Abstract
Adaptation to hypoxia depends on a conserved α/β heterodimeric transcription factor called Hypoxia Inducible Factor (HIF), whose α-subunit is regulated by oxygen through different concurrent mechanisms. In this study, we have identified the RNA binding protein dMusashi, as a negative regulator of the fly HIF homologue Sima. Genetic interaction assays suggested that dMusashi participates of the HIF pathway, and molecular studies carried out in Drosophila cell cultures showed that dMusashi recognizes a Musashi Binding Element in the 3′ UTR of the HIFα transcript, thereby mediating its translational repression in normoxia. In hypoxic conditions dMusashi is downregulated, lifting HIFα repression and contributing to trigger HIF-dependent gene expression. Analysis performed in mouse brains revealed that murine Msi1 protein physically interacts with HIF-1α transcript, suggesting that the regulation of HIF by Msi might be conserved in mammalian systems. Thus, Musashi is a novel regulator of HIF that inhibits responses to hypoxia specifically when oxygen is available.
INTRODUCTION
Animals can adapt to variations of oxygen levels by modifying their transcription profile. Oxygen-dependent gene expression is regulated mostly by the Hypoxia Inducible Factor (HIF), an evolutionary conserved heterodimeric transcription factor, whose α and β-subunits belong to the basic-Helix-Loop-Helix-PAS (bHLH-PAS) protein family (1). While the HIFβ subunit is constitutively expressed, HIFα expression is regulated primarily at the level of protein stability (2). HIFα is rapidly degraded in normoxia and stabilized in hypoxia, being its degradation dependent on the hydroxylation of key prolyl residues localized in the HIFα oxygen-dependent degradation domain (3,4). Hydroxylation of these prolines is mediated by specific prolyl-4-hydroxylases, termed PHDs, that utilise molecular oxygen as a co-substrate for catalysis, and are hence considered oxygen sensors (5,6). The bHLH-PAS proteins Similar (Sima) and Tango (Tgo) are respectively the Drosophila HIFα and HIFβ homologs (7), while the fatiga gene encodes the Drosophila PHD isoforms that control Sima stability in an oxygen dependent manner (8,9). The Drosophila HIF system has been shown to control adaptation to hypoxia in vivo through mechanisms identical to those operating in mammalian systems (10).
The Musashi (Msi) family of RNA binding proteins is an evolutionarily conserved group of proteins that regulate translation of target mRNAs by binding to consensus sequences, termed Musashi Binding Elements (MBEs), at their 3′ untranslated region (3′ UTR) (11–15). Musashi proteins have clear roles in stem cell maintenance and cell fate determination across the metazoan lineage (16,17). Two Msi paralogs, Msi1 and Msi2, are present in vertebrate species, and a few of their mRNA targets have been so far identified (17). These include the Notch inhibitor Numb (18), the cell cycle regulator CDKN1A/p21 (19), the neural microtubule-associated protein Doublecortin (20), the multidomain tumor suppressor protein Adenomatous Polyposis Coli -APC- (21), the Notch ligand Jagged1 (15), the phosphatase PTEN (22), the integral membrane protein Tetraspanin 3 (23) and the meiotic regulator c-mos in Xenopus laevis (24). In Drosophila melanogaster, a single musashi orthologous gene occurs (dmsi) which is known to mediate translational repression of the transcription factor Tramtrack69 (Ttk69), thereby controlling asymmetric cell divisions during adult sensory organ differentiation and photoreceptor differentiation during eye development (11,25,26). dMsi is also required for male germ line stem cell maintenance, although the mRNA target in this context is unknown (27).
In this study we show that dMsi is a novel inhibitor of HIF-dependent responses to hypoxia in Drosophila. dMsi recognizes a MBE within the 3′ UTR of sima mRNA and mediates its translational repression in normoxic conditions. dMsi is downregulated in hypoxia, lifting Sima repression and contributing to trigger HIF-dependent gene expression. We provide evidence that HIF regulation by Msi might be conserved in mammals.
MATERIALS AND METHODS
Identification of Musashi binding elements
To identify RNA regulatory motifs in sima or HIFα 3′ UTRs, we used the computational platform RegRNA2.0 (http://regrna2.mbc.nctu.edu.tw/index.html). Several Musashi Binding Elements were inferred through this analysis, and we focused on those conserved between species.
Fly strains
Flies were reared on a cornmeal-yeast-sucrose medium at 25°C. All the strains used in this study have been previously described. These fly lines were: HRE-LacZ (7), msi1 (25), msiDf(3R)6203 (Bloomington stock number 7682), fga9 and sima07607 (8), dSRF-Gal4 (28). The following stocks were from the Vienna Drosophila RNAi Center: UAS-msi RNAi (VDRC #44895), UAS-sima RNAi (VDRC #106504).
Cell culture
Semi-adherent Schneider (S2R+) Drosophila cells were maintained in Schneider Drosophila medium (Sigma) supplemented with Penicillin (50 U/ml, Invitrogen), Streptomycin (50 ug/ml, Invitrogen) and 10% FBS (Invitrogen) at 25°C in 25 cm2 T-flasks (Greiner). Synthesis of dsRNA and RNA interference treatments in S2R+ cells were performed as previously described (29).
Plasmids, transfection and Luciferase assays
For transient transfection experiments in S2R+ cells, we employed previously characterized vectors: pAC-LacZ, HRE-LucFF, pAC-LucRen (30) and pAC-Msi (11). All vectors generated in this manuscript employ the copper-inducible pMT/V5-His plasmid (Invitrogen) as the backbone vector. To obtain pMT-Luciferase Renilla (pMT-LucRen), LucRen coding sequence from pRL-SV40 vector (Promega) was directionally cloned into pMT/V5-His using HindIII/XbaI. pMT-Luciferase Firefly reporter construct (pMT-LucFF) was obtained by subcloning the coding sequence of LucFF from pGL3 vector (Promega) into EcoRI/XbaI sites of pMT/V5-His. All 3′ UTRs used here were obtained by PCR of cDNA obtained from Drosophila embryos and subsequently cloned into the XbaI/ApaI restriction sites of pMT-LucFF. The employed primers are as follows,
3′ UTR adh
Fw: 5′-GCTCTAGAGAAGTGATACTCCCAAAAAA-3′
Rv: 5′-GCCATTGGGCCCATCATAGGAAAATGAATTGC-3′
3′ UTR ttk69
Fw: 5′-GCTCTAGATCTCTGGGCACCTCACACCAAG-3′
Rv: 5′-GCCATTGGGCCCGAGTGTTTTTTGCATTGTGTATTT-3′
3′ UTR sima
Fw: 5′-GCTCTAGAATTACCAGTACCTTAGCATGCA-3′
Rv: 5′-GCCATTGGGCCCCAAAAACTTTTTTTCTCGTCACAGC-3′
The point mutations in the MBE of sima 3′ UTR (3′ UTR sima MBEmut) were introduced by nested PCR with the additional primers:
Fw: 5′-CACACTTGAATAGTTTTCTTCCCATGTTAACTGCC-3′
Rv: 5′-GGCAGTTAACATGGGAAGAAAACTATTCAAGTGTG-3′
For transfection experiments, 350.000 cells per well were plated in 24-well plate (Grenier) and 0.3 μg of total plasmid DNA were transfected employing Effectene transfection reagent (Qiagen). All pMT-LucFF-3′ UTR constructs were co-transfected (1:1) with a Renilla luciferase plasmid (pMT-LucRen) to normalize transfection efficiency. Expression of all pMT-Luc reporters was induced 24h after transfection by addition of 0.7 mM CuSO4 for 7 h. Luciferase activity was measured by using the Dual-Glo Luciferase Assay System (Promega) following the instructions of the manufacturer and measured in a Veritas Microplate Luminometer (Turner BioSystems).
Reverse transcription and qPCR (RT-qPCR)
Total RNA from S2R+ cells and fly embryos exposed to different treatments was isolated using 500 μl of Trizol reagent (Invitrogen). One μg of total RNA, measured with a NanoDrop 1000 spectrophotometer (Thermo Scientific), was used as template for complementary DNA (cDNA) synthesis, using SuperScript III First Strand Synthesis System for RT-PCR (Invitrogen). Quantitative PCR reactions were conducted employing a 1/30 dilution of cDNA sample, SYBRGreen, ROX reference dye and Taq DNA polymerase (Invitrogen) in a Mx3005P real time PCR device (Stratagene). The annealing temperature was 60°C and the elongation time at 72°C was 60 s. Relative mRNA abundances were estimated employing internal standard curves with a PCR efficiency of 100 ± 10% for each set of primer in each experiment. The MxPro qPCR software was used to analyze the data. The Ribosome protein Large 29 (RpL29) gene was used as normalizer. The primers utilized were:
firefly luciferase:
Fw: 5′-CATAGAACTGCCTGCGTGAG-3′ / Rv: 5′-ACCGTGATGGAATGGAACAA-3′
Renilla luciferase:
Fw: 5′-AAGTTCGTCGTCCAACATTATC / Rv: 5′-GGCACCTTCAACAATAGCATT-3′
rpl29:
Fw: 5′-GAACAAGAAGGCCCATCGTA / Rv: 5′-AGTAAACAGGCTTTGGCTTGC-3′
lactate-dehydrogrenase (ldh):
Fw: 5′-GTGTGACATCCGTGGTCAAG / Rv: 5′-CTACGATCCGTGGCATCTTT-3′
fgaB:
Fw: 5′-CACCCTTTCTCTGCACAACA / Rv: 5′-TGTCCAAAAGTTCCCGAAAG-3′
spermine oxidase:
Fw: 5′-GCATGGTTGGAGGATGTCTT / Rv: 5′-TCTGGGATTTTCCACCTCAG-3′
sequoia:
Fw: 5′-TCGCAGTACACCTTCACGAC / Rv: 5′-AGCAGCTCGTTCTTCAGCTC-3′
sima:
Fw: 5′-AGCCCAATCTGCCGCCAACC / Rv: 5′-TGGAGGCCAGGTGGTGGGAC-3′
SDS-PAGE and immunoblotting
Protein extracts from S2R+ cells or embryos (stage E14 to stage E17) were prepared in RIPA buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.1% (v/v) SDS, 0.5% (w/v) sodium deoxycholate and 1% Triton X-100) with the addition of proteinase inhibitory cocktail (Invitrogen) and kept at 4°C. 25–50 μg of total extracts were loaded on a 6–11% polyacrylamide gel, subjected to electrophoresis and then blotted onto nitrocellulose membrane (Bio-Rad). Thereafter, membranes were blocked for 1 h at room temperature with 5% nonfat milk or BSA in TBS with 0.1% Tween 20 (TBS-T) and incubated overnight with rabbit anti-Sima (29); rat anti-dMsi 3A5 (26) or mouse anti-tubulin (Invitrogen) in 5% nonfat milk in TBS-T. The secondary antibodies used were HRP conjugated (1/5000, Jackson ImmunoResearch). Immunoblots were developed with the ECL prime detection reagent (Amersham).
β-Galactosidase activity
For X-gal stainings, late-stage embryos were dechorionated in bleach for 1 min, incubated with heptane for 5 min, fixed 20 min at room temperature in glutaraldehyde 0,5% in PBS and washed in PT (0.1% Triton X-100 in PBS). Tissues were incubated in 500 μl of staining solution (5 mM K4Fe2+, 5 mM K4Fe3+ and 0.2% of 5-bromo-4 chloro-3 indolyl β-d-galactopyranoside in PT) at 37°C and the colorimetric reaction was monitored. Reactions were stopped by several washes with PT and recorded with a Nomarski Olympus BX-60 microscope.
RNA Immunoprecipitation (RIP)
S2R+ cells were harvested and lysed in the following extraction buffer (50 mM Tris–HCl pH 7.5, 1% (v/v) NP-40, 0.5% (w/v) sodium deoxycholate, 0.05% (w/v) SDS, 1 mM EDTA, 150 mM NaCl) containing complete protease inhibitor cocktail and RNasin (Promega). Extracts were sonicated with a Bioruptor at high amplitude with three 30-s bursts and insoluble material was precipitated. Supernatant was precleared with GammaBind G sepharose beads (GE Healthcare) for 30 min at 4°C before addition of anti-dMsi or non-immune IgG for incubation overnight. Complexes were immunoprecipitated with GammaBind G sepharose beads for 1 h and after three washes with the above extraction buffer; RNA was extracted with phenol–chloroform. cDNA was prepared from one-half of the RNA from anti-dMsi or IgG RIPs using 10-mer random primers. For every RNA fragment analyzed, each sample was quantified from three independent RIPs. The cDNA and no–reverse transcription control were analyzed by qPCR with the following primers:
adh
Fw: 5′-AGATAAATGGGAGCGGCAGG-3′; Rv: 5′-GTGCAATTCCTCCGCAATCC-3′
ttk69
Fw: 5′-GTTAATCCCGGGTCTGGGTC-3′; Rv: 5′-GATGTTACGGGGAACGGTGT-3′
sima
Fw: 5′-CGAATGGCGAAGGTGAAC-3′; Rv: 5′-CTTGGCTGCTTGGGTTTG-3′
CLIP-RT-qPCR assay
CLIP-RT-qPCR assays were performed as described previously (31) with modifications. Mouse embryonic brains were UV cross-linked at 254nm (UV-B) with 400 mJ/cm2 three times, lysed in PXL buffer; and immunoprecipitated for 2 h at 4°C with 4 μg polyclonal anti-mMsi1 or rabbit normal IgG antibodies bound to Dynabeads Protein G (Invitrogen). IPed lysates were washed with PXL, high salt wash buffer and PNK buffer twice respectively to completely remove indirect protein–protein interactions. To purify proteins directly bound to RNA, the complexes on beads were digested with proteinase K (Roche), followed by RNA isolation after phenol/chloroform extraction. RT reaction was performed as described in iScript cDNA Synthesis kit (BioRad). Quantitative RT-PCR was performed using Thunderbird Syber qPCR mix (Toyobo) on the Step-one-plus Real time PCR system (Life Technologies). CLIP-qrt-PCR enrichments were normalized by quantifying relative levels of gapdh mRNA, which is not a target of mMsi1. The following primers were used:
gapdh
Fw: 5′-AGGTCGGTGTGAACGGATTTG-3′; Rv: 5′-TGTAGACCATGTAGTTGAGGTCA-3′
HIF-1α
Fw: 5′-TGGAAGGTATGTGGCATTTATTTGG-3′; Rv: 5′-CAGAGGGACTGTTTTGAGTTGGT-3′
Epas1(HIF-2α)
Fw: 5′-GTGTGACAGTCCCAGGAGAGAAG-3′; Rv: 5′-TAGCGGCAACAGCACACAC-3′
HIF-3α
Fw: 5′-TACCTTATTCACCCCTCTTTGGA-3′; Rv: 5′-AGCCAGGACAATTTTTCCGGT-3′
Dcx
Fw: 5′-TTTCAGGAGCAAAACTCTTCAGG-3′; Rv: 5′-TTCTGTTTGGCAGTGAGAGCA-3′
Analysis of developmental phenotypes
For pupariation analysis, 50 synchronized first-instar larvae were placed in standard food and the number of pupae was measured every 24 h daily until day 8 after larval eclosion. To evaluate the maximal third-instar larval size, 25 synchronized first-instar larvae were placed in standard food and visually monitored throughout their development two times per day. To measure the maximal size reached, 25 third-instar wandering larvae were imaged per genotype. The volume of each larva was calculated from the area measured from photographs using ImageJ.
Quantification of tracheal phenotypes
Branching quantification was performed as previously reported (28). Briefly, first-instar larvae were placed in fresh vials, at a density of 25 individuals per vial and let them develop to third instar. Wandering larvae were ether anesthetized and ramifications of terminal cells of the third segment dorsal tracheal branch were counted and photographed using bright-field microscopy.
Hypoxia treatment
Hypoxia was applied in a Forma Scientific 3131 incubator, by regulating the proportions of oxygen and nitrogen at 25°C.
Statistical analysis
All the statistical analysis were performed using InfoStat version 2009 (Grupo InfoStat, FCA, Universidad Nacional de Córdoba, Argentina). In all cases normality and variance homogeneity were tested with the Shapiro-Wilk test and Levene's test, respectively. For all graphs, the error bars represent the standard error of the mean (SEM) of at least three independent experiments. The experimental groups with different letters indicate statistically significant differences. P<0.05 was considered statistically significant.
RESULTS
dMusashi behaves as a negative regulator of HIF/Sima
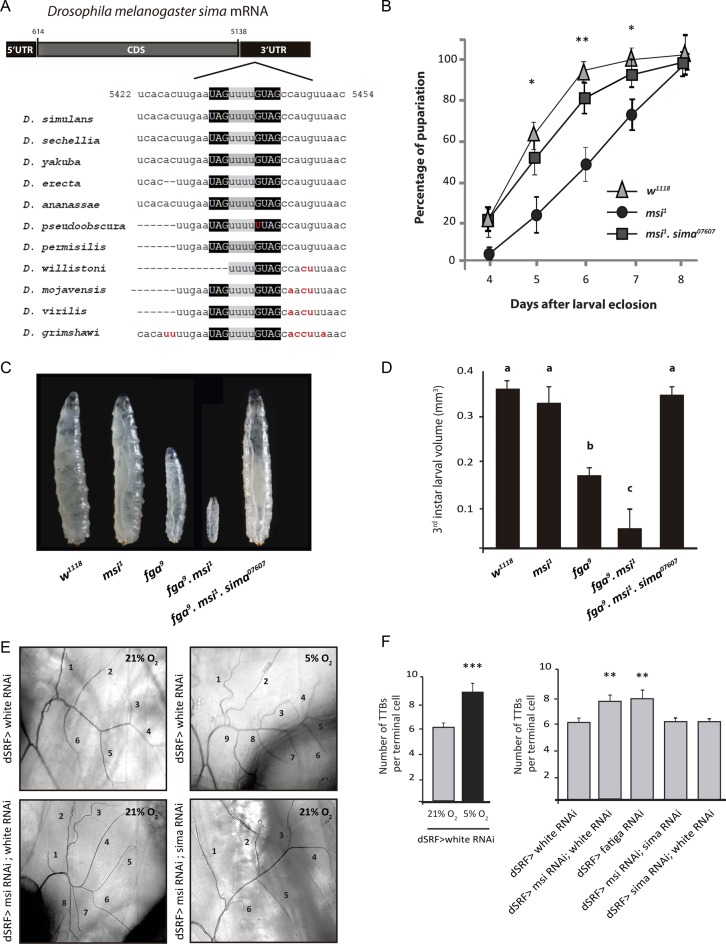
Bioinformatic analysis of sima 3′ UTR revealed the occurrence of a dMsi binding element (MBE) conserved across species of the Drosophilidae family (see Materials and Methods; Figure 1A). This phylogenetic sequence conservation in sima 3′ UTR suggests that dMsi plays a role in Drosophila HIFα regulation. To begin analyzing this possibility, we performed genetic interaction assays to assess if dMsi contributes to developmental phenotypes that are known to depend of the HIF pathway in Drosophila larvae. We have previously reported that hypomorphic mutants of fatiga (fga9), the main negative regulator of Sima, exhibit delayed pupariation and larval growth impairment (8), and that in fga9 sima07607double homozygous larvae normal growth and developmental timing are restored, indicating that an excess of Sima accounts for these developmental phenotypes. To begin exploring if dMsi negatively regulates Sima, we analyzed pupariation timing of dmusashi loss-of-function homozygous animals (msi1). Compared to control siblings, msi1 mutants show a 1–2 days pupariation delay (Figure 1B), which is reverted in msi1 sima07607 double homozygotes (Figure 1B), indicating that the effect of dMsi loss of function is due to Sima accumulation. These results suggest that dMsi may be a negative regulator of Sima. To further investigate the possibility that dMsi inhibits Sima, we analyzed genetic interactions between msi1 and fga9 mutants. The final size of msi1 third instar mutant larvae is indistinguishable from that of controls, while the size of fga9 homozygous larvae is clearly reduced (Figure 1C and D and (8)). Noteworthy, msi1 fga9 double homozygous third instar larvae fail to pupariate and are remarkably smaller than fga9 single mutants (Figure 1C and D), although they can remain alive for up to 2 weeks. Strikingly, msi1 fga9 sima07607 triple homozygous larvae undergo pupariation normally, attaining a final size similar to that of wild type controls (Figure 1C and D), suggesting that dMusashi cooperates with Fatiga in inhibiting the Sima pathway.
Figure 1.
dMusashi loss of function provokes Sima-dependent growth defects and enhances tracheal sprouting. (A) Layout of the Drosophila melanogaster sima transcript (FBtr0344374). A predicted Musashi binding element (MBE) occurs in the 3′ UTR of all the species analyzed of the Drosophilidae family. Minimal binding sequences for Msi (UAG and GUAG) are highlighted in black boxes, while the linker region between them is shown in grey. The nucleotides non-conserved between species are shown in red. (B) musashi homozygous mutants (msi1) display prolonged larval development, resulting in delayed pupariation in comparison to their wild type siblings (w1118), while in msi1, sima07607 double homozygotes, normal pupariation timing is restored (error bars represent SEM; n = 3 with a minimum of 60 individuals analyzed per genotype at each point of the curve; *P < 0.05 and **P ≤ 0.01; Student's t test compared with msi1). (C) Whereas musashi (msi1) homozygous mutants and wild type third instar larvae attain similar size before entering pupariation, fga9 homozygotes are smaller. fga9 msi1 double homozygous mutants reach the third larval instar but are remarkably smaller than fga9 homozygotes and never pupariate. In fga9 msi1 sima07607 triple homozygous third instar larvae, normal growth is rescued. (D) Quantification of final body volume of third instar larvae in the experiment depicted in panel C. Error bars represent SEM (n = 3 with a minimum of 25 larvae analyzed per genotype in each experiment; different letters indicate statistical differences with a P < 0.05 in a one-way ANOVA with a Bonferroni post-hoc test). (E) Photographs of the morphology of a terminal cell of the third dorsal branch of a third instar larva in different genotypes or oxygen concentrations are shown. Numbers indicate individual subcellular extensions of more than 1 μm diameter, the ‘‘thick terminal branches’’ (TTBs). Thinner terminal branches may ramify from TTBs but these were not counted. Ramification of terminal cells is enhanced in hypoxic (5% O2 16 h) wild type control larvae (dSRF> white RNAi), as well as in individuals with dMsi loss of function in these cells (dSRF > msiRNAi; white RNAi) maintained in normoxia. Sima knockdown, along with dMsi silencing, leads to rescue of the normal ramification pattern. (F) Quantification of the results shown in panel E. Error bars represent SEM (n = 3 with a minimum of 25 larvae analyzed per genotype in each experiment; **P ≤ 0.01; ***P ≤ 0.001; Student's t test).
Tracheal terminal cells of Drosophila third instar larvae are plastic and respond to hypoxia by projecting terminal branches in a Sima dependent manner (28), so we investigated if dMsi modulates tracheal sprouting. As we have previously shown, the number of terminal branches with more than 1μm diameter (‘thick terminal branches’, TTBs) of the dorsal branch of the third segment of third instar larvae is a sensitive parameter to quantify terminal tracheal branching after physiological or genetic interventions (Figure 1E and F left panel; (28)). We used a UAS-msi RNAi line that effectively downregulates dMsi protein levels (Supplementary Figure S1), expressed under control of the terminal cell-specific driver dSRF-Gal4 in normoxic larvae, and observed a significant increase in the number of TTBs in comparison to controls expressing an unrelated RNAi (Figure 1E and F right panel). To investigate if this tracheal ramification increase depends on Sima, we coexpressed msi RNAi along with a sima RNAi, and observed a complete reversion of the phenotype, with the number of TTBs being restored to wild type levels (Figure 1E and F right panel). These results indicate that dMusashi depletion can induce Sima-dependent tracheal terminal sprouting, which is a physiological response to hypoxia in Drosophila.
dMusashi controls Sima protein levels
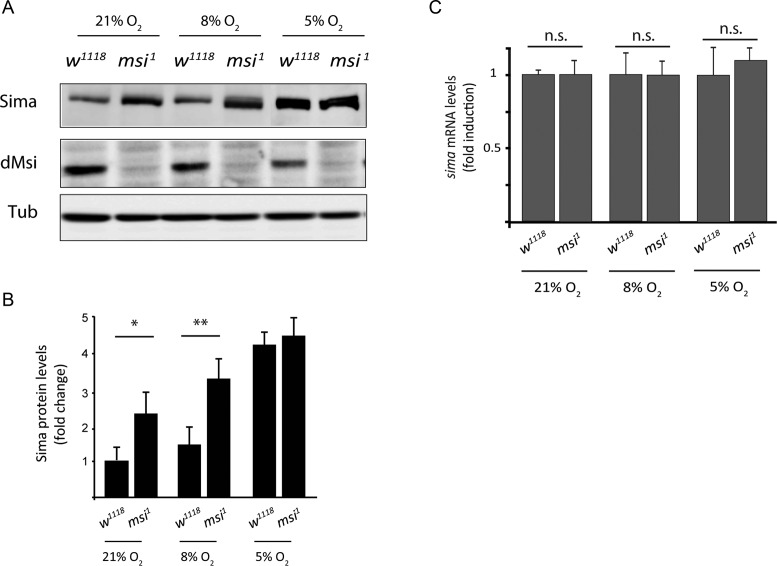
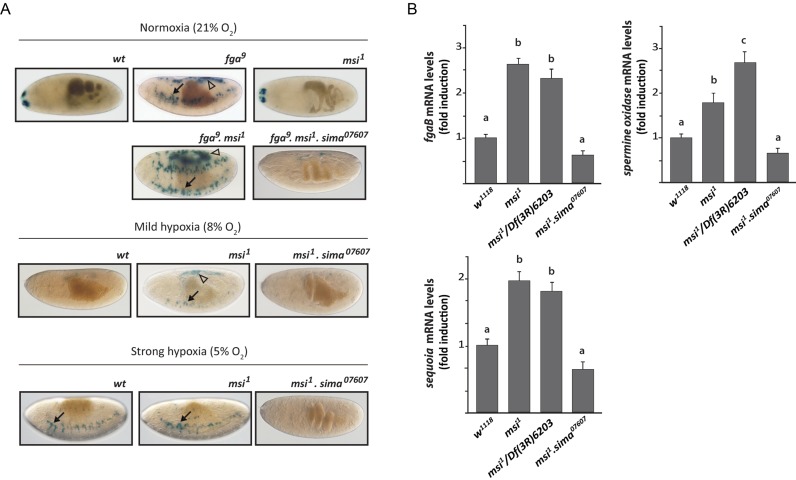
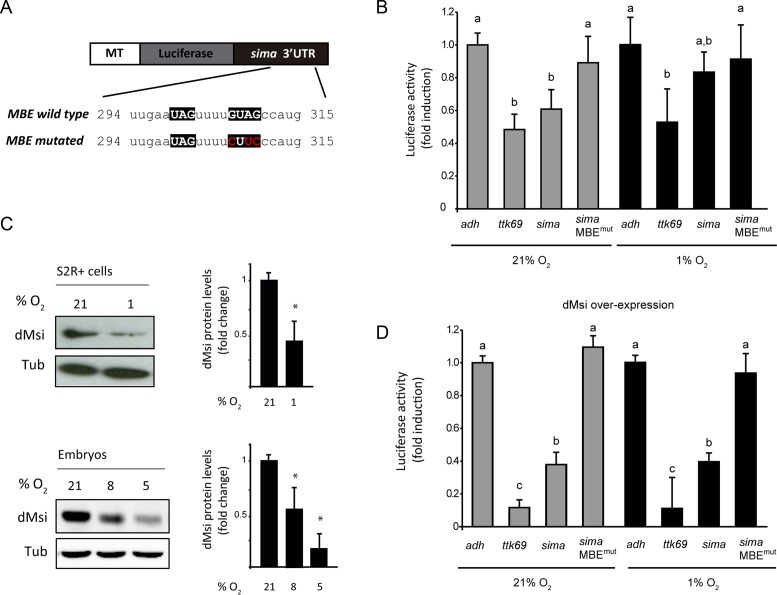
To investigate if dMusashi directly controls Sima protein abundance, we performed western blot analysis with anti-Sima antibodies of extracts of msi1 homozygous embryos in comparison to wild type controls in normoxia at 8% O2, an hypoxic condition previously shown to trigger mild HIF-dependent responses in Drosophila embryos (7), and at 5% O2, a condition that induces maximal HIF-dependent transcription in Drosophila in vivo (7). As previously reported, Sima accumulates in wild type embryos exposed to hypoxia, and interestingly, Sima protein levels are also increased in dmsi mutants, either in normoxia or in mild hypoxia (8% of O2) as compared to wild type controls (Figure 2A and B). In strong hypoxic conditions (5% of O2), no accumulation of Sima protein is detected in comparison with the wild type (Figure 2A and B). sima transcript levels are unaffected in all cases (Figure 2C), suggesting that dMsi downregulates Sima at a post-transcriptional level. Next, we studied if dMsi plays a role in the regulation of HIF-dependent transcription, by analyzing the expression of a hypoxia-inducible transcriptional reporter (HRE-LacZ) in transgenic fly lines (7). As previously reported, HRE-LacZ is silent in wild type embryos in normoxia (21%O2) or mild hypoxia (8% O2), and induced in strong hypoxic conditions (5% O2) (Figure 3A); in fga mutant embryos, the reporter is strongly expressed already in normoxia (Figure 3A) (7,8). Interestingly, in dmsi null mutants, expression of the reporter can be observed already in mild hypoxia (Figure 3A), suggesting that dMsi negatively regulates Sima dependent transcription. Consistent with this notion, in msi1 fga9 double mutants, expression of the reporter in normoxia is stronger than in embryos mutant for fga only (Figure 3A). As expected, no expression in any condition is observed in embryos carrying the sima07607 loss-of-function allele in combination with the msi1 mutation or with msi1 and fga9 mutations simultaneously (Figure 3A). To confirm that dmsi loss of function enhances Sima dependent transcription, we analyzed in msi1 mutant larvae the expression of three different endogenous genes that are upregulated in hypoxia in a Sima-dependent manner (Figure 3B and Supplementary Figure S2). Increased expression of all three target genes is observed, even in normoxic conditions, both in msi1 homozygous larvae and in larvae heterozygous for msi1 and the Df(3R)6203 chromosomal deficiency that covers the dmsi locus (Figure 3B). Expression of Sima target genes is restored to wild-type normoxic levels in msi1 sima07607 double homozygous larvae (Figure 3B), indicating that the enhanced expression of Sima target genes in dmsi loss-of-function larvae is due to overaccumulation of Sima. Noteworthy, the extent of upregulation of HIF target gene expression in msi1 mutant normoxic larvae (2–3-fold; Figure 3B), and the extent of their upregulation in wild type larvae exposed to hypoxia (Supplementary Figure S2) is similar. This comparison strengthens the notion that the Sima-dependent transcriptional response to hypoxia is constitutively activated in msi1 mutants. Consistent with the above results, in cultured S2R+ cells treated with msi dsRNA, strong enhancement of Sima-dependent transcription is observed in normoxia, while in hypoxia (1% O2) this enhancement is milder (Supplementary Figure S3). Altogether, these results indicate that dMsi mediates reduction of Sima protein in normoxic conditions, thereby restricting Sima-dependent transcription.
Figure 2.
dMusashi downregulates Sima protein levels. (A) Western blot showing that Sima protein levels increase in musashi1 homozygous mutant late-stage embryos (msi1) in comparison to wild type controls (w1118) at different oxygen concentrations. Sima accumulates in wild type embryos (w1118) in hypoxia, while in msi1 mutants, Sima accumulates in normoxia or mild hypoxia (8% O2 5 h), but not in strong hypoxia (5% O2 5 h), when compared with wild type controls. Tubulin was used as loading control. (B) Quantification of Sima protein levels from the western blot shown in panel A. Error bars represent SEM (n = 3, *P < 0.05; **P ≤ 0.01; Student's t test). (C) sima mRNA levels were analyzed by RT-qPCR from embryos in the same conditions as in panels A and B in normoxia or hypoxia. sima transcript levels are not significantly different between wild type and mutant individuals in any condition. Rpl29 mRNA was used for normalization. n.s.: values not significantly different, Student's t-test.
Figure 3.
dMusashi downregulates Sima dependent transcription. (A) Expression of the Sima-inducible reporter HRE-LacZ in wild type, fga9or msi1 mutant embryos maintained in normoxia or exposed to two different hypoxic conditions; arrows indicate groups of tracheal cells that express the reporter with maximal sensitivity, and arrowheads indicate groups of dorsal cells where the reporter expression is more variable but occurs consistently when HIF-dependent transcription is especially strong. In wild type embryos, the reporter is only expressed in strong hypoxia (5% O2 5h), while in fga9 mutants expression can be detected even in normoxia (21% O2), and in msi1 mutants it is already detectable in mild hypoxia (8% O2 5 h); fga9 msi1 double homozygous mutants display overall enhanced expression of the reporter in normoxia in comparison to fga9 single mutants. (B) Expression of three Sima target genes was analyzed by RT-qPCR in wild type, musashi loss-of-function (msi1 homozygous mutants or larvae with the heteroallelic combination msi1/Df(3R)6203) or in msi1 sima07607double homozygous mutant third instar larvae maintained in normoxia. In both msi loss-of-function backgrounds (msi1 or msi1/Df(3R)6203), expression of the three Sima target genes, fgaB, sequoia and spermidine oxidase, is higher than in wild type controls or in msi1 sima07607double mutants. Error bars represent SEM (n = 3; different letters indicate statistical differences with a P < 0.05 in a one-way ANOVA with a Bonferroni post-hoc test).
dMusashi binds to sima mRNA and represses translation
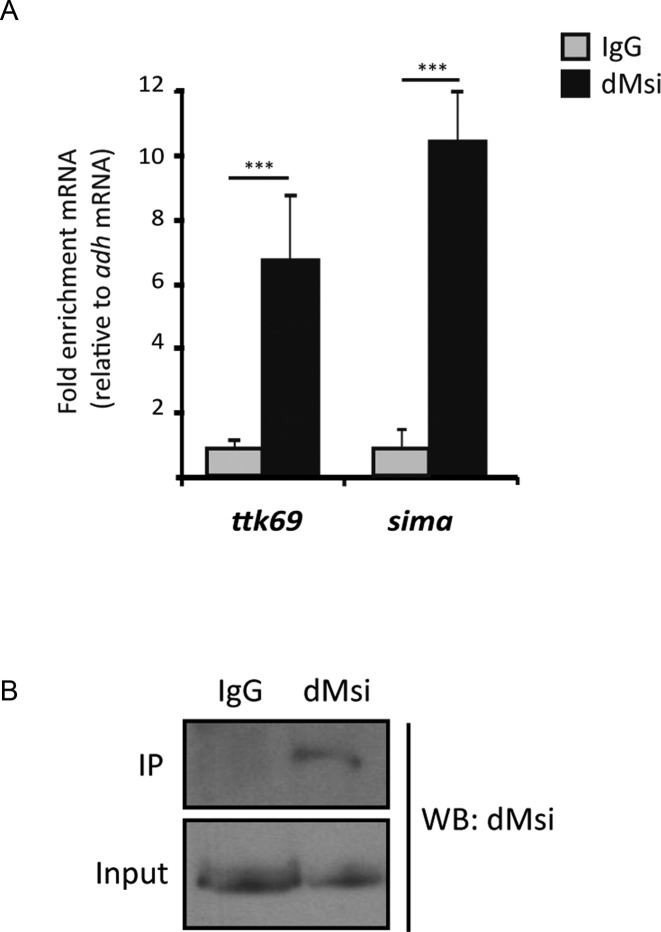
As a next step, we sought to explore the mechanism by which dMsi represses Sima. Given the occurrence of a MBE at the 3′ UTR of sima mRNA, we analyzed if dMsi protein can bind the sima transcript. RNA immunoprecipitation (RIP) analysis with an anti-dMsi antibody indicates specific binding to sima mRNA, which occurs with an efficiency comparable to binding of dMsi to ttk69 mRNA (Figure 4A and B), a previously reported dMsi target. Next, we investigated if the sima 3′ UTR, that contains a MBE, can mediate repression of a luciferase reporter in S2R+ cells. In cells maintained in normoxia, sima 3′ UTR exerts strong repression of luciferase reporter activity, which is comparable to the repression exerted by the ttk69 3′ UTR used as a positive control (Figure 5A and B) (11). The repressive capacity of sima 3′ UTR is largely abolished when the MBE is mutagenized (Figure 5A and B), suggesting that repression is mediated by dMsi. Remarkably, the sima 3′ UTR is less efficient in repressing luciferase activity in cells exposed to hypoxia (1% O2 for 20 h) (Figure 5B), which is consistent with a sharp reduction of dMsi protein levels (Figure 5C) in hypoxia. dMsi transcript levels do not vary in hypoxic conditions, suggesting that dMsi regulation by oxygen is post-transcriptional (Supplementary Figure S4). To get additional evidence that dMsi is responsible for the repression of luciferase activity mediated by sima 3′ UTR, we induced dMsi overexpression by cotransfecting cells with an actin-dMsi plasmid along with the 3′ UTR luciferase reporters (Supplementary Figure S5). Strong enhancement of the repression of both sima 3′ UTR and ttk69 3′ UTR luciferase reporters occurs in these cells in normoxia, while again, no repression is observed with the reporter in which the sima 3′ UTR was mutagenized at the MBE (Figure 5D). These results confirm that dMsi mediates the repression exerted by sima 3′ UTR. Noteworthy, over-expression of artificially high levels of dMsi protein in these cells, overrides the downregulation of dMsi protein levels provoked by the hypoxia treatment (Supplementary Figure S5). Consistent with this, oxygen dependence of sima 3′ UTR repressive capacity is lost completely, as repression is equally potent in normoxia and hypoxia (Figure 5D). RT-qPCR of luciferase transcript levels revealed no differences between constructs bearing different 3′ UTRs (Supplementary Figure S6), suggesting that Musashi represses Sima at a translational level, which is consistent with its well-established function as a translational repressor (17).
Figure 4.
dMusashi binds Sima mRNA. RNA immunoprecipitation (RIP) was performed from extracts of normoxic S2R+ cell cultures using an anti-dMusashi antibody or a pre-immune (IgG) serum. (A) Immunoprecipitated sima mRNA was quantified by RT-qPCR, along with ttk69 mRNA (positive control) and adh mRNA (that lacks Musashi Binding Elements) for normalization. dMsi binds sima mRNA with an efficiency comparable to that of ttk69 mRNA. (B) Western blot showing dMsi protein levels in whole cell extracts (input), and after IP; the protein is immunoprecipitated with the anti-dMsi antibody (dMsi) but not with the pre-immune serum (IgG). Error bars represent SEM (n = 3, ***P ≤ 0.001; Student's t test).
Figure 5.
dMusashi represses Sima translation in normoxia. (A) Schematic representation of firefly luciferase reporter constructs containing wild type or mutagenized versions of the sima 3′ UTR. Mutations (shown in red) have been generated within the Musashi Binding Element (MBE) by site-directed mutagenesis. The reporters were transiently transfected in S2R+ cells and expressed under control of a copper-inducible metallothionein promoter (MT). (B) S2R+ cells transfected with luciferase reporters containing the alcohol dehydrogenase (adh) 3′ UTR (negative control), tramtrack69 (ttk69) 3′ UTR (positive control), or sima 3′ UTR (either wild type or mutagenized, MBE mut) were maintained in normoxia or exposed to hypoxia (1% O2 20 h), and luciferase activity of cell lysates was measured, using a copper-inducible Renilla luciferase construct for normalization. In normoxic but not in hypoxic cells, sima 3′ UTR represses luciferase activity to a similar extent to ttk69 3′ UTR; sima 3′ UTR mutagenized in the MBE loses completely its repressive activity. (C) Western blot analysis of dMsi protein levels in S2R+ cells in normoxia or hypoxia (1% O2 20 h), as well as in embryos maintained in normoxia (21% O2) or exposed to hypoxia (8% or 5% O2 for 5h). Both in cell culture and in vivo, dMsi protein levels are sharply reduced in hypoxia. Tubulin was used as a loading control. Error bars represent SEM (n = 3, *P < 0.05; **P ≤ 0.01; Student's t test). (D) Cells transfected with the same set of reporters as in panel B) were co-transfected with a dMsi overexpression vector (pAc-dMsi) and maintained in normoxia or exposed to hypoxia (1% O2 20 h). dMsi overexpression enhances the repression exerted by ttk69 or sima 3′ UTRs irrespective of oxygen levels (compare with panel B), while the mutagenized version of sima 3′ UTR loses its repressive capacity. Luciferase activity is depicted as fold induction respect to the activity of the adh 3′ UTR reporter (negative control). In (B) and (D), error bars represent SEM (n = 3; different letters indicate statistical differences with a P < 0.05 in a one-way ANOVA with a Bonferroni post-hoc test).
The Msi-HIF axis in mammals
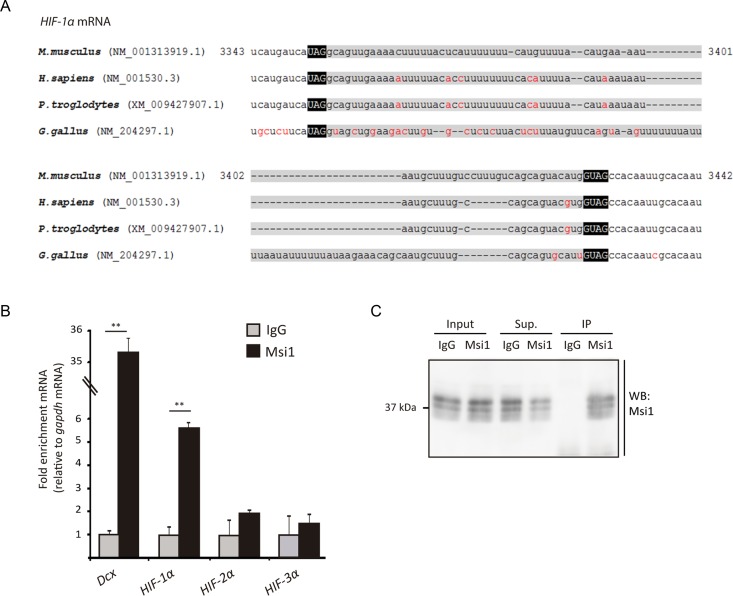
Having established that Msi regulates HIF in the Drosophila system, we asked whether this regulation might be conserved in mammals. Bioinformatic analysis of mammalian HIFα 3′ UTRs (see Materials and Methods) revealed the occurrence of a putative MBE in HIF-1α 3′ UTR (Figure 6A), but not in the 3′ UTRs of HIF-2α or HIF-3α. Noteworthy, the sequence is evolutionary conserved and conservation is not limited to the UAGxnGUAG bipartite Msi-binding consensus sequence (13), but rather, substantial conservation is also observed upstream to the UAG 5′ box and downstream to the 3′ GUAG box, as well as in the linker region (Figure 6A). These observations prompted us to analyze if mammalian Msi1 protein binds HIFα transcripts in a mammalian system. We carried out CLIP-based RT-qPCR assays (31) to analyze possible interactions in mouse embryonic brains (E14.5), using the doublecortin (Dcx) mRNA as a positive control (20). We performed UV-crosslinking on tissue samples, and prepared an extract (see Materials and Methods), which was subjected to immunoprecipitation with an anti-Msi1 antibody, followed by RT-qPCR analysis of HIF-1α, HIF-2α and HIF-3α transcripts. Brain extracts were prepared in stringent buffer conditions to minimize protein-protein non-covalent interactions and indirect protein-RNA interactions. As shown in Figure 6B and C, Msi1 physically interacts with HIF-1α mRNA but not with HIF-2α or HIF-3α transcripts. These observations suggest that Msi1 might potentially regulate HIF-1α translation in the mouse brain, as we have shown above for the Drosophila homologous proteins.
Figure 6.
Mammalian Msi1 interacts with HIF-1α mRNA. (A) Sequence alignment of HIF-1α 3′ UTRs of vertebrate species is shown. The bipartite Musashi Binding Element (MBE) is highlighted in black boxes. Note that sequence conservation is not limited to the MBE but includes extensive portions of the linker between the two boxes, as well as regions upstream to the 5′ UAG box and the 3′ GUAG box. Nucleotides shown in red color are not conserved between species. (B) Murine Msi1 (Msi1) interacts with HIF-1α mRNA in vivo; CLIP-RT-qPCR assays were performed from extracts of mouse embryonic brains using an anti-Msi1 or control antibody (IgG). Msi1-bound mRNAs was quantified by RT-qPCR, using the gapdh transcript for normalization. Msi1 protein interacts with HIF-1α but not with HIF-2α or HIF-3α mRNA (n = 3; error bars represent standard deviation). Asterisks indicate P < 0.01 by Student's t test. (C) Western blot showing dMsi protein levels in whole cell extracts (input), and after IP, in the supernatant (Sup.) or in the pellet (IP); the protein can be immunoprecipitated with the anti-dMsi antibody (dMsi) but not with the pre-immune serum (IgG).
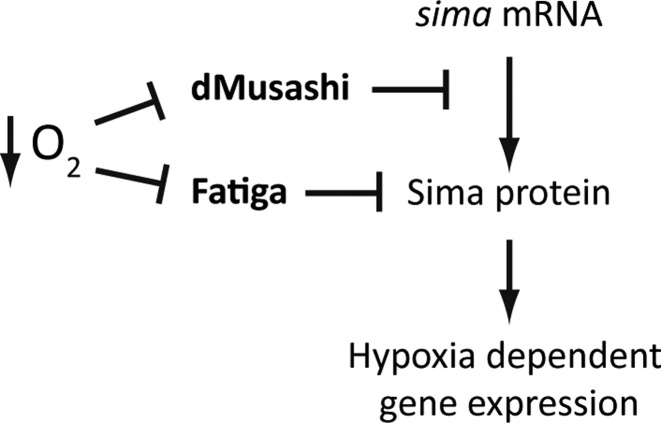
Thus, we have shown in the Drosophila system that dMusashi inhibits HIF dependent transcriptional responses in normoxia by repressing sima mRNA translation. In hypoxia, dMusashi is downregulated and sima translational repression is lifted, contributing to activation of HIF-dependent transcription and adaptation to hypoxia (Figure 7). We also found that in mouse embryonic brains, Msi1 physically interacts with the HIF-1α transcript suggesting that Msi-HIF functional interactions might be conserved in mammalian systems.
Figure 7.
Model for the regulation of Sima by dMusashi. The prolyl-4-hydroxylase Fatiga downregulates Sima protein levels by promoting its proteasomal degradation, while dMusashi represses sima mRNA translation by binding a Musashi Binding Element within the 3′ UTR. dMsi protein levels are reduced in hypoxia, allowing Sima dependent transcription.
DISCUSSION
In this work, we have established a role of the RNA binding protein dMusashi in the regulation of mRNA translation of the hypoxia inducible factor alpha subunit (HIFα). The Drosophila HIF alpha-subunit Sima is regulated primarily by oxygen-dependent proteasomal degradation (7,8). The Proline 850, localized in the oxygen-dependent degradation domain, is hydroxylated by the prolyl-4-hydroxylase Fatiga and degraded at the 26S proteasome. Sima oxygen-dependent subcellular localization is another important regulatory step: In hypoxia the protein accumulates in the nucleus, while following reoxygenation, Sima is exported very rapidly to the cytoplasm (2-5 min) by the nuclear export receptor CRM1 (32). Sima nuclear export is also dependent on Fatiga-dependent hydroxylation of Proline 850 (33).
The current work led to the identification of the translational repressor dMusashi as a novel regulator of Sima in Drosophila, which adds another layer of complexity to the control of transcriptional responses to hypoxia. We have shown both in cell culture and in vivo that dMsi is an inhibitor of HIF in normoxia, which operates by binding a Musashi Binding Element (MBE) in the Sima 3′ UTR and inhibits its translation. In hypoxia dMsi levels are strongly reduced, resulting in suppression of the inhibitory effect. Consistent with these observations, in msi mutants HIF-dependent transcription and biological outcomes, including sprouting of the tracheal system, are enhanced. We have provided evidence that Msi–dependent regulation of the HIF system might be conserved at least within higher eukaryotes, as Msi1 protein physically interacts with the HIF-1α transcript in embryonic mouse brains. Defining if HIF-1α regulation by Msi1 occurs in mammals is undoubtedly of interest, and requires further research.
Previous studies on mammalian HIF regulation have focused mostly on oxygen-dependent proteasomal degradation of its α-subunit (3–6,34), and transcriptional co-activator recruitment (35–37), while mechanisms supporting transcriptional (38–40) or translational control (41) are less well-defined. Nonetheless, it is clear that multiple mechanisms that mediate HIF translational regulation occur, contributing substantially to the regulation of responses to hypoxia. One of such mechanisms is the switch from CAP-dependent to CAP-independent (internal ribosomal entry site (IRES)-dependent) mRNA translation. In hypoxia, general CAP-dependent translation is largely suppressed, following Target of Rapamycin (TOR) inhibition, and activation of the eukaryotic translation initiation factor 4 binding proteins (4E-BPs), which bind 4E thereby preventing formation of the initiation complex (42,43). In hypoxia, HIF-1α translation becomes dependent on an IRES localized within the 5′ UTR of its mRNA, thereby escaping general translational inhibition (44). Besides its role in the regulation of CAP-dependent translation through 4E-BPs, TOR is required for HIFα translation, and thereby for triggering the hypoxic response. TOR mediates S6 kinase phosphorylation, which is in turn necessary for HIFα translation in hypoxia (30,45–47).
Another layer of oxygen-dependent regulation of HIFα is the one exerted by RNA binding proteins or micro-RNAs that bind its 5′ or 3′ UTRs, controlling mRNA stability or translation (49). HuR is an RNA binding protein that mediates potent enhancement of HIFα mRNA translation in hypoxia in human cervical carcinoma cells (48). Even though the mechanisms by which hypoxia activates HuR function to potentiate HIFα mRNA translation are so far unclear, it is known that they involve its binding to U or AU-rich sequences at the 5′ UTR. While HuR operates on HIFα 5′ UTR, another RNA binding protein, the polypyrimidine tract-binding protein (PTB) binds its 3′ UTR enhancing translation as well (48). The ability of PTB to enhance HIFα translation depends on HuR, supporting the notion that PTB and HuR cooperate with each other to promote this enhancement (48,49). The T-cell intracellular antigen-1 (TIA-1) and its related protein TIAR have also been reported to regulate HIFα translation. TIA-1 and TIAR are upregulated in hypoxia in a model of rat brain ischemia (50), and form stress granules mediating HIF repression (51). The effect is conveyed by AU-rich elements present at the 3′ UTR of HIF-1α mRNA (51). It remains to be established if HuR, PTB or TIA-1/TIAR homologs are involved in HIF regulation in the Drosophila system, and if so, to what extent they functionally interact with dMsi.
The potential biological implications of HIF regulation by the translational repressor Musashi, reported in this study, are appealing. Msi is highly expressed in several types of tumours, being required for maintenance of the undifferentiated state in aggressive leukemias (52–54); in most types of cancer its expression correlates with poor prognosis (15,55–58). Msi in addition participates in asymmetric cell divisions, epithelial-mesenchymal transitions (EMTs) and is a key determinant of stem cell maintenance in diverse organs and cellular contexts, including cancer stem cells (15,17). Remarkably, the biological processes regulated by HIF overlap with those controlled by Musashi: HIF promotes EMT and metastatic phenotypes in human cells and in mice (59); HIF plays a role in stem cell homeostasis in the hematopoietic lineage (60); and finally, HIF plays a pivotal role in tumorigenesis (61,62). Further research is required to determine if the functional relationship of HIF and Musashi described in this work plays a role in one or more of the above biological processes in which the two proteins suggestively overlap.
Supplementary Material
Acknowledgments
We wish to thank colleagues that contributed with fly strains, as well as to the Bloomington Drosophila Stock Center and the Vienna Drosophila Resource Centre for fly stocks; Andrés Liceri for preparing fly culture media, and members of the Wappner's laboratory for insightful comments and discussion.
Author contributions: APB, MJK, JMA, BP, DB-O, LG, ES and MY performed the experiments and analyzed the data; HK and HO contributed to the generation of anti-dMsi antibodies; APB and PW conceived the project; PW, APB, HO and AS wrote the manuscript.
Footnotes
Present address: Julieta M. Acevedo, German Cancer Research Center (DKFZ), Heidelberg, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Wellcome Trust [WT087675MA]; Agencia de Promoción Científica y Tecnológica [PICT2012 No. 0214, PICT 2014 No. 0649]. Funding for open access charge: Agencia de Promoción Científica y Tecnológica [PICT 2014 No. 0649].
Conflict of interest statement. None declared.
REFERENCES
- 1.Ratcliffe P.J. Oxygen sensing and hypoxia signalling pathways in animals: the implications of physiology for cancer. J. Physiol. 2013;591:2027–2042. doi: 10.1113/jphysiol.2013.251470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maxwell P.H., Wiesener M.S., Chang G.W., Clifford S.C., Vaux E.C., Cockman M.E., Wykoff C.C., Pugh C.W., Maher E.R., Ratcliffe P.J. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 3.Jaakkola P., Mole D.R., Tian Y.M., Wilson M.I., Gielbert J., Gaskell S.J., von Kriegsheim A., Hebestreit H.F., Mukherji M., Schofield C.J., et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 4.Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J.M., Lane W.S., Kaelin W.G., Jr HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 5.Epstein A.C., Gleadle J.M., McNeill L.A., Hewitson K.S., O'Rourke J., Mole D.R., Mukherji M., Metzen E., Wilson M.I., Dhanda A., et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 6.Bruick R.K., McKnight S.L. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 7.Lavista-Llanos S., Centanin L., Irisarri M., Russo D.M., Gleadle J.M., Bocca S.N., Muzzopappa M., Ratcliffe P.J., Wappner P. Control of the hypoxic response in Drosophila melanogaster by the basic helix-loop-helix PAS protein similar. Mol. Cell. Biol. 2002;22:6842–6853. doi: 10.1128/MCB.22.19.6842-6853.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centanin L., Ratcliffe P.J., Wappner P. Reversion of lethality and growth defects in Fatiga oxygen-sensor mutant flies by loss of hypoxia-inducible factor-alpha/Sima. EMBO Rep. 2005;6:1070–1075. doi: 10.1038/sj.embor.7400528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Acevedo J.M., Centanin L., Dekanty A., Wappner P. Oxygen sensing in Drosophila: multiple isoforms of the prolyl hydroxylase fatiga have different capacity to regulate HIFalpha/Sima. PLoS One. 2010;5:e12390. doi: 10.1371/journal.pone.0012390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romero N.M., Dekanty A., Wappner P. Cellular and developmental adaptations to hypoxia: a Drosophila perspective. Methods Enzymol. 2007;435:123–144. doi: 10.1016/S0076-6879(07)35007-6. [DOI] [PubMed] [Google Scholar]
- 11.Okabe M., Imai T., Kurusu M., Hiromi Y., Okano H. Translational repression determines a neuronal potential in Drosophila asymmetric cell division. Nature. 2001;411:94–98. doi: 10.1038/35075094. [DOI] [PubMed] [Google Scholar]
- 12.Okano H., Imai T., Okabe M. Musashi: a translational regulator of cell fate. J. Cell Sci. 2002;115:1355–1359. doi: 10.1242/jcs.115.7.1355. [DOI] [PubMed] [Google Scholar]
- 13.Ohyama T., Nagata T., Tsuda K., Kobayashi N., Imai T., Okano H., Yamazaki T., Katahira M. Structure of Musashi1 in a complex with target RNA: the role of aromatic stacking interactions. Nucleic Acids Res. 2012;40:3218–3231. doi: 10.1093/nar/gkr1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zearfoss N.R., Deveau L.M., Clingman C.C., Schmidt E., Johnson E.S., Massi F., Ryder S.P. A conserved three-nucleotide core motif defines Musashi RNA binding specificity. J. Biol. Chem. 2014;289:35530–35541. doi: 10.1074/jbc.M114.597112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katz Y., Li F., Lambert N.J., Sokol E.S., Tam W.L., Cheng A.W., Airoldi E.M., Lengner C.J., Gupta P.B., Yu Z., et al. Musashi proteins are post-transcriptional regulators of the epithelial-luminal cell state. eLife. 2014;3:e03915. doi: 10.7554/eLife.03915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sutherland J.M., McLaughlin E.A., Hime G.R., Siddall N.A. The Musashi family of RNA binding proteins: master regulators of multiple stem cell populations. Adv. Exp. Med. Biol. 2013;786:233–245. doi: 10.1007/978-94-007-6621-1_13. [DOI] [PubMed] [Google Scholar]
- 17.Fox R.G., Park F.D., Koechlein C.S., Kritzik M., Reya T. Musashi signaling in stem cells and cancer. Annu. Rev. Cell Dev. Biol. 2015;31:249–267. doi: 10.1146/annurev-cellbio-100814-125446. [DOI] [PubMed] [Google Scholar]
- 18.Imai T., Tokunaga A., Yoshida T., Hashimoto M., Mikoshiba K., Weinmaster G., Nakafuku M., Okano H. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol. Cell. Biol. 2001;21:3888–3900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Battelli C., Nikopoulos G.N., Mitchell J.G., Verdi J.M. The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21WAF-1. Mol. Cell. Neurosci. 2006;31:85–96. doi: 10.1016/j.mcn.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Horisawa K., Imai T., Okano H., Yanagawa H. 3′-Untranslated region of doublecortin mRNA is a binding target of the Musashi1 RNA-binding protein. FEBS Lett. 2009;583:2429–2434. doi: 10.1016/j.febslet.2009.06.045. [DOI] [PubMed] [Google Scholar]
- 21.Spears E., Neufeld K.L. Novel double-negative feedback loop between adenomatous polyposis coli and Musashi1 in colon epithelia. J. Biol. Chem. 2011;286:4946–4950. doi: 10.1074/jbc.C110.205922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muto J., Imai T., Ogawa D., Nishimoto Y., Okada Y., Mabuchi Y., Kawase T., Iwanami A., Mischel P.S., Saya H., et al. RNA-binding protein Musashi1 modulates glioma cell growth through the post-transcriptional regulation of Notch and PI3 kinase/Akt signaling pathways. PLoS One. 2012;7:e33431. doi: 10.1371/journal.pone.0033431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwon H.Y., Bajaj J., Ito T., Blevins A., Konuma T., Weeks J., Lytle N.K., Koechlein C.S., Rizzieri D., Chuah C., et al. Tetraspanin 3 Is Required for the Development and Propagation of Acute Myelogenous Leukemia. Cell Stem Cell. 2015;17:152–164. doi: 10.1016/j.stem.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Charlesworth A., Wilczynska A., Thampi P., Cox L.L., MacNicol A.M. Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation. EMBO J. 2006;25:2792–2801. doi: 10.1038/sj.emboj.7601159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakamura M., Okano H., Blendy J.A., Montell C. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron. 1994;13:67–81. doi: 10.1016/0896-6273(94)90460-x. [DOI] [PubMed] [Google Scholar]
- 26.Hirota Y., Okabe M., Imai T., Kurusu M., Yamamoto A., Miyao S., Nakamura M., Sawamoto K., Okano H. Musashi and seven in absentia downregulate Tramtrack through distinct mechanisms in Drosophila eye development. Mech. Dev. 1999;87:93–101. doi: 10.1016/s0925-4773(99)00143-4. [DOI] [PubMed] [Google Scholar]
- 27.Siddall N.A., McLaughlin E.A., Marriner N.L., Hime G.R. The RNA-binding protein Musashi is required intrinsically to maintain stem cell identity. Proc. Natl. Acad. Sci. U.S.A. 2006;103:8402–8407. doi: 10.1073/pnas.0600906103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Centanin L., Dekanty A., Romero N., Irisarri M., Gorr T.A., Wappner P. Cell autonomy of HIF effects in Drosophila: tracheal cells sense hypoxia and induce terminal branch sprouting. Dev. Cell. 2008;14:547–558. doi: 10.1016/j.devcel.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 29.Dekanty A., Romero N.M., Bertolin A.P., Thomas M.G., Leishman C.C., Perez-Perri J.I., Boccaccio G.L., Wappner P. Drosophila genome-wide RNAi screen identifies multiple regulators of HIF-dependent transcription in hypoxia. PLoS Genet. 2010;6:e1000994. doi: 10.1371/journal.pgen.1000994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dekanty A., Lavista-Llanos S., Irisarri M., Oldham S., Wappner P. The insulin-PI3K/TOR pathway induces a HIF-dependent transcriptional response in Drosophila by promoting nuclear localization of HIF-alpha/Sima. J. Cell. Sci. 2005;118:5431–5441. doi: 10.1242/jcs.02648. [DOI] [PubMed] [Google Scholar]
- 31.Ule J., Jensen K.B., Ruggiu M., Mele A., Ule A., Darnell R.B. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 32.Romero N.M., Irisarri M., Roth P., Cauerhff A., Samakovlis C., Wappner P. Regulation of the Drosophila hypoxia-inducible factor alpha Sima by CRM1-dependent nuclear export. Mol. Cell. Biol. 2008;28:3410–3423. doi: 10.1128/MCB.01027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Irisarri M., Lavista-Llanos S., Romero N.M., Centanin L., Dekanty A., Wappner P. Central role of the oxygen-dependent degradation domain of Drosophila HIFalpha/Sima in oxygen-dependent nuclear export. Mol. Biol. Cell. 2009;20:3878–3887. doi: 10.1091/mbc.E09-01-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaelin W.G., Jr, Ratcliffe P.J. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 35.Stolze I.P., Tian Y.M., Appelhoff R.J., Turley H., Wykoff C.C., Gleadle J.M., Ratcliffe P.J. Genetic analysis of the role of the asparaginyl hydroxylase factor inhibiting hypoxia-inducible factor (FIH) in regulating hypoxia-inducible factor (HIF) transcriptional target genes [corrected] J. Biol. Chem. 2004;279:42719–42725. doi: 10.1074/jbc.M406713200. [DOI] [PubMed] [Google Scholar]
- 36.Mahon P.C., Hirota K., Semenza G.L. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001;15:2675–2686. doi: 10.1101/gad.924501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galbraith M.D., Allen M.A., Bensard C.L., Wang X., Schwinn M.K., Qin B., Long H.W., Daniels D.L., Hahn W.C., Dowell R.D., et al. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell. 2013;153:1327–1339. doi: 10.1016/j.cell.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Uden P., Kenneth N.S., Webster R., Muller H.A., Mudie S., Rocha S. Evolutionary conserved regulation of HIF-1beta by NF-kappaB. PLoS Genet. 2011;7:e1001285. doi: 10.1371/journal.pgen.1001285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Belaiba R.S., Bonello S., Zahringer C., Schmidt S., Hess J., Kietzmann T., Gorlach A. Hypoxia up-regulates hypoxia-inducible factor-1alpha transcription by involving phosphatidylinositol 3-kinase and nuclear factor kappaB in pulmonary artery smooth muscle cells. Mol. Biol. Cell. 2007;18:4691–4697. doi: 10.1091/mbc.E07-04-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rius J., Guma M., Schachtrup C., Akassoglou K., Zinkernagel A.S., Nizet V., Johnson R.S., Haddad G.G., Karin M. NF-kappaB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1alpha. Nature. 2008;453:807–811. doi: 10.1038/nature06905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masuda K., Abdelmohsen K., Gorospe M. RNA-binding proteins implicated in the hypoxic response. J. Cell. Mol. Med. 2009;13:2759–2769. doi: 10.1111/j.1582-4934.2009.00842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clemens M.J. Translational regulation in cell stress and apoptosis. roles of the eIF4E binding proteins. J. Cell. Mol. Med. 2001;5:221–239. doi: 10.1111/j.1582-4934.2001.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu L., Cash T.P., Jones R.G., Keith B., Thompson C.B., Simon M.C. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol. Cell. 2006;21:521–531. doi: 10.1016/j.molcel.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lang K.J., Kappel A., Goodall G.J. Hypoxia-inducible factor-1alpha mRNA contains an internal ribosome entry site that allows efficient translation during normoxia and hypoxia. Mol. Biol. Cell. 2002;13:1792–1801. doi: 10.1091/mbc.02-02-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hudson C.C., Liu M., Chiang G.G., Otterness D.M., Loomis D.C., Kaper F., Giaccia A.J., Abraham R.T. Regulation of hypoxia-inducible factor 1alpha expression and function by the mammalian target of rapamycin. Mol. Cell. Biol. 2002;22:7004–7014. doi: 10.1128/MCB.22.20.7004-7014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brugarolas J.B., Vazquez F., Reddy A., Sellers W.R., Kaelin W.G., Jr TSC2 regulates VEGF through mTOR-dependent and -independent pathways. Cancer Cell. 2003;4:147–158. doi: 10.1016/s1535-6108(03)00187-9. [DOI] [PubMed] [Google Scholar]
- 47.Brugarolas J., Kaelin W.G., Jr Dysregulation of HIF and VEGF is a unifying feature of the familial hamartoma syndromes. Cancer Cell. 2004;6:7–10. doi: 10.1016/j.ccr.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 48.Galban S., Kuwano Y., Pullmann R., Jr, Martindale J.L., Kim H.H., Lal A., Abdelmohsen K., Yang X., Dang Y., Liu J.O., et al. RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1alpha. Mol. Cell. Biol. 2008;28:93–107. doi: 10.1128/MCB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gorospe M., Tominaga K., Wu X., Fahling M., Ivan M. Post-transcriptional control of the hypoxic response by RNA-binding proteins and microRNAs. Front. Mol. Neurosci. 2011;4:7. doi: 10.3389/fnmol.2011.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin K., Li W., Nagayama T., He X., Sinor A.D., Chang J., Mao X., Graham S.H., Simon R.P., Greenberg D.A. Expression of the RNA-binding protein TIAR is increased in neurons after ischemic cerebral injury. J. Neurosci. Res. 2000;59:767–774. doi: 10.1002/(SICI)1097-4547(20000315)59:6<767::AID-JNR9>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 51.Gottschald O.R., Malec V., Krasteva G., Hasan D., Kamlah F., Herold S., Rose F., Seeger W., Hanze J. TIAR and TIA-1 mRNA-binding proteins co-aggregate under conditions of rapid oxygen decline and extreme hypoxia and suppress the HIF-1alpha pathway. J. Mol. Cell Biol. 2010;2:345–356. doi: 10.1093/jmcb/mjq032. [DOI] [PubMed] [Google Scholar]
- 52.Ito T., Kwon H.Y., Zimdahl B., Congdon K.L., Blum J., Lento W.E., Zhao C., Lagoo A., Gerrard G., Foroni L., et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466:765–768. doi: 10.1038/nature09171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kharas M.G., Lengner C.J., Al-Shahrour F., Bullinger L., Ball B., Zaidi S., Morgan K., Tam W., Paktinat M., Okabe R., et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat. Med. 2010;16:903–908. doi: 10.1038/nm.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park S.M., Gonen M., Vu L., Minuesa G., Tivnan P., Barlowe T.S., Taggart J., Lu Y., Deering R.P., Hacohen N., et al. Musashi2 sustains the mixed-lineage leukemia-driven stem cell regulatory program. J. Clin. Invest. 2015;125:1286–1298. doi: 10.1172/JCI78440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Sousa Abreu R., Sanchez-Diaz P.C., Vogel C., Burns S.C., Ko D., Burton T.L., Vo D.T., Chennasamudaram S., Le S.Y., Shapiro B.A., et al. Genomic analyses of musashi1 downstream targets show a strong association with cancer-related processes. J. Biol. Chem. 2009;284:12125–12135. doi: 10.1074/jbc.M809605200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Byers R.J., Currie T., Tholouli E., Rodig S.J., Kutok J.L. MSI2 protein expression predicts unfavorable outcome in acute myeloid leukemia. Blood. 2011;118:2857–2867. doi: 10.1182/blood-2011-04-346767. [DOI] [PubMed] [Google Scholar]
- 57.Vo D.T., Subramaniam D., Remke M., Burton T.L., Uren P.J., Gelfond J.A., de Sousa Abreu R., Burns S.C., Qiao M., Suresh U., et al. The RNA-binding protein Musashi1 affects medulloblastoma growth via a network of cancer-related genes and is an indicator of poor prognosis. Am. J. Pathol. 2012;181:1762–1772. doi: 10.1016/j.ajpath.2012.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Glazer R.I., Vo D.T., Penalva L.O. Musashi1: an RBP with versatile functions in normal and cancer stem cells. Front. Biosci. (Landmark Ed.) 2012;17:54–64. doi: 10.2741/3915. [DOI] [PubMed] [Google Scholar]
- 59.Yang M.H., Wu M.Z., Chiou S.H., Chen P.M., Chang S.Y., Liu C.J., Teng S.C., Wu K.J. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat. Cell. Biol. 2008;10:295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 60.Lee K.E., Simon M.C. From stem cells to cancer stem cells: HIF takes the stage. Curr. Opin. Cell Biol. 2012;24:232–235. doi: 10.1016/j.ceb.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 61.Keith B., Simon M.C. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129:465–472. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keith B., Johnson R.S., Simon M.C. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat. Rev. Cancer. 2012;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.