Significance
Pre-mRNA splicing generates protein diversity, is involved in the regulation of cellular differentiation, and can be epigenetically regulated by histone modifications. Chromatin binding proteins, which recognize histone modifications, recruit splicing regulators to methylated histones around tissue-specific splicing regions and regulate pre-mRNA splicing. However, the interplay of epigenetic factors and the splicing machinery during spermatogenesis remains unclear. Here, we show that epigenetic regulation of pre-mRNA splicing is required for spermatogenesis and male fertility. Thus, novel splicing diversity is important for spermatogenesis, and defects in this system may trigger disease.
Keywords: infertility, fertility defects, splicing defects, epigenetics, spermiogenesis
Abstract
Splicing can be epigenetically regulated and involved in cellular differentiation in somatic cells, but the interplay of epigenetic factors and the splicing machinery during spermatogenesis remains unclear. To study these interactions in vivo, we generated a germline deletion of MORF-related gene on chromosome 15 (MRG15), a multifunctional chromatin organizer that binds to methylated histone H3 lysine 36 (H3K36) in introns of transcriptionally active genes and has been implicated in regulation of histone acetylation, homology-directed DNA repair, and alternative splicing in somatic cells. Conditional KO (cKO) males lacking MRG15 in the germline are sterile secondary to spermatogenic arrest at the round spermatid stage. There were no significant alterations in meiotic division and histone acetylation. Specific mRNA sequences disappeared from 66 germ cell-expressed genes in the absence of MRG15, and specific intronic sequences were retained in mRNAs of 4 genes in the MRG15 cKO testes. In particular, introns were retained in mRNAs encoding the transition proteins that replace histones during sperm chromatin condensation. In round spermatids, MRG15 colocalizes with splicing factors PTBP1 and PTBP2 at H3K36me3 sites between the exons and single intron of transition nuclear protein 2 (Tnp2). Thus, our results reveal that MRG15 is essential for pre-mRNA splicing during spermatogenesis and that epigenetic regulation of pre-mRNA splicing by histone modification could be useful to understand not only spermatogenesis but also, epigenetic disorders underlying male infertile patients.
Spermatogenesis is a complex process involving several biological events and dramatic changes of chromatin structure. Male germ cells undergo stem cell self-renewal, mitotic divisions in spermatogonial proliferation, genomic rearrangement by meiotic homologous recombination at the spermatocyte stage, and morphological changes of round spermatids into elongated spermatids to form mature spermatozoa (1–3). During spermiogenesis, nucleosomal histone proteins are replaced with transition nuclear proteins (TNPs) and subsequently, protamines, the major nucleosomal proteins in spermatozoa. Moreover, epigenetic modifications, such as histone methylation, dramatically change throughout spermatogenesis (3, 4).
Pre-mRNA splicing generates protein diversity and is involved in the regulation of cellular differentiation (5, 6). Recent advances have shown that histone modifications regulate alternative splicing through recruitment of splicing regulators via chromatin binding proteins, such as MORF-related gene on chromosome 15 (MRG15) (7). Histone H3 lysine 36 (H3K36) is methylated proximal to tissue-specific splicing regions (8–12). MRG15 specifically recognizes the methylated H3K36 and recruits polypyrimidine tract binding protein (PTB) at intronic splicing silencer elements near an exon to suppress exon insertions into mRNA (7). MRG15 is also a component of histone acetyltransferase (HAT) and histone deacetylase (HDAC) complexes and regulates transcription by balancing histone acetylation (7, 13–21). During spermatogenesis, before histones are replaced with transition proteins and protamines, histone H4 is highly acetylated, and the acetylation of histones is required for histone removal. In addition to regulation of pre-mRNA splicing and histone acetylation, MRG15 contributes to homology-directed DNA repair. Although MRG15 functions in multiple biological processes, the roles of MRG15 during spermatogenesis are unknown, because Mrg15 null mice are embryonic lethal (22).
In this report, we have deleted MRG15 specifically in postnatal male germ cells and analyzed the role of MRG15 during spermatogenesis. Spermatogenesis in the Mrg15 null testis arrests at the round spermatid stage without affecting meiotic division and histone acetylation. MRG15 contributes to epigenetic regulation of pre-mRNA splicing of Tnp2, one of the reasons for the arrest of spermatogenesis in the Mrg15 null germ cells.
Results
MRG15 Is Required for Postmeiotic Spermatogenesis.
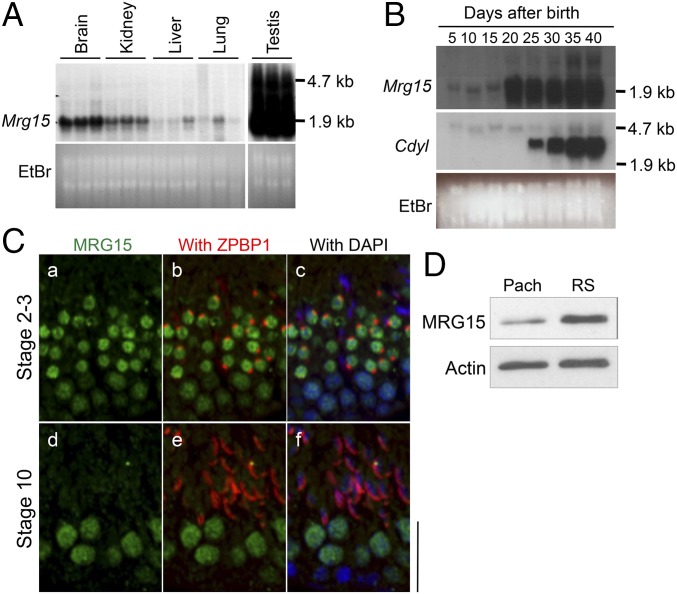
MRG15 mRNA levels are extremely high in testis, and expression initiates at 20 d after birth, when late-stage spermatocytes differentiate into round spermatids (Fig. 1 A and B). MRG15 protein localizes in spermatocytes, and its expression is highest in round spermatids during spermatogenesis (Fig. 1 C and D). To address the in vivo postnatal roles of MRG15 during spermatogenesis, an MRG15-floxed allele was generated in ES cells, and mice carrying this allele were crossed with Stra8-Cre mice to achieve germline-specific deletion (SI Appendix, Fig. S1) (23). Germline-specific Mrg15 null males have smaller testes and are sterile compared with control males (Fig. 2 A–C). In the Mrg15 null testes, spermatogenesis is arrested at the round spermatid stage, abnormal multinucleated cells are abundant, and there are no detectable mature sperm with condensed nuclei (Fig. 2D). Whereas spermatogonia and spermatocytes are histologically normal in Mrg15 null testes, spermatogenesis is arrested before round spermatid step 7, with significant apoptotic cell death (Fig. 2E and SI Appendix, Fig. S2). These findings suggest that MRG15 functions in round spermatids or late-stage spermatocytes for progression of spermatogenesis.
Fig. 1.
Spatiotemporal expression of MRG15 in the testis. (A and B) Transcripts of Mrg15 were examined by Northern blot analysis (A) in multiple tissues and (B) during postnatal development of the testis. Expression of chromodomain protein, Y chromosome-like (Cdyl) and/or ethidium bromide (EtBr) staining were performed as controls. The testis lanes in A were separately exposed to avoid strong radioactive signal from the testis lanes from invading adjacent lanes. (C) Immunolocalization of MRG15 in the testes. MRG15 (green), ZPBP1 (red), and DAPI (blue) localization in (a–c) stages 2 and 3 and (d–f) stage 10 seminiferous epithelium is shown. Seminiferous stages were determined by patterns of an acrosome staining of ZPBP1. (Scale bar: 10 µm.) (D) Western blot analysis of MRG15 in purified pachytene spermatocytes (Pach) and round spermatids (RS). Expression analysis of actin was performed as a control.
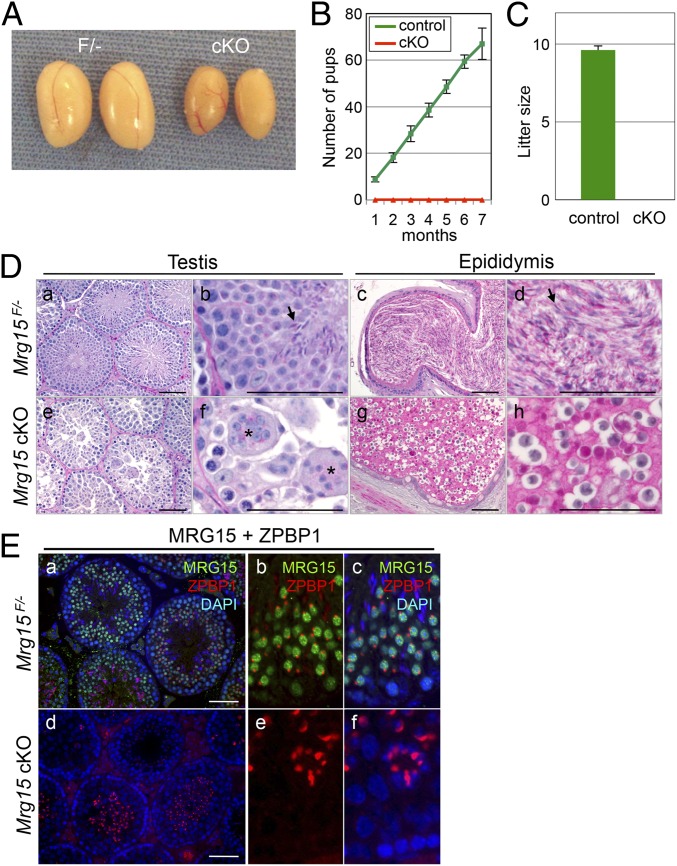
Fig. 2.
Infertile phenotype of germ cell-specific MRG15 null mice. (A) Gross images of Mrg15F− (F/−) and Mrg15F/−:Stra8-Cre+ (cKO) testes at 10 wk of age. (B) Average number of pups produced by F/− (control) and cKO males over 6 mo of breeding (n = 10 per genotype). (C) Average litter size produced by control and cKO males. (D) Histological analyses of (a, b, e, and f) testes and (c, d, g, and h) epididymis of (a–d) control and (e–h) cKO at 10 wk of age. Arrows and asterisks indicate mature sperm and abnormal multinucleated cells, respectively. (Scale bar: 50 µm.) (E) Validation of seminiferous stage of spermatogenetic arrest by lack of MRG15. MRG15 (green), ZPBP1 (red), and DAPI (blue) in (a–c) control and (d–f) cKO are shown. (Scale bar: 50 µm.)
MRG15 Is Not Required for Meiotic Progression.
Because MRG15 plays an essential role during homology-directed DNA damage repair, SCP1, γH2AX, and ubiquitylated H2A were analyzed in spermatocytes to examine whether MRG15 is required for meiotic homologous recombination (20, 21, 24). However, there were no significant differences in localizations of meiosis regulating proteins between control and Mrg15 null testes (SI Appendix, Figs. S3 and S4A). H3K36me3 and H4K20me3, both of which are recognized by MRG15, were not changed by lack of MRG15, indicating that MRG15 is not essential for meiotic homologous recombination (SI Appendix, Fig. S3) (25, 26).
Spermatogenesis in Mrg15 Null Testes Arrests Before Hyperacetylation of Histone H4.
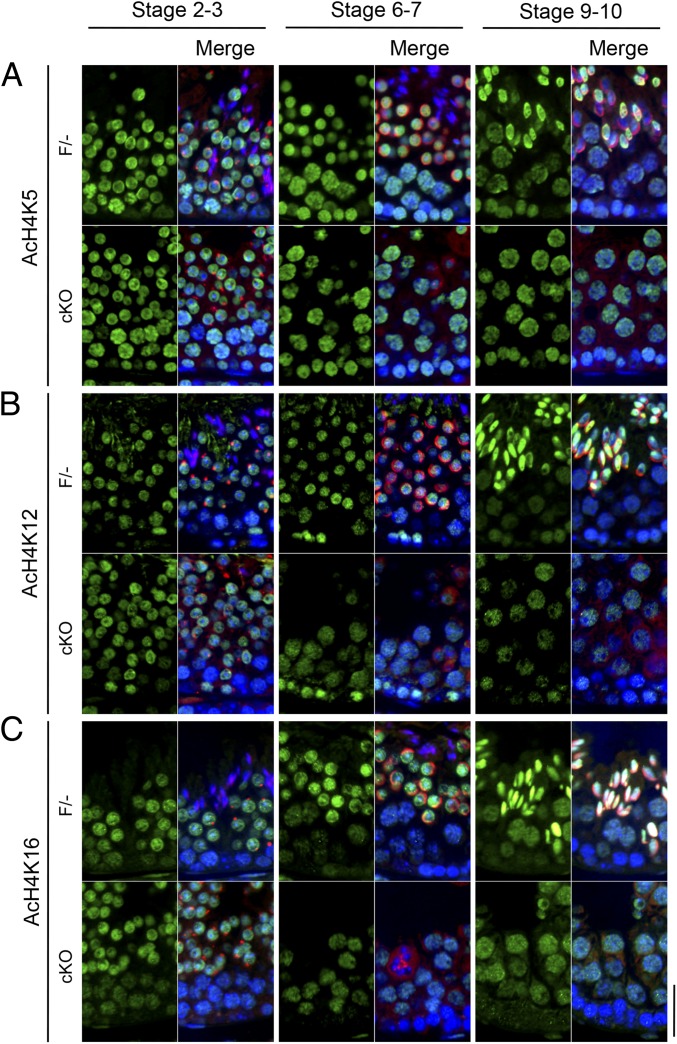
During spermatogenesis, histone H4 is highly acetylated before replacement of nucleosomal proteins from histones to TNPs to protamines (27). Because MRG15 is found in both HAT and HDAC complexes (13–21), MRG15 may be essential for postmeiotic events. To investigate whether MRG15 is involved in histone acetylation in postmeiotic spermatogenesis, acetylated histones were analyzed. Although acetylated histone H3 is highly localized in elongated spermatids in control testes, acetylated histone H3 levels in round spermatids are not significantly different between control and MRG15 conditional KO (cKO) testes (SI Appendix, Fig. S4B). CDYL, a histone H4 acetyltransferase that recognizes methylated H3K9 and is expressed in the testes after postnatal day 25 (Fig. 1B), is detectable in elongating spermatids in control testes but not in MRG15 cKO testes, probably because spermatogenesis is arrested before the elongating spermatid stage (SI Appendix, Fig. S4C) (28). When acetylation of lysines 5, 8, 12, and 16 of histone H4 was analyzed, these acetylated forms were reduced in the Mrg15 null testes (Fig. 3 and SI Appendix, Fig. S5). However, high accumulation of acetylation on each residue was observed in elongating spermatids of control testes, and these germ cells are no longer present in Mrg15 null testes (Fig. 3 and SI Appendix, Fig. S5A). When the amounts of histone H4 acetylation on each residue were compared in spermatid fractions between 22-d-old WT testes, in which spermatogenesis has developed to the round spermatid stage, and adult Mrg15 null testes, there were no significant differences (SI Appendix, Fig. S5B). Because spermatogenesis in Mrg15 null testes arrests at the round spermatid stage before histone H4 is hyperacetylated, it is difficult to determine if MRG15 is necessary for replacement of nucleosomal proteins.
Fig. 3.
Analyses of histone acetylation in MRG15 null spermatogenesis. Immunolocalization of acetylated histones in seminiferous cycle. Immunolocalization of (A) AcH4K5, (B) AcH4K12, and (C) AcH4K16 in stages 2 and 3, 6 and 7, and 9 and 10 of seminiferous cycles of both F/− and cKO are shown. Blue, DAPI; green, acetylated histones; red, ZPBP1. (Scale bar: 50 µm.)
Histone Modification Regulates Splicing of Tnp2 by MRG15.
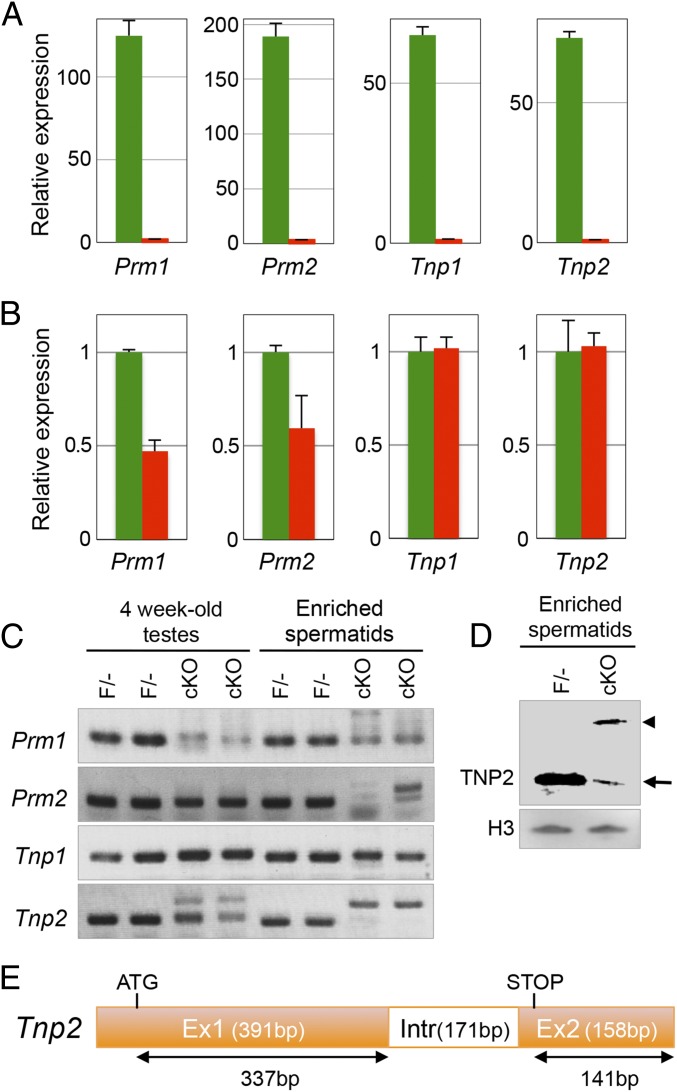
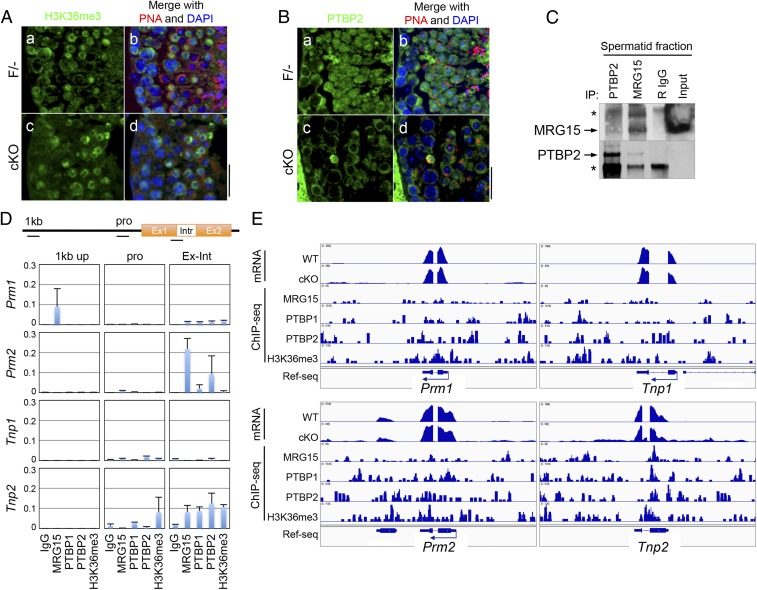
To study these processes in more molecular detail, expression of protamines (Prm1 and Prm2) and Tnps (Tnp1 and Tnp2) in testes and spermatids was analyzed in WT and Mrg15 null spermatid fraction. Expression levels of both Prm and Tnp genes were significantly reduced in the Mrg15 null adult testes, possibly because the expression of these proteins is the highest in latter stages of spermiogenesis, which are absent in the Mrg15 null testes (Fig. 4A). To further interpret our data, expression of the Prm and Tnp genes was compared between spermatids fractions isolated from 22-d-old WT and Mrg15 null testes, in which spermatogenesis has progressed to the early stage of round spermatids. Whereas expression levels of Prm1 and Prm2 are decreased in the absence of MRG15, Tnp1 and Tnp2 expression levels are not changed by Mrg15 deletion (Fig. 4B). However, the Tnp2 PCR products were larger in size in the absence of MRG15, and the shifts in size of the larger bands were exactly the size of the Tnp2 intron sequences (Fig. 4C). Larger bands in the absence of MRG15 were also detected in the Prm2 amplification in the spermatid-enriched fraction, although the larger bands were not observed in 4-wk-old testes. When expression of TNP2 protein was analyzed in the spermatid fraction, the amount of TNP2 (16 kDa) was extremely low in the absence of MRG15 compared with control testis, and a larger TNP2 protein was detected specifically in Mrg15 null spermatids (Fig. 4D), suggesting that splicing of the Tnp2 gene (Fig. 4E) could be regulated by MRG15.
Fig. 4.
Splicing defects observed in the absence of MRG15. (A and B) Quantitative expressions of Prm1, Prm2, Tnp1, and Tnp2 were examined by quantitative RT-PCR in (A) testes and (B) spermatid fractions of 22-d-old WT (green) and MRG15 cKO (red) mice. Hprt was used as an internal control. (C) Validation of the size of the PCR products of Prm1, Prm2, Tnp1, and Tnp2 in 4-wk-old testes and spermatid fractions of F/− and cKO males by electrophoresis. (D) Detection of TNP2 protein in F/− and cKO spermatids. Whole-protein extracts of crude spermatid fractions of F/− and cKO were separated by electrophoresis and probed with anti-TNP2 antibody. Arrow and arrowhead indicate a predicted size of TNP2 and a larger size of intron retained TNP2, respectively. Histone H3 was performed as control. (E) Structure of Tnp2 gene. Tnp2 mRNA consists of two exons (391 and 158 bp) and one intron (171 bp). The ORF of Tnp2 is 354 bp. When the intron is retained in the Tnp2 mRNA, the predicted length is 525 bp.
Because MRG15 regulates alternative splicing with recognition of methylated H3K36 and cooperation of PTB proteins, localization of methylated H3K36 and interaction between MRG15 and PTB proteins were analyzed. Methylated H3K36 is detected in a variety of male germ cells and observed at high levels in the nucleus of both control and Mrg15 null round spermatids (Fig. 5A), suggesting that H3K36 methylation is not affected by lack of MRG15. MRG15 interacts with PTBP2 in the WT spermatid fractions, and PTBP2 localizes to round spermatids similar to MRG15 and methylated H3K36 (Figs. 5 A–C). In the absence of MRG15, PTBP2 is excluded from the nuclei of round spermatids, suggesting that recruitment of PTBP2 to the splicing machinery is defective in Mrg15 null round spermatids (Fig. 5B). To further verify the recruitment of PTBP2 to splicing elements by MRG15, localization of MRG15 and PTBP proteins as well as H3K36me3 was analyzed by ChIP. We discovered specific colocalization of MRG15 and PTBP proteins between exons and introns of the Tnp2 gene but did not discover specific colocalization on either the 1-kb upstream region or the promoter region of the Tnp2 gene or the Prm1, Prm2, and Tnp1 genes (Fig. 5D).
Fig. 5.
MRG15 and PTBP2 cooperatively regulate splicing of Tnp2 gene. (A) Accumulation of H3K36me3 in the testes. (a and c) H3K36me3 (green) and (b and d) peanut agglutinin (PNA; red) were visualized in the (a and b) F/− and (c and d) cKO testes. (B) Immunolocalization of PTBP2. (a and c) PTBP2 (green) and (b and d) PNA (red) were stained in the (a and b) F/− and (c and d) cKO testes. (Scale bar: 50 µm.) (C) Coimmunoprecipitation of MRG15 and PTBP2 in WT spermatid fraction. Arrows indicate the bands of each protein. IP, immunoprecipitation; R, rabbit; *, IgG. (D) ChIP analyses of the protamine and transition protein genes. Quantitative PCR using primers located on 1-kb upstream, promoter, and exon–intron junction of Prm1, Prm2, Tnp1, and Tnp2 (each location is shown in Upper) was performed in ChIPs with indicated antibodies. Results are shown as relative amplification against input genome. (E) Colocalization of retained intron and MRG15 complexes. Genome browser views show the alignment of the read sequences of mRNA and ChIP with MRG15, PTBP1, PTBP2, and H3K36me3 on the mouse genomic regions of Prm1, Prm2, Tnp1, and Tnp2 of WT spermatids. Ref-seq, reference sequence.
To investigate a more widespread link between splicing defects and the lack of MRG15 and MRG15-containing protein complexes, we performed whole-mRNA sequencing and ChIP sequencing (ChIP-seq) against H3K36me3, MRG15, PTBP1, and PTBP2 using enriched spermatids collected from 22- to 25-d-old testes. We found that specific mRNA sequences disappeared from 66 germ cell-expressed genes in the absence of MRG15 and that specific intronic sequences were retained in mRNAs of 4 genes in the MRG15 cKO testes (Tables 1 and 2). In the Mrg15 null spermatids, colocalization of MRG15 complexes and H3K36me3 near splicing-defective regions was observed in 61 genes that had skipped sequences and all 4 genes that had retained sequences (Tables 1 and 2). Strong colocalization of MRG15, PTBP proteins, and H3K36me3 at the junction between exon and intron of the Tnp2 gene was confirmed by the ChIP-seq experiments (Fig. 5E). Although weak colocalization of the MRG15 complex and H3K36me3 was detected at the junction between exon and intron of Prm2, there was only a slight intron retention of the Prm2 transcript detected in deep sequencing results (Fig. 5E). Because the intron of Tnp2 was retained in its precursor mRNA, other genes, in which an intron sequence remained in their mRNAs in the absence of MRG15, were further investigated. Among four genes that had retained sequences, only Tnp2 retained the whole intron sequence, whereas Mfap5 and Nmnat3 retained a part of the intronic sequences, and Tpm1 retained an exon that was not expressed in MRG15-positive spermatids. We confirmed splicing defects of three genes in the absence of MRG15 and also, weak colocalization of MRG15, PTBPs, and H3K36me3 around each defective locus using mRNA sequencing and ChIP-seq (SI Appendix, Fig. S6). The results at least indicate that MRG15 recognizes H3K36me3 and recruits PTBP2 to the splicing machinery of the Tnp2 gene, and this protein complex regulates pre-mRNA splicing of Tnp2 cooperatively (Fig. 6).
Table 1.
Genes that have skipped sequences in the absence of MRG15
| Gene | Chromosome | Start | Stop | Gene ontology term (biological process) | Gene ontology term (cellular) | Ref. |
| Ankrd44 | Chr1 | 54,784,415 | 54,786,577 | — | — | |
| Ccdc93 | Chr1 | 123,383,243 | 123,387,698 | Golgi to plasma membrane transport | Endosome | |
| Mtap2 | Chr1 | 66,448,096 | 66,466,708 | Microtubule bundle formation | Cytoskeleton | 29 |
| Plekha6 | Chr1 | 135,142,720 | 135,155,585 | — | — | |
| Ptpn4 | Chr1 | 121,680,167 | 121,698,899 | Protein dephosphorylation | Cytoskeleton | |
| Spata17 | Chr1 | 188,964,376 | 188,998,253 | — | Cytoplasm | 35 |
| Ccdc34 | Chr2 | 109,872,597 | 109,880,730 | — | — | |
| Frmd5 | Chr2 | 121,417,499 | 121,632,465 | — | Cytoskeleton | |
| Pfkfb3 | Chr2 | 11,401,999 | 11,403,497 | Fructose 2,6-bisphosphate metabolism | Nucleoplasm | 46 |
| Zhx3 | Chr2 | 160,658,155 | 160,698,526 | Transcription, DNA templated | Nucleus | |
| Nbea | Chr3 | 55,514,662 | 55,521,698 | Protein targeting | Trans-Golgi network | |
| Pdzk1 | Chr3 | 96,634,080 | 96,654,150 | Calnitine transport | Plasma membrane | |
| Postn | Chr3 | 54,181,529 | 54,187,309 | Cell adhesion | Extracellular region | |
| Sgms2 | Chr3 | 131,045,379 | 131,047,507 | Lipid metabolic process | Golgi apparatus | |
| Aof2 | Chr4 | 136,110,316 | 136,111,087 | Transcription, DNA templated | Nuclear chromatin | |
| Eif4g3 | Chr4 | 137,702,289 | 137,707,496 | Translation | — | 30 |
| Map3k7 | Chr4 | 32,097,397 | 32,102,913 | MAPK cascade | Plasma membrane | |
| Pef1 | Chr4 | 129,798,686 | 129,802,306 | Proteolysis | Cytoplasm | |
| Slc9a1 | Chr4 | 132,972,414 | 132,973,814 | Ion transport | Plasma membrane | |
| Ssbp3 | Chr4 | 106,703,697 | 106,709,540 | Transcription, DNA templated | Nucleus | |
| Tssk3 | Chr4 | 129,166,977 | 129,167,705 | Spermatogenesis | Intracellular | |
| 1700023E05Rik | Chr5 | 77,445,664 | 77,471,120 | — | — | |
| 4931409K22Rik* | Chr5 | 24,056,624 | 24,057,778 | — | — | |
| Prom1 | Chr5 | 44,454,424 | 44,485,633 | Retina-layer formation | Extracellular space | |
| Cpa5 | Chr6 | 30,561,444 | 30,562,506 | Proteolysis | Extracellular region | |
| Vgll4 | Chr6 | 114,814,098 | 114,840,634 | Transcription, DNA templated | Nucleus | |
| Vhlh | Chr6 | 113,574,373 | 113,578,085 | Transcription, DNA templated | Nucleus | |
| Eftud1 | Chr7 | 89,846,528 | 89,899,301 | Translation | Intracellular | |
| Igf2* | Chr7 | 149,841,722 | 149,844,268 | Protein phosphorylation | Extracellular region | |
| Vasp | Chr7 | 19,843,269 | 19,844,151 | Actin cytoskeleton organization | Cytoskeleton | |
| Dynlrb2 | Chr8 | 119,038,866 | 119,039,579 | Microtubule-based movement | Cytoskeleton | |
| Psd3 | Chr8 | 70,314,841 | 70,341,846 | ARF protein signal transduction | Plasma membrane | |
| Stox2 | Chr8 | 48,288,628 | 48,437,384 | — | — | |
| Abhd14b | Chr9 | 106,352,563 | 106,353,723 | Transcription, DNA templated | Nucleus | |
| Ccdc33* | Chr9 | 57,878,228 | 57,879,421 | Spermatogenesis | Peroxisome | |
| Dixdc1 | Chr9 | 50,519,078 | 50,535,890 | Wnt signaling pathway | Cytoplasm | |
| Ncam1 | Chr9 | 49,365,344 | 49,372,973 | Cell adhesion | Plasma membrane | |
| Nptn | Chr9 | 58,491,610 | 58,498,303 | Cell adhesion | Plasma membrane | |
| Osbpl10 | Chr9 | 115,085,201 | 115,116,631 | Lipid metabolic process | Cytoskeleton | |
| Pkm2 | Chr9 | 59,519,881 | 59,523,374 | Glycolytic process | Mitochondrion | |
| Usp2 | Chr9 | 43,875,304 | 43,883,448 | Protein deubiquitination | Nucleus | 31 |
| Hbs1l | Chr10 | 21,056,882 | 21,061,519 | Translation | Intracellular | |
| Mtap7 | Chr10 | 19,868,887 | 19,950,589 | Microtubule cytoskeleton organization | Cytoskeleton | 32 |
| 4933404M19Rik | Chr11 | 78,017,700 | 78,018,341 | — | — | |
| Ace | Chr11 | 105,832,798 | 105,833,330 | Proteolysis | Extracellular region | 33 |
| Akap1 | Chr11 | 88,707,180 | 88,725,710 | Regulation of PKA signaling | Mitochondrion | |
| Ccdc46 | Chr11 | 108,392,780 | 108,431,601 | Receptor localization to synapse | Cytoskeleton | |
| Tex14 | Chr11 | 87,356,880 | 87,362,933 | Mitotic nuclear division | Kinetochore | 34 |
| 4930579E17Rik | Chr12 | 37,274,548 | 37,414,835 | Axon guidance | — | |
| Ankrd9* | Chr12 | 112,215,759 | 112,216,157 | Hydrolase activity | — | |
| Spata7 | Chr12 | 99,870,248 | 99,872,433 | Response to stimulus | Cytoskeleton | |
| 2010111I01Rik | Chr13 | 63,383,567 | 63,398,165 | Proteolysis | Cytoplasm | |
| Bmp6 | Chr13 | 38,438,457 | 38,561,482 | BMP signaling pathway | Extracellular region | |
| Cdc14b | Chr13 | 64,297,984 | 64,306,578 | DNA repair | Nucleus | 38 |
| 4933401F05Rik | Chr14 | 64,700,689 | 64,704,147 | Proteolysis | Membrane | |
| Diap3 | Chr14 | 87,172,510 | 87,210,142 | Actin cytoskeleton organization | Nucleus | |
| Apol7b | Chr15 | 77,258,240 | 77,277,809 | — | — | |
| Dab2 | Chr15 | 6,249,900 | 6,366,805 | Endocytosis | Plasma membrane | |
| Gm628 | Chr15 | 73,623,216 | 73,629,309 | — | — | |
| Lpp | Chr16 | 24,681,958 | 24,761,678 | Cell adhesion | Cytoplasm | |
| AW554918 | Chr18 | 25,362,478 | 25,448,439 | — | — | |
| Sncaip | Chr18 | 53,067,129 | 53,074,960 | Cell death | Cytoplasm | |
| Tcf4 | Chr18 | 69,802,670 | 69,810,875 | Transcription, DNA templated | Nucleus | |
| 6030443O07Rik | Chr19 | 45,046,176 | 45,047,623 | DNA repair | Nucleus | |
| Sorbs1 | Chr19 | 40,374,012 | 40,386,158 | Actin filament organization | Cytoskeleton | |
| Rpgr* | ChrX | 9,743,403 | 9,755,396 | Cilium assembly | Cytoskeleton | 36 |
These genes have skipped sequences but do not show colocalization of H3K36me3 and MRG15 complexes.
Table 2.
Genes with retained sequences in Mrg15 cKO but not in WT spermatids
| Gene | Chromosome | Start | Stop | Intron no. | Full name-location |
| Mfap5 | Chr6 | 122,471,947 | 122,474,504 | 6 | Mfap5_chr6_122471947_122474504_6 |
| Nmnat3 | Chr9 | 98,304,033 | 98,310,495 | 4 | Nmnat3_chr9_98304033_98310495_4 |
| Tpm1 | Chr9 | 66,895,719 | 66,896,825 | 1 | Tpm1_chr9_66895719_66896825_1 |
| Tnp2 | Chr16 | 10,788,187 | 10,788,357 | 1 | Tnp2_chr16_10788187_10788357_1 |
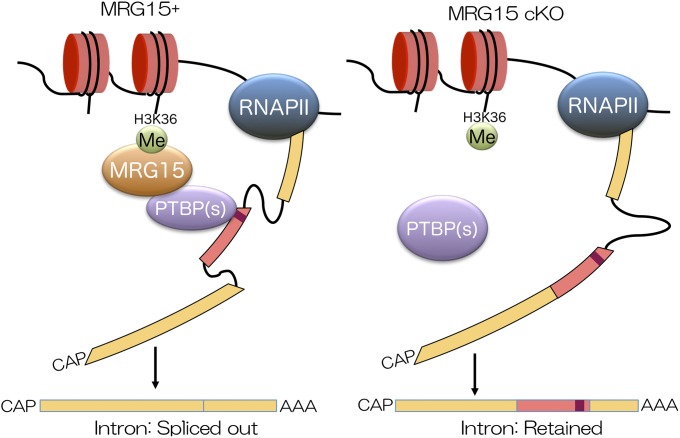
Fig. 6.
Proposed model for how MRG15 regulates pre-mRNA splicing of germ cell genes, such as Tnp2. AAA, poly A tail of mRNA; CAP, 5′ cap structure of mRNA; Me, methylation; RNAPII, RNA polymerase II.
In addition, at least Mtap2, Eif4g3, Usp2, Mtap7, Ace, and Tex14 are known to play a role in male fertility (29–34), and overexpression of Spata17 and Rpgr leads to male infertility (35, 36). Therefore, the infertility in the Mrg15 cKO mice could be secondary to alterations in the levels of these proteins. For example, our group has described the essential roles of TEX14 in male fertility and formation of intercellular bridges (34). The lack of TEX14 leads to the loss of intercellular bridges and the failure of proliferation and differentiation of spermatogonia (34, 37). Although the roles of TEX14 and intercellular bridges during postmeiotic process still remain to be elucidated, abnormal TEX14 might be a cause of the defects in spermatogenesis in the absence of Mrg15. Furthermore, Mtap2, Spata17, Frmd5, Vasp, Dynlrb2, Osbpl10, Mtap7, Ccdc46, Spata7, Cdc14b, Sorbs1, and Rpgr could be important for postmeiotic spermatogenic progression and might also be involved in the defects in the Mrg15 null germ cells (Table 1) (38); these proteins are associated with microtubules and cytoskeleton, and suppression of their levels could be involved in the formation of multinuclear round spermatids and the failure to form mature sperm.
Discussion
One of the biological events in which MRG15 is involved is homology-directed DNA repair after DNA damage (21). Many proteins that control homology-directed DNA repair, such as RAD51, BRCA1, BRCA2, and PALB2, are also involved in the regulation of meiotic homologous recombination (39–42). When MRG15 is down-regulated in somatic cells, localization of RAD51, BRCA2, and PALB2 to DNA damage sites is suppressed (21). However, the formation of RAD51 foci was not affected by the deletion of MRG15. Although localization of PALB2 to DNA damage sites can be regulated by BRCA1 independently with MRG15 and depletion of PALB2 leads to loss of RAD51 focus formation in the DNA damage sites of somatic cells, inhibition of the interaction between PALB2 and BRCA1 does not affect formation of RAD51 foci in meiotic double-strand breaks, even if it results in male sterility, likely secondary to defect in sex chromosome synapsis (42). Because there were no significant defects in meiosis Mrg15 null testes, MRG15 is apparently not required for meiotic homologous recombination.
Herein, we found splicing failures in multiple genes in the absence of MRG15 and showed that histone modification regulates splicing via chromatin binding proteins, such as MRG15 (7) (Fig. 6). These splicing forms might produce different products and have different functions from the typical annotated sequences. The results suggest that absence of a spliced form of a gene could be important for progression of spermatogenesis. When an abnormal splicing form is expressed in WT spermatids, the gene product could have different functions compared with the original product. In contrast, the gene product could lose its own function or exert an antagonistic effect on the original product when it is expressed in the Mrg15 null spermatids. In this study, we focused on the abnormally spliced mRNA, in which intronic sequences remained in the absence of MRG15. Among four genes with retained sequences, strong interaction between MRG15 complexes and abnormal splicing was found in the Tnp2 gene, whereas weak interaction was found in the other three genes: Mtap5, Nmnat3, and Tpm1. Of note, the retained sequence of Tpm1 in the absence of MRG15 was derived from an exon but not derived from an intron, indicating that MRG15 complexes could regulate alternatively splicing of Tpm1. In addition, skipped exon 3 of Tpm1 is a known target of PTBP (43). Interestingly, there was slight and weak colocalization of MRG15–PTBPs complexes around introns of Prm1 and Prm2, respectively, despite that H3K36me3 was strongly accumulated at the junction between exon and intron of Prm1 and Prm2. Some other proteins or modifications might participate in the regulation of splicing events of Tpm1, Prm1, and Prm2 mRNAs. Because MRG15 can interact with HAT, HDAC complexes, and other histone modifications, such as methylated H4K20, localization of MRG15 on the genome may be regulated by not only H3K36me3 but also, other histone modifications and/or protein complexes (13–21, 26). PTBPs are known to antagonize exon definition; therefore, those retained sequences could be recognized as an exon on the absence of MRG15 (44). Although detailed molecular mechanisms underlying epigenetic regulation of specific sequence retention still remain to be elucidated, amounts of MRG15–PTBPs complexes localized in the regulatory region seem to be correlated with ratios of splicing defects in the lack of MRG15 (Figs. 4C and 5E). However, many abnormally spliced mRNAs, in which exon sequences were skipped by the lack of MRG15, were identified. MRG15–PTBPs complexes could repress splicing of those skipped sequences in the absence of MRG15. Because Tnp2 null mice are not sterile, defects in other genes must be involved in the phenotype of the Mrg15 cKO mice (45). The most apparent defect in Mrg15 null spermatogenesis was spermatogenic arrest at the round spermatid stage. The epididymides of the Mrg15 cKO mice were filled with round spermatids, suggesting that the absence of MRG15 might induce splicing defects in the genes, which were essential for round spermatids to proceed to the elongation step. Moreover, multinuclear round spermatids were frequently detected in testes lacking MRG15, indicating that some genes, which were essential for cell division, are altered by the lack of MRG15. Among 61 genes with MRG15-regulated intron skipped sequences, 9 genes are categorized as cytoskeleton-associated genes, and 9 genes are categorized as microtubule, actin, or cytoskeletal protein binding genes (Table 1). Particularly, Mtap7 is localized in the spermatid manchette, and Mtap7 mutant mice are sterile because of deformation of spermatid nuclei. Another cytoskeletal-associated gene, Mtap2, is required for spermatocytes to exit meiotic prophase I via the G2/MI transition. Although the role of MTAP2 in round spermatids is not known, abnormal MTAP2 could be a cause of the formation of multinuclear round spermatids. CDC14B, which is a dual-specificity phosphatase, could also be involved in the formation of multinuclear round spermatids, because it can bind, bundle, and stabilize microtubules independent of its phosphatase activity (38). However, PFKFB3 could be involved in formation of mature sperm; PFKFB3 is present only in immature sperm and replaced with PFKFB4 during maturation of sperm (46). Thus, splicing defects of multiple genes regulated by MRG15–PTBPs complexes could cause the defects in spermatogenesis in the absence of Mrg15. Additional characterization as well as biological analyses of each abnormal splicing form and ChIP-seq using pure spermatids may uncover additional spermatogenic phenotypes in the Mrg15 null testes.
Germline-specific deletion of Ptbp2 results in male sterility secondary to defects in alternative splicing (47). In the Ptbp2 null mice, spermatogenesis also arrests at the round spermatid stage, and there are multinucleated cells as observed in the Mrg15 null germ cells. These results suggest that MRG15 and PTBP2 share target genes and regulate pre-mRNA splicing cooperatively. However, splicing defects that were found in the absence of PTBP2 were not altered in the Mrg15 null spermatids, suggesting that MRG15 and PTBP2 share some but not all splicing events (SI Appendix, Fig. S6) (47).
A large part of splicing forms could be regulated by other histone modifications and/or chromatin binding proteins but could not be regulated by H3K36me3 and MRG15. In fact, there are several complexes that include chromatin binding proteins (which recognize other histone modifications) and a splicing factor, such as H3K9me3/HP1a/hnRNPs, H3K4me3/CHD1/U2 snRNP, or acetylated H3/GCN5/U2 snRNP (5, 48–51). Splicing regulation via an MRG15–PTBP2 complex may also be physiologically relevant to the brain, because PTBP2 expression is restricted to testis and brain, and MRG15 is ubiquitously expressed (52–54). Moreover, regulation of splicing might be detectable in the hematopoietic lineage, because PTBP3 expression is restricted in hematopoietic cells (55). Thus, splicing regulation may be involved in other tissues, and tissue specificity might be regulated by not only the combination of chromatin binding proteins and splicing regulators present but also, regulation of histone modification. Alternative splicing forms and splicing regulation generate protein diversity; therefore, defects in this system may trigger disease.
Materials and Methods
Generation of Germ Cell-Specific Mrg15 Null Mice and Fertility Analysis.
Details regarding the targeting construct and generation of conditional null allele of MRG15 are in SI Appendix, SI Materials and Methods. A germline-specific deletion of Mrg15 was produced by mating with Stra8-Cre mice provided by Robert E. Braun, Jackson Laboratory, Bar Harbor, ME (23). For fertility analysis, 6-wk-old control (Mrg15F/−:Stra8-Cre−) and Mrg15 cKO (Mrg15F/−:Stra8-Cre+) male littermates were individually bred to WT females. The numbers of litters and pups born per litter were monitored over a 6-mo period. All mouse experiments were performed on a C57BL/6J:129S5 hybrid background in accordance with protocols approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine and the Experimental Animal Care Committee of Kyushu University.
Northern Blot Analysis and Semiquantitative and Quantitative RT-PCR.
Total RNA from mouse adult tissue samples and developing testes samples and cultured and isolated cell samples were extracted and subjected to Northern blot, semiquantitative RT-PCR, and quantitative PCR. Detailed methods and the primer sequences used in the experiments can be found in SI Appendix, SI Materials and Methods and Table S1, respectively.
Histology, Immunofluorescence, and TUNEL Staining.
For testis histology, 5-μm sections were stained with periodic acid-Schiff reagent and counterstained with hematoxylin. For immunofluorescence of tissue sections, paraformaldehyde-fixed sections were retrieved by microwave and then, incubated with primary antibodies overnight at 4 °C followed by Alexa 488- and Alexa 594-conjugated secondary antibodies (Invitrogen) for 1 h at room temperature. Fluorescent sections were mounted with VECTASHIELD containing DAPI (VECTOR Laboratories). To detect apoptotic cells, DNA fragmentation was analyzed by the ApopTag Plus Fluorescein In Situ Apoptosis Detection Kit (Millipore) according to the manufacturer’s instruction. Details regarding antibodies are included in SI Appendix, SI Materials and Methods.
Chromosome Analysis.
Chromosome spread analysis was performed as previously described (56). Antibody information is included in SI Appendix, SI Materials and Methods.
ChIP.
Approximately 10 million spermatids were cross-linked and sonicated until DNA was fragmented to an average length 200–500 bp. Control immunoprecipitation was performed with normal rabbit IgG. Immunoprecipitated DNA and input DNA were analyzed by PCR using the primers listed in SI Appendix, Table S1 or sequenced by Illumina HiSeq1500. Details regarding ChIP and antibodies used can be found in SI Appendix, SI Materials and Methods.
RNA Deep Sequencing and ChIP-Seq Analyses.
Details regarding RNA deep sequencing, ChIP sequencing, and their analyses are in SI Appendix, SI Materials and Methods. RNA sequencing and ChIP-seq data have been deposited in the DDBJ (DNA Data Bank of Japan) Sequence Read Archive under accession numbers DRA004783 and DRA004778, respectively.
Supplementary Material
Acknowledgments
We thank Dr. Olivia M. Pereira-Smith for her support and suggestions and Drs. Julio M. Castaneda and John W. Nelson for critical review of the manuscript. We also thank Dr. Robert E. Braun for the gift of the Stra8-Cre mice and Megumi Furukawa and Tomona Hayashi for their technical assistance. We thank the Genomic and RNA Profiling Core, Baylor College of Medicine for RNA deep sequencing; and the Research Institute for Information Technology, Kyushu University for ChIP-seq. This work was supported, in part, by the Japan Society for the Promotion of Sciences Grant-in-Aid for Young Scientists Grant 26712026 (to N.I.), Grant 15K21217 (to T.I.), Grant-in-Aid for Scientific Research (KAKENHI) Grant 26114506 (to T.I.), the Takeda Science Foundation (N.I.), Interdisciplinary Programs in Education and Projects in Research Development in Kyushu University (N.I.), and Eunice Kennedy Shriver National Institute of Child Health and Human Development/NIH Cooperative Agreement U01-HD076508 (to M.M.M.) as part of the Cooperative Program in Male Contraception.
Footnotes
The authors declare no conflict of interest.
Data deposition: The RNA sequencing and ChIP sequencing data have been deposited in the DDBJ Sequence Read Archive (DRA; accession nos. DRA004783 and DRA004778, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1611995113/-/DCSupplemental.
References
- 1.Oatley JM, Brinster RL. Regulation of spermatogonial stem cell self-renewal in mammals. Annu Rev Cell Dev Biol. 2008;24:263–286. doi: 10.1146/annurev.cellbio.24.110707.175355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kota SK, Feil R. Epigenetic transitions in germ cell development and meiosis. Dev Cell. 2010;19(5):675–686. doi: 10.1016/j.devcel.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Rathke C, Baarends WM, Awe S, Renkawitz-Pohl R. Chromatin dynamics during spermiogenesis. Biochim Biophys Acta. 2014;1839(3):155–168. doi: 10.1016/j.bbagrm.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 4.Payne C, Braun RE. Histone lysine trimethylation exhibits a distinct perinuclear distribution in Plzf-expressing spermatogonia. Dev Biol. 2006;293(2):461–472. doi: 10.1016/j.ydbio.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144(1):16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet. 2011;12(10):715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luco RF, et al. Regulation of alternative splicing by histone modifications. Science. 2010;327(5968):996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 2009;19(10):1732–1741. doi: 10.1101/gr.092353.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolasinska-Zwierz P, et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41(3):376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol. 2009;16(9):990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 11.Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36(2):245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tilgner H, et al. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol. 2009;16(9):996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- 13.Chen M, Tominaga K, Pereira-Smith OM. Emerging role of the MORF/MRG gene family in various biological processes, including aging. Ann N Y Acad Sci. 2010;1197:134–141. doi: 10.1111/j.1749-6632.2010.05197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yochum GS, Ayer DE. Role for the mortality factors MORF4, MRGX, and MRG15 in transcriptional repression via associations with Pf1, mSin3A, and Transducin-Like Enhancer of Split. Mol Cell Biol. 2002;22(22):7868–7876. doi: 10.1128/MCB.22.22.7868-7876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai Y, et al. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J Biol Chem. 2003;278(44):42733–42736. doi: 10.1074/jbc.C300389200. [DOI] [PubMed] [Google Scholar]
- 16.Doyon Y, Selleck W, Lane WS, Tan S, Côté J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24(5):1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayakawa T, et al. RBP2 is an MRG15 complex component and down-regulates intragenic histone H3 lysine 4 methylation. Genes Cells. 2007;12(6):811–826. doi: 10.1111/j.1365-2443.2007.01089.x. [DOI] [PubMed] [Google Scholar]
- 18.Sardiu ME, et al. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc Natl Acad Sci USA. 2008;105(5):1454–1459. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kusch T, et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306(5704):2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 20.Sy SMH, Huen MSY, Chen J. MRG15 is a novel PALB2-interacting factor involved in homologous recombination. J Biol Chem. 2009;284(32):21127–21131. doi: 10.1074/jbc.C109.023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayakawa T, et al. MRG15 binds directly to PALB2 and stimulates homology-directed repair of chromosomal breaks. J Cell Sci. 2010;123(Pt 7):1124–1130. doi: 10.1242/jcs.060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tominaga K, et al. MRG15 regulates embryonic development and cell proliferation. Mol Cell Biol. 2005;25(8):2924–2937. doi: 10.1128/MCB.25.8.2924-2937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sadate-Ngatchou PI, Payne CJ, Dearth AT, Braun RE. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis. 2008;46(12):738–742. doi: 10.1002/dvg.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia SN, Kirtane BM, Podlutsky AJ, Pereira-Smith OM, Tominaga K. Mrg15 null and heterozygous mouse embryonic fibroblasts exhibit DNA-repair defects post exposure to gamma ionizing radiation. FEBS Lett. 2007;581(27):5275–5281. doi: 10.1016/j.febslet.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun B, et al. Molecular basis of the interaction of Saccharomyces cerevisiae Eaf3 chromo domain with methylated H3K36. J Biol Chem. 2008;283(52):36504–36512. doi: 10.1074/jbc.M806564200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore SA, Ferhatoglu Y, Jia Y, Al-Jiab RA, Scott MJ. Structural and biochemical studies on the chromo-barrel domain of male specific lethal 3 (MSL3) reveal a binding preference for mono- or dimethyllysine 20 on histone H4. J Biol Chem. 2010;285(52):40879–40890. doi: 10.1074/jbc.M110.134312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meistrich ML, Trostle-Weige PK, Lin R, Bhatnagar YM, Allis CD. Highly acetylated H4 is associated with histone displacement in rat spermatids. Mol Reprod Dev. 1992;31(3):170–181. doi: 10.1002/mrd.1080310303. [DOI] [PubMed] [Google Scholar]
- 28.Lahn BT, et al. Previously uncharacterized histone acetyltransferases implicated in mammalian spermatogenesis. Proc Natl Acad Sci USA. 2002;99(13):8707–8712. doi: 10.1073/pnas.082248899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun F, Handel MA. A mutation in Mtap2 is associated with arrest of mammalian spermatocytes before the first meiotic division. Genes (Basel) 2011;2(1):21–35. doi: 10.3390/genes2010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun F, Palmer K, Handel MA. Mutation of Eif4g3, encoding a eukaryotic translation initiation factor, causes male infertility and meiotic arrest of mouse spermatocytes. Development. 2010;137(10):1699–1707. doi: 10.1242/dev.043125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bedard N, et al. Mice lacking the USP2 deubiquitinating enzyme have severe male subfertility associated with defects in fertilization and sperm motility. Biol Reprod. 2011;85(3):594–604. doi: 10.1095/biolreprod.110.088542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Komada M, McLean DJ, Griswold MD, Russell LD, Soriano P. E-MAP-115, encoding a microtubule-associated protein, is a retinoic acid-inducible gene required for spermatogenesis. Genes Dev. 2000;14(11):1332–1342. [PMC free article] [PubMed] [Google Scholar]
- 33.Hagaman JR, et al. Angiotensin-converting enzyme and male fertility. Proc Natl Acad Sci USA. 1998;95(5):2552–2557. doi: 10.1073/pnas.95.5.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greenbaum MP, et al. TEX14 is essential for intercellular bridges and fertility in male mice. Proc Natl Acad Sci USA. 2006;103(13):4982–4987. doi: 10.1073/pnas.0505123103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nie DS, Liu Y, Juan H, Yang X. Overexpression of human SPATA17 protein induces germ cell apoptosis in transgenic male mice. Mol Biol Rep. 2013;40(2):1905–1910. doi: 10.1007/s11033-012-2246-z. [DOI] [PubMed] [Google Scholar]
- 36.Brunner S, et al. Overexpression of RPGR leads to male infertility in mice due to defects in flagellar assembly. Biol Reprod. 2008;79(4):608–617. doi: 10.1095/biolreprod.107.067454. [DOI] [PubMed] [Google Scholar]
- 37.Iwamori N, Iwamori T, Matzuk MM. Characterization of spermatogonial stem cells lacking intercellular bridges and genetic replacement of a mutation in spermatogonial stem cells. PLoS One. 2012;7(6):e38914. doi: 10.1371/journal.pone.0038914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cho HP, et al. The dual-specificity phosphatase CDC14B bundles and stabilizes microtubules. Mol Cell Biol. 2005;25(11):4541–4551. doi: 10.1128/MCB.25.11.4541-4551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plug AW, Xu J, Reddy G, Golub EI, Ashley T. Presynaptic association of Rad51 protein with selected sites in meiotic chromatin. Proc Natl Acad Sci USA. 1996;93(12):5920–5924. doi: 10.1073/pnas.93.12.5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scully R, et al. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88(2):265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 41.Sharan SK, et al. BRCA2 deficiency in mice leads to meiotic impairment and infertility. Development. 2004;131(1):131–142. doi: 10.1242/dev.00888. [DOI] [PubMed] [Google Scholar]
- 42.Simhadri S, et al. Male fertility defect associated with disrupted BRCA1-PALB2 interaction in mice. J Biol Chem. 2014;289(35):24617–24629. doi: 10.1074/jbc.M114.566141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gooding C, et al. MBNL1 and PTB cooperate to repress splicing of Tpm1 exon 3. Nucleic Acids Res. 2013;41(9):4765–4782. doi: 10.1093/nar/gkt168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma S, Kohlstaedt LA, Damianov A, Rio DC, Black DL. Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nat Struct Mol Biol. 2008;15(2):183–191. doi: 10.1038/nsmb.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao M, et al. Targeted disruption of the transition protein 2 gene affects sperm chromatin structure and reduces fertility in mice. Mol Cell Biol. 2001;21(21):7243–7255. doi: 10.1128/MCB.21.21.7243-7255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gómez M, et al. Switches in 6-phosphofructo-2-kinase isoenzyme expression during rat sperm maturation. Biochem Biophys Res Commun. 2009;387(2):330–335. doi: 10.1016/j.bbrc.2009.07.021. [DOI] [PubMed] [Google Scholar]
- 47.Zagore LL, et al. RNA binding protein Ptbp2 is essential for male germ cell development. Mol Cell Biol. 2015;35(23):4030–4042. doi: 10.1128/MCB.00676-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sims RJ, 3rd, et al. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28(4):665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gunderson FQ, Johnson TL. Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly. PLoS Genet. 2009;5(10):e1000682. doi: 10.1371/journal.pgen.1000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Piacentini L, et al. Heterochromatin protein 1 (HP1a) positively regulates euchromatic gene expression through RNA transcript association and interaction with hnRNPs in Drosophila. PLoS Genet. 2009;5(10):e1000670. doi: 10.1371/journal.pgen.1000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loomis RJ, et al. Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol Cell. 2009;33(4):450–461. doi: 10.1016/j.molcel.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Polydorides AD, Okano HJ, Yang YY, Stefani G, Darnell RB. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc Natl Acad Sci USA. 2000;97(12):6350–6355. doi: 10.1073/pnas.110128397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Markovtsov V, et al. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol Cell Biol. 2000;20(20):7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu M, Hecht NB. Polypyrimidine tract binding protein 2 stabilizes phosphoglycerate kinase 2 mRNA in murine male germ cells by binding to its 3'UTR. Biol Reprod. 2007;76(6):1025–1033. doi: 10.1095/biolreprod.107.060079. [DOI] [PubMed] [Google Scholar]
- 55.Yamamoto H, Tsukahara K, Kanaoka Y, Jinno S, Okayama H. Isolation of a mammalian homologue of a fission yeast differentiation regulator. Mol Cell Biol. 1999;19(5):3829–3841. doi: 10.1128/mcb.19.5.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iwamori N, Zhao M, Meistrich ML, Matzuk MM. The testis-enriched histone demethylase, KDM4D, regulates methylation of histone H3 lysine 9 during spermatogenesis in the mouse but is dispensable for fertility. Biol Reprod. 2011;84(6):1225–1234. doi: 10.1095/biolreprod.110.088955. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.