Abstract
Coordinated regulation of gene expression that involves activation of lineage specific genes and repression of pluripotency genes drives differentiation of embryonic stem cells (ESC). For complete repression of pluripotency genes during ESC differentiation, chromatin at their enhancers is silenced by the activity of the Lsd1-Mi2/NuRD complex. The mechanism/s that regulate DNA methylation at these enhancers are largely unknown. Here, we investigated the affect of the Lsd1-Mi2/NuRD complex on the dynamic regulatory switch that induces the local interaction of histone tails with the Dnmt3 ATRX-DNMT3-DNMT3L (ADD) domain, thus promoting DNA methylation at the enhancers of a subset of pluripotency genes. This is supported by previous structural studies showing a specific interaction between Dnmt3-ADD domain with H3K4 unmethylated histone tails that is disrupted by histone H3K4 methylation and histone acetylation. Our data suggest that Dnmt3a activity is triggered by Lsd1-Mi2/NuRD-mediated histone deacetylation and demethylation at these pluripotency gene enhancers when they are inactivated during mouse ESC differentiation. Using Dnmt3 knockout ESCs and the inhibitors of Lsd1 and p300 histone modifying enzymes during differentiation of E14Tg2A and ZHBTc4 ESCs, our study systematically reveals this mechanism and establishes that Dnmt3a is both reader and effector of the epigenetic state at these target sites.
INTRODUCTION
DNA methylation is established by de novo methyltransferases (MTases) Dnmt3a and 3b and is maintained through replication by Dnmt1 that copies the methylation pattern from parent to daughter strand (1,2). In contrast to maintenance methylation, the mechanism(s) that determine the specificity of de novo methylation are not completely elucidated. Biochemical models proposed to explain the specificity of de novo DNA methylation include the intrinsic ability of Dnmt3a and 3b to find their target location (3–7) and the role of interacting factors such as DNA and chromatin binding proteins (8–11). Further, both direct and indirect relationships have been identified between histone tail modifications and DNA methylation (12–16). Interaction of HP1, a reader protein of histone H3K9 trimethylation (H3K9me3), with Dnmt3b leads to DNA methylation at pericentromeric repeats in mouse embryonic stem cells (ESCs) (17). H3K9me3 also has been shown to direct DNA methylation in plants (18) and in Neurospora (19). Structural and biochemical studies have shown a direct interaction between the PHD-like ATRX-DNMT3-DNMT3L (ADD) domain of de novo DNA MTases with an unmethylated H3K4 histone peptide leading to the catalytic activation of Dnmt3a. This interaction was blocked by H3K4 methylation and histone acetylation (20–23). These observations, taken together with the genome-wide studies showing the specific presence of H3K4me3 at unmethylated CpG rich regulatory regions, led to a model in which H3K4 methylation at regulatory elements protects them from DNA methylation by Dnmt3 enzymes (24,25). In a recent study, this model was tested by mutating Dnmt3a to render its ADD domain insensitive to H3K4 methylation. This Dnmt3a mutant was aberrantly targeted to active H3K4me3 promoters of a subset of developmental genes that led to a decrease in their gene expression and altered ESC differentiation (26). However, when the cell state changes during ESC differentiation and gene repression is induced, whether histone demethylation of H3K4 at some regulatory elements can locally recruit Dnmt3 enzymes has not been addressed. Therefore, we investigated a regulatory role of the interaction of Dnmt3-ADD domain with demethylated histone tails in site-specific DNA methylation during ESC differentiation.
In response to signals of differentiation, ESCs initiate global changes in gene expression that are forged by alterations in the epigenetic state of the regulatory elements of various cohorts of genes. Among these, pluripotency genes (PpGs) are downregulated and at their promoters and enhancers acquire a repressed chromatin state that includes loss of histone H3K4 methylation and gain of DNA methylation. The interaction of Dnmt3a with the G9A histone methyltransferase has been shown to promote DNA methylation at the PpG promoters post differentiation of ESCs (9,10). However, little is known about how DNA methylation is regulated at PpG enhancers. Histone H3K4me1 (monomethylation) and H3K27Ac (acetylation) are prototypical epigenetic modifications for active enhancers (27). Compared to demethylation of histone H3K4me3 at promoters, which is proposed to require the combined activity of Kdm5/Jarid family and Lsd1 demethylases, demethylation of H3K4me1 at enhancers only requires Lsd1 activity (28–31). During differentiation of ESCs, Lsd1 associates with the Mi2/NuRD deacetylase complex specifically at PpG enhancers. The activity of this repressive complex is triggered by dissociation of the OSN (Oct4/Sox2/Nanog/HATp300) coactivator complex resulting in enhancer repression by histone deacetylation and demethylation, which is critical for PpG repression (31). Based on these observations we speculated that deacetylated and demethylated H3K4 histone tails at PpG enhancers may locally activate Dnmt3 enzymes via an interaction with their ADD domains, thus conferring a unique regulatory potential to this series of epigenetic events. Our studies here provide evidence for the timing, location and role of the interaction of the Dnmt3 ADD domain with demethylated histone tails in regulating site-specific DNA methylation at a subset of PpG enhancers during ESC differentiation.
MATERIALS AND METHODS
ESC culture and differentiation method
E14Tg2A (WT) ESCs were maintained in media containing LIF and induced to differentiate by LIF withdrawal and followed by retinoic acid addition (32). ZHBTc4 cells were maintained in media with LIF and induced to differentiate by simultaneous withdrawal of LIF and addition of doxycycline (31). Treatment of the Lsd1 inhibitors pargyline (3 mM) (Sigma, P8013), tranylcypromine TCP (1 mM) (Millipore, 616431) and the p300 inhibitor C646 (400 nM) (Sigma, SML0002) were performed as described (31). Low passage Dnmt3KO ESC were obtained from Dr Taiping Chen, MD Anderson and ZHBTc4 were obtained from Dr Dana Levasseur, University of Iowa. Single (KO) and double knockout (DKO) cells for Dnmt3a and/or Dnmt3b, as well as the transgenic Myc-ADD and Myc-Vector expressing cell lines were grown and induced to differentiate similar to E14Tg2A ESCs. For all experiments, ESC (all lines) were expanded by passaging twice after which differentiation was induced. At various time points post induction, a third of cells were harvested for preparation of DNA and RNA and rest were crosslinked to prepare chromatin. For all experiments this procedure was repeated at least 2 times for sample collection.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed using crosslinked chromatin from ESCs and embryoid bodies. Cells were crosslinked with 1% formaldehyde in Crosslinking buffer (0.1 M NaCl, 1 mM EDTA, 0.5 mM EGTA and 50 mM HEPES pH 8) for 5 min, then quenched by adding glycine to 150 mM. The nuclei were isolated and chromatin was sheared using a Covaris E210 device, according to manufacturer's protocols. Eight micrograms of sonicated chromatin was added to protein A and protein G magnetic beads (Life Technologies, 10002D, 10004D) conjugated with desired antibody overnight. After washing the beads, bound chromatin was eluted and purified DNA was used for quantitative PCR (qPCR). DNA samples were quantified using a NanoDrop 3300 fluorospectrometer. qPCR was then performed using equal amounts of IN (input) and IP (immunoprecipitated sample) DNA. Percent input was calculated as follows: 2∧(Cq(IN)-Cq(IP))X100. Cq is the quantification cycle as calculated by the Biorad CFX Manager 3.1 based on the MIQE guidelines (33). Standard deviations represent at least 2 technical and 2 biological replicates. For the all the experiments, the significance of the average change in the enrichment across all loci was calculated by Wilcoxon matched pairs signed rank test using GraphPad Prism and P-values were ≤0.035. For Myc-ADD ChIP, the percent enrichment for the ADD domain was determined by subtracting the percent enrichment obtained for the vector control. Additional information is available in the Supplementary Methods section for ChIP. See Supplementary Table S4 for primers used. A detailed ChIP protocol is in Supplementary Materials.
DNA methylation-dependent qPCR assay (MD-qPCR)
Genomic DNA was harvested using a standard phenol:chloroform isolation, followed by ethanol precipitation. DNA from various samples was digested initially with CviQI (NEB, R0639L) overnight at 25°C. Purified samples were subjected to digestion by MspJI (NEB, R0166L) overnight at 37°C. Digested DNA was purified and quantified by PicoGreen according to manufacturer's protocol (Life Technologies, P11495) using the NanoDrop 3300 fluorospectrometer. Quantitative PCR was performed by using equal amount of DNA for each sample. Change in DNA methylation is represented by relative fold change in the Cq values as follows: 2∧[Cq(U)-Cq(I)], where Cq(U) is the Cq for the undifferentiated (ESC) sample, and Cq(I) represents a sample from a given time point post differentiation. Cq is the quantification cycle as calculated by the Biorad CFX Manager 3.1 based on the MIQE guidelines (Minimum Information for publication of Quantitative Real-Time PCR Experiments) (33). The primers used for DNA methylation-dependent qPCR analysis were the same as used for ChIP. Standard deviations represent at least 2 technical and 2 biological replicates.
Quantitative RT-PCR
RNA was isolated by using TRIzol (Invitrogen, 15596026) according to manufacturer's protocol, and purified after DNAse treatment (Roche, 04716728001). RT-qPCR (Real-Time quantitaive PCR) was performed by using Verso One-Step RT-qPCR kits (Thermo Scientific, AB-4104A). The data analysis was performed by calculating delta Cq normalized to Gapdh expression and represented as change relative to undifferentiated (ESC) expression, which was set to 0. Cq is the quantification cycle as calculated by the Biorad CFX Manager 3.1 based on the MIQE guidelines (33). Change in expression is reported in a log2 scale and the average data is calculated for 8 PpG. Standard deviations represent at least 2 technical and 2 biological replicates. The variance in average data is represented by SEM (standard error of the mean) with n = 8. The SD (standard deviation), SEM determination and P-value were calculated using GraphPad Prism using Student's paired t-test. See Supplementary Table S4 for primers used.
Microscopy
Alkaline phosphatase staining was performed using a kit (Sigma, AB0300) according to the manufacturers protocol. Cells were imaged under a Zeiss microscope at 20X objective. Immunofluorescence experiments were performed according to Whyte et al., 2012 (31). Antibodies used for immunofluorescence include: SSEA-1 (Millipore, MAB430) 1:2000 and AlexaFluor 555 nm (Life Technologies, A21422) 1:1000. Brightfield imaging was done with a Nikon Ts microscope with a 20X objective.
Bisulfite sequencing
Bisulfite sequencing was performed by using the EpiTect Fast Bisulfite Conversion Kit (Qiagen, 59802) according to the manufacturer's protocol. Bisulfite-converted DNA was amplified using nested primers according to the published methods (34). Products from the inner PCR were gel purified (Qiagen, 28704) and used to generate a barcoded library for Mi-Seq. Barcoded amplicons for each region were purified and pooled to a final concentration of 8 pM. Illumina MiSeq 500 cycle run generated paired-end 250 base reads. The reads were then mapped by Bowtie2 and analyzed by Bismark for DNA methylation. Instances of methylated and unmethylated CpG were quantified and summed to an overall percent methylation for each enhancer region with standard deviations. An average percent methylation for all the enhancers was computed and the variance is represented by SEM with n = 6. See Supplementary Table S2 for number of reads for each sample and site, and Supplementary Table S3 for the percent CpG methylation by region for each sample. See Supplementary Table S4 for primers used for bisulfite sequencing.
ChIP-Western and co-immunoprecipitation
For the ChIP-Western, crosslinked chromatin was processed as described in the ChIP protocol. For the detection of proteins in ChIP samples, elution was performed with Laemmli's loading buffer. For co-immunoprecipitation (Co-IP), the nuclear extract was prepared according to manufacturer's protocol (Active Motif, 40010) except that DNase was added for the release of chromatin-associated proteins. The Co-IP was performed using Dynabead M280 (Life Technologies, 11201D) conjugated with 5 μg antibody and with 50 μg of nuclear extract according to the manufacturer's protocol. Antibodies used include: anti-Dnmt3a (Active Motif, 39206), anti-Chd4 (Abcam, ab72418), anti-Hdac1 (Abcam, ab 7028), anti-Lsd1 (Abcam, ab17721).
RESULTS
Dnmt3a methylates a subset of PpG enhancers
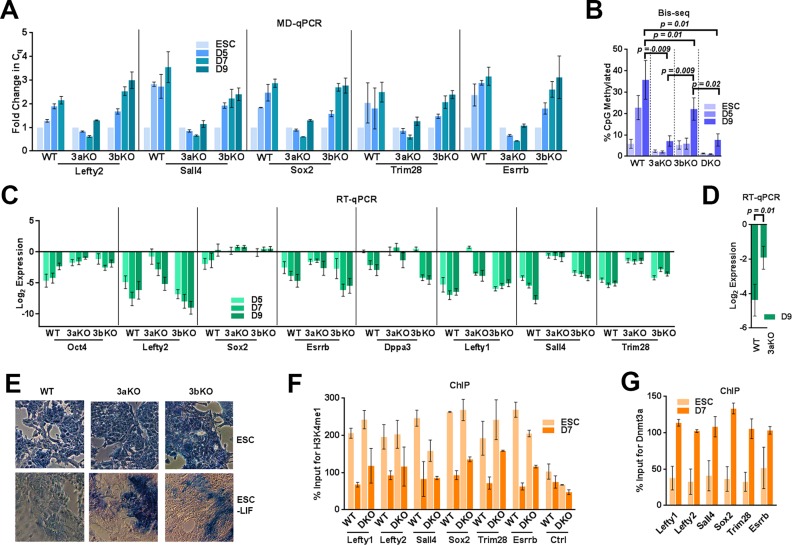
Dnmt3a and 3b have been previously reported to methylate PpG promoters (35) but their role in methylating PpG enhancers has not been fully defined. We tested the role of Dnmt3 enzymes in the methylation of the known enhancers of six commonly studied key PpGs; Lefty1, Lefty2, Esrrb, Sall4, Sox2, Trim 28 during ESC differentiation (36). We differentiated WT and Dnmt3a−/− 3b−/- DKO cells (Supplementary Figures S1 and S2A) and compared DNA methylation levels at these PpG enhancers (37). We used a methylation-dependent restriction method (MD-qPCR) in which the restriction enzyme MspJI cleaves methylated DNA (Supplementary Figure S2B) (38). A single methylation event will result in digestion by MspJI and loss of PCR amplification (39). The qPCR analysis showed an increase in Cq value representing an increase in the DNA methylation at PpG enhancers during differentiation of WT cells, which was completely absent in the DKOs (Supplementary Figure S2C). We further analyzed the DNA methylation at these PpG enhancers by bisulfite sequencing (Bis-seq) (Supplementary Tables S1 and S2) which confirmed the results obtained by MD-qPCR (Figure 1B and Supplementary Table S3). These data support the function of Dnmt3 enzymes in methylating these sites de novo during differentiation. Dnmt3a and 3b have distinct as well as redundant targets during early development (37). To determine whether both enzymes are involved in methylation of these PpG enhancers, we used Dnmt3a−/− and Dnmt3b−/− ESCs (3aKO, 3bKO) (Supplementary Figure S1), and induced them to differentiate as illustrated in Supplementary Figure S2A. Analysis using MD-qPCR and Bis-seq showed that DNA methylation at PpG enhancers in differentiating 3aKO cells was comparable to that in DKO cells, whereas in differentiating 3bKO cells, DNA methylation gradually increased to more than 50% of WT (Figure 1A and B). These data argue for a primary role of Dnmt3a in establishing DNA methylation at these PpG enhancers. The delayed and lower methylation in 3bKO cells suggests that either the activity of Dnmt3a is not fully operational in the absence of Dnmt3b or the propagation and/or maintenance of DNA methylation established by Dnmt3a requires Dnmt3b activity suggesting that combined activity of Dnmt3a and 3b is required to complete DNA methylation at these sites (37,40,41).
Figure 1.
Dnmt3a is the principal methyltransferase for PpG enhancer methylation. ESC: undifferentiated embryonic stem cells and D3–D9: Days post-induction of differentiation WT: Wild Type, 3aKO: Dnmt3a-/-, 3bKO: Dnmt3b-/-, DKO: Dnmt3a-/- Dnmt3b-/-. DNA methylation analysis of the PpG enhancers by (A) MD-qPCR and by (B) bisulfite sequencing (Bis-seq). For MD-qPCR, genomic DNA was digested with the restriction enzyme MspJI that cuts methylated DNA, followed by qPCR at PpG enhancers (illustration in Supplementary Figure S2B). An increase in the Cq (quantification cycle) represents the gain in DNA methylation. For Bis-seq (B) bisulfite-treated gDNA was used to determine the extent of CpG methylation at the PpG enhancers shown in A on a high throughput sequencing platform (MiSeq) and the data were analyzed using Bismark software. The number of CpGs, reads analyzed for each enhancer and percent CpG methylation by site in all cell lines and treatments are given in Supplementary Tables S1, S2, S3 and Materials and Methods. (C) Gene expression analysis of PpGs by RT-qPCR in WT and Dnmt3 KO cells; undifferentiated and post induction of differentiation. Gene expression is normalized to Gapdh and represented as a relative change to gene expression in ESC. (D) Average gene expression change across all 8 PpGs D9 post differentiation in Dnmt3aKO compared to WT. P-values are derived from Student's paired t-test (E) Alkaline phosphatase staining (blue) for pluripotency in ESCs and differentiated cells D9 post-induction, where the presence of stain indicates pluripotency. (F and G) ChIP-qPCR was used to determine percent enrichment of (F)H3K4me1 in WT and DKO cells (G) Dnmt3a in WT cells at PpG enhancers pre- and post-induction of differentiation. For A, C, F and G average and SD are shown for each gene.
Next we compared the changes in the expression of these PpGs in WT and Dnmt3 KO cells post induction of differentiation. Relative to gene expression changes post induction of differentiation in WT, we observed that more PpGs showed a partial gene repression in 3aKO and DKO cells compared to 3bKO cells (Figure 1C, Supplementary Figure S2D). The average effect on the repression of all the PpGs post differentiation in Dnmt3aKO was significant when compared to the WT (Figure 1D). This suggests a common role of Dnmt3a in the repression of these PpG during differentiation. Both Dnmt3a and 3b KO ESC differentiated into embryoid bodies (EBs), however, we noticed they had aberrant size and morphology (Supplementary Figure S2E). Compared to WT and 3bKO cells, the 3aKO cells also maintained a higher level of alkaline phosphatase (AP) expression post differentiation, a marker of pluripotency (Figure 1E). However, the germ layer specific genes were activated in both 3aKO and 3bKO cells post induction of differentiation (Supplementary Figure S2F, S2G), indicating only a partial defect in their differentiation potential.
Although we cannot exclude the possibility that in the 3aKO cells, the absence of DNA methylation at other Dnmt3a targets could affect the PpG repression, this observation resembles the published effects of Lsd1 inhibition on PpG repression. During ESC differentiation, Lsd1-Mi2/NuRD removes histone H3K4me1 and H3K27Ac from PpG enhancers and the demethylation activity of Lsd1 was shown to be critical for complete repression of PpGs (31). Therefore, we tested the Lsd1 activity at the PpG enhancers in DKO cells post induction of differentiation by ChIP of histone H3K4me1. Comparable levels of H3K4me1 were observed in DKO and WT cells, which were similarly reduced post induction, showing that Lsd1 activity is unaltered at PpG enhancers in DKO cells (Figure 1F). These results suggest Dnmt3a may be a potential downstream epigenetic effector of Lsd1-Mi2/NuRD activity and could contribute to the complete repression of these PpGs. We propose that during ESC differentiation, deacetylation and demethylation by the Lsd1-Mi2/NuRD complex primes the histone tails to interact with the Dnmt3a-ADD domain. This hypothesis is further supported by ChIP analysis showing an increased enrichment of Dnmt3a at PpG enhancers in differentiated cells (Figure 1G).
Given that several studies have described the relationship between histone deacetylation and DNA methylation (16), we investigated the contribution of Lsd1 activity in regulating DNA methylation by Dnmt3a at PpG enhancers during ESC differentiation.
Lsd1-mediated demethylation triggers Dnmt3a activity at PpG enhancers
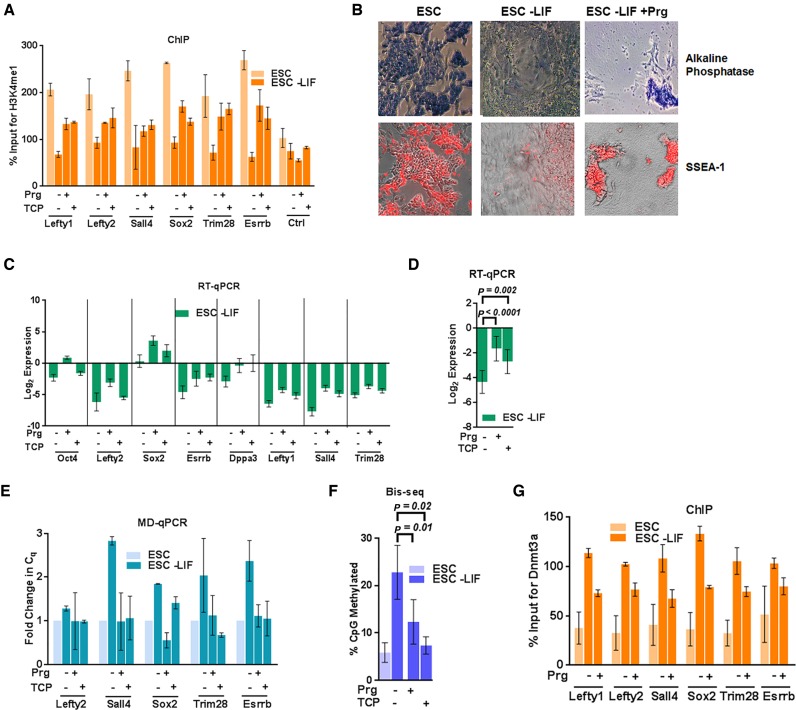
In contrast to other histone demethylases, Lsd1 uniquely catalyzes an FAD-dependent oxidation reaction to remove H3K4me1. It is specifically inhibited by the monoamine oxidase inhibitors pargyline (Prg) and tranylcypromine (TCP). Both of these inhibitors have been earlier demonstrated to inhibit Lsd1 activity and PpG repression during ESC differentiation (31,42,43). With the use of these inhibitors, we asked whether Dnmt3a activity is regulated by Lsd1-mediated histone demethylation at the examined PpG enhancers. The speculation that the Lsd1 inhibition will suppress Dnmt3a activity is supported by fluorescence polarization and isothermal titration calorimetry experiments, which show that H3K4me1 decreases the interaction of Dnmt3-ADD with H3 tails by 18- and 2-fold, respectively (21,26). Compared to H3K4me3, although H3K4me1 partially blocked this interaction in vitro, we anticipated the possibility of a significant effect in vivo. To test our hypothesis, ESC differentiation was induced in the presence of Prg or TCP (Supplementary Figure S3A). ChIP of H3K4me1 at a subset of the enhancers of PpGs confirmed the suppression of Lsd1 activity in inhibitor-treated, induced cells (Figure 2A). Consistent with earlier studies, treatment of Lsd1 inhibitors during differentiation lead to cell death (31,44), but the surviving cells retain pluripotency as shown by SSEA-1 and AP staining (Figure 2B). We also observed weak PpG repression and no significant change in the activation of germ layer-specific genes in Lsd1 inhibitor-treated induced cells (Figure 2C, D, Supplementary Figure S3B and S3C) confirming the previously known role of Lsd1 in regulation of PpG repression during ESC differentiation. Since the measurable increase in DNA methylation takes places 4–5d post differentiation in untreated WT cells (Figure 1A and B), we determined the effect of Lsd1 inhibition on the establishment of DNA methylation by analyzing these early time points using MD-qPCR and Bis-seq. Gain of DNA methylation at these PpG enhancers during differentiation was restricted in cells treated with Lsd1 inhibitors, suggesting a link between Lsd1 activity and DNA methylation at these PpG enhancers (Figure 2E and F). Consistent with these results, ChIP analysis showed that the enrichment of Dnmt3a at the PpG enhancers is reduced in Prg-treated induced cells (Figure 2G).
Figure 2.
Lsd1 inhibition restricts establishment of DNA methylation at PpG enhancers by Dnmt3a. ESC: undifferentiated embryonic stem cells, and ESC-LIF: Cells induced to differentiate. (A) ChIP-qPCR was used to determine the percent enrichments of H3K4me1 at the PpG enhancers in ESC and cells 7 days post- induction of differentiation in absence and presence of Lsd1 inhibitors, Prg: Pargyline, TCP: Tranylcypromine. (B) Alkaline phosphatase staining (blue) and SSEA-1 immunofluorescence (red) in ESCs and cells 9 days post induction of differentiation in absence and presence of pargyline, where positive staining indicates pluripotency. (C) Gene expression analysis of PpGs using RT-qPCR in ESCs and cells 9 days post induction of differentiation in absence and presence of Lsd1 inhibitors. Gene expression is normalized to Gapdh and represented relative to ESC. (D) Average expression change with SEM across all loci shown in C; (E) DNA methylation analysis of the PpG enhancers in ESCs and cells 5 days post induction of differentiation in absence and presence of pargyline and TCP was assayed by MD-qPCR as in Figure 1A. (F) Bis-seq was used to determine the extent of CpG methylation (details same as Figure 1B) and the data are the average and SEM of 6 PpG enhancers shown in E. (G) Percent enrichment of Dnmt3a using ChIP-qPCR at PpG enhancers in ESC and cells 7 days post-induction of differentiation in absence and presence of Lsd1 inhibitors. For D and F, P-values are derived from Student's paired t-test using GraphPad Prism. For A, C, E and G average and SD are shown for each gene.
Effect of the master regulator Oct4 on gain of DNA methylation at PpG enhancers
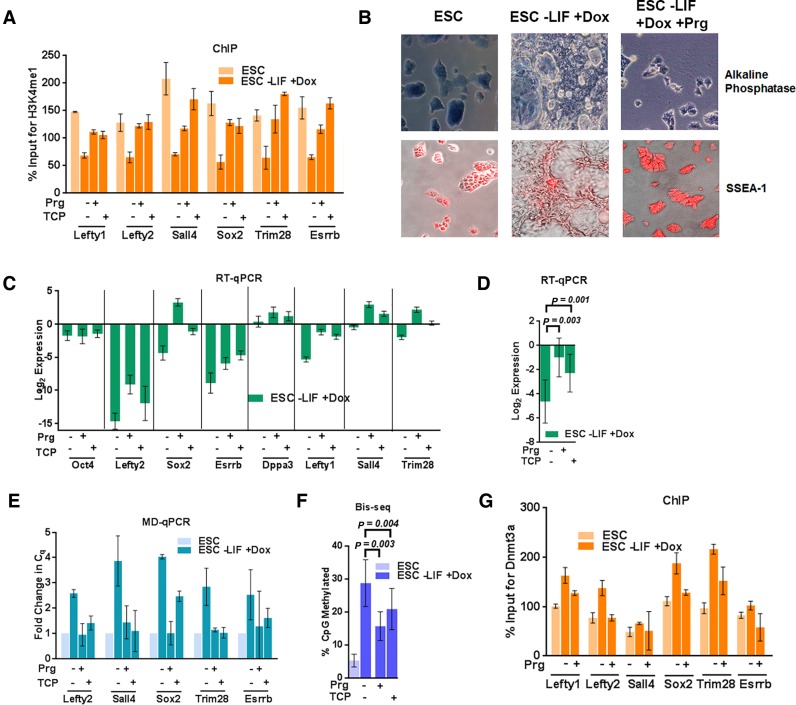
Because Lsd1 inhibitors block differentiation of ESCs, the gain of DNA methylation at PpG enhancers may be affected by other protective mechanisms that include binding of the master transcriptional regulator of PpGs, Oct4 to these PpG enhancers. Gene expression analysis of the Prg-treated and induced cells showed a higher expression of Oct4 (Figure 2C). We reasoned that in these conditions, high Oct4 expression may result in its continued binding to PpG regulatory elements, thus overriding the effect of epigenetic regulators in the repression pathway. To uncouple the effect of Oct4-reinforced transcriptional activation from Lsd1 regulation of DNA methylation, we used the transgenic ESC line, ZHBTc4 (Z), in which doxycycline treatment induces Oct4 repression and consequent cell differentiation into trophectoderm as shown by an increase in the expression of Cdx2 gene (Supplementary Figure S3D and S3E) (45). Similar to their effect in WT ESCs, in Z cells Lsd1 inhibitors prevented complete histone H3K4me1 demethylation (Figure 3A), impeded differentiation (Figure 3B) and caused incomplete repression of PpGs (Figure 3C, D and Supplementary Figure S3F). Further, compared to control differentiating Z cells, DNA methylation in treated cells was significantly reduced (Figure 3E–F) demonstrating that in the absence of Oct4 expression, inhibition of Lsd1 activity is sufficient to impede DNA methylation by Dnmt3a at PpG enhancers post induction of differentiation. ChIP analysis showed that similar to E14T cells, the enrichment of Dnmt3a at the PpG enhancers is reduced in Prg-treated induced cells (Figure 3G). The effect of Lsd1 inhibition on DNA methylation was not due to reduced expression of Lsd1, Dnmt3a, 3b or Dnmt1 (Supplementary Figure S4A–S4B) and did not affect global DNA methylation levels as evidenced by the integrity of genomic methylation at the repetitive elements, which is comparable to untreated differentiating ESCs (Supplementary Figure S4C). Taken together, our results support a functional interaction between the Lsd1-dependent histone H3K4 demethylation and Dnmt3a-catalyzed DNA methylation at these examined PpG enhancers.
Figure 3.
Lsd1 inhibition affects DNA methylation at PpG enhancers in absence of Oct4 expression. ESC: undifferentiated embryonic stem cells, and ESC -LIF +Dox: Cells induced to differentiate by Doxycycline treatment. (A) Percent enrichments of H3K4me1 using ChIP-qPCR at the PpG enhancers in ESC and cells 4 days post-induction of differentiation normalized to a negative control region in absence and presence of Lsd1 inhibitors, Prg: Pargyline, TCP: Tranylcypromine. (B) Alkaline phosphatase (blue stain) and SSEA-1 immunofluorescence staining (red stain) for pluripotency in ESCs and cells 8 days post-induction of differentiation in absence and presence of Prg, where positive staining indicates pluripotency. (C) Gene expression of PpGs by RT-qPCR in ZHBTc4 cells untreated and treated with Lsd1 inhibitors for 8 days post induction of differentiation. Gene expression is normalized to Gapdh and represented relative to ESC. (D) Average expression change with SEM across all loci shown in C; (E) DNA methylation analysis of the PpG enhancers in ESCs and cells 4 days post-induction of differentiation in absence and presence of pargyline and TCP was assayed by MD-qPCR as in Figure 1A. (F) Bis-seq was used to determine the extent of CpG methylation (details same as Figure 1B) and the data are the average and SEM of 6 PpG enhancers shown in E. (G) Percent enrichment of Dnmt3a using ChIP-qPCR at PpG enhancers from cells pre- and 4 days post-induction of differentiation in presence and absence of prg. P-values are derived from Student's paired t-test using GraphPad Prism. For A, C, E and G average and SD are shown for each gene.
Histone deacetylation by Mi2/NuRD and DNA methylation at PpG enhancers
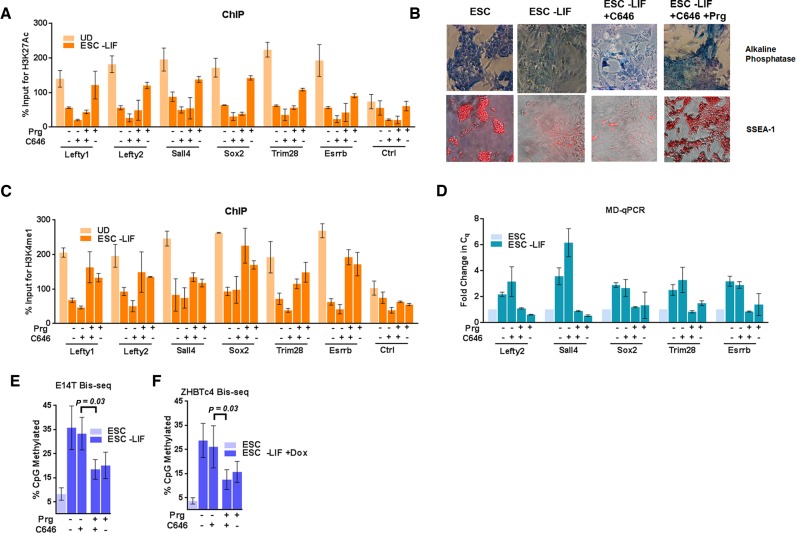
Previous studies have shown that in ESCs, Lsd1 is part of the Mi2/NuRD complex (31,46–48). During ESC differentiation, dissociation of the OSN coactivator complex (Oct4/Sox2/Nanog-HATp300) from the PpG enhancers triggers the histone deacetylase activity of the Mi2/NuRD complex, which is critical for Lsd1-mediated histone demethylation. Since the acetylation of histones is known to inhibit the interaction of unmethylated histone tails with the Dnmt3a-ADD domain (22,49), we asked if Dnmt3a activity at PpG enhancers was blocked by the disruption of the histone deacetylase activity of the Mi2/NuRD complex in Lsd1 inhibitor treated cells. We first verified histone deacetylation during differentiation at a subset of PpG enhancers by ChIP of histone H3K27Ac. Surprisingly, deacetylation was partially inhibited in Prg-treated cells (Figure 4A) (27,50), which may be a consequence of the recursive nature of epigenetic processes (51–55). Therefore, to distinguish the potential effect of remnant histone acetylation from that of histone methylation on Dnmt3a activity in Prg-treated cells, we induced differentiation of ESCs in the presence of the p300 HAT inhibitor C646, in addition to the treatment with Prg (Supplementary Figure S5A) (56). Treatment of cells with C646 did not affect the differentiation, however, treatment with Prg together with C646 impeded differentiation as shown by AP and SSEA-1 staining (Figure 4B). The decrease of histone acetylation at PpG enhancers in C646-treated differentiated cells (Figure 4A) is expected to be permissive for Lsd1 activity and DNA methylation by Dnmt3a. Consistent with this, we observed a decrease in H3K4 methylation and an increase in DNA methylation in C646 treated cells (Figure 4C, D and E). Simultaneous treatment with Prg and C646 can therefore be used to determine the direct impact of histone demethylation on Dnmt3a activity (Figure 4C). When ESCs were induced to differentiate in the presence of both C646 and Prg, gain of DNA methylation at these PpG enhancers was strongly inhibited (Figure 4D and E), even though the histone acetylation was similarly reduced in C646 treated cells with and without Prg (Figure 4A).
Figure 4.
Effect of histone deacetylation on DNA methylation at PpG enhancers. ESC: undifferentiated embryonic stem cells, and ESC -LIF: E14T cells induced to differentiate, ESC -LIF+Dox: ZHBTc4 cells induced to differentiate. (A) ChIP-qPCR was used to determine the percent enrichments of H3K27ac at the PpG enhancers in ESC and cells 7 days post-induction of differentiation in absence and presence of Prg and/or C646, Prg: Pargyline, C646: p300 Histone acetyltransferase inhibitor. (B) Alkaline phosphatase staining (blue) and SSEA-1 immunofluorescence (red) in ESCs and cells 9 days post induction of differentiation in absence and presence of C646 and C646 with pargyline, where positive staining indicates pluripotency. (C) ChIP-qPCR was used to determine the percent enrichments of H3K4me1 at the PpG enhancers in ESC and cells 7 days post-induction of differentiation in absence and presence of Prg and/or C646. (D) DNA methylation analysis of the PpG enhancers in ESCs and cells post induction of differentiation in absence and presence of Prg and/or C646 was assayed by MD-qPCR as in Figure 1A. (E and F) Bis-seq was used to determine the extent of CpG methylation at these enhancers in (E) E14T cells or (F) ZHBTc4 cells and the data were analyzed using Bismark software (details same as Figure 1B) and the data are the average and SEM of 6 PpG enhancers shown in D. P-values are derived from Student's paired t-test. For A, C and D average and SD are shown for each gene.
A similar effect on the gain of DNA methylation at the PpG enhancers was seen in Z cells when they were induced to differentiate by doxycycline treatment in the presence of Prg and/or C646 (Figure 4F). Use of Z cells in these experiments uncouples the protective effects of both Oct4 and histone acetylation from the regulatory role of Lsd1-mediated histone demethylation on Dnmt3a-catalyzed DNA methylation at PpG enhancers. Although Dnmt3a could have other targets during differentiation, its coordinated activity with Lsd1 is likely to be specific to PpG enhancers, since the Lsd1 activity was shown to be largely recruited at the PpG enhancers during ESC differentiation (31). Taken together, these data establish a dominant effect of Lsd1 inhibition on the mechanism that regulates the establishment of DNA methylation at a subset of PpG enhancers during ESC differentiation and suggest a functional link between Lsd1 activity and DNA methylation by Dnmt3a.
Interaction of Dnmt3a with Lsd1-Mi2/NuRD complex
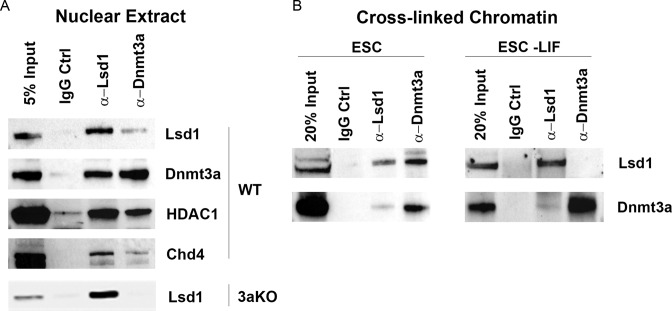
Previous studies have shown that Lsd1 and Mi2/NuRD form a complex in undifferentiated ESCs and associate with PpG enhancers while they are in the active state. The enzymatic activity of the Lsd1-Mi2/NuRD complex is, however, only triggered upon dissociation of the OSN complex during ESC differentiation (31,47). Therefore, we tested the interaction of Dnmt3a with the Lsd1-Mi2/NuRD complex both in the undifferentiated ESCs and in differentiated cells. Reciprocal Co-IP from ESC nuclear extracts and from crosslinked chromatin using Lsd1 or Dnmt3a antibodies revealed the presence of Dnmt3a along with known components of the Lsd1-Mi2/NuRD complex (Figure 5A and B). Similar Co-IP experiments to determine the presence of Dnmt3b in Lsd1-Mi2/NuRD complex showed no specific interaction, which may also be due to low specificity of the available antibodies. Compared to that in undifferentiated ESCs, a weaker or no interaction of Dnmt3a with the Lsd1 complex was observed in crosslinked chromatin from differentiated cells (Supplementary Figure S5B). This suggests that post differentiation, Lsd1-Mi2/NuRD activity allows the subsequent interaction of Dnmt3a-ADD with histone tails, after which the interaction of Dnmt3a with the Lsd1-Mi2/NuRD complex may not persist and may not be required. Lsd1-facilitated interaction of Dnmt3a with histone tails is supported by the results from Dnmt3a ChIP (Figure 1G) that showed a higher enrichment of Dnmt3a at these PpG enhancers post differentiation, which is reduced by Lsd1 inhibitor treatment (Figures 2G and 3G). This higher enrichment of Dnmt3a post differentiation is because its interaction with demethylated histone tails could facilitate its crosslinking to chromatin. Together, these data suggest a model in which the active enhancers of PpGs are poised for repression by the presence of a proximally located repressive Lsd1-Mi2/NuRD/Dnmt3a complex, which is activated as cells begin to differentiate (Figure 6C).
Figure 5.
Dnmt3a is associated with the Lsd1-Mi2/NuRD complex. ESC: undifferentiated embryonic stem cells, ESC-LIF: Days post induction of differentiation, 3aKO: Dnmt3a knockout. (A) Nuclear extract was prepared from WT and 3aKO ESCs and used in Co-IP (co-immunoprecipitation) performed with anti-Dnmt3a or anti-Lsd1 antibody and control IgG. The immune complexes and input (5% of nuclear extract used in CoIP) were blotted to probe for Lsd1-Mi2/NuRD subunits (Lsd1, Hdac1 and Chd4) and Dnmt3a (shown in the right). Co-IP using 3aKO nuclear extract serves as a control. (B) Crosslinked chromatin from ESC and cells 7 days post differentiation was used for a ChIP-Western performed with anti-Dnmt3a or anti-Lsd1. The immune complexes and the input (20%) was blotted to probe for Lsd1 and Dnmt3a.
Figure 6.
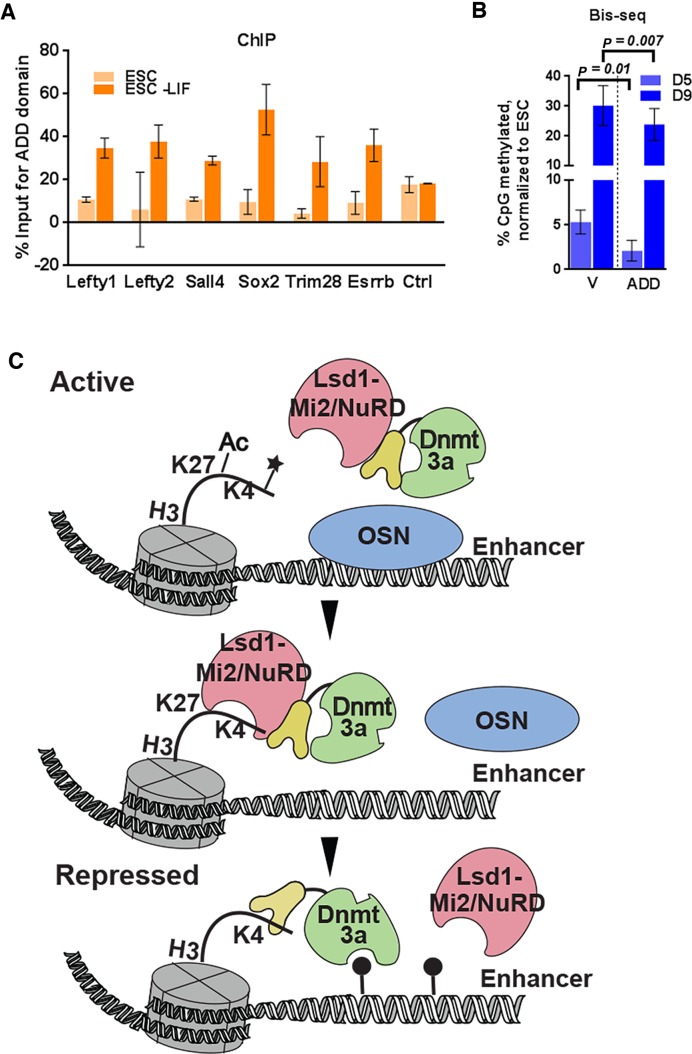
DNA methylation at PpG enhancers is facilitated by histone H3 and Dnmt3a-ADD interaction. ESC: undifferentiated embryonic stem cells, ESC-LIF: Cells induced to differentiate, D5, D9: Days post-induction of differentiation, V: empty Myc vector control and ADD: Myc-3a ADD expressing cells. (A) ChIP-qPCR was used to determine the percent enrichments of Myc-Dnmt3a ADD at the PpG enhancers in transgenic ESC lines expressing Myc Dnmt3a-ADD domain pre- and 9 days post-induction of differentiation; the percent enrichment is normalized by subtracting the percent enrichment of the empty vector control from the percent enrichment of ADD expressing cells. Average and SD is shown for each gene. (B) DNA methylation at PpG enhancers shown in A in transgenic ESC lines expressing Myc empty vector or Myc Dnmt3a-ADD domain 5 and 9 days post induction of differentiation was assayed by Bis-seq (details same as Figure 1B). The percent methylation for each cell line was were normalized to that in the respective ESC state. P-values are derived from Student's paired t-test. (C) Model illustrating the epigenetic switch at PpG enhancers that activates Dnmt3a through histone-ADD interaction. In their active state, PpGs enhancers are bound by the OSN coactivator complex and the chromatin is acetylated and methylated. Upon differentiation, dissociation of the coactivator complex triggers the activity of the Lsd1-Mi2/NuRD complex, which removes H3K27Ac and H3K4me1 at PpG enhancers. This in turn activates Dnmt3a through the interaction of its ADD domain with the newly generated H3K4me0 histone tails permitting site-specific DNA methylation. Ac: Acetylation; *: histone methylation; closed circle: DNA methylation.
ADD domain–histone interaction guides PpG enhancer DNA methylation
Our data suggest that during ESC differentiation Lsd1-Mi2/NuRD activity at a subset of PpG enhancers induces a local interaction between histone H3K4me0 and the Dnmt3a-ADD domain, thus facilitating de novo DNA methylation at these sites. To investigate the role of the interaction of the Dnmt3-ADD domain with H3K4me0 histone tails in PpG enhancer DNA methylation, we created transgenic ESCs overexpressing the Myc-epitope tagged-ADD domain of Dnmt3a (Supplementary Figure S5C). We confirmed the pluripotency of these lines by SSEA and AP staining (Supplementary Figure S5D) and induced them to differentiate. We reasoned that overexpression of this domain can potentially compete with endogenous Dnmt3a to interact with the demethylated histone tails at the examined PpG enhancers, thereby affecting enhancer DNA methylation during differentiation. ChIP analysis showed the binding of the recombinant Myc-ADD domain at these PpG enhancers post differentiation (Figure 6A). Indeed, accumulation of DNA methylation at these PpG enhancers in the Myc-ADD expressing cells was significantly reduced both at D5 and D9 post differentiation compared to the control cells (Figure 6B). We noticed a more substantial difference in DNA methylation at D5 post induction that narrowed as differentiation proceeded. This is because the Myc-ADD competes with the binding and not the activity of endogenous Dnmt3a; therefore, the DNA methylation deposited for every Dnmt3a binding event could be further propagated by an Lsd1 independent mechanism of Dnmt1 or by Dnmt3b. A partial gain in DNA methylation at these PpG enhancers in 3bKO cells supports this premise (Figure 1A and B). We further examined the potential effect of Myc-ADD on global DNA methylation which showed no change in DNA methylation at Line1 elements and minor satellite repeats (Supplementary Figure S5E). A significant inhibition of PpG enhancer DNA methylation by Myc-ADD is consistent with the model that the interaction of the Dnmt3a-ADD domain with demethylated histone tails at PpG enhancers regulates the establishment of de novo DNA methylation during the early phase of differentiation.
Taken together our data suggest that the histone editing activity of the Lsd1-Mi2/NuRD complex at a subset of PpG enhancers regulates the epigenetic switch to locally activate Dnmt3a, leading to site specific DNA methylation and enhancer repression (Figure 6C).
DISCUSSION
Current investigations have advanced our understanding of the contribution of epigenetic mechanisms in the regulation of pluripotency and cellular differentiation in normal and diseased states. However, the mechanisms that regulate the interplay between the readers and writers of epigenetic marks and their subsequent impact on gene expression have not been fully elucidated. In this study, we identified a functional role for Dnmt3a as a reader and effector in a queue of epigenetic events, where local histone deacetylation and demethylation by Lsd1-Mi2/NuRD complex allows the interaction and specific activation of Dnmt3a at a subset of PpG enhancers.
Earlier investigations on the regulation of DNA methylation at PpG regulatory elements have shown that both Dnmt3a and 3b are required for de novo methylation of the promoters of Oct4 and Nanog (35) and the interaction between G9a histone methyltransferase and Dnmt3a is required for DNA methylation at the Oct4 promoter (9,10,57,58). Other studies have shown that G9a may not be required for DNA methylation at the Oct4 enhancer (32,59). Our experiments using ESCs with targeted deletions of Dnmt3a and 3b (37) together with ChIP experiments first identify Dnmt3a as the primary MTase required for methylation of a subset of PpG enhancers. A supporting role of Dnmt3b in this process is suggested by delayed and partial gain of DNA methylation post differentiation in Dnmt3bKO ESCs. Further, our data suggest that DNA methylation at these PpG enhancers may contribute to the stable repression of PpGs during differentiation. Compared to WT cells, repression of some PpGs in Dnmt3aKO cells was partially affected suggesting that Dnmt3a-mediated DNA methylation is one of several processes that together promote complete repression of PpGs (Figure 1C). Additionally, in Dnmt3aKO cells, there was little or no effect on the activation of lineage specific genes during differentiation (Supplementary Figure S2F). Since a combination of PpG repression and lineage-specific gene activation is required to drive differentiation, these deficiencies may be insufficient to block development in utero thus explaining the ability of the Dnmt3a KO mice to reach term although they are born runted and die in less than 4 weeks after birth (37). Consistent with this argument we observed that 3aKO ESCs make embryoid bodies, albeit small (Supplementary Figure S2E), indicating that they can differentiate even though the PpG are not fully repressed.
Our data suggest that the activity of Dnmt3a at these PpG enhancers is regulated by the upstream Lsd1-Mi2/NuRD activity, which facilitates the binding of Dnmt3a-ADD domain to histone tails post differentiation. The inhibitory effect of the recombinant Myc-ADD domain on DNA methylation of these PpG enhancers confirms the mechanistic role of the interaction between the Dnmt3a-ADD domain and demethylated histone tails (H3K4me0) in regional specificity of Dnmt3a activity. Previous studies showing that Lsd1-Mi2/NuRD activity is localized at the PpG enhancers (31) suggest that this mechanism might specifically regulate the DNA methylation at these sites facilitated by the interaction of Dnmt3a with the Lsd1-Mi2/NuRD complex. Although the histone modification H3K4me0 is widely distributed in the mammalian genome, DNA methylation of these sites in ESCs and differentiated cells is largely maintained through mechanisms that are independent of Lsd1 activity and presence of the histone H3K4me0 modification (60–62) which is supported by our data showing no change in DNA methylation at the repetitive elements in Lsd1 inhibitor treated cells.
Previous studies have shown that continuous passage of Lsd1 KO ESCs leads to a progressive loss of DNA methylation at repetitive elements genome-wide due to loss of Dnmt1 protein (44). Under our experimental conditions, Lsd1 inhibition during induction of differentiation had no effect on Dnmt1 protein levels (Supplementary Figure S4B) or on global DNA methylation (Supplementary Figure S4C). In our studies, we exposed ESCs to Lsd1 inhibitors only 6 h prior to induction of differentiation and induced them for 8–9 days in the presence of these inhibitors. Therefore, most of the inhibitor treatment was post induction of differentiation in contrast to earlier studies where Lsd1 KO ESC were maintained and passaged in the pluripotent state, leading to reduced stability of Dnmt1 protein. This argues for the potential influence of the ESC state on the mechanism regulating Dnmt1 stability. Further, the near absence of DNA methylation in differentiated 3aKO cells suggests that the presence of Dnmt1 cannot compensate for the absence of Dnmt3a in establishing methylation at the PpG enhancers (Figure 1A and B). Taken together, our data suggest that inhibition of DNA methylation at PpG enhancers in Lsd1 inhibitor-treated cells is likely due to its effect on Dnmt3a activity rather than the loss of Dnmt1.
In this study, we also addressed the potential impact of the known protective mechanisms, which include the transcription factor Oct4 and histone acetylation, on gain of DNA methylation at these PpG enhancers in Lsd1 inhibitor treated cells. Use of Z cells allowed us to control the potential counteracting effects of the coactivator Oct4. Similar to E14T cells, although Lsd1 inhibition significantly impeded the gain of DNA methylation in Z cells, comparing Figure 2 to Figure 3, we indeed noticed that the effect on DNA methylation is less robust in Z cells. Use of C646 (a HAT inhibitor) and Prg during differentiation uncoupled the effect of histone acetylation and histone methylation on deposition of DNA methylation. We conclude that under these experimental conditions, deacetylation of histones is not sufficient to recruit Dnmt3a activity in the presence of H3K4me1 at PpG enhancers. However, this does not exclude the inhibitory effect of histone acetylation on the interaction of the Dnmt3a-ADD with histone tails when these enhancers are in the active state. We anticipate that in the ESC state, when PpG enhancers are active, histone acetylation together with histone methylation block the interaction of the Dnmt3a-ADD domain with histone H3 tails.
Recent structural studies have shown that the ADD domain of Dnmt3a autoinhibits its catalytic activity by sterically blocking its DNA binding domain and the interaction of the ADD domain with unmethylated histone tails relieves this autoinhibition (23). Our Co-IP and ChIP-Western experiments show that Dnmt3a associates with the Lsd1-Mi2/NuRD complex in undifferentiated ESCs. Based on these data, we speculate that in ESCs Dnmt3a associates with the Lsd1-Mi2/NuRD complex in its auto-inhibited state, poised to be activated during ESC differentiation. As differentiation proceeds, the Lsd1-Mi2/NuRD complex deacetylates H3K27 and demethylates H3K4 residues and the resulting ‘edited’ histone H3 tails interact with Dnmt3a-ADD domain and locally activate the enzyme.
These observations may also have potential implications for regulation of DNA methylation of other genomic elements with varying degrees of histone H3K4 methylation and histone acetylation, thus instructively creating tissue specific patterns of DNA methylation during differentiation. Therefore, it would be interesting to see if this represents a common mechanism for enhancer-mediated regulation of other genes controlling cell identity. For example, both Dnmt3a and Lsd1 are known to regulate hematopoietic stem cell (HSC) differentiation. Lsd1 mediated repression of the regulatory elements of hematopoietic stem and progenitor cell (HSPC) genes is required to fully silence these genes for proper hematopoietic maturation (63), and loss of Dnmt3a in HSCs results in the retention of multipotency gene expression during differentiation (64). It is possible that a similar epigenetic crosstalk mechanism may regulate enhancer repression and stable silencing of multipotency genes, thus maintaining the fidelity of differentiation during hematopoiesis. Additionally, several recent studies have enumerated the role of enhancer-mediated regulation of oncogenes in various cancers (65). Thus, our studies may have implications for the mechanisms that contribute to aberrant gene expression in various human diseases.
Supplementary Material
Acknowledgments
The authors thank Dr. Joe Ogas for helpful discussions, Dr. Sandra Rossie for reading the manuscript, Dr. Taiping Chen for providing Dnmt3KO ESC's and Dr. Dana Levasseur for ZHBTc4 cells.
Contributions : C.J.P., L.A., M.H., R.R. and H.G. performed the experiments. P.S.M. analyzed the Bis-seq data. C.J.P. and H.G. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Institutional Start up Fund from the College of Agriculture and the Department of Biochemistry, Purdue University. The authors gratefully acknowledge the DNA sequencing core facility and support from Purdue University Center for Cancer Research, NIH grant P30 CA023168, Graduate Research fellowship for LA supported by the Saudi Arabian Cultural Mission. Funding for open access charge: Institutional Start up Fund from College of Agriculture and the Department of Biochemistry, Purdue University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Goll M.G., Bestor T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 2.Cedar H., Bergman Y. Programming of DNA methylation patterns. Annu. Rev. Biochem. 2012;81:97–117. doi: 10.1146/annurev-biochem-052610-091920. [DOI] [PubMed] [Google Scholar]
- 3.Gowher H., Jeltsch A. Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: the enzyme modifies DNA in a non-processive manner and also methylates non-CpG [correction of non-CpA] sites. J. Mol. Biol. 2001;309:1201–1208. doi: 10.1006/jmbi.2001.4710. [DOI] [PubMed] [Google Scholar]
- 4.Holz-Schietinger C., Matje D.M., Harrison M.F., Reich N.O. Oligomerization of DNMT3A controls the mechanism of de novo DNA methylation. J. Biol. Chem. 2011;286:41479–41488. doi: 10.1074/jbc.M111.284687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Handa V., Jeltsch A. Profound flanking sequence preference of Dnmt3a and Dnmt3b mammalian DNA methyltransferases shape the human epigenome. J. Mol. Biol. 2005;348:1103–1112. doi: 10.1016/j.jmb.2005.02.044. [DOI] [PubMed] [Google Scholar]
- 6.Jia D., Jurkowska R.Z., Zhang X., Jeltsch A., Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gowher H., Jeltsch A. Molecular enzymology of the catalytic domains of the Dnmt3a and Dnmt3b DNA methyltransferases. J. Biol. Chem. 2002;277:20409–20414. doi: 10.1074/jbc.M202148200. [DOI] [PubMed] [Google Scholar]
- 8.Denis H., Ndlovu M.N., Fuks F. Regulation of mammalian DNA methyltransferases: a route to new mechanisms. EMBO Rep. 2011;12:647–656. doi: 10.1038/embor.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Epsztejn-Litman S., Feldman N., Abu-Remaileh M., Shufaro Y., Gerson A., Ueda J., Deplus R., Fuks F., Shinkai Y., Cedar H., et al. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat. Struct. Mol. Biol. 2008;15:1176–1183. doi: 10.1038/nsmb.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feldman N., Gerson A., Fang J., Li E., Zhang Y., Shinkai Y., Cedar H., Bergman Y. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat. Cell Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- 11.Dennis K., Fan T., Geiman T., Yan Q., Muegge K. Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev. 2001;15:2940–2944. doi: 10.1101/gad.929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones P.L., Veenstra G.J., Wade P.A., Vermaak D., Kass S.U., Landsberger N., Strouboulis J., Wolffe A.P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 13.Nan X., Ng H.H., Johnson C.A., Laherty C.D., Turner B.M., Eisenman R.N., Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 14.Thomson J.P., Skene P.J., Selfridge J., Clouaire T., Guy J., Webb S., Kerr A.R., Deaton A., Andrews R., James K.D., et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clouaire T., Webb S., Skene P., Illingworth R., Kerr A., Andrews R., Lee J.H., Skalnik D., Bird A. Cfp1 integrates both CpG content and gene activity for accurate H3K4me3 deposition in embryonic stem cells. Genes Dev. 2012;26:1714–1728. doi: 10.1101/gad.194209.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cedar H., Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 17.Lehnertz B., Ueda Y., Derijck A.A., Braunschweig U., Perez-Burgos L., Kubicek S., Chen T., Li E., Jenuwein T., Peters A.H. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 18.Jackson J.P., Lindroth A.M., Cao X., Jacobsen S.E. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 19.Tamaru H., Selker E.U. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- 20.Otani J., Nankumo T., Arita K., Inamoto S., Ariyoshi M., Shirakawa M. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 2009;10:1235–1241. doi: 10.1038/embor.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ooi S.K., Qiu C., Bernstein E., Li K., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S.P., Allis C.D., et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li B.Z., Huang Z., Cui Q.Y., Song X.H., Du L., Jeltsch A., Chen P., Li G., Li E., Xu G.L. Histone tails regulate DNA methylation by allosterically activating de novo methyltransferase. Cell Res. 2011;21:1172–1181. doi: 10.1038/cr.2011.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo X., Wang L., Li J., Ding Z., Xiao J., Yin X., He S., Shi P., Dong L., Li G., et al. Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature. 2015;517:640–644. doi: 10.1038/nature13899. [DOI] [PubMed] [Google Scholar]
- 24.Meissner A., Mikkelsen T.S., Gu H., Wernig M., Hanna J., Sivachenko A., Zhang X., Bernstein B.E., Nusbaum C., Jaffe D.B., et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weber M., Hellmann I., Stadler M.B., Ramos L., Paabo S., Rebhan M., Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 26.Noh K.M., Wang H., Kim H.R., Wenderski W., Fang F., Li C.H., Dewell S., Hughes S.H., Melnick A.M., Patel D.J., et al. Engineering of a histone-recognition domain in Dnmt3a alters the epigenetic landscape and phenotypic features of mouse ESCs. Mol. Cell. 2015;59:89–103. doi: 10.1016/j.molcel.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S.A., Flynn R.A., Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez-Bigas N., Kisiel T.A., Dewaal D.C., Holmes K.B., Volkert T.L., Gupta S., Love J., Murray H.L., Young R.A., Benevolenskaya E.V. Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Mol. Cell. 2008;31:520–530. doi: 10.1016/j.molcel.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng J.C., Valouev A., Swigut T., Zhang J., Zhao Y., Sidow A., Wysocka J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitz S.U., Albert M., Malatesta M., Morey L., Johansen J.V., Bak M., Tommerup N., Abarrategui I., Helin K. Jarid1b targets genes regulating development and is involved in neural differentiation. EMBO J. 2011;30:4586–4600. doi: 10.1038/emboj.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whyte W.A., Bilodeau S., Orlando D.A., Hoke H.A., Frampton G.M., Foster C.T., Cowley S.M., Young R.A. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Athanasiadou R., de Sousa D., Myant K., Merusi C., Stancheva I., Bird A. Targeting of de novo DNA methylation throughout the Oct-4 gene regulatory region in differentiating embryonic stem cells. PloS One. 2010;5:e9937. doi: 10.1371/journal.pone.0009937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 34.Tremblay K.D., Duran K.L., Bartolomei M.S. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol. Cell. Biol. 1997;17:4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J.Y., Pu M.T., Hirasawa R., Li B.Z., Huang Y.N., Zeng R., Jing N.H., Chen T., Li E., Sasaki H., et al. Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol. Cell. Biol. 2007;27:8748–8759. doi: 10.1128/MCB.01380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young R.A. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okano M., Bell D.W., Haber D.A., Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 38.Cohen-Karni D., Xu D., Apone L., Fomenkov A., Sun Z., Davis P.J., Kinney S.R., Yamada-Mabuchi M., Xu S.Y., Davis T., et al. The MspJI family of modification-dependent restriction endonucleases for epigenetic studies. Proc. Natl. Acad. Sci. U.S.A. 2011;108:11040–11045. doi: 10.1073/pnas.1018448108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng Y., Cohen-Karni D., Xu D., Chin H.G., Wilson G., Pradhan S., Roberts R.J. A unique family of Mrr-like modification-dependent restriction endonucleases. Nucleic Acids Res. 2010;38:5527–5534. doi: 10.1093/nar/gkq327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang G., Chan M.F., Tomigahara Y., Tsai Y.C., Gonzales F.A., Li E., Laird P.W., Jones P.A. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol. Cell. Biol. 2002;22:480–491. doi: 10.1128/MCB.22.2.480-491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen T., Ueda Y., Dodge J.E., Wang Z., Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 43.Sun G., Alzayady K., Stewart R., Ye P., Yang S., Li W., Shi Y. Histone demethylase LSD1 regulates neural stem cell proliferation. Mol. Cell. Biol. 2010;30:1997–2005. doi: 10.1128/MCB.01116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J., Hevi S., Kurash J.K., Lei H., Gay F., Bajko J., Su H., Sun W., Chang H., Xu G., et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 45.Niwa H., Miyazaki J., Smith A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 46.Amente S., Lania L., Majello B. The histone LSD1 demethylase in stemness and cancer transcription programs. Biochim. et Biophys. Acta. 2013;1829:981–986. doi: 10.1016/j.bbagrm.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 47.Forneris F., Binda C., Vanoni M.A., Battaglioli E., Mattevi A. Human histone demethylase LSD1 reads the histone code. J. Biol. Chem. 2005;280:41360–41365. doi: 10.1074/jbc.M509549200. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y., Zhang H., Chen Y., Sun Y., Yang F., Yu W., Liang J., Sun L., Yang X., Shi L., et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138:660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y., Jurkowska R., Soeroes S., Rajavelu A., Dhayalan A., Bock I., Rathert P., Brandt O., Reinhardt R., Fischle W., et al. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res. 2010;38:4246–4253. doi: 10.1093/nar/gkq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karmodiya K., Krebs A.R., Oulad-Abdelghani M., Kimura H., Tora L. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics. 2012;13:424–442. doi: 10.1186/1471-2164-13-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang P.H., Plass C., Chen C.S. Effects of histone deacetylase inhibitors on modulating H3K4 methylation marks - a novel cross-talk mechanism between histone-modifying enzymes. Mol. Cell. Pharmacol. 2011;3:39–43. [PMC free article] [PubMed] [Google Scholar]
- 52.Huang Y., Vasilatos S.N., Boric L., Shaw P.G., Davidson N.E. Inhibitors of histone demethylation and histone deacetylation cooperate in regulating gene expression and inhibiting growth in human breast cancer cells. Breast Cancer Res. Treat. 2012;131:777–789. doi: 10.1007/s10549-011-1480-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vasilatos S.N., Katz T.A., Oesterreich S., Wan Y., Davidson N.E., Huang Y. Crosstalk between lysine-specific demethylase 1 (LSD1) and histone deacetylases mediates antineoplastic efficacy of HDAC inhibitors in human breast cancer cells. Carcinogenesis. 2013;34:1196–1207. doi: 10.1093/carcin/bgt033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yin F., Lan R., Zhang X., Zhu L., Chen F., Xu Z., Liu Y., Ye T., Sun H., Lu F., et al. LSD1 regulates pluripotency of embryonic stem/carcinoma cells through histone deacetylase 1-mediated deacetylation of histone H4 at lysine 16. Mol. Cell. Biol. 2014;34:158–179. doi: 10.1128/MCB.00631-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee M.G., Wynder C., Bochar D.A., Hakimi M.A., Cooch N., Shiekhattar R. Functional interplay between histone demethylase and deacetylase enzymes. Mol. Cell. Biol. 2006;26:6395–6402. doi: 10.1128/MCB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bowers E.M., Yan G., Mukherjee C., Orry A., Wang L., Holbert M.A., Crump N.T., Hazzalin C.A., Liszczak G., Yuan H., et al. Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chem. Biol. 2010;17:471–482. doi: 10.1016/j.chembiol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bittencourt D., Lee B.H., Gao L., Gerke D.S., Stallcup M.R. Role of distinct surfaces of the G9a ankyrin repeat domain in histone and DNA methylation during embryonic stem cell self-renewal and differentiation. Epigenet. Chromatin. 2014;7:27–39. doi: 10.1186/1756-8935-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang Y., Sun L., Kokura K., Horton J.R., Fukuda M., Espejo A., Izumi V., Koomen J.M., Bedford M.T., Zhang X., et al. MPP8 mediates the interactions between DNA methyltransferase Dnmt3a and H3K9 methyltransferase GLP/G9a. Nat. Commun. 2011;2:533–543. doi: 10.1038/ncomms1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharma S., Gerke D.S., Han H.F., Jeong S., Stallcup M.R., Jones P.A., Liang G. Lysine methyltransferase G9a is not required for DNMT3A/3B anchoring to methylated nucleosomes and maintenance of DNA methylation in somatic cells. Epigenet. Chromatin. 2012;5:3–15. doi: 10.1186/1756-8935-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bostick M., Kim J.K., Esteve P.O., Clark A., Pradhan S., Jacobsen S.E. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 61.Sharif J., Muto M., Takebayashi S., Suetake I., Iwamatsu A., Endo T.A., Shinga J., Mizutani-Koseki Y., Toyoda T., Okamura K., et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 62.Karagianni P., Amazit L., Qin J., Wong J. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol. Cell. Biol. 2008;28:705–717. doi: 10.1128/MCB.01598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kerenyi M.A., Shao Z., Hsu Y.J., Guo G., Luc S., O'Brien K., Fujiwara Y., Peng C., Nguyen M., Orkin S.H. Histone demethylase Lsd1 represses hematopoietic stem and progenitor cell signatures during blood cell maturation. eLife. 2013;2:e00633. doi: 10.7554/eLife.00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Challen G.A., Sun D., Jeong M., Luo M., Jelinek J., Berg J.S., Bock C., Vasanthakumar A., Gu H., Xi Y., et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat. Genet. 2012;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kron K.J., Bailey S.D., Lupien M. Enhancer alterations in cancer: a source for a cell identity crisis. Genome Med. 2014;6:77–89. doi: 10.1186/s13073-014-0077-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.