Significance
Trillions of bacteria live in the primate gut, contributing to metabolism, immune system development, and pathogen resistance. Perturbations to these bacteria are associated with metabolic and autoimmune human diseases that are prevalent in Westernized societies. Herein, we measured gut microbial communities and diet in multiple primate species living in the wild, in a sanctuary, and in full captivity. We found that captivity and loss of dietary fiber in nonhuman primates are associated with loss of native gut microbiota and convergence toward the modern human microbiome, suggesting that parallel processes may be driving recent loss of core microbial biodiversity in humans.
Keywords: human microbiome, primate microbiome, dietary fiber, dysbiosis, microbial ecology
Abstract
The primate gastrointestinal tract is home to trillions of bacteria, whose composition is associated with numerous metabolic, autoimmune, and infectious human diseases. Although there is increasing evidence that modern and Westernized societies are associated with dramatic loss of natural human gut microbiome diversity, the causes and consequences of such loss are challenging to study. Here we use nonhuman primates (NHPs) as a model system for studying the effects of emigration and lifestyle disruption on the human gut microbiome. Using 16S rRNA gene sequencing in two model NHP species, we show that although different primate species have distinctive signature microbiota in the wild, in captivity they lose their native microbes and become colonized with Prevotella and Bacteroides, the dominant genera in the modern human gut microbiome. We confirm that captive individuals from eight other NHP species in a different zoo show the same pattern of convergence, and that semicaptive primates housed in a sanctuary represent an intermediate microbiome state between wild and captive. Using deep shotgun sequencing, chemical dietary analysis, and chloroplast relative abundance, we show that decreasing dietary fiber and plant content are associated with the captive primate microbiome. Finally, in a meta-analysis including published human data, we show that captivity has a parallel effect on the NHP gut microbiome to that of Westernization in humans. These results demonstrate that captivity and lifestyle disruption cause primates to lose native microbiota and converge along an axis toward the modern human microbiome.
Humans with metabolic disorders, autoimmune and inflammatory conditions affecting the intestines, colorectal cancer, and infectious diseases often exhibit dysbiosis, a state of microbial imbalance (1). Petersen and Round described three categories of dysbiosis: loss of beneficial microbial organisms, expansion of pathobionts or potentially harmful microorganisms, and loss of overall microbial diversity, all of which can occur simultaneously (2). In addition to the evident effect of lifestyle on the pathophysiology of many diseases (3), there is mounting evidence that an intimate interplay exists between the gut microbiota and the development of diseases, including obesity (4–6), Crohn’s disease and ulcerative colitis (7), diabetes (8–10), nonalcoholic fatty liver disease (11), Kwashiorkor (12), and many others. Thus, understanding how lifestyle affects the development and maintenance of the gut microbiome is an important health issue.
Specific environmental and genetic factors have been linked to the development of dysbiosis, including antibiotic use, stress, geography, race, host genetics, and diet (13, 14). Of these factors, diet, gastrointestinal motility, and medication history most strongly shape the gut microbiome (13, 15–19). Furthermore, previous studies examining dietary patterns suggest a strong association between Western lifestyle and dysbiosis (3, 13, 20), especially considering that in Westernized countries, diet-related chronic diseases are the largest contributors to morbidity and mortality (3, 21), affecting more than 50% of the population. Dysbiosis in Westernized countries is thought to be mainly a result of diet, as the Western diet is evolutionarily discordant from the diet of ancestral humans (3, 21) and tends to be high in fat and animal protein (e.g., red meat), high in sugar, and low in plant-based fiber (3, 13, 20). The consequences for humans of living in a state of evolutionary discordance with the native primate diet may include development of disease, increased morbidity and mortality, and reduction in reproductive success (3, 21).
Nonhuman primates (NHPs) are the most biologically relevant research animal models for humans, as they are our closest living relatives. In this study, we sought to explore the relationship between lifestyle and the gut microbiome in NHPs in an effort to better understand primate conservation and the etiology of modern human dysbiosis. In a unique study design, we examined the microbial communities of two NHP species, the red-shanked douc (Pygathrix nemaeus) and the mantled howling monkey (Alouatta palliata), in different individuals living in captive, semicaptive, and wild conditions. We also examined an additional novel captive NHP population comprising eight different species, in addition to previously published human populations living in the United States (i.e., Western) and in developing nations (i.e., non-Western). Both red-shanked doucs and mantled howling monkeys are folivorous NHPs, consuming a diet that is nutritionally poor and difficult to digest compared with diets consumed by nonfolivores. These species are rarely housed in captivity, in part because of the challenge of replicating their wild diets. Our ability to sample from captive, semicaptive, and wild populations from the same species gave us a unique opportunity to study the relationship between lifestyle and disturbance of the native gut microbiota in primates.
Results
Captivity Reproducibly Alters the Primate Microbiome.
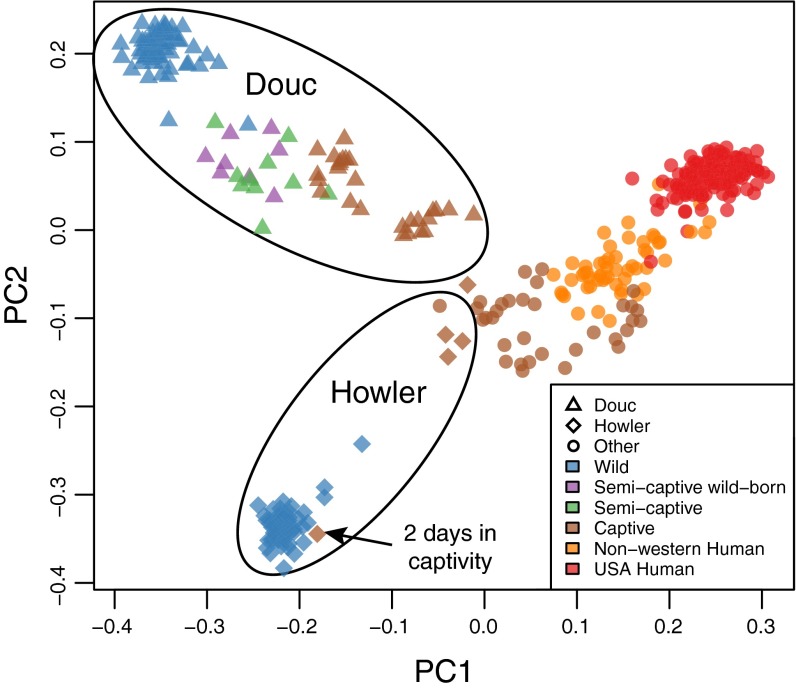
We performed amplicon sequencing of the V4 region of the 16S rRNA gene on fecal samples collected from captive and wild red-shanked doucs (n = 93) and captive and wild mantled howling monkeys (n = 56). We obtained samples from wild doucs in Vietnam and from captive doucs in two zoos in different continents (Southeast Asia, United States). Captive and wild howler samples were collected in Costa Rica. We first examined microbiome composition differences according to captivity status. To determine whether significant differences in gut microbiomes were present between captive and wild populations, we calculated unweighted UniFrac distances between all samples (22). This distance metric has been effective previously for distinguishing both highly divergent and subtly divergent microbial ecosystems (23). Examination of a principal coordinates analysis plot revealed that although the gut microbiomes of wild NHP populations (doucs and howlers) are highly divergent, captivity causes them to converge toward the same composition (Fig. 1 and SI Appendix, Fig. S1). This is true despite the highly distinct diet, gut physiology, and geographical location of the two species, and is true across three independent zoos in three countries. We use unweighted UniFrac distances throughout our analysis, as they provide much better clustering of our experimental data by population than weighted UniFrac or Bray Curtis distances (SI Appendix, Fig. S2). This indicates that the clustering is likely driven by presence or absence of key taxa in different populations, rather than by shifts in the ratios of dominant members of the microbiota.
Fig. 1.
The captive primate microbiome converges toward the modern human microbiome. Principal coordinates plot of unweighted UniFrac distances showing ecological distance between gut microbial communities in wild, semicaptive (from a sanctuary), and captive NHPs, as well as non-Westernized humans, and humans living in the United States (i.e., Westernized). All samples were obtained with the same protocol for V4 16S rRNA sequencing. Although in wild populations the douc and howler microbiomes are highly distinctive, captivity causes them to converge toward the same composition. Semicaptive doucs (green) fall in between wild and captive doucs along the same axis of convergence. The axis of convergence continues toward non-Westernized human populations (Malawi and Venezuela), and finally to the modern US human microbiome. One howler individual resembling wild howlers was sampled after only 2 d in captivity, indicating that the transition to the captive microbiome state requires more than 2 d. Semicaptive doucs born in the wild have similar microbiomes to their captivity-born counterparts, indicating that transition to captivity from the wild is sufficient to produce the captivity-related microbiome.
To confirm this pattern of microbiome convergence, we collected samples from an additional captive population comprising 33 individuals from eight different species, housed at a single zoological institution in the United States (n = 33), different from the US institution housing the sampled doucs. We found that the novel captive population had experienced a similar trend of convergence toward the same captive microbiome state (Fig. 1). To further test the hypothesis that this convergence toward the captive microbiome was a result of the major disruptions to diet and lifestyle associated with captivity, we obtained microbiome samples from 18 individual doucs housed at a primate sanctuary in Vietnam. These “semicaptive” animals are exposed to a less severe form of captivity than the fully captive animals. They are provided with diets consisting mostly of local plants obtained daily from the jungle by caregivers, but these plants do not represent the full diversity of plants the animals would normally eat in the wild, the animals are kept in large caged enclosures, and the animals also have increased exposure to humans relative to wild animals. However, they are not given antibiotics or other medicines and are not fed the manufactured or refined diets typical of zoos. One of the distinguishing features of our analysis of these animals is that they belong to the same species (red-shanked douc) as the wild and captive individuals to which we are comparing them. Notably, we found these animals to have an intermediate level of disruption to the gut microbiota, falling approximately in between the wild doucs and the captive doucs in our distance-based analysis (Fig. 1), suggesting that the level of severity of dietary and lifestyle disruption is associated with the level of disruption to the native gut microbiota. Analysis-of-similarities (ANOSIM) statistical analysis confirmed that NHP gut microbial communities grouped by captivity status (ANOSIM R = 0.69; P = 0.001), and a random-forest classifier obtained 100% cross-validation accuracy at discriminating wild douc, wild howler, semicaptive, and captive groups, with the sole exception of the recently captured howler monkey, which classified with the wild howlers (Fig. 2 and SI Appendix, Figs. S3 and S4). These analyses confirmed that the wild primate populations had unique signature microbiota that were lost in captivity. An additional meta-analysis including data previously published by Muegge et al. also confirmed that captive primates lose their native microbial biodiversity and converge toward a similar perturbed state (SI Appendix, Fig. S5) (17).
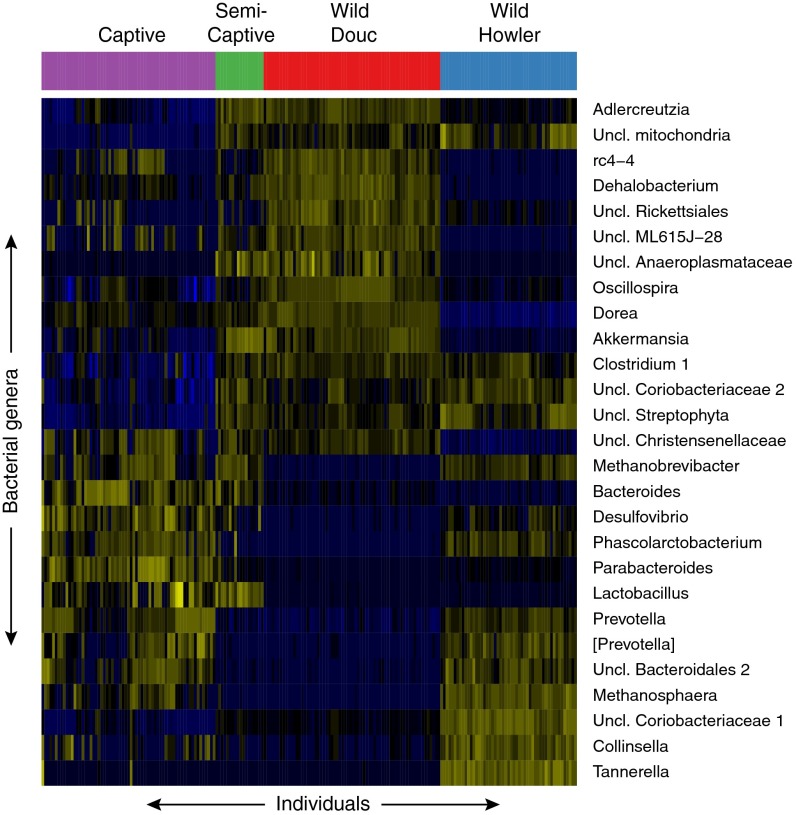
Fig. 2.
Discriminative taxa by species and captivity status. Highly predictive taxa for discriminating the captive, semicaptive, wild douc, and wild howler groups. Predictive taxa included are those with a 0.001 mean decrease in accuracy when removed as determined by the random forests classifier. The classifier was able to correctly predict the group label for unseen samples for all but one sample (99.6% accuracy). Semicaptive individuals show an overlap between some of the wild douc genera and some of the captive animal genera. Captive NHPs contain large blocks of bacterial genera that are absent from wild NHPs, and vice versa, indicating a massive shift in taxonomic composition between the populations. Although wild howlers tended to overlap more with captive NHPs than did the wild doucs, both the doucs and howlers lose most of their native signature gut bacteria in captivity.
The Captive Primate Microbiome Is Characterized by Loss of Diversity.
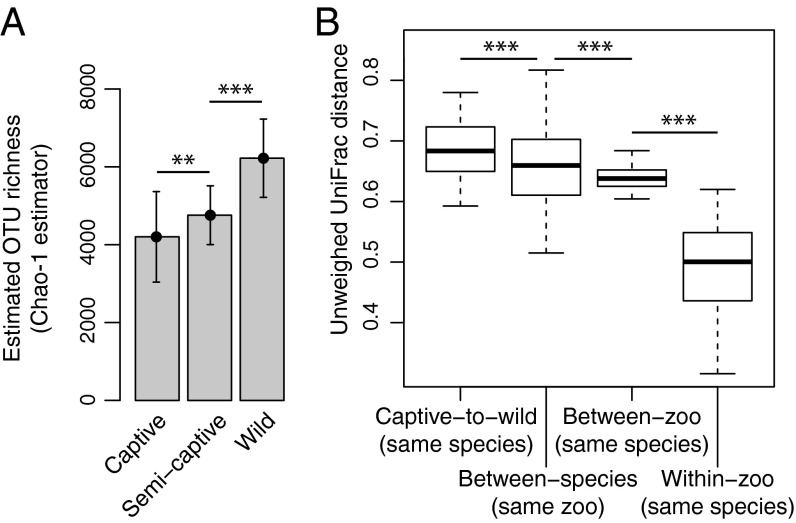
To investigate gut microbial diversity, including previously unknown taxa, we performed open-reference operational taxonomic units (OTUs) picking on the NHP samples. We tested for significant differences in within-individual biodiversity between primates in different living conditions. Adding to previously published results describing decreased NHP diversity in captivity (24), we found that gut microbial diversity decreased in accordance with the severity of captivity, with the highest alpha diversity observed in NHPs living under the most natural conditions (i.e., wild), the lowest alpha diversity seen in the NHPs living under the most unnatural conditions (i.e., captive), and an intermediate number of OTUs seen in the semicaptive individuals (t test wild vs. semicaptive, P = 2.1 × 10−21; semicaptive vs. captive, P = 1.9 × 10−5) (Fig. 3A). Rarefaction curves showed a similar trend (SI Appendix, Fig. S6).
Fig. 3.
Captivity reduces native primate microbiota. (A) Bar plot of mean and spread of gut microbial biodiversity, as measured by the number of species-like OTUs in the gut microbiome, of wild, semicaptive, and captive douc populations of NHPs, using the Chao1 estimator of total OTU richness. This indicates a significant loss of biodiversity from wild populations to semicaptive populations, and again from semicaptive to captive. Error bars indicate SD, and asterisks denote significance at *P < 0.05, **P < 0.01, and ***P < 0.001. (B) Standard box plot of microbiome variation (unweighted UniFrac distance) explained by different experimental factors, showing that captivity in general is associated with a greater change in microbiome state than variation in host species, zoological institution, or individual.
Factors Influencing Captive Primate Microbiome Convergence.
Several major lifestyle changes known to influence the gut microbiome are associated both with captivity in NHPs and with modernization in humans. These changes include dietary shifts, disease, changes in geography, and exposure to modern medicine and hygiene, including antibiotics. We performed several analyses to determine whether these factors, or host genetics, are associated with convergence toward humans and loss of microbiome diversity in captive primates.
First, we examined the relative effects of geography and location on microbiome variation. Including the additional 33 samples from the second US zoo, our samples span four different captive facilities in three different countries, all demonstrating the same trend of convergence toward the modern human microbiome in captive primates. We found that, although there is a strong effect of geography and location on the microbiota of the captive individuals, with interzoo microbiome distances being significantly greater than intrazoo microbiome distances, there is an even greater difference between captive and wild douc microbiomes (Fig. 3B) (permutation test on unweighted UniFrac distances; P < 0.001). We also note that the two captive douc populations are in very divergent geographical locations (United States and Southeast Asia), yet their microbiomes are more similar to each other than to the wild doucs. We confirmed in a meta-analysis, including samples from additional species in additional zoos (17), that although captive animals do cluster together with animals from the same zoo, all zoo animals tend to be more similar to each other, even between zoos, than they are to the wild animals (SI Appendix, Fig. S5). In addition, we note that although the captive and wild howler populations have approximately the same geographical location, we observed significant microbiome perturbations in the captive howlers. Thus, we find that geography and location alone are not sufficient to explain the observed changes in captive NHP microbiomes.
We also considered the effects of host genetics on the microbiome as a potential confounder. However, an important additional component of our study design is that we have controlled for host genetics at the species level by obtaining samples from wild and captive individuals within the same species, replicated across two different species (howler and douc). Although this does not control for within-species individual genetic variation that may cause dysbiosis (14), microbiome variation between captive individuals of the same species is smaller than variation between captive and wild individuals from the same species (Fig. 3B). Interestingly, we do find a correlation between host phylogeny and host microbiome variation within a given zoological institution (permutation test, P < 0.0001) (SI Appendix, Fig. S7), as noted previously in wild NHPs (25). However, this association is potentially confounded by diet and is less strong than the associations of microbiome state with zoological institution and captivity in general. Thus, we find that host genetics, although likely affecting the primate microbiome, has a much smaller effect on the microbiome than does captivity.
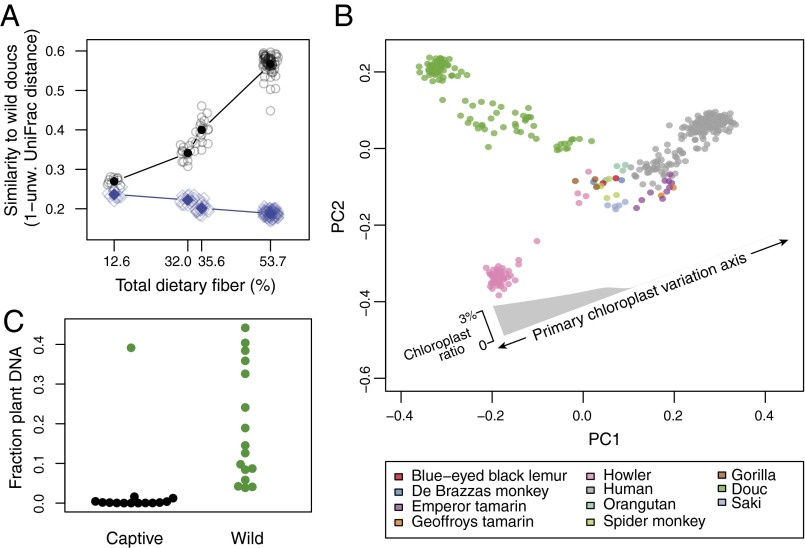
We followed several lines of study to examine the effects of dietary changes on the captive primate microbiome and found evidence that dietary perturbation is likely a primary driver of captive primate microbiome composition. Recent studies in humans and mice have supported the hypothesis that loss of natural dietary fiber causes a loss of native microbial diversity and a shift toward the modern Westernized human microbiome (26, 27). To quantify dietary perturbation in a subpopulation of wild, captive, and semicaptive doucs, we collected samples of typical plants and other dietary components for chemical analysis. We tracked wild doucs over the course of ∼9 mo and observed them feeding on 57 different plant species, whereas the semicaptive doucs were offered 43 different plant species over the course of 1 y. In contrast to the high dietary diversity (i.e., number of plant species) consumed by the wild and semicaptive doucs, the captive doucs were fed far fewer plant species, and thus consumed a much less diverse diet. Specifically, the Southeast Asia zoo doucs were offered ∼15 plant species and the US zoo doucs were offered only one plant species over the course of 1 year. We measured crude protein, crude fat, soluble sugars, acid detergent fiber, and neutral detergent fiber, in addition to several minerals, in the typical diets of each of these populations (wild, semicaptive, captive in Asia, captive in the United States). For neutral detergent fiber content in the wild and semicaptive douc diets, we used previously measured values from Ulibarri (28) and Otto (29), respectively (SI Appendix, Table S1). Although we do not have specific dietary chemical analysis for each individual animal in these populations, dietary content tends to be homogeneous within these captive and wild primate groups (see SI Appendix for detailed discussion of captive primate diet homogeneity). We then tested the hypothesis that decreased total neutral detergent fiber is associated with loss of native microbiota in captive primates and convergence away from the wild douc microbiome (ANOVA, P = 1.4 × 10−74). We found that populations consuming high-fiber diets had microbiomes more similar to those of wild doucs, and populations consuming low-fiber diets had microbiomes more similar to those of modern humans (Fig. 4A). Functional pathways predicted using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) (30) indicated that wild NHP (notably douc) microbiomes possessed increased metabolism of pyruvate, butanoate, glycerophospholipid, and propanoate (random forests feature importance score > 0.01), consistent with increased plant fiber degradation (SI Appendix, Fig. S8). However, these predicted functional profiles should be regarded as suggestive only because of the lack of accounting for unknown species and functional variability within OTU clusters.
Fig. 4.
Dietary fiber and gut microbiome in NHPs. (A) Mean ecological similarity of a given individual douc’s microbiome to all wild douc microbiomes, plotted against that individual’s estimated dietary fiber content (black); same, but showing mean ecological similarity to humans (blue). Doucs consuming more fiber more closely resemble wild doucs in their microbiome; doucs consuming less fiber more closely resemble humans. (B) The same samples plotted in Fig. 1, colored by host species. Also shown is the primary axis of correlation of chloroplast relative abundance, a proxy measurement for dietary raw plant content, with sample positions. A smoothed local regression curve for chloroplast ratio (i.e., relative abundance) along the primary axis of variation shows decreasing raw plant consumption from wild, to semicaptive, to captive primates, with almost no raw plant consumption in the humans or US captive primates. (C) Fraction of whole-genome shotgun data aligning at 97% identity to known plant genomes for 14 captive individuals (9 douc, 5 howler) and 16 wild individuals (8 douc, 8 howler). Wild individuals have a high fraction of plant DNA in their stool; captive individuals have almost none, with the exception of a single outlier individual who was recently rescued from the wild for treatment of electrical burns (also an outlier in Fig. 1).
We also estimated total raw plant dietary content for all NHPs and human subjects, using chloroplast sequences observed in the 16S amplicon sequencing data. We found that chloroplast content was substantial in wild populations (∼3% of all amplicon sequences, on average), but that chloroplast content decreased in semicaptive and captive animals and was nearly completely absent from the human samples and US-based captive primate samples (Fig. 4B). Finally, we performed deep shotgun metagenomic sequencing (28.2 ± 6.6 million sequences per sample) on a subset of 30 captive and wild howlers and doucs and aligned the sequences to known plant reference genomes at 97% identity. Although the existing plant reference genomes were not the same species as those being consumed by the wild primates, we nonetheless identified a large proportion of fecal DNA sequences in the wild primates that were homologous to regions of known cultivated plant genomes, ranging up to ∼40% of all observed sequences. In contrast, we observed almost no plant DNA in the captive samples, with the one exception of an individual howler monkey that had been rescued for electrical burn treatment within the last 48 h who had levels similar to those of the wild animals (Mann–Whitney U test, P < 0.0001) (Fig. 4C). This same animal was also an outlier in the principal coordinates analysis, clustering with the wild howler monkeys (Fig. 1). Plant DNA in captive NHP microbiomes also represented lower dietary plant alpha diversity than in wild NHP microbiomes (Mann–Whitney U test of Shannon index, P < 0.0001) (SI Appendix, Fig. S9).
We considered whether transition to captivity can produce major microbiome perturbations in an adult individual. We obtained birth location records for the semicaptive douc individuals and determined that approximately half of them were born in the wild (eight of 18). This provided us with an opportunity to test the hypothesis that transition to captivity has caused the observed disruption to the natural microbiota in the adult individuals. If the within-individual transition from wild to semicaptive caused only part of the observed disruption to the gut microbiota, then we would expect the wild-born sanctuary-housed doucs to resemble the wild doucs more closely than do those born in the sanctuary. However, we found that wild-born doucs were no closer to wild doucs than were the captive-born doucs (unweighted UniFrac permutation test, P = 0.663) (Fig. 1). This supports the hypothesis that transition to captivity can cause the observed microbiome disruption in these animals and indicates that a substantial part of the captivity-related microbiome perturbation can be acquired within an individual’s lifetime. This may have implications in the study of migration-related dysbiosis in humans.
We examined medical records for differences in antibiotic use and disease between individuals that could be affecting or affected by the microbiome. Sadly, five of the nine captive douc individuals we sampled died of gastrointestinal-related diseases including wasting and gastroenteritis within the following year. We tested whether individuals who died had microbiomes more divergent from the wild douc microbiome than their captive counterparts within the same zoo. We observed a positive trend, but there were only five deceased and four living individuals, and the trend was not significant (t test, P = 0.35) (SI Appendix, Fig. S10). This suggests there may be an association between the captive primate microbiome and disease, but the findings are not conclusive and require larger sample size for proper testing.
Finally, we determined that 11 of the 33 animals sampled from the second US zoo had never been prescribed antibiotics in their lives, based on their complete medical records. In contrast, the remaining 22 individuals had received an average of 4.6 ± 4.0 courses throughout life. We used this information to test whether the microbiomes of the 22 individuals that had received antibiotics more closely resembled modern human microbiomes, but they did not (t test, P = 0.5507), nor were they lower in diversity or less similar to wild primate microbiomes (SI Appendix, Fig. S11). This finding suggests that antibiotics are not a strong driver of the convergence of the captive primate microbiome toward the modern human microbiome.
Captivity in Primates Partially Parallels Modernization in Humans.
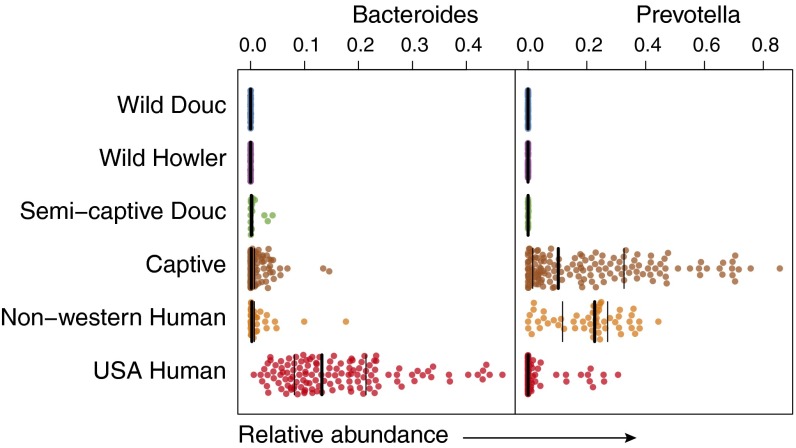
Using standardized methodologies across studies (i.e., sequencing of the V4 region), we were able to conduct a meta-analysis combining NHP populations with previously published samples from adult humans living in both Westernized (United States, n = 129) and non-Westernized (Malawi and Venezuela, n = 21, 34, respectively) countries (23). This analysis demonstrated that the axis of convergence seen in Fig. 1 continues from wild to captive NHPs, and then toward non-Westernized humans, and finally to Westernized humans, suggesting that a similar loss of signature biodiversity seen in captive NHPs has taken place as humans adapted to modern society. Interestingly, we observed higher relative abundances in captive primates of Bacteroides and Prevotella, the two dominant human gut microbiome genera compared with in wild and semicaptive NHPs (Fig. 5). In addition, a higher relative abundance of Bacteroides was seen in Westernized humans compared with non-Westernized humans. The high relative abundance of Bacteroides seen in both captive NHPs and Westernized humans is suggestive that captivity moves captive NHPs in the same direction along the Bacteroides gradient as does Westernization in humans.
Fig. 5.
Captive primates acquire Bacteroides and Prevotella, the dominant genera in the modern human gut microbiome. A bee swarm plot of the arc-sine square root relative abundance of bacterial genera Bacteroides and Prevotella, the two dominant modern human gut microbiome genera, shown in wild, semicaptive, and captive NHPs, as well as in non-Westernized and Westernized humans. As populations shift toward the captive state, their microbiomes become colonized by dominant human gut bacterial genera.
Discussion
Our analysis of species-matched wild, semicaptive, and captive individuals in two different species (red-shanked douc and mantled howler monkey) demonstrates that NHPs lose substantial portions of their signature microbiota in captivity and that they become colonized by human-associated gut bacterial genera Bacteroides and Prevotella. Species in both Bacteroides and Prevotella are capable of polysaccharide degradation, suggesting their emergence may be associated with a shift in diversity or types of dietary polysaccharides, rather than with overall loss of dietary fiber. In an additional captive NHP population, represented by 33 individuals across eight species at an independent zoo, we found that captive rearing caused these individuals to converge toward the same perturbed microbiome state as the captive doucs and howlers. Thus, we found that multiple populations of captive primates converged along an axis leading away from the wild primate microbiome state and toward the modern human microbiome state.
It is important to note that we do not know whether the microbiome perturbations we observe contribute to captive primate disease or are merely a consequence of gastrointestinal disease caused by other factors. However, perturbation of the gut microbiome has known causal and predictive roles in human gastrointestinal diseases, particularly in predisposing individuals to infection (31–33). This suggests there may be a broader role for maintaining keystone gut microbial genetic and functional diversity in the conservation of NHPs, although further research is needed to determine which microbiome perturbations represent dysbiosis, and which are merely benign or even beneficial adaptations to changes in lifestyle and diet. Previous studies have shown that changes in diet are directly associated with shifts in gut microbial community structure (18, 20, 34). By leveraging our study design and zoological medical records, we were able to rule out geography, host genetics, antibiotics exposure, and birth in captivity as the primary determinants of the captive primate microbiome. In contrast, chemical dietary analysis, deep shotgun sequencing, and tracking of fecal chloroplast ratios pointed to loss of dietary plant fiber as a primary driver of captive primate microbiome perturbation.
Recent studies have shown that modern humans have lost a substantial portion of their natural microbial diversity (34–36). Given the massive loss of gut microbiome diversity in captive primates in this study, captive NHPs may provide an informative model for understanding the effects of modernization and mass human migration on the development of human diseases linked to diet and the microbiome, such as obesity and diabetes. Our meta-analysis including Westernized and non-Westernized human microbiomes suggests that loss of dietary fiber may be driving loss of core microbial biodiversity in humans and captive primates alike.
Materials and Methods
Study Subjects and Sample Material.
Fecal samples (n = 251) were collected opportunistically immediately after defecation from two wild, one semicaptive, and three captive NHP populations located around the globe between 2009 and 2013. Ten different NHP species were sampled in this study, including the Red-shanked douc (Pygathrix nemaeus), Mantled howling monkey (Alouatta palliata), Western lowland gorilla (Gorilla gorilla gorilla), Sumatran orangutan (Pongo abelii), De Brazza’s monkey (Cercopithecus neglectus), Black-handed spider monkey (Ateles geoffroyi), White-faced saki (Pithecia pithecia), Blue-eyed black lemur (Eulemur flavifrons), Emperor tamarin (Saguinus imperator), and Geoffroy’s tamarin (Saguinus geoffroyi). A detailed breakdown of population groups is included in the SI Appendix.
The research in this study complied with protocols approved through the University of Minnesota Institutional Animal Care and Use Committee and adhered to the legal requirements of the countries in which it was conducted.
Bacterial 16S rDNA Extraction, Amplification, and Sequencing.
The bacterial 16S rRNA gene was extracted and amplified using primers 515F and 806R, which flanked the V4 hypervariable region of bacterial 16S rRNAs, following the Earth Microbiome Project protocol (37). Amplicons were sequenced on an Illumina MiSeq in 2 × 250 paired-end mode.
Data Analysis.
Raw forward-read sequences were analyzed with QIIME 1.8.0 pipeline (38) with standard settings, but keeping those sequences with 150 bp < length < 1,000 bp and mean quality score > 25. Reverse reads were discarded because of low average sequencing quality (SI Appendix, Fig. S12). Preprocessed sequences were clustered at 97% nucleotide sequence similarity level, using closed reference OTU picking against the Greengenes 13_8 reference database (39) for the meta-analyses ,including published human data, and using open-reference OTU picking for the analysis of alpha diversity in NHPs. All the sequences were rarefied to 14,100 reads per sample for the downstream analysis. Taxonomy assignment, diversity analysis, and principal coordinate analysis were obtained through QIIME with default settings and using custom R scripts. Published data used in our meta-analysis (17, 23, 40, 41) were analyzed similarly. Microbiome-based phylogenies for SI Appendix, Fig. S7 were generated using the ape package in R (42).
Supplementary Material
Acknowledgments
We thank Dr. Michael Murtaugh and Dr. Herbert Covert for critically reading the manuscript and providing feedback; Dr. Lisa Corewyn, Dr. Chris Vinyard, Dr. Susan Williams, Dr. Mark Teaford, and Dr. Marta Salas, who were vital in the collection of individualized fecal samples from the howlers at La Pacifica in Costa Rica; our field assistants, Ai Nguyen Tam and Nguyen Dat, for their help with sample collection and processing in Vietnam; the Endangered Primate Rescue Center, Philadelphia Zoo, Singapore Zoo, and Como Zoo for providing fecal samples from semicaptive and captive nonhuman primates; Tran Van Luong, Nguyen Van Bay, and Nguyen Manh Tien for their permission to work in Son Tra Nature Reserve and for their continued support and help; Kieu Thi Kinh and Thai Van Quang for help in obtaining the research permits; the Department of Forest Protection, the Danang University, and the Son Tra Nature Reserve for granting the research permits; and Christina Valeri and James Collins at the University of Minnesota Veterinary Diagnostic Laboratory for their assistance with acquiring and maintaining shipping permits. This research was funded in part by the Morris Animal Foundation through a Veterinary Student Scholars grant; the University of Minnesota through a College of Veterinary Medicine Travel grant; Merial/NIH through a Summers Scholars grant; the Margot Marsh Biodiversity Foundation; the Mohamed bin Zayed Species Conservation Fund; and the National Institutes of Health through a PharmacoNeuroImmunology Fellowship (NIH/National Institute on Drug Abuse T32 DA007097-32).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the European Bioinformatics Institute database (project no. PRJEB11414).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1521835113/-/DCSupplemental.
References
- 1.Yang Y, Jobin C. Microbial imbalance and intestinal pathologies: Connections and contributions. Dis Model Mech. 2014;7(10):1131–1142. doi: 10.1242/dmm.016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petersen C, Round JL. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol. 2014;16(7):1024–1033. doi: 10.1111/cmi.12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hold GL. Western lifestyle: A ‘master’ manipulator of the intestinal microbiota? Gut. 2014;63(1):5–6. doi: 10.1136/gutjnl-2013-304969. [DOI] [PubMed] [Google Scholar]
- 4.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 5.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3(4):213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gevers D, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15(3):382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giongo A, et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011;5(1):82–91. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown CT, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One. 2011;6(10):e25792. doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boerner BP, Sarvetnick NE. Type 1 diabetes: Role of intestinal microbiome in humans and mice. Ann N Y Acad Sci. 2011;1243:103–118. doi: 10.1111/j.1749-6632.2011.06340.x. [DOI] [PubMed] [Google Scholar]
- 11.Henao-Mejia J, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482(7384):179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith MI, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339(6119):548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawrelak JA, Myers SP. The causes of intestinal dysbiosis: A review. Altern Med Rev. 2004;9(2):180–197. [PubMed] [Google Scholar]
- 14.Knights D, et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 2014;6(12):107. doi: 10.1186/s13073-014-0107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hildebrandt MA, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137(5):1716–1724. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muegge BD, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332(6032):970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falony G, et al. Population-level analysis of gut microbiome variation. Science. 2016;352(6285):560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 19.Zhernakova A, et al. LifeLines cohort study Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352(6285):565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Medina M, et al. Western diet induces dysbiosis with increased E coli in CEABAC10 mice, alters host barrier function favouring AIEC colonisation. Gut. 2014;63(1):116–124. doi: 10.1136/gutjnl-2012-304119. [DOI] [PubMed] [Google Scholar]
- 21.Cordain L, et al. Origins and evolution of the Western diet: Health implications for the 21st century. Am J Clin Nutr. 2005;81(2):341–354. doi: 10.1093/ajcn.81.2.341. [DOI] [PubMed] [Google Scholar]
- 22.Lozupone C, Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uenishi G, et al. Molecular analyses of the intestinal microbiota of chimpanzees in the wild and in captivity. Am J Primatol. 2007;69(4):367–376. doi: 10.1002/ajp.20351. [DOI] [PubMed] [Google Scholar]
- 25.Ochman H, et al. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol. 2010;8(11):e1000546. doi: 10.1371/journal.pbio.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sonnenburg ED, et al. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529(7585):212–215. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Keefe SJD, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun. 2015;6:6342. doi: 10.1038/ncomms7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ulibarri LR. 2013. The socioecology of red-shanked doucs (Pygathrix nemaeus) in Son Tra Nature Reserve, Vietnam. PhD dissertation (University of Colorado, Boulder)
- 29.Otto C. 2005. Food intake, nutrient intake, and food selection in captive and semi-free douc langurs. PhD dissertation (University of Cologne, Cologne)
- 30.Langille MGI, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31(9):814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weingarden A, et al. Dynamic changes in short- and long-term bacterial composition following fecal microbiota transplantation for recurrent Clostridium difficile infection. Microbiome. 2015;3:10. doi: 10.1186/s40168-015-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montassier E, et al. Pretreatment gut microbiome predicts chemotherapy-related bloodstream infection. Genome Med. 2016;8(1):49. doi: 10.1186/s13073-016-0301-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujimura KE, Slusher NA, Cabana MD, Lynch SV. Role of the gut microbiota in defining human health. Expert Rev Anti Infect Ther. 2010;8(4):435–454. doi: 10.1586/eri.10.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martínez I, et al. The gut microbiota of rural Papua New Guineans: Composition, diversity patterns, and ecological processes. Cell Reports. 2015;11(4):527–538. doi: 10.1016/j.celrep.2015.03.049. [DOI] [PubMed] [Google Scholar]
- 35.Moeller AH, et al. Rapid changes in the gut microbiome during human evolution. Proc Natl Acad Sci USA. 2014;111(46):16431–16435. doi: 10.1073/pnas.1419136111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clemente JC, et al. The microbiome of uncontacted Amerindians. Sci Adv. 2015;1(3):e1500183. doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilbert JA, et al. Meeting report: The terabase metagenomics workshop and the vision of an Earth microbiome project. Stand Genomic Sci. 2010;3(3):243–248. doi: 10.4056/sigs.1433550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rollo F, Ermini L, Luciani S, Marota I, Olivieri C. Studies on the preservation of the intestinal microbiota’s DNA in human mummies from cold environments. Med Secoli. 2006;18(3):725–740. [PubMed] [Google Scholar]
- 41.Tito RY, et al. Insights from characterizing extinct human gut microbiomes. PLoS One. 2012;7(12):e51146. doi: 10.1371/journal.pone.0051146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20(2):289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.