Significance
The structure and function of membrane-bound chemoreceptor arrays in Bacteria and Archaea are well understood. The chemoreceptors form trimers-of-dimers that are organized into large, hexagonally packed arrays by rings of the histidine kinase CheA and the adaptor protein CheW. Even though many chemotactic prokaryotes are predicted to have additional, purely cytoplasmic chemoreceptor arrays, their structure and function remain poorly understood. We investigated the structure of the cytoplasmic array in the human pathogen Vibrio cholerae and discovered a receptor, DosM, with an unusual architecture. This chemoreceptor contains two signaling domains and is essential for the formation of cytoplasmic arrays. Furthermore, we show that DosM structurally stabilizes the cytoplasmic arrays.
Keywords: chemotaxis, chemoreceptor arrays, Vibrio cholerae, electron cryotomography, microscopy
Abstract
Nearly all motile bacterial cells use a highly sensitive and adaptable sensory system to detect changes in nutrient concentrations in the environment and guide their movements toward attractants and away from repellents. The best-studied bacterial chemoreceptor arrays are membrane-bound. Many motile bacteria contain one or more additional, sometimes purely cytoplasmic, chemoreceptor systems. Vibrio cholerae contains three chemotaxis clusters (I, II, and III). Here, using electron cryotomography, we explore V. cholerae’s cytoplasmic chemoreceptor array and establish that it is formed by proteins from cluster I. We further identify a chemoreceptor with an unusual domain architecture, DosM, which is essential for formation of the cytoplasmic arrays. DosM contains two signaling domains and spans the two-layered cytoplasmic arrays. Finally, we present evidence suggesting that this type of receptor is important for the structural stability of the cytoplasmic array.
Most motile bacteria move toward favorable environments through a process called chemotaxis. The molecular basis of this behavior is best understood in Escherichia coli, where transmembrane methyl-accepting chemotaxis proteins (MCPs, or chemoreceptors) form large arrays at the cell pole. The chemoreceptors bind attractants or repellents in the periplasm (1–3) and relay signals to histidine kinases (CheA) in the cytoplasm (4). When activated, CheA first autophosphorylates and then transfers the phosphoryl group to the response regulators CheY and CheB. Phosphorylated CheY binds to the flagellar motor, changing the direction of flagellar rotation. This allows the cells to switch from swimming forward smoothly (so-called “runs”) to tumbling randomly. Changes in the duration and frequency of run and tumble phases drive a biased random walk that moves the cells toward favorable environments (5). The other response regulator, CheB, is a methylesterase, which, in conjunction with the constitutively active methyltransferase CheR, tunes the sensitivity of the system by changing the methylation state of the chemoreceptors (6–8).
Although there is only one chemotaxis system in E. coli, most chemotactic bacterial and archaeal species have multiple systems (9). For instance, Rhodobacter sphaeroides has three chemotaxis systems encoded in three distinct clusters of genes, one of which (CheOp2) encodes two receptors without predicted transmembrane domains, TlpT and TlpC (10). Analysis of fluorescently tagged TlpT and TlpC revealed that they localize in foci around midcell. The foci split before cell division and are segregated to the midcell position within the future daughter cells. The chromosome-associated ParA-like ATPase PpfA controls the localization and segregation of the foci through an interaction with the N terminus of TlpT in a ParB-like manner (11).
Using electron cryotomography (ECT), we discovered that these cytoplasmic foci in R. sphaerodes are ordered arrays of chemoreceptors with the same basic architecture as membrane-bound arrays: trimers-of-dimers linked by rings of CheA and the adaptor protein CheW (12–14). Such cytoplasmic arrays are not unique to R. sphaeroides, but are also present in other organisms such as V. cholerae. We have shown previously that when V. cholerae cells are starved in late stationary phase, in addition to the membrane-bound chemoreceptor array that has always been present in this species in our studies (15), a second, cytoplasmic array appears (16). We also showed that cytoplasmic arrays exist in the archaeon Methanoregula formicicum (17), demonstrating that this alternative architecture is used across kingdoms. The main difference between membrane-bound and cytoplasmic arrays, however, is that in cytoplasmic chemoreceptor arrays, two layers of receptors are stacked head-to-head, sandwiched between two layers of CheA and CheW (16).
Although the genes for the core chemotaxis proteins CheA, CheW, CheR, and CheB are consistently clustered, chemoreceptor genes are often distributed throughout the genome. However, work of the Studdert laboratory has shown that only chemoreceptors of the same length can join in a single array (18), and this information can be used to identify which class of chemoreceptors forms any particular array, as seen in a cryotomogram (15). In species with more than one cluster of core chemotaxis genes and more than one type of array, an important question is whether different core proteins mix in different arrays or whether each cluster drives assembly of its own array. How these associations might affect signaling is also unknown; however, experimental evidence in the literature argues against the intermixing of chemotaxis proteins from different clusters (19).
V. cholerae contains three clusters of core chemotaxis genes and 43 chemoreceptor genes scattered throughout the genome. In this study, we show that the formation of the cytoplasmic arrays in V. cholerae is independent of cluster II and cluster III proteins, but dependent on two proteins expressed from cluster I. We also identify a chemoreceptor with an unusual domain architecture (20) that is essential for the formation of the cytoplasmic arrays: VC1403, hereinafter referred to as DosM (double signaling-domain MCP). We further show that this unusual receptor stabilizes the arrays by spanning across the two-layered cytoplasmic arrays.
Results and Discussion
Cluster II Proteins Are Responsible for the Membrane-Bound Array That Drives Chemotaxis.
Here and in previous work (15), we found by ECT that when V. cholerae is grown to exponential phase in rich medium such as LB, the cells contain a single membrane-bound chemoreceptor array. It is also known that cluster II proteins are expressed in those conditions, and that cluster II is the sole operon essential for chemotactic behavior under all conditions tested so far (21). In contrast, cluster I components are induced under low-oxygen conditions (22), and cluster III components are induced by the major stress-related sigma factor RpoS (23) inside hosts (during infection), in stationary phase, and during carbon starvation. The localization of cluster I proteins also has been reported to be dependent on microaerophilic conditions (24). Because the membrane-bound array is the only array consistently seen under conditions in which cells are known to be chemotactic (21), we conclude they are composed of cluster II proteins.
In strong support of this conclusion, bioinformatic analysis has suggested that only the cluster II system is expected to interact with 40H receptors (9). Based on the match of the predicted length of these receptors and the actual length of the receptors in the membrane-bound array as seen on ECT, we previously concluded that the membrane-bound array contains 40H receptors (15). At least one chemoreceptor of this class has been identified to sense amino acids and control chemotaxis (VC2161, also known as McpX or Mlp24) (25). Thus, because it has been shown that cluster I and III genes can be deleted and leave cells fully chemotactic (i.e, no observed phenotype) (21), under conditions conductive to chemotaxis, cluster II genes must support a membrane-bound array of 40H receptors like McpX.
Formation of Cytoplasmic Arrays in V. cholerae Depends on Cluster I, but Not on Cluster III.
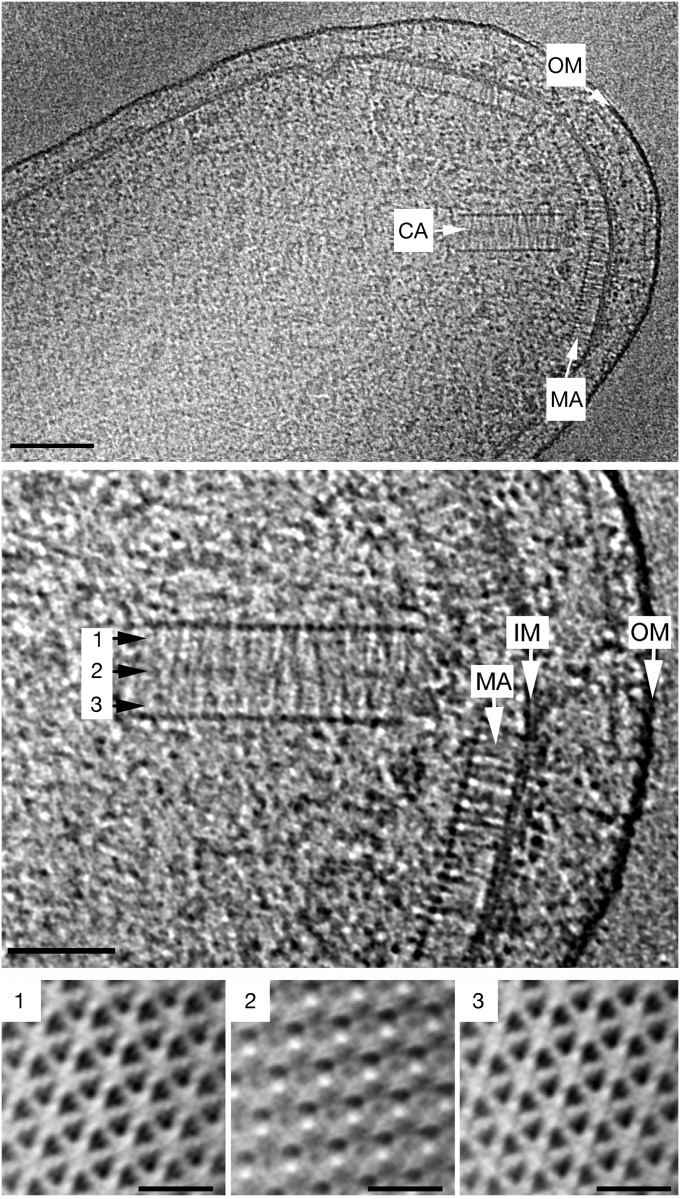
We again used ECT to test the dependency of cytoplasmic arrays on clusters I and III. In wild type (WT) cells starved in late stationary phase, cytoplasmic arrays were seen in 5 of 30 tomograms (17%) (Table 1 and Fig. 1, Top and Middle). As reported previously, the arrays exhibited the double-baseplate architecture, and unlike the cytoplasmic arrays in R. sphaeroides, the V. cholerae cytoplasmic arrays were flat (Fig. 1, Top and Middle) (16). In a deletion mutant missing both the CheW and the CheA proteins encoded by cluster III (C6706Z-∆vca1093 ∆vca1094 ∆vca1095), cytoplasmic arrays were seen in 7 of 29 tomograms (Table 1). In contrast, no cytoplasmic arrays were present (0 of 38 tomograms; Table 1) in the deletion mutant missing both CheA and CheW from cluster I (PM22, Δvc1397 and Δvc1402). The mutant with a deletion of only CheA from cluster I (PM15, Δvc1397) still exhibited cytoplasmic arrays, but less frequently than the WT (2 of 20 tomograms; Table 1). We conclude that the CheW0 of cluster I (VC1402) is essential for cytoplasmic array formation in V. cholerae. Although formally it is still possible that the CheA proteins from cluster II or cluster III are substituting in this special case, given that others have observed (by fluorescence microscopy) that membrane-bound chemoreceptor arrays can form in E. coli even in the complete absence of CheA (26), and given the homologous structures and roles of CheA and CheW in the baseplate rings, we strongly favor the interpretation that the cytoplasmic arrays in the V. cholerae mutant missing cluster I CheA (VC1397) are formed by all-cluster-I-CheW0 (VC1402) rings.
Table 1.
Cytosolic arrays detected by ECT in WT, cluster I, and cluster III deletion mutants
| Deletion | No. of tomograms | No. of cytosolic arrays |
| WT | 30 | 5 |
| CheWs and CheA (cluster III) | 29 | 7 |
| ∆vca1093 ∆vca1094 ∆vca1095 | ||
| CheA (cluster I) | 20 | 2 |
| Δvc1397 | ||
| CheA and CheW (cluster I) | 38 | 0 |
| Δvc1397 and Δvc1402 | ||
| DosM | 29 | 0 |
| Δvc1403 |
Fig. 1.
Electron tomography of cytoplasmic chemoreceptor arrays in V. cholera. (Top) Overview of a V. cholerae cell pole containing two structurally distinct chemoreceptor array varieties, a membrane-bound array (MA) and a cytoplasmic array (CA). (Scale bar: 100 nm.) (Middle) Magnified view of the membrane-bound and cytoplasmic arrays. (Scale bar: 50 nm.) (Bottom) Cross-sections of a subvolume average of a cytoplasmic array. The location of the cross-sections corresponds to the positions indicated in the side view of the array shown in the middle panel. (Scale bars: 20 nm.) OM, outer membrane; IM, inner membrane.
The Chemoreceptor DosM Is Necessary for Cytoplasmic Cluster Formation.
The genome of V. cholerae encodes 43 MCPs. Five of these MCPs (VC0098, DosM, VC1406, VCA0864, and VCA1092) lack transmembrane domains and thus are predicted to be purely cytoplasmic (27). One of these receptors, DosM, is encoded in cluster I. This receptor belongs to the signaling domain class 44H (28) and is the only receptor in the genome predicted to have two signaling domains (20). No cytoplasmic arrays were observed in a deletion strain lacking this receptor (PM16, ΔdosM VC1403) (0 of 29 tomograms; Table 1). We conclude that this receptor is essential for formation of the cytoplasmic arrays.
DosM Spans the Cytoplasmic Array.
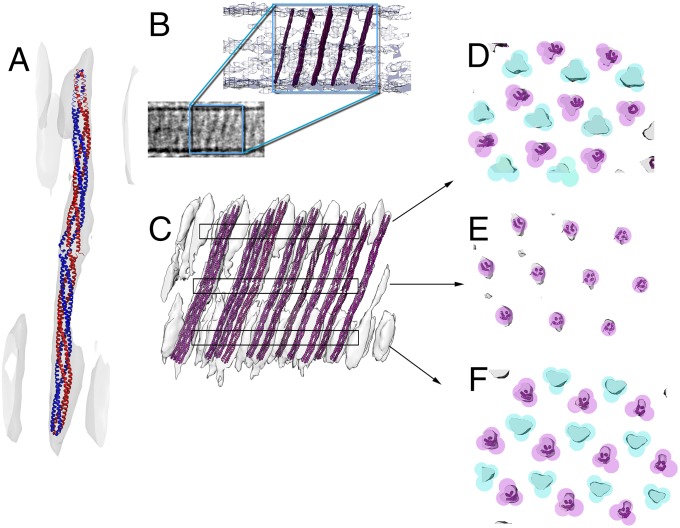
The two CheA/CheW base plates in the V. cholerae cytoplasmic arrays are 35 nm apart. Subvolume averaging within a higher-resolution tomogram of a V. cholerae cytoplasmic array revealed a unique structural element: continuous pillar-like electron densities extending between the base plates (Fig. 2). Although long and thin like the other chemoreceptor densities, the pillars were recognizable because their densities spanned the gap between layers at the position of every other trimer-of-dimer (Figs. 1, Bottom Middle, and 2D). Because the distance between the base plates exceeds the estimated physical length of any of the single-signal domain receptors present in the genome (15), we hypothesized that this long, pillar-like density represents the unusual receptor DosM, whose two signaling domains point in opposite directions. The subtomogram averages show DosM in trimer formation with other cytoplasmic receptors (Fig. 2 D and F) at both signaling tips.
Fig. 2.
MDFF of the homology model of the double signaling domain chemoreceptor DosM. (A) The homology model of a DosM receptor dimer (blue/red) was fit into the pillar-like electron density of the subvolume average using MDFF. (B) Several copies of the homology model of DosM were superimposed onto the electron density from the unaveraged electron tomogram, preserving both distribution and tilt angles from the fit into the subvolume average map shown in C. (C) Fitting of the homology model into the pillar-like electron densities of the subvolume average. (D–F) Cross-section of the areas marked in C, showing the locations of DosM correlating with every other trimer-of-dimer in the lattice. Each colored circle indicates a dimer; turquoise indicates dimers that are part of a trimer that is lacking DosM, and magenta indicates dimers that are DosM or are part of a DosM-containing trimer.
Four potential chemoreceptors lack transmembrane domains that could be part of the cytoplasmic arrays: VC0098, VC1406, VCA0864, and VCA1092. Along with DosM, at least one of these other receptors might be necessary for complex formation. The realization that DosM spans the cytoplasmic complex is in contrast to a recent hypothesis based on the analysis of a single-chain Tsr chemoreceptor construct. This modified E. coli receptor contains a double signaling domain, and the results suggest that in this case, the two signaling domains directly interact with each other (29). We speculate that this structural difference is the result of the transmembrane domain in the Tsr construct that forces the signaling domains to interact with one another, whereas the lack of transmembrane domain in DosM prevents direct interaction of the two signaling domains favoring the linear architecture shown in the tomograms.
To test this idea, we built a homology model of a single signaling domain tip of a DosM receptor dimer based on the 44H Thermotoga maritima TM1143 structure. We then duplicated it head to head to form a double signaling-domain DosM dimer. Finally, we used molecular dynamics flexible fitting (MDFF) (30) to fit this model into the pillar-like electron density; it fit well in terms of both length and width (Fig. 2A). Because this particular subtomogram average was derived from an array oriented with the base plate planes perpendicular to the electron beam (and thus suffered from poorer resolution in the direction of the pillar length owing to the “missing wedge” effect; ref. 31), we also fit the receptor dimer model into a cytoplasmic array that had been imaged with the base plate planes parallel to the electron beam (Fig. 2B). Placing the model in the array with the same angle as determined in the first fitting, we confirmed that the length of the model matched the distance between the base plates (Figs. 2 B and C). Although similar to other MCP densities, DosM pillar densities are unique in being continuous across the midplane of the array (side views) (Fig. 2E). DosM pillars occupied every other trimer-of-dimer position (Fig. 2 D and F).
Cytosolic chemoreceptors with a double signaling domain are not unique to V. cholerae, but also can be found in other bacterial species that possess the F9 class of chemotaxis systems (9). The F9 system is one of more than a dozen identified chemotaxis system classes and is characterized by the gene order (MCP-CheW-CheB…CheR-CheY-CheA), the presence of a double signaling-domain MCP that belongs to the 44H receptor class, and a concatenated triple CheW (9). The function of F9 systems (like this one in V. cholerae) remains unknown, but it appears that at least part of the role of DosM-like receptors here is to stabilize the two-layer structure. In R. sphaeroides and M. formicicum, both of which lack double signaling-domain receptors, cytoplasmic arrays are curved, and those in R. sphaeroides disassemble when cells are lysed in the absence of molecular-crowding agents (16). Double signaling-domain receptors may promote array flatness.
Fluorescence Microscopy Confirms That DosM Is Required for the Assembly of Stable Cluster I Foci.
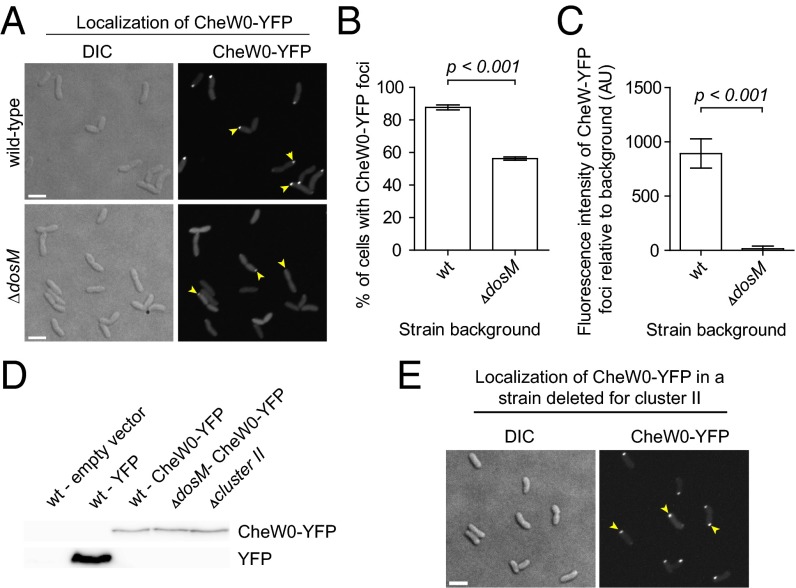
To further test the importance of DosM on cytoplasmic array formation, we fluorescently tagged the CheW protein from cluster I (VC1402, also called CheW0, which we have already identified using ECT as essential for cytoplasmic array formation) with YFP (CheW0-YFP). In WT cells, CheW0-YFP localized in distinct foci at the cell poles in 88% of cells grown to early stationary phase (n = 254) (Fig. 3 A and B). Note that although they are not membrane-bound, the V. cholerae cytoplasmic arrays nevertheless were always seen near the cell pole in cryotomograms (Fig. 1). In contrast, in a deletion mutant missing DosM, only 56% of cells showed polar foci of CheW0-YFP (n = 282) (Fig. 3 A and B), and their intensity was ∼60-fold lower than that in WT cells (Fig. 3C), thus indicating that cluster I signaling arrays in the absence of DosM are significantly smaller than those forming in WT cells. As a control, Western blot analysis using anti-YFP antibodies showed that the same amount of CheW0-YFP was produced in WT and ΔdosM backgrounds (Fig. 3D), indicating that the observed effect on CheW0-YFP foci intensity was a change in localization, not in abundance.
Fig. 3.
MCP DosM is required for the formation and stability of cluster I chemotactic signaling arrays. (A) Fluorescence micrograph showing the intracellular localization of CheW0-YFP in V. cholerae WT and ΔdosM strains. Yellow arrowheads indicate polar CheW0-YFP foci. (B) Bar graphs depicting the percentage of V. cholerae WT and ΔdosM cells with foci of CheW0-YFP. (C) Bar graphs representing the fluorescence intensity of of CheW0-YFP foci relative to the cytosolic fluorescence signal in WT and ΔdosM V. cholerae cells. (D) Immunoblot using anti-GFP antibodies (which also recognize YFP) on various strains of V. cholerae expressing either YFP or CheW0-YFP. As a negative control, a strain not expressing any YFP is included (lane 1). (E) Fluorescence microscopy showing the intracellular localization of CheW0-YFP in a V. cholerae strain deleted for the entire chemotaxis cluster II operon. Yellow arrowheads indicate polar CheW0-YFP foci. (Scale bar: 3 µm.)
Because no cytoplasmic arrays were seen in 29 DosM mutant cells, we assume that these weak foci of fluorescence arose either from tiny cytoplasmic arrays of just a few chemoreceptors, which were not noticed in the cryotomograms, or some kind of CheW0-YFP aggregates that collect near the cell pole. Another possible interpretation is that under these conditions, some CheW0-YFP is incorporated into the membrane-bound arrays. Taken together, the foregoing data indicate that DosM is required for both proper formation of cluster I signaling arrays and the actual size of the arrays.
In a strain in which cluster II genes were deleted entirely (strain SR28, Δvc2059, Δvc2060, Δvc2061, Δvc2062, Δvc2063, Δvc2064, and Δvc2065), CheW0-YFP foci were observed in 79% of cells (n = 299), a level similar to that found in a WT strain (Fig. 3E). If strong CheW0-YFP foci do indeed represent cytoplasmic arrays, then this observation is consistent with our previous conclusion that cluster II proteins form the membrane-bound arrays, and further reveals that, like cluster III proteins (ECT data), cluster II proteins are not needed to form the cytoplasmic arrays.
In summary, regarding the question of whether different chemotaxis gene clusters are responsible for different arrays, we have shown that two cluster I proteins, CheW0 (VC1402) and DosM (VC1403), are essential for the cytoplasmic arrays of V. cholerae. We also have localized the unusual DosM molecules within the arrays and have shown that cluster I CheW0-YFP foci do not depend on cluster II genes (fluorescent light microscopy data). Furthermore, we have shown that cytoplasmic arrays do not depend on cluster III genes (ECT data). Thus, cluster I proteins are responsible for the cytoplasmic arrays, which form without the need for the products of cluster II or III genes. Previously published work and additional data presented here show that cluster II proteins give rise the membrane-bound arrays, and that these arrays are the ones that support chemotaxis under the conditions tested so far. The structures formed by cluster III proteins remain unclear. Our most striking finding is that DosM molecules span across and stabilize the two-layered structure of the cytoplasmic arrays.
Materials and Methods
Strains and Growth Conditions.
All V. cholerae strains used were derived from the El Tor clinical isolate C6706. E. coli strain SM10λpir was used for conjugative transfer of plasmid DNA into V. cholera (32). Construction of V. cholerae deletion mutants was performed with standard allele-exchange techniques (33) using derivatives of plasmid pCVD442. Strain PM7 (Δvca1093 Δvca1094 Δvca1095) was constructed using pCVD442 derivative pPM045, strain PM15 (Δvc1397) was constructed using pCVD442 derivative pPM041. Strain PM16 (ΔdosM) was constructed using pCVD442 derivative pPM049, and strain PM22 (Δvc1397 Δvc1402) was constructed by consecutive deletions of vc1397 and vc1402 using pCVD442 derivatives pPM041 and pPM042, respectively.
Construction of Plasmids.
Plasmid pPM041 was constructed by PCR amplification of the upstream and downstream regions of vc1397 using primer pairs 317/318 and 319/320, respectively, and using C6706 chromosomal DNA as a template. In a third PCR, the two flanking regions were fused using the initial two products as a template and primer pair 317/320. This product was digested with XbaI and inserted into the equivalent site of pCVD442 to generate plasmid pPM041.
Plasmid pPM042 was constructed by PCR amplification of the upstream and downstream regions of vc1402 using primer pairs 322/323 and 324/325, respectively, and C6706 chromosomal DNA as a template. In a third PCR, the two flanking regions were fused using the initial two products as a template and primer pair 322/325. This product was digested with XbaI and inserted into the equivalent site of pCVD442 to generate plasmid pPM042.
Plasmid pPM045 was constructed by PCR amplification of the upstream region of vca1093 and the downstream region of vca1095 using primer pairs 337/338 and 339/340, respectively, and C6706 chromosomal DNA as a template. In a third PCR, the two flanking regions were fused using the initial two products as a template and primer pair 337/340. This product was digested with XbaI and inserted into the equivalent site of pCVD442 to generate plasmid pPM045.
Plasmid pPM049 was constructed by PCR amplification of the upstream and downstream regions of dosM using primer pairs 327/328 and 329/330, respectively, and C6706 chromosomal DNA as a template. In a third PCR, the two flanking regions were fused using the initial two products as a template and primer pair 327/330. This product was digested with XmaI and inserted into the equivalent site of pCVD442 to generate plasmid pPM049.
Plasmid pPM064 was constructed by PCR amplification of vc1402 and yfp with primer pairs 562/563 and 564/565, using C6706 chromosomal DNA and plasmid pMF390 as templates, respectively. In a third PCR, the two regions were fused using the initial two products as a template and primer pair 562/565. This product was digested with KpnI and SphI and inserted into the equivalent sites of plasmid pBAD33 to generate plasmid pPM064. The list of plasmids and primers used in this work is summarized in Tables 2 and 3.
Table 2.
Strains and plasmids
| Strain/plasmid name | Relevant genotype/description | Ref. |
| V. cholerae C6706 | lacZ- | |
| PM7 | C6706 lacZ- ∆vca1093 ∆vca1094 ∆vca1095 | This work |
| PM15 | C6706 lacZ- ∆vc1397 | This work |
| pM16 | C6706 lacZ- ∆dosM | This work |
| PM22 | C6706 lacZ- ∆vc1397 ∆vc1402 | This work |
| SR28 | lacZ- ΔcheY3 ΔcheZ ΔcheA2 ΔcheB2 ΔparC ΔparP ΔcheW1 | (24) |
| E. coli SM10λpir | KmR, thi-1, thr, leu, tonA, lacY, supE, recA::RP4-2-Tc::Mu, λpir | |
| pCVD442 | (34) | |
| pBAD33 | PBAD:: | (36) |
| pMF390 | PBAD::yfp | (37) |
| pPM064 | PBAD::vc1402-yfp | This work |
| pPM041 | Plasmid for deletion of vc1397 | This work |
| pPM042 | Plasmid for deletion of vc1402 | This work |
| pPM045 | Plasmid for deletion of vca1093, vca1094, vca1095 | This work |
| pPM049 | Plasmid for deletion of dosM | This work |
Table 3.
Primers
| Primer name | Primer sequence |
| 317 | ccccctctagaaacctcaacaaccgagcagaag |
| 318 | ctcgatgatgaagttttgcattattg |
| 319 | caataatgcaaaacttcatcatcgagactgaggaggtagtaaaacatgc |
| 320 | ccccctctagatacctgggagaacttgttgaatc |
| 322 | ccccctctagaacctgaatgaaatggcgaatattg |
| 323 | ttcgttgtctacaagggcttcac |
| 324 | gtgaagcccttgtagacaacgaaatggctgcaaacgatatggatag |
| 325 | ccccctctagatttcacgggttgttgcagatgg |
| 327 | ccccccccgggtcacgttccatgtcacatgcc |
| 328 | actggtagtgatatgtttcttgag |
| 329 | ctcaagaaacatatcactaccagtattgccgccgttgccgatgaa |
| 330 | ccccccccgggatctcgttcttcatcgatcacatc |
| 337 | ccccctctagaaattggctaatccctcctaaactc |
| 338 | aatcttgcgcagttgttccatatc |
| 339 | gatatggaacaactgcgcaagattcgcttaagcaccactgccgaa |
| 340 | ccccctctagacatcatcaaattcgtcgtcatgc |
| 562 | cccccggtaccatgaccagtgaagcccttgtag |
| 563 | cagctcctcgcccttgctcaccatgacatcctcgagctcgctatccatatcgtttgcagcca |
| 564 | atggtgagcaagggcgagga |
| 565 | cccccgcatgctcacttgtacagctcgtccatgcc |
Fluorescence Microscopy.
Fluorescence microscopy was done essentially as described previously (23, 34), with a few modifications. For microscopy, 200 µL of an overnight culture of V. cholerae harboring plasmid pPM064 for expression of CheW0-YFP was added to 10 mL of LB medium supplemented with 200 µg/mL streptomycin and 5 µg/mL chloramphenicol, followed by incubation at 30 °C with shaking. At an OD600 of 0.1, l-arabinose was added to a final concentration of 0.1% wt/vol to induce expression of CheW0-YFP. Cultures were then incubated until they reached an OD600 of 0.8, at which point samples were collected for microscopy. Simultaneously, samples were collected for immunoblot experiments to determine the cellular levels of CheW0-YFP. Microscopy was performed using a Nikon Eclipse Ti inverted Andor spinning-disk confocal microscope equipped with a 100× lens and an Andor Zyla sCMOS cooled camera. Images of WT and mutant strains were obtained using identical laser intensities and exposure times.
Microscopy images were analyzed using ImageJ (https://imagej.nih.gov/ij/), and the percentage of cells with polar foci was enumerated by hand, using the cell counter plug-in for ImageJ. To measure the intensity of CheW0-YFP foci relative to the cytosolic signal, the average fluorescence intensity was measured for equal areas of the polar foci and the cytosol. The value for the cytosolic signal was then subtracted from that of the polar focus, and the remaining fluorescence intensity was then plotted for each strain.
Immunoblot Analysis.
Samples acquired for immunoblot analysis during microscopy experiments were normalized for OD600. Samples were then analyzed by SDS/PAGE, followed by immunoblotting using JL-8 anti-GFP antibodies (Clontech) to detect CheW0-YFP levels. The JL-8 anti-GFP antibody also recognizes YFP. In addition, V. cholerae strains harboring an empty expression vector (pBAD33) or a vector for expression of YFP were included as negative and positive controls for YFP detection, respectively. Sample collection for the negative and positive controls was performed as described for the samples used for fluorescence microscopy.
ECT.
For EM experiments, cells were grown for 24 h in LB medium at 30 °C with shaking, after which 150 µL of cell suspension was diluted into 2 mL of Ca-Hepes buffer and grown for an additional 16 h at 30 °C with shaking. Cells were prepared on EM grids for ECT as described previously (16). Images were collected using either an FEI G2 300-keV field emission gun microscope or a FEI TITAN Krios 300-keV field emission gun microscope equipped with correction for lens aberration. Both microscopes were equipped with Gatan imaging filters and K2 summit counting electron-detector cameras (Gatan). Data were collected with the UCSFtomo software (37), using a cumulative dose of ∼160 e/A2 or less for each individual tilt series. Image processing, including CTF correction, frame alignment, and SIRT reconstruction, was done using the IMOD software package (38). Subvolume averaging was done using PEET software (39), and visualization was done with UCSF Chimera (40).
Bioinformatics.
To determine the signaling domains in the DosM sequence, we identified the limits of the MCPsignal (PF00015) Pfam (41) protein domain model with HMMER3 (42). To determine the heptad class of each domain, we used hidden Markov models provided previously (28, 43).
To build the homology model, we used the 44H, T. maritima chemoreceptor TM1143 structure (PDB ID code 2CH7) (44). Because there is no structure of chemoreceptors with two signaling domains available at present, we used VMD (45) to construct a PDB file with two structures of the chemoreceptor TM1403, linearly aligned but with the tips pointing in opposite directions. We built a corresponding sequence fasta file by duplicating the sequence of the crystal structure. Using BLAST 2.2.29+ (46), we selected close homologs of DosM (E-value <10E-30) from the MiST database (27) and added the composed sequence of duplicated TM1143 to build a multiple sequence alignment (MSA), using the l-INS-I algorithm from the MAFFT package (47). Using the sequences from DosM and double TM1143 from the MSA and the double TM1403 model structure, we built 200 homology models with MODELER 9.15 (48). The selected MODELER relevant parameters are as follows: max_var_interations = 1,000, library_schedule = autosched.slow, md_leve = refine.slow, repeat_optimization = 100, and max_molpdf = 1e6. We used MODELER’s DOPE and DOPEHR to select the top 10 best homology models, and on inspection of each model using PROCHECK (49), we selected the model with the fewest stereochemical problems. The model was hydrated and neutralized with KCl ions and superimposed manually to the electron-density map from the tomograms, using VMD. Finally, we used the NAMD2 (50) and MDFF plug-in for VMD (30) with default parameters to fit the DosM homology model into the electron-density map. The structure used in the figures was obtained after 50 ns of simulation.
Acknowledgments
We thank Drs. Zhiheng Yu, Jason de la Cruz, Chuan Hong, and Rick Huang for microscopy support at the Howard Hughes Medical Institute Janelia Research Campus. This work was supported by National Institute of General Medical Sciences Grant GM101425 (to G.J.J.) and by the Max Planck Society (S.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The electron cryotomography subtomogram average density map reported in this paper has been deposited in the Electron Microscopy Data Bank, www.emdatabank.org (accession no. EMD-3398).
References
- 1.Tam R, Saier MHJ., Jr Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol Rev. 1993;57(2):320–346. doi: 10.1128/mr.57.2.320-346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Milburn MV, et al. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 1991;254(5036):1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- 3.Englert DL, Adase CA, Jayaraman A, Manson MD. Repellent taxis in response to nickel ion requires neither Ni2+ transport nor the periplasmic NikA binding protein. J Bacteriol. 2010;192(10):2633–2637. doi: 10.1128/JB.00854-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: High-performance signaling in networked arrays. Trends Biochem Sci. 2008;33(1):9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turner L, Ryu WS, Berg HC. Real-time imaging of fluorescent flagellar filaments. J Bacteriol. 2000;182(10):2793–2801. doi: 10.1128/jb.182.10.2793-2801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toews ML, Goy MF, Springer MS, Adler J. Attractants and repellents control demethylation of methylated chemotaxis proteins in Escherichia coli. Proc Natl Acad Sci USA. 1979;76(11):5544–5548. doi: 10.1073/pnas.76.11.5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lupas A, Stock J. Phosphorylation of an N-terminal regulatory domain activates the CheB methylesterase in bacterial chemotaxis. J Biol Chem. 1989;264(29):17337–17342. [PubMed] [Google Scholar]
- 8.Kleene SJ, Hobson AC, Adler J. Attractants and repellents influence methylation and demethylation of methyl-accepting chemotaxis proteins in an extract of Escherichia coli. Proc Natl Acad Sci USA. 1979;76(12):6309–6313. doi: 10.1073/pnas.76.12.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wuichet K, Zhulin IB. Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal. 2010;3(128):ra50. doi: 10.1126/scisignal.2000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porter SL, Wadhams GH, Armitage JP. Rhodobacter sphaeroides: Complexity in chemotactic signalling. Trends Microbiol. 2008;16(6):251–260. doi: 10.1016/j.tim.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Roberts MAJ, Wadhams GH, Hadfield KA, Tickner S, Armitage JP. ParA-like protein uses nonspecific chromosomal DNA binding to partition protein complexes. Proc Natl Acad Sci USA. 2012;109(17):6698–6703. doi: 10.1073/pnas.1114000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, et al. Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc Natl Acad Sci USA. 2012;109(23):E1481–E1488. doi: 10.1073/pnas.1200781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, et al. The 3.2-Å resolution structure of a receptor: CheA:CheW signaling complex defines overlapping binding sites and key residue interactions within bacterial chemosensory arrays. Biochemistry. 2013;52(22):3852–3865. doi: 10.1021/bi400383e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Briegel A, et al. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc Natl Acad Sci USA. 2012;109(10):3766–3771. doi: 10.1073/pnas.1115719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Briegel A, et al. Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci USA. 2009;106(40):17181–17186. doi: 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Briegel A, et al. Structure of bacterial cytoplasmic chemoreceptor arrays and implications for chemotactic signaling. eLife. 2014;3:e02151. doi: 10.7554/eLife.02151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Briegel A, et al. Structural conservation of chemotaxis machinery across Archaea and Bacteria. Environ Microbiol Rep. 2015;7(3):414–419. doi: 10.1111/1758-2229.12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrera Seitz MK, Frank V, Massazza DA, Vaknin A, Studdert CA. Bacterial chemoreceptors of different length classes signal independently. Mol Microbiol. 2014;93(4):814–822. doi: 10.1111/mmi.12700. [DOI] [PubMed] [Google Scholar]
- 19.Güvener ZT, Tifrea DF, Harwood CS. Two different Pseudomonas aeruginosa chemosensory signal transduction complexes localize to cell poles and form and remould in stationary phase. Mol Microbiol. 2006;61(1):106–118. doi: 10.1111/j.1365-2958.2006.05218.x. [DOI] [PubMed] [Google Scholar]
- 20.Lilburn TG, Gu J, Cai H, Wang Y. Comparative genomics of the family Vibrionaceae reveals the wide distribution of genes encoding virulence-associated proteins. BMC Genomics. 2010;11:369. doi: 10.1186/1471-2164-11-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gosink KK, Kobayashi R, Kawagishi I, Häse CC. Analyses of the roles of the three cheA homologs in chemotaxis of Vibrio cholerae. J Bacteriol. 2002;184(6):1767–1771. doi: 10.1128/JB.184.6.1767-1771.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kan B, et al. Proteome comparison of Vibrio cholerae cultured in aerobic and anaerobic conditions. Proteomics. 2004;4(10):3061–3067. doi: 10.1002/pmic.200400944. [DOI] [PubMed] [Google Scholar]
- 23.Ringgaard S, Hubbard T, Mandlik A, Davis BM, Waldor MK. RpoS and quorum sensing control expression and polar localization of Vibrio cholerae chemotaxis cluster III proteins in vitro and in vivo. Mol Microbiol. 2015;97(4):660–675. doi: 10.1111/mmi.13053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hiremath G, et al. Hypoxia-induced localization of chemotaxis-related signaling proteins in Vibrio cholerae. Mol Microbiol. 2015;95(5):780–790. doi: 10.1111/mmi.12887. [DOI] [PubMed] [Google Scholar]
- 25.Nishiyama S, et al. Mlp24 (McpX) of Vibrio cholerae implicated in pathogenicity functions as a chemoreceptor for multiple amino acids. Infect Immun. 2012;80(9):3170–3178. doi: 10.1128/IAI.00039-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kentner D, Thiem S, Hildenbeutel M, Sourjik V. Determinants of chemoreceptor cluster formation in Escherichia coli. Mol Microbiol. 2006;61(2):407–417. doi: 10.1111/j.1365-2958.2006.05250.x. [DOI] [PubMed] [Google Scholar]
- 27.Ulrich LE, Zhulin IB. The MiST2 database: A comprehensive genomics resource on microbial signal transduction. Nucleic Acids Res. 2010;38(Database Issue):D401–D407. doi: 10.1093/nar/gkp940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alexander RP, Zhulin IB. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc Natl Acad Sci USA. 2007;104(8):2885–2890. doi: 10.1073/pnas.0609359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mowery P, Ames P, Reiser RH, Parkinson JS. Chemotactic signaling by single-chain chemoreceptors. PLoS One. 2015;10(12):e0145267. doi: 10.1371/journal.pone.0145267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K. Molecular dynamics flexible fitting: A practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods. 2009;49(2):174–180. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gan L, Jensen GJ. Electron tomography of cells. Q Rev Biophys. 2012;45(1):27–56. doi: 10.1017/S0033583511000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: Osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170(6):2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donnenberg MS, Kaper JB. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59(12):4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ringgaard S, et al. ParP prevents dissociation of CheA from chemotactic signaling arrays and tethers them to a polar anchor. Proc Natl Acad Sci USA. 2014;111(2):E255–E264. doi: 10.1073/pnas.1315722111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177(14):4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamaichi Y, Fogel MA, Waldor MK. par genes and the pathology of chromosome loss in Vibrio cholerae. Proc Natl Acad Sci USA. 2007;104(2):630–635. doi: 10.1073/pnas.0608341104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng SQ, et al. UCSF tomography: An integrated software suite for real-time electron microscopic tomographic data collection, alignment, and reconstruction. J Struct Biol. 2007;157(1):138–147. doi: 10.1016/j.jsb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 38.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116(1):71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 39.Nicastro D, et al. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313(5789):944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- 40.Pettersen EF, et al. UCSF Chimera: A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 41.Finn RD, et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016;44(D1):D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wheeler TJ, et al. Dfam: A database of repetitive DNA based on profile hidden Markov models. Nucleic Acids Res. 2013;41(Database Issue):D70–D82. doi: 10.1093/nar/gks1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14(9):755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 44.Park S-Y, et al. Reconstruction of the chemotaxis receptor-kinase assembly. Nat Struct Mol Biol. 2006;13(5):400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- 45.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graphics. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 46.Camacho C, et al. BLAST+: Architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Webb B, Sali A. 2014. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics 54:5.6.1–5.6.32.
- 49.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 50.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]