Significance
During chronic viral infection, CD8+ T cells are gradually deprived of their principal effector functions and irreversibly loose their plasticity to develop into memory populations, precluding the establishment of long-lasting protective immunity. Relevant host-derived factors directing this T-cell exhaustion process have remained elusive. Growing evidence suggests that the cytokine milieu dramatically impacts the outcome of chronic viral infection. However, it is unclear if cytokines directly promote CD8+ T-cell exhaustion and, if so, which specific cytokines are involved in this process. Here we demonstrate a critical role for two highly related cytokines, IL-2 and IL-15, in the terminal differentiation of highly exhausted CD8+ T cells and the lack of immunological memory observed during chronic viral infection.
Keywords: CD8 T cell, IL-2, IL-15, exhaustion, memory T cell
Abstract
Exhaustion of CD8+ T cells severely impedes the adaptive immune response to chronic viral infections. Despite major advances in our understanding of the molecular regulation of exhaustion, the cytokines that directly control this process during chronicity remain unknown. We demonstrate a direct impact of IL-2 and IL-15, two common gamma-chain–dependent cytokines, on CD8+ T-cell exhaustion. Common to both cytokine receptors, the IL-2 receptor β (IL2Rβ) chain is selectively maintained on CD8+ T cells during chronic lymphocytic choriomeningitis virus and hepatitis C virus infections. Its expression correlates with exhaustion severity and identifies terminally exhausted CD8+ T cells both in mice and humans. Genetic ablation of the IL2Rβ chain on CD8+ T cells restrains inhibitory receptor induction, in particular 2B4 and Tim-3; precludes terminal differentiation of highly defective PD-1hi effectors; and rescues memory T-cell development and responsiveness to IL-7–dependent signals. Together, we ascribe a previously unexpected role to IL-2 and IL-15 as instigators of CD8+ T-cell exhaustion during chronic viral infection.
CD8+ T cells are the cornerstone of antiviral immunity. Throughout their differentiation, CD8+ T cells integrate signals that define their cellular fate and functional capacities (1–3). Persistent infections with viruses such as HIV and hepatitis C (HCV) and B are associated with exhaustion of CD8+ T cells (4, 5). Exhaustion is characterized by progressive loss of effector functions (cytokine secretion, cytolytic potential, antigen-driven proliferation), increased cell death, and eventually physical deletion (6–9). This unresponsive state stems from increased expression of multiple inhibitory receptors on CD8+ T cells [including programmed death 1 (PD-1), lymphocyte-activation gene 3 (LAG-3), 2B4 (CD244), T-cell Ig and mucin domain-containing molecule-3 (Tim-3), CD160, and cytotoxic T-lymphocyte associated protein 4 (CTLA4)] that use distinct and nonredundant ways to suppress T-cell functions (10–12). Further, CD8+ T cells are gradually and irreversibly deprived of their developmental plasticity as they commit to exhaustion, restricting the cell-fate potential for memory development and precluding the establishment of long-lasting protective immunity (13–15).
Factors underlying T-cell exhaustion are not understood (5, 16). Antigen persistence is a culprit, but little is known about the intrinsic factors that program CD8+ T-cell differentiation to exhaustion. Certain transcription factors that regulate the differentiation of KLRG1hi IL7Rαlo short-lived effector cells (SLECs) during acute infection are also fundamental for sustaining the differentiation of highly exhausted T cells during chronic infection (1, 5). Despite being distinct effector subtypes (11, 13), both SLECs and exhausted CD8+ T cells have elevated and sustained expression of Blimp-1 (encoded by Prdm1) (11, 17, 18). Blimp-1 is indispensable for SLEC development and terminal differentiation in acute infection (17, 19), but is also pivotal to the establishment of exhaustion in chronic infection (18). Similarly, the T-box transcription factor Eomes, which matures SLECs with full effector functions (20–22), is indispensable to generate PD-1hi terminally exhausted cells in chronic settings (23). Thus, some of the cardinal transcriptional requirements for SLEC terminal differentiation during acute infection are also fundamental to directing the exhaustion of CD8+ T cells during chronic infections. Therefore, we hypothesize that determinants shaping the transcriptional program toward SLEC terminal differentiation during acute infections might also instigate CD8+ T-cell exhaustion in chronic viral infections.
During acute viral infection, SLEC terminal differentiation is partly controlled by the common gamma chain (γc)-dependent cytokines IL-2 and IL-15. Indeed, we have previously shown that these closely related cytokines that share common receptor components [i.e., IL-2 receptor β (IL2Rβ; CD122), IL2Rγ (CD132)] and signaling pathways (24) directly promote SLEC development in both primary and secondary responses to acute viral infection (25). Concomitantly, CD8+ T cells deficient for the IL2Rβ chain preferentially turn to KLRG1loIL7Rαhi memory precursor effector cells and convert rapidly to CD62LhiCD127hi central memory T cells (Tcm) (25). These results demonstrate that IL-2– and IL-15–dependent signals direct CD8+ T-cell lineage choices toward SLEC terminal differentiation. Importantly, IL-2 and IL-15 mediate this effect through the modulation of key transcription factors that are also pivotal during the establishment of CD8+ T-cell exhaustion (i.e., Blimp-1 and Eomes). Indeed, IL-2 directly induces Blimp-1 expression (26, 27), and both IL-2 and IL-15 trigger high levels of Eomes in CD8+ T cells in vitro (21, 28). These pieces of evidence strongly predict a role for IL-2 and IL-15 in CD8+ T-cell responses during a chronic viral infection. This role remains to be investigated.
Using a mouse model of chronic viral infection, lymphocytic choriomeningitis virus (LCMV) clone 13 (Cl-13), we observed selective maintenance of CD122, common to IL-2 and IL-15 receptors, on CD8+ T cells throughout antigen persistence. CD122 expression levels on CD8+ T cells correlated with severe exhaustion and clearly discriminated PD-1hi terminal effectors in LCMV-infected mice and humans with persistent HCV infection. Using adoptive transfer experiments of transgenic (Tg) P14 cells deficient for the IL2Rβ chain, we also observed that IL-2 and IL-15 directly regulated several aspects of CD8+ T-cell exhaustion. These cytokines controlled the expression of 2B4 and Tim-3 and augmented the levels of several other inhibitory receptors on CD8+ T cells, leading to higher dysfunction. Further, signaling through IL2Rβ instigated key transcriptional changes in CD8+ T cells that fostered the terminal differentiation of highly exhausted PD-1hi effectors while precluding memory development and IL-7–dependent signaling. Together, our results show a previously unexpected role for IL-2 and IL-15 in the deviation of CD8+ T cells toward terminal exhaustion during chronic viral infection.
Results
Increased and Sustained Expression of CD122 on CD8+ T Cells Marks Severe Exhaustion.
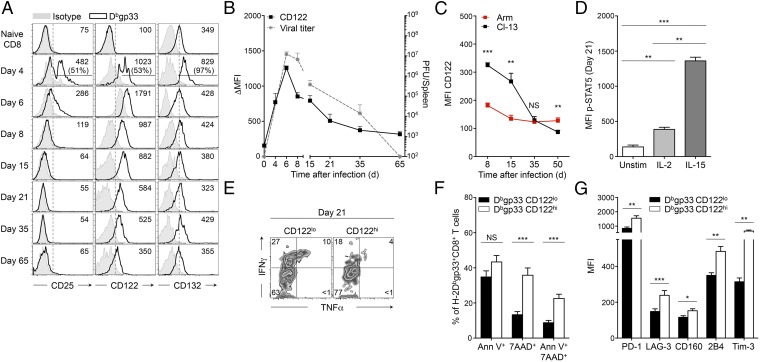
To delineate the impact of IL-2 and IL-15 on the CD8+ T-cell response to chronic viral infection, we first analyzed the expression kinetics of key IL-2 and IL-15 (IL-2/15) receptor components on H-2Db–restricted LCMV-specific CD8+ T cells throughout infection, focusing on the dominant epitope gp33-41 (Dbgp33). Following infection with LCMV Cl-13, CD25 (IL2Rα) expression was induced early but vanished after 6 d postinfection (p.i.) (Fig. 1A, Left), whereas CD132, the receptor chain shared by all cytokines of the common γc family, returned to baseline levels (Fig. 1A, Right and Fig. S1A). By contrast, CD122, exclusive to both IL-2 and IL-15, was expressed early and was sustained throughout the infection (Fig. 1A, Center), and its expression level declined gradually, mirroring the drop in viral titers (Fig. 1B). This association suggests that antigen persistence supports CD122 expression on CD8+ T cells. In line with this notion, the levels of CD122 were substantially higher at day 8 and 15 p.i. on chronically stimulated Dbgp33 CD8+ T cells than on Dbgp33 CD8+ T cells activated by the acute strain of LCMV, LCMV Armstrong (Arm), until day 30 p.i., when they reached comparable levels (Fig. 1C). Thereafter, CD122 expression continued to decline in mice previously infected with LCMV Cl-13, consistent with the inability of chronically stimulated cells to sustain CD122 expression after viral elimination (14, 15). Maintenance of the IL2Rβγ complex promoted signal transducer and activator of transcription 5 (STAT5) phosphorylation in response to IL-2 and, to a greater extent, to IL-15 at day 21 p.i., in agreement with previous reports (Fig. 1D) (29). In response to IL-2 and IL-15, CD8+ T cells chronically stimulated with Cl-13 expressed levels of phospho-STAT5 similar to those in their counterparts acutely infected with Arm at day 30 p.i. (Fig. S1B). These results suggested that CD122-dependent signals supported important physiological functions during antigen persistence. To understand these functions, we performed direct ex vivo comparative analysis between freshly isolated CD8+ T cells expressing high (CD122hi) or low (CD122lo) levels of IL2Rβ (see Fig. S1C for the gating strategy). We noted that a lower frequency of CD122hi Dbgp33-specific CD8+ T cells secreted IFNγ compared with CD122lo cells (22% versus 33%, P = 0.0086) (Fig. 1E and Fig. S1D). IFNγ/TNFα double producers were also less frequent within CD122hi effectors than in CD122lo cells (5% versus 9%, P = 0.0463); however, TNFα production was uniformly low (Fig. 1E and Fig. S1D). CD122hi effectors were also more apoptotic, as demonstrated by the higher frequency of 7-amino-actinomycin D (7AAD)+ and AnnexinV+/7AAD+ cells compared with CD122lo effectors (39% versus 13%, P < 0.0001, and 22% versus 9%, P < 0.0001, respectively) (Fig. 1F). This restricted cytokine profile and limited survival potential were suggestive of T-cell exhaustion. To correlate these findings with the exhaustion status of the cells, we assessed the expression of a panel of inhibitory receptors. On day 21 p.i., CD122hi effectors expressed higher levels of several inhibitory receptors including PD-1, LAG-3, CD160, 2B4, and Tim-3 than did CD122lo Dbgp33-specific T cells (Fig. 1G). Together, these results imply that high IL-2– and IL-15–dependent signaling may restrict T-cell function and survival through increased expression of inhibitory receptors, committing effector cells to severe exhaustion.
Fig. 1.
CD122 expression is maintained on CD8+ T cells and identifies severely exhausted cells. C57BL/6 (B6) mice were infected with LCMV Cl-13; analyses were performed in spleens on LCMV-specific H-2Dbgp33+CD8+ T cells at day 21 p.i., unless otherwise indicated. (A) Cell-surface expression of CD25, CD122, and CD132 at distinct time points after infection. Values in quadrants indicate MFI (and frequency of highly positive cells on Day 4); gray-filled histograms are isotype controls. (B) Cell-surface expression of CD122 [left y axis; filled squares represent Δ MFI (MFI minus isotype control MFI)] and viral titers (right y axis; dotted gray line) at indicated time points. (C) Cell-surface expression of CD122 on H-2Dbgp33+CD8+ T cells isolated at indicated time points from mice infected with LCMV Arm (acute infection; red squares) or LCMV Cl-13 (chronic infection; black squares). (D) Intracellular expression of p-STAT5 in activated (CD11a+) CD8+ T cells following stimulation with the indicated cytokines. Unstim, unstimulated control. (E) Intracellular production of IFNγ and TNFα in CD122lo H-2Dbgp33+CD8+ T cells (Left) and CD122hi H-2Dbgp33+CD8+ T cells (Right). Values indicate the frequency of cells in each quadrant. (F) Frequency of CD122lo H-2Dbgp33+CD8+ T cells (black bars) and CD122hi H-2Dbgp33+CD8+ T cells (white bars) positive for Annexin V and/or 7AAD. (G) Expression of the indicated inhibitory receptors on CD122lo H-2Dbgp33+CD8+ T cells (black bars) and CD122hi H-2Dbgp33+CD8+ T cells (white bars). Bars represent the MFI. *P < 0.05; **P < 0.005; ***P < 0.0005; NS P ≥ 0.05 (two-tailed unpaired Student’s t test). Data are pooled from two (C and G) or three (B and F) independent experiments with at least five mice per group or are representative of three independent experiments (A, D, and E) with similar results (n = 2 or 3 mice per group in each). Error bars in B–D, F, and G indicate mean ± SEM.
Fig. S1.
CD122 expression on CD8+ T cells identifies severely exhausted cells. C57BL/6 (B6) mice were infected with LCMV Cl-13; analyses were performed in spleens on day 21 p.i. unless otherwise indicated. (A) Cell-surface expression of CD132 (left y axis; filled squares represent Δ MFI) and viral titers (right y axis; dashed gray line) at the indicated time points. (B) Intracellular expression of p-STAT5 in activated (CD11a+) CD8+ T cells following stimulation with the indicated cytokines from animals infected with LCMV Arm (acute infection; red bars) or LCMV Cl-13 (chronic infection; black bars) at day 30 p.i.; Unstim, level in unstimulated control. (C) Gating strategy for CD122hi H-2Dbgp33+CD8+ T cells (upper gate) and CD122lo H-2Dbgp33+CD8+ T cells (lower gate). Gates are set above and below the center of the population. CD122int/lo cells include cells negative for CD122. Values indicate the frequency in each gate. (D) Frequency of CD122lo (black bars) and CD122hi (white bars) H-2Dbgp33+CD8+ T cells producing IFNγ, TNFα, or both IFNγ and TNFα simultaneously. *P < 0.05; **P < 0.005; ***P < 0.0005; NS P ≥ 0.05; two-tailed unpaired Student’s t test. Data are pooled from two (A) or three (D) independent experiments with four to eight mice per group or are representative of three (C) or four (B) independent experiments with similar results (two to three mice per group in each). Error bars in A, B, and D indicate mean ± SEM.
Elevated CD122 Expression on CD8+ T Cells Identifies PD-1hi Terminal Effectors in Mice Infected with LCMV Cl-13 and in Humans with Persistent HCV Infection.
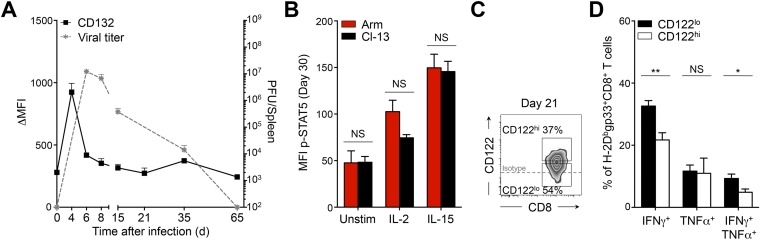
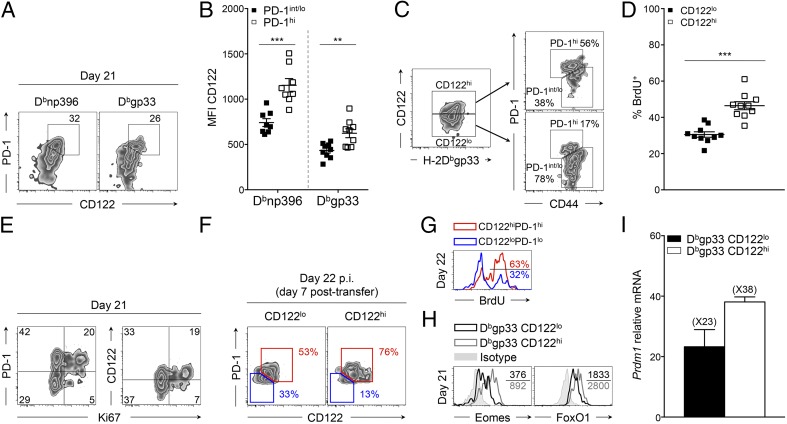
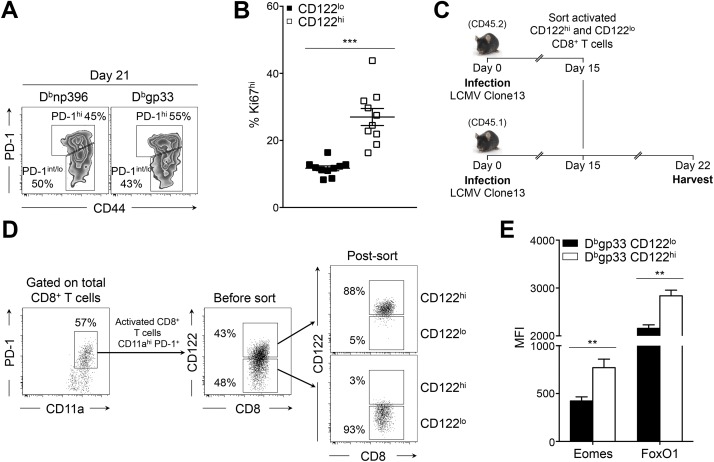
During chronic infection, it was demonstrated that PD-1int/lo progenitor cells serve as a reservoir for continuously replenishing a PD-1hi progeny population (23). Conversion into PD-1hi cells is terminal and is associated with enhanced exhaustion, increased expression of inhibitory receptors, loss of cytokine production capacity, and increased apoptosis (7, 10, 23). Because CD122hi cells presented these same characteristics (Fig. 1), we questioned the lineage relationship between CD122hi and PD-1hi cells. At day 21 p.i., we observed a correlation between CD122 and PD-1 expression on antigen-specific CD8+ T cells (Fig. 2A). PD-1hi cells expressed higher levels of CD122 than PD-1int/lo progenitors (see Fig. 2B and Fig. S2A for the gating strategy). The majority of CD122hi Dbgp33 CD8+ T cells were also PD-1hi progenies, and, inversely, CD122lo cells were largely composed of PD-1int/lo progenitors (Fig. 2C). Conversion from PD-1int/lo to PD-1hi terminal effectors and subsequent increase in PD-1 levels follows extensive proliferation (23). Similarly, CD122 levels increased with proliferation as most CD122hi Dbgp33 CD8+ T cells incorporated 5-bromo-2-deoxyuridine (BrdU) between day 15 and 30 p.i. and preferentially expressed the proliferative marker Ki67 (Fig. 2 D and E and Fig. S2B). To delineate the lineage relationship between CD122lo and CD122hi cells, we tested whether CD122lo cells were the progenitors of the CD122hi terminal effector population. To do so, CD122lo and CD122hi CD45.2+ cells sorted at day 15 p.i. were transferred into infection-matched CD45.1+ mice (Fig. S2 C and D). One week later (day 22 p.i.), 46 ± 4% of transferred CD122lo cells presented a CD122hi PD-1hi phenotype, whereas 74 ± 2% of CD122hi cells remained CD122hi PD-1hi (Fig. 2F). Conversion of transferred CD122lo cells into CD122hi PD-1hi effectors was accompanied by intense proliferation, as demonstrated by the incorporation of BrdU over 1 wk, whereas the majority of CD122lo PD-1lo cells were BrdU− (Fig. 2G). Differentiation into PD-1hi effectors is associated with specific transcriptional changes including elevated levels of Eomes, Blimp-1, and the Forkhead box protein O1 (FoxO1) (18, 23, 29). Accordingly, CD122hi Dbgp33 CD8+ T cells isolated at day 21 p.i. expressed higher levels of Eomes and FoxO1 than their CD122lo counterparts (Fig. 2H and Fig. S2E). Prdm1 mRNA levels also appeared higher in CD122hi cells, although the difference did not reach statistical significance (Fig. 2I). Together, these results define CD122 as an independent marker distinguishing the PD-1int/lo progenitors from the terminally exhausted PD-1hi progeny population and reveal CD122lo cells as the progenitors of the CD122hi PD-1hi population.
Fig. 2.
Lineage relationship between CD122hi cells and PD-1hi cells. Mice were infected and analyzed as in Fig. 1. (A) Cell-surface expression of CD122 and PD-1 on H-2Dbnp396+CD8+ T cells (Left) and H-2Dbgp33+CD8+ T cells (Right). Values indicate the frequency of CD122hiPD-1hi cells. (B) Expression of CD122 on PD-1int/lo (filled squares) and PD-1hi (open squares) H-2Dbnp396+ CD8+ T cells (Left) and H-2Dbgp33+ CD8+ T cells (Right). (C) Expression of PD-1 and CD44 on CD122hi (Upper Right) and CD122lo (Lower Right) H-2Dbgp33+CD8+ T cells. Values indicate the frequency of PD-1hiCD44int and PD-1int/loCD44hi cells in each panel. (D) Frequency of CD122lo (filled squares) and CD122hi (open squares) H-2Dbgp33+CD8+ T cells that incorporated BrdU between day 15 and 30 p.i. (E) Cell-surface expression of CD122, PD-1, and intracellular Ki67 in H-2Dbgp33+CD8+ T cells. Values indicate the frequency of cells in each gate. (F) Cell-surface expression of CD122 and PD-1 on CD122lo (Left) and CD122hi (Right) transferred cells (CD45.2+) at day 22 p.i. (7 d posttransfer). Values indicate the frequency of CD122hiPD-1hi (red) and CD122loPD-1lo (blue) cells. (G) BrdU incorporation (day 15–22 p.i.) by CD122hiPD-1hi CD45.2+ cells (red histogram) and CD122loPD-1lo CD45.2+ cells (blue histogram) recovered from mice adoptively transferred with CD122lo (CD45.2+) cells. Values indicate the frequency of positive cells. (H) Intracellular Eomes and FoxO1 expression in CD122lo (black histograms) and CD122hi (gray histograms) H-2Dbgp33+CD8+ T cells. Values indicate the MFI; gray-filled histograms are isotype controls. (I) Expression of Prdm1 assessed by quantitative RT-QPCR analysis in sorted CD122lo (black bar) and CD122hi (white bar) H-2Dbgp33+CD8+ T cells. Values indicate the fold increase relative to naive P14 CD8+ T cells. **P < 0.005; ***P < 0.0005; two-tailed unpaired Student’s t test. Data are pooled from two (I) or three (B) independent experiments with at least eight mice per group or are representative of one (G), two (F and H), or three (A and C) independent experiments with similar results (n = 2 or 3 mice per group). Data in D and E are representative of one experiment with 10 mice per group. Error bars in B, D, E, and I indicate mean ± SEM.
Fig. S2.
CD122hi cells share functional and transcriptional features with PD-1hi cells. Mice were infected and analyzed as in Fig. S1. (A) Gating strategy for PD-1hi (upper gates) and PD-1int/lo (lower gates) H-2Dbnp396+ CD8+ T cells (Left) and H-2Dbgp33+ CD8+ T cells (Right). Values indicate the frequency of cells in each gate. (B) Cumulative frequency of CD122lo H-2Dbgp33+CD8+ T cells (filled squares) and CD122hi H-2Dbgp33+CD8+ T cells (open squares) expressing high levels of Ki67. Data are representative of one experiment with 10 mice per group. (C and D) Experimental approach (C) and sorting strategy (D) for adoptive transfer experiments of CD122lo and CD122hi cells. Splenocytes were harvested from CD45.2+ donor mice at day 15 p.i. Activated CD11a+ PD-1+ CD8+ T cells were sorted based on high (CD122hi) or intermediate/low (CD122lo) expression of CD122 and were infused i.v. in distinct CD45.1+ infection-matched recipient mice. Recipient mice were given BrdU daily from day 15–22 p.i., and CD45.2+ transferred cells were harvested at day 22 and analyzed for surface markers and BrdU incorporation. (E) Intracellular Eomes and FoxO1 expression in CD122lo H-2Dbgp33+CD8+ T cells (black bars) and CD122hi H-2Dbgp33+CD8+ T cells (white bars). **P < 0.005; ***P < 0.0005; two-tailed unpaired Student’s t test. Data are pooled from two independent experiments with at least six mice per group (E) or are representative of three independent experiments with similar results (n = 2 or 3 mice per group) (A). Error bars in B and E indicate mean ± SEM.
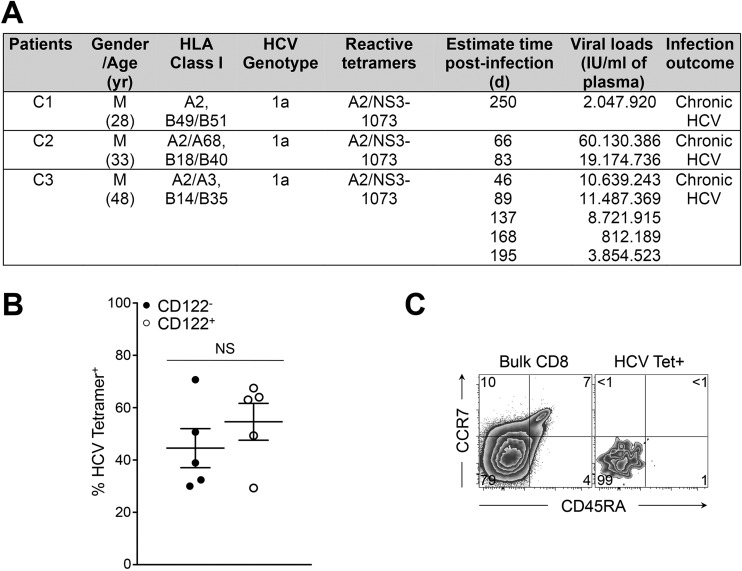
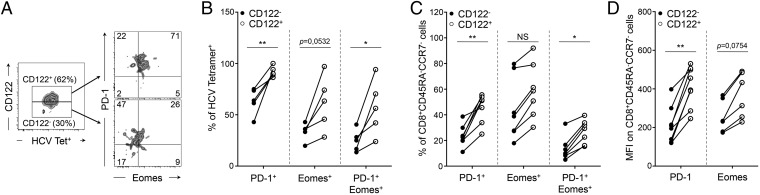
To validate and extend our observations to human chronic viral infection, we performed similar analysis on peripheral blood samples obtained at different stages p.i. from individuals who developed persistent HCV infection (Fig. S3A). In these patients we detected a substantial fraction of peripheral HCV-specific CD8+ T cells expressing CD122 (54.6 ± 7%) (Fig. S3B). Interestingly, CD122+ HCV-specific CD8+ T cells were enriched in cells expressing PD-1, Eomes, or both markers (Fig. 3 A and B), indicating that these effectors were more exhausted and terminally differentiated (23). Concomitantly, the frequency of cells expressing these markers was reduced in CD122− HCV-specific CD8+ T cells (Fig. 3 A and B). Similar patterns were observed when we gated bulk CD8+ T cells with an effector memory CD45RAloCCR7lo phenotype, which constitutes the majority of effector cells in HCV-infected patients (Fig. 3 C and D and Fig. S3C). Taken together, our results demonstrate that CD122 is a relevant biological marker of CD8+ T-cell exhaustion and terminal differentiation in both mice and humans.
Fig. S3.
Chronically infected patients with HCV. PBMCs were collected during the first year of HCV infection from individuals who had high viral titers and had developed persistent viremia. (A) Patients' characteristics. (B) Frequency of CD122− HCV-specific CD8+ T cells (filled circles) and CD122+ HCV-specific CD8+ T cells (open circles). (C) Expression of CCR7 and CD45RA on total CD8+ T cells (Left) and HCV-specific CD8+ T cells (Right). Values indicate the frequency of cells in each gate. Data are representative of five to eight samples from three infected patients processed independently.
Fig. 3.
CD122 identifies exhausted cells in individuals infected with HCV. PBMCs were collected during the first year of HCV infection from individuals who had high viral titers and developed persistent viremia as described in Fig. S3A. (A) Expression of extracellular PD-1 and intracellular Eomes in CD122+ (Upper Right) and CD122− (Lower Right) HCV-specific CD8+ T cells. Gates for CD122+ and CD122− cells are set based on fluorescence minus one (FMO) controls. Values indicate the frequency of cells in each quadrant. (B) Frequency of CD122− (filled circles) and CD122+ (open circles) HCV-specific CD8+ T cells positive for the indicated molecules. (C) Frequency of CD122− (filled circles) and CD122+ (open circles) CD45RA−CCR7− effector memory CD8+ T cells positive for the indicated molecules. (D) Expression of PD-1 and Eomes in CD122− (filled circles) and CD122+ (open circles) CD45RA−CCR7− effector memory CD8+ T cells represented by their MFI. *P < 0.05; **P < 0.005; NS P ≥ 0.05; two-tailed unpaired Student’s t test. Data are pooled from five to eight distinct time points from two to three infected patients processed independently.
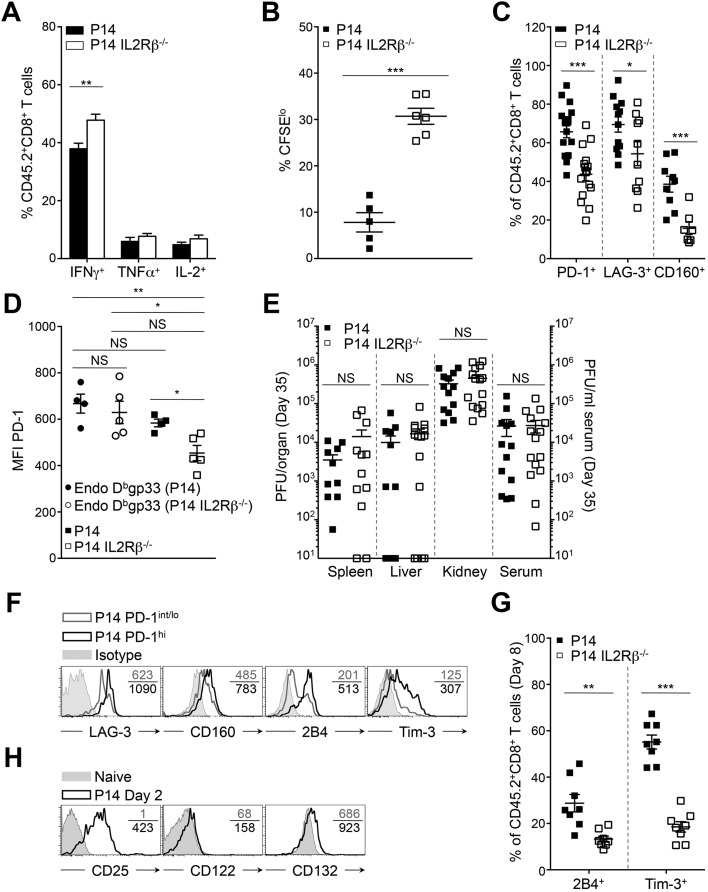
Deleting CD122 on CD8+ T Cells Abrogates Their Differentiation into PD-1hi Exhausted Cells.
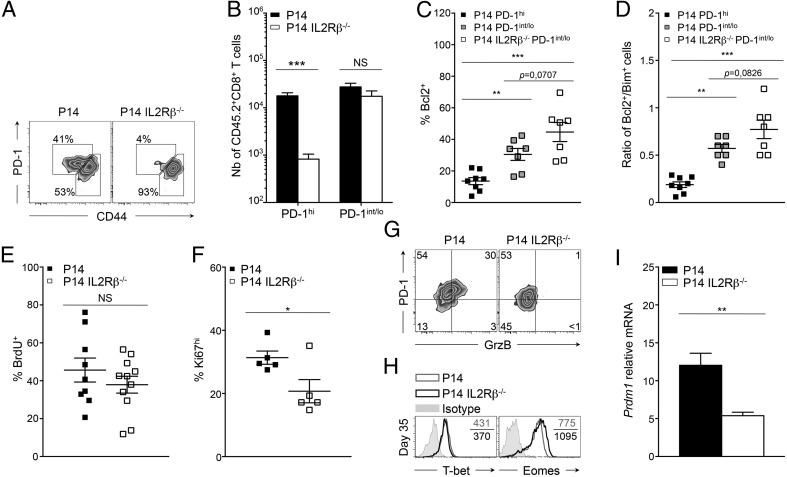
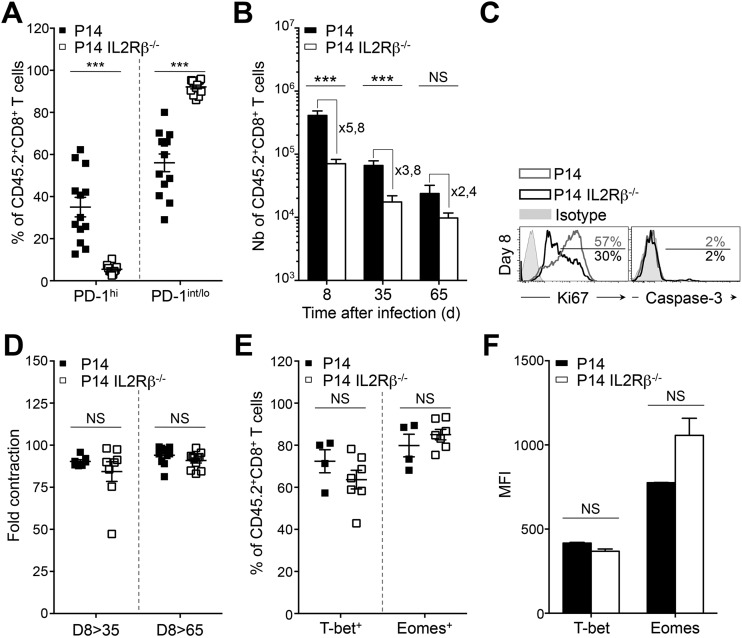
To elucidate the cell-intrinsic significance of increased CD122 levels during PD-1hi conversion, we performed adoptive transfer of IL2Rβ-deficient P14 (P14 IL2Rβ−/−) cells or IL2Rβ-sufficient controls (P14 cells) into distinct C57BL/6 hosts 20 h before infection with LCMV Cl-13 (25). As expected, P14 cells could be identified in the spleen as two distinct populations on day 35 p.i.: PD-1int/lo progenitors (PD-1int/loCD44hi; 2.8 × 104 cells) and PD-1hi progenies (PD-1hiCD44int/lo; 1.8 × 104 cells) (Fig. 4 A and B and Fig. S4A) (7, 23). Strikingly, PD-1hi effectors were nearly undetectable in the absence of IL2Rβ (8 × 102 cells), whereas the number of PD-1int/lo cells was unaffected (1.8 × 104 cells) (Fig. 4 A and B and Fig. S4A). Absence of PD-1hi cells correlated with a 3.8-fold reduction in the absolute number of P14 IL2Rβ−/− cells compared with P14 controls at day 35 p.i. (Fig. S4B). This salient difference in cell numbers resulted mainly from reduced proliferation of IL2Rβ−/− cells at day 8 p.i. rather than from a survival defect, as demonstrated by the lower frequency of Ki67hi cells in the absence of IL2Rβ but equally low expression of active caspase-3 (Fig. S4C). The fold difference in absolute numbers did not increase but rather decreased between day 8 and 35 p.i., and both groups presented similar contraction phases (Fig. S4 B and D). We and others have previously demonstrated that IL2Rβ-dependent signals improved the survival of terminal effectors during the contraction phase of an acute viral infection in a B-cell lymphoma 2 (Bcl2)-dependent manner (24, 25, 30, 31). Only 16 ± 8% of P14 PD-1hi cells, which present the highest levels of CD122 (Fig. 2B), expressed Bcl2, as compared with 30 ± 10% of P14 PD-1int/lo cells (Fig. 4C). The absence of IL2Rβ further increased the proportion of cells expressing Bcl2 in PD-1int/lo cells (45 ± 6%) (Fig. 4C). This increase resulted in a superior ratio of P14 IL2Rβ−/− PD-1int/lo cells expressing Bcl2 over Bim as compared with either P14 PD-1hi or PD-1int/lo cells (Fig. 4D). Collectively, these data demonstrate that IL2Rβ-dependent signals are essential for the development of PD-1hi cells rather than for their maintenance and further hamper the survival of the PD-1int/lo progenitor pool. Because PD-1hi development is the result of extensive proliferative events (23), we then questioned whether the defective development into PD-1hi cells seen in the absence of IL2Rβ was caused by altered proliferation during viral persistence. To test this hypothesis, mice were given daily BrdU starting at day 15 p.i. At day 30 p.i., P14 and P14 IL2Rβ−/− CD8+ T cells had incorporated equivalent amounts of BrdU (Fig. 4E). However, the immediate proliferation rate of P14 controls was superior to P14 IL2Rβ−/− cells as assessed by direct Ki67 staining at day 30 p.i. (31 ± 2% versus 21% ± 4% respectively) (Fig. 4F). Although 21% of P14 IL2Rβ−/− cells underwent proliferation, they never acquired granzyme B expression, a cytotoxic molecule known to increase with PD-1hi terminal differentiation (Fig. 4G) (23). These findings indicate that IL2Rβ-dependent signals sustain the immediate proliferation of CD8+ T cells during progression to chronic infection but also affect intrinsic differentiation pathways critical for the generation of terminally exhausted PD-1hi cells. To understand how IL2Rβ-dependent signals impacted the differentiation of PD-1hi progenies, we evaluated known transcription factors involved in the differentiation of PD-1hi terminal effectors (23). Unexpectedly, levels of T-bet were nearly equivalent in both groups of cells, whereas Eomes levels were slightly increased in P14 IL2Rβ−/− cells, although the increase did not reach statistical significance (Fig. 4H and Fig. S4 E and F). However, we detected a marked reduction in the levels of Prdm1 mRNA (the gene encoding Blimp-1) in P14 IL2Rβ−/− cells at day 35 p.i. compared with P14 controls (Fig. 4I). These results point to a critical role for IL-2 and IL-15 in driving the lineage choice to PD-1hi cells by promoting the expression of Blimp-1, a key transcription factor associated with CD8+ T-cell terminal differentiation and exhaustion (17–19).
Fig. 4.
IL2Rβ deficiency abrogates PD-1hi terminal differentiation. Tg P14 or P14 IL2Rβ−/− (CD45.2) cells were adoptively transferred into recipient mice (CD45.1.2) before LCMV Cl-13 infection. Tg cells (CD45.2) were compared in the spleen at day 35 p.i. (A) Representative dot plots of PD-1hi and PD-1int/lo P14 cells (Left) and P14 IL2Rβ−/− cells (Right). Values indicate the frequency of cells in each gate. (B) Absolute numbers of PD-1hi (Left) and PD-1int/lo (Right) splenic cells from P14 (black bars) and P14 IL2Rβ−/− (white bars) chimeric mice. (C) Frequency of cells expressing Bcl2 in P14 PD-1hi cells (filled squares), P14 PD-1int/lo cells (gray squares), and P14 IL2Rβ−/− PD-1int/lo cells (open squares). (D) Frequency of Bcl2+ cells over frequency of Bim+ cells in P14 PD-1hi cells (filled squares), P14 PD-1int/lo cells (gray squares), and P14 IL2Rβ−/− PD-1int/lo cells (open squares). (E) Frequency of P14 cells (filled squares) and P14 IL2Rβ−/− cells (open squares) that incorporated BrdU between day15 and 30 p.i. (F) Frequency of Ki67hi P14 cells (filled squares) and P14 IL2Rβ−/− cells (open squares). Data represent one experiment with five mice per group. (G) Expression of PD-1 and intracellular granzyme B in P14 cells (Left) and P14 IL2Rβ−/− cells (Right). Values indicate the frequency of cells in each gate. (H) Intracellular T-bet and Eomes expression in P14 cells (gray histograms) and P14 IL2Rβ−/− cells (black histograms). Values indicate the MFI; gray-filled curves are isotype controls. (I) Prdm1 levels assessed by quantitative RT-QPCR analysis in sorted P14 cells (black bar) and P14 IL2Rβ−/− cells (white bar) relative to naive P14 cells. *P < 0.05; **P < 0.005; ***P < 0.0005; NS P ≥ 0.05; two-tailed unpaired Student’s t test. Data are pooled from two (C, D, and I) or three (B and E) independent experiments with at least seven mice per group or are representative of two (G and H) or three (A) independent experiments with similar results (n = 2–4 mice per group in each). Error bars in B–F and I indicate mean ± SEM.
Fig. S4.
IL2Rβ deficiency abrogates PD-1hi terminal differentiation. Tg P14 or P14 IL2Rβ−/− (CD45.2) CD8+ T cells, specific for the H-2Dbgp33 epitope of LCMV, were adoptively transferred into recipient mice (CD45.1.2) before LCMV Cl-13 infection. Development of Tg CD8+CD45.2+ PD-1hi and PD-1int/lo cells in the two groups of chimeric mice was compared at day 35 p.i. (A) Cumulative frequencies of PD-1hi (Left) and PD-1int/lo (Right) P14 cells (filled squares) and P14 IL2Rβ−/− cells (open squares). (B) Absolute numbers of CD8+CD45.2+ P14 splenic cells (black bars) and P14 IL2Rβ−/− splenic cells (white bars) calculated based on H-2Dbgp33 tetramer and CD45.2 congenic marker staining at the indicated time points. Values indicate the fold difference in the absolute numbers of P14 cells over P14 IL2Rβ−/− cells at that precise time point. (C) Intracellular expression of Ki67 (Left) and active-caspase 3 (Right) in CD8+CD45.2+ P14 cells (gray histograms) and P14 IL2Rβ−/− cells (black histograms) at day 8 p.i. Values indicate the frequency of Ki67hi or active caspase-3+ cells. (D) Fold contraction in the number of P14 cells (filled squares) and P14 IL2Rβ−/− cells (open squares) in the indicated time frames. (E and F) Frequency (E) and MFI (F) of intracellular T-bet and Eomes in P14 cells (filled squares in E and black bars in F) and P14 IL2Rβ−/− cells (open squares in E and white bars in F) at day 35 p.i. ***P < 0.0005; NS P ≥ 0.05; two-tailed unpaired Student’s t test. Data are pooled from two (E and F), three (A and D), or four (B) independent experiments with 3–10 mice per group or are representative of three (C) independent experiments with similar results (n = 3 mice per group). Error bars in A, B, and D–F indicate mean ± SEM.
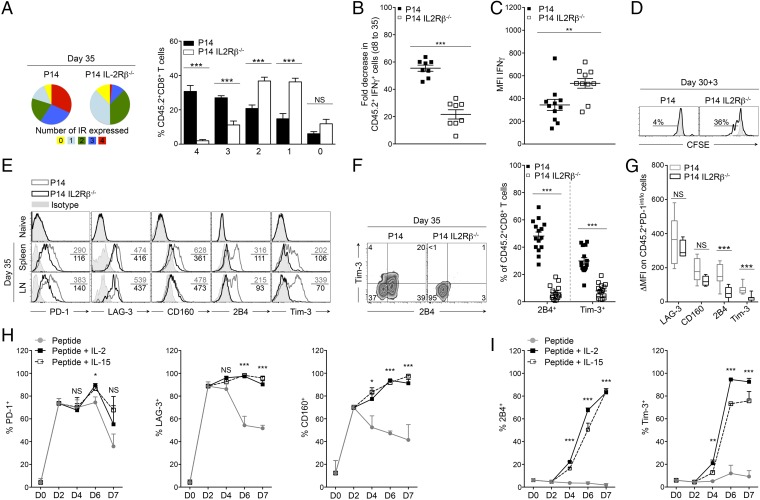
Lack of IL2Rβ-Dependent Signaling Restrains the Development of Highly Exhausted Effector Cells and Blocks 2B4 and Tim-3 Induction on CD8+ T Cells.
To confirm the importance of IL2Rβ-dependent signals on the development of highly exhausted CD8+ T cells, we analyzed the composition of the CD8+ T-cell pool on day 35 p.i. First we measured the frequency of highly dysfunctional P14 and P14 IL2Rβ−/− cells coexpressing multiple inhibitory receptors (10). We observed that IL2Rβ deficiency enhanced the quality of the effector pool by favoring the accumulation of less exhausted CD8+ T cells that coexpressed only one or two inhibitory receptors (Fig. 5A) (10). Conversely, most P14 controls were found to express three or four inhibitory receptors simultaneously, indicating more pronounced exhaustion (10). In IL2Rβ−/− cells, this observation correlated both with a preserved capacity to produce IFNγ and increased IFNγ production on a per-cell basis (as indicated by the mean fluorescence intensity, MFI) (Fig. 5 B and C and Fig. S5A) and also with the ability to proliferate in response to antigenic stimulation (Fig. 5D and Fig. S5B), two fundamental characteristics lost by exhausted T cells (6, 8). Together, these results indicated that IL2Rβ deficiency protected effector CD8+ T cells from severe exhaustion. When analyzed individually, we also observed that IL2Rβ deficiency was associated with a major reduction in the expression of all tested inhibitory receptors (PD-1, LAG-3, CD160, 2B4, and Tim-3), with the exception of CD160, which was also reduced on P14 cells from the lymph nodes (Fig. 5E and Fig. S5C). Most striking was the nearly complete abolition of 2B4 and Tim-3 expression on IL2Rβ−/− cells (Fig. 5 E and F). These observations did not reflect discrepancies between the two groups of mice, because endogenous Dbgp33 (CD45.1.2) cells of mice adoptively transferred with either P14 or P14 IL2Rβ−/− cells expressed equivalent levels of PD-1 (Fig. S5D). Moreover, both groups had similar viral titers by day 35 p.i. (Fig. S5E). Because PD-1hi cells are known to express higher levels of inhibitory receptors (Fig. S5F and ref. 10), we questioned whether the differences we observed between the two groups might reflect the defective development of PD-1hi cells in the absence of IL2Rβ. We compared the expression of LAG-3, CD160, 2B4, and Tim-3 in the PD-1int/lo compartment of P14 and P14 IL2Rβ−/− cells. Highly significant differences in 2B4 and Tim-3 expression were observed in the two groups, whereas LAG-3 and CD160 expression were comparable (Fig. 5G). The restricted expression of 2B4 and Tim-3 was detectable at earlier time points in the absence of IL2Rβ (Fig. S5G), excluding a potential selective loss of this population during chronicity. These results imply that the increase in LAG-3 and CD160 expression resulted from an efficient conversion in PD-1hi progenies, whereas IL-2 and IL-15 directly controlled 2B4 and Tim-3 expression. To confirm this finding, we designed an in vitro assay to assess the impact of IL-2 and IL-15 on the induction of inhibitory receptors while keeping constant the antigenic stimulation throughout the assay. IL-2 or IL-15 was added on day 2 of the culture, a time when IL-2/15 receptor components were efficiently induced (Fig. S5H). Adequate T-cell receptor (TCR) stimulation was sufficient to trigger high levels of PD-1, LAG-3, and CD160 on P14 cells at day 2 of the culture, and IL-2 and IL-15 maintained their expression thereafter (Fig. 5H). In contrast, 2B4 and Tim-3 expression was strictly dependent on the addition of IL-2 or IL-15 to the culture medium (Fig. 5I). Our findings provide evidence that IL-2 and IL-15 use both direct and indirect routes to regulate CD8+ T-cell exhaustion. First, IL-2 and IL-15 directly and redundantly promote 2B4 and Tim-3 expression. Second, these cytokines control the terminal differentiation of PD-1hi effectors and the subsequent increase in LAG-3 and CD160 levels. These data point to the critical role of IL-2 and IL-15 in the development of severe T-cell exhaustion during chronic viral infection.
Fig. 5.
IL2Rβ-dependent signals regulate the levels of inhibitory receptor expression. (A–G) P14 or P14 IL2Rβ−/− chimeric mice were generated and analyzed as described in Fig. 4. (A) Frequency of P14 cells (left pie chart and black bars) and P14 IL2Rβ−/− cells (right pie chart and white bars) coexpressing PD-1, LAG-3, 2B4, and CD160. Numbers 1–4 represent the number of inhibitory receptors expressed simultaneously. (B) Fold decrease in the frequency of P14 cells (filled squares) and P14 IL2Rβ−/− cells (open squares) producing IFNγ between days 8 and 35 p.i. (C) Production of IFNγ (as shown by MFI) in CD8+CD45.2+ P14 cells (filled squares) and P14 IL2Rβ−/− cells (open squares). (D) Proliferation of P14 cells (Left) and P14 IL2Rβ−/− cells (Right) in response to gp33 stimulation. Proliferation is represented as CFSE dilution over 3 d with values indicating the frequency of CFSEint cells. Gray-filled histograms are unstimulated controls. (E) Expression of inhibitory receptors on P14 cells (gray histograms) and P14 IL2Rβ−/− cells (black histograms). Values indicate the MFI; gray-filled histograms are isotype controls. (F, Left) Cell-surface expression of 2B4 and Tim-3 on P14 cells and P14 IL2Rβ−/− cells. Values indicate the frequency of cells in each quadrant. (Right) Cumulative frequencies of 2B4+ and Tim-3+ P14 cells (filled squares) and P14 IL2Rβ−/− cells (open squares). (G) Expression of inhibitory receptors on PD-1int/lo P14 cells (gray boxes) and P14 IL2Rβ−/− cells (black boxes) represented by their Δ MFI. (H and I) Frequency of P14 cells expressing the indicated inhibitory receptors during coculture with gp33 peptide alone (gray lines) or with additional IL-2 (black lines) or IL-15 (dashed lines). Data are pooled from five independent experiments performed in triplicate. *P < 0.05; **P < 0.005; ***P < 0.0005; NS P ≥ 0.05; two-tailed unpaired Student’s t test (A–G) or one-way ANOVA non parametric tests; Dunnett’s t (G and H). Data are pooled from two (A), three (B, C, and G), or four (F) independent experiments with 5–16 mice per group or are representative of two (D) or five (E) independent experiments with similar results (n = 2–5 mice per group in each). Error bars in A–C and F–I indicate mean ± SEM.
Fig. S5.
IL2Rβ-dependent signals regulate the level of inhibitory receptor expression and the associated cellular dysfunctions. P14 or P14 IL2Rβ−/− chimeric mice were generated as described in Fig. S4, and cells were analyzed at day 35 p.i. (A) Intracellular production of the indicated cytokines by CD8+CD45.2+ P14 cells (black bars) and P14 IL2Rβ−/− cells (white bars). (B) Cumulative frequency of P14 cells (filled squares) and P14 IL2Rβ−/− cells (open squares) proliferating in response to gp33 peptide stimulation, as assessed by a CFSE dilution assay. (C) Cumulative frequencies of PD-1+ (Left), LAG-3+ (Center), and CD160+ (Right) P14 cells (filled squares) and P14 IL2Rβ−/− cells (open squares) at day 35 p.i. (D) Cumulative cell-surface expression of PD-1 on endogenous CD45.1.2+H-2Dbgp33+CD8+ T cells from chimeric mice transferred with P14 cells (filled circles) or P14 IL2Rβ−/− cells (open circles). Tg P14 (filled squares) and Tg P14 IL2Rβ−/− (open squares) CD45.2+H-2Dbgp33+CD8+ T cells are also represented. (E) Viral titers in the indicated organs (left axis) and in the serum (right axis) at day 35 p.i. from P14 (filled squares) and P14 IL2Rβ−/− (open squares) chimeric mice. (F) Expression of the indicated inhibitory receptors on P14 PD-1int/lo cells (gray histograms) and P14 PD-1hi cells (black histograms). Values indicate the MFI; gray-filled histograms are isotype controls. (G) Cumulative frequencies of 2B4+ (Left) and Tim-3+ (Right) P14 cells (filled squares) and P14 IL2Rβ−/− cells (open squares) at day 8 p.i. (H) Expression of CD25, CD122, and CD132 on P14 cells after 2 d of in vitro stimulation with dendritic cells loaded with gp33-41. Values indicate the MFI; gray-filled histograms are isotype controls. *P < 0.05; **P < 0.005; ***P < 0.0005; NS P ≥ 0.05; two-tailed unpaired Student’s t test. Data are pooled from one (D), two (B), three (A, E, and G), or five (C) independent experiments with 4–18 mice per group or are representative of one (H) or three (F) independent experiments with similar results (n = 2–4 mice per group). Error bars in A–E and G indicate mean ± SEM.
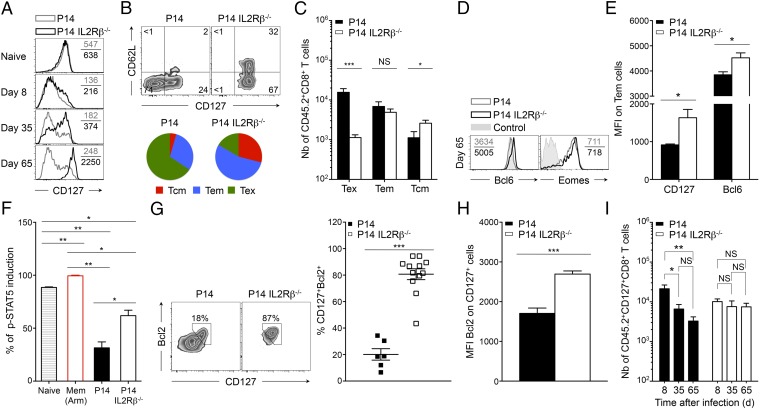
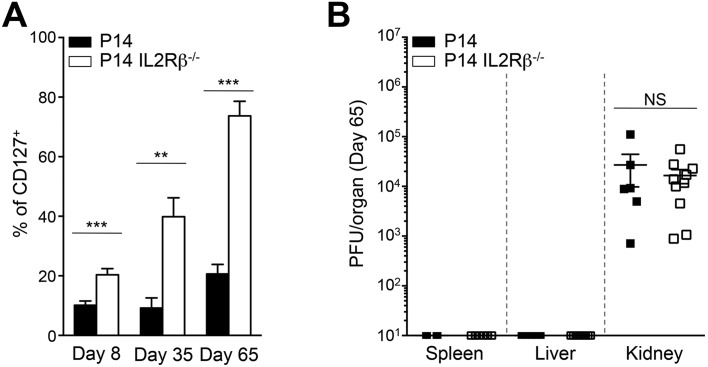
IL2Rβ Deficiency Restores CD8+ Memory T-Cell Development and IL-7–Dependent Signaling.
Resolution of an acute infection allows the differentiation of a CD8 memory T-cell pool that persists and protects the host from reinfection. During chronic viral infection, however, CD8 memory T-cell development is completely blocked (13–15). The factors involved in this arrested memory differentiation remain unclear, and very few immunomodulatory methods used to limit exhaustion have been shown to rescue CD8 memory T cells. Strikingly, we noted that abrogation of IL2Rβ-dependent signals led to the rapid reexpression of the cardinal memory marker CD127 from day 8 p.i. onwards (Fig. 6A). At that time point the frequency of CD127+ P14 IL2Rβ−/− cells already exceeded that of P14 controls and increased rapidly thereafter (Fig. S6A). This finding suggested that IL2Rβ deficiency caused an early differentiation bias, allowing a substantial fraction of effector CD8+ T cells to escape exhaustion and conserve memory potential. Indeed, at day 65 p.i., when the virus was eliminated from most tissues except for the kidneys (Fig. S6B), 74 ± 5% of P14 IL2Rβ−/− cells expressed CD127, as compared with 21 ± 3% of P14 controls in which CD127 expression remained repressed, in line with previous reports (14, 15) (Fig. 6A and Fig. S6A). We found that 27 ± 2% of P14 IL2Rβ−/− memory T cells (2.6 × 103 cells) efficiently converted to the CD127highCD62Lhigh Tcm phenotype (Fig. 6 B and C), whereas in P14 controls this population barely developed and was significantly inferior numerically (1.2 × 103 cells), with 65% ± 4% of the cells remaining CD127negCD62Lneg (1.6 × 104 cells), a state we refer to as “exhausted T cells” (Tex) (Fig. 6 B and C). Rescued Tcm development in P14 IL2Rβ−/− cells correlated with increased expression of the Tcm-associated transcription factor Bcl6 (32, 33), whereas the levels of Eomes remained equivalent to those in P14 controls (Fig. 6D). Although equivalent numbers of CD127highCD62Lneg T effector memory cells (Tem) were generated with or without of IL2Rβ (Fig. 6C), these cells expressed higher levels of both CD127 and Bcl6 in the absence of IL2Rβ compared with their P14 counterparts (Fig. 6E). Collectively, these results indicate that both the quality and the quantity of the memory T-cell pool generated in the absence of IL2Rβ-dependent signals are greatly enhanced. We show that antigen persistence associated with chronic viral infections does not impede the capacity of CD8+ T cells to differentiate into memory T cells, as previously thought. Rather, we demonstrate that IL-2 and IL-15 are the main mediators of this arrested developmental process during viral persistence. Conventional memory T cells provide long-lasting protection and persist in the host through their ability to respond to the homeostatic cytokine IL-7 (34, 35). We thus tested if enhanced memory generation in the absence of IL2Rβ was sufficient to restore responsiveness to IL-7–dependent signals. We first assessed STAT5 phosphorylation in response to IL-7 and showed that at day 65 P14 IL2Rβ−/− memory T cells efficiently triggered STAT5 phosphorylation compared with P14 controls, although not to the same levels as conventional memory T cells (Fig. 6F). However, the majority of P14 IL2Rβ−/− memory T cells reexpressed the prosurvival molecule Bcl2, a known target of IL-7, and its expression strictly overlapped with that of CD127 (Fig. 6G). Conversely, few P14 control cells coexpressed CD127 and Bcl2, and the level of Bcl2 in CD127+ cells always remained inferior to that in their IL2Rβ−/− counterparts (Fig. 6 G and H). These results indicate that lack of IL2Rβ-dependent signaling reinstated responsiveness to IL-7–mediated homeostatic signals (14, 15), leading to the generation of a stable population of CD127+ IL2Rβ−/− memory T cells over time, in contrast to P14 controls (Fig. 6I). Together, our results strongly demonstrate that IL-2 and IL-15 are the main repressors of memory CD8+ T-cell development during chronic viral infection. The absence of these signals restored Tcm development as well as responsiveness to IL-7–dependent signals and subsequent Bcl2 expression. This finding indicates that chronically stimulated CD8+ T cells are able to overcome the typical antigen addiction that comes with exhaustion and can restore a functional IL-7–STAT5–Bcl2 survival axis when IL2Rβ-dependent signals are interrupted.
Fig. 6.
IL2Rβ deficiency restores CD8+ memory T-cell development. P14 or P14 IL2Rβ−/− chimeric mice were generated as described in Fig. 4. Cells were analyzed in the spleen at day 65 p.i. unless otherwise indicated. (A) CD127 expression on P14 cells (gray histograms) and P14 IL2Rβ−/− cells (black histograms) at the indicated time points. Values indicate the MFI. (B, Upper) Contour plots show the expression of CD62L and CD127 on P14 cells (Left) and P14 IL2Rβ−/− cells (Right). Values in each quadrant indicate the frequency. (Lower) Pie charts show the cumulative frequency of Tcm (CD62LhiCD127hi; red), Tem (CD62LloCD127hi; blue), and Tex (CD62LloCD127lo; green) cells in P14 cells (Left) and P14 IL2Rβ−/− cells (Right). (C) Absolute numbers of Tex (CD62LloCD127lo) (Left), Tem (CD62LloCD127hi (Center), and Tcm (CD62LhiCD127hi) (Right) cells in P14 cells (filled bars) and P14 IL2Rβ−/− cells (open bars). (D) Intracellular expression of Bcl6 and Eomes in P14 cells (gray histograms) and P14 IL2Rβ−/− cells (black histograms). Values indicate the MFI; gray-filled histograms are isotype controls (Eomes) and TCRβ− cells (Bcl6). (E) Expression of CD127 (Left) and Bcl6 (Right) on P14 Tem cells (black bars) and P14 IL2Rβ−/− Tem cells (white bars). (F) Induction of p-STAT5 in P14 cells (black bar) and P14 IL2Rβ−/− cells (white bar) following stimulation with IL-7. Naive P14 cells are represented by the striped bar; antigen-specific memory cells are represented by the red bar. (G, Left) Expression of CD127 and Bcl2 in P14 cells and P14 IL2Rβ−/− cells. Values in contour plots indicate the frequency of double-positive cells. (Right) Cumulative frequency of CD127+Bcl2+ cells in P14 cells (filled squares) and P14 IL2Rβ−/− cells (open squares). (H) Expression of Bcl2 in CD127+ P14 cells (black bar) and P14 IL2Rβ−/− cells (white bar). (I) Absolute numbers of CD127+ P14 cells (Left) and CD127+ P14 IL2Rβ−/− cells (Right) at the indicated time points. *P < 0.05; **P < 0.005; ***P < 0.0005; NS P ≥ 0.05; two-tailed unpaired Student’s t test. Data are pooled from two (G and H) or three (C and I) independent experiments with 6–17 mice per group or are representative of two (D and F) or three (A, B, and E) independent experiments with similar results (n = 2–7 mice per group). Error bars in C and E–I indicate mean ± SEM.
Fig. S6.
IL2Rβ deficiency restores CD127 expression. P14 or P14 IL2Rβ−/− chimeric mice were generated as described in Fig. S4. (A) Frequency of CD127+ P14 cells (black bars) and P14 IL2Rβ−/− cells (white bars) at the indicated time points. (B) Viral titers in the indicated organs at day 65 p.i. from P14 (filled squares) and P14 IL2Rβ−/− (open squares) chimeric mice. **P < 0.005; ***P < 0.0005; NS P ≥ 0.05; two-tailed unpaired Student’s t test. Data are pooled from two (B) or three (A) independent experiments with 5–10 mice per group. Error bars indicate mean ± SEM.
Discussion
The physiological impact of IL-2 and IL-15, two key cytokines involved in the differentiation of CD8+ effector T cells (25, 36), has not been thoroughly evaluated during chronic infections. Here we demonstrate a critical role for these two cytokines in the development of severe CD8+ T-cell exhaustion and the associated abrogation of memory T-cell development. We provide evidence for preferential maintenance of CD122, the IL2Rβ chain common to IL-2 and IL-15, on CD8+ T cells during chronic viral infection in both humans and mice. We show that IL-2– and IL-15–dependent signals direct key aspects of CD8+ T-cell exhaustion including (i) the terminal differentiation of highly exhausted PD-1hi cells, (ii) the expression of multiple inhibitory receptors, in particular 2B4 and Tim3, at the surface of the cells and the associated cellular dysfunctions, and (iii) the arrested CD8 memory T-cell differentiation observed during chronic viral infection. These observations demonstrate a direct role of cytokines, in this case IL-2 and IL-15, in instigating CD8+ T-cell exhaustion during chronic viral infection.
Of primary importance, we identified distinct subsets of exhausted CD8+ T cells expressing high (CD122hi) or low (CD122lo) levels of the IL2Rβ chain, the latter being the progenitors of the former. These subsets had distinct patterns of exhaustion and selectively marked PD-1hi and PD-1int/lo effectors, respectively (7, 18, 23). Hence, we defined CD122 as a reliable biological marker of severe exhaustion and terminal differentiation in both humans and mice with chronic viral infections. Moreover, our observations expanded the understanding of the dynamics involved in the conversion from PD-1int/lo progenitors to PD-1hi terminal effectors. Previous studies demonstrated that the development of PD-1hi effectors was dependent on proliferative events driven by antigen and Eomes (23). Here, IL2Rβ-deficient CD8+ T cells expressed normal or even slightly increased Eomes levels and proliferated during chronicity, albeit at a lower rate than control cells, but did not terminally differentiate into PD-1hi progenies. This result underscored another level of regulation in which IL-2– and IL-15–dependent signals drove the terminal differentiation of PD-1hi effectors, likely through the induction of Blimp-1. This statement is in line with the essential role of this transcriptional repressor in directing CD8+ T-cell terminal differentiation in acute infection models and severe exhaustion in chronic settings (17–19). We propose that conversion to PD-1hi effectors is a two-step process in which antigen first drives the proliferation of PD-1int/lo progenitors. Then, IL-2 and IL-15 sustain this proliferation and cooperate to perfect PD-1hi terminal maturation and exhaustion. This process defines a key role for IL-2 and IL-15 in directing cell-fate decision to PD-1hi terminal differentiation during chronic viral infection.
We further provided evidence for IL2Rβ-dependent signals in supporting TCR-dependent PD-1, LAG-3, and CD160 expression during chronic antigenic stimulation. In addition, these signals are mandatory for the induction of 2B4 and Tim-3 expression at the surface of the cells in conjunction with TCR signals. These results present evidence that distinct regulatory pathways induce inhibitory receptor expression. Previous studies support the dichotomy observed in the regulation of inhibitory receptors (18, 37). Indeed, T-bet directly repressed PD-1 and decreased LAG-3 and CD160 expression on CD8+ T cells while promoting Tim-3 and 2B4 to a lesser extent (37). Blimp-1 deficiency in CD8+ T cells prevented 2B4 induction, but PD-1, LAG-3, and CD160 were less affected (18). These results point to specific inherent signals for the induction of distinct inhibitory receptors at the surface of the cells, fine-tuning the level of inhibition that is needed in different contexts. We unveil a previously unexpected immunoregulatory role for IL-2 and IL-15 in governing the induction of a specific pattern of inhibitory receptors (i.e., 2B4 and Tim-3) on CD8+ T cells during chronic viral infection.
Our data strongly indicate that IL-2 and IL-15 cooperate to sustain the exhaustion of CD8+ T cells during chronic infections. Indeed, despite the early loss of CD25 expression on CD8+ effectors during chronic stimulation, exhausted CD8+ T cells integrate both IL-2– and IL-15–dependent signals at day 21 p.i., likely through the IL2Rβγ complex (38). However, in a setting in which IL-2 production is severely hindered (6, 39), IL-15 phosphorylated STAT5 at day 21 of infection more efficiently than did IL-2, pointing to an increased sensitivity of exhausted T cells to IL-15–dependent signals. IL-2 and IL-15 further supported 2B4 and Tim-3 expression on CD8+ T cells in vitro. Collectively, these results are in agreement with the notion that IL-2 and IL-15 induce similar transcriptional programs in CD8+ T cells (24) and imply that IL-2 and IL-15 act in concert to direct the differentiation of exhausted cells during chronic viral infection. Determining the exact timeframe during which IL-2 and IL-15 program and regulate this process will be an important question to address in the future.
Finally, we demonstrate that IL-2 and IL-15 are responsible for the progressive and irreversible loss of memory development potential that marks CD8+ T cells during chronic viral infection (13–15). IL2Rβ deficiency on CD8+ T cells allowed CD127 reexpression and Tcm development despite viral persistence and reinstated a potent CD127–Stat5–Bcl2 survival axis. Transcriptionally, memory resurgence in IL2Rβ-deficient CD8+ T cells resulted from an imbalance favoring the expression of Bcl6 over Blimp-1, two transcription factors promoting central memory development and terminal differentiation, respectively (17, 32, 40). Similarly, in acute infection settings, CD25−/− and CD122−/− CD8+ T cells rapidly turned to memory precursor effector cells and preferentially developed into Tcm cells through the regulation of the same transcription factors (21, 25, 36, 41). However, the impact of IL-2 and IL-15 in chronic settings is far more dramatic, because it leads to the complete abrogation of memory development. This interesting observation indicates that, although chronic antigenic stimulation is a prerequisite to CD8+ memory T-cell defects (14, 15), IL-2– and IL-15–dependent signals direct the arrested memory differentiation of CD8+ T cells during chronic viral infection.
The lack of memory T-cell development and the intense conversion to PD-1hi terminal effectors during chronic viral infection has dramatic consequences for the long-term outcome of the host. In fact, PD-1hi cells present critical survival defects and have shorter lifespans than PD-1int/lo progenitors (7, 10, 23). These terminal effectors barely proliferate to antigenic stimulation and mediate a poor immune protection upon secondary challenge despite having better killing potential (7, 23). More importantly, prolonged and elevated viral loads in mice and humans continuously enforce the transition to PD-1hi cells, leading to a gradual erosion of the progenitor pool and ultimately to the loss of T-cell immunity (23). The demonstration that IL2Rβ deficiency halts PD-1hi conversion, supports the survival of PD-1int/lo progenitors, and promotes memory T-cell development paves the way to new therapeutic approaches aiming to restore T-cell immunity during chronic viral infection. It identifies salient therapeutic opportunities to limit poor outcomes associated with the loss of CD8+ memory T cells in patients with chronic viral infections by interfering with IL2Rβ-dependent signals. Sustaining an IL-7–Stat5–Bcl2 survival axis would provide considerable survival advantage to antiviral T cells following treatment-induced viral decline and would provide an interesting complementary approach to anti-PDL1 therapies, because a limitation of such treatment is its ineffectiveness in rescuing terminally differentiated PD-1hi cells that represent the majority of the CD8+ T-cell pool (7, 23). Hence, limiting the conversion to PD-1hi cells (i.e., by blocking IL-2– and/or IL-15–dependent signals) might increase the proportion of PD-1int/lo progenitors that are highly responsive to αPDL1 therapy and thereby improve treatment efficacy. Globally, our findings provide promising therapeutic perspectives for the treatment of chronic viral infection.
Here we identified previously unknown immunosuppressive functions for IL-2 and IL-15 on CD8+ T-cell responses during chronic viral infection. These effects include the induction of specific inhibitory receptors, the enforced terminal differentiation of CD8+ effectors, and the suppression of memory potential. These findings provide the next crucial steps to developing innovative therapeutic measures to prevent the dramatic loss of immunity that comes with T-cell exhaustion during chronic viral infection.
Materials and Methods
Mice, Virus, and Infection.
Six-week-old C57BL/6 (B6) mice were obtained from The Jackson Laboratory. CD45.2+ P14 Tg mice bearing the H-2Dbgp33-41–specific TCR were kindly provided by A. Freitas, Institut Pasteur, Paris. RAG2−/− IL2Rβ-deficient P14 Tg mice (CD45.2+) were obtained by breeding P14 mice with RAG2−/− IL2Rβ−/− mice (both strains from The Jackson Laboratory) and were previously shown to generate a monoclonal population of naive CD8+ T cells (25). Adoptive transfer recipients (B6 CD45.1.2) were obtained by breeding Tg− littermates (CD45.2 mice) with CD45.1 B6.SJL mice (The Jackson Laboratory) to ensure genetic compatibility between donor and recipient mice, avoiding rejection of transferred cells. P14 and P14 IL2Rβ−/− chimeric mice were obtained by infusing splenocytes from 6- to 9-wk-old Tg mice containing 104 P14 IL2Rβ+/− or P14 IL2Rβ−/− (CD45.2) cells into 6- to 10-wk-old recipient mice (CD45.1/2) 20 h before infection. To confirm the adoptive transfer of naive cells, the activation status (CD69, CD25, CD62L, Ly6C) of Tg cells was verified before each experiment. LCMV Arm and LCMV Cl-13, kindly provided by Rolf M. Zinkernagel, Zurich University Hospital, Zurich, were produced on BHK-21 and L929 cells, respectively, and were titrated by plaque assays on MC57G cells as previously described (42, 43). Mice were infected i.p with 2 × 105 pfu of LCMV Arm or i.v with 2 × 106 pfu of LCMV Cl-13 to generate chronic infection. All donor and virus-free recipient mice were housed in specific pathogen-free facilities at the Sainte-Justine University Hospital Research Center. Infectious experiments were conducted in accordance with Canadian Council on Animal Care guidelines and were approved by the Institutional Committee for Animal Care in Research of the Sainte-Justine University Hospital Research Center and the Institutional Committee for Animal Care of the Experimental Biological Center of the Institut Armand Frappier, where the experiments were performed.
Study Subjects.
HCV acutely infected subjects were recruited among high-risk HCV-seronegative injection drug users participating in the Montreal Hepatitis C Cohort at the Saint-Luc Hospital of the University of Montreal Health Center as previously described (44). Blood samples were processed and analyzed as previously described (45). The institutional ethics committee at the University of Montreal Hospital Research Center approved the human study (SL05.014). All participants signed informed consent forms upon enrollment, and experiments were performed in accordance with the Declaration of Helsinki.
Flow Cytometry, Intracellular Staining, and Cell Sorting.
Spleen and lymph nodes (axillary and inguinal) harvested from infected mice were filtered on a 100-μM nylon mesh and were treated with NH4Cl for erythrocyte removal. For all experiments, dead cells were stained with fixable LIVE/DEAD Aqua (Thermo Fisher) and were excluded from the analysis. Cells were Fc-blocked (BD), and extracellular staining was performed in 50–100 μL of PBS with 2% (vol/vol) FBS for 20 min on ice before fixation. For cytokine release assays, splenocytes were restimulated for 4 h with cognate gp33 peptide (0.1 mM) in the presence of GolgiStop (BD). Cells then were fixed and permeabilized using the Cytofix/Cytoperm kit (BD) and were stained for IFNγ, TNFα, and IL-2. Intracellular Granzyme B and active caspase 3 were assessed directly ex vivo using the same BD Cytofix/Cytoperm kit (BD). Transcription factors and intracellular proteins were detected using the Foxp3 Fixation/Permeabilization kit (eBioscience). To allow Bim detection, an additional 1-h staining was performed with a PE-conjugated donkey anti-rabbit secondary antibody (Jackson ImmunoResearch). Annexin V/7AAD stainings were realized using the Apoptosis Detection Kit I (BD) in accordance with the manufacturer’s protocols. Phosphorylated STAT5 was detected as follows. Briefly, splenocytes from P14 and P14 IL2Rβ−/− transferred mice were rested for 2–4 h in nude medium at 37 °C as previously described (29). Cells then were stained with LIVE/DEAD Aqua before 30 min of restimulation with IL-2, IL-15 (20 ng/mL) (R&D), or IL-7 (5 ng/mL) (PeproTech) in complete RPMI medium. Splenocytes then were fixed with methanol-free 4% (wt/vol) formaldehyde, permeabilized with 90% (vol/vol) ice-cold methanol, and stained for 1 h at room temperature with surface and p-STAT5 antibodies in staining buffer (BD). Anti-mouse antibodies were purchased from eBioscience [CD8, TCRβ, CD45.1, CD45.2, CD127, CD62L, CD25, CD122, LAG-3, CD160, 2B4, Tim-3 (RMT3-23 clone), CD69, Ly6C, Bcl2, Ki67, Eomes, and Bcl6], BD Bioscience (active-Caspase 3, CD44, CD132, IFNγ, TNFα, IL-2), BioLegend [PD-1 (RMP1-30 clone) and Tim-3 (RMT3-23 clone)], Santa Cruz Biotechnology (T-bet), Thermo Fisher (Granzyme B), and Cell Signaling Technology (p-STAT5, FoxO1, and Bim). H-2Dbgp33 tetramers were kindly provided by F. Lemaître, Institut Pasteur, Paris, and were coupled to ultra-avidin-R-phycoerythrin (Leinco). H-2Dbnp396 tetramers were homemade and coupled to streptavidin R phycoerythrin (Thermo Fisher). Human peripheral blood mononuclear cell (PBMC) samples were stained with MHC class I tetramers carrying the HLA-A2–restricted NS3-1073 epitope (A2/NS3-1073) (NIH Tetramer Core Facility, Emory University, Atlanta, GA). Anti-human antibodies were purchased from BD Bioscience (CD3, CD8, CCR7, CD45RA, CD122, and PD-1) and eBioscience (Eomes). All data were acquired on an LSR FORTESSA II (BD) and were analyzed with FlowJo v9.7.6 (Tree Star). For quantitative PCR analysis, cells were sorted on an FACS Aria II (BD) based on Aqua−CD8+CD44+Tet+ for endogenous responses and additional gating on CD45.2+CD45.1− congenic markers for transferred cells. Purity was routinely >95%.
Adoptive Transfers of CD122hi and CD122lo Subsets.
CD45.2+ mice were infected with LCMV Cl-13, and spleens were harvested at day 15 p.i. Activated CD11a+ PD-1+ CD8+ T cells were sorted based on their differential expression of CD122. CD122hi and CD122lo cells (1 × 106 each) were infused i.v. into infection-matched CD45.1+ recipients and analyzed 1 wk later.
BrdU Treatment.
Mice were i.p. injected daily with 1 mg of BrdU (Sigma-Aldrich) between day 15 and day 30 p.i. Harvested splenocytes were stained with extracellular markers, and BrdU incorporation was measured with the BrdU Flow Kit (BD) in accordance with the manufacturer’s protocol.
In Vitro Stimulations.
For proliferative assay, splenocytes from infected mice at day 35 p.i. were labeled with carboxyfluorescein succinimidyl ester (CFSE) (2 μM) and were cultured with H-2Db–restricted gp33 peptide (0.1 μM) in complete Roswell Park Memorial Institute (RPMI) medium. Proliferation of CD45.2+ P14 and P14 IL2Rβ−/− cells was assessed 72 h later by CFSE dilution. Inhibitory receptor induction was performed by coculturing (ratio 1:1) naive P14 cells with gp33-pulsed DCs (0.1 μM) isolated by magnetic separation with CD11c MicroBeads (Miltenyi). At days 2, 4, and 6 of coculture, P14 cells were numbered, normalized, and cultured back in triplicates in RPMI medium complemented with gp33 peptide (1 nM) alone or in combination with IL-2 (R&D) or IL-15 (R&D) at 10 U/mL.
Quantitative PCR.
Endogenous H-2Dbgp33 and adoptively transferred P14 and P14 IL2Rβ−/− cells were sorted directly in TRIzol (Invitrogen). Total RNA was precipitated by adding chloroform (Sigma-Aldrich), washed with 70% (vol/vol) ethanol, and purified using the RNeasy Micro Kit (Qiagen) according to the manufacturer’s protocol. cDNA was generated with the SuperScript Vilo cDNA Synthesis Kit (Thermo Fisher), and quantitative PCR was realized in two steps using the Brilliant II SYBR Green qRT-PCR Low ROX master mix (Agilent Technologies) on an Mx 3000p quantitative RT-PCR system (Stratagene). PCRs were performed in triplicate for mouse Blimp-1 (Prdm1) (forward, 5′-ACA CAC AGG AGA GAA GCC ACA TGA-3′ and reverse, 5′-TCG AAG GTG GGT CTT GAG ATT GCT-3′) and hypoxanthine guanine phosphoribosyl transferase as internal control (forward, 5′-CTC CTC AGA CCG CTT TTT GC-3′ and reverse, 5′-TAA CCT GGT TCA TCA TCG CTA ATC-3′). For all experiments, mRNA levels are expressed as a fold increase compared with naive P14.
Statistical Analyses.
Statistical significance was determined by a standard Student’s t test or one-way ANOVA nonparametric tests (Dunnett’s t) using ABI Prism 6. Significance was set as *P < 0.05, **P < 0.005, and ***P < 0.0005.
Acknowledgments
We thank A. Freitas for P14 mice; Rolf M. Zinkernagel for LCMV virus; F. Lemaître for gp33 tetramers; F. Le Deist for flow cytometry and cell sorting; V. Abadie for insightful comments; V. Abadie, L. Barreiro, E. Kritikou, and J. Kashul for critical reading of the manuscript; C. Mathieu, E. Tihic, N. Cotta-Grand, E. Tarrab, S. Condotta, and M. Richer for technical assistance; and all members of the animal facilities at Sainte-Justine University Hospital Research Center and the Experimental Biological Centre at Institut Armand Frappier for mouse care. H.D. is supported by Canadian Institutes of Health Research (CIHR) Grant MOP-130469, the Fonds de Recherche du Québec–Santé (FRQS), the Leukemia and Lymphoma Society of Canada, the Cole Foundation, clinician scientists salary awards from the FRQS and the Canadian Child Health Clinician Scientist Program. A.L. received support from CHIR Grant MOP-89797 and holds the Jeanne and J.-Louis Lévesque Research Chair in Immunovirology from the J.-Louis Lévesque Foundation. N.H.S. is supported by CIHR Grant MOP-133680, the FRQS AIDS and Infectious Diseases Network, and an FRQS Chercheurs Boursier-Senior Award. J-C.B. has research scholarships from the University of Montreal, the Sainte-Justine University Hospital Research Center Foundation, and the American Association of Immunologists.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604256113/-/DCSupplemental.
References
- 1.Gray SM, Kaech SM, Staron MM. The interface between transcriptional and epigenetic control of effector and memory CD8⁺ T-cell differentiation. Immunol Rev. 2014;261(1):157–168. doi: 10.1111/imr.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang N, Bevan MJ. CD8(+) T cells: Foot soldiers of the immune system. Immunity. 2011;35(2):161–168. doi: 10.1016/j.immuni.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Decaluwe H, et al. Gamma(c) deficiency precludes CD8+ T cell memory despite formation of potent T cell effectors. Proc Natl Acad Sci USA. 2010;107(20):9311–9316. doi: 10.1073/pnas.0913729107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zehn D, Wherry EJ. Immune memory and exhaustion: Clinically relevant lessons from the LCMV model. Adv Exp Med Biol. 2015;850:137–152. doi: 10.1007/978-3-319-15774-0_10. [DOI] [PubMed] [Google Scholar]
- 5.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15(8):486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol. 2003;77(8):4911–4927. doi: 10.1128/JVI.77.8.4911-4927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackburn SD, Shin H, Freeman GJ, Wherry EJ. Selective expansion of a subset of exhausted CD8 T cells by alphaPD-L1 blockade. Proc Natl Acad Sci USA. 2008;105(39):15016–15021. doi: 10.1073/pnas.0801497105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barber DL, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439(7077):682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 9.Jin HT, et al. Cooperation of Tim-3 and PD-1 in CD8 T-cell exhaustion during chronic viral infection. Proc Natl Acad Sci USA. 2010;107(33):14733–14738. doi: 10.1073/pnas.1009731107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blackburn SD, et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol. 2009;10(1):29–37. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wherry EJ, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27(4):670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Odorizzi PM, Wherry EJ. Inhibitory receptors on lymphocytes: Insights from infections. J Immunol. 2012;188(7):2957–2965. doi: 10.4049/jimmunol.1100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Angelosanto JM, Blackburn SD, Crawford A, Wherry EJ. Progressive loss of memory T cell potential and commitment to exhaustion during chronic viral infection. J Virol. 2012;86(15):8161–8170. doi: 10.1128/JVI.00889-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wherry EJ, Barber DL, Kaech SM, Blattman JN, Ahmed R. Antigen-independent memory CD8 T cells do not develop during chronic viral infection. Proc Natl Acad Sci USA. 2004;101(45):16004–16009. doi: 10.1073/pnas.0407192101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin H, Blackburn SD, Blattman JN, Wherry EJ. Viral antigen and extensive division maintain virus-specific CD8 T cells during chronic infection. J Exp Med. 2007;204(4):941–949. doi: 10.1084/jem.20061937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beltra JC, Decaluwe H. Cytokines and persistent viral infections. Cytokine. 2016;82:4–15. doi: 10.1016/j.cyto.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Rutishauser RL, et al. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31(2):296–308. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shin H, et al. A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity. 2009;31(2):309–320. doi: 10.1016/j.immuni.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kallies A, Xin A, Belz GT, Nutt SL. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity. 2009;31(2):283–295. doi: 10.1016/j.immuni.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 20.Banerjee A, et al. Cutting edge: The transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J Immunol. 2010;185(9):4988–4992. doi: 10.4049/jimmunol.1002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pipkin ME, et al. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 2010;32(1):79–90. doi: 10.1016/j.immuni.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belz GT, Masson F. Interleukin-2 tickles T cell memory. Immunity. 2010;32(1):7–9. doi: 10.1016/j.immuni.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 23.Paley MA, et al. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science. 2012;338(6111):1220–1225. doi: 10.1126/science.1229620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ring AM, et al. Mechanistic and structural insight into the functional dichotomy between IL-2 and IL-15. Nat Immunol. 2012;13(12):1187–1195. doi: 10.1038/ni.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathieu C, et al. IL-2 and IL-15 regulate CD8+ memory T-cell differentiation but are dispensable for protective recall responses. Eur J Immunol. 2015;45(12):3324–3338. doi: 10.1002/eji.201546000. [DOI] [PubMed] [Google Scholar]
- 26.Gong D, Malek TR. Cytokine-dependent Blimp-1 expression in activated T cells inhibits IL-2 production. J Immunol. 2007;178(1):242–252. doi: 10.4049/jimmunol.178.1.242. [DOI] [PubMed] [Google Scholar]
- 27.Xin A, et al. A molecular threshold for effector CD8(+) T cell differentiation controlled by transcription factors Blimp-1 and T-bet. Nat Immunol. 2016;17(4):422–432. doi: 10.1038/ni.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinrichs CS, et al. IL-2 and IL-21 confer opposing differentiation programs to CD8+ T cells for adoptive immunotherapy. Blood. 2008;111(11):5326–5333. doi: 10.1182/blood-2007-09-113050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Staron MM, et al. The transcription factor FoxO1 sustains expression of the inhibitory receptor PD-1 and survival of antiviral CD8(+) T cells during chronic infection. Immunity. 2014;41(5):802–814. doi: 10.1016/j.immuni.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yajima T, et al. IL-15 regulates CD8+ T cell contraction during primary infection. J Immunol. 2006;176(1):507–515. doi: 10.4049/jimmunol.176.1.507. [DOI] [PubMed] [Google Scholar]
- 31.Sanjabi S, Mosaheb MM, Flavell RA. Opposing effects of TGF-beta and IL-15 cytokines control the number of short-lived effector CD8+ T cells. Immunity. 2009;31(1):131–144. doi: 10.1016/j.immuni.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ichii H, Sakamoto A, Kuroda Y, Tokuhisa T. Bcl6 acts as an amplifier for the generation and proliferative capacity of central memory CD8+ T cells. J Immunol. 2004;173(2):883–891. doi: 10.4049/jimmunol.173.2.883. [DOI] [PubMed] [Google Scholar]
- 33.Ichii H, et al. Role for Bcl-6 in the generation and maintenance of memory CD8+ T cells. Nat Immunol. 2002;3(6):558–563. doi: 10.1038/ni802. [DOI] [PubMed] [Google Scholar]
- 34.Schluns KS, Kieper WC, Jameson SC, Lefrançois L. Interleukin-7 mediates the homeostasis of naïve and memory CD8 T cells in vivo. Nat Immunol. 2000;1(5):426–432. doi: 10.1038/80868. [DOI] [PubMed] [Google Scholar]
- 35.Kaech SM, et al. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol. 2003;4(12):1191–1198. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 36.Mitchell DM, Ravkov EV, Williams MA. Distinct roles for IL-2 and IL-15 in the differentiation and survival of CD8+ effector and memory T cells. J Immunol. 2010;184(12):6719–6730. doi: 10.4049/jimmunol.0904089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kao C, et al. Transcription factor T-bet represses expression of the inhibitory receptor PD-1 and sustains virus-specific CD8+ T cell responses during chronic infection. Nat Immunol. 2011;12(7):663–671. doi: 10.1038/ni.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura Y, et al. Heterodimerization of the IL-2 receptor beta- and gamma-chain cytoplasmic domains is required for signalling. Nature. 1994;369(6478):330–333. doi: 10.1038/369330a0. [DOI] [PubMed] [Google Scholar]
- 39.Brooks DG, Teyton L, Oldstone MB, McGavern DB. Intrinsic functional dysregulation of CD4 T cells occurs rapidly following persistent viral infection. J Virol. 2005;79(16):10514–10527. doi: 10.1128/JVI.79.16.10514-10527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui W, Kaech SM. Generation of effector CD8+ T cells and their conversion to memory T cells. Immunol Rev. 2010;236:151–166. doi: 10.1111/j.1600-065X.2010.00926.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Obar JJ, et al. CD4+ T cell regulation of CD25 expression controls development of short-lived effector CD8+ T cells in primary and secondary responses. Proc Natl Acad Sci USA. 2010;107(1):193–198. doi: 10.1073/pnas.0909945107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dutko FJ, Oldstone MB. Genomic and biological variation among commonly used lymphocytic choriomeningitis virus strains. J Gen Virol. 1983;64(Pt 8):1689–1698. doi: 10.1099/0022-1317-64-8-1689. [DOI] [PubMed] [Google Scholar]
- 43.Battegay M, et al. Quantification of lymphocytic choriomeningitis virus with an immunological focus assay in 24- or 96-well plates. J Virol Methods. 1991;33(1-2):191–198. doi: 10.1016/0166-0934(91)90018-u. [DOI] [PubMed] [Google Scholar]
- 44.Grebely J, et al. InC Study Group Cohort profile: The international collaboration of incident HIV and hepatitis C in injecting cohorts (InC3) study. Int J Epidemiol. 2013;42(6):1649–1659. doi: 10.1093/ije/dys167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kared H, Fabre T, Bédard N, Bruneau J, Shoukry NH. Galectin-9 and IL-21 mediate cross-regulation between Th17 and Treg cells during acute hepatitis C. PLoS Pathog. 2013;9(6):e1003422. doi: 10.1371/journal.ppat.1003422. [DOI] [PMC free article] [PubMed] [Google Scholar]