Significance
The Na+/I− symporter (NIS) uses the Na+ electrochemical gradient to actively transport I− into the thyroid with a 2 Na+:1 I− stoichiometry. In NIS and related transporters, one of the two Na+ binding sites, Na2, includes a Ser and a Thr residue. We determined that, besides these residues, four other amino acids participate in binding Na+ at the Na2 site, at different stages of the transport cycle. These residues are conserved throughout the entirety of solute carrier family 5 (SLC5), to which NIS belongs. Replacing any of these residues has a marked effect on transport and on the dynamics of NIS. These results not only deepen our understanding of the mechanism of transport by NIS but are also relevant to the study of other Na+-driven cotransporters.
Keywords: Na+/I− symporter, NIS, Na+-driven cotransporters, Na+ binding site, protein dynamics
Abstract
The sodium/iodide symporter (NIS) mediates active I− transport in the thyroid—the first step in thyroid hormone biosynthesis—with a 2 Na+: 1 I− stoichiometry. The two Na+ binding sites (Na1 and Na2) and the I− binding site interact allosterically: when Na+ binds to a Na+ site, the affinity of NIS for the other Na+ and for I− increases significantly. In all Na+-dependent transporters with the same fold as NIS, the side chains of two residues, S353 and T354 (NIS numbering), were identified as the Na+ ligands at Na2. To understand the cooperativity between the substrates, we investigated the coordination at the Na2 site. We determined that four other residues—S66, D191, Q194, and Q263—are also involved in Na+ coordination at this site. Experiments in whole cells demonstrated that these four residues participate in transport by NIS: mutations at these positions result in proteins that, although expressed at the plasma membrane, transport little or no I−. These residues are conserved throughout the entire SLC5 family, to which NIS belongs, suggesting that they serve a similar function in the other transporters. Our findings also suggest that the increase in affinity that each site displays when an ion binds to another site may result from changes in the dynamics of the transporter. These mechanistic insights deepen our understanding not only of NIS but also of other transporters, including many that, like NIS, are of great medical relevance.
The sodium/iodide symporter (NIS), a member of solute carrier family 5 (SLC5), is the key plasma membrane protein that mediates active iodide (I−) uptake in the thyroid gland, the first step in the biosynthesis of the thyroid hormones (THs), of which iodine is an essential constituent (1). The extensive characterization of NIS at the molecular level began with the 1996 isolation by our group of the cDNA that encodes NIS (2). NIS couples the “uphill” inward transport of I− against its electrochemical gradient to the “downhill” inward translocation of Na+ down its electrochemical gradient, which is generated by the Na+/K+ ATPase (1, 3). I− transport mediated by NIS is electrogenic, with a 2 Na+: 1 I− stoichiometry (4). Other anions, such as perrhenate (ReO4−) (5) and the environmental pollutant perchlorate (ClO4−) (6), are also transported by NIS, but, surprisingly, with an electroneutral (1 Na+: 1 anion) stoichiometry. Besides the thyroid, NIS mediates active I− transport in several other tissues, including the lactating breast (7), salivary glands (7, 8), stomach (9), and small intestine (10, 11). The function of NIS appears to be an evolutionary adaptation to the scant amount of I− in the environment (12, 13).
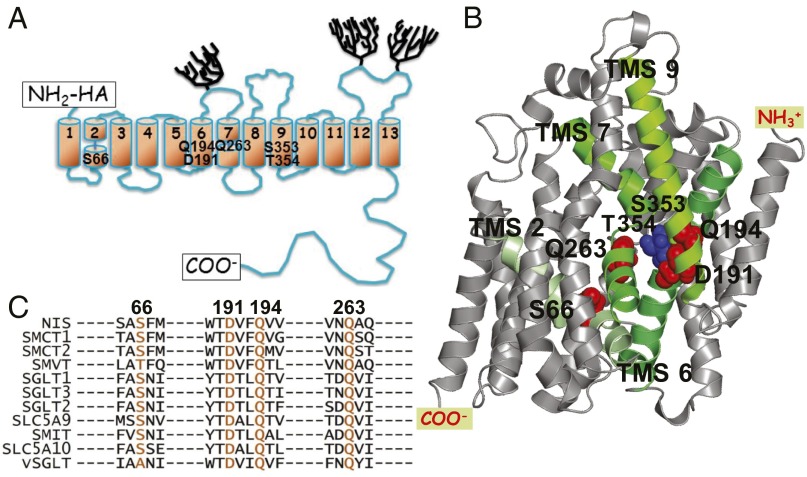
Amphipathy and helix propensity analyses suggest that NIS traverses the plasma membrane 13 times. Experimental testing has confirmed this topology and revealed that the N terminus faces the extracellular milieu and the long C terminus the cytosol (14). NIS is glycosylated at three positions—N225, N489, and N502 (15) (Fig. 1A)—but glycosylation is not essential for NIS function (15). We have generated a NIS homology model (5, 16, 17) (Fig. 1B) based on the structure in the inwardly facing conformation of another member of the SLC5 family (18), the Na+/galactose cotransporter of Vibrio parahaemolyticus (vSGLT). NIS and vSGLT share 32% sequence identity and 64% similarity (between core residues 50 and 476). In this NIS homology model, the Na2 site of NIS is between the unwound region of transmembrane segment 2 (TMS2) and TMS9. Trigonal bipyramidal coordination of Na+ is effected by the β-OH groups of S353 and T354 and the main chain –COs of A65, M68, and G350 (Fig. S1A). We recently suggested (12) that, in the case of NIS, the release of the ions into the cytoplasm after the transition from the outwardly to the inwardly facing conformation can take place without a change in the affinity of the symporter for the ions: the change in ion concentrations between the external milieu and the cytoplasm is sufficient to trigger the release of the ions. If this is the case, the coordination of the ions bound to NIS probably involves the same residues in both conformations. Mechanisms in which the affinities for some of the transported substrates are similar in the outwardly and the inwardly open conformations have been reported for the bacterial lactose permease (19) and GltPh (20). Here, we use our NIS homology model to predict and interpret the effect of mutations on transport by NIS even though, naturally, this process involves both the outwardly and the inwardly open conformations.
Fig. 1.
NIS models and SLC5 family alignment. (A) NIS secondary structure model. Numbered cylinders represent the transmembrane segments (TMSs); trees, the carbohydrates; and rectangles, the carboxy terminus and the HA tag at the amino terminus. The positions of the residues determined to be significant for Na+ coordination at the Na2 site are identified by residue name. (B) NIS 3D homology model (5). Green TMSs are those containing the residues that interact with Na+ at the Na2 site. (C) The residues named in A are highly conserved in the SLC5 family. The residues in the other members of the family that correspond to the NIS residues of interest are shown in orange. Alignment was generated using ClustalW2 (www.ebi.ac.uk/Tools/msa/clustalw2/).
Fig. S1.
Na+ coordination at the Na2 site and interactions between residues. Bipyramidal trigonal coordination of Na+ at the Na2 site is effected by the –OHs of S353 and T354 and the main chain –COs of A65, M68, and G350. The positions of S66, D191, Q194, and Q263 are shown (A). Q194 interacts with D191 and S353 (B). Q263 interacts with atoms in the main chain of V266 (C), M258 (D), and Y259 (E) in TMS7 and with S66 at the Na2 site (F). Distances (in Å) between atoms of interest are shown.
Mutations in the SLC5A5 gene (21), the gene that encodes NIS, cause congenital iodide transport defect (ITD), a rare autosomal recessive condition that results from NIS being either inactive (22–24) or not targeted to the plasma membrane (16, 17, 25, 26). The resulting impairment in I− uptake in the thyroid leads to congenital hypothyroidism, which, unless treated early in life by TH administration, causes goiter and even mental retardation. Study of the 14 ITD-causing NIS mutations identified to date (27–36) has been instrumental in the identification of residues key for NIS transport activity (5, 16, 17, 22–25, 37, 38). For example, the analysis of T354P revealed the role in NIS function of TMS9, where this mutation is located (23). TMS9 contains the most amino acids with OH groups at the β-carbon of any NIS TMS, and many of these residues are conserved in all the members of the SLC5 family. Five residues in TMS9—T351, S353, T354, S356, and T357—are critical for NIS function because they participate in Na+ binding and/or translocation, a conclusion supported by the high-resolution structure (1.65 Å) of the Aquifex aeolicus Na+-dependent leucine transporter (LeuT) (39), in which the T354 and S355 side-chain oxygens coordinate Na+ at the Na2 site. Because these residues correspond to NIS S353 and T354, we proposed that, although the sequence identity between LeuT and NIS is extremely low (∼12%), NIS may have the same fold as LeuT (37).
The crystal structures (39–42), and computational studies (43), of several Na+-driven transporters have revealed that they share the LeuT fold. Some of these transporters, including NIS, use two Na+ ions. Others use only one; in those, the Na+ binding site is in a position structurally equivalent to that of the Na2 site of LeuT. These structural findings, together with converging biochemical data, have led to the proposal that the Na2 binding site is conserved in all Na+-coupled transporters with the LeuT fold (44, 45).
Many of the important questions about the use of the two ions have been investigated, particularly by employing LeuT as a model. The answers to several of these questions, however—such as what the binding order is—remain controversial (46–50). One topic of particular interest is the allosteric interaction between the two sites. Surprisingly, the thermodynamic and structural determinants of this interaction have not been thoroughly investigated, despite its importance. In NIS, our analysis of whole-cell transport data carried out using a statistical thermodynamics formalism found that binding to one of the sites significantly increases the affinity for Na+ of the other (12). Experiments using 22Na+ in a scintillation proximity assay (SPA) also showed strong cooperativity between the two sites (51). Furthermore, the cooperativity is lost when residues S353 and T354—Na+ ligands at the Na2 site—are replaced by alanine (51). The effect of these mutations suggests that the cooperativity between the sites involves, at least in part, some of the residues that form the Na2 site. As allosteric interactions require that the protein have access to multiple conformations, it is important to determine whether other residues besides S353 and T354 participate in the coordination of Na+ at the Na2 site.
In this paper, we describe the identification of residues involved in Na+ binding at the Na2 site in NIS, a mammalian transporter—including residues not previously identified as participating in Na+ binding in any transporter belonging to this family. Initial clues about which residues might participate in coordinating Na+ at the Na2 site were sought using short molecular dynamics (MD) simulations.
As the MD simulations (52) involved short runs, no attempt was made to identify conformational transitions between the outwardly and inwardly facing conformations, their onset, or the binding and unbinding of the ions. The MD simulations identified direct Na+ ligands and residues that contribute to the coordination of the Na+ by contacting other liganding residues. In addition to correctly identifying residues S353 and T354 as Na+ ligands at the Na2 site, in agreement with our previously reported biochemical results (37), the MD simulations suggested that residues S66, D191, Q194, and Q263 also participate in Na+ coordination at this site (52). These suggestions were thoroughly tested experimentally: each residue was replaced with many other amino acids and the I− transport activity of the resulting NIS mutants measured. Substitutions at all six positions significantly impaired transport by NIS. To interpret these experimental observations, we carried out MD simulations in which some of these residues were replaced by some of the same amino acids whose effects we investigated experimentally. These simulations suggested that replacing any of the four residues mentioned above affects the overall coordination by the other residues.
Results
Ion-Binding Cavities.
Of the cavities identified by the programs Cavenv, Voidoo, and fpocket in the NIS homology model (Materials and Methods), one corresponds to the Na2 site conserved in all transporters that share the LeuT fold (see below). The other was the large cavity that holds the substrate in the structures of LeuT, vSGLT, and BetP. We already reported the effects on I− and ReO4− transport of substitutions of residues lining this large cavity (5). Na+ and I− were placed in different positions within the large cavity by themselves or in pairs with and without Na+ at the Na2 site and short MD simulations were run and used to identify residues that get within a 3-Å radius of the Na+ ion and/or a 4-Å radius of the I−. The binding site for each ion was considered to encompass the volume the ion was found in 90% of the frames. The detailed positions of the ions and their coordination were dependent on whether the sites of the other ions were occupied. Residues that coordinate I− under the different site occupancies include F67, Q72, Q94, M258, and S416, and those in the Na1 site are F67, Q72, Q94, and L289. Because the thrust of this paper relates to the Na2 site, further experimental validation of the proposed Na1 site and I− coordinating residues is beyond the scope of this work.
Identification of Residues That Coordinate Na+ at the Na2 Site.
We cast a wide net for possible binders of Na+ at the Na2 site using a distance cutoff of 5 Å—i.e., any residue of which at least one atom was ≤5 Å from the Na+ at the Na2 site in any frame was tagged as a possible ligand. Analysis of the fraction of the frames in which these residues were within coordinating distance of the ion enabled us to identify six residues as the main contributors to binding Na+ to this site. Two of them (S353 and T354) were already known to be ligands (37), and the remaining four—S66, D191, Q194, and Q263—were newly identified as ligands (Fig. 2). The participation of these six residues in coordinating Na+ at the Na2 site was analyzed using three different criteria. The frequency with which the residues appear at a given distance from the Na+ (histograms; Fig. 2 A–F) reveals that the coordination of the ion at the Na2 site depends strongly on whether the sites for the other two ions are occupied. These data are also summarized in Fig. 2 G and H. The fraction of all occurrences in which the distance between the residue of interest and the Na+ at the Na2 site is between 0 and 3 Å (Fig. 2G; integrated probabilities) shows that two of the residues, Q194 and Q263, do not contact the Na+ directly. That these residues do nonetheless participate is demonstrated by the integrated probabilities between 3 and 5 Å (Fig. 2H), which show that both residues remain in the second coordination sphere when all three ions are bound. Q194 is also in this position with high probability when only the two Na+ ions are bound. The two conditions (0 ≤ dNa ≤ 3 Å and 3 Å ≤ dNa ≤ 5 Å) correspond to two possible states that comprise a large number of conformations of each of the residues in the ensemble of conformations sampled. The probabilities (Pi) of these states can be used to estimate the free energy [ΔG = −RTln(Pi)]. Free energies computed in this manner (Fig. 2 G and H) represent the excess ΔG of conformations of the residues in the ensemble that occur with low frequency. The limiting values are ΔG = 0 for conformations that are part of the ground state and ΔG > 5.24 kcal/mol for conformations that are not observed in the simulation.
Fig. 2.
Coordination of the Na+ at the Na2 site. (A–F) The distance between the Na+ at the Na2 site and each residue shown was measured when only the Na2 site was occupied (blue), Na2 and Na1 were occupied (red), or Na2, Na1, and I− were all occupied (black). The x axis represents the distance between the Na+ and the atom in the residue closest to it. The y axis represents the fraction of the 5,000 frames in which the Na+ at the Na2 site was found at the indicated distance from the indicated residue. Each Inset shows two distances between the Na+ at the Na2 site and the side-chain oxygen when the three ions were present (black line). For Q263, only one distance is shown, corresponding to the observed single peak. Each Inset also shows the position of S353. (G) Summary of the integrated frequencies and free energies of residues 0–3 Å from the Na+ at the Na2 site. (H) Summary of the integrated frequencies and free energies of residues 3–5 Å from the Na+ at the Na2 site.
Binding of Its Ion Substrates Affects the Overall Fluctuations of NIS.
The effects of ion binding on the overall dynamics of NIS were studied using the elastic network model (ENM) formalism. Because the software currently available does not consider the effect of bound ions on the dynamics of the protein (53), we modified the code to increase the effect of small changes in the structure, including those induced by ion binding and mutations, on the low-frequency modes. In the original software, only Cα-Cα distances are considered in determining which residues are connected in the elastic network. This program does not identify residues that coordinate ions, and underestimates or even fails to detect the effects of mutations. Our modification to the software consists in using the distances between all atoms in determining which Cα atoms are connected to each other (off-diagonal terms) and to the ions, which are considered one-atom residues (Materials and Methods). The matrix, however, still only considers Cα and ion positions, and its size is equal to the number of amino acids plus the number of ions. The distance cutoff used (5 Å) was much lower than that used in the conventional algorithm. As a result of our modifications, the off-diagonal terms reflect changes in the side chains introduced by the mutations and the presence of the bound ions. Fluctuation profiles were obtained by generating random linear combinations of the six lowest-frequency vibrational modes to bring out the effects of the low-frequency/high-amplitude motions. We recently demonstrated that binding of one Na+ increases the affinity for Na+ of the other binding site. Also, Na+ binding increases the affinity of NIS for I− by a factor of ∼10 (12). These changes must reflect modifications in the conformation and the dynamics of the protein.
As expected, in the absence of bound substrates, the protein shows greater fluctuations, especially in the loops connecting TMSs (Fig. 3). When Na+ is placed in the Na1 site, there is a marked reduction in fluctuations along the entire protein. Binding of Na+ to the Na2 site results in a lesser reduction; the reduction is more pronounced in the regions between TMSs 6 and 7 and TMSs 8 and 9. When Na+ is bound to both sites, the fluctuations are similar to those with only the Na2 site occupied (Fig. 3). When all three sites (the Na1, Na2, and I− sites) are occupied, there is a reduction in the fluctuations, equivalent to that observed when only the Na1 site is occupied. In summary, the fluctuations are highly dependent on the occupancy of the substrate sites: Na2 ∼ Na1 + Na2 > Na1 ∼ Na1 + Na2 + I−. These findings suggest that interactions between the ion binding sites involve changes in the overall dynamics of the protein brought about by ion binding. These changes may be responsible for the changes in affinity that occur at one substrate site when the other sites are occupied (12).
Fig. 3.
Fluctuations of NIS from ENM calculations. NIS fluctuation profiles were obtained by generating random linear combinations of the first six vibrational modes. Fluctuations are shown for NIS with no substrates (black) and NIS with the Na1 (red); Na2 (blue); Na1 and Na2 (green); and Na1, Na2, and I− (pink) sites occupied. Numbered rectangles represent TMSs.
Substitutions of S66, D191, Q194, and Q263 Markedly Affect NIS Function.
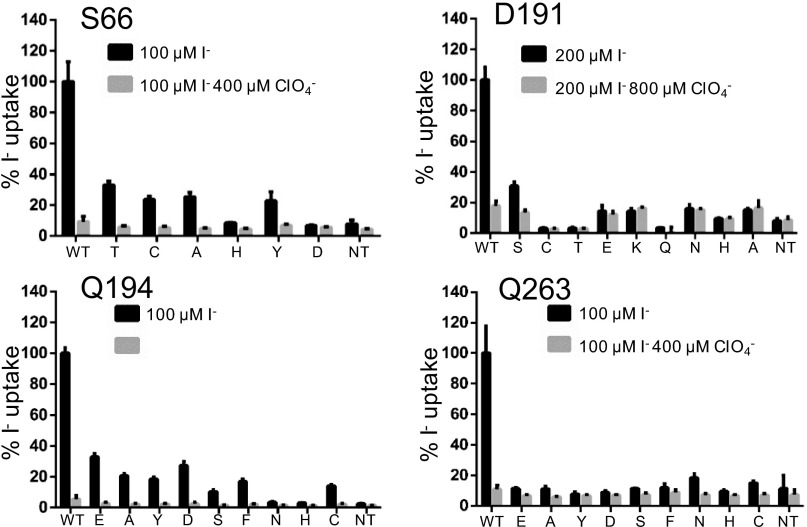
The role on NIS function of residues identified computationally as involved in the coordination of Na+ at the Na2 site—S66, D191, Q194, and Q263—was investigated by carrying out transport assays in whole cells using NIS mutants with several substitutions at each of these positions (Fig. 4 and Fig. S2) (residues S353 and T354 had been investigated before; ref. 37). All mutants were properly targeted to the plasma membrane, as determined by flow cytometry under nonpermeabilized conditions using an antibody against the extracellular amino terminus hemagglutinin (HA) tag epitope (Fig. 1A and Fig. S3), and/or by cell surface biotinylation followed by Western blot analysis (Fig. S4). Steady-state I− uptake experiments were carried out at subsaturating concentrations of I− (20 µM) (Fig. 4), and at near-saturating concentrations (100 or 200 µM I−; Fig. S2).
Fig. 4.
NIS-mediated I− uptake at steady state. cDNA constructs coding for NIS mutants with the indicated residues replaced were transfected into COS7 or HEK cells, and I− uptake mediated by these mutants was measured at 20 µM I− and at 140 mM Na+ for 30 min with or without the NIS-specific inhibitor ClO4−. Results are expressed as % of WT NIS I− uptake ± SE. Values represent averages of the results from three different experiments, each of which was carried out in triplicate. NT, nontransfected cells.
Fig. S2.
I− uptake at steady state. I− uptake was measured at the saturating concentrations of 100 or 200 µM I− and at 140 mM Na+ for 30 min with or without the NIS-specific inhibitor ClO4−. Results are expressed as % of WT NIS I− uptake ± SE. Values represent averages of the results from three different experiments, each of which was carried out in triplicate.
Fig. S3.
All NIS mutants are properly targeted to the plasma membrane. Nontransfected HEK cells and HEK cells transfected with WT NIS or the indicated NIS mutants were incubated under nonpermeabilized conditions with an anti-HA antibody that recognizes the extracellular N terminus HA epitope and analyzed by flow cytometry. The x axes show the intensity of the fluorescence of each single cell; these values were arrived at using the forward or the side scatter, parameters, respectively, representing the size and the complexity of the cells and shown on the y axes. For each acquisition, NT cells were used as a reference to identify the negative cells and determine the percentage of cells expressing WT NIS or the indicated NIS mutants.
Fig. S4.
All NIS mutants are properly targeted to the plasma membrane. Nontransfected HEK cells and HEK cells transfected with WT NIS or the indicated NIS mutants [(A) S66, (B) Q194, (C) Q263] were biotinylated under nonpermeabilized conditions and immunoblotted with anti-NIS Abs (Upper) or anti-Na+/K+ ATPase Abs, as a loading control (Lower).
Replacements of S66 with Thr, Cys, Ala, His, Tyr, and Asp had distinct effects on NIS activity. S66T and S66A NIS were the most active, retaining ∼40% the activity of WT NIS; S66C and Y retained ∼20%; and S66H and D were completely inactive (Fig. 4 and Fig. S2). Replacing D191 with Cys, Thr, Lys, Gln, His, or Ala caused a total loss of activity; replacing it with Ser, in contrast, produced a mutant that displayed ∼20% the activity of WT NIS at 20 µM (Fig. 4) or even 200 µM I− (Fig. S2). Substitutions at positions 194 and 263 produced mutant proteins with markedly reduced function. Q194D and E NIS were the most active mutants: each retained ∼30% the activity of WT NIS; Q194A, Y, S, F, and C retained ∼15–20%; and Q194H and N were completely inactive (Fig. 4 and Fig. S2). Q263E, Y, F, D, and H NIS were completely inactive at both 20 and even 100 µM I−, whereas Q263A and S exhibited ≤10%, and Q263N and C exhibited ∼20% the activity of WT NIS (Fig. 4 and Fig. S2).
The Q194E and Y and Q263N NIS Mutants Have a Lowered Apparent Affinity for Na+.
Kinetic analyses of I− transport in COS-7 or HEK cells expressing the S66T, D191S, Q194E, Q194Y, or Q263N NIS mutants were carried out at varying concentrations of either I− or Na+. S66T and D191S have a Km for I− similar to that of WT NIS (30-40 µM at 140 mM Na+) and a Km for Na+ similar to that of WT NIS (40–60 mM at 100–200 µM I−) (Fig. S5). Interestingly, Q194E and Y and Q263N NIS have a higher Km for Na+ (∼120 mM) than WT NIS (Fig. 5), suggesting a key role played by Q194 and Q263 in Na+ binding at the Na2 site.
Fig. S5.
Kinetic analysis of S66T and D191S NIS. Initial rates of I− uptake (2-min time points) at varying concentrations of I− and 140 mM Na+ (A and C) and at varying concentrations of Na+ and a constant concentration of I− (100 µM for S66T NIS and 200 µM for D191S NIS) (B and D) were determined for WT and S66T NIS (A and B) and D191S NIS (C and D) as described in Materials and Methods. Background from the values obtained from nontransfected cells was subtracted from each data point. KmI− and KmNa+ values are given in the tables. Results are expressed as pmol/µg DNA ± SD. Graphs show data from experiments carried out at least two times in triplicate.
Fig. 5.
Kinetic analysis of Q194 E and Y and Q263N NIS and effect of varying pH. Initial rates of I− uptake (2-min time points or 6-min time points for Q263N) were determined at 140 mM Na+ and varying concentrations of I− (A and C) and at varying concentrations of Na+ and 50 µM I−, for Q194 E and Y, or 200 µM I−, for Q263N (B and D). KmI− and KmNa+ values are indicated in E. Absolute values of WT NIS I− uptake in a representative experiment at higher I− or Na+ concentrations were, respectively, 128 ± 10 and 90 ± 5 pmol/µg DNA. (F) I− uptake at 5 min was assessed in HEK cells expressing WT or Q194 E, D, or H NIS at 40 µM I− and at increasing pH values. Absolute values of WT NIS I− uptake in a representative experiment at pH 6, 7.4, and 8.2 were, respectively, 132 ± 5, 180 ± 5, and 185 ± 5 pmol/µg DNA. Results are expressed as % of WT NIS I− uptake ± SE. Values represent averages of the results from two or three different experiments, each of which was carried out in triplicate.
An Uncharged Residue Is Required at Position 194.
To investigate whether or not the charge of the side chain in Q194D and E was responsible for the small reduction in activity, we measured I− uptake at three different pHs—6.0, 7.4, and 8.2—and compared the results to those obtained with WT NIS under the same conditions (Fig. 5F). At pH 6, the mutants displayed ∼45% the activity of WT NIS, whereas at pH 7.4, it was ∼20%, and at pH 8.2, <15% (Fig. 5F). These results suggest that neutralizing the negative charge of Asp and Glu at position 194 removes the major difference between Gln and the substituted residues. The opposite effect was observed with Q194H. At pH 6.0 or 7.4, there was no I− uptake (as in nontransfected cells). At pH 8.2, however, Q194H NIS displayed low but significant transport (Fig. 5F), underscoring the similarity between neutral His and Gln.
Discussion
The architecture of the Na+ binding site known as Na2 is highly conserved among Na+-dependent transporters with the LeuT fold (39), and it is to this site that Na+ binds in Na+-dependent transporters with a 1 Na+: 1 substrate stoichiometry. The first LeuT 3D structure showed that Na+ is coordinated at the Na2 binding site by two amino acid side chains with an –OH at the β-carbon (the other oxygens are main-chain carbonyls) (39). The NIS residues corresponding to these LeuT residues are S353 and T354 (37). In different transporters, individual or double replacement of the β-OH-residues by Ala at the Na2 site can have markedly different consequences (37, 43, 45, 48, 54–56).
Binding of any of the ions to empty NIS increases the affinity of the protein for the other ions (12). The ENM analysis shows that ion binding changes the dynamics of the protein (Fig. 3). This result suggests a mechanism for the increase in apparent affinity that may not involve a conformational change in the traditional sense—i.e., one between two distinct ground states. The decrease in the magnitude of the structural fluctuations revealed by the ENM analysis may instead involve an increase in the protein populations that are conformationally poised to bind the other ions. Effects of this type—i.e., allosteric effects in the absence of a conformational change between two largely different structures—have been proposed to occur in several other systems (57–59).
Our analysis of the MD trajectories suggested that, besides S353 and T354, four other residues, S66, D191, Q194, and Q263, also take part in Na+ coordination at the Na2 site (first and second coordination spheres). Alignment of SLC5 family members shows that these residues are highly conserved (Fig. 1C), whereas residues in the region corresponding to TMS3 of NIS—which we have proposed is involved in substrate specificity and stoichiometry—are not highly conserved. The latter region, together with other nonconserved residues facing the same cavity, provides anion specificity (5).
The importance of S66, D191, Q194, and Q263 in NIS transport was determined by measuring the activity of mutants with amino acid substitutions at these positions. Although replacing these residues did not affect the plasma membrane targeting of the resulting mutants (Figs. S3 and S4), it markedly affected their ability to transport I− (Fig. 4 and Fig. S2).
D191 is key for NIS activity. All amino acid substitutions at this position result in loss of activity except D191S, which produces a functional mutant that, however, has only 20% the activity of WT NIS (Fig. 4). Studies of other SLC5 family members have suggested that the equivalent residue in those proteins may also participate in the transport cycle. MD simulations of vSGLT (60, 61) showed that, when Na+ is released from its binding site, it moves closer to D189 (which corresponds to D191 of NIS). When the corresponding residue in hSGLT1 (the human Na+/glucose cotransporter), D204, was replaced with E, C, or N, the resulting mutants retained some activity (62). In NIS, the effects were more dramatic: the corresponding NIS mutants were completely inactive (Fig. 4 and Fig. S2).
Position 191 has a stringent requirement for Asp in NIS. The residue is a direct ligand of the Na+ at the Na2 site when either Na+ is bound to the Na2 site or both Na1 and Na2 have ions bound to them (Fig. 2 B and G). When I− binds, the residue leaves the coordination sphere of the ion (Fig. 2 B and G) and becomes part of the second coordination sphere (Fig. 2H). These functions require an amino acid that is an excellent Na+ ligand, and a good H-bond acceptor. Glu, despite having these characteristics, may be too long and hence fail to coordinate the Na+. Lys not only is too long but has a positive charge. Asn, Gln, Cys, Ala, and His are not good Na+ ligands, and Gln and His are probably also too long. Ser and Thr do frequently coordinate Na+, but only Ser yields a (partially) active NIS molecule. These two amino acids behave differently because Thr, a β-branched residue, can adopt only a limited number of side chain conformations, especially when located in a helix. D191S NIS is less active than the WT probably because of the loss of the negative charge and because the interaction with the Na+ is weakened owing to the failure of the shorter serine side chain to fully reach a liganding position. The presence of Ser at position 191 dramatically affects the overall coordination of Na+ at the Na2 site, impairing transport but the Km of D191S for Na+ is not altered (Fig. S5D). This behavior may be due to the change in coordination in the mutant: Na+ at the Na2 site is closer to S353 and T354 (Fig. 6 C, G, and H) than to the side chain of residue 191. In short simulations, the –COO− of the side chain of D191 forms a hydrogen bond with the amino group of the side chain amide of Q194 (Fig. S1B), which is located in the same TMS. This interaction seems to be essential for NIS function. The hypothesis that D191 and Q194 interact in the course of transport by NIS is consistent with the results obtained with PutP (63).
Fig. 6.
Coordination of Na+ at the Na2 site in WT NIS and NIS mutants. (A–F) MD simulations were carried out with WT NIS or the indicated NIS mutants with the Na2, Na1, and I− sites occupied. The residual activity of each NIS mutant is given in parentheses as a % of WT NIS activity. Ep and E, respectively, refer to the protonated and nonprotonated forms of glutamate. (G) Summary of the integrated frequencies and free energies of residues 0–3 Å from the Na+ at the Na2 site. (H) Summary of the integrated frequencies and free energies of residues 3–5 Å from the Na+ at the Na2 site.
The biochemical data point to S66 as one of the key residues in NIS-mediated I− uptake (Fig. 4). S66 coordinates Na+ at the Na2 site when only this site is occupied and when all three ion sites have ions bound to them (Fig. 2 A and G). When only the two Na+ sites are occupied, S66 oscillates between coordinating the Na+ directly and interacting as a second sphere ligand (Fig. 2 A, G, and H). The equivalent position in PutP is also important (64, 65). In NIS, the Na+ at the Na2 site interacts with the –CO of the main chain and with the Oβ of the S66 side chain (Fig. 7D). The most active of the S66 mutants, S66T NIS, retains 40% of the activity of the WT even though it coordinates Na2 significantly differently. In the conformations permitted to the Thr in an α-helix, T66 does not coordinate the Na+ (Fig. 6 B and G). Instead, it moves to the second coordination sphere (Fig. 6H), where it stabilizes other coordinating residues. In S66T NIS, which has a Km for Na+ similar to that of WT NIS (Fig. S5B), S353 and T354 are at distances from the Na+ at the Na2 site similar to the corresponding distances in WT NIS, whereas D191 and Q194 are closer to, and T66 farther away from, the Na+ at the Na2 site than in WT NIS (Fig. 6 B, G, and H), suggesting a different but still effective coordination.
Fig. 7.
Na2 network. (A) Correlation of the distances between residues whose movements are associated with coordination of the Na+ at the Na2 site. The correlation coefficients of the distances of the two residues from the Na+ at the Na2 site shown in column 1 are given in column 2. Two residues that replace each other in Na+ coordination have a negative correlation value (orange); two residues that jointly approach or move away from Na+ have a positive correlation value (light green). The average distance between each of the residues and the Na+ and the corresponding SD are shown in columns 3 and 4. Correlations between residue displacements are summarized. (B) Coordination of the Na+ at the Na2 site mediated by S353 and T354 when Na1 and Na2 are occupied. (C–E) Representative frames corresponding to the farther-away peak for T354, D191, and S66. (C) When Na1, Na2, and I− are occupied, the Na+ at the Na2 site moves far away from S353 and T354, and S66 coordinates the Na+. (D) When T354 is not coordinating the Na+ at the Na2 site, D191 and S66 are close enough to the Na+ to hold it. (E) When T354 and S66 are coordinating the Na+, Q263 is too far away to be a Na+ ligand. Note the movement of the side chain of S66 (compare with C).
Replacing Q194 with other residues yields mutants that transport 15–30% as much I− as the WT, except for Q194N and H NIS, which are both completely inactive (Fig. 4). Kinetic analysis of Q194E and Y NIS revealed that Q194 is involved in Na+ binding, as the Km for Na+ of these mutants was approximately twice that of WT NIS (Fig. 5E). The pH dependence of I− uptake by Q194E indicates that protonation of the –COO− of the glutamate side chain leads to a considerable increase in activity. (The same is true for Q194D.) A pH effect is also observed with the Q194H mutant: deprotonation of the imidazole results in low but still significant activity, probably because a deprotonated His can play a role similar to that of Gln (Fig. 5F). MD simulations with the protonated Q194Ep suggest that the conformation of Q263 is most affected (Fig. 6H), but the lower apparent affinity of Q194Ep NIS for Na+ may be due to an increased distance in this mutant between T354 and the Na+ at the Na2 site (3.8 Å), which is greater than the corresponding distance in WT NIS (∼2.2 Å) (Fig. 6D; see also Fig. 6 G and H). When E194 is not protonated (Q194E−), more substantial changes occur: only T354 and S66 remain at a coordination distance from Na2 (Fig. 6 E, G, and H).
As mentioned above, Q263N NIS had a higher Km for Na+ than WT NIS even at 200 µM I−. Q263 is in the unwound region of TMS7, and its side chain forms H-bonds with the main chain groups of V266, M258, and Y259 (Fig. S1 C–E). Moreover, Q263 interacts with S66, and even though in our MD simulations Q263 remains far from the Na+ at the Na2 site (second coordination sphere; Fig. 2H), this residue helps maintain the binding conformation of S66 (Fig. S1F). In NIS, replacements of Q263 severely affect I− transport. Even the most active 263 mutant, Q263N NIS, retained only ∼20% the activity of WT NIS. Q263N NIS showed marked changes in Na2 coordination, as only T354 and S66 remain within liganding distance of the Na+ at the Na2 site (Fig. 6 F and G), consistent with the higher Km for Na+ of Q263N (three times that of WT NIS). The residues corresponding to Q263 of NIS have never been characterized in other SLC5 family members.
Surprisingly, S353 was found to be close to Na+ at the Na2 site only when I− was not bound, suggesting that in the inwardly open conformation this Na+ is the first to be released [as has been proposed for the multihydrophobic amino acid transporter (MhsT) (40)]. On the other hand, NIS T354 coordinates Na+ at the Na2 site, along with S66, when all ions are bound (Fig. 2 A, F, and G), indicating that T354 continues to ligate Na+ during the conformational changes that lead to substrate translocation.
The experimental affinity for Na+ is low and the coordination is dynamic—as expected for the coordination of an alkali metal. Accordingly, under some substrate-binding conditions, two distances between S66, D191, and T354 and the Na+ at Na2 were frequently observed (Fig. 2 A, B, F, G, and H), indicating that the side chains adopt two different stable conformations, only one of which participates in the coordination. If Na+ is to remain bound at the Na2 site when S353 adopts a nonliganding conformation, other residues must compensate for the loss of binding. When one residue replaces another, as its distance from Na+ decreases, the distance between the other residue and Na+ increases (yielding a negative correlation coefficient; Fig. 7A). Two residues that jointly approach or move away from the Na+ at the Na2 site have a positive correlation coefficient (Fig. 7A). Interestingly, these conformations are observed even in these short simulations, indicating that Na+ coordination at this site is indeed highly dynamic.
When D191 is far from the Na+ at the Na2 site, the site’s coordination is taken over by the Oβ of T354 and the –CO of the S66 main chain (Fig. 7C). When T354 is far from Na+, the coordination of this cation at the Na2 site is mediated by S66, through its Oβ, and by D191 (Fig. 7D); but when the Na+ at the Na2 site is closest to the Q263 side chain, T354 and S66 mediate the coordination (Fig. 7E).
The data presented here shed light on the relative affinities of the Na1 and Na2 sites and their cooperativity—i.e., how binding to one site affects the apparent affinity for Na+ of the other. Although S353 and T354 of NIS do not directly coordinate Na+ at Na2 when only Na2 is occupied (Fig. 2), substitutions at these positions have a dramatic effect on the transport activity of the protein (37, 51). Both these residues, however, coordinate Na+ at the Na2 site when the Na1 site is occupied. Far from being surprising, this observation leads to an important mechanistic conclusion: binding of Na+ to the Na1 site increases the affinity for Na+ of the Na2 site by allowing residues S353 and T354 to coordinate Na+ at the Na2 site—either through a defined conformational change or by reducing the magnitude of the fluctuations, thereby increasing the population of the binding-competent conformations. In support of this conclusion, SPA Na+ binding data on the S353A/T354A mutant showed that the mutant NIS has two affinities—one in the range of 1–2 mM and the other in the range of 90–120 mM (51). This finding can be interpreted as indicating that, in the double mutant, the cooperative interactions are lost, yielding a high-affinity Na1 site and a disrupted Na2 site (51).
Conclusions
Residues S353 and T354 and S66, D191, Q194, and Q263 participate in coordinating Na+ at Na2. The results of our biochemical experiments designed on the basis of our observations from the short MD simulations are summarized in Fig. 8. This analysis suggests that, when all three ions are bound to NIS, S353 drifts away from the Na2 site, aiding in releasing the Na+. At this stage, T354 still coordinates Na+, together with S66 and D191. Once Na+ is released into the cytoplasm, the Na2 site may be stabilized by the S353–Q194 interaction (Fig. S1B). Q263, located in the unwound region of TMS7, may function as a sensor for the release of Na+ from the Na2 site, and direct the ensuing conformational changes in TMS7 to bring about the release of Na+ from the Na1 site and I− from the I− site.
Fig. 8.
Schematic representation of Na+ coordination at the Na2 site. (Upper) Na+ coordination at the Na2 site of WT NIS, when the Na2 (blue box); Na2 and Na1 (red box); and Na2, Na1, and I− (black box) sites are occupied. (Lower) Na+ coordination at the Na2 site of NIS mutants, with all three ions bound. Blue circles show substituted amino acids. Residual activity is given in parentheses as % of WT NIS activity. Double red lines represent distances ≤2.5 Å; green lines, distances >2.6 and ≤3.9 Å; and violet lines, distances ≥4 Å. Ep and E−, respectively, refer to the protonated and nonprotonated forms of glutamate.
Along with our direct observations of the effects of the mutations, the data suggest that once Na+ binds to Na1, the affinity for Na+ of Na2 increases, making binding of the second Na+ more favorable. Given these affinities, most molecules will—even in the absence of an obligatory binding order—bind Na+ first to Na1 and then to Na2, unless there are very different kinetic barriers for binding to the two sites; binding in the opposite order is possible but will be infrequent.
Materials and Methods
Cell Culture and Transfections.
COS-7 and HEK 293T cell lines obtained from the American Type Culture Collection were cultured in DMEM media (Life Technologies) supplemented with 10% FBS (Gemini BioProducts), 10 mM glutamine, 100 U/mL penicillin, and 100 mg/mL streptomycin (Cellgro). Cells were transiently transfected with 4 µg NIS cDNAs per 10-cm plate using Lipofectamine/Plus reagent (Life Technologies) following the manufacturer’s instructions. Cells were split 24 h after transfection and analyzed by FACS to determine the transfection efficiency (25). I− uptake activity, immunoblot, and surface biotinylation were assayed 48 h after transfection.
Site-Directed Mutagenesis.
Generation of S66, D191, Q194, and Q263 mutants was performed using as a template HA-tagged rat NIS cloned into pcDNA3.1. A PCR was performed with primers carrying the specific codon using Kod Hot Start DNA polymerase (Novagen). The template plasmid was digested with DpnI (NEB), and the PCR product was phosphorylated with T4 PNK (NEB), ligated with T4 DNA ligase (NEB), and transformed in TOP10. All constructs were sequenced to verify specific nucleotide substitutions.
Cell Surface Biotinylation.
Biotinylation of cell surface proteins was performed as previously described (24). Briefly, transfected cells were incubated with 1 mg/mL of the membrane-impermeable biotin reagent sulfo-NHS-SS-biotin (Pierce Chemical), which covalently interacts with extracellular primary amines. Cells were lysed, and biotinylated proteins were precipitated overnight with streptavidin-coated beads (Pierce Chemical). Beads were washed, and adsorbed proteins were eluted with sample buffer containing 10 mM DTT at 75 °C for 5 min and analyzed by immunoblot.
Immunoblotting.
SDS/PAGE, electrotransference to PVDF membranes, and immunoblotting were carried out as previously described (14). Membranes were incubated with 4 nM of an affinity-purified polyclonal anti-rat NIS Ab directed against 16 residues of the cytosolic NIS carboxyl terminus. Equal loading was assessed by stripping and reprobing the same blot with 0.5 µg/mL monoclonal anti-Na+/K+ ATPase α-subunit Ab (Affinity BioReagents). HRP-linked secondary anti-mouse and anti-rabbit Abs were from Jackson ImmunoResearch and Amersham Biosciences, respectively. Proteins were visualized using the enhanced chemiluminescence Western blot detection system (Amersham Biosciences).
Flow Cytometry.
Paraformaldehyde-fixed cells were incubated with 0.1 µg/mL anti-HA (YPYDVPDYA) antibody (Roche Applied Science) in PBS containing 0.2% BSA (16). The antibody interacts with the extracellular HA epitope in nonpermeablilized cells. After washing, cells were incubated with 50 nM of Alexa-488–conjugated goat anti-mouse antibody (Life Technologies). The fluorescence of 104 cells per tube was assayed in FACSCalibur flow cytometer (BD Bio-sciences). Data were analyzed with FlowJo software (Tree Star).
I− Transport.
Transfected cells were washed twice with HBSS [140 mM NaCl, 5.4 mM KCl, 1.3 mM CaCl2, 0.4 mM MgSO4, 0.5 mM MgCl2, 0.4 mM Na2HPO4, 0.44 mM KH2PO4, 5.55 mM glucose, and 10 mM Hepes (pH 7.5)] as previously described (66). For steady-state experiments, cells were incubated with HBSS containing 20, 100, or 200 µM KI supplemented with carrier-free Na125I (specific activity 50 mCi/mmol). Incubations proceeded at 37 °C for 30 min in a humidified atmosphere. For I−-dependent kinetic analysis, cells were incubated in HBSS buffer containing the I− concentrations indicated (1.2–600 µM) for 2 min (except where otherwise indicated). For Na+-dependent kinetic analysis, cells were incubated in HBSS buffer containing 50, 100, or 200 µM I− and the indicated Na+ concentrations (0–260 mM) for 2 min (except where otherwise indicated). Accumulated 125I− was released with ice-cold ethanol and then quantified in a Cobra II Gamma Counter (Packard Bioscience). DNA was determined by the diphenylamine method after trichloroacetic acid precipitation (10). Iodide uptake was expressed as picomoles of I− per microgramg DNA. In all flux experiments, the activity was standardized by NIS expression at the cell surface analyzed by FACS under nonpermeabilized conditions. Initial rate data were analyzed by nonlinear regression using the following equations: v = (Vmax × [I−])/(Km + [I−]) for I−-dependent kinetic and v = (Vmax × [I−]n)/(Km + [I−]n) for Na+-dependent kinetic. Background obtained in nontransfected cells was subtracted.
Identification of Ion-Binding Sites.
The final model of NIS was analyzed for cavities that might correspond to ion-binding sites using the programs Voidoo (Uppsala Software Factory), Cavenv (CCP4 suite), and fpocket (67). Besides the Na2 site, the program identified an additional major cavity that structurally coincides with the locations of the vSGLT and LeuT substrates previously determined by X-ray crystallography. We then tested for potential binding sites for Na+ and I− by randomly selecting different locations in the major cavity and running short (10 ns) MD simulations. In the first set of simulations, we used one ion type at a time, but because binding can be cooperative, we also ran simulations using combinations of ions. Each run comprised 20 simulations with the ions at different initial positions. Only the last 8 ns of each simulation was used to determine which residues were involved in binding the ions. We considered only those simulations in which the ions were bound to the protein and discarded those in which the ions diffused away from the NIS cavity.
MD Simulations.
Constant number of atoms, volume, and temperature (NVT) MD simulations were carried out with CHARMM 35b1 (52) using the CHARMM 27 force field parameters with implicit solvent. The Generalized Born using Molecular Volume (GBMV) model was used to account for the influence of the membrane (68–71). The system was minimized using conjugate gradients and the adapted-basis Newton–Raphson (ABNR) method, for 4,000 steps each. The system was then gradually heated from 100 to 300 K over the course of a 100-ps simulation, followed by equilibration at 300 K (for 100 ps) and data collection for 500 ps in the NVT ensemble. The cutoff distance for nonbonded Coulomb and Lennard–Jones interactions was set to 12 Å, with a switching function at 10 Å. Simulations were carried out with an integration time step of 1 fs using the SHAKE algorithm for bonds involving H-atoms. Data were analyzed using VMD and programs written in-house. Results were validated for WT NIS with explicit water and a POPC membrane using GROMACS 5.0 (72, 73). The initial water molecules and the bilayer were built using the web-based automated builder CHARMM-GUI (74–76). The protein model used was the same as that used in the implicit solvent simulation. The NIS model was wrapped in a 1.8-nm-thick POPC lipid bilayer (220 lipid molecules), and the membrane-protein system was solvated with a 2-nm-thick layer of TIP3 water on each side of the lipid (22,205 TIP3 water molecules), which shapes the model into a cubic box. The explicit solvent system has five net positive charges and was neutralized by replacing 125 TIP3 water molecules with 60 Na+ ions and 65 Cl− ions (0.15 mM NaCl). Ten Cl− ions were manually replaced with 10 I− ions, which made the I− concentration 0.02 mM. Two Na+ ions and one I− ion were manually aligned to the proposed binding sites using the molecular visualization tool VMD (77). The CHARMM 36 force field was used in the simulation, and the topology was generated automatically while CHARMM-GUI built the model. The energy was minimized using the steepest descent method.
The reference temperature was set to 310 K, and the system was coupled to an external heat bath using a Berendsen thermostat and was equilibrated in six stages. The time step was set to 1 fs during the first three stages and 2 fs during the following three stages. The first and second stages consisted in a 50-ps-long NVT equilibration, and the third involved a 25-ps-long constant number of atoms, pressure, and temperature (NPT) equilibration. The last three stages consisted in a 2.1-ns-long NPT equilibration. The equilibrated system was extended into a 100-ns-long MD production simulation using a Nose–Hoover thermostat. The LINear Constant Solvent (LINCS) algorithm was used to constrain bonds, and the Particle Mesh Ewald algorithm was used to treat long-range electrostatic interactions during equilibration of the system and MD production runs (72–77).
The frequency with which the coordinating residues are present at a given distance from the Na+ at the Na2 site (Figs. 2 and 6) provides information about the free energy cost of having the residue in a conformation compatible with liganding the ion. Using the term Nclose to denote the number of occurrences of a coordinating residue at a liganding distance (i.e., d ≤ 3.0 Å; see ref. 78) and Nfar to denote the number of occurrences of a coordinating residue too far away to bind the ion, the integrated probability Pi of the residue being in a liganding position is given by Pi = Nclose/Nclose + Nfar and the free energy cost of adopting the binding conformation is ΔG = −RTln(Nclose/Nclose + Nfar). For residues that remain in the liganding conformation, ΔG is zero, and for those adopting a liganding conformation only part of the time, ΔG will be greater than zero. Obviously, as in the histograms, the values will differ between the WT and mutants and among the mutants. To capture residues that contribute to Na+ binding by interacting with and positioning liganding residues, we did the same calculations counting the occurrences of the selected residues between 3 and 5 Å from the Na+. The data for the WT with Na+ at Na2, with Na+ at Na2 and Na1, and with the three ions bound are shown in Fig. 2. Data for the WT and mutants in the presence of all three ions are shown as part of Fig. 6.
Normal Mode Analysis.
Normal mode analysis calculations were done with a version of the Gaussian network model (GNM) program (79–81) modified to consider distances between all atoms in determining which Cαs are connected in the network and to include the ion substrates. The nodes in the networks represented all of the Cα atoms plus the bound ions. To determine which nodes were connected (off-diagonal terms), a double loop was added to the program for each residue pair to determine the distances between all atoms in the two residues. Two Cαs were considered connected if any two atoms in the residues were closer than a cutoff distance (Rc = 5 Å) much shorter than that used in the original program. For programming purposes, the ions were treated as one-atom residues. For the GNM program, the structures were energy minimized as described above for the MD simulations.
Fluctuation profiles were estimated as follows: for each position, we combined the contributions of the eigenvectors of the first six modes using random numbers αk (Σαk = 1) (to bring out the effects of the low-frequency/high-amplitude motions). This calculation was repeated 1,000 times with different random numbers, and the 1,000 values for each individual Cα atom in the chain were averaged (82). We showed that the calculation converges when more than 500 values are averaged. In addition, very similar profiles were obtained when the first 12 eigenvectors were used.
Note Added in Proof.
While this paper was being reviewed, Adelman et al. (83) published a paper reporting the identification of residues in the Na+ site of vSGLT corresponding to some of the residues discussed here.
Acknowledgments
We thank Dr. A. Lau for critical reading of the manuscript and valuable discussions, and the reviewers for suggestions that significantly improved the analysis and presentation of the data. This study was supported by the NIH Grants DK-41544 (to N.C.) and GM-114250 (to N.C. and L.M.A.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1607231113/-/DCSupplemental.
References
- 1.Carrasco N. Iodide transport in the thyroid gland. Biochim Biophys Acta. 1993;1154(1):65–82. doi: 10.1016/0304-4157(93)90017-i. [DOI] [PubMed] [Google Scholar]
- 2.Dai G, Levy O, Carrasco N. Cloning and characterization of the thyroid iodide transporter. Nature. 1996;379(6564):458–460. doi: 10.1038/379458a0. [DOI] [PubMed] [Google Scholar]
- 3.De La Vieja A, Dohan O, Levy O, Carrasco N. Molecular analysis of the sodium/iodide symporter: Impact on thyroid and extrathyroid pathophysiology. Physiol Rev. 2000;80(3):1083–1105. doi: 10.1152/physrev.2000.80.3.1083. [DOI] [PubMed] [Google Scholar]
- 4.Eskandari S, et al. Thyroid Na+/I− symporter. Mechanism, stoichiometry, and specificity. J Biol Chem. 1997;272(43):27230–27238. doi: 10.1074/jbc.272.43.27230. [DOI] [PubMed] [Google Scholar]
- 5.Paroder-Belenitsky M, et al. Mechanism of anion selectivity and stoichiometry of the Na+/I− symporter (NIS) Proc Natl Acad Sci USA. 2011;108(44):17933–17938. doi: 10.1073/pnas.1108278108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dohán O, et al. The Na+/I− symporter (NIS) mediates electroneutral active transport of the environmental pollutant perchlorate. Proc Natl Acad Sci USA. 2007;104(51):20250–20255. doi: 10.1073/pnas.0707207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tazebay UH, et al. The mammary gland iodide transporter is expressed during lactation and in breast cancer. Nat Med. 2000;6(8):871–878. doi: 10.1038/78630. [DOI] [PubMed] [Google Scholar]
- 8.Venturi S, Venturi M. Iodine in evolution of salivary glands and in oral health. Nutr Health. 2009;20(2):119–134. doi: 10.1177/026010600902000204. [DOI] [PubMed] [Google Scholar]
- 9.Altorjay A, et al. Expression of the Na+/I− symporter (NIS) is markedly decreased or absent in gastric cancer and intestinal metaplastic mucosa of Barrett esophagus. BMC Cancer. 2007;7:5. doi: 10.1186/1471-2407-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicola JP, et al. The Na+/I− symporter mediates active iodide uptake in the intestine. Am J Physiol Cell Physiol. 2009;296(4):C654–C662. doi: 10.1152/ajpcell.00509.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicola JP, Reyna-Neyra A, Carrasco N, Masini-Repiso AM. Dietary iodide controls its own absorption through post-transcriptional regulation of the intestinal Na+/I− symporter. J Physiol. 2012;590(23):6013–6026. doi: 10.1113/jphysiol.2012.241307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicola JP, Carrasco N, Amzel LM. Physiological sodium concentrations enhance the iodide affinity of the Na+/I− symporter. Nat Commun. 2014;5:3948. doi: 10.1038/ncomms4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Portulano C, Paroder-Belenitsky M, Carrasco N. The Na+/I- symporter (NIS): Mechanism and medical impact. Endocr Rev. 2014;35(1):106–149. doi: 10.1210/er.2012-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy O, et al. Characterization of the thyroid Na+/I− symporter with an anti-COOH terminus antibody. Proc Natl Acad Sci USA. 1997;94(11):5568–5573. doi: 10.1073/pnas.94.11.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levy O, et al. N-linked glycosylation of the thyroid Na+/I− symporter (NIS). Implications for its secondary structure model. J Biol Chem. 1998;273(35):22657–22663. doi: 10.1074/jbc.273.35.22657. [DOI] [PubMed] [Google Scholar]
- 16.Li W, Nicola JP, Amzel LM, Carrasco N. Asn441 plays a key role in folding and function of the Na+/I− symporter (NIS) FASEB J. 2013;27(8):3229–3238. doi: 10.1096/fj.13-229138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paroder V, Nicola JP, Ginter CS, Carrasco N. The iodide-transport-defect-causing mutation R124H: A δ-amino group at position 124 is critical for maturation and trafficking of the Na+/I− symporter. J Cell Sci. 2013;126(Pt 15):3305–3313. doi: 10.1242/jcs.120246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faham S, et al. The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science. 2008;321(5890):810–814. doi: 10.1126/science.1160406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guan L, Kaback HR. Binding affinity of lactose permease is not altered by the H+ electrochemical gradient. Proc Natl Acad Sci USA. 2004;101(33):12148–12152. doi: 10.1073/pnas.0404936101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reyes N, Oh S, Boudker O. Binding thermodynamics of a glutamate transporter homolog. Nat Struct Mol Biol. 2013;20(5):634–640. doi: 10.1038/nsmb.2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riedel C, Dohán O, De la Vieja A, Ginter CS, Carrasco N. Journey of the iodide transporter NIS: From its molecular identification to its clinical role in cancer. Trends Biochem Sci. 2001;26(8):490–496. doi: 10.1016/s0968-0004(01)01904-1. [DOI] [PubMed] [Google Scholar]
- 22.Dohán O, Gavrielides MV, Ginter C, Amzel LM, Carrasco N. Na+/I− symporter activity requires a small and uncharged amino acid residue at position 395. Mol Endocrinol. 2002;16(8):1893–1902. doi: 10.1210/me.2002-0071. [DOI] [PubMed] [Google Scholar]
- 23.Levy O, Ginter CS, De la Vieja A, Levy D, Carrasco N. Identification of a structural requirement for thyroid Na+/I− symporter (NIS) function from analysis of a mutation that causes human congenital hypothyroidism. FEBS Lett. 1998;429(1):36–40. doi: 10.1016/s0014-5793(98)00522-5. [DOI] [PubMed] [Google Scholar]
- 24.De La Vieja A, Ginter CS, Carrasco N. The Q267E mutation in the sodium/iodide symporter (NIS) causes congenital iodide transport defect (ITD) by decreasing the NIS turnover number. J Cell Sci. 2004;117(Pt 5):677–687. doi: 10.1242/jcs.00898. [DOI] [PubMed] [Google Scholar]
- 25.De la Vieja A, Ginter CS, Carrasco N. Molecular analysis of a congenital iodide transport defect: G543E impairs maturation and trafficking of the Na+/I− symporter. Mol Endocrinol. 2005;19(11):2847–2858. doi: 10.1210/me.2005-0162. [DOI] [PubMed] [Google Scholar]
- 26.Nicola JP, et al. Sodium/iodide symporter mutant V270E causes stunted growth but no cognitive deficiency. J Clin Endocrinol Metab. 2015;100(10):E1353–E1361. doi: 10.1210/jc.2015-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujiwara H. Congenital hypothyroidism caused by a mutation in the Na+/l−symporter. Nat Genet. 1997;17(1):122. doi: 10.1038/ng0997-122a. [DOI] [PubMed] [Google Scholar]
- 28.Fujiwara H, et al. Recurrent T354P mutation of the Na+/I− symporter in patients with iodide transport defect. J Clin Endocrinol Metab. 1998;83(8):2940–2943. doi: 10.1210/jcem.83.8.5029. [DOI] [PubMed] [Google Scholar]
- 29.Fujiwara H, et al. A novel V59E missense mutation in the sodium iodide symporter gene in a family with iodide transport defect. Thyroid. 2000;10(6):471–474. doi: 10.1089/thy.2000.10.471. [DOI] [PubMed] [Google Scholar]
- 30.Kosugi S, Bhayana S, Dean HJ. A novel mutation in the sodium/iodide symporter gene in the largest family with iodide transport defect. J Clin Endocrinol Metab. 1999;84(9):3248–3253. doi: 10.1210/jcem.84.9.5971. [DOI] [PubMed] [Google Scholar]
- 31.Kosugi S, Inoue S, Matsuda A, Jhiang SM. Novel, missense and loss-of-function mutations in the sodium/iodide symporter gene causing iodide transport defect in three Japanese patients. J Clin Endocrinol Metab. 1998;83(9):3373–3376. doi: 10.1210/jcem.83.9.5245. [DOI] [PubMed] [Google Scholar]
- 32.Kosugi S, Okamoto H, Tamada A, Sanchez-Franco F. A novel peculiar mutation in the sodium/iodide symporter gene in spanish siblings with iodide transport defect. J Clin Endocrinol Metab. 2002;87(8):3830–3836. doi: 10.1210/jcem.87.8.8767. [DOI] [PubMed] [Google Scholar]
- 33.Kosugi S, et al. High prevalence of T354P sodium/iodide symporter gene mutation in Japanese patients with iodide transport defect who have heterogeneous clinical pictures. J Clin Endocrinol Metab. 1998;83(11):4123–4129. doi: 10.1210/jcem.83.11.5229. [DOI] [PubMed] [Google Scholar]
- 34.Matsuda A, Kosugi S. A homozygous missense mutation of the sodium/iodide symporter gene causing iodide transport defect. J Clin Endocrinol Metab. 1997;82(12):3966–3971. doi: 10.1210/jcem.82.12.4425. [DOI] [PubMed] [Google Scholar]
- 35.Pohlenz J, et al. Hypothyroidism in a Brazilian kindred due to iodide trapping defect caused by a homozygous mutation in the sodium/iodide symporter gene. Biochem Biophys Res Commun. 1997;240(2):488–491. doi: 10.1006/bbrc.1997.7594. [DOI] [PubMed] [Google Scholar]
- 36.Pohlenz J, et al. Congenital hypothyroidism due to mutations in the sodium/iodide symporter. Identification of a nonsense mutation producing a downstream cryptic 3′ splice site. J Clin Invest. 1998;101(5):1028–1035. doi: 10.1172/JCI1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De la Vieja A, Reed MD, Ginter CS, Carrasco N. Amino acid residues in transmembrane segment IX of the Na+/I− symporter play a role in its Na+ dependence and are critical for transport activity. J Biol Chem. 2007;282(35):25290–25298. doi: 10.1074/jbc.M700147200. [DOI] [PubMed] [Google Scholar]
- 38.Reed-Tsur MD, De la Vieja A, Ginter CS, Carrasco N. Molecular characterization of V59E NIS, a Na+/I- symporter mutant that causes congenital I- transport defect. Endocrinology. 2008;149(6):3077–3084. doi: 10.1210/en.2008-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl−dependent neurotransmitter transporters. Nature. 2005;437(7056):215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 40.Malinauskaite L, et al. A mechanism for intracellular release of Na+ by neurotransmitter/sodium symporters. Nat Struct Mol Biol. 2014;21(11):1006–1012. doi: 10.1038/nsmb.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weyand S, et al. Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science. 2008;322(5902):709–713. doi: 10.1126/science.1164440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ressl S, Terwisscha van Scheltinga AC, Vonrhein C, Ott V, Ziegler C. Molecular basis of transport and regulation in the Na+/betaine symporter BetP. Nature. 2009;458(7234):47–52. doi: 10.1038/nature07819. [DOI] [PubMed] [Google Scholar]
- 43.Khafizov K, et al. Investigation of the sodium-binding sites in the sodium-coupled betaine transporter BetP. Proc Natl Acad Sci USA. 2012;109(44):E3035–E3044. doi: 10.1073/pnas.1209039109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krishnamurthy H, Piscitelli CL, Gouaux E. Unlocking the molecular secrets of sodium-coupled transporters. Nature. 2009;459(7245):347–355. doi: 10.1038/nature08143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z, Albers T, Fiumera HL, Gameiro A, Grewer C. A conserved Na+ binding site of the sodium-coupled neutral amino acid transporter 2 (SNAT2) J Biol Chem. 2009;284(37):25314–25323. doi: 10.1074/jbc.M109.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loo DD, Jiang X, Gorraitz E, Hirayama BA, Wright EM. Functional identification and characterization of sodium binding sites in Na symporters. Proc Natl Acad Sci USA. 2013;110(47):E4557–E4566. doi: 10.1073/pnas.1319218110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meinild AK, Forster IC. Using lithium to probe sequential cation interactions with GAT1. Am J Physiol Cell Physiol. 2012;302(11):C1661–C1675. doi: 10.1152/ajpcell.00446.2011. [DOI] [PubMed] [Google Scholar]
- 48.Zhou Y, Zomot E, Kanner BI. Identification of a lithium interaction site in the gamma-aminobutyric acid (GABA) transporter GAT-1. J Biol Chem. 2006;281(31):22092–22099. doi: 10.1074/jbc.M602319200. [DOI] [PubMed] [Google Scholar]
- 49.Zomot E, Gur M, Bahar I. Microseconds simulations reveal a new sodium-binding site and the mechanism of sodium-coupled substrate uptake by LeuT. J Biol Chem. 2015;290(1):544–555. doi: 10.1074/jbc.M114.617555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao C, et al. Ion-controlled conformational dynamics in the outward-open transition from an occluded state of LeuT. Biophys J. 2012;103(5):878–888. doi: 10.1016/j.bpj.2012.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ravera S, Quick M, Nicola JP, Carrasco N, Amzel LM. Beyond non-integer Hill coefficients: A novel approach to analyzing binding data, applied to Na+-driven transporters. J Gen Physiol. 2015;145(6):555–563. doi: 10.1085/jgp.201511365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brooks BR, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bahar I, Wallqvist A, Covell DG, Jernigan RL. Correlation between native-state hydrogen exchange and cooperative residue fluctuations from a simple model. Biochemistry. 1998;37(4):1067–1075. doi: 10.1021/bi9720641. [DOI] [PubMed] [Google Scholar]
- 54.Hilger D, Böhm M, Hackmann A, Jung H. Role of Ser-340 and Thr-341 in transmembrane domain IX of the Na+/proline transporter PutP of Escherichia coli in ligand binding and transport. J Biol Chem. 2008;283(8):4921–4929. doi: 10.1074/jbc.M706741200. [DOI] [PubMed] [Google Scholar]
- 55.Krishnamurthy H, Gouaux E. X-ray structures of LeuT in substrate-free outward-open and apo inward-open states. Nature. 2012;481(7382):469–474. doi: 10.1038/nature10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sala-Rabanal M, et al. Bridging the gap between structure and kinetics of human SGLT1. Am J Physiol Cell Physiol. 2012;302(9):C1293–C1305. doi: 10.1152/ajpcell.00397.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McLeish TC, Rodgers TL, Wilson MR. Allostery without conformation change: Modelling protein dynamics at multiple scales. Phys Biol. 2013;10(5):056004. doi: 10.1088/1478-3975/10/5/056004. [DOI] [PubMed] [Google Scholar]
- 58.Seckler JM, Barkley MD, Wintrode PL. Allosteric suppression of HIV-1 reverse transcriptase structural dynamics upon inhibitor binding. Biophys J. 2011;100(1):144–153. doi: 10.1016/j.bpj.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cooper A, Dryden DT. Allostery without conformational change. A plausible model. Eur Biophys J. 1984;11(2):103–109. doi: 10.1007/BF00276625. [DOI] [PubMed] [Google Scholar]
- 60.Li J, Tajkhorshid E. Ion-releasing state of a secondary membrane transporter. Biophys J. 2009;97(11):L29–L31. doi: 10.1016/j.bpj.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zomot E, Bahar I. The sodium/galactose symporter crystal structure is a dynamic, not so occluded state. Mol Biosyst. 2010;6(6):1040–1046. doi: 10.1039/b927492h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quick M, Loo DD, Wright EM. Neutralization of a conserved amino acid residue in the human Na+/glucose transporter (hSGLT1) generates a glucose-gated H+ channel. J Biol Chem. 2001;276(3):1728–1734. doi: 10.1074/jbc.M005521200. [DOI] [PubMed] [Google Scholar]
- 63.Amin A, Ando T, Saijo S, Yamato I. Role of Asp187 and Gln190 in the Na+/proline symporter (PutP) of Escherichia coli. J Biochem. 2011;150(4):395–402. doi: 10.1093/jb/mvr062. [DOI] [PubMed] [Google Scholar]
- 64.Pirch T, Landmeier S, Jung H. Transmembrane domain II of the Na+/proline transporter PutP of Escherichia coli forms part of a conformationally flexible, cytoplasmic exposed aqueous cavity within the membrane. J Biol Chem. 2003;278(44):42942–42949. doi: 10.1074/jbc.M308253200. [DOI] [PubMed] [Google Scholar]
- 65.Pirch T, Quick M, Nietschke M, Langkamp M, Jung H. Sites important for Na+ and substrate binding in the Na+/proline transporter of Escherichia coli, a member of the Na+/solute symporter family. J Biol Chem. 2002;277(11):8790–8796. doi: 10.1074/jbc.M111008200. [DOI] [PubMed] [Google Scholar]
- 66.Purtell K, et al. The KCNQ1-KCNE2 K⁺ channel is required for adequate thyroid I− uptake. FASEB J. 2012;26(8):3252–3259. doi: 10.1096/fj.12-206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Le Guilloux V, Schmidtke P, Tuffery P. Fpocket: An open source platform for ligand pocket detection. BMC Bioinformatics. 2009;10:168. doi: 10.1186/1471-2105-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feig M. Implicit membrane models for membrane protein simulation. Methods Mol Biol. 2008;443:181–196. doi: 10.1007/978-1-59745-177-2_10. [DOI] [PubMed] [Google Scholar]
- 69.Im W, Feig M, Brooks CL., 3rd An implicit membrane generalized born theory for the study of structure, stability, and interactions of membrane proteins. Biophys J. 2003;85(5):2900–2918. doi: 10.1016/S0006-3495(03)74712-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Im W, Lee MS, Brooks CL., 3rd Generalized born model with a simple smoothing function. J Comput Chem. 2003;24(14):1691–1702. doi: 10.1002/jcc.10321. [DOI] [PubMed] [Google Scholar]
- 71.Lee MS, Feig M, Salsbury FR, Jr, Brooks CL., 3rd New analytic approximation to the standard molecular volume definition and its application to generalized Born calculations. J Comput Chem. 2003;24(11):1348–1356. doi: 10.1002/jcc.10272. [DOI] [PubMed] [Google Scholar]
- 72.Berendsen HJC, Van der Spoel D, Van Drunen R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput Phys Commun. 1995;91(1–3):43–56. [Google Scholar]
- 73.Abraham MJ, et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. [Google Scholar]
- 74.Jo S, Kim T, Im W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS One. 2007;2(9):e880. doi: 10.1371/journal.pone.0000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J Comput Chem. 2008;29(11):1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 76.Lee J, et al. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J Chem Theory Comput. 2015;12(1):405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 78.Varma S, Rempe SB. Coordination numbers of alkali metal ions in aqueous solutions. Biophys Chem. 2006;124(3):192–199. doi: 10.1016/j.bpc.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 79.Atilgan AR, et al. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys J. 2001;80(1):505–515. doi: 10.1016/S0006-3495(01)76033-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bahar I, Atilgan AR, Erman B. Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential. Fold Des. 1997;2(3):173–181. doi: 10.1016/S1359-0278(97)00024-2. [DOI] [PubMed] [Google Scholar]
- 81.Tirion MM. Large amplitude elastic motions in proteins from a single-parameter, atomic analysis. Phys Rev Lett. 1996;77(9):1905–1908. doi: 10.1103/PhysRevLett.77.1905. [DOI] [PubMed] [Google Scholar]
- 82.Echeverria I, Liu Y, Gabelli SB, Amzel LM. Oncogenic mutations weaken the interactions that stabilize the p110α-p85α heterodimer in phosphatidylinositol 3-kinase α. FEBS J. 2015;282(18):3528–3542. doi: 10.1111/febs.13365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Adelman JL, et al. Stochastic steps in secondary active sugar transport. Proc Natl Acad Sci USA. 2016;113(27):E3960–E3966. doi: 10.1073/pnas.1525378113. [DOI] [PMC free article] [PubMed] [Google Scholar]