Abstract
All-trans Retinoic acid (RA) and its derivatives are potent therapeutics for immunological functions including wound repair. However, the molecular mechanism of RA modulation in innate immunity is poorly understood, especially in macrophages. We found that topical application of RA significantly improves wound healing and that RA and IL-4 synergistically activate Arg1, a critical gene for tissue repair, in M2 polarized macrophages. This involves feed forward regulation of Raldh2, a rate-limiting enzyme for RA biosynthesis, and requires Med25 to coordinate RAR, STAT6 and chromatin remodeler, Brg1 to remodel the +1 nucleosome of Arg1 for transcription initiation. By recruiting elongation factor TFIIS, Med25 also facilitates transcriptional initiation-elongation coupling. This study uncovers synergistic activation of Arg1 by RA and IL-4 in M2 macrophages that involves feed forward regulation of RA synthesis and dual functions of Med25 in nucleosome remodeling and transcription initiation-elongation coupling that underlies robust modulatory activity of RA in innate immunity.
INTRODUCTION
All-trans Retinoic acid (RA), the principal active ingredient of vitamin A, plays crucial roles in a wide spectrum of biological processes, such as development, vision, reproduction, cell proliferation/differentiation and immune function (1–4). It is one of the most potent therapeutics for diseases like acute promyelocytic leukemia, inflammatory diseases, psoriasis and wound repair for its anti-tumor and potent immune-modulatory effects (5,6). Studies have shown that retinoids can enhance wound healing in preoperative use, but clinical application for tissue repair has remained contradictory due to toxicity elicited by their prolonged use (7,8). This has been suggested to involve an alteration in arginine metabolism (9). The molecular mechanism by which RA affects wound healing remains unclear.
Nutritional studies have established that RA deficiency is frequently associated with elevated susceptibility to infectious diseases, and that RA is important to the development and functions of multiple immune cells such as dendritic cells, T cells and B cells (10–12). RA is particularly critical for promoting mucosal immunity by inducing gut-homing effector T cells and IgA-producing B cells. RA is also known to regulate Foxp3+ regulatory T cell and Th17 effector T cell differentiation (13–15). Endogenously, RA can be synthesized in various cell types particularly antigen presenting cells (APCs), such as macrophages and dendritic cells. APCs are principal players in innate immunity, and are active in RA biosynthesis (16,17). These cells express rate-limiting enzymes in RA biosynthesis, retinal dehydrogenases (Raldhs) that catalyze the conversion of retinal to RA. In APCs, RA is known to regulate Raldhs gene expression in conjunction with inputs from pro- or anti-inflammatory cytokines (18–20), resulting in functional amplification of immune responses. However, the physiological relevance remains to be elucidated.
RA binds to nuclear RA receptors (RARs) that act as transcription factors and interact with co-regulators to regulate target gene transcription. The Mediator complex is a multi-unit co-regulator that recruits RNA Pol II and basal transcriptional machinery (21,22). Certain subunits, such as, MED1, MED17 and MED25, are known as RAR or RXR-interacting proteins (23–25). IL-4, an anti-inflammatory cytokine, binds to its receptor, IL-4R, which is then auto-phosphorylated and activates JAK1/3. The phosphorylated JAK1/3 recruits and phosphorylates signal transducer and activator of transcription 6 (STAT6), which is then translocated to the nucleus as a homodimer and acts as a master transcription activator for M2 macrophage gene expression (26). It is believed that signal integration between RA and various cytokines contributes to RA's potent actions in immune modulation (17–19).
Macrophages are the principal innate immune cells and play a central role in inflammation, host defense and resolution of inflammation. Upon activation, macrophages exhibit, mainly, classical, pro-inflammatory activation (M1) or alternative, anti-inflammatory activation (M2) (27). M1 macrophages are characterized by high levels of pro-inflammatory cytokines, reactive nitrogen and oxygen intermediates, and harbor strong microbicidal and tumoricidal activities. M2 macrophages are involved in the resolution of inflammation and parasite containment and promote wound healing, tissue remodeling and fibrosis. Arginase-1 (Arg1) is a cytosolic enzyme which catalyzes arginine hydrolysis to urea and ornithine (28). The production of urea allows excess nitrogen to be removed from the body, and ornithine can be used to generate polyamines and proline, which is important for collagen synthesis and wound healing. As such, Arg1 is a prototypic marker for M2 activation, and its gene regulation involves multiple transcription factors (STAT6, C/EBPβ, KLF4, PU.1 and nuclear receptors RAR, RXR and PPARγ) (29). Among these, a cytokine responsive enhancer for STAT6 binding, located 3 kb upstream (30) and an RA response element located 1 kb upstream of the proximal promoter (19) are most noticeable. But how these hormones/cytokines coordinately regulate Arg1 expression on the chromatin level is unclear.
In this study, we found that RA and IL-4 synergistically facilitate wound healing in mice and dampen inflammatory responses. On the genome level, the synergism occurs rapidly and specifically, which is to robustly activate Arg1 expression in M2 macrophage. This involves RA feed forward regulation of Raldh2 and dual functions of Med25. Mechanistically, Med25 coordinates the remodeling of a specific nucleosome spanning the transcription initiation site of Arg1 gene and facilitates elongation by recruiting transcription elongation factor TFIIS. Feed forward regulation of Raldh2 provides endogenous RA source to sustain prolonged Arg1 activation that is required for wound healing.
MATERIALS AND METHODS
Reagents
Reagent sources: all-trans RA (Sigma, R-2625), 0.1% Tretinoin cream (Boynton Health Service in University of Minnesota), IL-4 (Cell signaling #5208), AGN 193109 (pan-RAR antagonist, Santa Cruz CAS 171746-21-7), LPS (lipopolysaccharide, Sigma, L4391). Antibodies for STAT6 (M-20, sc-981), RAR (M-454, sc-773), RARβ (C-19, sc-552), RNA Pol II (sc-899), Brg1 (H-88, sc-10768), TFIIS (B-6, sc-393520), β-actin (C4, sc-47778) are from Santa Cruz. Antibodies for MED25 (Aviva systems biology, OAAB00209), H3.3 (Abcam, ab62642), H3k36me3 (Active motif, 61101), Arginase-1 (Cell signaling, #9819) are from indicated companies.
Animals
All studies were carried out using male C57BL/6J mice from The Jackson Laboratory and maintained in the animal facility of University of Minnesota. Animal studies were approved by the University of Minnesota Institutional Animal Care and Use Committee.
In vivo wound healing assay
As described in (31), cutaneous wounds were made on both sides of the shaved mice back (2 wounds per animal, n = 6) with a 5 mm round biopsy punch under anesthesia. Wound size was recorded daily, and reagents (control cream or 0.1% Tretinoin cream) were applied on wounds until day 5. Wound size were measured and analyzed by Image J.
Cell culture, lentivirus production and transduction
RAW264.7 cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% antibiotics. STAT6 KD and Med25 KD lentivirus were produced in 293TN cells using lentivirus packing system (System Biosciences). Lentivirus constructs and package plasmids were transfected using Lipofectamine 2000 (Thermo Fisher Scientific) and incubated for 3 days. Cell culture medium containing virus soup was collected and concentrated with Lenti-X Concentrator (Clontech) overnight. Viruses were centrifuged and mixed with fresh medium, stored in −80° freezer. STAT6 and Med25 shRNA lentivirus constructs were purchased from Genomic Center at the University of Minnesota:
-Full hairpin sequence for STAT6 KD:
#1-CCGGGCCACCTTATGATCTTGGAATCTCGAGATTCCAAGATCATAAGGTGGCTTTTTG
#2- CCGGCGGCTGATCATTGGCTTTATTCTCGAGAATAAAGCCAATGATCAGCCGTTTTTG
#3-CCGGCCACAGTCCATCCACTCATTTCTCGAGAAATGAGTGGATGGACTGTGGTTTTTG
#4-CCGGCGGTTCAGATGCTTTCTGTTACTCGAGTAACAGAAAGCATCTGAACCGTTTTTG
-Full hairpin sequence for Med25 KD:
#1-TGCTGTTGACAGTGAGCGACCAGGTCATCACCAACCACAATAGTGAAGCCACAGATGT
ATTGTGGTTGGTGATGACCTGGCTGCCTACTGCCTCGGA
#2-TGCTGTTGACAGTGAGCGCCCGGAACTCAAGAATGGTTCATAGTGAAGCCACAGATG
TATGAACCATTCTTGAGTTCCGGATGCCTACTGCCTCGGA
#3-TGCTGTTGACAGTGAGCGACTCAGGCTCTCTGCAGACCAATAGTGAAGCCACAGATGT
ATTGGTCTGCAGAGAGCCTGAGGTGCCTACTGCCTCGGA
#4-TGCTGTTGACAGTGAGCGCCTCAAGAATGGTTCAGTTCCATAGTGAAGCCACAGATGT
ATGGAACTGAACCATTCTTGAGTTGCCTACTGCCTCGGA
RNA isolation, gene expression analyses and Immunoblotting
Total RNA was isolated using TRIzol (Invitrogen). Reverse transcription (RT) was performed by using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative real-time PCR (qPCR) was performed with SYBR enzyme mix (Applied Biosystems). Each gene expression experiment was performed in triplicate. Expression levels were normalized to β-actin mRNA level. For the immunoblotting, cells were lysed in radioimmunoprecipitation assay (RIPA) buffer. Protein was quantified with Bradford method and separated by 10% SDS-PAGE gel.
Chromatin-immunoprecipitation (ChIP) assay
RAW264.7 cells were cross-linked with 1% formaldehyde. Cell pellets were lysed with NP40-lysis buffer. Nuclear extracts were sonicated and immunoprecipitated with antibodies and protein A/G beads overnight. Beads were washed, and DNA-protein complex was eluted from beads. Following decrosslinking, DNA was isolated and DNA enrichment was analyzed by qPCR with primer sets for the indicated regions of Arg1 gene.
-Arg1 enhancer STAT6 binding site: (F) TGAACAGGCTGTAGCCAACA, (R) AGCACCCTCAACCCAAAGTG,
-Arg1 RARE: (F) CAGGTTGCAGAAGAATCGAA, (R) TGACACTCTGCTGGTGTGTAGA,
-Arg1 +1 Nucleosome: (F) GAAAAAGATGTGCCCTCTGT, (R) AGAGAGACCCAAGGTCGCCG.
Nucleosome positioning assay
As described in (32), RAW264.7 cells nuclear extracts were digested with 30U of MNase at 37°C for 30 min, and digested genomic DNA was purified. The specific DNA regions were amplified with 20 primer sets by using qPCR. The nucleosome positions were determined by the ratio of digested/undigested DNA. Each primer set amplified about 100 bp product with an average of ∼60 bp overlap to achieve mononucleosome resolution. Data represent average gene amplification signal protected against MNase digestion from three biological replicates.
The measurement of pre-mRNA synthesis
As modified from (33), RAW264.7 cells were treated with DRB (5,6-Dichloro-1-β-D-ribofuranosylbenzimidazole, Sigma D-1916) for 3 h, and cells were treated with IL-4 or IL-4/RA. Cells were collected every 10 min after the stimulation. Total RNA was extracted with High Pure RNA isolation kit (Roche), and newly synthesized pre-mRNAs were measured with primer sets that can detect Arg1 intron 7-exon 8 by using qRT-PCR.
In vitro arginase activity
RAW264.7 cells were lysed with RIPA buffer, and arginase activity from each sample was measured according to manufacturer's instruction (Sigma, MAK112). Arginase catalyzes arginine to urea and ornithine, and the urea reacts with the substrate to generate a colored product, which is proportional to the arginase activity. The arginase activity of a sample was determined by measuring the absorbance at 430 nm.
Statistical analysis
Experiments were performed at least three times and results were presented as means ± SD. Student's t-test was used. P values of 0.05 or less were considered statistically significant (*P < 0.05; **P < 0.01; ***P < 0.001).
RESULTS
RA enhances wound healing in mice and promotes anti-inflammatory macrophage activation by synergizing with IL-4 to activate Arg1 expression
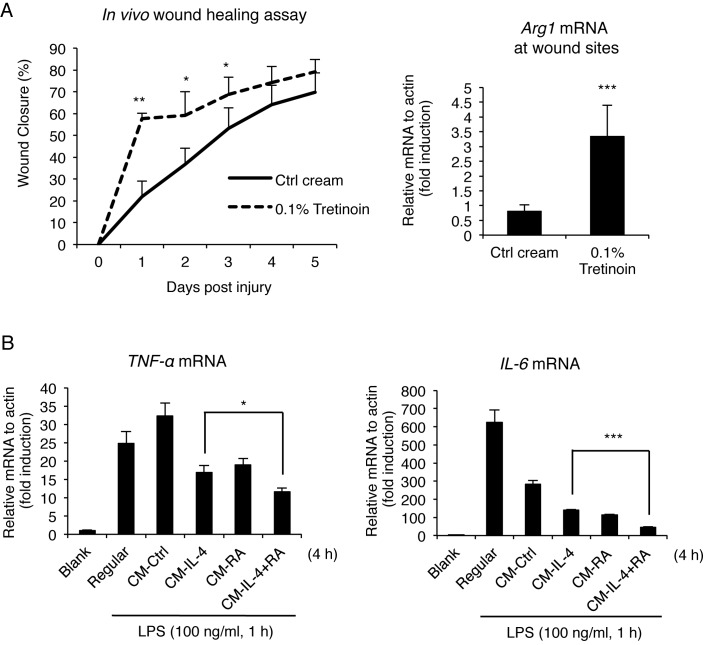
Although RA can regulate both pro- and anti-inflammatory genes in APCs, in general, RA signaling ultimately establishes an anti-inflammatory environment (34). We first employed a wound-healing model in the mouse to determine how RA affects the healing process, which involves, mainly, the anti-inflammatory (alternative, or M2, macrophage activation) response. Wounds were created on both sides of the back of the mouse, and monitored for 5 days with or without topical RA application (0.1% Tretinoin cream). As shown in Figure 1A, RA-treated mice exhibited significantly improved wound closure (57.7%) as compared to control mice (21.9%) as early as 1 day after wound creation. During wound healing, alternative (M2) macrophage activation is crucial following initial inflammatory (classical, or M1 macrophage activation) response (35). Therefore, we examined Arg1 mRNA expression, a key M2 marker, in wounded tissues. Indeed, Arg1 mRNA level was significantly elevated in RA cream-treated animals, indicating increased M2 activation by RA treatment (Figure 1A right). M2 activation is typically triggered by anti-inflammatory cytokines such as IL-4 and IL-13 (28). To examine whether RA and anti-inflammatory cytokines individually or combinatorially affect gene expression in macrophage activation, we profiled gene expression in cultured macrophages during pro- and anti-inflammatory responses (Figures 1B and 2A).
Figure 1.
RA enhances wound healing and represses inflammatory responses. (A) In vivo wound healing assay. The wounds were created and treated with a control or 0.1% Tretinoin cream for 5 days. The size of wound was monitored daily by recording wound closure (%) (n = 6 per each group) (left). qRT-PCR analyses of Arg1 mRNA at wound sites (right). Each wound tissue was collected at day 5 for mRNA extraction. (B) qRT-PCR analyses of LPS-induced (100 ng/ml, 1 h) pro-inflammatory cytokine gene (TNF-αand IL-6) mRNA in RAW264.7 cells treated with conditioned medium (CM, 4 h) collected from IL-4, RA or IL-4/RA-treated (6 h) RAW264.7 cells. Cells were treated with IL-4 (10 ng/ml) and/or RA (100 nM) in this figure. Data are representative of three experimental repeats (mean ± s.d), and Student's t-test (n = 3) was used (*P < 0.05; **P < 0.01; ***P < 0.001).
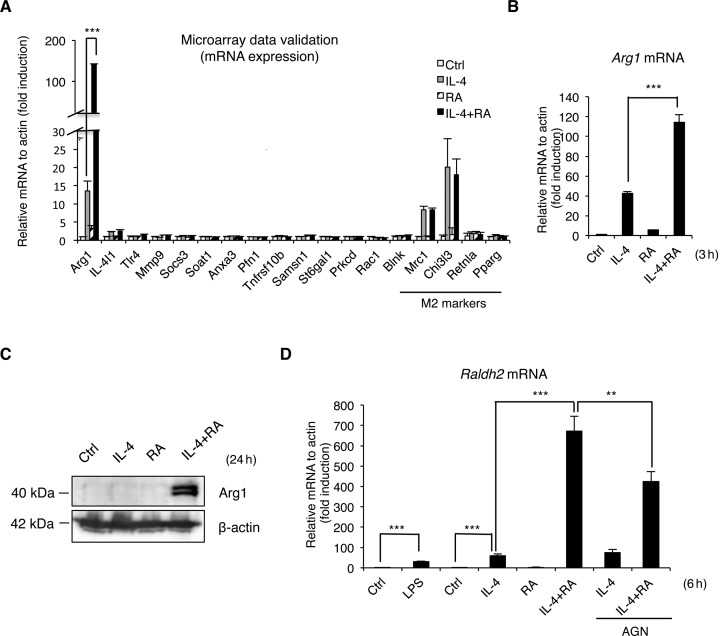
Figure 2.
RA promotes anti-inflammatory macrophage activation by synergizing with IL-4 to activate Arg1 expression. (A) qRT-PCR for genes selected from microarray analysis of RAW264.7 cells treated with IL-4, RA or IL-4/RA for 3 h. (B) qRT-PCR analyses of Arg1 mRNA in mouse peritoneal macrophages stimulated with IL-4, RA or IL-4/RA for 3 h. (C) Western blot analyses of Arg1 protein from the extract of RAW264.7 cells treated with IL-4, RA or IL-4/RA for 24 h. (D) qRT-PCR analyses of Raldh2 mRNA in mouse bone marrow-derived macrophages treated with indicated ligands and cytokines for 6 h. Cells were treated with IL-4 (10 ng/ml) and/or RA (100 nM) in this figure. Data are representative of three experimental repeats (mean ± s.d), and Student's t-test (n = 3) was used (*P < 0.05; **P < 0.01; ***P < 0.001).
It appeared that, for LPS-stimulated macrophages (for pro-inflammatory M1 response), supplement with conditioned medium (CM) derived from IL-4/RA co-treated RAW264.7 macrophages (M2 macrophage) dampened their expression of inflammatory cytokine genes such as TNF-α and IL-6 genes (Figure 1B). These results demonstrate the suppression of inflammation by RA/IL-4 stimulated M2 (anti-inflammatory) macrophage activation. To comprehensively examine changes in gene expression during the anti-inflammatory phase, which is most critical to wound healing, we first employed microarrays to profile genes synergistically induced by IL-4/RA co-treatment. These were further validated quantitatively by qRT-PCR (Figure 2A). Very interestingly, only the Arg1 gene exhibited a robust synergistic induction by IL-4/RA (about 140-fold higher than control treatment) in M2 activation. This result revealed that IL-4 and RA synergize to robustly activate Arg1 gene, a gene most critical to tissue repair, which can lead to an important functional outcome, i.e. enhanced wound healing. Arg1 gene regulation by IL-4, RA or IL-4/RA co-treatment was further validated in experiments using primary macrophages (Figure 2B). Interestingly, Arg1 activation by IL-4/RA co-treatment was sustained over a period of 24 h (Supplementary Figure S1A). Western blot data confirmed synergistic elevation in Arg1 protein level stimulated by IL-4/RA co-treatment (Figure 2C). Sustained Arg1 expression would require continuous supply of RA in M2 polarized macrophages, which is supported by the finding of significant activation of Raldh2, a rate-limiting enzyme for RA biosynthesis, as demonstrated using primary bone marrow-derived macrophages (BMDM). For a comparison (Figure 2D), we found that LPS enhanced Raldh2 expression for a 31-fold in M1 macrophages, but IL-4 and RA co-treatment robustly (for a 671-fold) induced its expression in M2 macrophages. This synergism was reduced by AGN (a pan RAR antagonist), suggesting activation elicited by a canonical, RAR-mediated event. It appeared that synergistic activation of Raldh2 by IL-4 and RA was sustained over 24 h (data not shown), indicating that this positive feed forward mechanism could be critical for maintaining enhanced Arg1 activation for a longer period during the healing process. These data show robust induction of Arg1 by RA and IL-4 co-treatment in M2 macrophages, which involves RA's feed forward regulation of Raldh2 that contributes to sustained Arg1 expression and enhanced wound healing.
STAT6 and RAR are required for the synergistic effect of IL-4 and RA on Arg1 expression
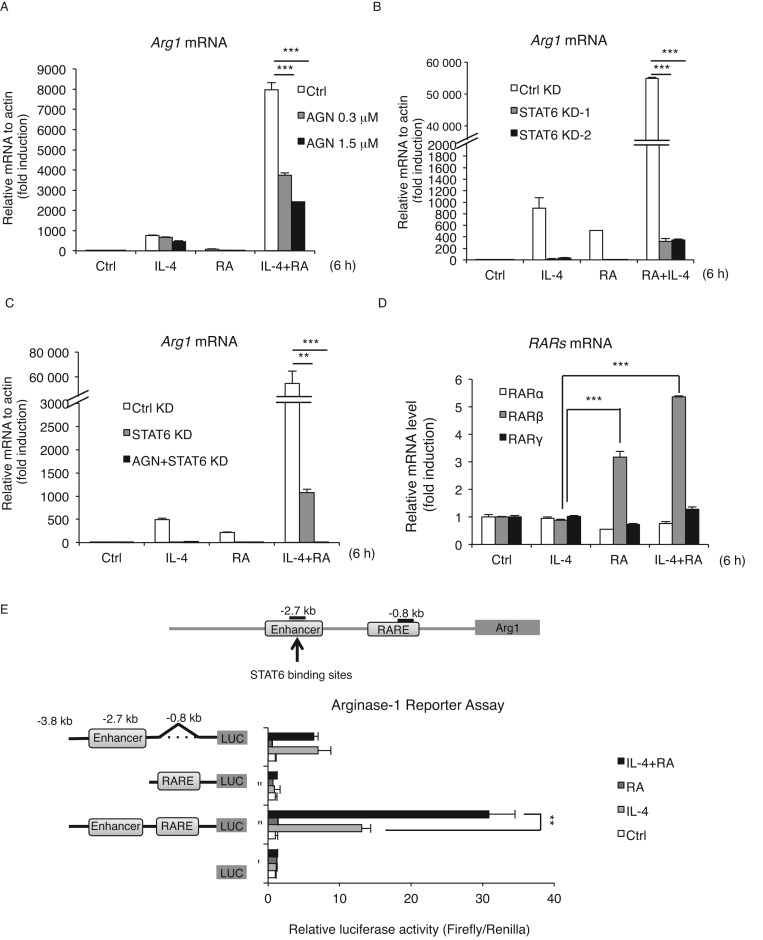
To determine whether synergistic activation of Arg1 gene by RA and IL-4 was mediated by canonical pathways, i.e. through the actions of RAR and STAT6, we employed both pharmacological and genetic approaches. RAW264.7 macrophages were pre-treated with a RAR pan-antagonist, AGN to block the actions of RARs (Figure 3A), which indeed dampened the synergistic induction of Arg1 expression in a concentration-dependent manner. The synergism was also significantly reduced in two independent STAT6 stably knockdown (STAT6 KD-1,2) clones as compared to control knockdown (Ctrl KD) clones (Figure 3B and Supplementary Figure S1B). Furthermore, a combination of AGN pre-treatment and STAT6 KD almost completely abolished the induction of Arg1 gene by IL-4 and RA (Figure 3C). Interestingly, among the three RAR isoforms (RARα, β, γ), only RARβ responded to RA or RA/IL-4 treatment in this cellular context (Figure 3D), indicating that RARβ may be the one mediating RA activation of Arg1 gene in macrophages. To determine the targets on the Arg1 gene promoter responding to RA and IL-4, we employed a luciferase reporter system to dissect STAT6 binding sites in the upstream enhancer (for responding to IL-4) and RARE near the promoter (for responding to RA). As shown in Figure 3E, the reporter construct containing both the IL-4-responsive enhancer and the RARE showed a synergistic effect of IL-4 and RA whereas constructs deleted in either the enhancer or the RARE exhibited no synergistic effect. Therefore, the canonical signaling pathways of IL-4 and RA, mediated through their corresponding genetic elements (STAT6 binding site and RARE), are responsible for the synergistic activation of Arg1 in IL-4 stimulated M2 macrophages co-treated with RA.
Figure 3.
STAT6 and RAR are required for the synergistic effect of IL-4 and RA on Arg1 activation. (A–C) qRT-PCR analyses of (A) Arg1 mRNA expression in RAW264.7 cells treated with IL-4, RA or IL-4/RA for 6 h with or without AGN (RAR pan-antagonist, 300 nM) pre-treatment for 30 min (B) Control knockdown (Ctrl KD) or STAT6 knockdown (STAT6 KD) cells treated with IL-4, RA or IL-4/RA for 6 h (C) Control, STAT6 KD or AGN-pretreated STAT6 KD cells treated with IL-4, RA or IL-4/RA for 6 h. (D) qRT-PCR analyses of RAR isoforms mRNA expression in RAW264.7 cells treated with IL-4, RA or IL-4/RA for 6 h. (E) Arg1 gene luciferase reporter assay deleted in enhancer (STAT6-binding sites, −2.7 kb), RARE (RAR-response element, −0.8 kb) or both in RAW264.7 cells treated with IL-4, RA or IL-4/RA 24 h. Cells were treated with IL-4 (10 ng/ml) and/or RA (100 nM) in this figure. Data are representative of three experimental repeats (mean ± s.d), and Student's t-test (n = 3) was used (*P < 0.05; **P < 0.01; ***P < 0.001).
Med25 plays dual functional roles in synergistic activation of Arg1 by IL-4 and RA, involving remodeling +1 nucleosome for transcription initiation and enhancing transcription elongation
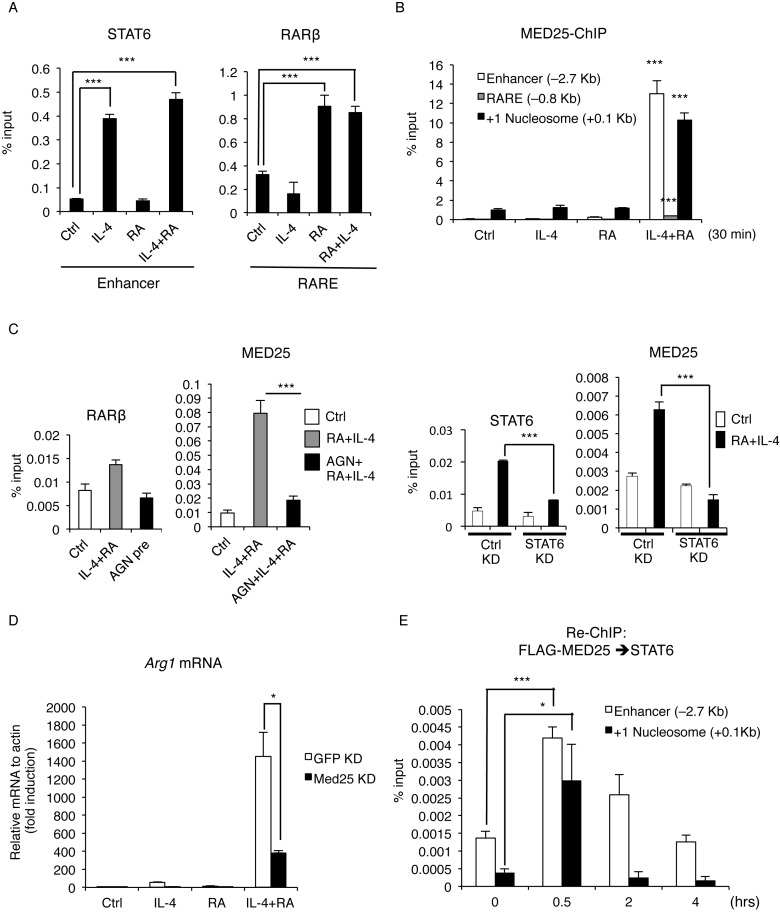
To examine the molecular events mediating the robust synergism of IL-4 and RA on Arg1 gene, we first validated that RARβ and STAT6 were indeed recruited to the endogenous chromatin regions of Arg1 gene upon RA/IL-4 co-treatment using chromatin immunoprecipitation (ChIP) assay (Figure 4A). The data showed similar levels of recruitment of RARβ and STAT6 by RA/IL-4 co-treatment as compared to single treatment, ruling out the possibility of enhanced transcription factor recruitment by co-treatment. Given that the enhancer and RARE are separated by a relatively long (∼2 kb) distance, we speculated a role for the Mediator complex that might act to bridge RARE and the enhancer. It appeared that among several reported RAR-interacting Mediator components, only Med25 recruitment was increased on the enhancer and the +1 nucleosome regions in IL-4/RA co-stimulated cells (Figure 4B). Since Med25 recruitment occurred after the recruitment of STAT6 and RARβ (Supplementary Figure S2A), Med25 is likely to be recruited by RARβ or STAT6. This was confirmed in the two following findings: IL-4/RA-induced Med25 enrichment to the +1 nucleosome region was dampened by AGN-pretreatment (Figure 4C, left), and its recruitment to the enhancer region was reduced by STAT6 KD (Figure 4C, right). Finally, the functional role for Med25 was validated in the experiments using loss-of-function (Med25 KD) (Figure 4D) and gain-of-function (over-expression, Supplementary Figure S3A) approaches.
Figure 4.
STAT6 and RARβ synergistically recruit Med25 upon IL-4 and RA treatment to enhance Arg1 transcription. (A) ChIP (Chromatin immunoprecipitation) assay of STAT6 and RARβ in RAW264.7 cells treated with IL-4, RA or IL-4/RA for 30 min. STAT6 and RARβ enrichment on enhancer and RARE, respectively, was measured by qPCR. (B) ChIP assay of Med25 on Arg1 enhancer, RARE or +1 nucleosome region in IL-4, RA or IL-4/RA-treated RAW264.7 cells. (P value: compared with ctrl or single treatment) (C) ChIP assay of RARβ or Med25 on RARE or +1 nucleosome, respectively, upon IL-4 and RA treatment with or without AGN (300 nM) pre-treatment for 30 min (left). ChIP assay of STAT6 or Med25 on the enhancer in Ctrl KD or STAT6 KD RAW264.7 cells upon IL-4 and RA treatment (right). (D) qRT-PCR analyses of Arg1 mRNA expression in Ctrl KD (GFP KD) or Med25 KD RAW264.7 cells upon IL-4, RA or IL-4/RA treatment. (E) Re-ChIP assay of FLAG-MED25 and STAT6 on the enhancer and the +1 nucleosome of Arg1 at indicated time-points after IL-4 and RA treatment. Cells were treated with IL-4 (10 ng/ml) and/or RA (100 nM) in this figure. Data are representative of three experimental repeats (mean ± s.d), and Student's t-test (n = 3) was used (*P < 0.05; **P < 0.01; ***P < 0.001). See also Supplementary Figure S3.
Med25 belongs to the Mediator complex that recruits general transcription machinery including RNA Pol II to the target promoter. To investigate whether Med25 may also regulate other M2 genes, we monitored M2 markers’ mRNA expression patterns in control KD and Med25 KD macrophages upon IL-4 stimulation. As shown in Supplementary Figure S3B, most M2 genes were not affected by Med25 KD, such as Chi3l3, Retnla and Pparg, except for Mrc1 gene. Thus, RA/IL-4 synergism during M2 activation, which requires the Med25, seems to occur only on selected M2 genes, such as the Arg1 gene. Interestingly, MED1, also a RAR-interacting Mediator component (23), is not involved in the synergistic induction of Arg1 by IL-4 and RA (Supplementary Figure S3C), suggesting a unique role for Med25 in mediating synergism of RA and IL-4 on Arg1 gene. Sequential ChIP (Re-ChIP) assay revealed that Med25 forms a complex with STAT6, most evidently on the +1 nucleosome region and the enhancer region (Figure 4E); whereas STAT6 and RARβ complex formation occurred mainly on the enhancer region (Supplementary Figure S4A). These data show that Med25, recruited by RARβ and STAT6, is required for the synergistic activation of Arg1 gene by IL-4 and RA through enhanced transcription factor-Mediator complex formation on the initiation site covered by the first (+1) nucleosome. Thus, transcription factors/cofactors recruitment on the Arg1 gene regulatory region occurs sequentially, with each transcription factor first recruited to its own corresponding DNA element upon stimulation, which then alters the recruitment of other co-activators such as the Mediator complex to transcription initiation site to stimulate transcription initiation.
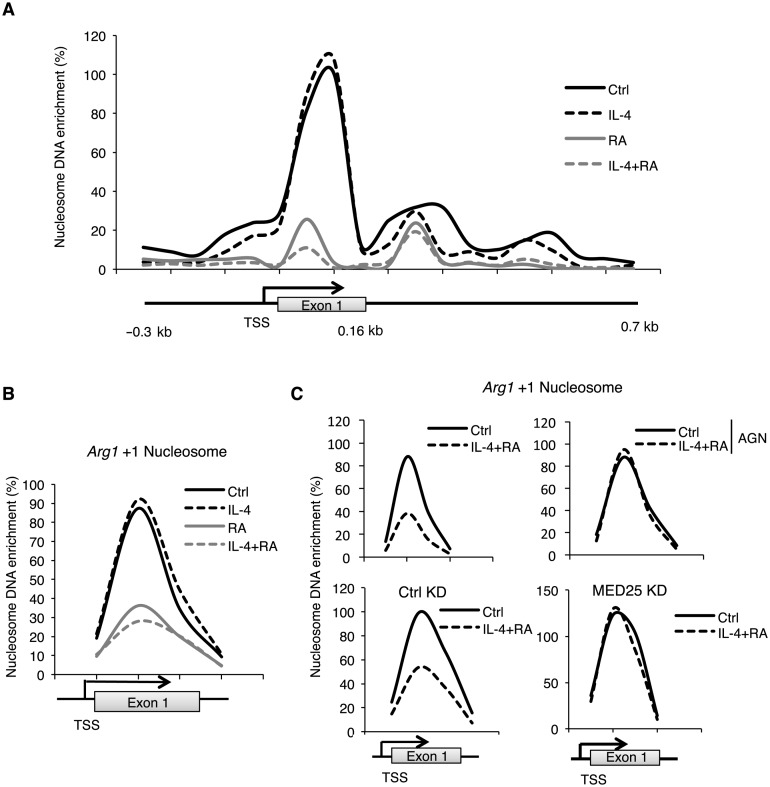
It is known that nucleosomes spanning the gene promoter region affect RNA pol II recruitment for transcription initiation (36). To survey nucleosome occupancy on Arg1 gene, we employed MNase-digestion to generate mono-nucleosomes and scanned nucleosome occupancy on the Arg1 gene regulatory regions (nucleosome array, Figure 5A). Interestingly, the intensity of the nucleosome spanning exon 1 region was most robustly reduced in cells stimulated by RA or IL-4/RA. This result suggests that this +1 nucleosome is destabilized mainly by RA treatment (Figure 5B). Consistently, this effect was blocked by AGN pre-treatment (Figure 5C, upper). In Med25 KD stable clones, we found that Med25 KD also mainly affected this +1 nucleosome (Figure 5C, bottom). We then determined which of the two ATP-dependent chromatin remodelers, Brg1 and Brm, was involved and found that the recruitment of Brg1, but not Brm, on +1 nucleosome region was significantly enhanced upon IL-4 and RA co-treatment. Further, this was dampened by Med25 KD (Figure 6A, left, Supplementary Figure S4B). Together, the data show that IL-4/RA stimulation destabilizes, specifically, the +1 nucleosome, which involves RAR- and STAT6-recruited Med25 and its subsequent recruitment of the ATPase chromatin remodeler Brg1.
Figure 5.
Med25 mediates synergistic activation of Arg1 by IL-4 and RA via remodeling +1 nucleosome. (A) Nucleosome scanning on the Arg1 promoter, the +1 nucleosome and the gene body region in IL-4, RA or IL-4/RA-treated RAW264.7 cells (30 min). (B) The +1 nucleosome occupancy immediately down-stream of Arg1 gene's TSS in RAW264.7 cells treated with IL-4, RA or IL-4/RA. (C) The +1 nucleosome occupancy in RAW264.7 cells with (upper right) or without (upper left) AGN (300 nM) pre-treatment for 30 min, and that in Ctrl KD (bottom left) or Med25 KD (bottom right) RAW264.7 cells. Cells were treated with IL-4 (10 ng/ml) and/or RA (100 nM) in this figure. Data are representative of three experimental repeats.
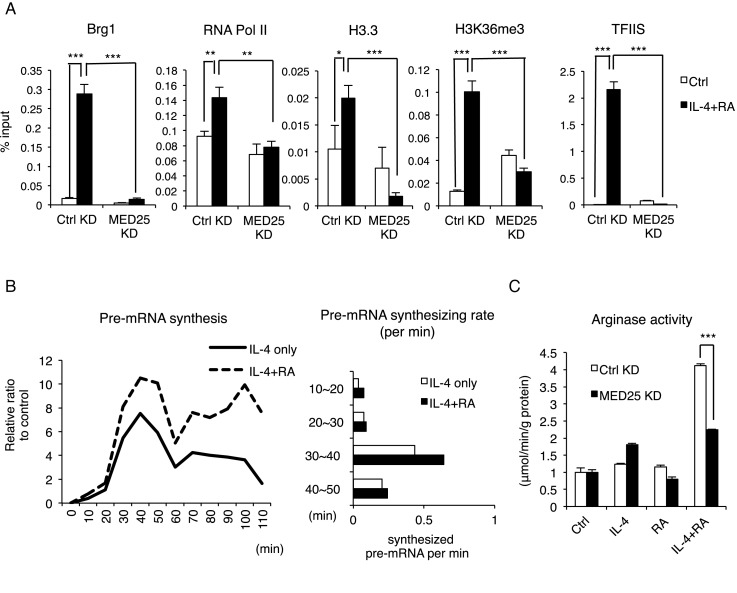
Figure 6.
IL-4 and RA synergistically induce Med25 recruitment for transcription initiation-elongation coupling on Arg1 gene. (A) ChIP assay of Brg1, RNA Pol II, H3.3, H3K36me3 and TFIIS on the +1 nucleosome in Ctrl KD or Med25 KD RAW264.7 cells with or without IL-4 and RA treatment for 30 min. (B) The kinetics of Arg1 pre-mature mRNA synthesis. Total RNAs were isolated to monitor Arg1 pre-mRNA level determined with primers spanning intron 7 and exon 8 regions in qRT-PCR (left). Pre-mRNA synthesizing rate was deduced through measuring the differences in relative ratios of synthesized pre-mRNA per min (right) (C) In vitro arginase activity assayed in Ctrl KD or Med25 KD RAW264.7 cells following IL-4, RA or IL-4/RA treatment for 24 h. Cells were treated with IL-4 (10 ng/ml) and/or RA (100 nM) in this figure. Data are representative of three experimental repeats (mean ± s.d), and Student's t-test (n = 3) was used (*P < 0.05; **P < 0.01; ***P < 0.001).
Among various steps of the transcription process, we found that Med25 KD reduced the recruitment of RNA Pol II, a major component of transcriptional pre-initiation complex, and the expected histone variants/post-translational modifications (PTM) (Figure 6A, second left). Among the histone variants, H3.3, known as an active histone variant with H2A.Z (37), was most dramatically augmented by IL-4/RA co-treatment, which was reversed by Med25 KD (Figure 6A, middle). Surprisingly, H3K36me3 enrichment, a histone marker related to transcription elongation (38), was also significantly enhanced upon IL-4/RA co-treatment, which was also abolished by Med25 KD (Figure 6A, second right). This result suggests an extended Med25 function in transcription elongation, which was supported by the finding of enhanced recruitment of TFIIS, an elongation complex component and an inducer of catalytic mode of promoter-bound Pol II (39). Importantly, this was also abolished by Med25 KD (Figure 6A. right). To further substantiate the synergistic effects of RA/IL-4 on transcription elongation of Arg1, we monitored newly synthesized pre-mRNA of Arg1 every 10 min by removing pre-existing pre-mRNA through blocking transcription with 5,6-Dichloro-1-β-D-ribofuranosylbenzimidazole (DRB), a RNA Pol II-dependent elongation inhibitor, for 3 h before the stimulation (Figure 6B, left).(33). Primers spanning intron 7 and exon 8 regions were used in qRT-PCR to quantify pre-mRNA accumulation. Indeed, the relative level of pre-mRNA in cells co-stimulated with IL-4 and RA was much higher than when stimulated with IL-4 alone, and the difference was readily evident in 30 min (Figure 6B, right). These data indicate that co-stimulation of IL-4 and RA accelerates RNA Pol II release from pausing for transcription initiation and also enhances progression from initiation to elongation possibly by Med25-dependent recruitment of elongation factors such as TFIIS (as shown in Figure 6A). Finally, we validated the functional role of Med25 in IL-4/RA's synergistic activation of Arg1 enzyme activity as shown in Figure 6C.
DISCUSSION
Vitamin A deficiency has long been associated with delayed wound healing and delayed recovery following inflammation (2). Therefore, retinoids have been very useful in dermatology. Preoperative use of retinoids to enhance wound healing has been clinically documented, which involves stimulation of collagen synthesis, epithelialization and angiogenesis (8,40–42). Our current study unambiguously demonstrates the molecular and cellular bases underlying a robust synergism of RA and anti-inflammatory cytokine IL-4 on Arg1, a gene critical to the wound healing processes. Importantly, this involves RA feed forward regulation of Raldh2 and requires dual functions of Med25, which accelerates nucleosome remodeling for transcription initiation and enhances transcription elongation.
There are two distinct isoforms of arginase in mammalian cells. Arginase 1 (encoded by Arg1) is a widely expressed cytosolic enzyme with a principal role in the hepatic urea cycle; whereas arginase 2 (encoded by Arg2) is a mitochondrial enzyme with limited expression only in certain organs such as kidney, brain and liver. Importantly, Arg1 has been known to play important roles in anti-inflammation, tumor immunity, fibrosis and immunosuppression-related diseases mainly because of its activity in regulating L-arginine metabolism in immune cells including macrophages (43–45). As such, controlling Arg1 expression levels through the synergism of RA with anti-inflammatory cytokine IL-4 can provide a potential therapeutic strategy in treating inflammatory diseases that may be further exploited clinically.
Interestingly, the synergism of IL-4 and RA co-treatment yielded a 4-fold increase in Arg1 enzyme activity, whereas the effect on Arg1 mRNA levels was detected for hundreds- to thousands-fold higher than control treatment. This suggests that while the enhanced capacity to produce Arg1 enzyme is critical for wound healing, there are additional regulations needed to control the level of this enzyme activity in the very dynamic process of wound healing, such as by regulation at translational/post-translational levels. This requires further investigation in future studies. In our animal wound healing experiments (Figure 1 and Supplementary Figure S1), elevation of the Arg1 mRNA level, stimulated by RA/IL-4 co-treatment, was sustained for longer than 1 day (Supplementary Figure S1 shows sustained elevation after 24 h, and our unpublished data show sustained elevation can be as long as 3 days). In order to monitor direct effects of RA and IL-4 on endogenous Raldh2 expression, we have routinely measured Raldh2 mRNA levels at early time points, such as 6 h post-stimulation (Figure 2D); however, it appears that Raldh2 regulation by RA can last as long as 24 h (data not shown). Presumably, this is to ensure the endogenous supply of RA, which would be required for prolonged activation of Arg1 gene during wound healing without pharmacological intervention. An interesting question arises as to the dampening of this feed forward regulation of Raldh2 by RA in a physiological context. Since efficient Raldh2 activation in M2 macrophages requires IL-4 stimulation, reduction in immune cytokines, including IL-4, in the wounded tissues as the healing process occurs would likely provide a break to prevent continuous activation of Raldh2 gene later after healing has completed and RA is no longer needed.
MED25 was originally identified as a PTOV1 relative (46) and is also identical to ARC92/ACID1, which is a direct target of the activator VP16 (47). While the Mediator complex is highly conserved from yeast to humans (48), There may be an evolutionary advantage of MED25 in higher eukaryotes given it is not found in Caenorhabditis elegans or yeast. Although MED25 has similar features with MED1 in its domains and the action on nuclear receptor (NR)-mediated transcription, MED25 possesses distinct functions separated from MED1 upon different environmental signals in different cells (25,49). Our data provide evidence for unique functional roles for Med25, such as in regulating Arg1 gene expression in M2 macrophage that is critically needed for wound healing.
In gene transcription, the formation of pre-initiation complex (PIC) and the initiation steps were considered to be most critical. However, it has become increasingly clear that post-recruitment steps, including RNA Pol II release from promoter-proximal pausing and transcription elongation, are also important in the control of transcriptional rate (50,51). Our data show elongation-related histone marker, H3K36me3 and elongation factor TFIIS are enhanced by IL-4 and RA co-stimulation, which were blocked by Med25 KD. This is in line with a recent study showing cooperation of the Mediator complex and TFIIS in altering intrinsic catalytic properties of RNA Pol II transit across the +1 nucleosome (52).
Supplementary Material
Acknowledgments
We thank R. Jasty, E. Gordon and B. Cheng for technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [R01DK54733, R01DK60521, R01DK54733-11S, RO1DK60521-12S1 to L.-N.W.]; Dean's Commitment and the Distinguished McKnight Professorship of University of Minnesota (to L.-N. W). Funding for open access charge: National Institutes of Health (NIH), USA [DK60521 and DK54733].
Conflict of interest statement. None declared.
REFERENCES
- 1.Clagett-Dame M., DeLuca H.F. The role of vitamin A in mammalian reproduction and embryonic development. Annu. Rev. Nutr. 2002;22:347–381. doi: 10.1146/annurev.nutr.22.010402.102745E. [DOI] [PubMed] [Google Scholar]
- 2.Sommer A. Vitamin a deficiency and clinical disease: an historical overview. J. Nutr. 2008;138:1835–1839. doi: 10.1093/jn/138.10.1835. [DOI] [PubMed] [Google Scholar]
- 3.Carman J.A., Pond L., Nashold F., Wassom D.L., Hayes C.E. Immunity to Trichinella spiralis infection in vitamin A-deficient mice. J. Exp. Med. 1992;175:111–120. doi: 10.1084/jem.175.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith S.M., Levy N.S., Hayes C.E. Impaired immunity in vitamin A-deficient mice. J. Nutr. 1987;117:857–865. doi: 10.1093/jn/117.5.857. [DOI] [PubMed] [Google Scholar]
- 5.Verma A.K. Retinoids in chemoprevention of cancer. J. Biol. Regul. Homeost. Agents. 2003;17:92–97. [PubMed] [Google Scholar]
- 6.Malkovský M., Doré C., Hunt R., Palmer L., Chandler P., Medawar P.B. Enhancement of specific antitumor immunity in mice fed a diet enriched in vitamin A acetate. Proc. Natl. Acad. Sci. U.S.A. 1983;80:6322–6326. doi: 10.1073/pnas.80.20.6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdelmalek M., Spencer J. Retinoids and wound healing. Dermatol. Surg. 2006;32:1219–1230. doi: 10.1111/j.1524-4725.2006.32280.x. [DOI] [PubMed] [Google Scholar]
- 8.Hunt T.K. Vitamin A and wound healing. J. Am. Acad. Dermatol. 1986;15:817–821. doi: 10.1016/s0190-9622(86)70238-7. [DOI] [PubMed] [Google Scholar]
- 9.Ulland A.E., Shearer J.D., Coulter C., Caldwell M.D. Altered wound arginine metabolism by corticosterone and retinoic acid. J. Surg. Res. 1997;70:84–88. doi: 10.1006/jsre.1997.5099. [DOI] [PubMed] [Google Scholar]
- 10.Hall J.A., Grainger J.R., Spencer S.P., Belkaid Y. The role of retinoic acid in tolerance and immunity. Immunity. 2011;35:13–22. doi: 10.1016/j.immuni.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ross A.C. Vitamin A and retinoic acid in T cell-related immunity. Am. J. Clin. Nutr. 2012;96:1166S–1172S. doi: 10.3945/ajcn.112.034637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ross A.C., Chen Q., Ma Y. Vitamin A and retinoic acid in the regulation of B-cell development and antibody production. Vitam. Horm. 2011;86:103–126. doi: 10.1016/B978-0-12-386960-9.00005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mora J.R., Iwata M., Eksteen B., Song S.-Y., Junt T., Senman B., Otipoby K.L., Yokota A., Takeuchi H., Ricciardi-Castagnoli P., et al. Generation of gut-homing IgA-secreting B cells by intestinal dendritic cells. Science. 2006;314:1157–1160. doi: 10.1126/science.1132742. [DOI] [PubMed] [Google Scholar]
- 14.Mucida D., Park Y., Kim G., Turovskaya O., Scott I., Kronenberg M., Cheroutre H. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317:256–260. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 15.Sun C.-M., Hall J.A., Blank R.B., Bouladoux N., Oukka M., Mora J.R., Belkaid Y. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J. Exp. Med. 2007;204:1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwata M., Hirakiyama A., Eshima Y., Kagechika H., Kato C., Song S.-Y. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 2004;21:527–538. doi: 10.1016/j.immuni.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 17.Broadhurst M.J., Leung J.M., Lim K.C., Girgis N.M., Gundra U.M., Fallon P.G., Premenko-Lanier M., McKerrow J.H., McCune J.M., Loke P. Upregulation of retinal dehydrogenase 2 in alternatively activated macrophages during retinoid-dependent type-2 immunity to helminth infection in mice. PLoS Pathog. 2012;8:e1002883. doi: 10.1371/journal.ppat.1002883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu B., Buttrick T., Bassil R., Zhu C., Olah M., Wu C., Xiao S., Orent W., Elyaman W., Khoury S.J. IL-4 and retinoic acid synergistically induce regulatory dendritic cells expressing Aldh1a2. J. Immunol. 2013;191:3139–3151. doi: 10.4049/jimmunol.1300329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang J., Thangamani S., Kim M.H., Ulrich B., Morris S.M., Kim C.H. Retinoic acid promotes the development of Arg1-expressing dendritic cells for the regulation of T-cell differentiation. Eur. J. Immunol. 2013;43:967–978. doi: 10.1002/eji.201242772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eriksen A.B., Berge T., Gustavsen M.W., Leikfoss I.S., Bos S.D., Spurkland A., Harbo H.F., Blomhoff H.K. Retinoic acid enhances the levels of IL-10 in TLR-stimulated B cells from patients with relapsing-remitting multiple sclerosis. J. Neuroimmunol. 2015;278:11–18. doi: 10.1016/j.jneuroim.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 21.Malik S., Roeder R.G. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taatjes D.J. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem. Sci. 2010;35:315–322. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan C.X., Ito M., Fondell J.D., Fu Z.Y., Roeder R.G. The TRAP220 component of a thyroid hormone receptor- associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc. Natl. Acad. Sci. U.S.A. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park J.M., Kim J.M., Kim L.K., Kim S.N., Kim-Ha J., Kim J.H., Kim Y.-J. Signal-induced transcriptional activation by Dif requires the dTRAP80 mediator module. Mol. Cell. Biol. 2003;23:1358–1367. doi: 10.1128/MCB.23.4.1358-1367.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee H.-K., Park U.-H., Kim E.-J., Um S.-J. MED25 is distinct from TRAP220/MED1 in cooperating with CBP for retinoid receptor activation. EMBO J. 2007;26:3545–3557. doi: 10.1038/sj.emboj.7601797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mikita T., Campbell D., Wu P., Williamson K., Schindler U. Requirements for interleukin-4-induced gene expression and functional characterization of Stat6. Mol. Cell. Biol. 1996;16:5811–5820. doi: 10.1128/mcb.16.10.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mantovani A., Sozzani S., Locati M., Allavena P., Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–555. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 28.Gordon S. Alternative activation of macrophages. Nat. Rev. Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 29.Pourcet B., Pineda-Torra I.X.S. Transcriptional regulation of macrophage arginase 1 expression and its role in atherosclerosis. Trends Cardiovasc. Med. 2013;23:143–152. doi: 10.1016/j.tcm.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Gray M.J., Poljakovic M., Kepka-Lenhart D., Morris S.M. Induction of arginase I transcription by IL-4 requires a composite DNA response element for STAT6 and C/EBPbeta. Gene. 2005;353:98–106. doi: 10.1016/j.gene.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 31.Lin Y.-W., Lee B., Liu P.-S., Wei L.-N. Receptor-interacting protein 140 Orchestrates the dynamics of macrophage M1/M2 polarization. J. Innate Immun. 2016;8:97–107. doi: 10.1159/000433539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu C.-Y., Feng X., Wei L.-N. Coordinated repressive chromatin-remodeling of Oct4 and Nanog genes in RA-induced differentiation of embryonic stem cells involves RIP140. Nucleic Acids Res. 2014;42:4306–4317. doi: 10.1093/nar/gku092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh J., Padgett R.A. Rate of in situ transcription and splicing in large human genes. Nat. Struct. Mol. Biol. 2009;16:1128–1133. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pino-Lagos K., Guo Y., Noelle R.J. Retinoic acid: A key player in immunity. BioFactors. 2010;36:430–436. doi: 10.1002/biof.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray P.J., Wynn T.A. Protective and pathogenic functions of macrophage subsets. Nat. Publishing Group. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fuda N.J., Ardehali M.B., Lis J.T. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber C.M., Henikoff S. Histone variants: dynamic punctuation in transcription. Genes Dev. 2014;28:672–682. doi: 10.1101/gad.238873.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wen H., Li Y., Xi Y., Jiang S., Stratton S., Peng D., Tanaka K., Ren Y., Xia Z., Wu J., et al. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature. 2014;508:263–268. doi: 10.1038/nature13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J., Guermah M., Roeder R.G. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell. 2010;140:491–503. doi: 10.1016/j.cell.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elias P.M. Epidermal effects of retinoids: supramolecular observations and clinical implications. J. Am. Acad. Dermatol. 1986;15:797–809. doi: 10.1016/s0190-9622(86)70236-3. [DOI] [PubMed] [Google Scholar]
- 41.Hevia O., Nemeth A.J., Taylor J.R. Tretinoin accelerates healing after trichloroacetic acid chemical peel. Arch. Dermatol. 1991;127:678–682. [PubMed] [Google Scholar]
- 42.Popp C., Kligman A.M., Stoudemayer T.J. Pretreatment of photoaged forearm skin with topical tretinoin accelerates healing of full-thickness wounds. Br. J. Dermatol. 1995;132:46–53. doi: 10.1111/j.1365-2133.1995.tb08623.x. [DOI] [PubMed] [Google Scholar]
- 43.Bronte V., Serafini P., Mazzoni A., Segal D.M., Zanovello P. L-arginine metabolism in myeloid cells controls T-lymphocyte functions. Trends Immunol. 2003;24:301–305. doi: 10.1016/s1471-4906(03)00132-7. [DOI] [PubMed] [Google Scholar]
- 44.Bronte V., Zanovello P. Regulation of immune responses by L-arginine metabolism. Nat. Rev. Immunol. 2005;5:641–654. doi: 10.1038/nri1668. [DOI] [PubMed] [Google Scholar]
- 45.Munder M. Arginase: an emerging key player in the mammalian immune system. Br. J. Pharmacol. 2009;158:638–651. doi: 10.1111/j.1476-5381.2009.00291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benedit P., Paciucci R., Thomson T.M., Valeri M., Nadal M., Càceres C., de Torres I., Estivill X., Lozano J.J., Morote J., et al. PTOV1, a novel protein overexpressed in prostate cancer containing a new class of protein homology blocks. Oncogene. 2001;20:1455–1464. doi: 10.1038/sj.onc.1204233. [DOI] [PubMed] [Google Scholar]
- 47.Mittler G., Stühler T., Santolin L., Uhlmann T., Kremmer E., Lottspeich F., Berti L., Meisterernst M. A novel docking site on Mediator is critical for activation by VP16 in mammalian cells. EMBO J. 2003;22:6494–6504. doi: 10.1093/emboj/cdg619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boube M., Joulia L., Cribbs D.L., Bourbon H.-M. Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell. 2002;110:143–151. doi: 10.1016/s0092-8674(02)00830-9. [DOI] [PubMed] [Google Scholar]
- 49.Rana R., Surapureddi S., Kam W., Ferguson S., Goldstein J.A. Med25 is required for RNA polymerase II recruitment to specific promoters, thus regulating xenobiotic and lipid metabolism in human liver. Mol. Cell. Biol. 2011;31:466–481. doi: 10.1128/MCB.00847-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jonkers I., Lis J.T. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015;16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu X., Kraus W.L., Bai X. Ready, pause, go: regulation of RNA polymerase II pausing and release by cellular signaling pathways. Trends Biochem. Sci. 2015;40:516–525. doi: 10.1016/j.tibs.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nock A., Ascano J.M., Barrero M.J., Malik S. Mediator-regulated transcription through the +1 nucleosome. Mol. Cell. 2012;48:837–848. doi: 10.1016/j.molcel.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.