Significance
Our work shows a remarkable synergy between physiological levels of vitamin C and 5-aza-CdR. The combination enhances the viral mimicry response to DNA methyltransferase inhibitors, including the upregulation of endogenous retroviruses in the dsRNA form and the induction of viral defense pathways. Because patients with hematological and other cancers are often markedly vitamin C deficient, the addition of vitamin C to treatment protocols may be a straightforward way to increase the clinical efficacy of such drugs in patients with myelodysplastic syndrome and leukemia.
Keywords: epigenetic therapy, DNA methyltransferase inhibitor, vitamin C, endogenous retrovirus, 5-hydroxymethylcytosine
Abstract
Vitamin C deficiency is found in patients with cancer and might complicate various therapy paradigms. Here we show how this deficiency may influence the use of DNA methyltransferase inhibitors (DNMTis) for treatment of hematological neoplasias. In vitro, when vitamin C is added at physiological levels to low doses of the DNMTi 5-aza-2′-deoxycytidine (5-aza-CdR), there is a synergistic inhibition of cancer-cell proliferation and increased apoptosis. These effects are associated with enhanced immune signals including increased expression of bidirectionally transcribed endogenous retrovirus (ERV) transcripts, increased cytosolic dsRNA, and activation of an IFN-inducing cellular response. This synergistic effect is likely the result of both passive DNA demethylation by DNMTi and active conversion of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) by ten–eleven translocation (TET) enzymes at LTR regions of ERVs, because vitamin C acts as a cofactor for TET proteins. In addition, TET2 knockout reduces the synergy between the two compounds. Furthermore, we show that many patients with hematological neoplasia are markedly vitamin C deficient. Thus, our data suggest that correction of vitamin C deficiency in patients with hematological and other cancers may improve responses to epigenetic therapy with DNMTis.
Alterations in DNA methylation are among the earliest and most common events during tumorigenesis (1). De novo methylation of CpG island promoters silences tumor suppressors in cancer and can be viewed as an alternative to genetic mutation in inactivating such important regulators. Because of its dynamic nature, DNA methylation is attractive as a therapeutic target for Food and Drug Administration-approved DNA methyltransferase inhibitors (DNMTis) such as 5-aza-2′-deoxycytidine (5-aza-CdR) or 5-azacytidine (5-aza-CR). These nucleosides are incorporated into the DNA of proliferating cells during S phase and inhibit cytosine methylation by trapping DNA methyltransferases (DNMTs) onto the DNA, leading to their proteolysis (2–4). Demethylation occurs via subsequent passive dilution during DNA replication; the eventual restoration of normal DNMT levels leads to remethylation of CpG sites (5, 6).
DNMTis have exhibited significant success in the treatment of hematological malignancies such as myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) (7–11). It has been proposed that the molecular mechanisms underlying such efficacy involve demethylation of tumor-suppressor gene promoters and oncogene bodies, thereby restoring more normal expression levels (1, 6). Recently, we found that inhibiting DNA methylation by 5-aza-CdR causes up-regulation of endogenous retroviruses (ERVs) and induces an IFN response, which may be responsible for apoptosis and sensitization to subsequent immune checkpoint therapy (12, 13). This response has been called “viral mimicry” because the cell responds as it would to an exogenous viral infection.
Only about 50% of patients with MDS or AML respond to 5-aza-CdR alone (14, 15), so recent clinical trials are exploring combinations with conventional chemotherapy or immune checkpoint therapy (12, 16, 17). To improve the clinical efficacy of DNMTis, we explored a combination of physiological levels of vitamin C with low-dose 5-aza-CdR. Vitamin C is a cofactor for the Fe(II) 2-oxoglutarate dioxygenase family, exemplified by the three ten–eleven translocation (TET) enzymes (18), which convert 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) (19, 20). In addition, 5hmC depletion in a variety of human cancers suggests decreased TET activity in cancer cells (21). The addition of vitamin C to ES cells promotes DNA demethylation through increased TET activity (18, 22). We hypothesized that combination treatment of 5-aza-CdR plus vitamin C would lead to improved efficacy against cancer cells, possibly by pharmacological inhibition of DNMTs by 5-aza-CdR and activation of TET enzymes by vitamin C.
Results
Vitamin C at Physiological Levels Enhances the Apoptotic Effects of 5-aza-CdR.
Virtually all culture media lack vitamin C. We therefore examined whether daily supplementation of vitamin C at physiological levels in five cancer cell lines would alter the outcome of DNMTi treatment. Previous studies showed that transient (24-h) exposure of proliferating cells to 5-aza-CdR at 20–300 nM leads to long-term growth inhibition that mimics the prolonged patient responses seen in clinical settings (23–25). We tested various concentrations of 5-aza-CdR and vitamin C alone and in combination (SI Appendix, Fig. S1A). With HCT116 colorectal carcinoma cells, 5-aza-CdR alone showed the expected dose-dependent toxicity between 75 and 600 nM, with about 65% inhibition of cell growth at the highest concentration. Vitamin C alone at concentrations up to 57 μM had little effect on cell growth but was toxic at 228 μM (SI Appendix, Fig. S1B), in line with recent studies of high vitamin C concentrations (125–2,000 μM) (26, 27). In our combination approach, vitamin C increased the effects of low doses of 5-aza-CdR, with 57 μM vitamin C almost doubling the growth inhibition. Using the Chou–Talalay method (28), we found that the two compounds indeed acted synergistically, rather than additively, to inhibit cancer cell growth over the physiological ranges of vitamin C in healthy individuals (26–84 μM) (SI Appendix, Fig. S1C).
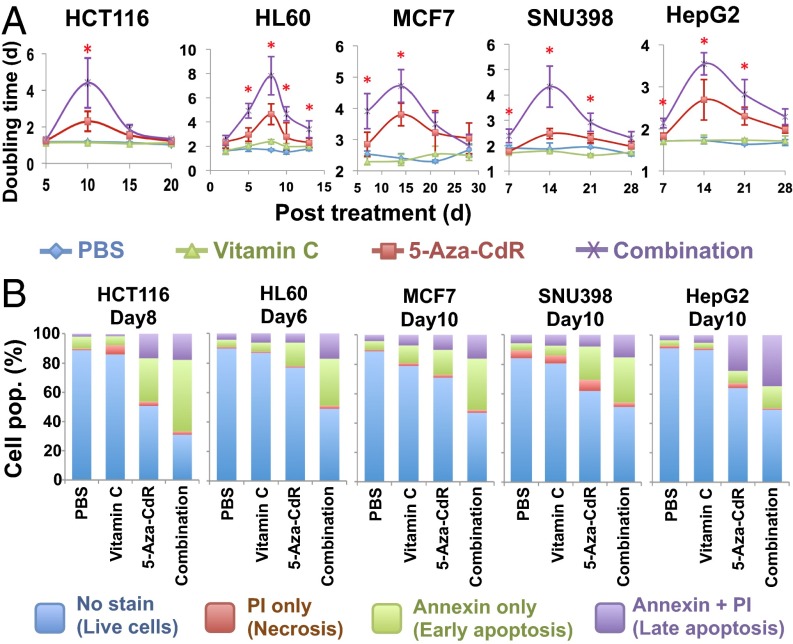
We selected four other cancer cell lines: HL60 (acute myeloid leukemia), MCF7 (breast carcinoma), and SNU398 and HepG2 (hepatocellular carcinomas) for further analysis. HCT116 and HL60 cells were treated with 300 nM 5-aza-CdR for 24 h, and MCF7, SNU398, and HepG2 cells were treated with three consecutive daily doses of 5-aza-CdR at 100 nM for 72 h to compensate for the fact that incorporation of the drug is dependent on cell-doubling time (5, 24). Prolonged increases in population-doubling times occurred in all cell lines, with peaks at 8–15 d after 5-aza-CdR treatment (Fig. 1A). The daily addition of vitamin C at 57 μM further lengthened the population-doubling times (Fig. 1A). The increased doubling time was correlated with enhanced apoptosis, as determined by annexin V measurements using flow cytometry (Fig. 1B). We observed a 1.5- to 2.2-fold increase in apoptosis with combination treatment relative to 5-aza-CdR treatment alone; daily vitamin C treatment alone increased apoptosis only slightly (Fig. 1B). Further analysis confirmed a synergy between vitamin C and 5-aza-CdR in inducing apoptosis in all these cell lines (SI Appendix, Fig. S1D).
Fig. 1.
The combination of 5-aza-CdR and physiological levels of vitamin C inhibits cell growth and enhances apoptosis. (A) Population-doubling time after treatment with PBS, vitamin C, 5-aza-CdR, or the combination. HCT116 and HL60 cells were treated with one dose of 300 nM 5-aza-CdR for 24 h; MCF7, SNU398, and HepG2 cells were treated with three consecutive daily doses of 5-aza-CdR at 100 nM, with drug withdrawal at 72 h. All cells were treated with daily doses of 57 μM vitamin C. Values are mean ± SD of three independent experiments. *P < 0.05 by paired Student’s t test comparing combination vs. 5-aza-CdR treatment. (B) Apoptosis analysis showing an increased percentage of apoptotic cells after combination treatment relative to either 5-aza-CdR or vitamin C alone. Cells were stained with annexin V-FITC and PI and were analyzed by flow cytometry.
Both 5-aza-CdR and vitamin C at high doses (in the micromolar and millimolar range, respectively) cause cytotoxicity by damaging DNA, as evidenced by a robust increase in phosphorylation of the histone variant H2AX at serine 139 (γ-H2AX) and an increase in p53 (27, 29). However, daily vitamin C alone at 57 μM did not increase the γ-H2AX or p53 levels in HCT116 cells (SI Appendix, Fig. S2), and 5-aza-CdR treatment increased them only slightly. Moreover, the combination treatment did not increase these levels further over 5-aza-CdR alone (SI Appendix, Fig. S2), suggesting that the apoptosis induced by combination treatment was not caused by DNA damage.
Combination Treatment of Vitamin C and 5-Aza-CdR Up-Regulates Endogenous Viral-Defense Genes.
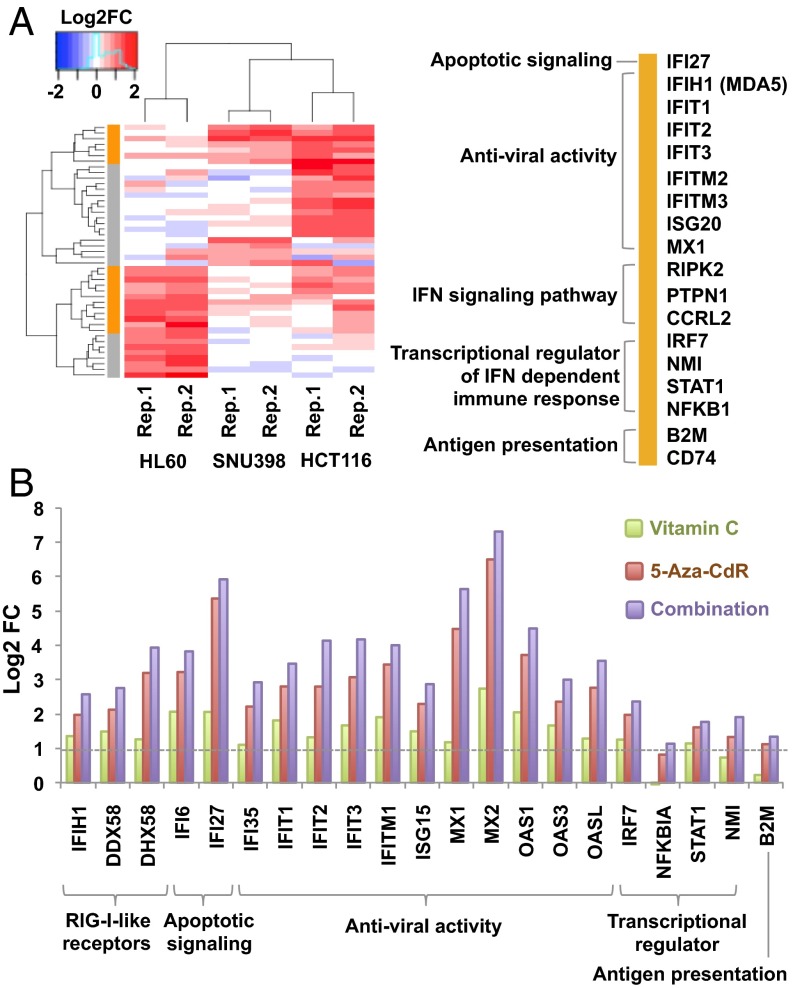
We next used gene-expression microarrays to screen for early changes in gene expression at day 5 after treatment, before the peak of apoptosis. Although few genes were down-regulated (SI Appendix, Fig. S3), genes responsive to IFN-α and -γ were consistently up-regulated by combination treatment (relative to 5-aza-CdR alone) in HCT116, HL60, and SNU398 cells (Fig. 2A and SI Appendix, Fig. S3). These up-regulated genes are involved in dsRNA recognition in the cytoplasm, participating in viral recognition and defense, IFN signaling, apoptotic signaling, and antigen presentation (Fig. 2A). We and others have shown that these viral defense-related genes can be up-regulated by 5-aza-CdR treatment and are responsible for inducing apoptosis in cancer cells (12, 13). Similarly, up-regulation of these genes by combination treatment may cause the increased apoptosis seen with combination treatment compared with 5-aza-CdR treatment alone.
Fig. 2.
Combination treatment of vitamin C and 5-aza-CdR up-regulates viral-defense genes. (A) Comparison of the expression of IFN-responsive genes upon combination treatment vs. 5-aza-CdR treatment in HCT116, HL60, and SNU398 cells. Treatment was as described in Fig. 1A. Transcripts were analyzed by microarray at 5 d after treatment. Orange indicates genes up-regulated in at least two cell lines (genes are listed at the right); gray denotes genes up-regulated in only one line. (B) A log2 plot of the expression of dsRNA defense genes after treatment with vitamin C, 5-aza-CdR, or their combination vs. untreated HCT116 cells. Values are calculated from fragments per kilobase per million fragments mapped and are averaged from two independent RNA-seq datasets.
We explored this possibility further by quantitation with RNA-sequencing (RNA-seq) in HCT116 cells. 5-Aza-CdR treatment alone up-regulated the viral-defense genes four- to 64-fold in HCT116 cells (Fig. 2B and SI Appendix, Fig. S4), as previously reported (12, 13, 24, 30). Daily vitamin C treatment alone increased the expression of these genes two- to fourfold relative to untreated cells (Fig. 2B and SI Appendix, Fig. S4), whereas the transcription of most housekeeping genes remained stable (SI Appendix, Fig. S5A). All the viral-defense genes were up-regulated further (up to 128-fold) by the combination treatment (Fig. 2B and SI Appendix, Fig. S4), but the expression of housekeeping genes was comparable to that under 5-aza-CdR treatment alone (SI Appendix, Fig. S5A). However, cytosolic sensors for DNA and Toll-like receptors that recognize viral DNA/RNA in endosomes were not up-regulated by any treatment (SI Appendix, Fig. S5B). Vitamin C and 5-aza-CdR therefore synergistically up-regulate the cytosolic viral RNA defense pathways, strengthening the viral mimicry state produced by DNMTi treatment alone (12, 13).
Combination Treatment of Vitamin C with 5-Aza-CdR Up-Regulates Endogenous Retroviruses in dsRNA Forms.
It is still unknown to what extent DNMTis up-regulate ERV transcripts genome-wide, which is key for triggering a viral-defense response. We analyzed the transcripts mapping uniquely to ERV loci (which are not covered by the expression array) in two replicates of the RNA-seq data for HCT116 cells after vitamin C, 5-aza-CdR, or combination treatment. The percentage of ERV transcripts relative to total reads did not change after vitamin C treatment but increased in cells treated with 5-aza-CdR and increased further with combination treatment (SI Appendix, Fig. S6A). The total reads for the ERV-1, ERVL, and ERVL-MaLR (mammalian apparent LTR retroposon) subfamilies were increased by both 5-aza-CdR and combination treatment, whereas the ERVK family was down-regulated relative to PBS controls (SI Appendix, Fig. S6B). Expression of other retrotransposons, such as the long interspersed nuclear elements (LINEs) and short interspersed nuclear elements (SINEs), was also increased by 5-aza-CdR and combination treatment; the expression levels of low-complexity repeats, simple repeats, tRNA, and small cytoplasmic RNAs remained stable (SI Appendix, Fig. S6A).
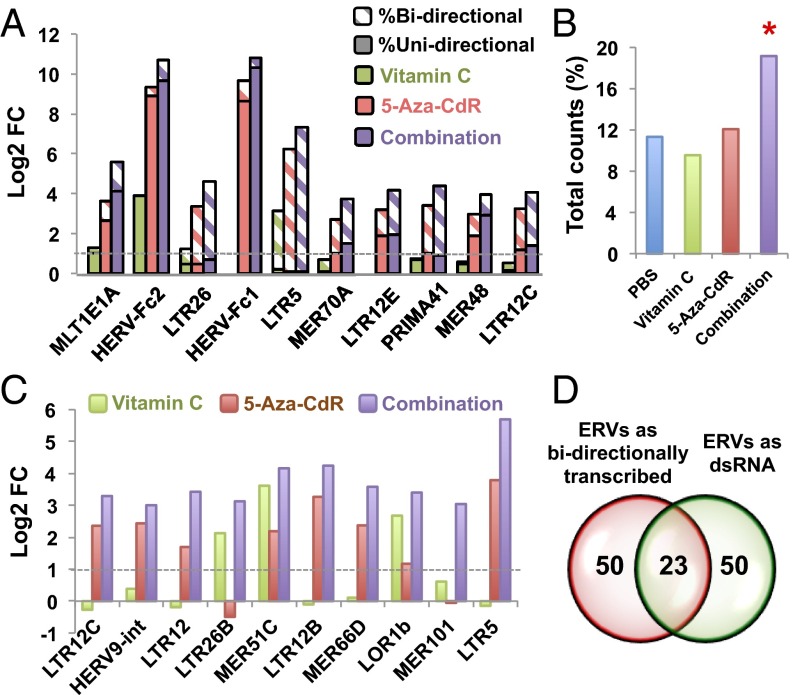
We then asked whether the up-regulated ERVs whose LTRs possess bidirectional promoter activities (31, 32) could form dsRNA secondary structures. Using directional RNA sequencing methods, we detected some unidirectionally transcribed ERVs (e.g., HERV-Fc1, HERV-Fc2, and MLT1E1A) as well as some transcribed bidirectionally (e.g., LTR5, LTR26, PRIMA41, and LTR12C) (Fig. 3A and SI Appendix, Fig. S6C). To test whether bidirectionally transcribed ERVs physically formed dsRNA secondary structures, we used the J2 antibody (33) specifically to pull down transcripts containing dsRNAs (Fig. 3 B–D). Remarkably, the combination treatment greatly increased the expression of ERVs as dsRNAs relative to 5-aza-CdR or vitamin C treatment alone (Fig. 3 B and C). In addition, the 50 most abundant bidirectionally transcribed ERVs largely overlapped with the ERVs that formed dsRNAs (Fig. 3D). LTR12C, which has more than 2,500 copies present in the human genome, was the most abundant dsRNA up-regulated by combination treatment in HCT116 cells (Fig. 3 A and C). Finally, transfection of polyinosinic:polycytidylic acid [poly(I:C)], which mimics the presence of dsRNA, up-regulated viral-defense genes in HCT116 cells to a similar extent as the combination treatment (SI Appendix, Fig. S7). Thus ERVs up-regulated as dsRNA by the combination treatment may be responsible for triggering a strong IFN response and for inducing apoptosis in cancer cells.
Fig. 3.
Combination treatment up-regulates endogenous retroviruses in both ssRNA and dsRNA forms in HCT116 cells. (A) Log2 fold change (FC) of transcripts uniquely mapped to individual ERV loci, comparing vitamin C, 5-aza-CdR, and combination treatment vs. untreated cells. Diagonal shading shows the percentage of bidirectional transcripts. (B) Percentage of double-stranded ERV (dsERV) counts relative to total reads in HCT116 cells after PBS, 5-aza-CdR, or combination treatment. *P < 0.05 by χ2 test for equality of proportions without continuity correction, compared with untreated cells. (C) Log2 fold change of dsERV levels comparing 5-aza-CdR and combination treatment (purple) relative to untreated cells. (D) The Venn diagram shows that ERVs that are bidirectionally transcribed largely overlap with ERVs that form dsRNA. The 50 most abundant bidirectionally transcribed ERVs and dsERVs in HCT116 cells after the combination treatment were determined by directional RNA-seq and dsRNA-seq, respectively.
Combination Treatment Promotes 5hmC Production at ERV LTRs.
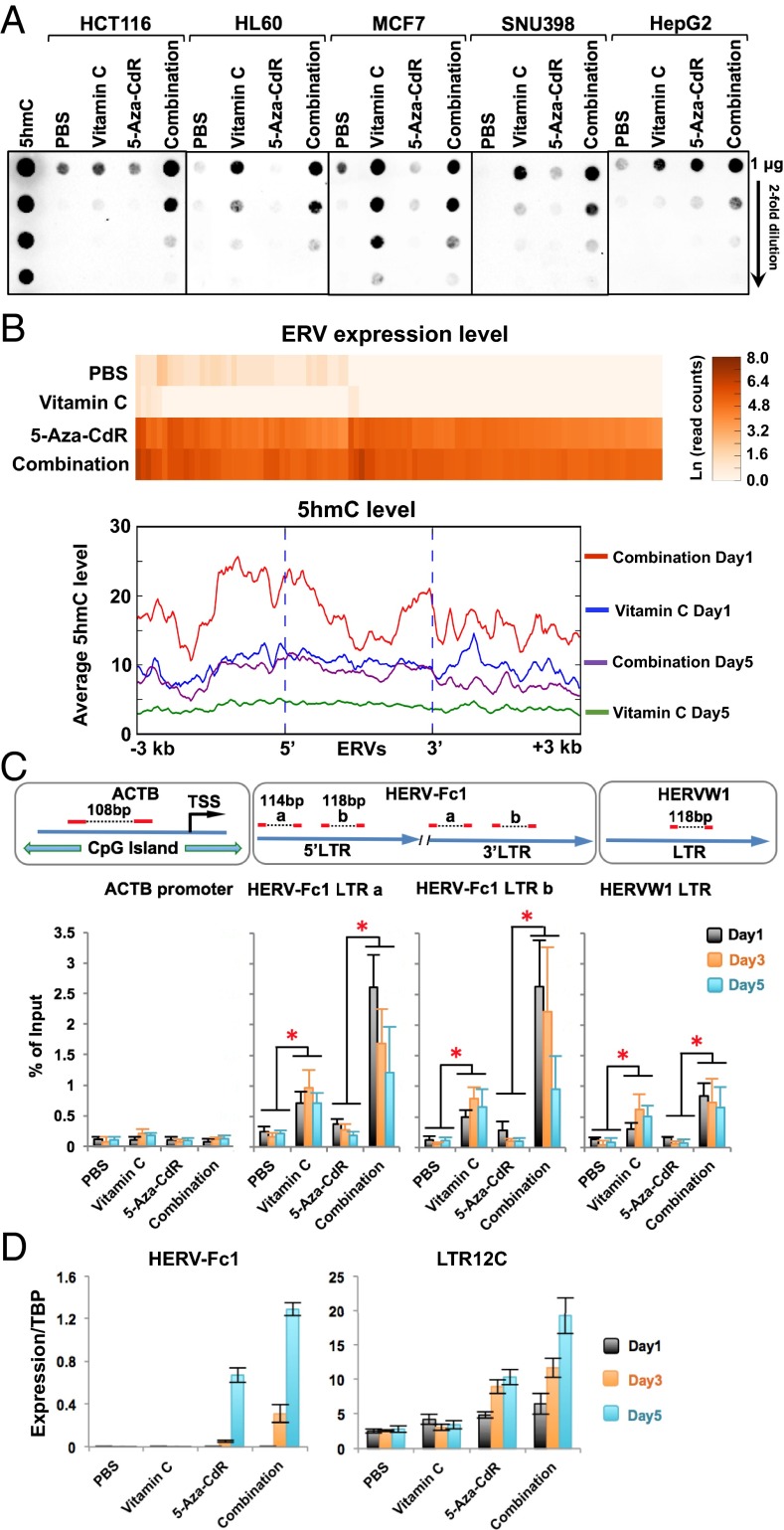
Because vitamin C is a cofactor for TET proteins (18, 22), we performed dot blot analysis with a 5hmC antibody to test whether vitamin C or combination treatment altered the global levels of 5hmC. Vitamin C treatment increased 5hmC in all five cell lines, with the biggest response seen in MCF7 cells (Fig. 4A). Combination treatment further increased 5hmC by about twofold relative to vitamin C treatment in all but MCF7 cells (Fig. 4A), in which combination treatment resulted in 5hmC levels comparable to those of vitamin C treatment alone (Fig. 4A). This result might be explained by the very high expression of TET2 and TET3 in the MCF7 cell line (SI Appendix, Fig. S8). In general, in most cell lines the expression of the three TET genes was unaffected by vitamin C, 5-aza-CdR, or the combination treatment (SI Appendix, Figs. S5C and S8). Therefore increased 5hmC by vitamin C and combination treatments was most likely caused by increased TET enzyme activity.
Fig. 4.
Combination treatment promotes 5hmC production at ERV LTRs. (A) Dot blot analysis of global 5hmC levels in cancer cells on day 5 after treatment with an antibody against 5hmC. (B) Genomewide analysis of 5hmC at up-regulated ERV loci in HCT116 cells on days 1 and 5 after vitamin C and combination treatment. The top 100 most up-regulated ERVs after combination treatment compared with untreated cells were identified by RNA-seq, and 5hmC enrichment and were analyzed using the Hydroxymethyl Collector Kit (Active Motif) followed by next-generation sequencing. (Upper) The heat map shows the expression levels of individual ERVs in the unit of natural log read counts. (Lower) The average base counts of 5hmC reads at all ERV loci and their ± 3-kb flanking region. (C, Lower) Quantification of 5hmC in HCT116 cells at LTRs of ERVs on days 1, 3, and 5 after treatment by qPCR. (Upper) Primer design. The promoter region of β-actin (ACTB) was used as a negative control. Values are presented as mean ± SD of five independent experiments. *P < 0.05 by Student’s t test. (D) Expression of HERV-Fc1 and LTR12C transcripts in HCT116 cells at days 1, 3, and 5 after treatment relative to the TATA-binding protein (TBP) levels by quantitative RT-PCR. Values are mean ± SD of three independent experiments.
Sodium bisulfite treatment is considered the gold standard for measuring DNA methylation, but it cannot distinguish between 5hmC and 5mC and therefore actually measures the sum of both modified bases. We examined the “modification” status (i.e., 5mC+5hmC) of DNA extracted before and after treatment on the Infinium HM450 DNA methylation BeadChip platform that is based on sodium bisulfite treatment. However, at 5 d after treatment we did not observe pronounced cytosine modification changes induced by vitamin C or by combination treatment as compared with 5-aza-CdR treatment (SI Appendix, Fig. S9A).
Because the HM450 BeadChip platform contains few repetitive elements such as ERVs, this result does not preclude localized decreases in cytosine modifications elsewhere in the genome. We next assessed dynamic 5hmC changes at ERV genomic loci, using biotin-based enrichment of 5hmC followed by sequencing on days 1 and 5 after treatment. The levels of 5hmC in untreated and 5-aza-CdR–treated cells were too low to allow successful sequencing. Notably, ERVs up-regulated by combination treatment gained high 5hmC levels on day 1 which were reduced by twofold on day 5 after the treatment (Fig. 4B and SI Appendix, Fig. S10). Vitamin C treatment also transiently increased the presence of 5hmC on day 1, but the levels were lower than seen with combination treatment (Fig. 4B and SI Appendix, Fig. S10). Quantitative PCR showed that the highest 5hmC levels appeared at the LTRs of HERV-Fc1 and HERVW1 on day 1 and gradually decreased on days 3 and 5 after combination treatment (Fig. 4C), whereas the expression of ERVs increased from day 1 to day 5 after the treatment (Fig. 4D). Reduction of 5hmC on day 5 may be caused by the loss of 5mC substrates after inhibition of DNMTs. These data also confirmed the global analysis shown in Fig. 4B. In contrast, 5hmC levels were increased only slightly by vitamin C treatment but did not change with 5-aza-CdR treatment (Fig. 4C). We next examined cytosine modification patterns in the HERV-Fc1 5′ LTR by bisulfite sequencing on day 5 after treatment (SI Appendix, Fig. S9B). No changes were induced by vitamin C or by combination treatment as compared with cells treated with 5-aza-CdR (SI Appendix, Fig. S9B). Further loss of cytosine modifications by combination treatment was not seen, suggesting that the presence of 5hmC at the ERV LTRs may directly facilitate the transient up-regulation of these ERVs.
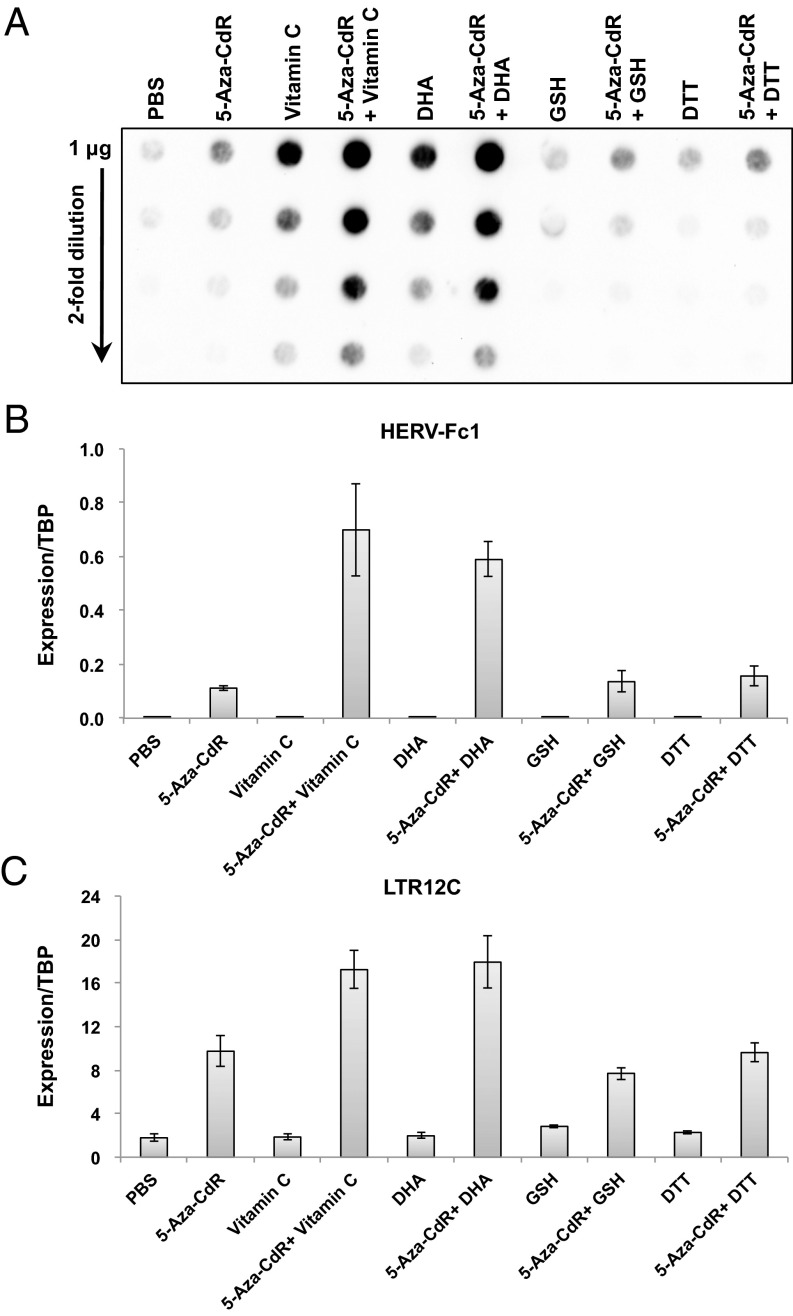
It is worth noting that the effects of vitamin C on 5hmC production and ERV up-regulation could also be achieved by treatment with dehydroascorbic acid (DHA) (Fig. 5), the oxidized form of vitamin C. Therefore, as previously reported (26), it is possible that vitamin C was oxidized to DHA before it was transported into the cells. On the other hand, other antioxidants such as glutathione (GSH) or DTT were unable to elicit the same effects (Fig. 5).
Fig. 5.
DHA elicits effects similar to those of vitamin C, but other antioxidants do not. (A) Dot blot analysis of global 5hmC levels with an antibody against 5hmC in HCT116 cells untreated or treated with 300 nM 5-aza-CdR for 24 h, daily doses of vitamin C or DHA at 57 μM, daily doses of GSH at 4.9 mM or DTT at 500 μM, and the combination of 5-aza-CdR with vitamin C, DHA, GSH, or DTT. (B and C) Expression levels of HERV-Fc1 (B) and LTR12C (C) by quantitative RT-PCR in HCT116 cells on day 5 after treatment. Values are presented as mean ± SD of three independent experiments.
TET2 Knockout Reduces the Expression of ERVs upon Combination Treatment.
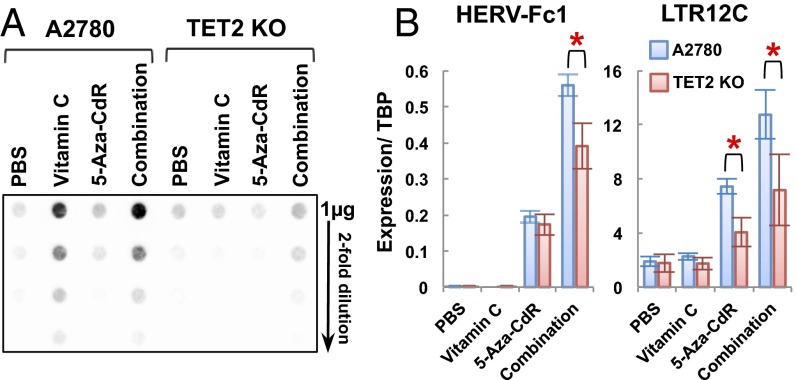
Because TETs are the only known enzymes that oxidize 5mC to 5hmC, we reasoned that the effects of vitamin C may be mediated by TET proteins. To test this hypothesis, we used a TET2-KO cell line established by CRISPR/Cas9 technology in A2780 human ovarian carcinoma cells (SI Appendix, Fig. S11). The global levels of 5hmC were about four- to eightfold lower in TET2-KO cells than in the wild-type A2780 cells after vitamin C or combination treatment (Fig. 6A), indicating that knocking out TET2 substantially decreases 5hmC levels in A2780 cells. Furthermore, we observed a significant reduction of HERV-Fc1 and LTR12C expression in TET2-KO cells compared with wild-type A2780 cells after combination treatment (Fig. 6B). Therefore the effects of vitamin C on ERV expression in A2780 cells are mediated in part by TET2 protein. Other TET proteins also might compensate for the loss of TET2, because an up-regulation of 5hmC and ERV expression was observed after combination treatment in the KO cells, although the effect was less than that seen in the wild-type A2780 cells (Fig. 6B). Interestingly, knocking out TET2 also reduces the expression of LTR12C after 5-aza-CdR treatment (Fig. 6B). It is possible that TET2 may still have weak activity in wild-type A2780 cells cultured in McCoy’s 5A medium with a low vitamin C concentration (2.9 μM) and may partly contribute to the effects of 5-aza-CdR alone.
Fig. 6.
TET2 is involved in ERV up-regulation upon combination treatment. (A) Dot blot analysis of global 5hmC levels in wild-type A2780 and TET2-KO cells after treatment. (B) Expression levels of HERV-Fc1 and LTR12C by quantitative RT-PCR in wild-type A2780 and TET2-KO cells on day 5 after treatment. Values are presented as mean ± SD of four independent experiments. *P < 0.05 by Student’s t test.
Patients with Hematological Neoplasia Are Often Markedly Vitamin C Deficient.
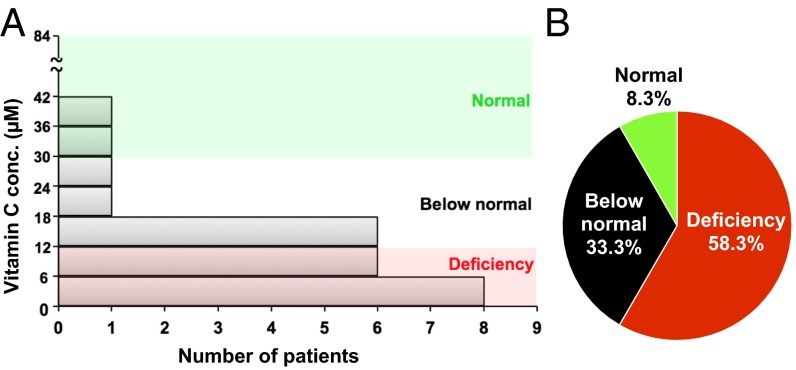
These results show that targeting the cancer DNA methylome by combining low-dose 5-aza-CdR and vitamin C stimulates the expression of ERVs, the induction of a cell-autonomous immune activation response, and increased apoptosis of cancer cells. The lack of vitamin C in a patient’s plasma might cause a poor response to DNMTi treatment in some patients. We therefore measured plasma concentrations of vitamin C in a small number of patients with miscellaneous hematologic malignancies. Strikingly, 58% of patients with hematological neoplasia who were not taking vitamin C supplements had severe vitamin C deficiency (serum concentration <11.4 μM, at which clinical features of scurvy may be manifested) (34), and 33% had vitamin C levels below the normal range (Fig. 7 and SI Appendix, Table S1). In this small sample, no specific pattern was observed with respect to patient age, disease state, or treatment status. In contrast, only 7.1% of the United States population was found to be deficient in vitamin C according to the 2003–2004 National Health and Nutrition Examination Survey (NHANES) (34). Vitamin C deficiency has been seen previously in patients with multiple types of cancer, including hematological malignancies (35–37). We predict that these patients might receive the most benefits from the combination treatment.
Fig. 7.
Plasma vitamin C concentrations in patients with hematopoietic malignancies. (A) The bar graph shows the distribution of plasma vitamin C levels in 24 patients with hematopoietic malignancies who were not taking vitamin C supplements. (B) The pie chart shows the percentage of patients with vitamin C deficiency (plasma level <11.4 μM), patients with levels below normal (11.4–26 μM), and patients with levels within the normal range (26–84 μM).
Discussion
Our work shows a remarkable synergy between an inhibitor of DNA methylation and a stimulator of the TET enzymes that catalyze the conversion of 5mC to 5hmC. We show that the combination establishes a viral mimicry response by the up-regulation of endogenous retroviruses, including dsRNA production, and induction of an innate immune response. These events are relevant to apoptosis and the targeting of cancer stem cells by DNMTis (12, 13). The addition of vitamin C to treatment protocols therefore may be a straightforward way to increase the clinical efficacy of such drugs in MDS and leukemia patients (7, 9–11, 24). Oral administration of vitamin C should be sufficient for the therapeutic strategy, because the concentrations reported in this study would not require i.v. administration.
Vitamin C is an essential nutrient for humans and has been reported to increase IFN levels in human cells upon virus infection (38). Gulo−/− mice, which are unable to synthesize vitamin C, exhibit a significant reduction in IFN-α/β production (39). In our study, daily treatment with vitamin C alone at physiological concentrations enhanced the expression of viral-defense genes relative to untreated cells. When combined with low-dose 5-aza-CdR, physiological concentrations of vitamin C synergistically inhibited cancer-cell growth and induced apoptosis. Such synergy was associated with increased ERV expression and dsRNA in treated cells. The mechanism of action differs from that of vitamin C at higher doses, which involves its pro-oxidant activity, including GSH inhibition, to generate reactive oxygen species (40). This activity has been shown to induce DNA damage and to enhance the sensitivities of myeloid malignancies, multiple myeloma, and cutaneous T-cell lymphoma to arsenic trioxide (41–44). It also can increase chemosensitivity of ovarian cancer cells (27) and selectively kill KRAS or BRAF mutant colorectal cancer cells by inhibiting GAPDH (26).
Our data suggest a mechanism involving both pharmacological knockdown of DNMTs by 5-aza-CdR (causing passive DNA demethylation) and the activation of TET proteins by vitamin C (inducing 5hmC production at LTRs of ERVs). The transcription of repetitive elements including ERVs is mainly silenced by DNA methylation (32, 45). Dnmt3a and/or Dnmt3b have been found to be specifically responsible for maintaining the methylation patterns of endogenous repetitive regions in mouse ES cells (46, 47). Therefore, pharmacological knockdown of DNMTs by 5-aza-CdR may allow TET proteins to be recruited to these regions when activated by vitamin C, oxidizing the 5mC residues into 5hmC and resulting in the up-regulation of repetitive-element expression. Consistent with this model, the highest level of 5hmC at the LTRs of both HERV-Fc1 and ERVW1 was found on day 1 after combination treatment (Fig. 4C), when most DNMTs have been degraded (4, 48). Indeed, this model is strengthened further by the fact that TET2 knockout reduces the synergistic effect of up-regulation of ERVs. The presence of 5hmC at the LTRs may be sufficient to allow transcription, since strong ERV expression was seen even though the overall cytosine modifications (5mC+5hmC) were unchanged after the combination treatment.
The finding that cancer patients are often vitamin C deficient and have less 5hmC in their tumors may suggest important roles for this vitamin in cancer prevention and treatment. In many types of cancers, 5hmC is greatly depleted or even lost completely (21). Furthermore, genetic mutations affecting enzymes regulating DNA methylation (DNMT3A/B, TET2, and IDH 1/2) contribute to altered 5mC/5hmC patterns, as observed in a subgroup of AMLs (21). Further study is warranted to determine whether vitamin C deficiency causes low 5hmC levels in cancers either with or without mutations in DNA methylation regulators. Based on preclinical studies, vitamin C deficiency in cancer patients also might complicate established therapeutic paradigms, because humans cannot synthesize vitamin C (39). In addition, most cells cultured in vitro are vitamin C deficient; this deficiency should be considered when interpreting the results of preclinical studies.
Our results show that supplementation with vitamin C can boost the outcome of DNMTi treatment of cancer cells in vitro. Clinical trials are needed to determine whether such supplements will enhance the therapeutic responses in patients treated with DNMTis. We also expect that the combination may provide a better response to immune checkpoint therapy, because high basal levels of viral-defense genes correlate with better clinical outcomes after anti-CTL4 immune checkpoint therapy (12, 49).
Materials and Methods
Participants.
Twenty-four patients diagnosed with hematological malignancies from Department of Hematology, Rigshospitalet, Copenhagen University Hospital, Denmark, were randomly selected for measurement of plasma vitamin C levels (details are listed in SI Appendix, Table S1). The study was approved by the Regional Ethical Committee, Capital Region of Denmark, and all participants provided written informed consent before enrollment. Detailed description of the materials and methods used in this study are provided in SI Appendix, SI Materials and Methods.
Cell Culture.
HCT116 (ATCC no. CCL-247), HL60 (ATCC no. CCL-240), MCF7 (ATCC no. HTB-22), SNU398 (ATCC no. CRL-2233), and HepG2 (ATCC no. HB-8065) cells were obtained from the American Type Culture Collection (ATCC; https://www.atcc.org/). A2780 and TET2-KO cells were obtained from the S.B.B. laboratory. Mutations contained in these cell lines are listed as Dataset S1. Information about cell culture, cell viability assays, and evaluation of combination effects are described in SI Appendix, SI Materials and Methods.
Drug Treatment and Measurement of Cell-Doubling Time.
HCT116 and HL60 cells were untreated or treated with a daily dose of 57 μM vitamin C, with 300 nM 5-aza-CdR for 24 h, or with the combination of daily vitamin C at 57 μM and transient exposure to 5-aza-CdR for 24 h. For MCF7, SNU398, and HepG2 cells, 5-aza-CdR treatment was performed with three daily consecutive doses at 100 nM (72-h exposure). Cell-doubling time was calculated as previously reported (13) and is described in SI Appendix, SI Materials and Methods.
Apoptosis Assay.
Cellular apoptosis was measured by annexin V and propidium iodide (PI) staining using the annexin V-FITC apoptosis detection kit (MBL) according to the manufacturer’s protocol. After staining, the cells were analyzed by a MoFlo Astrios Cell Sorter (Beckman Coulter, Inc.). Annexin V-FITC staining indicated apoptotic cells.
Microarray Gene-Expression Analysis.
Gene-expression analysis was performed at Sanford–Burnham Medical Institution, La Jolla, CA using the Illumina genome-wide expression BeadChip (HumanHT-12_V4.0_R1) (Illumina). Gene-expression data were normalized using the quantile normalization method with the lumi package, and differential expression analyses were performed using the limma package in R.
RNA-Seq.
For total RNA-seq with rRNA reduction and dsRNA sequencing, libraries were sequenced as single-end 75 bases on a NextSeq 500 instrument (Illumina) at the Van Andel Research Institute Genomics Core. For directional RNA-seq with rRNA reduction and strand specificity retained, libraries were prepared and sequenced as paired-end 50 bases on a HiSeq 2500 instrument (Illumina) at HudsonAlpha Institute for Biotechnology Genomic Services Laboratory. Details of J2 antibody pull-down, RNA-seq library preparation, and data processing are described in SI Appendix, SI Materials and Methods.
Dot Blot Analysis and Chemical Pulldown of 5hmC.
Dot blot analysis of 5hmC was performed as described previously (18). Chemical capture of 5hmC was performed using the Hydroxymethyl Collector Kit (Active Motif, 55013) according to the manufacturer’s protocol with minor modifications described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the Van Andel Research Institute (VARI) Flow Cytometry Core; the VARI Genomics Core; the HudsonAlpha Institute for Biotechnology Genomic Services Laboratory; the VARI Bioinformatics and Biostatistics Core; Zack Ramjan for technical assistance; and David Nadziejka (VARI) for technical editing of the article. This work was supported by National Cancer Institute Grant R01CA082422; by VARI through funding from the VARI–Stand Up To Cancer Epigenetics Dream Team (Stand Up To Cancer is a program of the Entertainment Industry Foundation, administered by the American Association for Cancer Research) (P.A.J., S.B.B., and K.G.); by the Vicky Joseph Cancer Research Fund (G.L.); by the Novo Nordisk Foundation and the Danish Cancer Society (K.G.); and by the Lundbeck Foundation (A.D.Ø.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited at the Gene Expression Omnibus (GEO), www.ncbi.nlm.nih.gov/geo/ (accession codes GSE77031, GSE77032, GSE77034, GSE77035, and GSE77036).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612262113/-/DCSupplemental.
References
- 1.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11(10):726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429(6990):457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 3.Sharma S, De Carvalho DD, Jeong S, Jones PA, Liang G. Nucleosomes containing methylated DNA stabilize DNA methyltransferases 3A/3B and ensure faithful epigenetic inheritance. PLoS Genet. 2011;7(2):e1001286. doi: 10.1371/journal.pgen.1001286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghoshal K, et al. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol. 2005;25(11):4727–4741. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Bender CM, et al. Roles of cell division and gene transcription in the methylation of CpG islands. Mol Cell Biol. 1999;19(10):6690–6698. doi: 10.1128/mcb.19.10.6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang X, et al. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26(4):577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kantarjian H, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: Results of a phase III randomized study. Cancer. 2006;106(8):1794–1803. doi: 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- 8.Kantarjian HM, et al. Multicenter, randomized, open-label, phase III trial of decitabine versus patient choice, with physician advice, of either supportive care or low-dose cytarabine for the treatment of older patients with newly diagnosed acute myeloid leukemia. J Clin Oncol. 2012;30(21):2670–2677. doi: 10.1200/JCO.2011.38.9429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenaux P, et al. International Vidaza High-Risk MDS Survival Study Group Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: A randomised, open-label, phase III study. Lancet Oncol. 2009;10(3):223–232. doi: 10.1016/S1470-2045(09)70003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fenaux P, et al. Azacitidine prolongs overall survival compared with conventional care regimens in elderly patients with low bone marrow blast count acute myeloid leukemia. J Clin Oncol. 2010;28(4):562–569. doi: 10.1200/JCO.2009.23.8329. [DOI] [PubMed] [Google Scholar]
- 11.Issa JP, et al. Safety and tolerability of guadecitabine (SGI-110) in patients with myelodysplastic syndrome and acute myeloid leukaemia: A multicentre, randomised, dose-escalation phase 1 study. Lancet Oncol. 2015;16(9):1099–1110. doi: 10.1016/S1470-2045(15)00038-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiappinelli KB, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162(5):974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roulois D, et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162(5):961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee YG, et al. Korean Society of Haematology AML/MDS working party Comparative analysis between azacitidine and decitabine for the treatment of myelodysplastic syndromes. Br J Haematol. 2013;161(3):339–347. doi: 10.1111/bjh.12256. [DOI] [PubMed] [Google Scholar]
- 15.Schecter J, Galili N, Raza A. MDS: Refining existing therapy through improved biologic insights. Blood Rev. 2012;26(2):73–80. doi: 10.1016/j.blre.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Balch C, Nephew KP. Epigenetic targeting therapies to overcome chemotherapy resistance. Adv Exp Med Biol. 2013;754:285–311. doi: 10.1007/978-1-4419-9967-2_14. [DOI] [PubMed] [Google Scholar]
- 17.Wrangle J, et al. Alterations of immune response of non-small cell lung cancer with azacytidine. Oncotarget. 2013;4(11):2067–2079. doi: 10.18632/oncotarget.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blaschke K, et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500(7461):222–226. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pastor WA, Aravind L, Rao A. TETonic shift: Biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14(6):341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfeifer GP, Kadam S, Jin SG. 5-hydroxymethylcytosine and its potential roles in development and cancer. Epigenetics Chromatin. 2013;6(1):10. doi: 10.1186/1756-8935-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung TL, et al. Vitamin C promotes widespread yet specific DNA demethylation of the epigenome in human embryonic stem cells. Stem Cells. 2010;28(10):1848–1855. doi: 10.1002/stem.493. [DOI] [PubMed] [Google Scholar]
- 23.Cashen AF, Shah AK, Todt L, Fisher N, DiPersio J. Pharmacokinetics of decitabine administered as a 3-h infusion to patients with acute myeloid leukemia (AML) or myelodysplastic syndrome (MDS) Cancer Chemother Pharmacol. 2008;61(5):759–766. doi: 10.1007/s00280-007-0531-7. [DOI] [PubMed] [Google Scholar]
- 24.Tsai HC, et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell. 2012;21(3):430–446. doi: 10.1016/j.ccr.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bender CM, Pao MM, Jones PA. Inhibition of DNA methylation by 5-aza-2′-deoxycytidine suppresses the growth of human tumor cell lines. Cancer Res. 1998;58(1):95–101. [PubMed] [Google Scholar]
- 26.Yun J, et al. Vitamin C selectively kills KRAS and BRAF mutant colorectal cancer cells by targeting GAPDH. Science. 2015;350(6266):1391–1396. doi: 10.1126/science.aaa5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma Y, et al. High-dose parenteral ascorbate enhanced chemosensitivity of ovarian cancer and reduced toxicity of chemotherapy. Sci Transl Med. 2014;6(222):222ra18. doi: 10.1126/scitranslmed.3007154. [DOI] [PubMed] [Google Scholar]
- 28.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58(3):621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 29.Karpf AR, Moore BC, Ririe TO, Jones DA. Activation of the p53 DNA damage response pathway after inhibition of DNA methyltransferase by 5-aza-2′-deoxycytidine. Mol Pharmacol. 2001;59(4):751–757. [PubMed] [Google Scholar]
- 30.Karpf AR, et al. Limited gene activation in tumor and normal epithelial cells treated with the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine. Mol Pharmacol. 2004;65(1):18–27. doi: 10.1124/mol.65.1.18. [DOI] [PubMed] [Google Scholar]
- 31.Dunn CA, Romanish MT, Gutierrez LE, van de Lagemaat LN, Mager DL. Transcription of two human genes from a bidirectional endogenous retrovirus promoter. Gene. 2006;366(2):335–342. doi: 10.1016/j.gene.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20(2):116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- 33.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80(10):5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schleicher RL, Carroll MD, Ford ES, Lacher DA. Serum vitamin C and the prevalence of vitamin C deficiency in the United States: 2003-2004 National Health and Nutrition Examination Survey (NHANES) Am J Clin Nutr. 2009;90(5):1252–1263. doi: 10.3945/ajcn.2008.27016. [DOI] [PubMed] [Google Scholar]
- 35.Mayland CR, Bennett MI, Allan K. Vitamin C deficiency in cancer patients. Palliat Med. 2005;19(1):17–20. doi: 10.1191/0269216305pm970oa. [DOI] [PubMed] [Google Scholar]
- 36.Huijskens MJ, Wodzig WK, Walczak M, Germeraad WT, Bos GM. Ascorbic acid serum levels are reduced in patients with hematological malignancies. Results Immunol. 2016;6:8–10. doi: 10.1016/j.rinim.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nannya Y, Shinohara A, Ichikawa M, Kurokawa M. Serial profile of vitamins and trace elements during the acute phase of allogeneic stem cell transplantation. Biol Blood Marrow Transplant. 2014;20(3):430–434. doi: 10.1016/j.bbmt.2013.12.554. [DOI] [PubMed] [Google Scholar]
- 38.Dahl H, Degré M. The effect of ascorbic acid on production of human interferon and the antiviral activity in vitro. Acta Pathol Microbiol Scand [B] 1976;84B(5):280–284. doi: 10.1111/j.1699-0463.1976.tb01938.x. [DOI] [PubMed] [Google Scholar]
- 39.Kim Y, et al. Vitamin C is an essential factor on the anti-viral immune responses through the production of interferon-alpha/beta at the Initial stage of influenza A virus (H3N2) infection. Immune Netw. 2013;13(2):70–74. doi: 10.4110/in.2013.13.2.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Linster CL, Van Schaftingen E. Vitamin C. Biosynthesis, recycling and degradation in mammals. FEBS J. 2007;274(1):1–22. doi: 10.1111/j.1742-4658.2006.05607.x. [DOI] [PubMed] [Google Scholar]
- 41.Amadori S, Fenaux P, Ludwig H, O’dwyer M, Sanz M. Use of arsenic trioxide in haematological malignancies: Insight into the clinical development of a novel agent. Curr Med Res Opin. 2005;21(3):403–411. doi: 10.1185/030079904X20349. [DOI] [PubMed] [Google Scholar]
- 42.Michel L, et al. Arsenic trioxide induces apoptosis of cutaneous T cell lymphoma cells: Evidence for a partially caspase-independent pathway and potentiation by ascorbic acid (vitamin C) J Invest Dermatol. 2003;121(4):881–893. doi: 10.1046/j.1523-1747.2003.12479.x. [DOI] [PubMed] [Google Scholar]
- 43.Hoffer LJ, et al. Phase I clinical trial of i.v. ascorbic acid in advanced malignancy. Ann Oncol. 2008;19(11):1969–1974. doi: 10.1093/annonc/mdn377. [DOI] [PubMed] [Google Scholar]
- 44.Bejanyan N, et al. A phase 2 trial of combination therapy with thalidomide, arsenic trioxide, dexamethasone, and ascorbic acid (TADA) in patients with overlap myelodysplastic/myeloproliferative neoplasms (MDS/MPN) or primary myelofibrosis (PMF) Cancer. 2012;118(16):3968–3976. doi: 10.1002/cncr.26741. [DOI] [PubMed] [Google Scholar]
- 45.Bestor TH, Tycko B. Creation of genomic methylation patterns. Nat Genet. 1996;12(4):363–367. doi: 10.1038/ng0496-363. [DOI] [PubMed] [Google Scholar]
- 46.Liang G, et al. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol Cell Biol. 2002;22(2):480–491. doi: 10.1128/MCB.22.2.480-491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10(11):805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patel K, et al. Targeting of 5-aza-2′-deoxycytidine residues by chromatin-associated DNMT1 induces proteasomal degradation of the free enzyme. Nucleic Acids Res. 2010;38(13):4313–4324. doi: 10.1093/nar/gkq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snyder A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.