Significance
The brassinosteroid (BR) signaling pathway has important functions in plant development, but the mechanisms that control its key regulator BR-INSENSITIVE 2 (BIN2) remain poorly understood. We found that the histone deacetylase HDA6 can interact with and deacetylate BIN2 to inhibit its activity, providing significant insights into the repression of BIN2 and the function of the histone deacetylase in modifying nonhistone proteins. We also found that this repression may be related to energy status in plants.
Keywords: HDA6, BIN2, brassinosteroid signaling, deacetylation, development
Abstract
Glycogen synthase kinase 3 (GSK3)-like kinases play important roles in brassinosteroid (BR), abscisic acid, and auxin signaling to regulate many aspects of plant development and stress responses. The Arabidopsis thaliana GSK3-like kinase BR-INSENSITIVE 2 (BIN2) acts as a key negative regulator in the BR signaling pathway, but the mechanisms regulating BIN2 function remain unclear. Here we report that the histone deacetylase HDA6 can interact with and deacetylate BIN2 to repress its kinase activity. The hda6 mutant showed a BR-repressed phenotype in the dark and was less sensitive to BR biosynthesis inhibitors. Genetic analysis indicated that HDA6 regulates BR signaling through BIN2. Furthermore, we identified K189 of BIN2 as an acetylated site, which can be deacetylated by HDA6 to influence BIN2 activity. Glucose can affect the acetylation level of BIN2 in plants, indicating a connection to cellular energy status. These findings provide significant insights into the regulation of GSK3-like kinases in plant growth and development.
Brassinosteroids (BRs), plant-specific steroid hormones, play important roles in development and stress responses (1). In the Arabidopsis thaliana BR signaling pathway, perception of BRs through the plasma membrane receptor BRASSINOSTEROID INSENSITIVE 1 (BRI1) (2) and coreceptor BRI1-ASSOCIATED KINASE 1 (BAK1) (3) results in release of the negative regulator BRI1 KINASE INHIBITOR 1 (BKI1) (4) from the plasma membrane. BRI1 can phosphorylate downstream kinases, such as BR SIGNALING KINASEs (BSKs) and CONSTITUTIVE DIFFERENTIAL GROWTH 1 (CDG1) (5). These kinases regulate the Kelch-repeat phosphatase BRI1 SUPPRESSOR 1 (BSU1), which may dephosphorylate BR-INSENSITIVE 2 (BIN2) to inhibit its kinase activity (6) and lead to the accumulation of the dephosphorylated BRI1-EMS-SUPPRESSOR 1 (BES1) and BRASSINAZOLE RESISTANT 1 (BZR1) (7–9). As the active form, dephosphorylated BES1 regulates BR-responsive gene expression, and the longer form, BES1-L, has stronger activity to promote BR signaling (10).
In the BR signaling pathway, the negative regulator BIN2 is one of 10 glycogen synthase kinase 3 (GSK3)-like kinases (GSK3s) encoded in the A. thaliana genome. BIN2 plays key roles in linking multiple signaling pathways to regulate many aspects of plant development (11–14). The gain-of-function mutant bin2-1 showed a BR-deficient phenotype, including dark green dwarf stature, epinastic round leaves, delayed flowering and senescence, male infertility, activated stress responses, and hypersensitivity to abscisic acid (ABA) (11, 15–17). BIN2 can also regulate auxin signaling by phosphorylating ARF2 to repress its DNA binding activity (18). In addition, BIN2 can enhance ABA signaling by phosphorylating SnRK2.2 and SnRK2.3 to promote their kinase activity (19). BIN2 and its homologs can phosphorylate EGL3 and TTG to promote root hair formation (20), phosphorylate PIF4 to prepare it for degradation in the regulation of hypocotyl elongation (21), and phosphorylate YDA to inhibit its phosphorylation of MAKK4 in the regulation of stomatal development (22).
The regulatory mechanisms by which BRs and other signals regulate BIN2 activity and stability remain unclear. Although in BR signaling it is reported that BIN2 can be regulated by dephosphorylation of the Y200 residue (6), the functions of many other phosphorylation sites remain unknown. In addition, overexpression of Y200 phosphatase BSU1 only partially rescued the BRI1 mutant bri1-116 (6), indicating that other mechanisms may also be involved in the regulation of BIN2 activity. Furthermore, BRs can regulate BIN2 degradation mediated by the 26S proteasome by unknown mechanisms (23). Besides BRs, other signals can also regulate BIN2 activity. For example, BIN2 can be recruited to the plasma membrane by OCTOPUS, a polar-localized plasma membrane-associated protein functioning in phloem differentiation; this relocalization inhibits the activity of BIN2 in the nucleus (24).
In mammals, GSK3-like kinases can be regulated by multiple modifications. Most substrates of GSK3s must be phosphorylated by other kinases to be recognized by the phospho-pocket of GSK3, which includes the R96, R180, and K205 amino acids (25). However, if the N-terminal Ser-9/21 residues of GSK3s are phosphorylated, the N terminus can bind to the pocket to inhibit GSK3 activity (26). Furthermore, phosphorylation of Tyr-279-GSK3α and Tyr-216-GSK3β is important for full activity of GSK3α and -β (26). Other sites on GSK3β, such as Ser-48, -389, and -390, can also be phosphorylated, but their functions remain unknown (27). Besides phosphorylation, other modifications also regulate GSK3 activities, such as cleavage by calpain and by matrix metalloproteinase 2 leading to activated GSK3 fragments, mono-ADP ribosylation leading to GSK3 inhibition, and citrullination leading to nuclear localization of GSK3 (27, 28). Recent work reported that mammalian GSK3β can be acetylated, and depletion of the deacetylase led to reduced phosphorylation and enhanced kinase activity of GSK3β (29). Lysine acetylation is an important posttranslational modification that neutralizes the positively charged lysine residue. Acetylation of many nonhistone proteins plays an important role in regulating biological processes such as the cell cycle, splicing, nuclear transport, metabolism, aging, and several diseases (30–34).
In this study, to explore the regulatory mechanisms of BIN2 in Arabidopsis, we used a mass spectrometry approach and BIN2-FLAG transgenic Arabidopsis to identify a number of BIN2-interacting proteins, including the deacetylase HDA6. We used various genetic and biochemical approaches to confirm that HDA6 can interact with BIN2. Interestingly, we did not find that BIN2 phosphorylates HDA6; rather, we found that HDA6 can deacetylate BIN2 to repress its kinase activity and enhance BR signaling in Arabidopsis. The BIN2 acetylation site is in the conserved phospho-binding pocket of GSK3-like kinases and plays an essential role in regulating BIN2 kinase activity, likely under conditions of energy limitation.
Results
HDA6 Interacts with the BIN2 Kinase.
BIN2 functions as a key negative regulator in the BR signaling pathway and plays an important role in linking many other signaling pathways (11, 19, 35). To identify substrates or regulators of BIN2 kinase, we immunoprecipitated BIN2-FLAG from 35S::BIN2-FLAG transgenic Arabidopsis plants. We then used mass spectrometry to identify a number of potential BIN2-interacting proteins (19), including HDA6. The expression of HDA6 and subcellular localization of HDA6 are very similar to those of BIN2 (36); also, the hda6 knockout mutants and bin2-1 gain-of-function mutant have similar phenotypes, such as late flowering (35, 37), sensitivity to ABA (16, 38), delayed senescence (35, 37), and more root hairs (20, 39). Therefore, we focused on HDA6 for further studies. To confirm the interaction of BIN2 with HDA6, we first used bimolecular fluorescence complementation (BiFC) in the pavement cells of Nicotiana benthamiana and found that BIN2 and HDA6 can interact in the cytoplasm and nucleus (Fig. 1A). Second, we purified the recombinant proteins HDA6-GST and BIN2-His and the kinase-dead BIN2K69R-His from Escherichia coli to conduct GST-pull down assays. We observed that HDA6-GST can directly interact with both forms of BIN2-His (Fig. 1B). Third, we created transgenic plants expressing HDA6-YFP and crossed them with BIN2-FLAG plants to perform coimmunoprecipitation (co-IP), which showed that HDA6-YFP can interact with BIN2-FLAG in planta (Fig. 1C). Taken together, these results suggested that BIN2 can interact with HDA6 in vitro and in vivo.
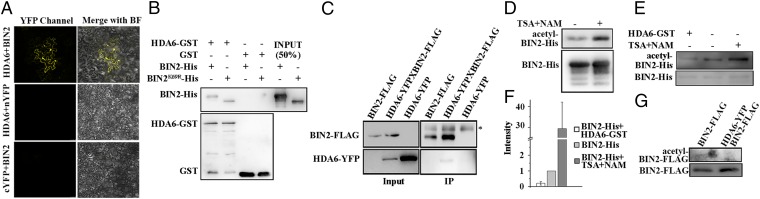
Fig. 1.
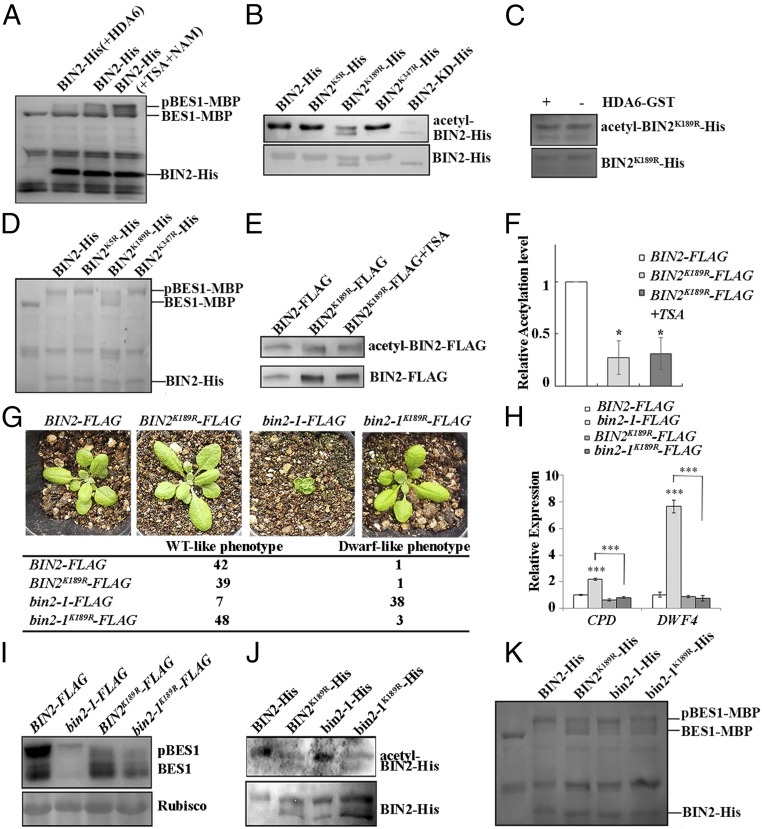
HDA6 interacts with and deacetylates BIN2. (A) HDA6 interacts with BIN2 in BiFC assays. The nYFP-BIN2 or nYFP and HDA6-cYFP or cYFP constructs were cotransformed into the pavement cells of N. benthamiana. BF, bright field; magnification, 20×. (B) HDA6-GST interacts with BIN2-His in GST-pull down assays. (C) HDA6-YFP coimmunoprecipitated by BIN2-FLAG in plants. The asterisk indicates a nonspecific band. (D) The recombinant BIN2-His protein purified from E. coli was acetylated. The E. coli strain containing the BIN2-His construct was cultured in LB medium with or without 5 nM TSA and 5 mM NAM. The acetylation level of the purified BIN2-His was detected with an anti–acetyl-Lys antibody. (E) BIN2-His can be deacetylated by HDA6 in vitro. The acetylation status of BIN2-His was determined with an anti–acetyl-Lys antibody. (F) Statistical analysis of the relative acetylation level of BIN2-His. (G) BIN2-FLAG is acetylated in plants. BIN2-FLAG and HDA6-YFP BIN2-FLAG (crossed from BIN2-FLAG and HDA6-YFP) plants were grown on soil for 2 wk. The acetylation level of the immunoprecipitated BIN2-FLAG protein was detected with an antiacetyl-Lys antibody. Error bars represent SE.
HDA6 Can Deacetylate BIN2.
To investigate the biochemical outcomes of the HDA6–BIN2 interaction, we first conducted in vitro kinase assays to test whether BIN2 can phosphorylate HDA6, because some HDACs in mammals, such as HDAC4, HDAC5, HDAC7, and HDAC9, can be phosphorylated to regulate their subcellular localization and shuttling between the nucleus and cytoplasm (40). However, we did not observe significant phosphorylation of HDA6 by BIN2 (Fig. S1A).
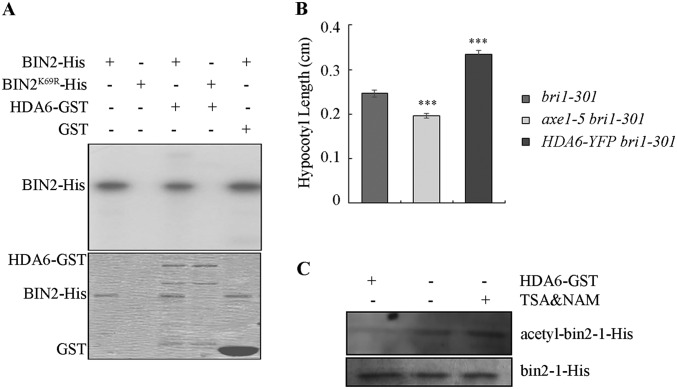
Fig. S1.
HDA6 regulates BR signaling through the BIN2 kinase. (A) BIN2 cannot phosphorylate HDA6. (B) Hypocotyl lengths of dark-grown bri1-301 (n = 25), axe1-5 bri1-301 (n = 25), and HDA6-YFP bri1-301 (n = 25) on medium containing 1 μM BRZ220. (C) HDA6 can deacetylate bin2-1 protein. Error bars represent SE. ***P < 0.001.
Mammalian GSK3β, a close homolog of BIN2, can be acetylated, and the deacetylation of GSK3β can promote dephosphorylation of Ser9 to activate GSK3β kinase activity (29). Therefore, we speculated that BIN2 may be modified by acetylation, and that HDA6 may deacetylate BIN2 to regulate its function. Thus, we purified BIN2-His recombinant protein from E. coli cultured with or without the deacetylation inhibitors trichostatin A (TSA) and β-nicotinamide (NAM). We detected the acetylation status of BIN2-His with an anti–acetyl-lysine antibody. We found that the acetylation level of BIN2-His from the cultures treated with TSA and NAM was higher than that of BIN2-His from the untreated culture (Fig. 1D), indicating that BIN2-His can be acetylated in E. coli. To test whether HDA6 can deacetylate BIN2 in E. coli, we transformed the E. coli strain containing the BIN2-His construct with the HDA6-GST construct. We found that the acetylation level of BIN2-His significantly decreased in the cells producing BIN2-His and HDA6-GST, compared with cells producing only BIN2-His (Fig. 1 E and F). To test whether HDA6 can deacetylate BIN2 in planta, we detected the acetylation level of BIN2-FLAG from transgenic plants harboring BIN2-FLAG alone or both BIN2-FLAG and HDA6-YFP, which we produced by crossing the HDA6-YFP line with BIN2-FLAG plants. We found that the acetylation level of BIN2-FLAG from the HDA6-YFP BIN2-FLAG plants was lower than that of BIN2-FLAG from the plants expressing only BIN2-FLAG (Fig. 1G). These experiments demonstrated that HDA6 can deacetylate BIN2 both in vitro and in vivo.
HDA6 Plays a Positive Role in BR Signaling.
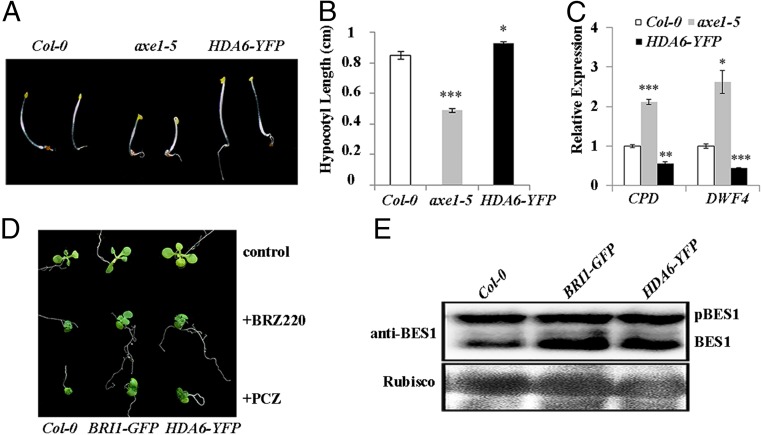
To explore the function of BIN2 deacetylation by HDA6, we conducted a series of biochemical and genetic experiments to test the effect of HDA6 on BIN2 function and the BR signaling outputs. First, we checked whether the HDA6 knockout mutant or the HDA6 overexpression line showed altered BR-related phenotypes. Using the hypocotyl elongation assay, we discovered that the axe1-5 mutant, which has an HDA6 loss-of-function mutation (41), had shorter hypocotyls compared with wild-type Col-0, and that the HDA6-YFP plants had longer hypocotyls (Fig. 2 A and B). We further measured the transcript levels of the BR-responsive genes CPD (CONSTITUTIVE PHOTOMORPHOGENIC DWARF) and DWF4 (DWARF4) by quantitative real-time RT-PCR (qRT-PCR) and found that their expression was up-regulated in the axe1-5 mutant and down-regulated in the HDA6-YFP line, compared with the wild-type (Fig. 2C). We also found that more dephosphorylated BES1 accumulated in the HDA6-YFP plants, similar to the BRI1-GFP overexpression line (Fig. 2E). Finally, we treated the HDA6-YFP seedlings with the BR synthesis inhibitors brassinozole 220 (BRZ220) or propiconazole and found that, like the BRI1-GFP overexpression line, the HDA6-YFP plants showed less growth inhibition than the wild-type plants (Fig. 2D). These data suggested that HDA6 plays a positive role in the BR signaling pathway.
Fig. 2.
HDA6 positively regulates BR signaling. (A) Dark-grown hypocotyl phenotypes of Col-0, the axe1-5 mutant, and the HDA6-YFP overexpression line. (B) Hypocotyl lengths of the dark-grown Col-0 (n = 25), axe1-5 (n = 25), and HDA6-YFP (n = 25). (C) Expression levels of the BR-responsive genes CPD and DWF4. Their expression level in Col-0 was defined as “1.” (D) The seedling phenotype of Col-0, BRI1-GFP, and HDA6-YFP plants grown on medium containing 1 μM BR synthetic inhibitor BRZ220 and propiconazole (PCZ). (E) Phosphorylation status of BES1 in Col-0, BRI1-GFP, and HDA6-YFP plants. BES1 was detected by an anti-BES1 antibody. Error bars represent SE. *P < 0.05, **P < 0.01, and ***P < 0.001.
HDA6 Regulates BR Signaling Through GSK3s.
To test whether the regulation of BR signaling by HDA6 acts through BIN2 and its homologs, we first used the hypocotyl elongation assay to examine the effect of the deacetylation inhibitor TSA on BRI1-GFP, bin2-3 bil1 bil2, and BES1-D plants. We found that the BRI1-GFP line, which overexpresses the BR receptor BRI1, was more sensitive to TSA compared with the wild-type. By contrast, the bin2-3 bil1 bil2 triple mutant, which knocks out BIN2 and its close homologs BIL1 and BIL2, and the BES1-D line, which is a gain-of-function mutant of BR transcription factors, were less sensitive to TSA in hypocotyl elongation compared with their wild-type counterparts (Fig. 3 A–C), indicating that HDA6 may function downstream of the BR receptor BRI1 and upstream or through BIN2 to modulate BR signaling.
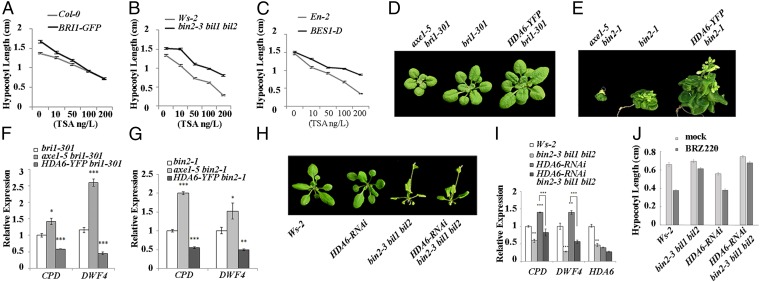
Fig. 3.
HDA6 regulates BR signaling through BIN2 and its homologs. (A–C) Hypocotyl length of Col-0 and the BRI1-GFP line (A), Ws-2 and bin2-3 bil1 bil2 plants (B), and En-2 and bes1-D plants (C). Plants were grown in the dark on medium containing different concentrations of TSA. (D) HDA6 partially rescued the dwarf phenotype of bri1-301. (E) HDA6 can partially suppress the dwarf phenotype of bin2-1. (F) Expression levels of the BR-responsive genes CPD and DWF4 in bri1-301, axe1-5 bri1-301, and HDA6-YFP bri1-301. Their expression in bri1-301 was defined as 1. (G) Expression levels of the BR-responsive genes CPD and DWF4 in bin2-1, axe1-5 bin 2-1, and HDA6-YFP bin 2-1. Their expression in bin-2-1 was defined as 1. (H) HDA6-RNAi cannot suppress the seedling phenotype of the bin2-3 bil1 bil2 triple mutant. (I) Expression levels of CPD, DWF4, and HDA6 in bin2-3 bil1 bil2 and HDA6-RNAi bin2-3 bil1 bil2 plants. Their expression level in Ws-2 was defined as 1. (J) HDA6-RNAi hardly affects hypocotyl elongation in the bin2-3 bil1 bil2 background growing on medium containing 1 μM BRZ220 in the dark. Error bars represent SE. *P < 0.05, **P < 0.01, and ***P < 0.001.
To further test whether the HDA6-enhanced BR signaling was mediated by BIN2 or other components in the BR signaling pathway, we conducted genetic analysis, generating double or multiple mutants of HDA6-related and BR-related mutants and overexpression lines. We first generated double mutants of axe1-5 or HDA6-YFP with the BR receptor mutant bri1-301. We found that the axe1-5 bri1-301 seedlings were smaller than the bri1-301 seedlings, and that the HDA6-YFP bri1-301 seedlings were larger than the bri1-301 seedlings (Fig. 3D). The expression of the BR-responsive genes CPD and DWF4 was enhanced in the axe1-5 bri1-301 double mutants but repressed in the HDA6-YFP bri1-301 double mutants, compared with bri1-301 (Fig. 3F). Because bri1-301 is a weak allele of bri1, to mimic a null allele of bri1, we grew bri1-301, axe1-5 bri1-301, and HDA6-YFP bri1-301 on medium containing a BR biosynthetic inhibitor, BRZ220, and measured the hypocotyl length of the dark-grown seedlings. We found that the hypocotyl length of axe1-5 bri1-301 is still shorter than that of bri1-301, and that the hypocotyl length of HDA6-YFP bri1-301 is still longer than that of bri1-301 (Fig. S1B). These results support that HDA6 acts downstream of BRI1 to modulate BR signaling. We further generated the axe1-5 bin2-1 double mutant and the HDA6-YFP bin2-1 line, and observed that the axe1-5 bin2-1 seedlings were smaller but the HDA6-YFP bin2-1 seedlings were larger than bin2-1 (Fig. 3E). Similarly, the expression of CPD and DWF4 in the axe1-5 bin2-1 plants was enhanced, compared with that in bin2-1, but was suppressed in HDA6-YFP bin2-1 plants (Fig. 3G).
Because bin2-1 is a gain-of-function mutant of BIN2, the above results indicate that HDA6 may function downstream of BIN2, or HDA6 may regulate the activity of the mutant BIN2 protein. To distinguish these possibilities, we further generated HDA6-RNAi lines in Ws-2 and crossed one line to the bin2-3 bil1 bil2 triple-knockout background to gain HDA6-RNAi bin2-3 bil1 bil2. We found that the HDA6-RNAi bin2-3 bil1 bil2 plants showed similar phenotypes in both seedling growth and CPD and DWF4 expression to bin2-3 bil1 bil2, compared with the HDA6-RNAi plants (Fig. 3 H and I), suggesting that without BIN2 and its homologs the effect of HDA6 on BR signaling was reduced. We also found that the HDA6-RNAi bin2-3 bil1 bil2 seedlings were less sensitive to the BR biosynthetic inhibitor BRZ in hypocotyl elongation inhibition than the HDA6-RNAi seedlings but similar to bin2-3 bil1 bil2 plants in the dark (Fig. 3J), suggesting that HDA6 regulates BR signaling largely through BIN2 and its homologs in the dark. Furthermore, we found that HDA6 can still deacetylate the bin2-1 protein (Fig. S1C). The bin2-3 bil1 bil2 mutant had lower HDA6 transcript levels than the wild-type Ws-2 (Fig. 3I), implying feedback inhibition of HDA6 expression by the GSK3-like kinase family. Taken together, these observations demonstrated that HDA6 enhances BR signaling through BIN2 and other GSK3-like kinases.
HDA6 Deacetylates the K189 Residue of BIN2 to Inhibit BIN2 Kinase Activity.
To investigate how HDA6 enhanced BR signaling, we tested whether BIN2 that had been deacetylated by HDA6 had altered kinase activity. We used the phosphorylation status of BES1 as a biochemical marker to evaluate whether HDA6 influences the kinase activity of BIN2 (4, 42). Therefore, we conducted BES1-MBP phosphorylation assays using BIN2-His deacetylated by HDA6, normal BIN2-His, and hyperacetylated BIN2-His produced in E. coli treated with TSA and NAM. We found that the acetylated BIN2-His had the highest kinase activity, based on its ability to phosphorylate BES1-MBP, and that the deacetylated BIN2-His had the lowest kinase activity (Fig. 4A). Therefore, we concluded that HDA6 can deacetylate BIN2 to inhibit its kinase activity.
Fig. 4.
HDA6 deacetylates BIN2 to inhibit its kinase activity. (A) Acetylated BIN2 has higher kinase activity to phosphorylate BES1-MBP. (B) Acetylation levels of the mutated BIN2 proteins on the predicted acetylation residues. BIN2K189R showed reduced acetylation level and autophosphorylation activity compared with other forms of BIN2. BIN2K69R showed unphosphorylated BIN2. (C) Additional HDA6 did not change the acetylation level of BIN2K189R. (This acetylation band of BIN2K189R may be caused by nonspecific recognition of the antibody or acetylation of BIN2 by other enzymes on other residues.) (D) BES1-MBP phosphorylation by BIN2, BIN2K5R, BIN2K189R, and BIN2K347R. (E) Acetylation level of BIN2-FLAG and BIN2K189R-FLAG from plants with or without TSA treatments. (F) Statistical analysis of the relative acetylation level of BIN2-FLAG and BIN2K189R-FLAG from plants with or without TSA treatments. The acetylation level of BIN2-FLAG was defined as 1. (G) bin2-1K189R-FLAG overexpression rescued the dwarf phenotype of bin2-1-FLAG overexpression. (H) Expression levels of the BR-responsive genes CPD and DWF4 in BIN2-FLAG, BIN2K189R-FLAG, bin2-1-FLAG, and bin2-1K189R-FLAG plants. (I) BES1 phosphorylation status in BIN2-FLAG, BIN2K189R-FLAG, bin2-1-FLAG, and bin2-1K189R-FLAG plants detected by an anti-BES1 antibody. (J) Acetylation levels of BIN2-His, BIN2K189R-His, bin2-1-His, and bin2-1K189R-His proteins purified from E. coli. (K) Phosphorylation of BES1-MBP by BIN2-His, BIN2K189R-His, bin2-1-His, and bin2-1K189R-His protein. Error bars represent SE. *P < 0.05 and ***P < 0.001.
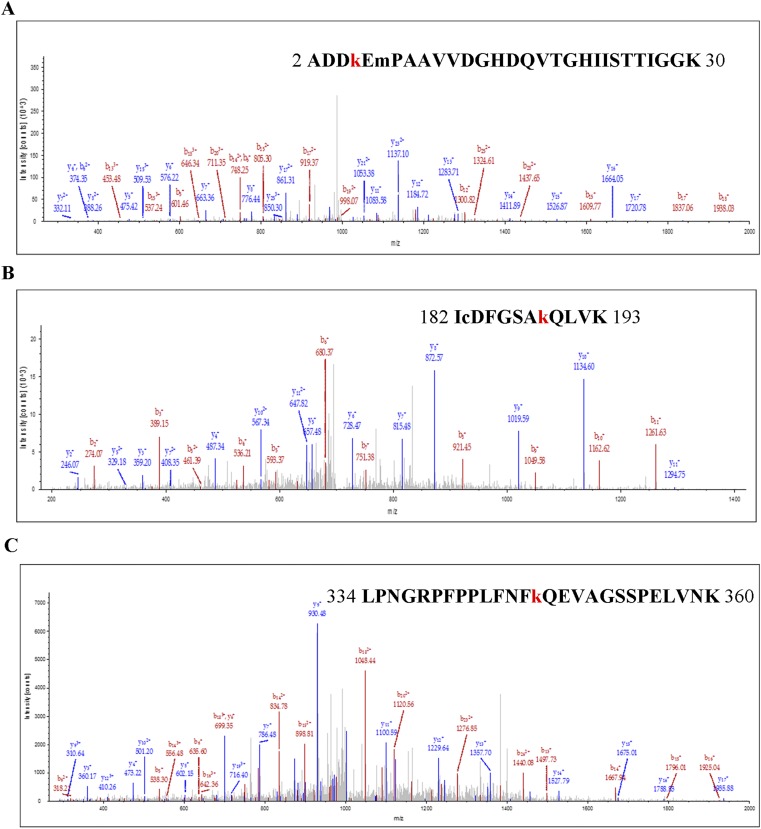
To understand which acetylated/deacetylated residues of BIN2 are important for its kinase activity, we used mass spectrometry to identify its potential acetylation sites using recombinant BIN2-His protein produced in E. coli treated with TSA and NAM. We found three potential acetylation sites, K5, K189, and K347, of BIN2 (Fig. S2). We mutated each K to R and produced these BIN2 mutant proteins in E. coli treated with TSA and NAM. The acetylation level of BIN2K189R-His substantially decreased compared with wild-type BIN2 (Fig. 4B), and BIN2K189R-His could not be deacetylated by HDA6 in vitro (Fig. 4C). Moreover, the kinase activity of BIN2K189R-His was dramatically reduced, based on BIN2K189R-His phosphorylation of BES1-MBP (Fig. 4D), indicating that K189 is a crucial site for BIN2 acetylation and activity. BIN2K189R-His also had decreased autophosphorylation activity (Fig. 4B), consistent with its decreased kinase activity.
Fig. S2.
Identification of the acetylation sites of BIN2 by LC-MS/MS. (A) K5 is a potential acetylation site of BIN2. (B) K189 is a potential acetylation site of BIN2. (C) K347 is a potential acetylation site of BIN2.
To further investigate whether the K189 site can be acetylated and has important functions in plants, we detected the acetylation level of BIN2-FLAG and BIN2K189R-FLAG with and without TSA treatments in the transgenic plants. The results showed that the acetylation level of BIN2K189R-FLAG was substantially lower than that of BIN2-FLAG, and TSA treatment did not significantly enhance the acetylation level of BIN2K189R-FLAG (Fig. 4 E and F), indicating that K189 is an important acetylation site in vivo. Previous studies showed that overexpression of the wild-type BIN2 in Arabidopsis did not lead to a strong BR-deficient phenotype, but that overexpression of the bin2-1 (BIN2E263K) protein can cause a severe BR-deficient phenotype, for unknown reasons (36, 43). To investigate whether BIN2K189R had altered activity in vivo, we made constructs to express bin2-1-FLAG and bin2-1K189R-FLAG and transferred these constructs into Ws-2. We speculated that if the acetylation of K189 is important for BIN2 kinase activity, the K189R mutation should cause a phenotype different from the dwarf phenotype shown by plants expressing bin2-1-FLAG. Indeed, our results showed that the bin2-1K189R-FLAG plants were similar to the wild-type, rather than the bin2-1-FLAG overexpression lines (Fig. 4G). In addition, the BR marker genes CPD and DWF4 and the BES1 phosphorylation status in the bin2-1K189R-FLAG plants were similar to that in the BIN2-FLAG and BIN2K189R-FLAG plants rather than that in the bin2-1-FLAG plants (Fig. 4 H and I), indicating that bin2-1K189R had reduced kinase activity. Furthermore, in vitro acetylation assays indicated that bin2-1K189R-His had lower acetylation levels (Fig. 4J) and lower kinase activity to phosphorylate BES1-MBP (Fig. 4K), compared with bin2-1-His. Therefore, we concluded that the acetylation site K189 of BIN2 is essential for its kinase activity in vivo.
Discussion
This study has provided several lines of evidence to support the idea that HDA6 plays a positive role in the BR signaling pathway. First, the HDA6 loss-of-function mutant axe1-5 and the HDA6 overexpression line had altered hypocotyl elongation phenotypes in the dark and altered expression of BR marker genes. Second, the triple knockout of bin2-3 bil1 bil2 and the bes1-D gain-of-function mutant showed reduced sensitivity to the deacetylase inhibitor TSA in hypocotyl elongation. Third, axe1-5 enhanced the dwarf phenotype of bri1-301, and the HDA6 overexpression line partially rescued its dwarf phenotype. Fourth, HDA6 can directly interact with and deacetylate BIN2 to repress its kinase activity.
We provide genetic and biochemical data to demonstrate that the deacetylation of the K189 residue of BIN2 by HDA6 is important to regulate BIN2 activity in Arabidopsis. The acetylated K189 of BIN2 is equivalent to K205 of GSK3β in mammals (11), which is one of three positively charged amino acids (R96, R180, and K205) constituting the binding pocket for a phosphoserine residue in GSK3β (25). BIN2 K189 is a novel acetylation site found in plants. Acetylation neutralizes this positively charged side chain of K189, likely to inhibit phospho-binding by BIN2. The K189R mutation nullified the dwarf phenotype of the bin2-1-FLAG transgene, suggesting that the acetylation of K189 is important for the kinase activity of BIN2. In many other kinases like CDK2 and ERK2, the residue equivalent to K189 is arginine rather than lysine (25); therefore, K189 acetylation may play a unique role in GSK3s. In animals, this pocket is occupied by the phosphorylated N terminus of GSK3β to take the place of the primed phosphorylated substrate to repress GSK3β activity (25). However, BIN2 has no priming phosphorylation or conserved N terminus to repress its activity in plants (23). Therefore, other phospho-residues may inhibit BIN2 activity by binding to the pocket after BIN2 deacetylation.
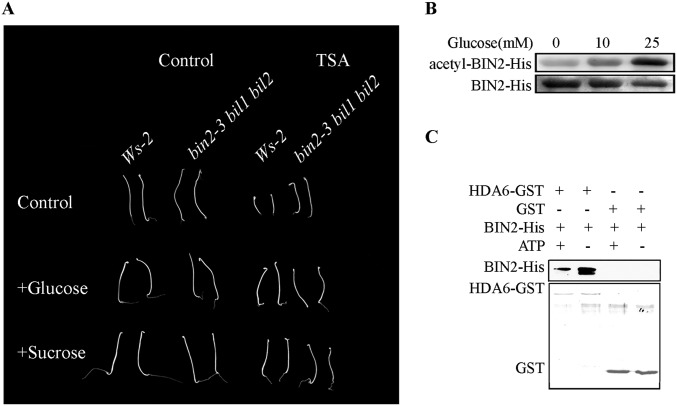
Protein acetylation, which plays a major role in metabolic regulation in mammalian systems, may also be important in regulating plant development under different energy conditions. Almost all of the important enzymes in several mammalian metabolic pathways, such as glycolysis, gluconeogenesis, the TCA cycle, the urea cycle, fatty acid metabolism, and glycogen metabolism, can be acetylated (31). Animal GSK3s are closely related to energy responses. For example, GSK3s were named due to their role in phosphorylating glycogen synthase to inhibit glycogen synthesis, which is closely related to energy status in mammals (12). In addition, under starvation conditions, GSK3s phosphorylate TSC2 to inhibit the mTOR pathway, which promotes protein and lipid synthesis (44), and, under energy-rich conditions, GSK3s phosphorylate and inhibit AMPK, which inhibits protein and fatty acid synthesis (45). Therefore, GSK3s act as critical players to regulate many anabolic and catabolic pathways in mammals (45). To test whether energy affects the acetylation of BIN2, we added glucose and sucrose to the medium to observe any changes of the BIN2 acetylation level and plant growth. We found that the short hypocotyl phenotype caused by TSA treatment can be rescued by additional glucose or sucrose in the growth medium (Fig. S3A), and the applied glucose can enhance BIN2 acetylation (Fig. S3B). We also found that additional ATP can reduce the interaction between HDA6 and BIN2 in vitro (Fig. S3C), which may cause the increase in BIN2 acetylation. Indeed, we only observed the BR-related phenotype of the HDA6 mutants and overexpression lines in sugar-free medium or in BR-deficient mutant backgrounds (Figs. 2A and 3 D–F and J). Therefore, the repression of BIN2 by HDA6-dependent acetylation may only occur under energy-limiting conditions.
Fig. S3.
Energy affects the acetylation of BIN2. (A) Seedling phenotype of Ws-2 and bin2-3 bil1 bil2 grown in the dark on medium with or without TSA and glucose or sucrose. (B) Glucose enhances BIN2-His acetylation. (C) ATP inhibits the interaction between HDA6 and BIN2.
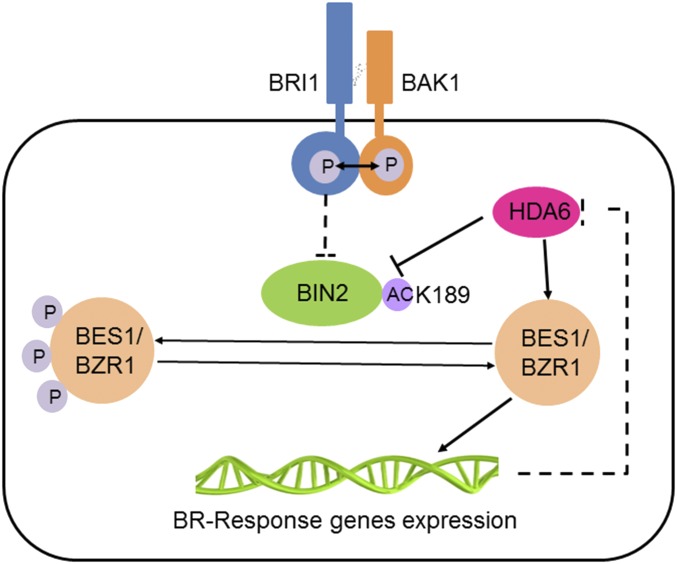
Based on previous work and our findings, we propose a model to illustrate how HDA6 regulates BR signaling through GSK3s in Arabidopsis. HDA6 can interact with BIN2 and deacetylate BIN2 on K189. The acetylated form of BIN2 has stronger activity to repress BR signaling. Under certain growth conditions (most likely those involving low energy levels), HDA6 is activated to deacetylate and inhibit BIN2; this enhances BR signaling. Meanwhile, BR signaling also represses HDA6 transcription in a feedback regulatory mechanism (Fig. S4). Besides the regulation of HDA6 to deacetylation BIN2, HDA6 may also function downstream of BIN2, because CPD/DWF4 expression was increased in HDA6-RNAi bin2-3 bil1 bil2 plants compared to bin2-3 bil1 bil2 (Fig. 3I, the fourth lane and the second lane of marker genes CPD and DWF4). We then tested the physical interaction between HDA6 and the downstream transcription factor BES1 and found that they can interact with each other (Fig. S5), suggesting that HDA6 may also directly regulate BES1. Further study is needed to investigate how HDA6 regulates BES1 activity. In addition, how energy affects the regulation of BIN2 acetylation status and activity in plants also requires further investigation.
Fig. S4.
Proposed model. BIN2, an important negative regulator in the BR signaling pathway, is regulated by upstream BR signals through phosphorylation. Phosphorylated and acetylated BIN2 has higher activity to phosphorylate the downstream transcription factors BES1 and BZR1 to inhibit BR signaling. Under energy-limited conditions, HDA6 interacts with BIN2 and deacetylates BIN2 on the K189 site to inhibit BIN2 activity and promote BR signaling. The expression of HDA6 can be feedback-inhibited, likely through BES1- and BZR1-mediated transcription regulation. HDA6 may also interact with BES1 to regulate its transcription activity.
Fig. S5.
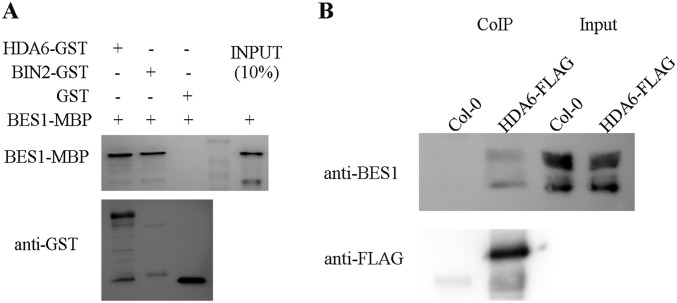
HDA6 interacts with BES1. (A) HDA6-GST interacts with BES1-MBP in GST-pull down assays. (B) BES1 interacts with HDA6-FLAG in plants.
Materials and Methods
Plant Materials and Growth Conditions.
A. thaliana plants for genetic analysis, RNA samples for genetic analysis, acetylation levels in vivo, and BES1 phosphorylation status were grown on 1/2× Murashige-Skoog medium for 9–10 d and then transferred to soil (sand:nutrient soil:roseite 9:3:1) to grow at 23 °C with a 16-h light/8-h dark cycle for an additional 2 wk. Hypocotyl length and RNA samples for qRT-PCR were from seedlings grown at 23 °C in the dark for 3 d. Plant materials used in this research are listed in Table S1.
Table S1.
Plant materials used in this research
| Col-0 background | Ws-2 background | En2 background |
| axe1-5 | bin2-3 bil1 bil2 | BES1-D |
| HDA6-YFP | HDA6-RNAi | |
| bri1-301 | HDA6-RNAi bin2-3 bil1 bil2 | |
| BIN2-FLAG | ||
| bin2-1 | BIN2K189R-FLAG | |
| axe1-5 bri1-301 | bin2-1-FLAG | |
| bin2-1K189R-FLAG | ||
| axe1-5 bin2-1 | ||
| HDA6-YFP bri1-301 | ||
| HDA6-YFP bin2-1 | ||
| BIN2-FLAG | ||
| HDA6-YFP BIN2-FLAG | ||
| BRI1-GFP |
Bimolecular Fluorescence Complementation.
HDA6-cYFP and BIN2-nYFP constructs in agrobacteria were injected into the leaves of N. benthamiana and grown for 2–3 d. HDA6-cYFP cotransfected with nYFP and BIN2-nYFP cotransfected with cYFP were used as controls. Pavement cells around the injected tobacco leaves were observed using a confocal laser-scanning microscope (Leica SP8).
In Vitro Pull-Down Assay.
Details of in vitro pull-down assay and co-IP assay can be found in SI Materials and Methods.
Detection of BIN2 Acetylation Status.
We detected BIN2 acetylation levels using an acetyl-lysine antibody (Cell Signaling Technology; 9681).
Gene Expression Analysis by Quantitative Real-Time PCR.
We extracted total RNA with an RNApre Plant Kit (Tiangen) and used a Reverse Transcriptase M-MLV Kit (TaKaRa) to generate first-strand cDNAs. cDNAs were combined with SYBR Master Mix (Invitrogen) for qRT-PCR, which was performed with an Eppendorf iCycler. Primers for qRT-PCR are listed in Table S2.
Table S2.
Primers used for quantitative RT-PCR
| Name | Directionality: sequence, 5′–3′ |
| U-BOX | Forward: TGCGCTGCCAGATAATT |
| (At5g15400) | Reverse: TGCTGCCCAACATCAGGTT |
| CPD | Forward: TCCTTGTGGGTCTAGTGTTTG |
| (At5g05690) | Reverse: TTGAACCATTGAAGCAGAAGAG |
| DWF4 | Forward: CATTGCTCTCGCTATCTTCTTC |
| (At3g50660) | Reverse: GACTCTCCTAGTTCCTTCTTGG |
| HDA6 | Forward: CCACAACTCCTAGTAATG |
| (At5g63110) | Reverse: CTCTCACTCAGAATCTCT |
Generation of Transgenic Arabidopsis RNAi Lines.
The HDA6-RNAi construct has been described in Cai et al. (19), and the primers used in this paper are listed in Table S3. The construct was transferred to Ws-2 plants to obtain HDA6-RNAi lines. Then, a line showing strong repression of HDA6 was chosen by qRT-PCR to cross to bin2-3 bil1 bil2 plants to obtain the HDA6-RNAi bin2-3 bil1 bil2 line.
Table S3.
Primers used for construction of HDA6-RNAi lines
| Name | Sequence, 5′–3′ |
| HDA6-I | gaTTGATTTTCTAACTTGGGCCTtctctcttttgtattcc |
| HDA6-II | gaAGGCCCAAGTTAGAAAATCAAtcaaagagaatcaatga |
| HDA6-III | gaAGACCCAAGTTAGTAAATCATtcacaggtcgtgatatg |
| HDA6-IV | gaATGATTTACTAACTTGGGTCTtctacatatatattcct |
In Vitro Phosphorylation Assay of BES1.
For phosphorylation assays, 2 μg of BES1-MBP and 0.5 μg of each form of BIN2-His were reacted in 1× kinase buffer (20 mM Tris, pH 7.4, 100 mM NaCl, 12 mM MgCl2, 1 mM DTT, and 1 mM ATP) at 37 °C for 30 min. Reactions were terminated by adding 5× SDS loading buffer and boiling at 95 °C for 10 min and then loaded onto SDS/PAGE.
Generation of Point Mutants of BIN2.
Details can be found in SI Materials and Methods.
Identification of the Acetylation Site of BIN2 by Liquid Chromatography/Tandem Mass Spectrometry.
E. coli cells were transformed with the BIN2-His vector for expressing BIN2 protein, and then 5 nM TSA and 5 mM NAM were added to the medium. Cells were harvested after 16 h of treatment and BIN2-His protein was purified. BIN2 protein was purified by SDS/PAGE and subjected to in-solution alkylation/tryptic digestion followed by liquid chromatography/tandem mass spectrometry (LC-MS/MS) as described by Cai et al. (19).
SI Materials and Methods
In Vitro Pull-Down Assay.
HDA6-GST, GST, BIN2-His, and BIN2K69R-His recombinant proteins were expressed in E. coli and purified with glutathione resin (GenScript) and TALON Metal Affinity Resin (Clontech). We tested whether HDA6-GST can interact with BIN2-His or BIN2K69R-His. Reactions were performed with 2× extraction buffer [100 mM Tris·HCl, pH 7.5, 300 mM NaCl, 2 mM EDTA, pH 8.0, 1% Triton X-100, 10% (vol/vol) glycerol, and protease inhibitor mixture (AMRESCO)] with 5 mM DTT for 1 h at 37 °C and incubated with glutathione resin for 2–3 h at 4 °C. Beads were washed with 2× extraction buffer several times and boiled with 1× SDS loading buffer at 95 °C for 10 min and loaded onto SDS polyacrylamide gels.
Co-IP Assay.
BIN2-FLAG, HDA6-YFP×BIN2-FLAG, and HDA6-YFP plants were all ground to fine powder in liquid nitrogen, and then equal volumes of 2× extraction buffer were added and incubated at 4 °C for 1–2 h to extract total protein. After being centrifuged at 13,400 × g, the supernatant was incubated with anti-FLAG M2 agarose gel (Sigma) for 3–4 h. The gel was washed three or four times with 2× extraction buffer and boiled with 1× SDS loading buffer at 95 °C for 10 min and loaded onto SDS polyacrylamide gels.
Generation of Point Mutants of BIN2.
Site-specific point mutants of three BIN2 constructs were created as follows. The BIN2K5R construct was generated by mutating the forward PCR primer to 5′-CGCGGATCCATGGCTGATGATAGGGAGATGCCTG-3′. The BIN2K189R construct was generated by overlap extension PCR according to Molecular Cloning (47), and the primers used for overlapping were BIN2-K189-F (forward), 5′-GGCAGTGCGAGACAGCTCGTT-3′; and BIN2-K189-R (reverse), 5′-AACGAGCTGTCTCGCACTGCC-3′. The BIN2K347R construct was generated by the “megaprimer” PCR method according to Ke and Madison (46), and the primer used to generate the megaprimer was BIN2-M-F (forward), 5′-TTCAACTTCAGACAAGAAGTAGCTG-3′, and BIN2-R (reverse), 5′-GCAGGTCGACAGTTCCAGATTGATTCAAGAAGCTT-3′.
Acknowledgments
We thank Prof. Y. Yin (Iowa State University) for providing the anti-BES1 antibody, and T. Asami (RIKEN) for providing BRZ220. This work was supported by Grants 91317302, 31271300, and 31430046 (to X.W.) of the National Natural Science Foundation of China; and Grant 2012CB114304 of the Ministry of Science and Technology of China (to X.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1521363113/-/DCSupplemental.
References
- 1.Yang CJ, Zhang C, Lu YN, Jin JQ, Wang XL. The mechanisms of brassinosteroids’ action: From signal transduction to plant development. Mol Plant. 2011;4(4):588–600. doi: 10.1093/mp/ssr020. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90(5):929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 3.Li J, et al. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110(2):213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Chory J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science. 2006;313(5790):1118–1122. doi: 10.1126/science.1127593. [DOI] [PubMed] [Google Scholar]
- 5.Kim TW, Guan S, Burlingame AL, Wang ZY. The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol Cell. 2011;43(4):561–571. doi: 10.1016/j.molcel.2011.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim TW, et al. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol. 2009;11(10):1254–1260. doi: 10.1038/ncb1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao J, et al. Two putative BIN2 substrates are nuclear components of brassinosteroid signaling. Plant Physiol. 2002;130(3):1221–1229. doi: 10.1104/pp.102.010918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin Y, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109(2):181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 9.He JX, Gendron JM, Yang Y, Li J, Wang ZY. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA. 2002;99(15):10185–10190. doi: 10.1073/pnas.152342599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang J, Zhang C, Wang X. A recently evolved isoform of the transcription factor BES1 promotes brassinosteroid signaling and development in Arabidopsis thaliana. Plant Cell. 2015;27(2):361–374. doi: 10.1105/tpc.114.133678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saidi Y, Hearn TJ, Coates JC. Function and evolution of ‘green’ GSK3/Shaggy-like kinases. Trends Plant Sci. 2012;17(1):39–46. doi: 10.1016/j.tplants.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Youn JH, Kim TW. Functional insights of plant GSK3-like kinases: Multi-taskers in diverse cellular signal transduction pathways. Mol Plant. 2015;8(4):552–565. doi: 10.1016/j.molp.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 13.Jonak C, Hirt H. Glycogen synthase kinase 3/SHAGGY-like kinases in plants: An emerging family with novel functions. Trends Plant Sci. 2002;7(10):457–461. doi: 10.1016/s1360-1385(02)02331-2. [DOI] [PubMed] [Google Scholar]
- 14.Qi X, Chanderbali AS, Wong GK, Soltis DE, Soltis PS. Phylogeny and evolutionary history of glycogen synthase kinase 3/SHAGGY-like kinase genes in land plants. BMC Evol Biol. 2013;13:143. doi: 10.1186/1471-2148-13-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Nam KH, Vafeados D, Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001;127(1):14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang S, Cai Z, Wang X. The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc Natl Acad Sci USA. 2009;106(11):4543–4548. doi: 10.1073/pnas.0900349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye Q, et al. Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proc Natl Acad Sci USA. 2010;107(13):6100–6105. doi: 10.1073/pnas.0912333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vert G, Walcher CL, Chory J, Nemhauser JL. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci USA. 2008;105(28):9829–9834. doi: 10.1073/pnas.0803996105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai Z, et al. GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc Natl Acad Sci USA. 2014;111(26):9651–9656. doi: 10.1073/pnas.1316717111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng Y, et al. Brassinosteroids control root epidermal cell fate via direct regulation of a MYB-bHLH-WD40 complex by GSK3-like kinases. Elife. 2014;3:e02525. doi: 10.7554/eLife.02525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernardo-García S, et al. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 2014;28(15):1681–1694. doi: 10.1101/gad.243675.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim TW, Michniewicz M, Bergmann DC, Wang ZY. Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature. 2012;482(7385):419–422. doi: 10.1038/nature10794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng P, Yan Z, Zhu Y, Li J. Regulation of the Arabidopsis GSK3-like kinase BRASSINOSTEROID-INSENSITIVE 2 through proteasome-mediated protein degradation. Mol Plant. 2008;1(2):338–346. doi: 10.1093/mp/ssn001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anne P, et al. OCTOPUS negatively regulates BIN2 to control phloem differentiation in Arabidopsis thaliana. Curr Biol. 2015;25(19):2584–2590. doi: 10.1016/j.cub.2015.08.033. [DOI] [PubMed] [Google Scholar]
- 25.Bax B, et al. The structure of phosphorylated GSK-3beta complexed with a peptide, FRATtide, that inhibits beta-catenin phosphorylation. Structure. 2001;9(12):1143–1152. doi: 10.1016/s0969-2126(01)00679-7. [DOI] [PubMed] [Google Scholar]
- 26.Frame S, Cohen P, Biondi RM. A common phosphate binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation. Mol Cell. 2001;7(6):1321–1327. doi: 10.1016/s1097-2765(01)00253-2. [DOI] [PubMed] [Google Scholar]
- 27.Beurel E, Grieco SF, Jope RS. Glycogen synthase kinase-3 (GSK3): Regulation, actions, and diseases. Pharmacol Ther. 2015;148:114–131. doi: 10.1016/j.pharmthera.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stadler SC, et al. Dysregulation of PAD4-mediated citrullination of nuclear GSK3β activates TGF-β signaling and induces epithelial-to-mesenchymal transition in breast cancer cells. Proc Natl Acad Sci USA. 2013;110(29):11851–11856. doi: 10.1073/pnas.1308362110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, et al. SIRT2 overexpression in hepatocellular carcinoma mediates epithelial to mesenchymal transition by protein kinase B/glycogen synthase kinase-3β/β-catenin signaling. Hepatology. 2013;57(6):2287–2298. doi: 10.1002/hep.26278. [DOI] [PubMed] [Google Scholar]
- 30.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 31.Zhao S, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327(5968):1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu JY, Lin YY, Zhu H, Chuang LM, Boeke JD. Protein acetylation and aging. Aging (Albany, NY) 2011;3(10):911–912. doi: 10.18632/aging.100398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iyer A, Fairlie DP, Brown L. Lysine acetylation in obesity, diabetes and metabolic disease. Immunol Cell Biol. 2012;90(1):39–46. doi: 10.1038/icb.2011.99. [DOI] [PubMed] [Google Scholar]
- 34.Xu W, Li Y, Liu C, Zhao S. Protein lysine acetylation guards metabolic homeostasis to fight against cancer. Oncogene. 2014;33(18):2279–2285. doi: 10.1038/onc.2013.163. [DOI] [PubMed] [Google Scholar]
- 35.Li J, Nam KH. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295(5558):1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- 36.Vert G, Chory J. Downstream nuclear events in brassinosteroid signalling. Nature. 2006;441(7089):96–100. doi: 10.1038/nature04681. [DOI] [PubMed] [Google Scholar]
- 37.Wu K, Zhang L, Zhou C, Yu CW, Chaikam V. HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J Exp Bot. 2008;59(2):225–234. doi: 10.1093/jxb/erm300. [DOI] [PubMed] [Google Scholar]
- 38.Luo M, et al. HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J Exp Bot. 2012;63(8):3297–3306. doi: 10.1093/jxb/ers059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Z, et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci USA. 2011;108(30):12539–12544. doi: 10.1073/pnas.1103959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parra M, Verdin E. Regulatory signal transduction pathways for class IIa histone deacetylases. Curr Opin Pharmacol. 2010;10(4):454–460. doi: 10.1016/j.coph.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 41.Murfett J, Wang XJ, Hagen G, Guilfoyle TJ. Identification of Arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. Plant Cell. 2001;13(5):1047–1061. doi: 10.1105/tpc.13.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang H, et al. Dual role of BKI1 and 14-3-3 s in brassinosteroid signaling to link receptor with transcription factors. Dev Cell. 2011;21(5):825–834. doi: 10.1016/j.devcel.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 43.Peng P, Zhao J, Zhu Y, Asami T, Li J. A direct docking mechanism for a plant GSK3-like kinase to phosphorylate its substrates. J Biol Chem. 2010;285(32):24646–24653. doi: 10.1074/jbc.M110.142547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inoki K, et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126(5):955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki T, et al. Inhibition of AMPK catabolic action by GSK3. Mol Cell. 2013;50(3):407–419. doi: 10.1016/j.molcel.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ke SH, Madison EL. Rapid and efficient site-directed mutagenesis by single-tube ‘megaprimer’ PCR method. Nucleic Acids Res. 1997;25(16):3371–3372. doi: 10.1093/nar/25.16.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Green MR, Sambrook J. Creating insertions or deletions using overlap extension PCR mutagenesis. In: Sambrook J, Russell DW, editors. Molecular Cloning: A Laboratory Manual. 4th Ed. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 2012. pp. 1059–1130. [Google Scholar]