Abstract
The range of commercially available array platforms and analysis software packages is expanding and their utility is improving, making reliable detection of copy-number variants (CNVs) relatively straightforward. Reliable interpretation of CNV data, however, is often difficult and requires expertise. With our knowledge of the human genome growing rapidly, applications for array testing continuously broadening, and the resolution of CNV detection increasing, this leads to great complexity in interpreting what can be daunting data. Correct CNV interpretation and optimal use of the genotype information provided by single-nucleotide polymorphism probes on an array depends largely on knowledge present in various resources. In addition to the availability of host laboratories’ own datasets and national registries, there are several public databases and Internet resources with genotype and phenotype information that can be used for array data interpretation. With so many resources now available, it is important to know which are fit-for-purpose in a diagnostic setting. We summarize the characteristics of the most commonly used Internet databases and resources, and propose a general data interpretation strategy that can be used for comparative hybridization, comparative intensity, and genotype-based array data.
Keywords: array, classification, CNV, database, data interpretation, diagnostic, genome wide
Introduction
Currently, all the laboratories that offer genome-wide array testing for diagnostic purposes use commercially available platforms in combination with a suitable software package to analyze data. These packages include predefined reference datasets and enable the use of several publicly available datasets through links in their software. Often, the user is able to include their own or national datasets by adding custom annotation tracks as well as additional data sources, all of which aid the array data interpretation. Correct interpretation of array data focuses on assessing the clinical relevance of the detected copy-number variant (CNV) events. In addition, arrays that contain single-nucleotide polymorphism (SNP) probes can both determine the copy-number state of each interrogated genomic sequence, and provide genotype information of the SNPs tested. This genotype information allows stretches of homozygosity to be detected, the parental origin of an aberration to be determined, and sample mix-ups to be identified. The SNP data can also help in identifying aneuploidy, which is often seen in cancer samples. The overall aim of using arrays for diagnostic testing is to optimally interpret the results in the shortest possible time, and to reach a clear and straightforward conclusion about whether a finding is clinically relevant to the patient.
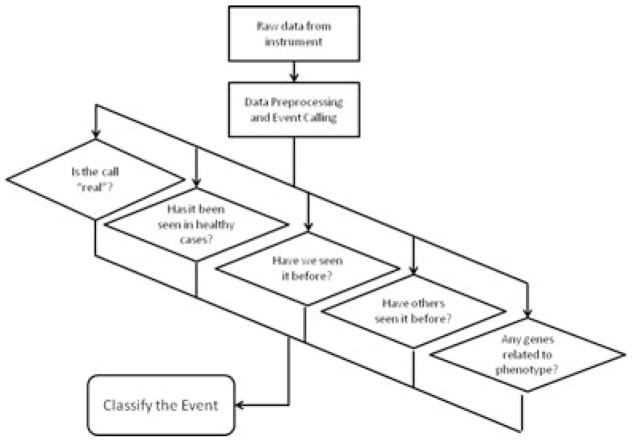
Although laboratories use different array platforms, reference DNA pools, and procedures for array-based comparative genomic hybridization (CGH) analysis, the general workflow for determining the relevance of each detected event to the observed phenotype can be summarized across most institutions as shown in Figure 1. The process starts by collecting raw intensity data from the microarray instrument and applying appropriate data preprocessing. This includes filtering out poorly performing probes, and platform-specific normalization and recentering, background correction, channel balancing (in the case of two dye arrays), and correction for systematic hybridization biases (e.g., %GC wave correction and fragmentation length). See Vermeesch et al. (2012, this issue) for more detailed information. The next step is to apply a CNV calling algorithm turning the probe intensities into comparative (“test over reference”) ratios to identify regions of possible CNV and, in the case of SNP arrays, allelic events. Once the CNVs have been detected, the user must classify these events in an accurate and efficient manner.
Figure 1.
Schematic representation of a general workflow to determine the relevance of an event detected by genome-wide array analysis to the observed phenotype of a patient.
We will briefly discuss the classification and interpretation of CNVs in constitutional diagnostics, whereas Simons et al. (2012, this issue) will deal with arrays in tumor diagnostics. We also provide an overview of the publicly available databases and resources, describing their main objectives and characteristics, as well as their potential limitations.
All the information about the resources laboratory specialists use for their array data interpretation is based on our own experiences or was gathered through a questionnaire and subsequent discussion at the international symposium on “Array in Daily Practice,” held in Amsterdam, the Netherlands, on May 27, 2011.
CNV Classification
The term copy-number variant or CNV is used to describe any change in copy number of a region of genomic sequence as a loss or gain relative to a control sample (from one or more control individuals) [Feuk et al., 2006]. For clinical use, every CNV detected needs to be interpreted [South and Brothman, 2011]. Some CNVs are common, whereas others are rare. Those that are common usually represent normal genomic variation or benign CNVs that are mostly not involved in disease risk. In some instances, a common CNV can represent a susceptibility locus [reviewed by Lee and Scherer, 2010]. CNVs that are rare are more likely to be penetrant for disease but, as with common variants, some will be benign variants specific to an individual or family, and the clinical relevance of other rare CNVs will be uncertain. In particular, rare CNVs are challenging to interpret and classify [Tsuchiya et al., 2009]. There are no generally established rules, but most laboratories classify the various CNVs into different categories using some or all of the CNV classifications listed in Table 1 [see also Vermeesch et al., (2012) this issue]. These classifications are largely based on those used in gene mutation analysis of the nucleotide changes detected, in particular of unclassified variants. Although the terms and abbreviations of CNV classifications may differ between laboratories, the respective implications are often the same. Using standard terms would facilitate communication between all those involved.
Table 1.
Classifications of Copy-Number Variants (CNVs) in the Human Genome
| CNV classification | Alternative terms |
|---|---|
| Benign CNV | Normal genomic variant |
| Likely benign CNV | |
| CNV of uncertain clinical relevance | Variant of uncertain significance (VOUS) |
| CNV of possible clinical relevance | High-susceptibility locus/risk factor/likely pathogenic variant |
| Clinically relevant CNV | Pathogenic variant |
When interpreting and classifying CNVs, it is essential to distinguish gains from losses, as the potential clinical consequences may differ significantly. Hence, it is crucial to compare gains with gains, and losses with losses [Conrad et al., 2010; Vermeesch et al., 2007]. Fortunately, the databases listed below have agreed to standardize the colors used to distinguish CNVs, depicting gains in blue and losses in red.
Public Internet Databases and Sources for Array Data Interpretation
Assigning a CNV to one of the classifications given in Table 1 is achieved by using many references and other datasets. These can be consulted sequentially, but a growing number of software packages offer semiautomated analysis using both public as well as local datasets, with the option to add and include additional datasets. Easy access to multiple data sources decreases reporting time significantly, but often extensive “manual” interpretation of one, or several, CNV(s) is still required to reach an exact conclusion for an array result. The main reason for this is that a CNV with the observed size may not have been reported before and hence adequate interpretation requires personal attention. The three interpretation steps (in any given order) are similar for most laboratories: (1) comparison with in-house, national, and international control datasets; (2) comparison with in-house and international affected individual datasets; and (3) gene content analysis and literature studies.
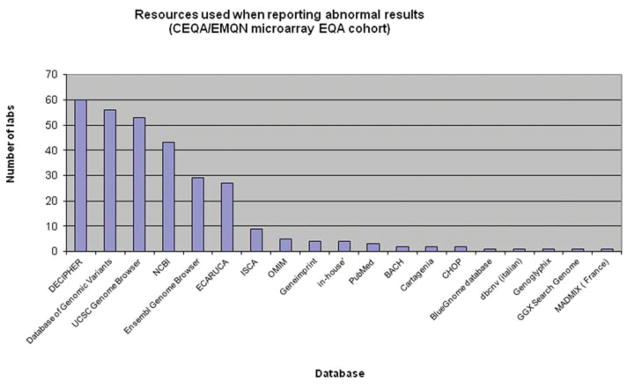
Laboratories use a variety of resources to interpret their array results (Fig. 2). A recent survey of European Molecular Genetics Quality Network and Cytogenetic European Quality Assessment microarray EQA participants showed the range of resources used by all 63 laboratories. In some cases, further resources need to be consulted for specific information, for example, where imprinting is suspected, a marker chromosome is identified or a mosaic aberration is encountered (Table 2).
Figure 2.
Graph showing the number of laboratories (Y-axis) using the various resources (X-axis) to interpret their array results.
Table 2.
Useful Internet Resources for Array Data Interpretation
| Name | URL | Main objective |
|---|---|---|
| COREMINE | www.coremine.com | COREMINE Medical is a product of the PubGene Company designed to be used by anyone seeking information on health, medicine, and biology. |
| ENDEAVOUR | www.esat.kuleuven.be/endeavour | ENDEAVOUR is a Web resource for the prioritization of candidate genes that uses a training set of genes known to be involved in a biological process of interest. |
| GECCO | http://sourceforge.net/projects/genomegecco/ | GeCCO (Genomic CNV Classification Objectively) is a bioinformatics tool for classifying copy number variants as either benign or pathogenic. |
| GeneImprint | www.geneimprint.com | GeneImprint is a portal into the burgeoning field of genomic imprinting, collecting relevant articles and reviews, press reports, video and audio lectures, and genetic information. |
| Genomic Oligoarray and SNP array evaluation tool v1.0 | www.ccs.miami.edu/cgi-bin/ROH/ROH_analysis_tool.cgi | This tool is designed to assist in the evaluation of genes, and subselections, including OMIM genes, OMIM genes annotated as associated with autosomal and/or autosomal recessive inherited phenotypes in runs of homozygosity (ROH), and chromosomal regions involved in microdeletions and microduplications. |
| GeneTests | www.ncbi.nlm.nih.gov/sites/GeneTests | Medical genetics information resource developed for physicians, other healthcare providers, and researchers. |
| Chromosomal Mosaicism | http://mosaicism.cfri.ca | A Website to provide information to patients, families, health care providers, students, and the general public on the unique conditions of chromosomal mosaicism. |
| Orphanet | www.orpha.net | Orphanet is the reference portal for information on rare diseases and orphan drugs, for all audiences. Orphanet’s aim is to help improve the diagnosis, care, and treatment of patients with rare diseases. |
| Small Supernumerary Marker Chromosomes Database | www.med.uni-jena.de/fish/sSMC/00START.htm | To collect all available case reports on small supernumerary marker chromosomes (sSMC) and provide detailed information for patients and medical professionals. |
| Unique | www.rarechromo.org | Unique is a source of information and support to families and individuals affected by any rare chromosome disorder and to the professionals who work with them. |
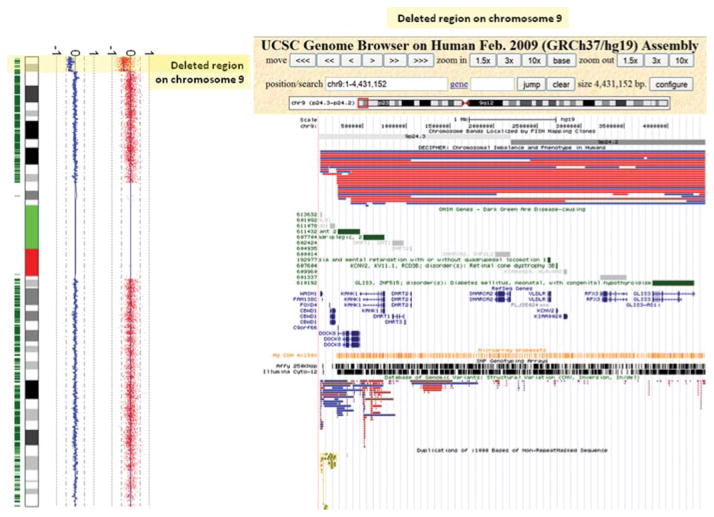
The majority of analytical software packages used to process data also provide visualization tools, showing the array data, genetic content (obtained from a recent version of the human genome reference sequence), probe distribution of the array platform, and often combining data from a number of public databases and sources. Sources such as PubMed, OMIM, and human genome browsers (UCSC, Ensembl) provide numerous valuable tracks of genome-oriented data. There are also specific databases that collect individual cases (e.g., DatabasE of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER) [Firth et al., 2009], European Cytogeneticists Association Register of Unbalanced Chromosome Aberrations (ECARUCA) [Feenstra et al., 2006], and International Standards for Cytogenomic Arrays (ISCA) [Kaminsky et al., 2011]) or control information (e.g., DGV [Church et al., 2010]), providing more genetic and phenotypic details, and information about the genomic region of interest (see Fig. 3). The tools and databases available for CNV interpretation can be divided into three categories. First, “in-house” repositories that are created to maintain cases processed by the laboratory itself. These range in complexity from an Excel spreadsheet to an enterprise-wide relational database fully integrated into the institution’s patient management database. The second type of database is a specialized repository with a relatively narrow objective or “theme.” Such a database collects individual case or control information regarding genetic and phenotypic details, for example, a collection of CNVs from a particular (control) population or a disease-specific database. Finally, the third type is the category of “data aggregators,” where the database is created by collating and organizing data from different sources. Queries are performed across the entire dataset allowing the user to display results based on numerous tracks. Each type of database and some of the Internet sources are reviewed below to highlight their objectives, characteristics, and limitations.
Figure 3.
Plot of chromosome 9 showing a terminal loss of the short arm detected by genome-wide SNP array analysis. The deleted region is shown in the UCSC Genome Browser and several tracks are selected to help in interpreting this loss and to determine its clinical relevance. From top to bottom, the following tracks were selected: chromosome bands, DECIPHER, OMIM genes, RefSeq genes, Microarray Probe sets, SNP Genotyping Arrays, DGV, and duplications.
In-House Databases
A number of laboratories have the resources to develop their own laboratory flow monitoring system (e.g., POEMA, designed and realized by RUNMC and www.rimarcable.nl) and interpretation strategies, involving databases holding genotype and phenotype information, prioritizing pipelines, and the semiautomatic compilation of diagnostic reports. Some private laboratories or companies provide access to their data only to customers; others use their large databases as a private resource for their own laboratory and effectively as a marketing tool. As more disease data becomes publicly available, for example, through DECIPHER, ECARUCA, and ISCA, the potential marketing value of these proprietary databases will decrease and at some point these may be released to the public.
Several commercial tools are available that provide in-house database capabilities aimed at improving the entire diagnostic workflow. These tools embed up-to-date, clinically relevant databases, and robust workflow automation tools to aid interpretation of variants (e.g., Cartagenia BENCH). Others aim to improve data interpretation by using several local and international datasets for a variety of array platforms (e.g., Nexus copy number). To maintain sales, commercial tools typically: (1) provide technical support and maintenance; (2) include training programmes; and (3) are created according to industry standards that follow software testing, validation, and certification protocols, which support laboratory accreditation. Like many of the noncommercial databases, (4) software companies interact with many different laboratories, leading to consensus approaches and best-of-breed tools, but furthermore, (v) companies have a financial incentive to continuously produce innovative software. Although the use of commercial software can be limited by financial constraints, such investments have the potential to improve the cost-effectiveness of a laboratory. An investment in a commercial tool set can help standardize a laboratory’s interpretative workflow; this leads to gains in speed, efficiency, and diagnostic confidence, allowing the laboratory to manage large numbers of cases and report findings confidently. The end result should be at least a break-even situation, and preferably a positive financial balance in the broadest sense.
Specialized CNV Databases
There are a number of common sources of CNV data that are employed during the CNV classification process. These consist of either sources of data on unaffected, potentially unaffected or control samples, or repositories providing an overview of samples with mainly affected individuals.
Datasets on Population and Family Control Individuals
Control datasets are important and useful resources for interpreting array data from patients. A number of laboratories that perform array analysis have accumulated array data from control individuals (i.e., volunteers, blood donors, etc.) as well as data on affected and unaffected parents from patients in whom array testing has been done. Some laboratories using the same array platform collaborate and collect their data in a national registry, thereby providing a larger, mutual control dataset than individual laboratories could achieve alone. In addition, numerous studies, such as HapMap, have examined control populations using a variety of different array platforms [Church et al., 2010]. The majority of these studies have been published and most have been collected in the Database of Genomic Variants (DGVs).
Database of Genomic Variants (http://projects.tcag.ca/variation/)
The DGV provides a useful catalogue of control data for studies aiming to correlate genomic variation with phenotypic data [Iafrate et al., 2004; Zhang et al., 2006]. It differs from other structural variation databases in that it focuses solely on variants identified in control samples. The database is continuously updated with new data from published research studies. All data included in DGV have, therefore, undergone peer review and informed consent is a requirement prior to publication. Only high-quality studies and data are included in this database, ensuring that the reliability of the CNV calls in the database is high. As the DGV includes data from large-scale CNV discovery efforts, it includes samples that were analyzed independently in multiple studies. These samples primarily represent controls from reference resource datasets commonly used in genetics research, such as the HapMap and Human Genome Diversity Panel cohorts, and other population controls; however, consistent documentation and standard sample nomenclature ensure that this redundancy is available to users.
DGV does not currently accept direct submission of new data. Instead, peer reviewed, published data are first submitted in a standardized format to either dbVar (National Center for Biotechnology Information [NCBI]) or DGVa (EBI) for accession and then passed on to DGV for review and curation by DGV staff [Church et al., 2010]. The data undergo rigorous testing using multiple methods to ensure their quality. High-quality studies that fulfill the criteria for inclusion in DGV are then selected and imported into DGV.
The objective of DGV is to provide a comprehensive summary of structural variation in the human genome. Structural variation is defined as genomic alterations that involve segments of DNA that are larger than 50 bp. This threshold was recently changed to reflect the development of better variation detection technologies and to adhere to the definition used by the 1000 Genomes Project and the archival structural variation databases (DGVa and dbVar) [Scherer et al., 2007]. All variants that are greater than 50 bp and less than 3 Mb (10 Mb for inversions) are included. Additional filters are also applied as well as the size restriction. Study-specific filters and instructions from authors can be applied to filter out low quality or spurious variants (i.e., variants described in >1 individual, or called by >1 algorithm could constitute a high-quality set of calls). Variants mapped to random or unknown chromosomes are excluded. If a variant is mapped to the Y chromosome in a female subject, or if a variant span gaps in the assembly, they are removed. Studies that report on both cases and controls and data pertaining to cases are excluded. A comparison with the regions associated with genomic disorders listed on DECIPHER is also performed to ensure that variants in control individuals do not coincide with known disease-causing variants. Thus, the content of the database only represents structural variation identified in control samples.
The DGV is freely accessible to any researcher, scientist, physician, or individual who wishes to search any data contained in it. The full contents of the database are also available for download. Although the database contains only data originally described in controls, this does not mean the database should be used as a substitute for running a control set with your patient samples. The database is meant to serve as a guide; it will provide information about whether there is a common variant in the region of interest, but just because a variant is annotated in the database does not mean that a similar variant cannot be disease causing in your patient sample. Explanations for such phenomena are given in the final section of this paper. Similarly, a lack of variants in a specific region of the database does not necessarily mean there are no common variants at that locus. Factors such as probe coverage and resolution differ significantly between platforms. Details on the DGV are summarized in Table 3.
Table 3.
Overview of Free Online Databases of CNVs in Controls (DGV) or Patients with Genotype–Phenotype Information
| DGV | DECIPHER | ECARUCA | ISCA | |
|---|---|---|---|---|
| Account holders | No accounts are created, freely accessible to all users. | Professionals in medical genetics only. | Professionals in medical genetics and families (patient portal; access to own data only). | ISCA Members |
| Access | No restrictions | Free public access to anonymized, consented data. Login upon registration for data entry and curation. |
Login upon registration. | Login upon registration. |
| Costs involved? | No | No | No | No |
| Number of account holders | 0 | 1,135 | >1,500 | >900 |
| Objectives of the database | To provide a comprehensive summary of structural variation in the human genome. The DGV provides a useful catalogue of control data for studies aiming to correlate genomic variation with phenotypic data. | Catalogue of pathogenic submicroscopic copy number variants and associated phenotype. | Reliable information on rare chromosome anomalies. | Leveraging high-quality clinical copy number data to create a CNV Atlas of the human genome. |
| Number cases | ||||
| Total | 11,941 controls | >11,300 | > 4,600 | >28,000 deidentified “calls-only” cases; approximately 8,000 of these cases have been collected under the opt-out method of consent and have associated raw data files available in dbGaP. |
| Consented | 11,941 | >5,300 (for free public access and browser display) | > 4,600 | |
| Number cases | ||||
| Prenatal cases | 0 | 0 | ~200 | ~4,000a |
| Postnatal cases | 11,941 | >11,300 | >4,400 | >28,000 postnatal |
| Number of aberrations | 101,923 (66,741 CNV; 34,229 InDels; 953 inversions | 31,148 | >6,200 | >13,000 |
| Aberrations | Primarily submicroscopic variants are included. Maximum size for CNVs is 3 Mb, while inversions up to 10 Mb are included and may be cytogenetically visible. | Some cases have benign CNVs listed as well. | Cytogenetically visible and submicroscopic imbalances. | Copy number variants identified via clinical constitutional microarray testing. |
| Karyotyping | 394 | Only clinically relevant findings are registered. | ||
| FISH | 2,457 | |||
| MLPA | 475 | |||
| QF-PCR | 1,271 | |||
| GW array | 31,148 | |||
| Number of cases with clinical features | 0 | 4,054 | >4,600; based on LDDB | >5,000 cases with ≥ one HPO term |
| Gender | Yes | Yes | Yes | |
| Age | At examination | At last examination | At time of testing | |
| Ethnicity | No | No | No | |
| Data curation | Peer reviewed data submitted, with additional data curation provided by DGV staff prior to data entry. | Account holders responsibility. | Quality control upon entry by the Database Management Team. | Submitting laboratories have the opportunity to curate their data against their own previous submissions and the ISCA curated dataset; expert curation committee periodically curates the entire dataset. |
| Visualization of database content in genome browser? | UCSC | UCSC | UCSC | UCSC |
| DGV genome browser | Ensembl DECIPHER | Ensembl | dbVar | |
To be submitted at end of prenatal grant.
DGV, Database of Genomic Variants; DECIPHER, DatabasE of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources ECARUCA, European Cytogeneticists Association Register of Unbalanced Chromosome Aberrations; ISCA, International Standards for Cytogenomic Arrays.
Datasets on Individuals with Disease
When a certain CNV has not been previously identified in one or more controls, the next step is to determine if the CNV has been detected previously in a patient. Laboratories are encouraged to store their own patient data in a database, including as much detail on their clinical features as possible. The likelihood that a CNV is clinically relevant increases when the same, or a similar CNV, has previously been detected in a patient with a similar phenotype. To allow phenotype comparison, it is essential that standard nomenclatures such as the Human Phenotype Ontology (HPO) are used for encoding clinical features. Riggs et al. elaborate on this in the upcoming Human Mutation special issue on phenotyping. If the local database does not show any “hits,” other databases with genotype–phenotype information are available and laboratories are advised to search for similar genetic aberrations. At present, there is at least one proprietary-owned database, the Genoglyphix Chromosome Aberration Database [Neill et al., 2010], and three freely available databases (DECIPHER, ECARUCA, and ISCA) that are most commonly used for array data interpretation. These last three databases are described below and summarized in Table 3.
DECIPHER (http://decipher.sanger.ac.uk/)
The DECIPHER is an interactive Web-based database that incorporates a suite of tools designed to aid the interpretation of submicroscopic chromosomal imbalances. The primary purposes of the DECIPHER project are as follows: (1) to increase medical and scientific knowledge about chromosomal microdeletions/duplications, (2) to improve medical care and genetic advice for individuals/families with submicroscopic chromosomal imbalances, and (3) to facilitate research into the study of genes that affect human development and health. DECIPHER enhances clinical diagnosis by retrieving information from a variety of bioinformatics resources relevant to the imbalance found in the patient. Known and predicted genes within an aberration are listed in the DECIPHER patient report, common copy-number changes in control populations are displayed, and genes of recognized clinical importance are highlighted. With the patient’s consent, positional genomic information together with a brief description of the associated phenotype becomes viewable without password protection, for example, via the DECIPHER track in Ensembl or the UCSC Genome Browser. The data in DECIPHER can be used both by clinicians advising patients with similar genomic findings and by researchers working on specific disorders or on the function of genes contained within an aberration. The DECIPHER consortium provides these data in good faith as a research tool and the database has facilitated and been cited in more than 200 publications.
DECIPHER contains a powerful search engine enabling the aggregate consented data to be rapidly searched by genomic location, phenotype term, cytogenetic band, or gene name. It incorporates innovative bioinformatics resources such as a predicted haploinsufficiency score for genes [Huang et al., 2010], a consensus CNV track displayed in four frequency bands, and context-sensitive link-outs to other data sources.
ECARUCA (www.ecaruca.net)
The ECARUCA is a Web-based database that contains cytogenetic and clinical data of patients with rare chromosome abnormalities, including microscopically visible aberrations, as well as microdeletions and duplications. ECARUCA collects the results of genetic and cytogenetic tests and the associated clinical features. The database can be queried on aberrations of chromosome regions according to the ISCN 2009 nomenclature [Shaffer et al., 2009], but also offers searching by base pair position. The submission of and search for clinical features are based on the strategy of the Winter–Baraitser Dysmorphology Database of the London Medical Databases (www.lmdatabases.com) [Winter and Baraitser, 1987, 2001]. ECARUCA is interactive, dynamic, and has possibilities to store cytogenetic, molecular, and clinical data for the long term. Currently, it contains more than 6,200, mainly unique, chromosomal aberrations detected by routine cytogenetic analysis, FISH, MLPA, and/or genome-wide array analysis in over 4,500 patients. It also includes nearly all the cases previously published by Schinzel (2001) with phenotypes associated with the great majority of cytogenetically visible, unbalanced chromosome abnormalities. In addition to this published set of data, all submitted data are curated by the ECARUCA daily management team, ensuring the up-to-date quality of the collection. Individual “parent accounts” allow parents to inform the ECARUCA team about the follow-up of their child. Thus, the ECARUCA database provides health care workers with accurate information on clinical aspects of rare chromosome disorders. In addition, detailed correlations between chromosome aberrations and their phenotypes are of invaluable help in localizing genes for intellectual disabilities (IDs) and congenital anomalies.
ECARUCA aims to be a database that is easily accessible for all account holders and it encourages both exchanges of information and technical knowledge. It aims to improve patient care and collaboration between genetic centres in the field of clinical cytogenetics. This free online database is one of the largest genetic registries with curated genetic and clinical information in the world.
ISCA (www.iscaconsortium.org)
The ISCA Consortium has established a publicly available database to leverage data from thousands of patients with developmental disabilities, congenital anomalies, and other phenotypes being screened by clinical laboratories, to accelerate our understanding of CNVs in the clinical population [Kaminsky et al., 2011].
Laboratories performing chromosome microarray testing on a clinical basis may submit to the database. Membership is free and open to the public. ISCA members may search the publicly available database, currently housed within the Database of Genomic Structural Variation (dbVar) at the NCBI (http://www.ncbi.nlm.nih.gov/dbvar/studies/nstd37/). The publicly available database currently contains over 13,000 CNVs identified from over 28,000 individuals, as well as information on the clinical interpretation of each CNV as determined by the submitting laboratory. In addition, a separate database with controlled access is housed within the Database of Genotypes and Phenotypes (dbGaP at NCBI) and contains raw data files from laboratories that have initiated the opt-out method of consent. To utilize these data, a researcher must have an institutional review board-approved protocol and apply for access through a Data Access Committee.
Phenotype information is also available within the public database for the subset of cases in which it was reported and this is displayed in HPO terms to allow for computational manipulation of the data and to avoid the complexities and variation of free text entries. The submission of clinical information is encouraged but not required. In an effort to encourage this, one-page phenotype forms (one for prenatal and one for postnatal characterization) have been developed to aid the collection of this information. Electronic versions of these forms, as well as the necessary tools to facilitate genotype and phenotype data submission from the laboratory and clinic to the registry, are made available by the ISCA consortium in collaboration with Cartagenia.
Data within the ISCA database are curated on several different levels. Periodically, the entire database is curated using an evidence-based protocol by a panel of expert reviewers. This group evaluates regions with discrepant calls using information gathered from, among other things, peer-reviewed literature, other large-scale case-control datasets, and expert opinion. Original calls are not changed, but curation decisions are reflected in a separate study in dbVar (http://www.ncbi.nlm.nih.gov/dbvar/studies/nstd45/) and on a separate track in UCSC for clarity.
Submitting laboratories are given the opportunity to curate their current submission against their past submissions to ensure intra-laboratory consistency. They are also given the opportunity to curate their submission against the ISCA-curated dataset. Submitters are required to acknowledge that they have reviewed the curation report, although they are not required to change any of their calls should they so choose.
Gene Content Analysis and Literature Studies
After consulting various datasets to classify a certain CNV, it is usually necessary to consult more resources to complement the array report with useful detailed information, for example, on the function of a specific gene in the aberrant region, potential candidate disease genes in stretches of homozygosity, or valuable clinical information for optimal patient care.
Data Aggregators
UCSC Genome Browser (http://genome.ucsc.edu/)
Human genome browsers are essential to quickly obtain relevant information of a certain genomic region in one view. By selecting one or several data tracks, information from various sources and sites can be shown and through direct and indirect links can relatively quickly and easily lead to more detailed information, from clinical information to experimental fundamental data. The UCSC Genome Browser [Fujita et al., 2011] is at present the most commonly used by diagnostic laboratories (Fig. 3) because it is accessible, stable, and it has links to all of the aforementioned databases, as well as data from the HapMap projects and segmental duplication data [Alkan et al., 2009]. Customized tracks can also be added. DGV, DECIPHER, and OMIM data tracks at UCSC are regularly updated. UCSC serves as a data aggregator for much publicly available data, but it currently has no provision for integrating nonconsented data.
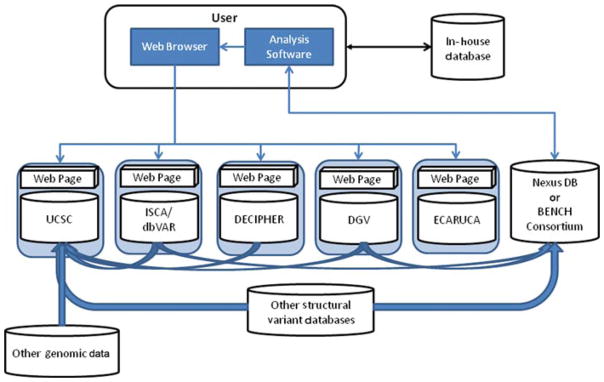
Other well-known sites that are routinely used in data interpretation (Fig. 2) are PubMed, OMIM, GeneImprint and CHOP, but also Unique, Orphanet, Genetests, GeneReviews, Google, ENDEAVOUR, and COREMINE (Table 2). If these resources are approached individually, the process becomes very time consuming. There is thus an urgent need for a fully integrated search engine that is able to address and search multiple databases and sources and report via one single login site. Some of the public databases (e.g., DECIPHER) and commercial products (e.g., Cartagenia BENCH and Nexus DB™) provide context-specific link-outs to other data sources, thereby offering some of the required utility (Fig. 4). Cartagenia is working on an open-access and noncommercial initiative that will allow querying multiple resources through a single portal.
Figure 4.
Schematic representation of possible connections between databases and Internet resources that can be used to optimize the quality and speed of array data interpretation. See text for details on the various resources.
Nexus DB™ (www.biodiscovery.com/software/nexus-db/)
Nexus DB™ (Biodiscovery, Inc, El Segundo, California) is a component of the Nexus Copy Number commercial software. It has been designed to serve the single purpose of storing CNV and loss of heterozygosity events from any platform and genome, and to be able to efficiently query any region for such events. Although Nexus DB™ is accessed globally via the Internet, it does not have a Web browser interface and relies on the genome browser integrated within Nexus Copy Number. As a data aggregator, CNV data from more than 35,000 cases are available to the users. These data are obtained from dbVar (ISCA dataset) as well as other publicly available sources. Nexus DB™ also provides a means for global collaboration and creation of special interest consortia (e.g., disease specific or regional) through secure data access and dynamic group creation mechanisms. A user of Nexus Copy Number looking at a single sample can compare, with a single click, the result of their samples against all of the samples in their local repository, as well as go to Nexus DB™ and search all of these sources. Various filtering options can be set so a user working with constitutional samples, for example, will not get aberrations reported for cancer samples.
Analytical Tools for Improving CNV Classification Efficiency
The CNV classification efficiency can be improved by applying computational methods and/or by annotation of the CNVs via data aggregation. Analytical approaches have been developed that utilize the sample phenotype information with the genomic content of the region of interest in order to provide additional guidance on the possible relevance of the event to the phenotype (e.g., deletion event in an area with a gene associated with neural development in a patient with ID). Two examples of such tools are GEnomic Classification of CNVs Objectively (GECCO) and the prioritization module within Cartagenia BENCH.
GEnomic Classification of CNVs Objectively
Using the aforementioned sources, the functional annotation of CNVs revealed distinct differences between CNVs associated with ID and those occurring in the general population. By using these annotation features, a classifier was developed to delineate ID-associated CNVs from CNVs seen in the general population. This bioinformatics tool is called GECCO (http://sourceforge.net/projects/genomegecco/) and measures each CNV for the presence and frequency of 13 genomic features, such as the density of repetitive elements within the CNV [Hehir-Kwa et al., 2010]. This classifier is able to complement existing clinical diagnostic workflows. The ability to predict the phenotypic effect of a CNV is particularly useful when parental samples are not available or when a rare, inherited CNV might contribute to the disease phenotype. Using the classifier, approximately 70% of rare, inherited CNVs and CNVs with unknown inheritance could be classified as either probably pathogenic or probably benign [Hehir-Kwa et al., 2010]. The GECCO classifier can also be used to independently confirm the pathogenicity of rare, de novo CNVs. Such tools give an estimation of pathogenicity, but should be used with care and consideration as they are based on a statistical calculation. It is important to be aware of potential false-positive or false-negative classifications. Adding the GECCO classifier to existing CNV interpretation methodologies, which are primarily based on frequency and inheritance, provides extra, objective information on the CNVs based on their genomic content.
Encompassing Database and Interpretation Platform for Clinical Routine Laboratory Workflow
Cartagenia (www.cartagenia.com)
Cartagenia BENCH is a software and database platform geared at interpreting genomic variation in routine diagnostics. It aims to automate the entire laboratory flow, from data intake to clinical variant interpretation to finished report. It has a strong clinical focus and automatically consults key databases and resources for interpretation.
First, BENCH allows the laboratory to build its own local variant and patient database, aggregates all relevant public databases and resources, and automates the laboratory workflow; analysis pipelines are set up and then saved for routine use, implementing a laboratory’s standard operating procedures for genetic variant filtering, classification, and interpretation in a single click. This significantly reduces hands on time.
Embedded databases include the laboratory’s internal variant database, DGV, CHOP, OMIM, RefSeq, UCSC and ENSEMBL datasets, PubMed literature analysis, custom gene and syndrome lists, DECIPHER, the ISCA CNV atlas, and so on and are kept up-to-date so that a counselor can send out laboratory reports based on accurate and recent information. Reports are generated automatically through customized laboratory and clinical templates.
Second, BENCH is also a rich phenotyping platform. The laboratory’s internal variant and patient database facilitates annotation of clinical features through user-friendly forms or through a physician portal, where referrers enter clinical detail electronically when they request an assay. Although these forms are based on the HPO, they avoid confronting counselors and clinicians with the complexity of phenotype vocabularies.
By integrating clinical features, BENCH will identify similar patients in the laboratory’s internal database as well as in external patient registries. Importantly, when clinical information is available for a patient, BENCH will assist variant interpretation through candidate-gene prioritization based on PubMed literature, automatically highlighting which variants might explain the patient’s phenotype. Besides automated genotype–phenotype correlation, the system facilitates advanced queries such as “are there other patients with this aberration that have the same or a similar heart defect?”
Third, the BENCH Consortium module facilitates the collaboration of consortia. These range from local or regional collaborations, to anonymous sharing of genotype and phenotype data within a national consortium (as set up in France and the Netherlands), to integration with international registries through single-click submission to DECIPHER, the ISCA CNV Atlas, and soon to ECARUCA.
Array Data Interpretation—The Conclusion
Using some or many of the aforementioned databases and resources will help determine whether a CNV or other significant array finding is considered (potentially) causative for the clinical phenotype of a patient and can be reported back to the requesting physician. Normal genomic variants or benign CNVs are not specified in the karyotype, but one has to be aware that these benign CNVs may sometimes (indirectly) cause or contribute to pathogenicity if:
there is a deletion on one allele and a mutated gene on the other allele [see Zhang et al., 2011 for an example];
the same deletion is present on both alleles, hence two benign heterozygote deletions generating a deleterious homozygous deletion;
each parent has a different, benign (heterozygous) deletion in the same gene, which, when both are inherited, causes a deleterious effect in the offspring (i.e., a compound heterozygote);
the region contains an imprinted gene possibly leading to differences in pathogenicity [Demars et al., 2011];
the CNV is on the X chromosome and inherited by a male offspring from an unaffected mother [De Leeuw et al., 2010; Ramocki et al., 2010];
the CNV is inherited from a mosaic carrier, who is not or only mildly affected [Willemsen et al., 2011];
the CNV occurs in combination with another CNV and together these lead to a pathogenic defect [Girirajan et al., 2010].
In any of the above circumstances, a benign CNV becomes pathogenic and must be mentioned in the karyotype with a detailed explanation in the array report.
Search and Submission of Valuable Data
New genetic as well as clinical information is available on a daily basis, and hence the current version of a certain software analysis package is soon outdated, in the sense that new comparative data may be available for a certain gene or genetic region. It is therefore crucial to consult up-to-date sources when one is not entirely certain about the meaning of a specific array finding. Databases with reliable genotype–phenotype information are crucial for geneticists, cytogeneticists, clinicians, and other medical professionals, as well as parents. They provide valuable structured clinical knowledge on (rare) chromosome imbalances that is often lacking in the literature, mostly due to a significant decline in the interest of scientific journals to publish case reports.
Submission to Databases and Organization of Data
The success of the databases largely depends on a constant flow of new findings based on up-to-date technologies. Submission or bulk upload of genetic data is essential and relatively easy for DECIPHER, ISCA, and ECARUCA, and their systems are continuously being optimized to improve these processes. A patient’s clinical information should include at least gender, age, and (basic) clinical features, and preferably be achieved by filling out a digital request form once, for both diagnostic purposes and potential submission to one of the databases. It is crucial for search and interpretation purposes to enter clinical features in a structured and unambiguous fashion. Unfortunately, the most broadly used international medical classification systems, International Classification of Diseases (ICD) and Systematized Nomenclature of Medicine (SNOMED), currently lack the levels of detail needed to code dysmorphic phenotypes. We therefore recommend using standardized terminology as provided by the HPO (www.human-phenotype-ontology.org), which is continuously expanding to meet clinical requirements and optimize the output of search strategies [Robinson et al., 2008, 2010]. See Supp. Table S1 for examples using HPO and ISCN. The ethnicity of an individual, either case or control, should preferably also be registered, as it is known that population-specific genomic variants exist.
Ideally, every professional using these resources for interpreting array data should have the discipline to contribute their own data. In practice, this is done for far too few cases. The main reasons for not submitting data vary, but are often lack of time, difficulty of obtaining informed consent, and limited or absent clinical information on the diagnostic request form. Simplifying the submission of genetic and clinical data from a local database to an international database by just a single “mouse click” is an ideal solution to improve this situation or, alternatively, enable data aggregation software to live query and combine data present in local databases instead of copying data to a central one. But even when submission is only “a mouse-click away” (as in the case of Nexus DB™ and Cartagenia BENCH), other limitations remain, such as lack of clinical details due to national laws and legislation protecting the privacy of an individual. The submission of genotypic and phenotypic data could be stimulated and improved by the introduction of “microattribution,” which shows who contributed the data, an approach described by Mons et al. [2011]. In this system, each data submission is treated as a mini-publication and adds to the scientist’s track record, submitting data to relevant databases in a manner complementary to the classic system of credits for publishing in a peer-reviewed scientific journal. Journals could require data to be submitted to one of the public databases prior to accepting a manuscript for publication.
Finally, there is more and more anecdotal evidence from patient groups that many patients are eager to update their phenotype information themselves. This is motivated by the desire to get into contact with other patients and share experiences and already occurs using social networks such as Facebook and disease-specific forums, where patients report in detail on their disease development and the success of interventions. We recommend tapping into these developments and putting the legal and IT frameworks in place for patients to add or systematically extend their own case reports and to facilitate interpatient contact.
One serious point of concern, whether for cases or controls, is to ensure a single registration per database and among databases because the scientific community must have accurate data and not an overrepresentation of repeated data. A unique, universal identifier should be employed by all repositories, but even this may not guarantee that individuals will not appear in more than one database.
Harmonization
It is useful to have multiple databases that provide accurate data. However, this does require other tools to function as “data aggregators,” whereby a user can search multiple sources for events/samples at a particular genomic locus, in a certain genome build. Conversion from one build to another should be easy and reliable. An example of this is the UCSC viewer, which, like other genome browsers, provides the user with a multitude of “tracks” that can be viewed, with each track representing data from different sources. Different people and applications prefer different user interface “front-ends” and having some level of choice between multiple viewers encourages continuous improvement. One major requirement for any viewer is the availability of data to aggregator sites. For example, the ISCA data are currently being deposited in dbVar and dbGaP at NCBI. The dbVar data can be publicly accessed and contains information about the call (start and stop positions, and basic phenotype). This is useful for aggregators. However, the data in DECIPHER and ECARUCA can be visualized in the UCSC genome browser, although the details are not directly viewable and require the host databases to be opened. It would be ideal for all repositories (holding data on either cases or controls) to provide an equivalent public level of data access without any personally identifiable information, such as detailed SNP data. Moreover, in addition to gender and age, and preferably also ethnicity, the clinical features observed in a patient should be listed in a uniform, structured way, conforming to a structured vocabulary such as HPO, to enable direct comparison and to determine the clinical consequences of a certain CNV or other genetic finding.
A Global Search Engine
The more genotype–phenotype information that becomes available to the medical/scientific community, the better geneticists can perform fast, reliable interpretation of array data now, and of whole genome sequencing data in the future. It is essential not only to enable fast and easy submission to these databases but also to accommodate a single search engine (data aggregation) that retrieves relevant information from different sources upon entry of a certain query. The majority of attendees at the symposium in Amsterdam agreed that all array data should be made publicly available, regardless of whether it comes from diagnostics or research, but these data should meet certain quality criteria. This is in line with current requirements for genetic data to be submitted to public repositories such as GEO, dbVar, or dbGaP. A minimal standard for queries should also be established by the software developers in this community, one that is available freely as “open source.” Local laboratory software developers as well as commercial software providers could then use this to connect to their databases in an easy way. Monitoring the quality of individual cases also remains a challenge, but the general opinion is that the quality of the submitted data, and, in particular, ensuring appropriate informed consent from the patient or legal representative is the submitter’s responsibility. The responsibility of the submitter to provide objective curation of data by experienced professionals is becoming increasingly important because many of the databases contain some incomplete, inaccurate, or confusing data such as patients with >100 unclassified CNVs. (Incomplete records are predominantly those on patients without clinical information.) In addition, different databases use different builds of the genome (NCBI35 [hg17], NCBI36 [hg18], and GRCh37 [hg19]), which is perceived as problematic when comparing data.
Conclusion
The collection of genetic data being made available to a larger audience is growing fast and current technical limitations will be relatively easily overcome in the near future. The most challenging part, however, remains obtaining and linking relevant clinical information to genetic observations in a structured way, to aid accurate data interpretation. Only by submitting and sharing their own data can the genetics community successfully search and interpret clinical data from patients with developmental disorders toward improving their health care worldwide.
Acknowledgments
Contract grant sponsor: DECIPHER is supported by the Wellcome Trust (grant number WT077008).
We would like to thank all participants of the “Array in daily practice: promises and pitfalls” symposium, held on May 27, 2011, in Amsterdam, the Netherlands. Part of the information for this paper was gathered during the symposium and based on participants’ answers to a questionnaire and their input during the workshop on “The interpretation of CNVs and the use of databases.” We thank Erin Riggs, Margie Manker, and Jeffrey R. MacDonald for their contribution and Jackie Senior for editorial advice.
Footnotes
Disclosure Statement: The authors declare no conflict of interest.
Additional Supporting Information may be found in the online version of this article.
References
- Alkan C, Kidd JM, Marques-Bonet T, Aksay G, Antonacci F, Hormozdiari F, Kitzman JO, Baker C, Malig M, Mutlu O, Sahinalp SC, Gibbs RA, Eichler EE. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. 2009;41:1061–1067. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church DM, Lappalainen I, Sneddon TP, Hinton J, Maguire M, Lopez J, Garner J, Paschall J, DiCuccio M, Yaschenko E, Scherer SW, Feuk L, Flicek P. Public data archives for genomic structural variation. Nat Genet. 2010;42:813–814. doi: 10.1038/ng1010-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, Fitzgerald T, Hu M, Ihm CH, Kristiansson K, Macarthur DG, Macdonald JR, Onyiah I, Pang AW, Robson S, Stirrups K, Valsesia A, Walter K, Wei J, Tyler-Smith C, Carter NP, Lee C, Scherer SW, Hurles ME Wellcome Trust Case Control Consortium. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–412. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demars J, Rossignol S, Netchine I, Lee KS, Shmela M, Faivre L, Weill J, Odent S, Azzi S, Callier P, Lucas J, Dubourg C, Andrieux J, Bouc YL, El-Osta A, Gicquel C. New insights into the pathogenesis of Beckwith-Wiedemann and Silver-Russell syndromes: contribution of small copy number variations to 11p15 imprinting defects. Hum Mutat. 2011;32:1171–1182. doi: 10.1002/humu.21558. [DOI] [PubMed] [Google Scholar]
- Feenstra I, Fang J, Koolen DA, Siezen A, Evans C, Winter RM, Lees MM, Riegel M, de Vries BBA, van Ravenswaaij CMA, Schinzel A. European cytogenetics association register of unbalanced chromosome aberrations (ECARUCA): an online database for rare chromosomal abnormalities. Eur J Med Genet. 2006;49:279–291. doi: 10.1016/j.ejmg.2005.10.131. [DOI] [PubMed] [Google Scholar]
- Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, Van Vooren S, Moreau Y, Pettett RM, Carter NP. DECIPHER: database of chromosomal imbalance and phenotype in humans using Ensembl resources. Am J Hum Genet. 2009;4:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, Diekhans M, Dreszer TR, Giardine BM, Harte RA, Hillman-Jackson J, Hsu F, Kirkup V, Kuhn RM, Learned K, Li CH, Meyer LR, Pohl A, Raney BJ, Rosenbloom KR, Smith KE, Haussler D, Kent WJ. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Rosenfeld JA, Cooper GM, Antonacci F, Siswara P, Itsara A, Vives L, Walsh T, McCarthy SE, Baker C, Mefford HC, Kidd JM, Browning SR, Browning BL, Dickel DE, Levy DL, Ballif BC, Platky K, Farber DM, Gowans GC, Wetherbee JJ, Asamoah A, Weaver DD, Mark PR, Dickerson J, Garg BP, Ellingwood SA, Smith R, Banks VC, Smith W, McDonald MT, Hoo JJ, French BN, Hudson C, Johnson JP, Ozmore JR, Moeschler JB, Surti U, Escobar LF, El-Khechen D, Gorski JL, Kussmann J, Salbert B, Lacassie Y, Biser A, McDonald-McGinn DM, Zackai EH, Deardorff MA, Shaikh TH, Haan E, Friend KL, Fichera M, Romano C, Gécz J, DeLisi LE, Sebat J, King MC, Shaffer LG, Eichler EE. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010;42:203–209. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehir-Kwa JY, Wieskamp N, Webber C, Pfundt R, Brunner HG, Gilissen C, de Vries BB, Ponting CP, Veltman JA. Accurate distinction of pathogenic from benign CNVs in mental retardation. PLoS Comput Biol. 2010;6:e1000752. doi: 10.1371/journal.pcbi.1000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N, Lee I, Marcotte EM, Hurles ME. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Kaminsky EB, Kaul V, Paschall J, Church DM, Bunke B, Kunig D, Moreno-De-Luca D, Moreno-De-Luca A, Mulle JG, Warren ST, Richard G, Compton JG, Fuller AE, Gliem TJ, Huang S, Collinson MN, Beal SJ, Ackley T, Pickering DL, Golden DM, Aston E, Whitby H, Shetty S, Rossi MR, Rudd MK, South ST, Brothman AR, Sanger WG, Iyer RK, Crolla JA, Thorland EC, Aradhya S, Ledbetter DH, Martin CL. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet Med. 2011;13:777–784. doi: 10.1097/GIM.0b013e31822c79f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Scherer SW. The clinical context of copy number variation in the human genome. Expert Rev Mol Med. 2010;12:e8. doi: 10.1017/S1462399410001390. [DOI] [PubMed] [Google Scholar]
- De Leeuw N, Bulk S, Green A, Jaeckle-Santos L, Baker LA, Zinn AR, Kleefstra T, van der Smagt JJ, Vianne Morgante AM, de Vries BB, van Bokhoven H, de Brouwer AP. UBE2A deficiency syndrome: mild to severe intellectual disability accompanied by seizures, absent speech, urogenital, and skin anomalies in male patients. Am J Med Genet A. 2010;152A:3084–3090. doi: 10.1002/ajmg.a.33743. [DOI] [PubMed] [Google Scholar]
- Mons B, van Haagen H, Chichester C, Hoen PB, den Dunnen JT, van Ommen G, van Mulligen E, Singh B, Hooft R, Roos M, Hammond J, Kiesel B, Giardine B, Velterop J, Groth P, Schultes E. The value of data. Nat Genet. 2011;43:281–283. doi: 10.1038/ng0411-281. [DOI] [PubMed] [Google Scholar]
- Neill NJ, Torchia BS, Bejjani BA, Shaffer LG, Ballif BC. Comparative analysis of copy number detection by whole-genome BAC and oligonucleotide array CGH. Mol Cytogenet. 2010;3:11. doi: 10.1186/1755-8166-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramocki MB, Tavyev YJ, Peters SU. The MECP2 duplication syndrome. Am J Med Genet A. 2010;152A:1079–1088. doi: 10.1002/ajmg.a.33184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs ER, Jackson L, Miller DT, Van Vooren S. Phenotypic information in genomic variant databases enhances clinical care and research: The international standards for cytogenomic arrays consortium experience. Hum Mutat. 2012;33:787–796. doi: 10.1002/humu.22052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PN, Köhler S, Bauer S, Seelow D, Horn D, Mundlos S. The Human phenotype ontology: a tool for annotating and analyzing human hereditary disease. Am J Hum Genet. 2008;83:610–615. doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PN, Mundlos S. The human phenotype ontology. Clin Genet. 2010;77:525–534. doi: 10.1111/j.1399-0004.2010.01436.x. [DOI] [PubMed] [Google Scholar]
- Scherer SW, Lee C, Birney E, Altshuler DM, Eichler EE, Carter NP, Hurles ME, Feuk L. Challenges and standards in integrating surveys of structural variation. Nat Genet. 2007;39:S7–S15. doi: 10.1038/ng2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinzel A. Catalogue of Unbalanced Chromosome Aberrations in Man. 2. Berlin: Walter de Gruyter; 2001. p. 966. [Google Scholar]
- Shaffer LG, Slovak ML, Campbell LJ. ISCN 2009: An International System for Human Cytogenetic Nomenclature (2009) Basel: S. Karger AG; 2009. p. 138. [Google Scholar]
- Simons A, Sikkema-Raddatz B, de Leeuw N, Konrad N, Hastings RJ, Schoumans J. Genome-wide arrays in routine diagnostics of haematological malignancies. Hum Mutat. 2012;33:941–948. doi: 10.1002/humu.22057. [DOI] [PubMed] [Google Scholar]
- South ST, Brothman AR. Clinical laboratory implementation of cytogenomic microarrays. Cytogenet Genome Res. 2011;135:203–211. doi: 10.1159/000331425. [DOI] [PubMed] [Google Scholar]
- Tsuchiya KD, Shaffer LG, Aradhya S, Gastier-Foster JM, Patel A, Rudd MK, Biggerstaff JS, Sanger WG, Schwartz S, Tepperberg JH, Thorland EC, Torchia BA, Brothman AR. Variability in interpreting and reporting copy number changes detected by array-based technology in clinical laboratories. Genet Med. 2009;11:866–873. doi: 10.1097/GIM.0b013e3181c0c3b0. [DOI] [PubMed] [Google Scholar]
- Vermeesch JR, Fiegler H, de Leeuw N, Szuhai K, Schoumans J, Ciccone R, Speleman F, Rauch A, Clayton-Smith J, van Ravenswaaij C, Sanlaville D, Patsalis PC, Firth H, Devriendt K, Zuffardi O. Guidelines for molecular karyotyping in constitutional genetic diagnosis. Eur J Hum Genet. 2007;15:1105–1114. doi: 10.1038/sj.ejhg.5201896. [DOI] [PubMed] [Google Scholar]
- Vermeesch JR, Brady PD, Sanlaville D, Kok K, Hastings RJ. Genome-wide arrays: quality criteria and platforms to be used in routine diagnostics. Hum Mutat. 2012;33:906–915. doi: 10.1002/humu.22076. [DOI] [PubMed] [Google Scholar]
- Willemsen MH, Beunders G, Callaghan M, de Leeuw N, Nillesen WM, Yntema HG, van Hagen JM, Nieuwint AW, Morrison N, Keijzers-Vloet ST, Hoischen A, Brunner HG, Tolmie J, Kleefstra T. Familial Kleefstra syndrome due to maternal somatic mosaicism for interstitial 9q34.3 microdeletions. Clin Genet. 2011;80:31–38. doi: 10.1111/j.1399-0004.2010.01607.x. [DOI] [PubMed] [Google Scholar]
- Winter RM, Baraitser M. The London dysmorphology database. J Med Genet. 1987;24:509–510. doi: 10.1136/jmg.24.8.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter RM, Baraitser M. London Dysmorphology Database, London Neurogenetics Database and Dysmorphology Photo Library on CD-ROM [Version 3] Oxford University Press; 2001. [Google Scholar]
- Zhang H, Gao J, Ye J, Gong Z, Gu X. Maternal origin of a de novo microdeletion spanning the ERCC6 gene in a classic form of the Cockayne syndrome. Eur J Med Genet. 2011;54:e389–e393. doi: 10.1016/j.ejmg.2011.03.012. [DOI] [PubMed] [Google Scholar]
- Zhang J, Feuk L, Duggan GE, Khaja R, Scherer SW. Development of bioinformatics resources for display and analysis of copy number and other structural variants in the human genome. Cytogenet Genome Res. 2006;115:205–214. doi: 10.1159/000095916. [DOI] [PubMed] [Google Scholar]