Modern diets and gut microbiome composition
Red-shanked doucs (Pygathrix nemaeus) on Son Tra Peninsula, Vietnam.
Rich in fat, sugar, and animal protein, the quintessential modern Western diet is often deficient in plant-derived fibers and thought to disrupt the delicate balance of human gut microbes. To explore the effects of diet on gut microbiomes, Jonathan Clayton et al. (pp. 10376–10381) sequenced DNA extracted from fecal samples from two primate species—red-shanked douc and mantled howling monkey—that were raised in zoos, sanctuaries, and nature, representing captive, semicaptive, and wild settings, respectively. Despite being raised on vastly divergent diets in zoos as far-flung as Southeast Asia and the United States, captive primates, unlike wild-reared individuals, displayed similar gut microbiome compositions to modern humans, including a predominance of Prevotella and Bacteroides species. Sanctuary-reared primates that were fed a plant-based diet that included some of the plants available to wild-reared primates displayed middling levels of microbiome diversity and disruption, compared with zoo-reared primates of the same species. Further, microbiome disruption was significantly associated with changes in dietary fiber content, but not with factors such as geographic location and antibiotic use, suggesting that diet largely influences microbiome composition in captive primates. According to the authors, the findings underscore the link between fiber-rich diets and gut microbiome diversity. — P.N.
Safe, opioid-like pain relief in monkeys
Opioid analgesics such as morphine, which targets mu opioid peptide (MOP) receptors, carry grave risks, including addiction and respiratory arrest due to abuse or overdose. Recent studies have found that nociceptin/orphanin FQ peptide (NOP) receptor agonists, which are used to block the addictive effects of drugs such as morphine, also interact with the MOP agonist buprenorphine to produce an analgesic effect. Huiping Ding et al. (pp. E5511–E5518) present the analgesic analog BU08028, a ligand that exhibits a receptor-binding profile similar to buprenorphine but with greater affinity and efficacy at NOP receptors. In a series of trials with monkeys, the authors demonstrate that BU08028 targets both MOP and NOP opioid receptors to produce full and lasting pain relief with no corresponding psychological addiction or physical dependence. Furthermore, the study shows that BU08028 does not depress respiratory function or cause cardiac arrest at 10 to 30 times the analgesic dose. Because monkey models faithfully recapitulate human opioid receptor function and drug effects, the findings suggest that mixed MOP/NOP agonists can lead to the development of effective analgesics in humans without risk of abuse and side effects of opioids, according to the authors. — T.J.
Phages in the human gut
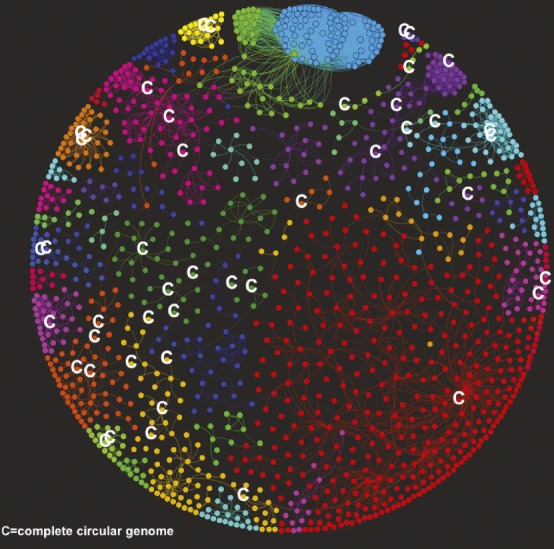
Taxonomic groups of bacteriophages identified in the human gut.
Researchers have previously suggested that most human gut microbial communities are comprised of similar microbial groups that perform comparable functions. However, few studies of healthy human gut microbiomes have focused on bacteriophages, which are viruses that infect bacteria. To investigate whether humans share bacteriophage communities, called phageomes, Pilar Manrique et al. (pp. 10400–10405) analyzed bacteriophage DNA from stool samples of two healthy people, ages 26 and 55, as well as a bacteriophage DNA dataset from a previous study of 62 healthy people and 102 people with ulcerative colitis or Crohn’s disease who lived in Boston, Chicago, or Cambridge, United Kingdom. The authors identified 23 bacteriophages shared by more than half of the 64 healthy people. Compared with healthy people, fewer individuals with gastrointestinal diseases carried the shared bacteriophages. Taxonomic network analyses identified 44 groups of bacteriophages in the healthy people, and more than half of the healthy people shared nine of these groups. According to the authors, common bacteriophage groups may be globally distributed and play a role in maintaining the structure and function of a healthy human gut microbiome. — L.C.



