Significance
Bacterial motility has been studied for many years, but much less is known about the flagellar system in archaea that provides motility. We have determined the structure of a flagellar-like filament from an archaeon using cryo-EM and can show how it has evolved combining domains from two different protein families. The mechanical properties of the filament are now explained from a computational analysis of the atomic model that we have been able to build. These results provide insights into how motile systems can arise by convergent evolution.
Keywords: archaea, flagellar filaments, helical polymers, cryo-EM
Abstract
The bacterial flagellar apparatus, which involves ∼40 different proteins, has been a model system for understanding motility and chemotaxis. The bacterial flagellar filament, largely composed of a single protein, flagellin, has been a model for understanding protein assembly. This system has no homology to the eukaryotic flagellum, in which the filament alone, composed of a microtubule-based axoneme, contains more than 400 different proteins. The archaeal flagellar system is simpler still, in some cases having ∼13 different proteins with a single flagellar filament protein. The archaeal flagellar system has no homology to the bacterial one and must have arisen by convergent evolution. However, it has been understood that the N-terminal domain of the archaeal flagellin is a homolog of the N-terminal domain of bacterial type IV pilin, showing once again how proteins can be repurposed in evolution for different functions. Using cryo-EM, we have been able to generate a nearly complete atomic model for a flagellar-like filament of the archaeon Ignicoccus hospitalis from a reconstruction at ∼4-Å resolution. We can now show that the archaeal flagellar filament contains a β-sandwich, previously seen in the FlaF protein that forms the anchor for the archaeal flagellar filament. In contrast to the bacterial flagellar filament, where the outer globular domains make no contact with each other and are not necessary for either assembly or motility, the archaeal flagellin outer domains make extensive contacts with each other that largely determine the interesting mechanical properties of these filaments, allowing these filaments to flex.
The bacterial flagellar system has been an object of intense study for many years (1–4). It has helped to elucidate issues of assembly, motility, and chemotaxis at a molecular level in a relatively simple system, typically containing ∼40 different proteins. It has also been the icon for creationists in the United States who deny evolution (5–7). The bacterial flagellar filament, largely composed of a single protein, flagellin, has been fascinating from a structural point of view. In an ideal helical homopolymer, all subunits (excluding those at ends) have identical environments, and the minimum energy conformation of such a filament is a straight rod. However, the rotation of a straight rod generates no thrust, and bacterial flagellar filaments supercoil so as to behave as an Archimedean screw when rotated. The explanation for this supercoiling (8–12) is based on the notion that protofilaments in the filament can exist in two states: long and short. The short protofilaments will form the inside of a supercoil, whereas the long protofilaments will be on the outside. Structural studies of the flagellar filament using X-ray crystallography, fiber diffraction, and cryo-EM have provided a detailed picture of the switching between these two states (13–18).
The proteins that form the bacterial flagellar system have no known homologs in eukaryotic cells. The eukaryotic flagellar, based on a microtubule-containing axoneme, is vastly more complicated. In fact, the current estimate for the number of different proteins in the axoneme is ∼425 (19). In contrast, the archaeal flagellar system appears simpler than the bacterial one and can contain as few as 13 different proteins (20). As with the eukaryotic flagellar system, the archaeal one does not have homology with the bacterial one and must have arisen by means of convergent evolution. In some archaea, the flagellar filament contains mainly one protein, whereas in other archaea, several related proteins are found (21). All of these archaeal flagellins contain an N-terminal domain that is a homolog of the N-terminal domain found in bacterial type IV pilin (T4P) (22), and all contain a short signal sequence at the extreme N terminus that is cleaved by a conserved peptidase to form the mature protein, similar to what exists in T4P. As with the T4P, the highly hydrophobic and conserved N-terminal α-helix exists as a transmembrane helix before the polymeric protein filament is formed. Thus, both T4P and archaeal flagellin are integral membrane proteins, assembling into a filament by a presumed common mechanism where subunits add on at the basal end. In contrast, bacterial flagellar filaments assemble in a completely different manner, where largely unfolded subunits diffuse through the hollow lumen of the filament to add on at the distal end (23).
In addition to the highly conserved N-terminal domain, bacterial T4Ps have a globular domain that shows much more sequence variation. However, in some bacterial pilins, this globular domain can be almost completely absent (24). In archaeal flagellins, however, no homology has yet been found outside of the N-terminal domain with any bacterial or eukaryotic proteins. We have previously described the structure of the Ignicoccus hospitalis Iho670 filament at 7.5-Å resolution (25). Because I. hospitalis has been shown to be nonmotile, these filaments were not called true flagellar filaments and have been called adhesion filaments (26). At a resolution of 7.5 Å, the N-terminal helix was clearly seen, establishing that it is packed differently in these adhesion filaments than in several different packings seen in bacterial T4P filaments (27–30). However, no information about the large globular domain was obtained at this limited resolution, and the sequence of Iho670 showed no homology with any other protein. We have now been able to take advantage of a direct electron detector to reconstruct by cryo-EM at ∼4-Å resolution the Iho670 filament. This reconstruction has allowed us to trace most of the protein chain and establish that the globular domain is a β-sandwich and has the same fold expected for true archaeal flagellins. The Iho670 filament is, thus, a flagellar-like filament and gives us insights at the atomic level into the interfaces that hold this filament together.
Results
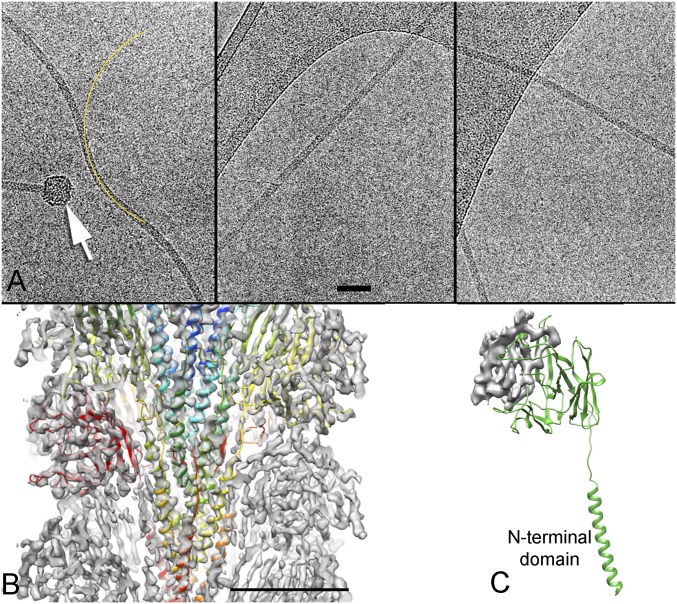
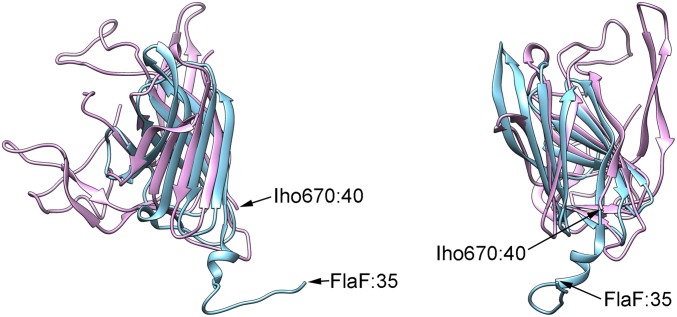
Because the preparation of Iho670 filaments was very dilute, we needed to image from the same grid for 10 d to collect ∼30,000 images, from which ∼300 micrographs were selected that contained one or more filaments (Fig. 1A). No difference was found between images collected on the 1st day and the 10th day, showing that contamination of the grid was not an issue over this long timespan. Using the iterative helical real space reconstruction (IHRSR) method (31), a 3D reconstruction was generated (Fig. 1B), which was estimated (Fig. S1) to have an overall resolution of ∼4 Å. However, the resolution was higher in the core and deteriorated at the very outside of the filament. The overall resolution was high enough (Fig. 2) that an atomic model could be built for most of the 303-residue sequence, except for a region on the periphery (residues 135–195). The model was built mostly automatically with minor manual adjustments using a unique protocol (Materials and Methods). The structure of one subunit consists of an N-terminal helix that is connected by a linker of 8 residues to the globular outer domain containing 264 residues (Fig. 1C). The linker region has very weak density, presumably because of the flexibility in this region. The globular domain contains a β-sandwich fold. The results for the final refined subunit model are shown in Table 1. A Dali server (32) search with our Iho670 structure found closest similarity (Z score = 6.6) to FlaF [Protein Data Bank (PDB) ID code 4P94] from Sulfolobus acidocaldarius (33), an archaeal flagellar protein that has recently been shown to contain an Ig-like β-sandwich in addition to the conserved N-terminal transmembrane helix (Fig. S2), with no other proteins having significant similarity. FlaF is not the actual FlaB flagellar filament protein in S. acidocaldarius but a homolog of FlaB that has been assumed to function as a stator protein that anchors the rotating flagellar filament to the archaeal cell envelope (33). We have compared the observed secondary structures of FlaF and Iho670 with those predicted for a number of true archaeal flagellins, such as FlaB and Mhun_3140 (34) (Fig. S3). This comparison shows that all of these proteins are expected to have a similar globular domain that is largely composed of β-strands.
Fig. 1.
Cryo-EM and 3D reconstruction of Iho670 filaments. (A) Images of frozen hydrated Iho670 filaments. The arrow points to a bulbous structure sometimes seen at one end of the filaments, likely the basal body (60). The yellow line is an arc with a radius of curvature of 1,500 Å. (Scale bar: 500 Å.) (B) A cutaway view of the 3D reconstruction, with a ribbon representation of the atomic model shown in a different color for each subunit. The core of the filament is entirely α-helical, with each subunit contributing a single highly hydrophobic helix. (Scale bar: 50 Å.) (C) A single subunit from the model. The gray density shown is the region where we were unable to build a unique model, accounting for residues 135–195.
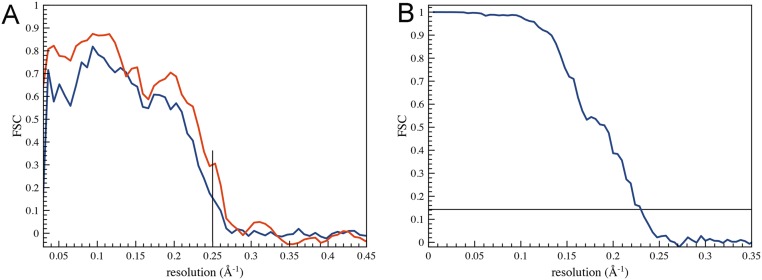
Fig. S1.
The resolution of the reconstruction has been estimated using FSC comparisons (A) between the map and the refined atomic model and (B) between two half-maps generated from independent nonoverlapping datasets. (A) The blue curve is from the comparison between the model and the full map (including the region containing 61 residues where no model has been built), whereas the red curve is from masking off both the model and the map, so that only the α-helical core is being compared. The vertical line suggests that the overall reconstruction is somewhat worse than 4.0 Å, whereas the inner core is somewhat better than 4.0 Å. In B, no masking of the poorly ordered outer region has been done, and the overall resolution using an FSC = 0.143 criterion is 4.3 Å.
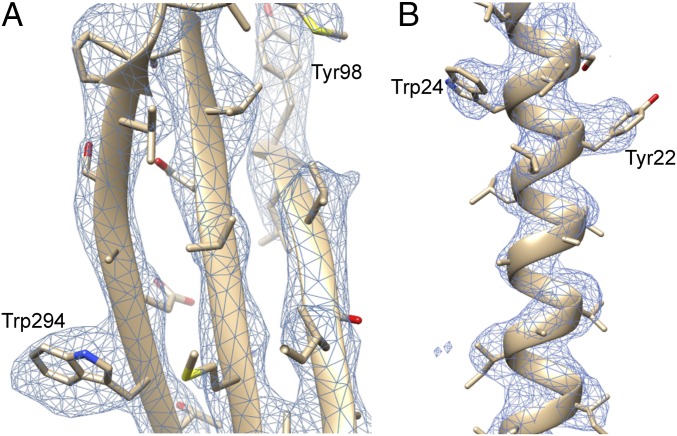
Fig. 2.
Secondary structure is clearly resolved. (A) A β-sheet with two bulky aromatic residues. The presence of such large side chains allows for an unambiguous threading of the sequence through the density map. (B) A portion of the N-terminal α-helix, again showing two clearly resolved bulky aromatics.
Table 1.
Refinement and model statistics
| Category | Result |
| Data collection | |
| No. of overlapping segments | 146,696 |
| No. of unique molecules | ∼300,000 |
| Pixel size (Å) | 1.08 |
| Defocus range (µm) | 0.5–3.5 |
| Voltage (kV) | 300 |
| Electron dose (e− Å−2) | 20 |
| Refinement | |
| Helical symmetry | 106.65°/5.45 Å |
| Resolution (Å) | 4.0 |
| Free shell (Å) | 4.3–4.0 |
| FSCavg (work)/FSCavg (free) | 0.81/0.45 |
| Cwork/Cfree | 0.81/0.43 |
| Rwork/Rfree | 31.8/48.5 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.0018 |
| Bond angles (°) | 0.47 |
| Model quality | |
| Molprobity score | 2.15 (100th percentile) |
| Clash score, all atoms | 7.38 (100th percentile) |
| Good rotamers (%) | 91.7 |
| EMRinger score | 3.42 |
| Ramachandran statistics | |
| Favored (%) | 96.2 |
| Outlier (%) | 1.7 |
Fig. S2.
A comparison between the globular domains of Iho670 (magenta) and FlaF (cyan; PDB ID code 4P94). The hydrophobic N-terminal domain of FlaF was removed before crystallization and is, thus, not available for this comparison, whereas the Iho670 model was truncated at residue 40. The two views are related by a rotation of 80° about the vertical axis.
Fig. S3.
The predicted α-helices (light blue) and β-strands (light red) for a number of archaeal flagellin-like proteins as well as the observed α-helices (dark red) and β-strands (dark blue) for Iho670 (in the text) and Sulfolobus acidocaldarius FlaF (PDB ID code 4P94). The secondary structure predictions were made using the Phyre2 server (61). The N-terminal domain of the FlaF protein was removed for crystallization (33), so that only the predicted secondary structure is available in that region. The sequences shown, from top to bottom, are Iho670 from Ignicoccus hospitalis (iho670; Uniprot A8AAA0), FlaF (flaf_sa; Uniprot Q4J9K8) and FlaB (flab_sa; Uniprot Q4J9K5) from Sulfolobus acidocaldarius; FlaB from Sulfolobus solfataricus (flab_ss; Uniprot Q97WB2); FlaB from Thermoplasma acidophilum (flab_ta; Uniprot Q9HKP4); flgB2 from Haloferax mediterranei (fglb2_hm; Uniprot I3R3X2); flgB2 from Halobacterium salinarum (flgb2_hs; Uniprot B0R4J0); and Mhun_3140 from Methanospirillum hungatei (mhun; Uniprot Q2FUM4). The alignment numbering at the top is arbitrary and does not correspond to any particular sequence.
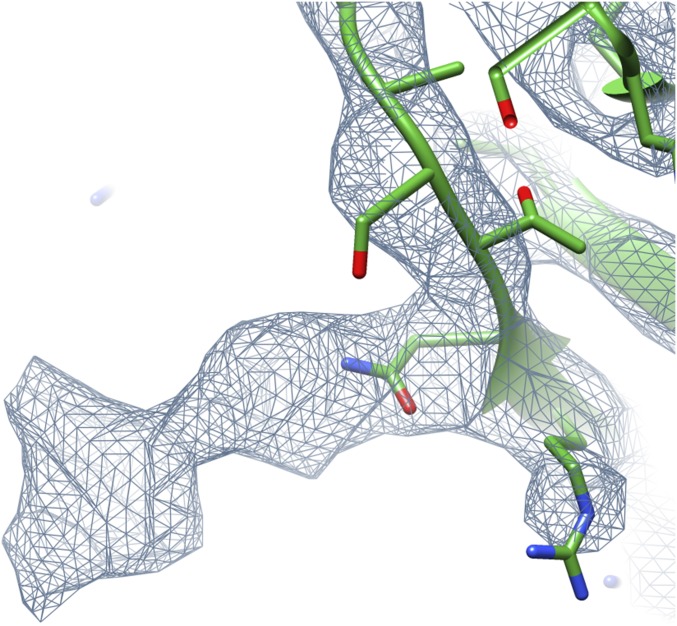
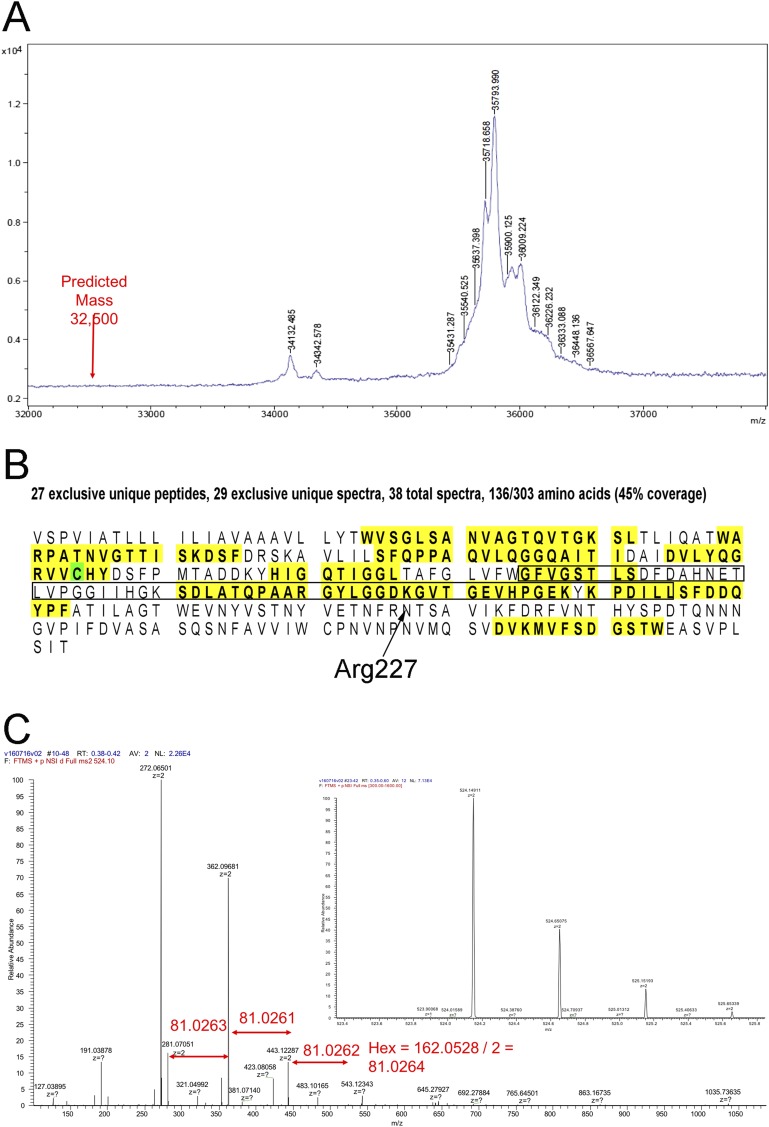
Interestingly, there is significant additional density in the reconstruction at Asn227 (Fig. S4) that suggested glycosylation. This finding would be consistent with previous observations (35, 36) that such glycosylation is widespread in archaeal flagellins, although for the I. hospitalis filament, it was not detected biochemically (26). To test this possibility, we did MS (Fig. S5), which showed that no unmodified protein was present and that the extra mass was from 1.5 to 3.5 kDa. The single largest species present (35,794 Da) corresponded to an additional 3.3 kDa above the predicted molecular mass. Fragmentation analysis of a number of ions showed that hexose molecules were being removed, establishing that some, if not most, of the additional mass was caused by glycosylation. Because the extra mass seen in Fig. S4 might account for only ∼0.5 kDa if filled with sugars, the additional glycosylation is most likely in the region of residues 135–195 that we were not able to trace [specifically in residues 143–160, where there was no peptide coverage (Fig. S5C)], providing additional explanation of why this density is poorly resolved. The modification of Asn227 is likely important for the stability of the flagellar filaments, because it occurs at a pocket between three subunits. There is also a potential intramolecular disulfide bond between Cys104 and Cys271, but the resolution of the map is not good enough to see if this bond actually exists.
Fig. S4.
A tube of extra density projects out from Asn227 into a pocket formed by three subunits. This extra density is consistent with glycosylation, and such glycosylation has been confirmed by MS (Fig. S5).
Fig. S5.
MS of Iho670. (A) MALDI-TOF mass analysis showing that the actual mass is from 1 to 4 kDa higher than that predicted from the protein sequence. The single largest species (35,794 Da) has a mass that is ∼3.3 kDa greater than expected. (B) Digestion of the protein (yellow) shows the unmodified peptides that were identified. Cys104 (green) was determined to be modified with a carbamidomethyl group (57 Da). The region that we were unable to trace (residues 135–195) in building an atomic model is shown within a box. (C) Fragmentation analysis of several ions showed that mass loss involved the removal of hexose, establishing that glycosylation was present.
Interface Analysis.
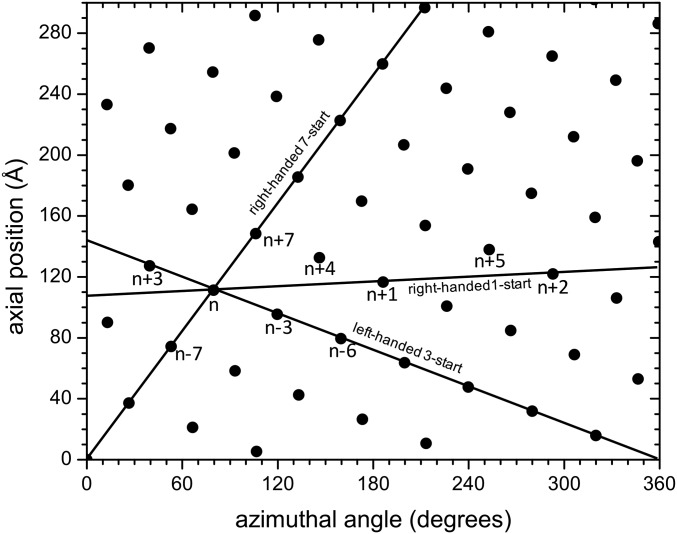
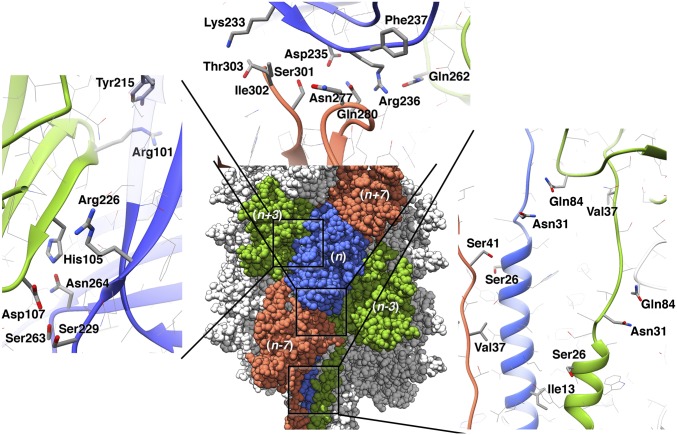
Although the model is missing 61 residues in the peripheral region, it is clear from the density that those residues are not involved in any interactions between the subunits. Therefore, the interfaces between neighboring subunits are fully described in the model and were analyzed using the Rosetta InterfaceAnalyzer as used in the work in ref. 37 (Table 2). For the globular domains, all interfaces are either along the right-handed seven-start helices or the left-handed three-start helices (Fig. 3). The left-handed three-start contacts are formed by subunit n with subunits n + 3 and n − 3, whereas the right-handed seven-start contacts are formed by subunit n with subunits n + 7 and n − 7. The subunit interface along the seven-start helices (Fig. 4) has a larger area, and its interaction energy is stronger than along the left-handed three-start helices. To identify residues in the interfaces that contribute strongly to the interaction, alanine scanning was performed using Rosetta (38, 39). The determined hotspot residues are listed in Table 2, and details of specific subunit interactions are shown in Fig. 4.
Table 2.
Results from the Rosetta InterfaceAnalyzer and Alanine Scanning
| Right-handed seven start | Left-handed three start | |
| No. of residues in interface | 20 | 44 |
| Buried interface area (Å2) | 1,044 | 1,623 |
| Hydrophobic interface area (Å2) | 514 | 754 |
| Polar interface area (Å2) | 550 | 931 |
| Interface energy (Rosetta units) | −5.92 | −7.50 |
| Alanine scanning hotspot residues | Ser26 | Ile13 |
| Val37 | Gln84 | |
| Ser41 | His105 | |
| Asp235 | Asp107 | |
| Arg236 | Tyr215 | |
| Gln280 | Arg226 | |
| Ser301 | Arg236 | |
| Ile302 | Gln262 |
Fig. 3.
The helical net of Iho670 using the conventions that the surface is unrolled and that we are looking from the outside. There is a right-handed one-start helix that passes through every subunit, and subunits are labeled along this helix (e.g., n − 1, n, n + 1, etc.). The outer globular domains only make significant contacts along the right-handed seven-start helices and the left-handed three-start helices, whereas the inner N-terminal helices also make contacts along the one-start helix and a four-start helix.
Fig. 4.
Details of the subunit–subunit interfaces, with residues highlighted that were identified as hotspots by the alanine scanning. A close-up view of the interface with the n + 3 subunit (blue and green) is shown in Left. (Upper) The n/n + 7 interface (blue and orange) involves the C terminus. Arg236 interacts with two subunits (n + 7 and n + 3). The major interactions of the helical core with subunits n + 3 and n + 7 involve hydrophobic contacts (Val37 and Ile13) as well as Ser–Ser hydrogen bonds.
The right-handed seven-start interface is stabilized by hydrogen bonds between Gln280 and Ser301 (n) with Asp235 (n + 7). Also, the C-terminal carboxyl group of Ile302 (n) forms a salt bridge with Lys233 (n + 7). There is a potential backbone hydrogen bond between the carbonyl group of Asn277 (n) and the amino group of Phe237 (n + 7). Arg236 was identified as a hotspot that is relevant for both interfaces and interacts with Gln262 (n − 3) and Gln280 (n − 7). The left-handed three-start interface is stabilized by a hydrogen bond between Asn264 and His105 (n + 3), and Asp107 (n + 3) forms hydrogen bonds with both Ser229 and Ser263 (n). In addition, there is potential hydrogen bonding between Try215 (n) and Arg101 (n + 3).
The hydrophobic core of the filament consists of a bundle of N-terminal helices. Each helix in this bundle contacts eight other helices: n − 7, n − 4, n − 3, n − 1, n + 1, n + 3, n + 4, and n + 7. Thus, each helix makes contacts along the one-, three-, four-, and seven-start helices in contrast to the globular domains that only make contacts along the three- and seven-start helices The results from the interface analysis of one N-terminal helix (residues 1–36) with its neighboring subunits are shown in Table 3. The alanine scanning suggests a particularly stabilizing role of Ile13 in this bundle. Val37 forms a hydrophobic interaction with the N-terminal helix of subunit n + 7, in particular with Leu20 and Leu21. Ser26 stabilizes the interaction of the N-terminal helix with the outer domain of a neighboring subunit via a hydrogen bond with Ser41 on subunit n − 7. Another important interaction of the N-terminal domain is formed by Gln84 in the globular domain, which makes contact with Asn31 of the neighboring subunit (n + 3) along the left-handed three-start helix.
Table 3.
Results from the Rosetta InterfaceAnalyzer for the helical core
| Interface of helical core n with | ||||||||
| n − 7 | n − 4 | n − 3 | n − 1 | n + 1 | n + 3 | n + 4 | n + 7 | |
| No. of residues in interface | 39 | 15 | 29 | 10 | 10 | 21 | 14 | 11 |
| Buried interface area (Å2) | 1,187 | 553 | 1,054 | 352 | 373 | 692 | 568 | 399 |
| Hydrophobic interface area (Å2) | 946 | 508 | 837 | 271 | 294 | 611 | 559 | 392 |
| Polar interface area (Å2) | 241 | 44 | 217 | 81 | 78 | 81 | 9 | 7 |
| Interface energy (Rosetta units) | −4.91 | −5.91 | −2.79 | −2.38 | −1.91 | −4.04 | −5.35 | −1.24 |
The N-terminal helix was cut from one subunit, and the interface with all neighboring (complete) subunits was calculated.
Motion Analysis.
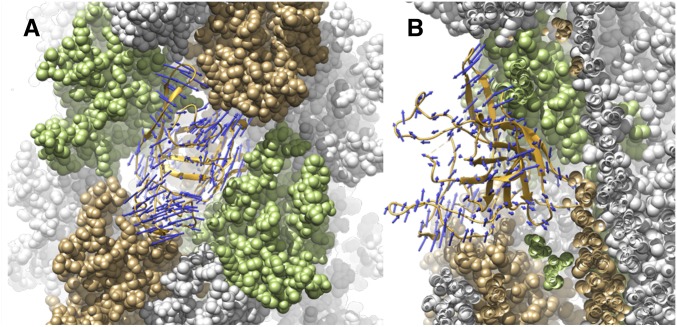
Although bacterial flagellar filaments can generate a fixed supercoil by means of switching between two subunit states (13), the absence of any long-pitch protofilaments in the Iho670 filament raises questions about whether similar switching could exist in archaeal flagellar filaments. However, both theoretical and experimental studies have suggested that helical waveforms can be induced at low Reynold’s number in rotating, semiflexible filaments, and the rotation of these waveforms will generate thrust (40–42). To study the potential conformational motions of the filament, we used the program CONCOORD (43) to generate an ensemble of 400 models. The average rmsd between subunits in this ensemble is ∼4 Å; however, we used only a short filament with 21 subunits, and therefore, the conformational variability could be overestimated, because the motion of a longer filament would be more restrained. The distribution of vertical separations between subunits in the ensemble had an SD of ∼3 Å. We focused on the interaction between the globular domains (because these would dominate the mechanical properties of the filament), and the structures generated in the CONCOORD ensemble were aligned onto a central subunit. This aligned subunit together with the directly contacting four neighbors (n − 3, n + 3, n − 7, and n + 7) were then subjected to principal component analysis. The first eigenvector shows an elongation along the right-handed seven-start helices coupled to a compression of the subunit along the left-handed three-start helices and vice versa (Fig. 5A and Movie S1). Because the left-handed three-start and right-handed seven-start helices have different pitches, compression along one with a similar amount of extension along the other does not cancel, and this motion along the first eigenvector will lead to an overall extension and compression of the filament.
Fig. 5.
Potential deformations of the filament from the CONCOORD analysis. (A) The first eigenvector shows an expansion of a subunit along the right-handed seven-start helix along with a compression along the left-handed three-start helix (and vice versa). This motion induces a change of the length of the filament (Movie S1). (B) The second eigenvector describes an opening and closing of the wedge-shaped subunits, which affect the bending of the filament (Movie S2).
The second eigenvector describes a motion that is related to the bending of the filament. The globular part of the subunit that points toward the core of the filament has a wedge shape (Fig. 5B and Movie S2). The second eigenvector mainly describes a change of the angle of this wedge; the smaller the angle, the more the filament bends.
The radius of curvature of the typical filament shown in Fig. 1A is ∼1,500 Å. The transition from a straight filament to one with such a curvature requires the spacing between subunits on the inside to be compressed and the spacing between subunits on the outside to be expanded by only about 1 Å at a helical radius of 35 Å (the radius of the strongest contacts), which is well within the fluctuations of 3 Å observed by the CONCOORD analysis.
Discussion
We had previously shown (25) that, although the Iho670 filaments contain an N-terminal domain that is a homolog of the N-terminal domain of bacterial T4P, the packing of these hydrophobic helices is different in Iho670 than in either Neisseria gonorrheae (27) or PulG (28, 29). We can now go farther and show that, although this region (residues 2–28) is completely α-helical in Iho670, the corresponding region in Neisseria meningitidis (and by extension, N. gonorrheae, which is identical in sequence in this region) is partially melted when the bacterial filament is formed (44). This observation shows that homology at the level of tertiary structure does not dictate conserved quaternary structure because of the sensitivity of quaternary interactions to small changes in sequence (45, 46). Thus, although we expect that the packing of the N-terminal helices will be conserved across many archaeal flagellar filaments, it is also possible that small sequence changes in this family of proteins may result in different arrangements.
The high-resolution structures obtained for the Salmonella flagellar filament (13–15, 17) arose from using flagellin mutants that failed to supercoil. In general, rotating such a straight and rigid mutant filament does not generate thrust, and bacteria with such a mutation are nonmotile (47). Because archaeal flagellar filaments do not have the long-pitch protofilaments found in bacterial flagellar filaments (48), it has been a mystery how supercoiling might be induced in these filaments. Theoretical and experimental studies of semiflexible filaments (40–42) in highly viscous media have suggested that thrust can be generated by the rotation of such filaments. Our atomic model for the flagellar-like filament from I. hospitalis allows us to predict some of the conformational dynamics of such a filament (Fig. 5), and the curvature seen in micrographs (Fig. 1A) is easily within the accessible predicted conformational fluctuations without requiring large conformational changes. Thus, our structure provides the basis for future studies to understand archaeal flagellar motility in atomic detail.
Materials and Methods
EM.
Samples were prepared as described (25) and applied to plasma-cleaned lacey carbon grids. These grids were verified using a Virobot Mark IV (FEI) and imaged on a Titan Krios operating at a voltage of 300 keV with a Falcon II direct electron detector. The magnification used yielded 1.08 Å per pixel. No dose fractionation was used, and the integrated images corresponded to a dose of ∼20 electrons per 1 Å2. Because the sample was very dilute and because most images did not contain any filaments, >30,000 images were collected, from which 302 were chosen that had one or more filaments. From these images, 220 were selected that showed a good contrast transfer function (CTF). The defocus was measured using CTFFIND3 (49). The phase reversals caused by the CTF were corrected by multiplying each image by the calculated CTF, which is a Wiener filter in the limit of a very poor signal-to-noise ratio. Long filament boxes were extracted using the e2helixboxer routine within EMAN2 (50), and most other operations used the SPIDER software package (51). Overlapping boxes 400 pixels long, with a shift of 10 pixels between adjacent boxes, were cut from the long boxes, yielding 172,230 segments. The IHRSR method (31) was used for the helical reconstruction, and the final volume was from 146,696 segments. The CTF was corrected by dividing the amplitudes of the reconstruction by the sum of the squared CTFs, because the images had been previously multiplied by the CTF twice (once by the microscope and once when phases were corrected).
MS.
Three 5-µL aliquots of the sample were taken for digestion with Lys-C, Asp-N, and chymotrypsin. Each aliquot was heated to 95 °C for 10 min. The sample was reduced with 10 mM DTT in 0.1 M ammonium bicarbonate at room temperature for 0.5 h. The sample was then alkylated with 50 mM iodoacetamide in 0.1 M ammonium bicarbonate at room temperature for 0.5 h. The sample was again heated to 95 °C for 10 min and then, digested overnight at 37 °C with 1 µg enzyme in 50 mM ammonium bicarbonate. The sample was acidified with acetic acid to stop digestion.
The liquid chromatography–MS system consisted of a Thermo Electron Velos Orbitrap ETD Mass Spectrometer System with an Easy Spray Ion Source connected to a Thermo 3-µm C18 Easy Spray Column (through precolumn); 300 fmol each digest was injected, and the peptides were eluted from the column by an acetonitrile/0.1 M acetic acid gradient at a flow rate of 0.25 µL/min over 0.5 h. The nanospray ion source was operated at 2.3 kV. The digest was analyzed using the rapid switching capability of the instrument acquiring a full-scan mass spectrum to determine peptide molecular masses followed by product ion spectra to determine amino acid sequence in sequential scans. The data were analyzed by database searching using the Sequest search algorithm against the known sequence.
Model Building.
The reconstructed density was sharpened using the VISDEM procedure (52) and then, filtered to 3.8 Å before the atomic model building. A map for a single subunit was segmented in Chimera (53) and used for initial model building and refinement of a single chain. To build the backbone trace, beads were placed using the tool dxbeadgen, which is part of the DireX package. The bead positions were refined using DireX, and some outliers were manually adjusted in Chimera. The connectivity was determined using e2pathwalker.py (54) from the EMAN2 package (50), where the density score was slightly modified to enforce connections to be within density. Because the peripheral region of the density had significantly lower resolution, a reliable backbone trace could not be obtained in that area. The final model is, therefore, missing 61 residues (142–202). The full atomic backbone model was built using a unique protocol. Briefly, a backbone fragment library (sequence independent) was built from seven-residue fragments by clustering of all seven-residue fragments from high-resolution X-ray structures mutated to alanine. The fragments from this library were then docked in the neighborhood of each bead position, and the best-fitting fragment was selected. These fragments were merged to yield a complete [all-alanine (all-ALA)] backbone.
The assignment of the sequence to the backbone was done by testing the fit of each amino acid at each residue position. From the resulting scores, a sequence profile was built, which was then aligned to the sequence using the Needleman–Wunsch algorithm to obtain an assignment. The all-ALA model was mutated according to the sequence assignment, and the full model was built using MODELLER, version 9 (55). As an additional test, all 326,668 sequences in SWISSPROT that are longer than 241 residues (the number of residues built in our model) were threaded onto the backbone of our model. For each sequence, a fit score was calculated. The best score obtained from the SWISSPROT sequences is 181.4, whereas the Iho670 sequence yielded a score of 232.3, which both confirms the high quality of the map and provides additional confidence in the correctness of the sequence assignment.
Additional model optimization was performed using the cryo-EM refinement protocol of Rosetta (56). The 20 best models were manually combined with Coot (57) to contain those parts of the models that fit best to the density. The final refinement of the single-chain model was done with CNS (crystallography and NMR System), and the results from this refinement are reported in Table 1. For cross-validation, only the Fourier shells 130.0–4.3 Å were used for the refinement, and the shells 4.3–4.0 Å were used for calculating the Cfree, Rfree, and Fourier shell correlation [FSC; FSCavg(free)] values as described (58). To include subunit interactions, a partial filament structure was built for 21 subunits. The refined subunit models were placed into the unmasked filament density by rigid body docking in Chimera. Rosetta was used to optimize rotamers at the interfaces, and those side chains for which no clear density was visible were adapted to these predicted rotamers. The EMRinger score (59) was calculated (Table 1) to assess the map to model agreement, and it was excellent.
The atomic model has been deposited in the PDB (ID code 5KYH), and the map has been deposited to the Electron Microscopy Data Bank (accession number EMD-8298).
Supplementary Material
Acknowledgments
We thank Michael Thomm, Harald Huber, Thomas Hader, and Konrad Eichinger for support in using the fermentation facility of the Archaea Centre Regensburg to grow Ignicoccus cells at large scale. This work was initially supported by Deutsche Forschungsgemeinschaft Grant WI 731/10-1 (to R.R. and R.W.) and also supported by NIH Grant EB001567 (to E.H.E.). The W. M. Keck Biomedical Mass Spectrometry Laboratory is funded by a grant from the University of Virginia’s School of Medicine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5KYH) and the Electron Microscopy Data Bank (EMDB accession no. EMD-8298).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1607756113/-/DCSupplemental.
References
- 1.Aizawa SI. What is essential for flagellar assembly? In: Jarrell KF, editor. Pili and Flagella: Current Research and Future Trends. Caister Academic Press; Poole, UK: 2009. pp. 91–98. [Google Scholar]
- 2.Toft C, Fares MA. The evolution of the flagellar assembly pathway in endosymbiotic bacterial genomes. Mol Biol Evol. 2008;25(9):2069–2076. doi: 10.1093/molbev/msn153. [DOI] [PubMed] [Google Scholar]
- 3.Larsen SH, Reader RW, Kort EN, Tso WW, Adler J. Change in direction of flagellar rotation is the basis of the chemotactic response in Escherichia coli. Nature. 1974;249(452):74–77. doi: 10.1038/249074a0. [DOI] [PubMed] [Google Scholar]
- 4.Berg HC, Anderson RA. Bacteria swim by rotating their flagellar filaments. Nature. 1973;245(5425):380–382. doi: 10.1038/245380a0. [DOI] [PubMed] [Google Scholar]
- 5.Miller KR. Deconstructing design: A strategy for defending science. Cold Spring Harb Symp Quant Biol. 2009;74:463–468. doi: 10.1101/sqb.2009.74.012. [DOI] [PubMed] [Google Scholar]
- 6.Forrest B, Gross PR. Creationism’s Trojan Horse: The Wedge of Intelligent Design. Oxford University Press; New York: 2004. [Google Scholar]
- 7.Egelman EH. Reducing irreducible complexity: Divergence of quaternary structure and function in macromolecular assemblies. Curr Opin Cell Biol. 2010;22(1):68–74. doi: 10.1016/j.ceb.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Kamiya R, Asakura S, Yamaguchi S. Formation of helical filaments by copolymerization of two types of ‘straight’ flagellins. Nature. 1980;286(5773):628–630. doi: 10.1038/286628a0. [DOI] [PubMed] [Google Scholar]
- 9.Calladine CR. Construction of bacterial flagella. Nature. 1975;255(5504):121–124. doi: 10.1038/255121a0. [DOI] [PubMed] [Google Scholar]
- 10.Kamiya R, Asakura S. Helical transformations of Salmonella flagella in vitro. J Mol Biol. 1976;106(1):167–186. doi: 10.1016/0022-2836(76)90306-5. [DOI] [PubMed] [Google Scholar]
- 11.Asakura S. Polymerization of flagellin and polymorphism of flagella. Adv Biophys. 1970;1:99–155. [PubMed] [Google Scholar]
- 12.Asakura S, Eguchi G, Iino T. Salmonella flagella: In vitro reconstruction and over-all shapes of flagellar filaments. J Mol Biol. 1966;16(2):302–316. doi: 10.1016/s0022-2836(66)80174-2. [DOI] [PubMed] [Google Scholar]
- 13.Maki-Yonekura S, Yonekura K, Namba K. Conformational change of flagellin for polymorphic supercoiling of the flagellar filament. Nat Struct Mol Biol. 2010;17(4):417–422. doi: 10.1038/nsmb.1774. [DOI] [PubMed] [Google Scholar]
- 14.Yonekura K, Maki-Yonekura S, Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424(6949):643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- 15.Samatey FA, et al. Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature. 2001;410(6826):331–337. doi: 10.1038/35066504. [DOI] [PubMed] [Google Scholar]
- 16.Hasegawa K, Yamashita I, Namba K. Quasi- and nonequivalence in the structure of bacterial flagellar filament. Biophys J. 1998;74(1):569–575. doi: 10.1016/S0006-3495(98)77815-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamashita I, et al. Structure and switching of bacterial flagellar filaments studied by X-ray fiber diffraction. Nat Struct Biol. 1998;5(2):125–132. doi: 10.1038/nsb0298-125. [DOI] [PubMed] [Google Scholar]
- 18.Mimori-Kiyosue Y, Vonderviszt F, Yamashita I, Fujiyoshi Y, Namba K. Direct interaction of flagellin termini essential for polymorphic ability of flagellar filament. Proc Natl Acad Sci USA. 1996;93(26):15108–15113. doi: 10.1073/pnas.93.26.15108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pazour GJ, Agrin N, Leszyk J, Witman GB. Proteomic analysis of a eukaryotic cilium. J Cell Biol. 2005;170(1):103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas NA, Bardy SL, Jarrell KF. The archaeal flagellum: A different kind of prokaryotic motility structure. FEMS Microbiol Rev. 2001;25(2):147–174. doi: 10.1111/j.1574-6976.2001.tb00575.x. [DOI] [PubMed] [Google Scholar]
- 21.Cohen-Krausz S, Trachtenberg S. The structure of the archeabacterial flagellar filament of the extreme halophile Halobacterium salinarum R1M1 and its relation to eubacterial flagellar filaments and type IV pili. J Mol Biol. 2002;321(3):383–395. doi: 10.1016/s0022-2836(02)00616-2. [DOI] [PubMed] [Google Scholar]
- 22.Bayley DP, Jarrell KF. Further evidence to suggest that archaeal flagella are related to bacterial type IV pili. J Mol Evol. 1998;46(3):370–373. [PubMed] [Google Scholar]
- 23.Evans LD, Poulter S, Terentjev EM, Hughes C, Fraser GM. A chain mechanism for flagellum growth. Nature. 2013;504(7479):287–290. doi: 10.1038/nature12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reardon PN, Mueller KT. Structure of the type IVa major pilin from the electrically conductive bacterial nanowires of Geobacter sulfurreducens. J Biol Chem. 2013;288(41):29260–29266. doi: 10.1074/jbc.M113.498527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu X, et al. Filaments from Ignicoccus hospitalis show diversity of packing in proteins containing N-terminal type IV pilin helices. J Mol Biol. 2012;422(2):274–281. doi: 10.1016/j.jmb.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Müller DW, et al. The Iho670 fibers of Ignicoccus hospitalis: A new type of archaeal cell surface appendage. J Bacteriol. 2009;191(20):6465–6468. doi: 10.1128/JB.00858-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Craig L, et al. Type IV pilus structure by cryo-electron microscopy and crystallography: Implications for pilus assembly and functions. Mol Cell. 2006;23(5):651–662. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Nivaskumar M, et al. Distinct docking and stabilization steps of the Pseudopilus conformational transition path suggest rotational assembly of type IV pilus-like fibers. Structure. 2014;22(5):685–696. doi: 10.1016/j.str.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campos M, Nilges M, Cisneros DA, Francetic O. Detailed structural and assembly model of the type II secretion pilus from sparse data. Proc Natl Acad Sci USA. 2010;107(29):13081–13086. doi: 10.1073/pnas.1001703107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Egelman EH, Craig L. Structure of the Vibrio cholerae Type IVb Pilus and stability comparison with the Neisseria gonorrhoeae type IVa pilus. J Mol Biol. 2012;418(1-2):47–64. doi: 10.1016/j.jmb.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85(4):225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 32.Holm L, Rosenstrom P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banerjee A, et al. FlaF is a β-sandwich protein that anchors the archaellum in the archaeal cell envelope by binding the S-layer protein. Structure. 2015;23(5):863–872. doi: 10.1016/j.str.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poweleit N. The structure of the Methanospirillum hungatei flagellum as determined by cryo electron microscopy. Biophys J. 2016;110(3):22a. [Google Scholar]
- 35.Ng SY, Chaban B, Jarrell KF. Archaeal flagella, bacterial flagella and type IV pili: A comparison of genes and posttranslational modifications. J Mol Microbiol Biotechnol. 2006;11(3-5):167–191. doi: 10.1159/000094053. [DOI] [PubMed] [Google Scholar]
- 36.Logan SM. Flagellar glycosylation - a new component of the motility repertoire? Microbiology. 2006;152(Pt 5):1249–1262. doi: 10.1099/mic.0.28735-0. [DOI] [PubMed] [Google Scholar]
- 37.Lewis SM, Kuhlman BA. Anchored design of protein-protein interfaces. PLoS One. 2011;6(6):e20872. doi: 10.1371/journal.pone.0020872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kortemme T, Baker D. A simple physical model for binding energy hot spots in protein-protein complexes. Proc Natl Acad Sci USA. 2002;99(22):14116–14121. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kortemme T, Kim DE, Baker D. Computational alanine scanning of protein-protein interfaces. Sci STKE. 2004;2004(219):pl2. doi: 10.1126/stke.2192004pl2. [DOI] [PubMed] [Google Scholar]
- 40.Coq N, Du Roure O, Marthelot J, Bartolo D, Fermigier M. Rotational dynamics of a soft filament: Wrapping transition and propulsive forces. Phys Fluids (1994) 2008;20(5):051703. [Google Scholar]
- 41.Wolgemuth CW, Powers TR, Goldstein RE. Twirling and whirling: Viscous dynamics of rotating elastic filaments. Phys Rev Lett. 2000;84(7):1623–1626. doi: 10.1103/PhysRevLett.84.1623. [DOI] [PubMed] [Google Scholar]
- 42.Tony SY, Lauga E, Hosoi A. Experimental investigations of elastic tail propulsion at low Reynolds number. Phys Fluids (1994) 2006;18(9):091701. [Google Scholar]
- 43.de Groot BL, et al. Prediction of protein conformational freedom from distance constraints. Proteins. 1997;29(2):240–251. doi: 10.1002/(sici)1097-0134(199710)29:2<240::aid-prot11>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 44.Kolappan S, Coureuil M, Yu X, Nassif X, Craig L, Egelman EH. Structure of the Neisseria meingitidis Type IV pilus. Nat Commun. 2016 doi: 10.1038/ncomms13015. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Egelman EH, et al. Structural plasticity of helical nanotubes based on coiled-coil assemblies. Structure. 2015;23(2):280–289. doi: 10.1016/j.str.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galkin VE, et al. Divergence of quaternary structures among bacterial flagellar filaments. Science. 2008;320(5874):382–385. doi: 10.1126/science.1155307. [DOI] [PubMed] [Google Scholar]
- 47.Hyman HC, Trachtenberg S. Point mutations that lock Salmonella typhimurium flagellar filaments in the straight right-handed and left-handed forms and their relation to filament superhelicity. J Mol Biol. 1991;220(1):79–88. doi: 10.1016/0022-2836(91)90382-g. [DOI] [PubMed] [Google Scholar]
- 48.Trachtenberg S, Galkin VE, Egelman EH. Refining the structure of the Halobacterium salinarum flagellar filament using the iterative helical real space reconstruction method: Insights into polymorphism. J Mol Biol. 2005;346(3):665–676. doi: 10.1016/j.jmb.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 49.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142(3):334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 50.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 51.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116(1):190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 52.Spiegel M, Duraisamy AK, Schröder GF. Improving the visualization of cryo-EM density reconstructions. J Struct Biol. 2015;191(2):207–213. doi: 10.1016/j.jsb.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 53.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 54.Baker MR, Rees I, Ludtke SJ, Chiu W, Baker ML. Constructing and validating initial Cα models from subnanometer resolution density maps with pathwalking. Structure. 2012;20(3):450–463. doi: 10.1016/j.str.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 56.DiMaio F, et al. Atomic-accuracy models from 4.5-Å cryo-electron microscopy data with density-guided iterative local refinement. Nat Methods. 2015;12(4):361–365. doi: 10.1038/nmeth.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 58.Falkner B, Schröder GF. Cross-validation in cryo-EM-based structural modeling. Proc Natl Acad Sci USA. 2013;110(22):8930–8935. doi: 10.1073/pnas.1119041110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barad BA, et al. EMRinger: Side chain-directed model and map validation for 3D cryo-electron microscopy. Nat Methods. 2015;12(10):943–946. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meyer C, Heimerl T, Wirth R, Klingl A, Rachel R. The Iho670 fibers of Ignicoccus hospitalis are anchored in the cell by a spherical structure located beneath the inner membrane. J Bacteriol. 2014;196(21):3807–3815. doi: 10.1128/JB.01861-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10(6):845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.