Abstract
Harmful oxidation of proteins, lipids and nucleic acids is observed when reactive oxygen species (ROS) are produced excessively and/or the antioxidant capacity is reduced, causing ‘oxidative stress’. Nuclear poly-ADP-ribose (PAR) formation is thought to be induced in response to oxidative DNA damage and to promote cell death under sustained oxidative stress conditions. However, what exactly triggers PAR induction in response to oxidative stress is incompletely understood. Using reverse phase protein array (RPPA) and in-depth analysis of key stress signaling components, we observed that PAR formation induced by H2O2 was mediated by the PLC/IP3R/Ca2+/PKCα signaling axis. Mechanistically, H2O2-induced PAR formation correlated with Ca2+-dependent DNA damage, which, however, was PKCα-independent. In contrast, PAR formation was completely lost upon knockdown of PKCα, suggesting that DNA damage alone was not sufficient for inducing PAR formation, but required a PKCα-dependent process. Intriguingly, the loss of PAR formation observed upon PKCα depletion was overcome when the chromatin structure-modifying protein HMGB1 was co-depleted with PKCα, suggesting that activation and nuclear translocation of PKCα releases the inhibitory effect of HMGB1 on PAR formation. Together, these results identify PKCα and HMGB1 as important co-regulators involved in H2O2-induced PAR formation, a finding that may have important relevance for oxidative stress-associated pathophysiological conditions.
INTRODUCTION
Reactive oxygen species (ROS) are a group of chemical species that contain at least one oxygen atom, but display stronger reactivity than molecular oxygen. ROS can typically arise from exogenous sources such as UVA or γ-irradiation, drugs, heavy metals (1–3), or from endogenous sources e.g. oxidative metabolism, apoptosis, bystander cells or enzymatic activity (4–7). When ROS are produced excessively or antioxidant capacity is reduced, indiscriminate oxidation of proteins, lipids and nucleic acid elicits harmful effects, known as ‘oxidative stress’. ROS as well as the more stable and less reactive by-product of ROS production, hydrogen peroxide (H2O2), are more than toxic products of respiratory burst, they are also effectors for a plethora of signaling pathways inducing innate and adaptive immune cell recruitment, cell proliferation, tissue healing, cell survival and apoptosis (8–11).
ADP-ribosylation is a post-translational protein modification that consists of mono- and poly-ADP-ribose (PAR) molecules covalently linked to specific residues of target proteins (12). The linear or branched PAR polymer can consist in vitro of up to 200–400 ADP-ribose moieties linked by O-glycosidic 1′-2′ ribose-ribose bonds. These modifications are synthesized by a subfamily of ADP-ribosyltransferases (ARTs), which use NAD+ as a substrate and belong to the ART diphtheria toxin-like (ARTD, originally PARP) family. In humans, the ARTD family is comprised of 18 members, which contain a characteristic catalytic ART domain conferring enzymatic activity (13). Most of the family members are mono-ARTs, while ARTD9 and ARTD13 are so far found to be inactive and ARTD1, ARTD2, ARTD5 and ARTD6 also catalyze poly-ADP-ribosylation.
The best-characterized ARTD family member is ARTD1 (originally PARP1), a 116 kDa nuclear enzyme consisting of an N-terminal DNA-binding domain (DBD), a central automodification domain (AMD) and a C-terminal catalytic domain (CAT) (14). ARTD1 automodifies itself at the AMD (15) and modifies other target proteins such as histones, transcription factors and DNA repair proteins, which points at an important function of ADP-ribosylation in epigenetics, transcriptional regulation and repair (16). Indeed, ADP-ribosylation is implicated in the regulation of a plethora of cellular processes, biological phenomena and medical conditions (14,16,17). ARTD1 has recently been termed a ‘cellular rheostat’, because it integrates different types and levels of stress signals (14). In response to mild or moderate stresses, it regulates transcription and DNA repair, while upon severe and sustained stress conditions, hyper-activation of ARTD1 leads to apoptosis or necrosis (18,19). Interestingly, a series of studies has shown that ARTD1 is not only automodified in the presence of DNA damages, but also by specific DNA structures such as cruciform hairpins (20). Moreover, ARTD1 activity can also be stimulated by polyamines (spermine) or core histones (H1 and H3), indicating that DNA-independent mechanisms can activate ARTD1 in vitro as well (21). The phosphorylation of H2AvSer137 can also stimulate ARTD1 activity, and the acetylation of H2ALys5 further enhances ARTD1 activity (22). The fact that single histones as well as modified histones stimulate PAR formation, suggests an important role of chromatin for the activation of ARTD1. However, by which mechanism chromatin activates PAR formation in vivo has not been elucidated previously. HMGB1 is a chromatin-associated protein that plays a role in the organization, sliding and incorporation of nucleosomes (23–25), as well as the compaction of chromatin (26). There is evidence that the nucleosome occupancy in cells lacking HMGB1 changes globally over the genome and that the DNA is more accessible to MNase digestion (27). Post-translational modifications of HMGB1 can lead to changes in its localization, as well as in its binding to DNA and various DNA structures (28–30), and thus to bend DNA and modify chromatin structure (24,31).
Cellular signaling pathways regulate ARTD1 activity also independently of DNA damage. For example, positive regulation of ARTD1 activity has been described for the extracellular signal-regulated kinase (ERK) (32–34) as well as for c-Jun N-terminal kinase (JNK) (35), while both positive and negative effects of protein kinase C (PKC) signaling in the regulation of ARTD1 have been reported (36–39). The activation of ARTD1 independent of DNA damage adds an additional layer to the traditional view that considers ARTD1 as part of the DNA damage response induced upon genotoxic or oxidative stress. Upon oxidative stress, ROS are believed to produce oxidative DNA damage and cause DNA strand breaks in the nucleus, which then strongly stimulates the enzymatic activity of ARTD1 and induces the formation of PAR (12). However, until now it has not been determined whether ARTD1 is activated by oxidative DNA damage in vivo or whether other pathways stimulate ADP-ribosylation in response to oxidative stress.
In this work, we deliberately interrupted the cellular signaling pathways induced early upon stimulation of cells with H2O2 to elucidate the molecular mechanisms involved in PAR formation. Using a systematic reverse phase protein array (RPPA) approach and in-depth molecular analysis of the key signaling components, we identified activation of the PLC/IP3R/Ca2+/PKCα signaling axis as a key regulator of PAR formation. Ca2+-dependent signaling induced DNA damage very rapidly (within a few minutes) that, however, was not sufficient to induce PAR formation, since knockdown of PKCα completely abolished PAR formation, but not DNA damage. Moreover, our results show that PKCα activation leads to the nuclear reduction of HMGB1, which otherwise negatively regulates PAR formation. Together these findings identify a major so far underappreciated mechanism, involving PKCα and HMGB1, by which H2O2 stimulates PAR formation.
MATERIALS AND METHODS
Cell culture, siRNA transfection, lysis and proliferation assay
MRC-5 and IMR-90 human lung fibroblasts (40,41) were obtained from the American Type Culture Collection (ATCC) and cultured in MEM (Invitrogen) supplemented with 5% penicillin/streptomycin (P/S) and 10% fetal calf serum (FCS). NIH/3T3 were obtained from ATCC and cultured in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 5% P/S and 10% FCS. Mouse embryonic fibroblasts (MEFs) were cultivated in supplemented DMEM. Cells were pre-incubated with inhibitors (stocks dissolved in dimethyl sulfoxide, used at 1:1000) for 1 h prior to H2O2 treatment in FCS-free media, which was also used as a vehicle for H2O2. All inhibitors were obtained from Enzo Life Sciences, except for Olaparib (AstraZeneca), PD98059 (Santa Cruz), IKK VII and KN-93 (Merck Millipore), and used at a final concentration as indicated in the figures or figure legends. To specifically reduce ARTD1, PKCα/δ, PKC pan, HMGB1, IP3R or PLCγ expression, 2 × 105 NIH/3T3, 1 × 105 MRC-5 cells or 2.5 × 104 MEFs were transfected using mouse siARTD1 (QIAGEN, SI02731428), siPKCα (QIAGEN, SI01388583), siPKCδ (QIAGEN, SI01388744), siHMGB1 (Microsynth, sense: 3′-CCGCTGAAAAGAGCAAGAAAA-5′), siIP3R1 (QIAGEN, SI01079092), siIP3R3 (QIAGEN, SI01079106), siPLCγ1 (QIAGEN, SI01380869) or siPLCγ2 (QIAGEN, SI02747381), or human siARTD1 (Microsynth, 3′-GGUGAUCGGUAGCAACAAA-5′), siPKC pan (Santa Cruz Biotechnology, sc-29449), siHMGB1 (QIAGEN, SI03650374), or siMock (QIAGEN, scrambled sequence) lacking significant homology to any known human or mouse gene sequence, with Lipofectamine RNAiMAX transfection reagent (Invitrogen) according to the manufacturer's instructions. Cells were treated with H2O2 3–4 days after transfection. Whole cell lysates were prepared with standard RIPA lysis buffer (50 mM Tris/pH 8, 400 mM NaCl, 0.5% NP-40, 1% deoxycholic acid, 0.1% sodium dodecyl sulphate supplemented with proteinase inhibitor cocktail (Roche), 10 mM β-glycerolphosphate, 1 mM NaF and 1 mM DTT) and total protein concentration determined using the standard Lowry method (42). Nuclear and cytosolic fractions were prepared as described by Dignam et al. (43). Cell viability was determined by the MTT assay (Sigma).
Reverse phase protein arrays (RPPA)
RPPA was performed as described (44,45). In brief, whole cell lysates were spotted onto hydrophobically coated Zeptosens Chips (Bayer Technology Services GmbH). Serially diluted lysates (100, 75, 50 and 25%) were arrayed in duplicates onto hydrophobic Zeptosens Chips using the Nanoplotter NP2.0 (GeSiM), followed by blocking in an ultrasonic nebulizer (ZeptoFOG, Bayer Technology Services GmbH). Antibody incubation with a target specific primary antibody and a signal generating fluorophore-labeled secondary antibody (Invitrogen), microarray data acquisition (ZeptoREADER, Bayer Technology Services GmbH) and data analysis (ZeptoVIEW version 3.1.0.2, Bayer Technology Services GmbH) were performed exactly as described (46). The fluorescence signals obtained from the eight lysate spots (100, 75, 50, 25% lysate amount in duplicates) were fitted using a signal/spot-quality weighted linear least squares fit (47,48) and the relative fluorescence intensity (RFI value) determined at the median protein concentration or modification. To correct for small variations in protein content, relative intensities were normalized to the signals of β-catenin, which itself did not show any significant variation (ANOVA, P < 0.05) in response to H2O2 over 10–60 min.
Significance and clustering analysis
To identify significant proteome changes in response to H2O2, relative fluorescence intensities were imported to MultiExperiment Viewer (MeV) version 4.6 (49). Relative fluorescence intensities were log2 transformed and normalized, before performing statistical analysis using one-way ANOVA as described in (50). The mean transformed fluorescence intensities for group 1 (biological duplicates of untreated_10 min), group 2 (biological duplicates of 0.5 mM_10 min), group 3 (biological duplicates of 0.5 mM_60 min) were compared using F-statistics with P < 0.05. For fold-change analysis, log2 transformed means of the biological replicates were normalized to the untreated sample (set as 1) and proteome changes were filtered (cut off set at 1× SD equal to log2 > 0.77/ ≤0.77 for 0.5 mM). Significant proteome changes in response to H2O2 were selected for clustering analysis (Supplementary Figure S2A), if significant by one-way ANOVA (P < 0.05). To correct for potential false negatives or positives, significance analysis was performed using fold change cut off with thresholds described above or performing a quality control analysis by visualization (V.C.) for clustering analysis as shown Supplementary Figure S2C. Similar profiles of the fold changes over time were identified by clustering analysis using the k-means clustering algorithm and default parameters of MeV (49). K-means based clustering analysis in Figure 1F comprises significant proteome changes, identified by ANOVA (n = 2, P < 0.05), fold change cut off (1 SD) and/or visual control (V.C.) and thus might slightly differ from the results obtained by k-means clustering using only ANOVA (n = 2, P < 0.05) in Supplementary Figure S2A.
Figure 1.
Sublethal H2O2 treatment activates cytoplasmic kinases. (A) PAR immunofluorescence (IF) intensity of MRC-5 cells treated with 0.01–2 mM H2O2 for 3–60 min or left untreated as arbitrary units (a.u.). (B) WT and ARTD1 knockout MEFs were treated with 0.5 mM H2O2 for 10 min. Nuclear extracts (NE) were prepared and 10 μg were incubated with [32P]NAD in an in vitro ADP-ribosylation reaction for 15 min at 30°C. (C) PAR IF analysis of MRC-5 cells transfected with siRNA against ARTD1 or mock, and treated with 0.1 mM H2O2 for 10 min or left untreated. Intensity (a.u.) × 10−5. (D) MEFs were incubated with EdU (10 μM) for 15 min prior to 0.1 mM H2O2 treatment for 10 min. Quantification of the mean PAR signal intensity per nucleus (a.u.) in G1, S and G2 cells (n = 4). No significant (n.s.) difference in H2O2-induced PAR formation between the different cell cycle stages by t-test. (E) Work flow for characterization of H2O2-induced proteome changes by reverse phase protein microarrays (RPPA). MRC-5 cells were treated with 0.5 mM H2O2, cells lysed after 10 or 60 min and whole cell lysates (from biological duplicates) serially diluted and spotted on Zeptosens Chips in technical duplicates. Target-specific primary (see list of antibodies in Supplementary Data) and fluorophore-labeled secondary antibodies were incubated with the lysates. Microarray data were acquired using ZeptoREADER and ZeptoVIEW. (F) Time profile clustering of 0.5 mM H2O2-induced proteome changes in MRC-5 cells (ANOVA, n = 2, P < 0.05) analyzed by k-means algorithm as described in ‘Material and Methods’ section, showing increased protein expression/modification (green) or repression/de-modification (red). Modifications are indicated in brackets. In some cases, different antibodies (recognizing different epitopes of the same protein or protein modification) were applied in the screen. To differentiate these antibodies, a three digit number was included in the description. In some instances, the experiment was performed with the same antibody, e.g. p53 (phosphor-Ser15) (2-02) more than once, therefore the designation (2-02)_3 and _4. The corresponding experiments _1 and _2 were excluded from the heat map, since they did not pass the stringent statistical analysis (ANOVA, n = 2, P < 0.05). (G) The data in panel F were plotted for each analyte in separate graphs grouped according to the cluster determined to better visualize the kinetic profile of the proteomic changes within each given group.
Pathway and network analysis
Gene pathway membership data were obtained from the protein interaction databases PID (51) and KEGG (52). A total of 200 and 211 pathways were obtained from PID and KEGG, respectively. A list of 93 pathways from PID and 63 from KEGG was obtained by mapping the analyzed proteins (unique IDs) to the pathways they play a role in and only selecting for pathways containing at least five of the analyzed proteins.
Fisher's exact test was performed to identify pathways significantly affected by proteins (or modifications) altered in response to H2O2 (significant by ANOVA and fold-change) using the R statistical framework (53). P-values were corrected for multiple testing using the Benjamini-Hochberg correction (54). We have used a false discovery rate (FDR)-corrected P-value cut off of 0.1 to identify pathways significantly affected by proteome changes.
Significant proteome changes, identified by statistical (ANOVA) and fold-change analysis (log2 cut off) were subjected to protein–protein interaction analysis using STRING (v 9.0) (55). Only interactions with a STRING score of at least 0.7 were further analyzed using Cytoscape (http://www.cytoscape.org/) (56).
Immunoblotting
For western blot analysis, proteins were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis and bands were visualized by either using horseradish peroxidase-conjugated antibodies (1:5’000, GE Healthcare) and ECL detection (GE Healthcare) or by using IR-Dye-conjugated antibodies (1:15’000, LI-COR) and detection by the Odyssey infrared imaging system (LI-COR). For quantification, bands were analyzed by ImageJ 1.46 (35) and the Odyssey imaging software (LI-COR).
Antibodies used for Western blotting were anti-ARTD1 (Santa Cruz), anti-PKCα (CST), anti-PKC-δ (CST), anti-Tubulin (1:10’000, Sigma), anti-HMGB1 (1:5’000 abcam), anti-H3 (1:5’000 abcam). If not stated otherwise, antibody dilution was 1:1’000.
Immunofluorescence (IF) microscopy
For PAR IF analysis 1.5 × 105 NIH/3T3 or 2.5 × 104 MEF, MRC-5, or IMR-90 cells were grown on coverslips overnight, prior to 1 h pre-incubation with inhibitors and subsequent H2O2 treatment. After the indicated time of treatment, cells were washed once with phosphate buffered saline (PBS), fixed with ice-cold methanol and acetic acid (3:1) for 10 min at 4°C, washed three times with PBS, blocked in 5% milk/0.05% Tween-20 in PBS for 30 min and stained immunohistochemically with primary mouse anti-poly(ADP-ribose) IgG (10H, made in-house), rabbit anti-poly(ADP-ribose) (Enzo), mouse anti-myc (Santa-Cruz) or rabbit anti-HMGB1 (abcam). Next, cells were washed three times with PBS before hybridization with a secondary antibody (Cy3 conjugated anti-mouse IgG, Jackson ImmunoResearch or Alexa Fluor 488 conjugated anti-rabbit IgG, Invitrogen). All antibodies were used at a dilution of 1:250. After incubation, cells were mounted on glass slides using DAPI-containing VECTASHIELD (Vector Labs) and images acquired using an inverted fluorescence microscope at 40×, oil immersion (Leica). Fluorescence intensities were quantified using ImageJ (v. 1.46r) or Imaris (v. 7.6.0, Bitplane) and equal set-up between the images and experiments.
Quantitative image-based cell cycle staging
For quantitative image-based cell cycle staging of MEFs, automated wide-field microscopy was performed as described (57,58) with the following modifications: Images were acquired on a Leica DMI 6000 inverted microscope equipped with a motorized stage, a Tri-band bandpass filter (DAPI/FITC/TX; BP387/11/BP 494/20/BP 575/20) and a 12-bit monochrome EMCCD camera (Leica DFC 350 FX, 1392 × 1040 pixels, 6.4 μm pixel size). Automated image acquisition under non-saturating conditions was performed using the Leica Matrix Screening Software. All images were imported to the Olympus ScanR Image Analysis Software Version 2.5.1, a dynamic background correction was applied and nuclei segmentation was performed using an integrated intensity-based detection module. Pulsed 5-ethynyl-2′-desoxyuridine (EdU) incorporation, PAR levels and DAPI intensities were measured. G1 cells were identified based on their low EdU and low DAPI content, S phase cells based on their high EdU content, and G2 cells based on their low EdU and high DAPI content.
In vitro kinase and ARTD1 automodification assay
For PKCδ kinase assay, 0.2 μl recombinant PKC catalytic subunit of the PKCδ isoform from rat brain (Sigma, P1609) and 1 μg ARTD1 full-length (fl.) or ARTD1 fragments (fr.) in PKCδ kinase buffer (120 mM Tris/pH 7.5, 40 mM MgCl2, 2 mM CaCl2, 2 mM DTT) and 20 nM ATP (Sigma) were spiked with 0.74 MBq gamma-labeled ATP (20 nM) and incubated for 30 min. For PKCα kinase assay, 100 ng recombinant PKCα (Enzo, BML-SE494-0005) was incubated in PKCα kinase buffer (25 mM MOPS/pH 7.2, 12.5 mM β-glycerophosphate, 25 mM MgCl2, 5 mM EGTA, 2 mM EDTA and 0.25 mM DTT) with 100 μM ATP (Sigma) spiked with 0.74 MBq gamma-labeled ATP (20 nM)/reaction and 1 μg recombinant ARTD1 fl., ARTD1 deletion fragments (ARTD1 fr.), HMGB1 or histone-mix (Roche) for 15 min. ARTD1 automodification assay contained PARP reaction buffer (50 mM Tris-HCl/pH 8, 4 mM MgCl2, 250 μM DTT, 1 μg pepstatin/bestatin/leupeptin), 100 μM or 100 nM unlabeled NAD (Sigma) spiked with 100 nM gamma-labeled NAD and in some cases 0.5 pmol of EcoRI dsDNA linker to induce ARTD1 activity in vitro.
Evaluation of DNA damage
DNA damage (double-strand breaks, single-strand breaks, abasic sites) in cells was determined using the Alkaline Comet Assay (Trevigen) according to the manufacturer's instructions. %DNA in tail was calculated using Comet Assay IV (Perceptive Instruments).
RESULTS
H2O2 rapidly induces nuclear PAR and in a cell cycle-independent manner
To study the signaling mechanisms by which oxidative stress induces PAR formation and to obtain kinetic and dose response information on this process, MRC-5 primary human fibroblasts were treated for various durations with increasing concentrations of H2O2 (0.01–2 mM), and PAR formation was analyzed by immunofluorescence microscopy (Figure 1A and Supplementary Figure S1A). These experiments revealed that H2O2 in a concentration-dependent manner transiently induced strong nuclear PAR formation already at 10 min after treatment, with levels returning to baseline after 60 min, and that maximum PAR formation was observed already at 0.1 mM H2O2. PAR formation coincided with activation of ARTD1, as shown by the automodification of ARTD1 in nuclei isolated from WT, but not ARTD1 knockout MEFs treated with H2O2 for 10 min and incubated with radioactively labeled NAD+ (Figure 1B). Moreover, knockdown of ARTD1 by siRNA confirmed that H2O2-induced PAR formation was predominantly mediated by ARTD1 (Figure 1C). To investigate the cell cycle stage dependency of H2O2-induced PAR formation, H2O2-treated MEFs were analyzed by quantitative image-based cell cycle staging. Nuclear PAR formation was induced by H2O2 to the same extent in all cell cycle stages (Figure 1D and Supplementary Figure S1B), indicating cell cycle-independent ARTD1 activation. To investigate the early H2O2-induced signaling events under sub-lethal conditions, cell viability was assessed 24 h after treatment of cells with H2O2. The results revealed that H2O2 at a concentration of 0.5 mM or below was sub-lethal under the tested condition (Supplementary Figure S1C).
H2O2 treatment induces dynamic proteomic changes
To investigate the early H2O2-induced signaling events that may regulate nuclear PAR formation, a proteomics screen using RPPA focusing on intracellular signaling cascades (i.e. kinases and their substrates) was performed on MRC-5 cells treated with a sublethal concentration of H2O2 (i.e. 0.5 mM) for either 10 or 60 min, respectively (Figure 1E and F; Supplementary Figure S2A–D). The two different time-points were chosen because strong PAR formation was observed at 10 min, while PAR was no longer detectable at 60 min after H2O2 treatment (Figure 1A and Supplementary Figure S1A). Significant changes in protein level or in post-translational modification in the RPPA analysis were determined by statistical analysis using ANOVA (P < 0.05, n = 2). Untreated samples were used as a control group (Figure 1F; Supplementary Figure S2A–D). The significant changes were clustered based on the temporal changes upon H2O2 treatment, taking all time points into account, using k-means clustering algorithm (Figure 1G; Supplementary Figure S2A–D). The majority of significant signal reductions concerned total protein levels, while increases in response to H2O2 treatment were often observed for phosphorylation events (Supplementary Figure S2D). Interestingly, early activation clusters 1–3 (10 min) comprised kinases exclusively found in the cytoplasm and known to be activated directly by either stress stimuli (p-p38, p-JNK) or growth factor signaling (ErbB-2, p-Akt). Ca2+ signaling transducers were also found among the early-induced proteome changes (p-PLCγ, p-PKC, p-CaMKII, p-CREB, p-PLA2) (Figure 1F and G; Supplementary Figure S2D). The majority of late activated signaling events (cluster 4, 60 min) included DNA damage response and cell cycle players such as p-aurora A, p-Chk1, p-cyclin D, p-p53, p-p27 and γH2AX (Figure 1F and G). Downregulated protein or phosphorylation levels (cluster 5) included several factors involved in cell death. However, the main hubs (more ≥ 20 interactions) in the protein–protein interaction network (Akt, p38, ERK2) were rather cytoplasmic signaling components (except for p53) mostly induced at the early time point (10 min), indicating cytoplasmic signaling events as the initial signaling events upon sublethal H2O2 (Supplementary Figure S2E). In summary, our systematic RPPA analyses of the early H2O2-induced proteome changes have identified several cytoplasmic signaling components. From the observed proteomic changes, the kinetics of several protein phosphorylations (e.g. of ERK1/2 and PKC substrates) strongly correlated with the kinetics of PAR formation, pointing at kinases as possible candidates that regulate PAR formation.
Activation of membrane-associated phospholipase C and Ca2+ signaling from the ER via IP3 are required for nuclear PAR formation
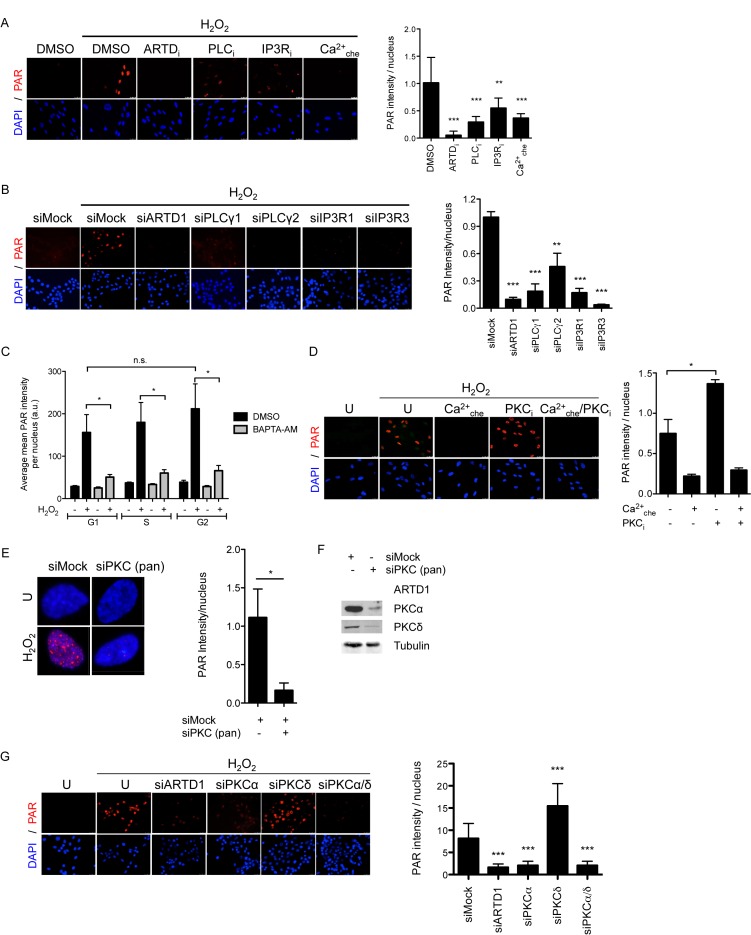
To further validate the H2O2-induced early signaling proteomic changes potentially responsible for PAR formation, a screen with small molecule inhibitors or siRNA for a sub-set of the identified early-activated cytoplasmic kinases was performed. Thus, human IMR-90 or mouse NIH/3T3 fibroblasts were either treated with inhibitors for IKK, AMPK, p38, JNK or MEK, or with siRNA against these proteins, and H2O2-induced PAR formation assessed by IF analysis or western blotting using the 10H anti-PAR antibody (Supplementary Figure S3A–F). Although their H2O2-induced activation kinetics correlated with the enhanced PAR formation, pharmacological inhibition or knockdown/knockout of any of these kinases did not, or only weakly, affect H2O2-induced PAR levels, suggesting that the above-mentioned pathways do not regulate PAR formation. The investigation was therefore extended to additional pathways that were explored with small molecular inhibitors for their role in PAR formation. One of the earliest changes observed by the RPPA (10 min) was enhanced phosphorylation of membrane-associated phospholipase C (PLCγ) (Figure 1F). To investigate a possible involvement of PLCγ in H2O2-induced PAR formation, IMR-90 fibroblasts were treated with the PLC inhibitor (PLCi) U-73122 (Figure 2A). Treatment of cells with PLCi before H2O2 treatment significantly reduced PAR formation, revealing that PLC contributes to PAR formation. PLC is known to stimulate the inositol triphosphate receptor (IP3R) at the ER through the synthesis of IP3. Thus, to determine whether this pathway is important for H2O2-induced PAR formation, IMR-90 fibroblasts were pre-incubated with IP3R inhibitor 2-APB (IP3Ri), which also led to significantly reduced H2O2-induced PAR formation. Importantly, the knockdown of PLCγ or IP3R by siRNA-mediated knockdown led to a very strong reduction in H2O2-induced PAR formation in NIH/3T3 fibroblasts (Figure 2B and Supplementary Figure S4A). Together, these results support the notion that PLC induces a signaling cascade through IP3 and subsequent stimulation of the IP3R at the ER membrane for the induction of PAR formation. Activation of IP3R is known to release Ca2+ from the ER. To investigate the role of Ca2+-signaling in H2O2-induced PAR formation, cells were pre-treated with the cell membrane-permeable intracellular Ca2+ chelator BAPTA-AM (Ca2+che). Indeed, pre-treatment of cells with BAPTA-AM significantly reduced H2O2-induced PAR formation assessed by immunofluorescence microscopy or immunoblot analysis (Figure 2A and Supplementary Figure S4B), indicating that Ca2+ signaling positively regulates PAR formation. Moreover, to rule out any cell cycle-specific Ca2+-dependency, H2O2-induced PAR formation in MEFs pre-incubated with BAPTA-AM were analyzed using quantitative image-based cell cycle staging. BAPTA-AM significantly inhibited nuclear PAR formation in all cell cycle phases, indicating that the Ca2+-dependency of PAR formation is cell cycle-independent (Figure 2C). BAPTA-AM did not block the very weak but detectable basal PAR levels in untreated cells, suggesting that basal PAR formation is not dependent on Ca2+ (Supplementary Figure S4C).
Figure 2.
Negative and positive modulation of ARTD1 activity by different PKC family members. (A) PAR IF analysis of IMR-90 pre-incubated with selected inhibitors or DMSO (control) prior to 0.1 mM H2O2 treatment for 10 min, or left untreated. Inhibitors against Ca2+ (BAPTA-AM, 10 μM), IP3R (2-APB, 100 μM), PLC (U-73122, 1 μM). Quantification of 10–20 nuclei analyzed per area (n = 10), intensity in arbitrary units × 10−5. (B) PAR IF analysis of NIH/3T3 upon treatment with 0.1 mM H2O2 for 10 min 48 h after siRNA transfection. Quantification of 20–40 nuclei analyzed per area (n = 3). (C) MEFs pre-treated with 10 μM BAPTA-AM or DMSO were incubated with EdU (10 μM) for 15 min, stimulated for 10 min with H2O2 (0.1 mM), before fixation and staining. Quantification of the mean PAR signal intensity per nucleus in G1, S and G2 cells, displayed as arbitrary units, n = 4. (D) PAR IF analysis of MRC-5 pre-incubated with 5 μM GF109203X (PKC inhibitor) or 10 μM BAPTA-AM (Ca2+ chelator) or left untreated (U) prior to 0.1 mM H2O2 for 10 min. Intensity in arbitrary units × 10−5. (E) PAR IF staining of MRC-5 cells transfected with siPKC (pan) prior to H2O2 treatment for 10 min (left). Quantification of PAR IF staining of MRC-5 cells transfected with siPKC (pan) prior to H2O2 treatment for 10 min (10–20 cells/condition, n = 5) (right). (F) Knockdown efficiency in e) was confirmed by immunoblotting with anti-ARTD1, anti-PKCα or anti-PKCδ using anti-tubulin as loading control. (G) PAR IF analysis of NIH/3T3 cells transfected with siPKCα and/or siPKCδ, siARTD1 (positive ctl) or siMock (negative control) prior to H2O2 (0.1 mM) treatment for 10 min (500 cells/condition, n = 2). Data represent mean +/− SD analyzed by t-test with *P < 0.05, **P < 0.01, ***P < 0.001, n.s. not significant.
PKCα controls H2O2-induced PAR formation
An early-altered potential downstream component observed in the RPPA analysis of the identified Ca2+-dependent signaling cascade, was the phosphorylation of PKC substrates. The PKC family of protein kinases consists of the conventional, Ca2+-dependent enzymes, as well as the novel and atypical Ca2+-independent kinases (59). In mammals, the PKC protein family consists of four conventional (PKCα, -βI/II and -γ), four novel (PKCδ, -ϵ, -η and -θ) and two atypical (PKCζ and -ι) isozymes (59). To investigate whether early-activated PKCs are involved in H2O2-induced PAR formation, MRC-5 cells were pre-incubated with the PKC pan inhibitor GF109203X (PKCi), which targets all isoforms. Interestingly, pan inhibition of PKCs led to enhanced H2O2-induced PAR formation (Figure 2D). Similar results were obtained with IMR-90 cells (Supplementary Figure S4D). However, the PKCi-mediated enhancement of H2O2-induced PAR formation was completely abolished by the Ca2+ chelator BAPTA-AM (Ca2+che), suggesting that Ca2+-dependent signaling dominates the negative regulatory effect mediated by PKC. To confirm PKC-dependent regulation of H2O2-induced PAR formation, MRC-5 cells were transfected with a pan siPKC against all PKC family members. In contrast to the PKC inhibitor effect, knockdown of PKC isoforms, including PKCα and PKCδ, led to a strong reduction in H2O2-induced PAR formation (Figure 2E and F), suggesting that different PKC family members may regulate PAR formation in opposing ways. Thus, to dissect the positive and negative regulatory effects observed by PKC inhibition and downregulation, NIH/3T3 cells were transfected with siPKCα and/or siPKCδ. In line with the PAR-reducing effect of BAPTA-AM, knockdown of the Ca2+-dependent isoform PKCα resulted in significantly lower H2O2-induced PAR formation as compared to control siRNA (Figure 2G and Supplementary Figure S4E). This effect was confirmed using PKCα knockout MEFs, which showed significantly less H2O2-induced PAR formation as compared to WT MEF (Supplementary Figure S4F and G). In contrast, knockdown of the Ca2+-independent isoform PKCδ resulted in enhanced PAR formation, comparable to that with the pan PKC inhibitor. Double knockdown of PKCα and PKCδ caused a reduction in PAR levels similar to that by knockdown of PKCα only, suggesting a dominant effect of PKCα over PKCδ on PAR formation (Figure 2G). Knockdown of the conventional PKC family members β and γ showed no effect on PAR formation (Supplementary Figure S4H). Together, these results demonstrate that while PKCδ rather attenuates H2O2-induced PAR formation, PKCα is required for H2O2-induced PAR formation.
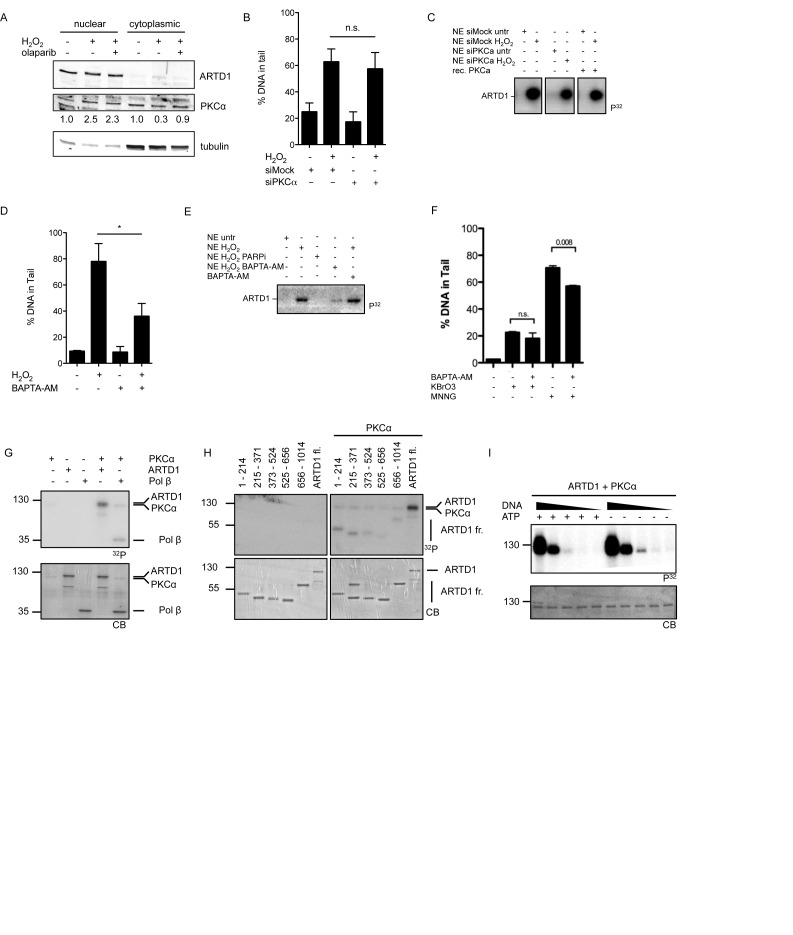
To further investigate the positive regulatory function of PKCα in H2O2-induced PAR formation and whether PKCα translocates to the nucleus in response to H2O2 to potentially regulate ARTD1 activity in this compartment, a nuclear/cytoplasmic separation of H2O2-treated MEFs was performed. Analysis of PKCα localization by immunoblotting showed that indeed 10 min after treatment of MEFs with H2O2, PKCα levels were ∼2.5-fold increased in the nucleus, with a corresponding decrease in the cytoplasm (Figure 3A), suggesting that upon H2O2 treatment a part of PKCα translocates to the nucleus.
Figure 3.
PKCα regulates PAR formation in a DNA break-independent manner. (A) MEFs pre-treated with 10 μM olaparib or left untreated prior to 0.5 mM H2O2 treatment for 10 min. Nuclear and cytoplasmic extracts were analyzed by immunoblotting. Quantification of PKCα levels was performed by densitometry, normalizing nuclear levels to ARTD1 and cytoplasmic levels to tubulin. (B) Alkaline comet assay of siPKCα transfected MEFs or siMock (negative control) prior to 0.1 mM H2O2 treatment for 10 min (100 nuclei analyzed/independent experiment, n = 2). (C) MEFs transfected with siPKCα or siMock were treated with 0.5 mM H2O2 for 10 min. NE were prepared and 10 μg incubated in an in vitro ADP-ribosylation assay using NAD (32P) in the presence or absence of recombinant PKCα for 15 min at 30°C. Loading controlled by Coomassie blue (CB) staining of the gel (lower panel). (D) Alkaline comet assay of MEFs pre-treated with 10 μM BAPTA-AM (Ca2+che) for 30 min or left untreated prior to 0.1 mM H2O2 treatment for 10 min (100 nuclei analyzed/independent experiment, n = 3. (E) MEFs were pre-incubated with either 10 μM olaparib, 10 μM BAPTA-AM or left untreated prior to 0.5 mM H2O2 for 10 min. NE were prepared and 10 μg incubated in an in vitro ADP-ribosylation assay using NAD (32P) in the presence or absence of BAPTA-AM for 15 min at 30°C. (F) Alkaline comet assay of NIH/3T3 cells pre-incubated with BAPTA (20 μM) or DMSO prior to KBrO3 (30 mM, 1 h) or MNNG (50 μM, 1 h) treatment (50 nuclei analyzed/independent experiment, n = 6). (G) PKCα kinase assay using ARTD1 full length and Pol β (positive control) as substrates and radiolabeled ATP (32P). (H) PKCα kinase assay using ARTD1 deletion fragments or full length ARTD1 as substrate and radiolabeled ATP (32P). (I) ADP-ribosylation assay using radiolabeled NAD (32P). PKCα was pre-incubated with ATP and/or ARTD1 for 30 min before adding PKC inhibitor (GF109203X, 5 μM), NAD (32P) and 5, 0.5, 0.05, 0.005 or 0 pmol of EcoRI linker (ds DNA) and incubated for 15 min at 30°C. Data in bar graphs represent mean +/− SD analyzed by t-test with *P < 0.05, **P < 0.01, n.s. not significant.
Ca2+-signaling induces DNA damage that is neither dependent on PKCα, OGG1 nor APE1
ARTD1 has been described to be activated by DNA damage (60). To confirm that the zinc fingers of ARTD1 are required for H2O2-induced PAR formation, ARTD1 knockout fibroblasts were genetically complemented with a mutant lacking zinc fingers 1 and 2, or a mutant lacking PAR forming activity (E988K). The results of these experiments revealed that the presence of zinc fingers and thus the binding to DNA is important for the ARTD1-mediated PAR formation in vivo (Supplementary Figure S5A). Interestingly, H2O2-induced PAR formation did not coincide with an increase in γH2A.X staining, which was observed only after 30 min, or 53BP1 foci formation, for which no increase could be observed within 60 min of H2O2 treatment (Supplementary Figure S5B). However, H2O2 in a dose and time-dependent manner did induce DNA damage as measured by DNA tail formation using the alkaline comet assay (Supplementary Figure S5C). Since ARTD1 is strongly activated by DNA damage, we tested whether PKCα-dependent signaling is responsible for the induction of the observed DNA damage and activation of ARTD1. MEF cells were treated with siPKCα, stimulated with H2O2 and DNA integrity assessed by the alkaline comet assay. H2O2 induced DNA tail formation comparably in cells treated with siPKCα or siMock (Figure 3B), suggesting that PKCα does not regulate PAR formation by affecting H2O2-induced DNA damage. Similarly, H2O2-induced activation of ARTD1 measured by automodification activity in nuclear extracts (NE) was not affected by siPKCα treatment (Figure 3C). Thus, since PAR formation was completely abrogated upon knockdown of PKCα, these experiments provide evidence that the induced DNA damage seems to be required but not sufficient to induce PAR formation by ARTD1. Intriguingly, treatment of cells with BAPTA-AM caused a marked reduction in H2O2-induced DNA tail formation (Figure 3D) and activation of ARTD1 (Figure 3E), suggesting that Ca2+-dependent signaling is important both for the observed DNA tail formation and ARTD1 activation. Interestingly, neither knockdown of the Ca2+-dependent DNA-glycosylase OGG1 nor APE1 affected PAR formation or induced DNA tail formation after 10 min of H2O2 (Supplementary Figure S5D–F), suggesting that the observed DNA damage may not be 8oxoG DNA modifications or abasic sites. To investigate whether the dependency on Ca2+ was specific for H2O2-induced DNA tail formation, 3T3 cells were treated for 1 h with KBrO3 or MNNG, after pre-incubation with BAPTA-AM or DMSO. The DNA damage induced by these agents was either not (KBrO3) or only moderately affected (MNNG) by treatment with BAPTA-AM, suggesting that the dependency on Ca2+ is particularly specific for H2O2-induced DNA tail formation and that H2O2 induces DNA damage by an as yet unidentified Ca2+-dependent mechanism (Figure 3F).
PKCα-mediated phosphorylation of ARTD1, histones or HMGB1 do not directly regulate ARTD1 enzymatic activity in vitro
Since PKCα was observed to translocate to the nucleus (Figure 3A) and to assess a direct regulation of ARTD1 activity through phosphorylation by PKCα in the nucleus, full-length ARTD1 was incubated with PKCα and radioactively labeled ATP in vitro. DNA polymerase beta (Polβ), known to be phosphorylated by PKC (61) was included as positive control in the reaction containing PKCα (Figure 3G). A radioactive modification was detected for ARTD1 in the presence of PKCα, indicating that PKCα is able to directly phosphorylate ARTD1 in vitro. To narrow down the ARTD1 phosphorylation site, different ARTD1 fragments were tested to be modified by PKCα. All tested fragments were phosphorylated by PKCα, although to a different extent, indicating that ARTD1 is phosphorylated indiscriminately at various sites across the whole protein (Figure 3H). To assess whether the enzymatic activity of ARTD1 is affected by PKCα-mediated phosphorylation, ARTD1 pre-incubated with PKCα in the presence or absence of ATP was subsequently incubated with radioactively labeled NAD+ and different amounts of DNA (Figure 3I). ARTD1 activity was essentially the same whether ARTD1 was pre-phosphorylated by PKCα or not, thus excluding direct PKCα-dependent phosphorylation of ARTD1 as the mechanism by which PAR formation is induced in a Ca2+-dependent manner upon H2O2 treatment and suggest an indirect mechanism by which PKCα activates ARTD1. Interestingly, comparable experiments with the Ca2+-independent PKCδ isoform revealed that PKCδ phosphorylates ARTD1 at the N-terminus (amino acids 1–214) and phosphorylation of ARTD1 by PKCδ inhibited DNA-induced PAR formation by ARTD1 in vitro (Supplementary Figure S6A and S6B), suggesting that the observed stimulatory effect of the PKCi on PAR formation might be due to inhibition of ARTD1 by PKCδ.
Considering that PKCα does neither influence ARTD1 activity in vitro nor in NE, but in the nucleus on the chromatin, other chromatin-associated PKCα targets could potentially positively influence PAR formation. To study the consequences of histone phosphorylation by PKCα, recombinant histones were incubated with PKCα and radioactively labeled ATP. A signal corresponding to phosphorylated core histones was observed, but showed no enhancing effect on ARTD1 autocatalytic activity, neither in the phosphorylated nor the non-phosphorylated state (Supplementary Figure S6C and D), indicating that PKCα does not mediate PAR formation by the phosphorylation of histones.
Another known PKCα target within the chromatin landscape that could potentially affect PAR formation is the ubiquitously expressed histone-like protein high mobility-group protein box 1 (HMGB1). To study the consequences of HMGB1 phosphorylation by PKCα, recombinant HMGB1 was incubated with PKCα and radioactively labeled ATP. A strong signal corresponding to phosphorylated HMGB1 was observed (Supplementary Figure S6E), confirming that HMGB1 is a target of PKCα in vitro. However, neither phosphorylated nor non-phosphorylated HMGB1 showed any influence on ARTD1 autocatalytic activity in vitro (Supplementary Figure S6F), suggesting that HMGB1 does not directly influence ARTD1 activity in a simple in vitro setting. However, these in vitro studies are not in the context of the nucleus and chromatin, which could be an important factor for the activity of ARTD1 and its potential modulation by HMGB1.
Chromatin-associated HMGB1 represses PAR formation in vivo and is evicted upon H2O2 treatment in a PKCα-dependent manner
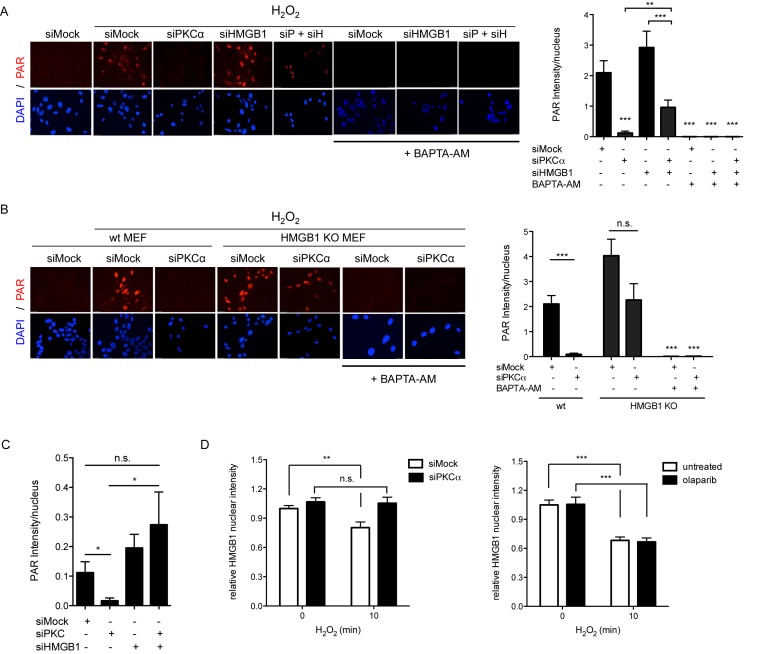
Since PKC-mediated phosphorylation of HMGB1 has been described to alter its DNA binding-affinity, as well as nuclear localization (29,62,63), we knocked down HMGB1 together with PKCα in WT MEF cells to further investigate a possible contribution of HMGB1 to PAR formation. Interestingly, knockdown of HMGB1 alone already enhanced H2O2-induced PAR formation, suggesting that the presence of HMGB1 represses PAR formation. Interestingly, double knockdown of HMGB1 and PKCα reversed the inhibitory effect of PKCα single knockdown on H2O2-induced PAR formation (Figure 4A and Supplementary Figure S6G). In HMGB1 knockout MEFs, the inhibitory effect on PAR formation by PKCα knockdown was also greatly attenuated (Figure 4B). This suggests that the presence of HMGB1 has an inhibitory effect on PAR formation, which is relieved by PKCα, thereby making PKCα a requirement for PAR formation upon H2O2 stimulation. To test whether the rescued PAR formation in PKCα/HMGB1 double knockdown MEFs is also dependent on Ca2+ signaling-induced DNA damage, MEFs were treated with BAPTA-AM after knockdown of HMGB1 alone or after HMGB1/PKCα double-knockdown (which showed the partial rescue) and PAR formation assessed after H2O2 treatment (Figure 4A and B). BAPTA-AM abrogated PAR formation also under these tested conditions, suggesting that Ca2+-dependent DNA damage is an important initiation step for H2O2-induced PAR formation.
Figure 4.
PKCα-dependent phosphorylation of HMGB1 is required for H2O2-induced PAR formation. (A) MEFs treated with scrambled siRNA or siRNA against HMGB1 and/or PKCα were incubated 10 min with 0.5 mM H2O2, in the absence or presence of BAPTA-AM (Ca2+che) and stained for PAR (left). Quantification of PAR intensity (right), 200 nuclei analyzed/experiment, n = 3. (B) HMGB1 knockout MEFs treated with scrambled siRNA or siRNA against PKCα were pretreated with 10 μM BAPTA-AM or left untreated, before 10 min treatment with 0.5 mM H2O2 and stained for PAR. Quantification of PAR intensity (right), 200 nuclei analyzed/experiment, n = 3. (C) Quantification of PAR intensity in MRC-5 cells treated with scrambled siRNA or siRNA against HMGB1 and/or PKC (pan), stained for PAR after 10 min of 0.5 mM H2O2 treatment (20 nuclei analyzed, n = 5). (D) Immunofluorescence analysis of nuclear HMGB1 in MEFs transfected with scrambled siRNA or siRNA against PKCα (left), or pretreated with 10 μM olaparib or DMSO (right), and treated with 0.5 mM H2O2 for 10 min, or left untreated. Data are mean +/− SD analyzed by t-test with *P < 0.05, **P < 0.01, ***P < 0.001, n.s. not significant.
To investigate whether HMGB1 also contributes to the effect of PKCα knockdown in human cells (cf. Figure 2E), the effect of PKCα knockdown (using siPKC) was tested in MRC-5 cells knocked-down for HMGB1 and compared to cells with proficient HMGB1 levels (Figure 4C and Supplementary Figure S6H). Also in this case, the attenuating effect of PKCα knockdown on PAR formation was reversed in cells depleted of HMGB1. These results provide strong evidence that the positive effect of PKCα on PAR formation is mediated in large parts through HMGB1 and that the effect is conserved.
To confirm that PKCα indeed also affects the chromatin association and cellular localization of HMGB1 upon H2O2 treatment, the nuclear levels of HMGB1 in H2O2-treated MEFs were examined by immunofluorescence. While decreased nuclear levels of HMGB1 could be detected in H2O2-treated as compared to non-treated MEFs, the nuclear levels of HMGB1 in MEFs with knocked down PKCα remained unaffected (Figure 4D), indicating that PKCα reduces HMGB1 nuclear levels. It has been reported that ARTD1 and PAR formation is important for the release of HMGB1 from the nucleus upon certain stimuli (64,65). However, the nuclear reduction of HMGB1 upon 10 min H2O2 treatment was not due to HMGB1 ADP-ribosylation, since inhibition of PAR formation by olaparib did not affect the HMGB1 levels (Figure 4D), suggesting that not PARylation by ARTD1, but rather PKCα is important for the reduction of nuclear HMGB1 levels upon 10 min H2O2 treatment.
DISCUSSION
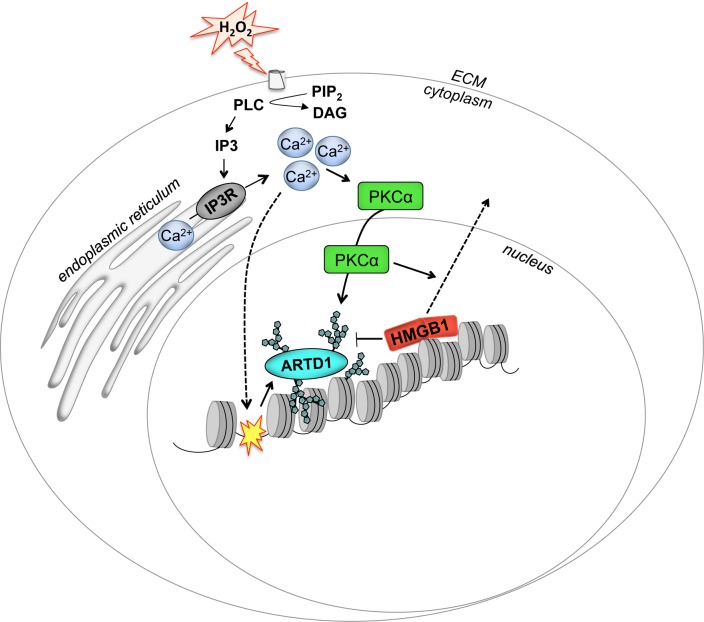
Our results indicate that H2O2 causes ARTD1-mediated PAR formation by inducing intracellular Ca2+ through PLC-generated IP3. Increased Ca2+ then increases PAR formation by two main molecular mechanisms: DNA damage and activation/nuclear translocation of PKCα, which induces chromatin eviction of HMGB1 to successfully induce full PAR formation in the nucleus (Figure 5).
Figure 5.
Schematic representation of the signaling pathways involved in H2O2-induced PAR formation. ARTD1 is activated by Ca2+ by two different mechanisms: (i) Ca2+-dependent formation of DNA strand breaks, as indicated by the curved dashed arrow and yellow star on the DNA and (ii) PKCα activation, translocation to the nucleus and subsequent reduction of nuclear HMGB1 (straight dashed arrow), which releases the repression of ARTD1.
H2O2-induced oxidative stress is a potent activator of nuclear PAR formation in different cell types, but a coherent picture of the molecular events that lead to ARTD1 activation and PAR formation has not existed so far. Although ARTD1 was suggested to be activated upon H2O2 treatment via the induction of oxidative DNA damage, a mechanism providing evidences for such a direct activation in vivo has not been reported previously and ample evidence suggests additional possibilities of controlling ARTD1 enzymatic activity, such as through phosphorylation of ARTD1 or the interaction with modified histones (22,34,35). To elucidate the signaling pathways activated during H2O2-induced oxidative stress, we applied a proteomics screen using RPPA focusing on the major intracellular signaling cascades (i.e. kinases and their substrates) in MRC-5 cells treated with a sublethal concentration of H2O2. Overall, H2O2 treatment principally caused decreases in protein signaling by diminishing total protein level, while producing increases in protein phosphorylation. This could be explained by H2O2-induced inhibition of translation, which causes a general reduction of the protein half-life (66). Moreover, this analysis revealed the kinetics of the H2O2-mediated activation of several pathways. The bioinformatics analysis of the samples treated with 0.5 mM did not significantly differ from samples treated with a lethal dose (i.e. 2 mM, data not shown), suggesting that the initiated signal pathways are the same, independent on the cell fate. Interestingly, validation of several pathways with inhibitors and siRNA revealed that many of the induced pathways did not, or only weakly, affect PAR formation, suggesting that these pathways do not regulate nuclear PAR formation. However, knockdown of PKCα significantly inhibited H2O2-induced PAR formation.
Signaling events upstream of PKCα, including Ca2+ release from the endoplasmatic reticulum and Ca2+-dependent signaling were confirmed to play a major role in H2O2-induced PAR formation. Interestingly, BAPTA-AM showed no effect on the PAR levels in untreated cells, suggesting that basal PAR formation is not dependent on Ca2+. Whether other PAR-inducing conditions depend on PKCα has to be further investigated. This finding is in agreement with studies that have implicated Ca2+ signaling in the activation of ARTD1 (67–69). Our findings revealed that Ca2+ release is initiated at the cellular plasma membrane through phospholipase C (PLC)-mediated signaling. This is in agreement with earlier publications reporting that phosphorylation and activation of PLC by a sulfhydryl oxidation-dependent mechanisms, which leads to increased IP3 synthesis and subsequent activation of the IP3 receptor, inducing the release of Ca2+ from intracellular stores (70,71). In addition, H2O2 treatment has also been reported to inactivate PTEN, which in turn increases cellular IP3 concentration (72). H2O2-induced Ca2+ mainly activated two different molecular processes that regulate nuclear PAR formation: the induction of DNA damage and the activation of PKCα, respectively. The exact nature of the observed Ca2+-induced DNA damage needs further investigation, since the comet assay used in these experiments detects both double-strand, and single-strand DNA breaks, and also abasic sites. Although we did observe a very strong correlation between Ca2+ release and the induction of DNA damage, we cannot assess the extent to which the induced DNA damage is required for PAR formation, since the molecular mechanism responsible for the induction of the observed damage is currently not known. Interestingly, neither knockdown of OGG1 nor APE1 affected PAR formation or DNA damage 10 min after H2O2, suggesting that the initial H2O2-induced DNA damage may not be at 8oxoG or abasic DNA sites. Importantly, since PAR formation but not DNA damage was completely abrogated upon knockdown of PKCα, DNA damage itself is required but not sufficient to induce PAR formation upon H2O2 treatment.
Ca2+ also activated the Ca2+-dependent PKC isoform PKCα, which upon H2O2-treatment translocated to the nucleus. Interestingly, PKCα has so far not been linked to the regulation of PAR metabolism and ARTD1 activity and was only studied as a factor affected by PARP inhibition (73). We found that PKCα was able to phosphorylate ARTD1, as previously shown for both PKCα and PKCβ (74). However, phosphorylation of ARTD1 by PKCα in vitro did not alter ARTD1 enzymatic activity. In contrast, H2O2 treatment also activated the Ca2+ independent PKC isoform PKCδ, which phosphorylated ARTD1 in vitro and reduced ARTD1-dependent DNA-induced PAR formation. This observation is in agreement with the data showing that inhibition of PKC signaling by the pan PKC inhibitor GF109203X led to a significant increase in H2O2-induced PAR formation. Indeed, PKC has been previously shown to phosphorylate ARTD1 in vitro (38) and its inhibition resulted in increased PAR induction upon alkylation stress (37), while there are also studies that have reported a positive regulation of ARTD1 by PKCδ in response to histamine (39,75). Thus, reported studies so far have identified two seemingly opposing (positively and negatively), Ca2+-dependent and -independent regulatory mechanisms for PAR formation upon H2O2-induced oxidative stress within the PKC family of proteins in human primary fibroblasts as well as in murine NIH/3T3 cells and MEFs. However, since the double knockdown of PKCα and PKCδ in our studies reduced PAR levels in the analyzed human and mouse fibroblast similar to that by knockdown of PKCα only, PKCα seems to play the dominant and more relevant role in PAR formation in the analyzed cell types.
H2O2 treatment of cells has been described to influence the chromatin structure, possibly dependent on Ca2+ signaling (76). The release of HMGB1 from chromatin and its translocation from the nucleus into the cytoplasm is inhibited by Ca2+ chelation (77), showing the importance of Ca2+ signaling in HMGB1 dissociation from chromatin. In HMGB1-deficient cells, we observed a stronger PAR formation in response to H2O2. Thus, there is a growing body of evidence that cellular signaling and chromatin-associated changes are involved in the activation of ARTD1 in a DNA damage-independent manner (78). These results suggest that the presence of HMGB1 in the nucleus represses PAR formation during H2O2-induced DNA damage, until the H2O2-stimulated Ca2+ release activates PKCα. This leads to decreased affinity of HMGB1 to the chromatin and subsequent nuclear release, revealing an intriguing interplay between PAR-stimulating and inhibiting mechanisms. We provide here evidence that PKCα interacts and phosphorylates HMGB1 in vitro, and likely the phosphorylation of HMGB1 or of other factors associated with HMGB1, leads to the reduction of HMGB1 protein levels in the nucleus. HMGB1 is known as an inflammatory signaling protein (79,80). Whether the reduction of HMGB1 in the nucleus is due to translocation to the cytosol and extracellular space or due to degradation, remains to be elucidated. Unfortunately, due to lack of phospho-specific HMGB1 antibodies and the transient nature of the interaction, several attempts with commercially available Ser/Thr phosphorylation antibodies and co-immunopreciptation failed to confirm the phosphorylation of HMGB1 in vivo or the interaction of PKCα with HMGB1 in an H2O2-dependent manner (data not shown). Since the phosphorylation of HMGB1 has also been suggested to affect its DNA binding affinity (62), its phosphorylation and effect on PAR formation could potentially also involve a change in DNA-binding and chromatin association. It has been suggested that PAR formation is important for the release of HMGB1 from the nucleus into the cytoplasm upon stress signaling, such as induced by LPS or MNNG (64,65,81). However, pre-incubation of cells with PARPi does not affect the nuclear levels of HMGB1 after 10 min of H2O2 treatment, suggesting that the release is phosphorylation-dependent, but ADP-ribosylation-independent. Interestingly, the regulatory effect of PKCα on PAR formation was strongly reduced in cells lacking HMGB1, suggesting that HMGB1 is the principle, although may not be the only PKCα target involved in controlling PAR formation. The impact of HMGB1 release on the chromatin structure, which chromatin domains (e.g. eu- or heterochromatin) are mainly affected, and how the release allows for PAR formation needs further investigation. Whether the derepression of ARTD1 activity, due to Ca2+-induced PKCα-dependent HMGB1 chromatin release includes additional proteins (e.g. H1 or HP1) or Ca2+-dependent processes in vivo remains to be determined.
In summary, our findings have identified the key players that determine H2O2-induced nuclear PAR formation, which involves parallel Ca2+-dependent signaling pathways: apart from Ca2+-induced DNA damage, the Ca2+-dependent activation of PKCα leads to its nuclear translocation, and subsequently, the eviction of HMGB1 from chromatin, allowing full PAR formation to take place. These results thus identify PKCα and HMGB1 as important regulators of the chromatin modulation involved in H2O2-induced PAR formation, a finding that may have important medical relevance for oxidative stress-associated pathophysiological conditions.
Supplementary Material
Acknowledgments
We are grateful to Roger Davies (University of Massachusetts Medical School) for providing JNK1/2 WT and JNK1/2-/- MEFs, Peter J. Parker (Francis Crick Institute, UK) for PKCα knockout MEFs and Marco E. Bianchi (San Raffaele University, Milano, Italy) for immortalized HMGB1 knockout MEFs. Markus Ehrat and Jan van Oostrum (Zeptosens—a division of Bayer (Schweiz) AG) are acknowledged for helpful input during the planning of the RPPA studies. Johann Grognux and Daniel Rechsteiner (Zeptosens) prepared the cell lysates for the Zeptosens Chips. Florian Freimoser and Stephan Christen (Department of Molecular Mechanisms of Disease, University of Zurich) provided valuable editorial assistance and critical input during the writing. We also thank Barbara van Loon, Nicole Grosse and Matthias Bosshard (Department of Molecular Mechanisms of Disease, University of Zurich) for help with the comet assay. The Center for Microscopy and Image Analysis at the University of Zurich is acknowledged for expert microscopy support.
Author contributions: A.A., A.B. and M.O.H. planned the experiments and wrote the manuscript. A.A. and A.B. performed and evaluated the data and M.O.H. supervised the study. J.T. supervised the RPPA study. F.T. and M.A. investigated cell cycle dependent PAR formation. N.K. performed the bioinformatics analysis and M.B. supervised the study. All authors reviewed the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss National Science Foundation [PP00P3_150690/1 to M.A.]; University of Zurich Association Research Talent Development Fund; University Research Priority Program ‘Integrative Human Physiology’ at the University of Zurich, the Swiss National Science Foundation Grant [310030B_138667; 310030_157019 to M.O.H., in part]; Oncosuisse grant [KLS 02396-02-2009]; Kanton of Zurich (to M.O.H.). Funding for open access charge: Kanton of Zurich.
Conflict of interest statement. J.T. was Zeptosens' Technology Manager at Bayer Technology Services GmbH and is currently Group Head for Process Analytical Technologies at the same company. The other authors declare no competing financial interests.
REFERENCES
- 1.Shokolenko I., Venediktova N., Bochkareva A., Wilson G.L., Alexeyev M.F. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 2009;37:2539–2548. doi: 10.1093/nar/gkp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoshida H., Nishikawa M., Kiyota T., Toyota H., Takakura Y. Increase in CpG DNA-induced inflammatory responses by DNA oxidation in macrophages and mice. Free Radic. Biol. Med. 2011;51:424–431. doi: 10.1016/j.freeradbiomed.2011.04.035. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Q., Itagaki K., Hauser C.J. Mitochondrial DNA is released by shock and activates neutrophils via P38 map kinase. Shock. 2010;34:55–59. doi: 10.1097/SHK.0b013e3181cd8c08. [DOI] [PubMed] [Google Scholar]
- 4.Brinkmann V., Zychlinsky A. Neutrophil extracellular traps: is immunity the second function of chromatin? J. Cell Biol. 2012;198:773–783. doi: 10.1083/jcb.201203170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins L.V., Hajizadeh S., Holme E., Jonsson I.M., Tarkowski A. Endogenously oxidized mitochondrial DNA induces in vivo and in vitro inflammatory responses. J. Leukoc. Biol. 2004;75:995–1000. doi: 10.1189/jlb.0703328. [DOI] [PubMed] [Google Scholar]
- 6.Saxena G., Chen J.Q., Shalev A. Intracellular shuttling and mitochondrial function of thioredoxin-interacting protein. J. Biol. Chem. 2010;285:3997–4005. doi: 10.1074/jbc.M109.034421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshihara E., Chen Z., Matsuo Y., Masutani H., Yodoi J. Thiol redox transitions by thioredoxin and thioredoxin-binding protein-2 in cell signaling. Method Enzymol. 2010;474:67–82. doi: 10.1016/S0076-6879(10)74005-2. [DOI] [PubMed] [Google Scholar]
- 8.Lane T., Flam B., Lockey R., Kolliputi N. TXNIP shuttling: missing link between oxidative stress and inflannmasome activation. Front Physiol. 2013;4:50. doi: 10.3389/fphys.2013.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kazak L., Reyes A., Holt I.J. Minimizing the damage: repair pathways keep mitochondrial DNA intact. Nat. Rev. Mol. Cell Biol. 2012;13:659–671. doi: 10.1038/nrm3439. [DOI] [PubMed] [Google Scholar]
- 10.Alexeyev M., Shokolenko I., Wilson G., LeDoux S. The maintenance of mitochondrial DNA integrity-critical analysis and update. Csh Perspect. Biol. 2013;5:a012641. doi: 10.1101/cshperspect.a012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki N., Kamataki A., Yamaki J., Homma Y. Characterization of circulating DNA in healthy human plasma. Clin. Chim. Acta. 2008;387:55–58. doi: 10.1016/j.cca.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Hassa P.O., Haenni S., Elser M., Hottiger M.O. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol. Mol. Biol. Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hottiger M.O., Hassa P.O., Lüscher B., Schüler H., Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Luo X., Kraus W.L. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26:417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altmeyer M., Messner S., Hassa P.O., Fey M., Hottiger M.O. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009;37:3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishnakumar R., Kraus W. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol. Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schiewer M.J., Goodwin J.F., Han S., Brenner J.C., Augello M.A., Dean J.L., Liu F., Planck J.L., Ravindranathan P., Chinnaiyan A.M., et al. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov. 2012;2:1134–1149. doi: 10.1158/2159-8290.CD-12-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kraus W.L., Hottiger M.O. PARP-1 and gene regulation: progress and puzzles. Mol. Aspects Med. 2013;34:1109–1123. doi: 10.1016/j.mam.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 19.David K.K., Andrabi S.A., Dawson T.M., Dawson V.L. Parthanatos, a messenger of death. Front. Biosci. 2009;14:1116–1128. doi: 10.2741/3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lonskaya I., Potaman V.N., Shlyakhtenko L.S., Oussatcheva E.A., Lyubchenko Y.L., Soldatenkov V.A. Regulation of poly(ADP-ribose) polymerase-1 by DNA structure-specific binding. J. Biol. Chem. 2005;280:17076–17083. doi: 10.1074/jbc.M413483200. [DOI] [PubMed] [Google Scholar]
- 21.Kun E., Kirsten E., Mendeleyev J., Ordahl C. Regulation of the enzymatic catalysis of poly(ADP-ribose) polymerase by dsDNA, polyamines, Mg2+, Ca2+, histones H1 and H3, and ATP. Biochemistry. 2004;43:210–216. doi: 10.1021/bi0301791. [DOI] [PubMed] [Google Scholar]
- 22.Thomas C.J., Kotova E., Andrake M., Adolf-Bryfogle J., Glaser R., Regnard C., Tulin A.V. Kinase-mediated changes in nucleosome conformation trigger chromatin decondensation via poly(ADP-Ribosyl)ation. Mol. Cell. 2014;53:831–842. doi: 10.1016/j.molcel.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osmanov T., Ugrinova I., Pasheva E. The chaperone like function of the nonhistone protein HMGB1. Biochem. Bioph. Res. Co. 2013;432:231–235. doi: 10.1016/j.bbrc.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 24.Joshi S.R., Sarpong Y.C., Peterson R.C., Scovell W.M. Nucleosome dynamics: HMGB1 relaxes canonical nucleosome structure to facilitate estrogen receptor binding. Nucleic Acids Res. 2012;40:10161–10171. doi: 10.1093/nar/gks815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonaldi T., Langst G., Strohner R., Becker P.B., Bianchi M.E. The DNA chaperone HMGB1 facilitates ACF/CHRAC-dependent nucleosome sliding. EMBO J. 2002;21:6865–6873. doi: 10.1093/emboj/cdf692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watson M., Stott K., Fischl H., Cato L., Thomas J.O. Characterization of the interaction between HMGB1 and H3-a possible means of positioning HMGB1 in chromatin. Nucleic Acids Res. 2014;42:848–859. doi: 10.1093/nar/gkt950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Celona B., Weiner A., Di Felice F., Mancuso F.M., Cesarini E., Rossi R.L., Gregory L., Baban D., Rossetti G., Grianti P., et al. Substantial histone reduction modulates genomewide nucleosomal occupancy and global transcriptional output. PLoS Biol. 2011;9:e1001086. doi: 10.1371/journal.pbio.1001086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ugrinova I., Pasheva E.A., Armengaud J., Pashev I.G. In vivo acetylation of HMG1 protein enhances its binding affinity to distorted DNA structures. Biochemistry. 2001;40:14655–14660. doi: 10.1021/bi0113364. [DOI] [PubMed] [Google Scholar]
- 29.Youn J.H., Shin J.S. Nucleocytoplasmic shuttling of HMGB1 is regulated by phosphorylation that redirects it toward secretion. J. Immunol. 2006;177:7889–7897. doi: 10.4049/jimmunol.177.11.7889. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Q., Wang Y. High mobility group proteins and their post-translational modifications. Biochim. Biophys. Acta. 2008;1784:1159–1166. doi: 10.1016/j.bbapap.2008.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paull T.T., Haykinson M.J., Johnson R.C. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 1993;7:1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- 32.Cohen-Armon M. PARP-1 activation in the ERK signaling pathway. Trends Pharmacol. Sci. 2007;28:556–560. doi: 10.1016/j.tips.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 33.Cohen-Armon M., Visochek L., Rozensal D., Kalal A., Geistrikh I., Klein R., Bendetz-Nezer S., Yao Z., Seger R. DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: a link to histone acetylation. Mol. Cell. 2007;25:297–308. doi: 10.1016/j.molcel.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 34.Kauppinen T., Chan W., Suh S., Wiggins A., Huang E., Swanson R. Direct phosphorylation and regulation of poly(ADP-ribose) polymerase-1 by extracellular signal-regulated kinases 1/2. Proc. Natl. Acad. Sci. U.S.A. 2006;103:7136–7141. doi: 10.1073/pnas.0508606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang S., Lin Y., Kim Y.S., Hande M.P., Liu Z.G., Shen H.M. c-Jun N-terminal kinase mediates hydrogen peroxide-induced cell death via sustained poly(ADP-ribose) polymerase-1 activation. Cell Death Differ. 2007;14:1001–1010. doi: 10.1038/sj.cdd.4402088. [DOI] [PubMed] [Google Scholar]
- 36.Bauer P.I., Farkas G., Buday L., Mikala G., Meszaros G., Kun E., Farago A. Inhibition of DNA binding by the phosphorylation of poly ADP-ribose polymerase protein catalysed by protein kinase C. Biochem. Biophys. Res. Commun. 1992;187:730–736. doi: 10.1016/0006-291x(92)91256-p. [DOI] [PubMed] [Google Scholar]
- 37.Hegedus C., Lakatos P., Olah G., Toth B.I., Gergely S., Szabo E., Biro T., Szabo C., Virag L. Protein kinase C protects from DNA damage-induced necrotic cell death by inhibiting poly(ADP-ribose) polymerase-1. FEBS Lett. 2008;582:1672–1678. doi: 10.1016/j.febslet.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka Y., Koide S.S., Yoshihara K., Kamiya T. Poly (ADP-ribose) synthetase is phosphorylated by protein kinase C in vitro. Biochem. Biophys. Res. Commun. 1987;148:709–717. doi: 10.1016/0006-291x(87)90934-x. [DOI] [PubMed] [Google Scholar]
- 39.Mizuguchi H., Terao T., Kitai M., Ikeda M., Yoshimura Y., Das A.K., Kitamura Y., Takeda N., Fukui H. Involvement of protein kinase Cdelta/extracellular signal-regulated kinase/poly(ADP-ribose) polymerase-1 (PARP-1) signaling pathway in histamine-induced up-regulation of histamine H1 receptor gene expression in HeLa cells. J. Biol. Chem. 2011;286:30542–30551. doi: 10.1074/jbc.M111.253104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacobs J.P., Jones C.M., Baille J.P. Characteristics of a human diploid cell designated MRC-5. Nature. 1970;227:168–170. doi: 10.1038/227168a0. [DOI] [PubMed] [Google Scholar]
- 41.Nichols W.W., Murphy D.G., Cristofalo V.J., Toji L.H., Greene A.E., Dwight S.A. Characterization of a new human diploid cell strain, IMR-90. Science. 1977;196:60–63. doi: 10.1126/science.841339. [DOI] [PubMed] [Google Scholar]
- 42.Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 43.Dignam J.D., Lebovitz R.M., Roeder R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pawlak M., Schick E., Bopp M.A., Schneider M.J., Oroszlan P., Ehrat M. Zeptosens' protein microarrays: a novel high performance microarray platform for low abundance protein analysis. Proteomics. 2002;2:383–393. doi: 10.1002/1615-9861(200204)2:4<383::AID-PROT383>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 45.Bluwstein A., Kumar N., Leger K., Traenkle J., Oostrum J., Rehrauer H., Baudis M., Hottiger M.O. PKC signaling prevents irradiation-induced apoptosis of primary human fibroblasts. Cell Death Dis. 2013;4:e498. doi: 10.1038/cddis.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voshol H., Ehrat M., Traenkle J., Bertrand E., van Oostrum J. Antibody-based proteomics: analysis of signaling networks using reverse protein arrays. FEBS J. 2009;276:6871–6879. doi: 10.1111/j.1742-4658.2009.07395.x. [DOI] [PubMed] [Google Scholar]
- 47.Lloyd S.P. Least squares quantization in PCM. IEEE Trans. Inf. Theory. 1982;28:128–137. [Google Scholar]
- 48.Bevington P.R. Data Reduction and Error Analysis for the Physical Sciences. 3rd edn. NY: McGraw-Hill; 2002. [Google Scholar]
- 49.Saeed A.I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 50.Zar J.H. Biostatistical Analysis. 5th edn. Upper Saddle River, NJ: Prentice Hall; 2009. [Google Scholar]
- 51.Schaefer C.F., Anthony K., Krupa S., Buchoff J., Day M., Hannay T., Buetow K.H. PID: the pathway interaction database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanehisa M., Goto S., Furumichi M., Tanabe M., Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ihaka R., Genleman R. R. A language for data analysis and graphics. J. Comput. Graph. Stat. 1996;5:299–314. [Google Scholar]
- 54.Reiner A., Yekutieli D., Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 55.Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Altmeyer M., Toledo L., Gudjonsson T., Grofte M., Rask M.B., Lukas C., Akimov V., Blagoev B., Bartek J., Lukas J. The chromatin scaffold protein SAFB1 renders chromatin permissive for DNA damage signaling. Mol. Cell. 2013;52:206–220. doi: 10.1016/j.molcel.2013.08.025. [DOI] [PubMed] [Google Scholar]
- 58.Toledo L.I., Altmeyer M., Rask M.B., Lukas C., Larsen D.H., Povlsen L.K., Bekker-Jensen S., Mailand N., Bartek J., Lukas J. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155:1088–1103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- 59.Newton A.C. Protein kinase C: poised to signal. Am. J. Physiol. Endocrinol. Metab. 2010;298:E395–E402. doi: 10.1152/ajpendo.00477.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ali A.A., Timinszky G., Arribas-Bosacoma R., Kozlowski M., Hassa P.O., Hassler M., Ladurner A.G., Pearl L.H., Oliver A.W. The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat. Struct. Mol. Biol. 2012;19:685–692. doi: 10.1038/nsmb.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tokui T., Inagaki M., Nishizawa K., Yatani R., Kusagawa M., Ajiro K., Nishimoto Y., Date T., Matsukage A. Inactivation of DNA-Polymerase Beta by Invitro Phosphorylation with Protein-Kinase-C. J. Biol. Chem. 1991;266:10820–10824. [PubMed] [Google Scholar]
- 62.Ugrinova I., Zlateva S., Pasheva E. The effect of PKC phosphorylation on the "architectural" properties of HMGB1 protein. Mol. Biol. Rep. 2012;39:9947–9953. doi: 10.1007/s11033-012-1863-x. [DOI] [PubMed] [Google Scholar]
- 63.Oh Y.J., Youn J.H., Ji Y., Lee S.E., Lim K.J., Choi J.E., Shin J.S. HMGB1 is phosphorylated by classical protein kinase C and is secreted by a calcium-dependent mechanism. J. Immunol. 2009;182:5800–5809. doi: 10.4049/jimmunol.0801873. [DOI] [PubMed] [Google Scholar]
- 64.Ditsworth D., Zong W.X., Thompson C.B. Activation of poly(ADP)-ribose polymerase (PARP-1) induces release of the pro-inflammatory mediator HMGB1 from the nucleus. J. Biol. Chem. 2007;282:17845–17854. doi: 10.1074/jbc.M701465200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang M., Liu L., Xie M., Sun X., Yu Y., Kang R., Yang L., Zhu S., Cao L., Tang D. Poly-ADP-ribosylation of HMGB1 regulates TNFSF10/TRAIL resistance through autophagy. Autophagy. 2015;11:214–224. doi: 10.4161/15548627.2014.994400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vogel C., Silva G.M., Marcotte E.M. Protein expression regulation under oxidative stress. Mol. Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.009217. M111 009217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ju B.-G., Solum D., Song E., Lee K.-J., Rose D., Glass C., Rosenfeld M. Activating the PARP-1 sensor component of the groucho/ TLE1 corepressor complex mediates a CaMKinase IIdelta-dependent neurogenic gene activation pathway. Cell. 2004;119:815–829. doi: 10.1016/j.cell.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 68.Midorikawa R., Takei Y., Hirokawa N. KIF4 motor regulates activity-dependent neuronal survival by suppressing PARP-1 enzymatic activity. Cell. 2006;125:371–383. doi: 10.1016/j.cell.2006.02.039. [DOI] [PubMed] [Google Scholar]
- 69.Bentle M., Reinicke K., Bey E., Spitz D., Boothman D. Calcium-dependent modulation of poly(ADP-ribose) polymerase-1 alters cellular metabolism and DNA repair. J. Biol. Chem. 2006;281:33684–33696. doi: 10.1074/jbc.M603678200. [DOI] [PubMed] [Google Scholar]
- 70.Sato H., Takeo T., Liu Q., Nakano K., Osanai T., Suga S., Wakui M., Wu J. Hydrogen peroxide mobilizes Ca2+ through two distinct mechanisms in rat hepatocytes. Acta Pharmacol. Sinica. 2009;30:78–89. doi: 10.1038/aps.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hong J.H., Moon S.J., Byun H.M., Kim M.S., Jo H., Bae Y.S., Lee S.I., Bootman M.D., Roderick H.L., Shin D.M., et al. Critical role of phospholipase Cgamma1 in the generation of H2O2-evoked [Ca2+]i oscillations in cultured rat cortical astrocytes. J. Biol. Chem. 2006;281:13057–13067. doi: 10.1074/jbc.M601726200. [DOI] [PubMed] [Google Scholar]
- 72.Lee S.R., Yang K.S., Kwon J., Lee C., Jeong W., Rhee S.G. Reversible inactivation of the tumor suppressor PTEN by H2O2. J. Biol. Chem. 2002;277:20336–20342. doi: 10.1074/jbc.M111899200. [DOI] [PubMed] [Google Scholar]
- 73.Bartha E., Solti I., Kereskai L., Lantos J., Plozer E., Magyar K., Szabados E., Kalai T., Hideg K., Halmosi R., et al. PARP inhibition delays transition of hypertensive cardiopathy to heart failure in spontaneously hypertensive rats. Cardiovasc. Res. 2009;83:501–510. doi: 10.1093/cvr/cvp144. [DOI] [PubMed] [Google Scholar]
- 74.Gagne J.P., Moreel X., Gagne P., Labelle Y., Droit A., Chevalier-Pare M., Bourassa S., McDonald D., Hendzel M.J., Prigent C., et al. Proteomic investigation of phosphorylation sites in poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase. J. Proteome Res. 2009;8:1014–1029. doi: 10.1021/pr800810n. [DOI] [PubMed] [Google Scholar]
- 75.Mizuguchi H., Miyagi K., Terao T., Sakamoto N., Yamawaki Y., Adachi T., Ono S., Sasaki Y., Yoshimura Y., Kitamura Y., et al. PMA-induced dissociation of Ku86 from the promoter causes transcriptional up-regulation of histamine H(1) receptor. Sci. Rep. 2012;2:916. doi: 10.1038/srep00916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Konat G.W. H2O2-induced higher order chromatin degradation: a novel mechanism of oxidative genotoxicity. J. Biosci. 2003;28:57–60. doi: 10.1007/BF02970132. [DOI] [PubMed] [Google Scholar]
- 77.Shin J.H., Lee H.K., Lee H.B., Jin Y., Lee J.K. Ethyl pyruvate inhibits HMGB1 phosphorylation and secretion in activated microglia and in the postischemic brain. Neurosci. Lett. 2014;558:159–163. doi: 10.1016/j.neulet.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 78.Burkle A., Virag L. Poly(ADP-ribose): PARadigms and PARadoxes. Mol. Aspects Med. 2013;34:1046–1065. doi: 10.1016/j.mam.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 79.Scaffidi P., Misteli T., Bianchi M.E. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–195. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- 80.Klune J.R., Dhupar R., Cardinal J., Billiar T.R., Tsung A. HMGB1: endogenous danger signaling. Mol. Med. 2008;14:476–484. doi: 10.2119/2008-00034.Klune. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Davis K., Banerjee S., Friggeri A., Bell C., Abraham E., Zerfaoui M. Poly(ADP-ribosyl)ation of high mobility group box 1 (HMGB1) protein enhances inhibition of efferocytosis. Mol. Med. 2012;18:359–369. doi: 10.2119/molmed.2011.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.