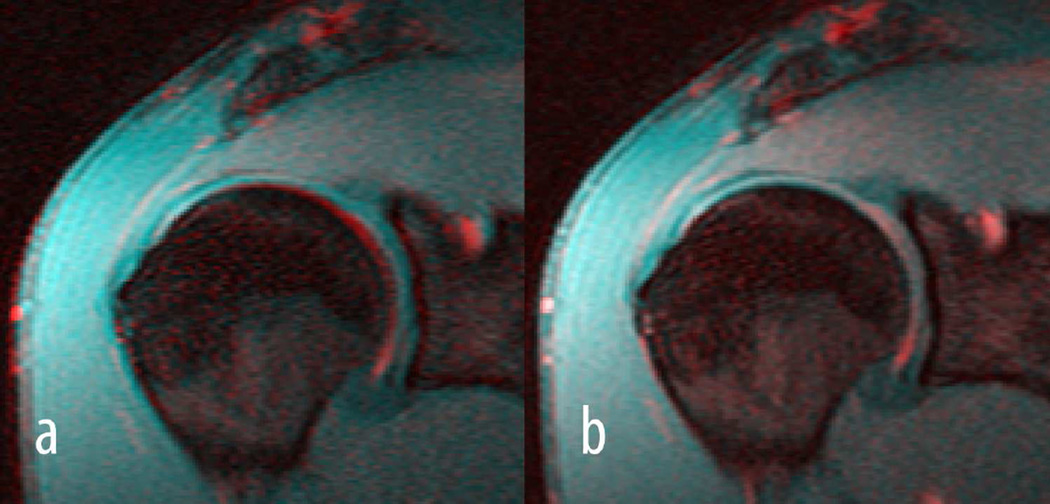
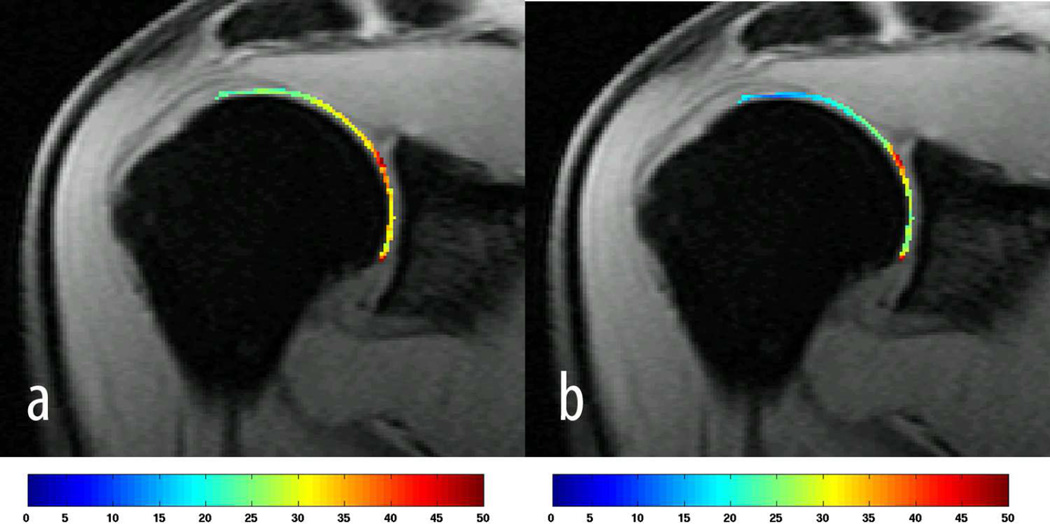
Figure 5 shows a representative example of motion correction where the fourth echo (colour coded in red) of a T2 acquisition was aligned to the first echo (colour coded in cyan), while Figure 6 demonstrates representative examples of colour-coded T1ρ and T2 maps. Results from T1ρ and T2 relaxation time measurements are summarized in Table 4 and Table 5, respectively. The RMS-CVs for T1ρ and T2 were 6.4% and 10.6%, respectively.
Figure 5.
Shown are colour-coded images of the fourth (red) and first (blue) echo of a T2 acquisition before registration a) and after registration b). In particular around the subchondral bone a clear misalignment can be seen between the two echoes in a) which disappeared after aligning the fourth to the first echo in b).
Figure 6.
A T1ρ map is shown in a) and a T2 map in b) from the same subject. The colour bars indicate T1ρ and T2 relaxation times in ms. Only the humeral head was segmented for T1ρ and T2 mapping.
Table 4.
Humeral cartilage T1ρ mean values and standard deviations for each subject.
| T1ρ [ms] | Baseline | Follow-up | ||
|---|---|---|---|---|
| Mean | StDev | Mean | StDev | |
| Subject 1 | 35.40 | 10.21 | 34.37 | 7.27 |
| Subject 2 | 32.78 | 21.76 | 30.74 | 13.53 |
| Subject 4 | 34.47 | 9.09 | 29.788 | 10.44 |
| Subject 5 | 33.38 | 11.42 | 35.48 | 7.64 |
| Subject 6 | 30.67 | 9.77 | 31.44 | 7.73 |
| Mean | 33.34 | 32.36 | ||
Table 5.
Humeral cartilage T2 mean values and standard deviations for each subject.
| T2 [ms] | Baseline | Follow-up | ||
|---|---|---|---|---|
| Mean | StDev | Mean | StDev | |
| Subject 1 | 20.93 | 9.74 | 21.2 | 9.11 |
| Subject 2 | 24.11 | 9.54 | 25.46 | 10.35 |
| Subject 4 | 24.47 | 7.35 | 25.64 | 11 |
| Subject 5 | 23.27 | 8.02 | 23.45 | 8.78 |
| Subject 6 | 17.8 | 6.49 | 21.09 | 6.86 |
| Mean | 22.12 | 23.37 | ||
Discussion
In this study, we have provided evidence that the MRI assessment of cartilage morphology and biochemical composition in-vivo is feasible and reproducible in healthy subjects. We have shown that the thin cartilage can be depicted in-vivo with good contrast using three state of the art pulse sequences with SPGR yielding the highest precision for cartilage thickness and volume in the humeral head. Furthermore, we have demonstrated the feasibility of assessing T1ρ and T2 relaxation times in the shoulder using a combined T1ρ-T2 pulse sequence featuring composite pulses as previously used [20]. The composite pulse was originally suggested by Dixon et al. [24] and has been shown to simultaneously correct for B0 and B1 field heterogeneities [21], which is in particular important for off-centre shoulder imaging. We have obtained T1ρ and T2 values similar to those previously reported for the knee [20], and high precision for T1ρ and modest precision for T2 was demonstrated.
Quantitative cartilage imaging in the shoulder is extremely challenging mainly due to the thin cartilage layer and off-centre imaging. Previously, Graichen et al. [18] investigated the feasibility of assessing shoulder cartilage morphology in human cadaveric specimens using MRI. This ex-vivo experiment allowed them to investigate the cartilage with high spatial resolution and a long imaging time of 19 minutes. They found a high agreement between the glenoid and humeral head cartilage volume and thickness measured by MRI and a water displacement method. Vanwanseele et al. [19] assessed the in-vivo precision of quantitative shoulder cartilage measurements in the humeral head after spinal cord injury using a 3D gradient echo sequence with selective water excitation with 1.5mm slice thickness and 0.273mm in-plane resolution in 11 minutes. They found a coefficient of variation of 4.5% for cartilage thickness measurements in the humeral head. They did not assess the cartilage of the glenoid. However, compared to our study they report higher thickness and volume values in a group of seven healthy volunteers (mean thickness 1.29mm and mean volume 4200mm3). This could be related to several reasons. The segmentation process is very challenging due to the extremely curved cartilage and partial volume effects. Part of the cartilage was consistently excluded to avoid volume average artefacts at the most anterior and posterior aspect of the cartilage and only the 7 central slices were segmented as seen in Figure 2. Thus, our approach was very conservative.
Our study aimed to assess cartilage morphology in both, the humeral head and the glenoid. Sequences were optimized for SNR, contrast and spatial resolution within a clinically feasible scan time shorter than 10 minutes. Highest precision was demonstrated for the SPGR sequence, followed by MERGE in the assessment of the humeral head. In contrast, the glenoid was more difficult to segment and showed lower precision values as expected. This was also reflected in the significant differences found for thickness and volume measures between the sequences for the glenoid at follow-up.
The MERGE sequence showed the lowest values for thickness and volume followed by FIESTA and SPGR. In general, contrast between cartilage and bone marrow was worse for FIESTA due to missing fat suppression, which is difficult to achieve for this type of pulse sequence without disturbing the steady state. SPGR and MERGE yielded similar contrast whereas MERGE was slightly more fluid sensitive.
We have also demonstrated that the assessment of biochemical information in the shoulder is feasible. T2 provided lower reproducibility values than T1ρ. This was primarily due to the combined acquisition method we used where all T1ρ TSLs are acquired first and subsequently all T2 echoes are acquired in the same scan and the first echo (TSL/TE=0) is shared by both [23]. We found substantial motion between the first echo and the last echoes for T2. Thus, we needed to register the different echoes, which significantly improved the precision, but interpolation of thick slices still could have an effect. The T2 scan was the last scan of the session and the subjects had difficulties to hold still. This was evident in subject 6 (Table 3) who had a much larger discrepancy between baseline and follow-up than all other subjects in T2. In contrast, the T1ρ values were very similar in that subject.
Our study has some limitations; the extremely curved surfaces of the humeral and glenoid cartilage, along with the non-orthogonal alignment of the images with the surface created prominent partial volume artefacts; however we consistently segmented the cartilage avoiding the most anterior and most posterior areas where this artefact was more severe. Another limitation is the lack of gold standard for the comparison of our measurements (arthrography or cadaveric specimens). We did not administrate intra-articular gadolinium, which would have allowed for more precise cartilage contour detection but would have been invasive.
In summary, we found very good precision using SPGR in the humerus for morphological assessment of cartilage. The glenoid was more difficult to segment and image in a clinically feasible scan time. We have also demonstrated the feasibility of biochemical assessment of shoulder cartilage using T1ρ and T2 mapping pulse sequences with good precision. We have found values, which are in good agreement with previously reported results in the knee. Thus, we conclude that morphological and biochemical assessment of shoulder cartilage is feasible in vivo within a clinically desired scan time.
Key Points.
The thin cartilage morphology can be assessed in the shoulder in-vivo.
Non-invasive biochemical assessment of shoulder cartilage is feasible in-vivo using MRI.
Acknowledgments
The authors would like to thank Dr. Xiaojuan Li and Dr. Cory Wyatt for helpful discussions. This work was supported by NIH/NIAMS R01AR057336. The scientific guarantor of this publication is Roland Krug. The authors of this manuscript declare relationships with the following companies: General Electrics. This study has received funding by NIH/NIAMS R01AR057336. No complex statistical methods were necessary for this paper. Institutional Review Board approval was obtained. Written informed consent was obtained from all subjects (patients) in this study. Methodology: experimental, performed at one institution.
References
- 1.Recht M, Bobic V, Burstein D, et al. Magnetic resonance imaging of articular cartilage. Clin Orthop Relat Res. 2001:S379–S396. doi: 10.1097/00003086-200110001-00035. [DOI] [PubMed] [Google Scholar]
- 2.Subburaj K, Souza RB, Stehling C, et al. Association of MR relaxation and cartilage deformation in knee osteoarthritis. J Orthop Res. 2012;30:919–926. doi: 10.1002/jor.22031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gold GE, Hargreaves BA, Stevens KJ, Beaulieu CF. Advanced magnetic resonance imaging of articular cartilage. Orthop Clin North Am. 2006;37:331–347. vi. doi: 10.1016/j.ocl.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Kornaat PR, Koo S, Andriacchi TP, Bloem JL, Gold GE. Comparison of quantitative cartilage measurements acquired on two 3.0T MRI systems from different manufacturers. J Magn Reson Imaging. 2006;23:770–773. doi: 10.1002/jmri.20561. [DOI] [PubMed] [Google Scholar]
- 5.Kornaat PR, Reeder SB, Koo S, et al. MR imaging of articular cartilage at 1.5T and 3.0T: comparison of SPGR and SSFP sequences. Osteoarthritis Cartilage. 2005;13:338–344. doi: 10.1016/j.joca.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Bolbos R, Benoit-Cattin H, Langlois JB, et al. Measurement of knee cartilage thickness using MRI: a reproducibility study in a meniscectomized guinea pig model of osteoarthritis. NMR Biomed. 2008;21:366–375. doi: 10.1002/nbm.1198. [DOI] [PubMed] [Google Scholar]
- 7.Carballido-Gamio J, Bauer J, Lee KY, Krause S, Majumdar S. Combined image processing techniques for characterization of MRI cartilage of the knee. Conf Proc IEEE Eng Med Biol Soc. 2005;3:3043–3046. doi: 10.1109/IEMBS.2005.1617116. [DOI] [PubMed] [Google Scholar]
- 8.Carballido-Gamio J, Bauer JS, Stahl R, et al. Inter-subject comparison of MRI knee cartilage thickness. Medical image analysis. 2008;12:120–135. doi: 10.1016/j.media.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juras V, Bohndorf K, Heule R, et al. A comparison of multi-echo spin-echo and triple-echo steady-state T2 mapping for in vivo evaluation of articular cartilage. Eur Radiol. 2015 doi: 10.1007/s00330-015-3979-6. 10.1007/s00330-015-3979-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Benjamin Ma C, Link TM, et al. In vivo T(1rho) and T(2) mapping of articular cartilage in osteoarthritis of the knee using 3 T MRI. Osteoarthritis Cartilage. 2007;15:789–797. doi: 10.1016/j.joca.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nozaki T, Kaneko Y, Yu HJ, et al. T1rho mapping of entire femoral cartilage using depth- and angle-dependent analysis. Eur Radiol. 2015 doi: 10.1007/s00330-015-3988-5. 10.1007/s00330-015-3988-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wyatt C, Kumar D, Subburaj K, et al. Cartilage T1rho and T2 Relaxation Times in Patients With Mild-to-Moderate Radiographic Hip Osteoarthritis. Arthritis Rheumatol. 2015;67:1548–1556. doi: 10.1002/art.39074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blanco FJ. Osteoarthritis year in review 2014: we need more biochemical biomarkers in qualification phase. Osteoarthritis Cartilage. 2014;22:2025–2032. doi: 10.1016/j.joca.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 14.Prasad AP, Nardo L, Schooler J, Joseph GB, Link TM. T(1)rho and T(2) relaxation times predict progression of knee osteoarthritis. Osteoarthritis Cartilage. 2013;21:69–76. doi: 10.1016/j.joca.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soslowsky LJ, Flatow EL, Bigliani LU, Mow VC. Articular geometry of the glenohumeral joint. Clin Orthop Relat Res. 1992:181–190. [PubMed] [Google Scholar]
- 16.Hodler J, Loredo RA, Longo C, Trudell D, Yu JS, Resnick D. Assessment of articular cartilage thickness of the humeral head: MR-anatomic correlation in cadavers. AJR American journal of roentgenology. 1995;165:615–620. doi: 10.2214/ajr.165.3.7645480. [DOI] [PubMed] [Google Scholar]
- 17.Yeh LR, Kwak S, Kim YS, et al. Evaluation of articular cartilage thickness of the humeral head and the glenoid fossa by MR arthrography: anatomic correlation in cadavers. Skeletal Radiol. 1998;27:500–504. doi: 10.1007/s002560050427. [DOI] [PubMed] [Google Scholar]
- 18.Graichen H, Jakob J, von Eisenhart-Rothe R, Englmeier KH, Reiser M, Eckstein F. Validation of cartilage volume and thickness measurements in the human shoulder with quantitative magnetic resonance imaging. Osteoarthritis Cartilage. 2003;11:475–482. doi: 10.1016/s1063-4584(03)00077-3. [DOI] [PubMed] [Google Scholar]
- 19.Vanwanseele B, Eckstein F, Hadwighorst H, Knecht H, Spaepen A, Stussi E. In vivo precision of quantitative shoulder cartilage measurements, and changes after spinal cord injury. Magn Reson Med. 2004;51:1026–1030. doi: 10.1002/mrm.20068. [DOI] [PubMed] [Google Scholar]
- 20.Li X, Wyatt C, Rivoire J, et al. Simultaneous acquisition of T1rho and T2 quantification in knee cartilage: repeatability and diurnal variation. J Magn Reson Imaging. 2014;39:1287–1293. doi: 10.1002/jmri.24253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen W, Takahashi A, Han E. Quantitative T(1)(rho) imaging using phase cycling for B0 and B1 field inhomogeneity compensation. Magn Reson Imaging. 2011;29:608–619. doi: 10.1016/j.mri.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 22.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307–310. [PubMed] [Google Scholar]
- 23.Glüer CC, Blake G, Blunt BA, Jergas M, Genant HK. Accurate Assessment of Precision Errors: How to measure the reproducibility of Bone Densitometry Techniques. Osteoporosis Int. 1995;5:262–270. doi: 10.1007/BF01774016. [DOI] [PubMed] [Google Scholar]
- 24.Dixon WT, Oshinski JN, Trudeau JD, Arnold BC, Pettigrew RI. Myocardial suppression in vivo by spin locking with composite pulses. Magn Reson Med. 1996;36:90–94. doi: 10.1002/mrm.1910360116. [DOI] [PubMed] [Google Scholar]