Abstract
Despite two decades of research, the major function of FBP-family KH domain proteins during animal development remains controversial. The literature is divided between RNA processing and transcriptional functions for these single stranded nucleic acid binding proteins. Using Drosophila, where the three mammalian FBP proteins (FBP1-3) are represented by one ortholog, Psi, we demonstrate the primary developmental role is control of cell and tissue growth. Co-IP-mass spectrometry positioned Psi in an interactome predominantly comprised of RNA Polymerase II (RNA Pol II) transcriptional machinery and we demonstrate Psi is a potent transcriptional activator. The most striking interaction was between Psi and the transcriptional mediator (MED) complex, a known sensor of signaling inputs. Moreover, genetic manipulation of MED activity modified Psi-dependent growth, which suggests Psi interacts with MED to integrate developmental growth signals. Our data suggest the key target of the Psi/MED network in controlling developmentally regulated tissue growth is the transcription factor MYC. As FBP1 has been implicated in controlling expression of the MYC oncogene, we predict interaction between MED and FBP1 might also have implications for cancer initiation and progression.
INTRODUCTION
The FBP-family of KH domain proteins were first described 20 years ago, however, the core function of these proteins in vivo remains unclear and controversial. KH motifs have been attributed RNA-binding functions (1,2), but can also engage single stranded DNA (ssDNA) with an affinity equal to RNA (at least in vitro) (3). Thus, there is a lack of consensus in the literature regarding the key physiological role of these single stranded nucleic acid binding factors. The weight of reports favour roles in RNA metabolism, with FBP proteins being ascribed multiple roles in RNA processing: both as negative and positive regulators of mRNA splicing, mRNA stability, mRNA export and mRNA translation (4). On the other hand the FBP1 protein has been implicated in modulating activated transcription via ssDNA binding and interaction with the general transcription factor complexes, particularly upstream of the promoter for the MYC oncogene (4–9).
Added to this confusion, mammalian genetics has not resolved the major molecular function of the FBP-family proteins most critical to development. In Drosophila the three FBP proteins (FBP1-3) are represented by one ortholog, P-element somatic inhibitor (Psi). Psi was originally ascribed the function of modulating splicing of transposable P-elements in Drosophila (1). However, given that P-elements are a recent addition to Drosophila melanogaster, only entering the genome within the past 50 years (10), this function cannot reflect evolutionary pressures. To clarify the primary role of the FBP family in development we took advantage of Drosophila genetics, and the lack of redundancy in these KH domain proteins, and here demonstrate that Psi is essential for cell and tissue growth.
Co-immunoprecipitation (Co-IP) mass spectrometry data (DPiM (11)) provided clues into possible mechanism(s) for growth control. Psi was predominantly detected in association with both core and gene-specific RNA Pol II transcriptional machinery; 63% of the 65 strongest Psi-interactors have been directly implicated in RNA Pol II activity. Strikingly, Psi was found in complex with most subunits of the transcriptional Mediator (MED) complex, which interacts with the RNA Pol II machinery to modulate transcription in all eukaryotes (12,13). Although the MED complex is required for most (if not all) RNA Pol II dependent transcription, the MED/CDK8 module can act as a sensor of developmental and environmental cues to activate specific transcriptional programs (14–20). Early studies in flies suggested that kohtalo and skuld, which encode Drosophila homologs of the MED12 and MED13 subunits of the kinase module, are essential for the transcription of Wg/Wnt and Notch pathway targets and, thus, required to establish compartment boundaries of the wing imaginal disc (16,21,22). More recently, a specific reduction in the expression of genes involved in wing margin formation was observed for MED26 null mutant wing disc clones (23). At the level of promoters, direct interplay between gene/tissue-specific Hox transcription factors and MED19 is essential for regulating expression of both embryonic and larval imaginal disc patterning genes (24). Here, we demonstrate that the interaction between Psi and the MED complex underlies Psi's essential function in cell and tissue growth.
The observation that Psi interacts with core RNA Polymerase II transcriptional machinery to maintain cell and tissue growth is of great interest given (i) the human Psi-related KH protein FBP1 has been implicated in modulating activated transcription from the MYC promoter (6,8,9,25–28), and (ii) the proficiency of the MYC transcription factor in driving cellular growth programs (29,30). These data do not exclude roles in RNA processing, but suggest dysregulation of dMYC transcription is key to phenotypic outcome, potentially as a consequence of the capacity of MYC to act as a context dependent global amplifier of elaborate developmental transcription programs (31,32). In the context of rapidly proliferating wing disc cells the major program of MYC-modulated transcription will include genes required for cell and tissue growth.
MATERIALS AND METHODS
Fly strains
Unless otherwise stated, the Drosophila strains were obtained from the Bloomington Stock Centre. We used two non-overlapping PsiRNAi lines: PsiRNAi 1 (chromosome 2) PsiRNAi 2 (chromosome 3). The UAS-Psi RNAi 1 line (V105135), UAS-Psi RNAi 2 line (V28990), UAS-CDK8 RNAi (V31264), UAS-CycC RNAi (V27937) and UAS-dMYC RNAi (V2948) lines were obtained from the Vienna Drosophila RNAi Center (33). UAS-MED18 RNAi from the Bloomington TRIP collection (BL42634). UAS-MED4 (767), UAS-MED7 (2735), UAS-MED17 (302), UAS-MED18 (2188), UAS-MED19 (1396), UAS-MED20 (2076), UAS-MED30 (1060), UAS-MED31 (988) were obtained from the DPiM transgenic resource.
Adult wing analysis
Adult wing size was determined for male wings imaged with Olympus SZ51 binocular at 4.5x magnification using Olympus DP20 camera. Wing size was measured in pixels for the area posterior to wing vein L5 using Photoshop software CS5. For wing hair counts adult male wings were imaged with Olympus BX 61 microscope at 20x magnification using the Olympus DP70 camera. Wing cell size was assessed using wing hair counts in a defined area (200 × 100 pixels) at the central region posterior of wing vein L5.
Co-IP and Western analysis
Co-IP for Drosophila was performed using 25 wild type 3rd instar larval heads dissociated in cold lysis buffer (50 mM Tris pH 7.5, 1.5 mM MgCl2, 125 mM NaCl, 0.2% NP40, 5% glycerol, 1x Protease inhibitor cocktail, Roche). Following homogenization, protein was collected by centrifugation at 12 000 rpm for 10 min at 4°C. The extract was pre-cleared by incubation with nProtein A Sepharose TM beads (GE Healthcare Life Science) for 1 h at 4°C with rotation and the supernatant collected by centrifugation at 12 000 rpm. Equal amounts of pre-cleared protein lysate were incubated with either guinea pig anti-MED17 (Gift from Michael Marr), mouse anti-Cdk8 (Abcam, ab52779) or anti-Psi (custom rabbit polyclonal antibody, Biomatik) antibodies overnight at 4°C. Beads were washed with lysis buffer 5 times and the eluent resolved using 10% SDS-PAGE/Western with anti-Psi antibody (1/1000) prior to detection with Li-Cor Odyssey IR detection.
Immunofluorescence
Larval imaginal tissues were fixed for 20 min in 4% paraformaldehyde (PFA) and blocked in 5% goat serum in Phosphate buffered saline PBS with 0.1% Tween (PBST) at room temperature for 1 h, followed by incubation overnight at 4°C with anti-Psi antibody (1/1000, rabbit polyclonal, Biomatik) and detection with fluorescently tagged secondary antibody. After placing in 80% glycerol, wing imaginal discs were dissected and imaged with the Zeiss Imager Z confocal microscope using Zen Meta software. Z-series with 1 μm sections were performed at 40x magnification. Fluorophores were imaged using band-pass filters to remove cross-detection between channels. Images were processed and prepared using Image J 1.43u, and Adobe Photoshop CS4 Version 11.0.2.
Luciferase reporter assays
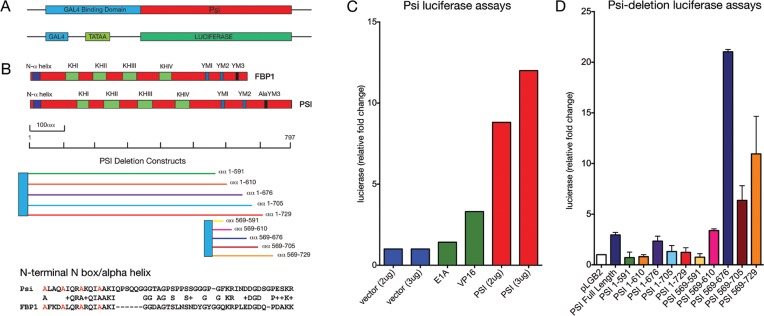
GAL4 fusion protein expression plasmids were generated for full-length Psi and Psi deletion constructs by cloning cDNA encoding the fragments (shown in Figure 6A) in frame with the GAL4 DNA binding domain in pSG424. The GAL4:VP16 and GAL4:E1a plasmids were used as positive controls for transactivation. The reporter plasmid (pLGB2) contains a minimal E1b TATAA (13 bp) and a single Gal4 site upstream of luciferase. Plasmids were transfected into 2 × 106 Hela cells (per well in a 6-well dish) using Lipofectamine, cells were harvested after 24–30 h, extracts prepared and assayed for luciferase. Assays were conducted as described previously (8,34).
Figure 6.
Psi activates transcription in vitro. (A) Design for GAL4-Psi fusion and the minimal promoter (GAL4-EB1 TATAA box) luciferase reporter fusion constructs. (B) Alignment of conserved domains for Psi and human FBP1, deletion constructs for Psi, amino acid sequence of the conserved N-terminal N-box. (C) Luciferase transcription activator assays for control E1a or VP16 activators compared with full-length Psi. (D) Luciferase activity following Psi deletion.
cDNA synthesis and q-RT PCR
RNA was isolated from equivalent numbers of wing imaginal discs (10 pairs for each genotype) using the Promega ReliaPrep RNA Cell miniprep system and eluted in 20 µl nuclease-free water. RNA purity and integrity was assessed using an automated electrophoresis system (2200 TapeStation, Agilent Technologies). A total of 6 µl of the eluted RNA samples were used for cDNA synthesis (Bioline Tetro cDNA Synthesis kit). qPCR was performed using KAPA SYBR FAST qPCR Master Mix (Geneworks). Each qPCR reaction contained 0.1 μl of cDNA, 5 μl KAPA SYBR Master Mix and 0.2 μM of each gene specific primer in a final volume of 10 μl. qPCR reactions (95°C for 2 min, followed by 40 cycles of: 90°C/5 s and 60°C/15 s) were run using the Viia7 Real-Time PCR System and Sequence Detection Systems in 384-well plates (Applied Biosystems). Amplicon specificity was verified by melt curve analysis. Average Ct values for three technical replicates were calculated for each sample. Multiple internal control genes were analysed for stability as described (35) and target gene expression was normalised to the geometric mean of cyp1 and tubulin, selected for having high expression and little sample-to-sample variability. Fold change was determined using the 2-ΔΔCT method.
The primers used were:
MYC - 5′ GTGGACGATGGTCCCAATTT 3′
5′ GGGATTTGTGGGTAGCTTCTT 3′;
PSI 5′ CGATGGCATCCCATTTGTTTGT 3′
5′ GGTGGTCAAGACTACTCGGC3′
Tubulin 5′ TGGGCCCGTCTGGACCACAA 3′
5′ TCGCCGTCACCGGAGTCCAT 3′
CYP1 – 5′ TCGGCAGCGGCATTTCAGAT 3′
5′ TGCACGCTGACGAAGCTAGG 3′
Cdk8 5′ GGACATGGACAATCCGGTGC 3′
5′ GCTTGTCTCCTTCCATTTCGC 3′
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were carried out essentially as described previously (36). For each ChIP sample, 30 larval heads were fixed in 4% PFA for 20 min. Larval heads were then mashed and chromatin sheared in 0.4% sodium dodecyl sulphate (SDS) using a Covaris S2 (10 min duration, 10% DUTY, 200 cycles per burst, Intensity 4, achieving average DNA fragment sizes 200–600 bp). ChIP was performed in IP buffer containing 0.1% SDS and 3 μg of antibody was used for each IP (anti-RNA Pol II phospho S5 antibody (ab5131), or anti-RNA Pol II phospho S2 (ab5095). Analysis was performed in triplicate using KAPA SYBR FAST qPCR Master Mix (Geneworks) on a Viia7 Real-Time PCR System and Sequence Detection Systems in 384-well plates (Applied Biosystems). To calculate the percentage of total DNA bound, non-immuno precipitated input samples from each condition were used as the qPCR reference for all qPCR reactions.
The primers for qPCR were:
MYC1 - 5′ GGCGATCGTTTCTGGCCTACGG 3′
5′ GCAGGCGCATTTGACTCGGC 3′;
MYC2 - 5′ ACTACTACTAACAACTGTCACAAGCCAAGT 3′
5′ TTTATGTATTTGCGCGGTTTTAAG 3′
MYC3 – 5′ TTCAAAATAGAATTTCTGGGAAAGGT 3′
5′ GCGGCCATGATCACTGATT 3′;
MYC4 - 5′ GGTTTTCCTTTTATGCCCTTG 3′
5′ CTATTAACCATTTGAACCCGAAATC 3′
MYC3′UTR - 5′ AGGGGTTAGAGTTTACGAGTGA 3′
5′ CCAAATCAAATCGCGCGGAA 3′
Statistical analysis
All statistical tests were performed with Graphpad Prism 6 using unpaired 2-tailed t-test with 95% confidence interval. In all figures error bars represent SEM and according to the Graphpad classification of significance points *(P = 0.01–0.05), **P = 0.001–0.01, ***P = 0.0001–0.001 and ****P < 0.0001
RESULTS
Psi is required for cell and tissue growth
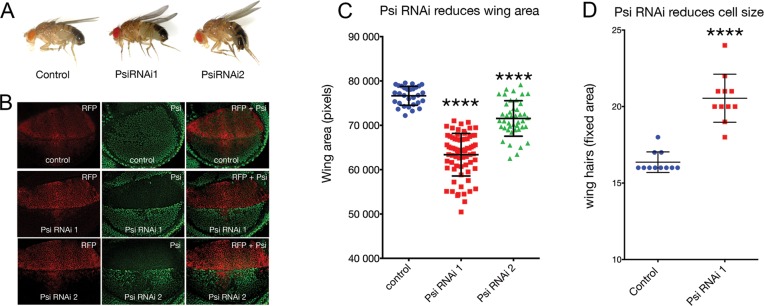
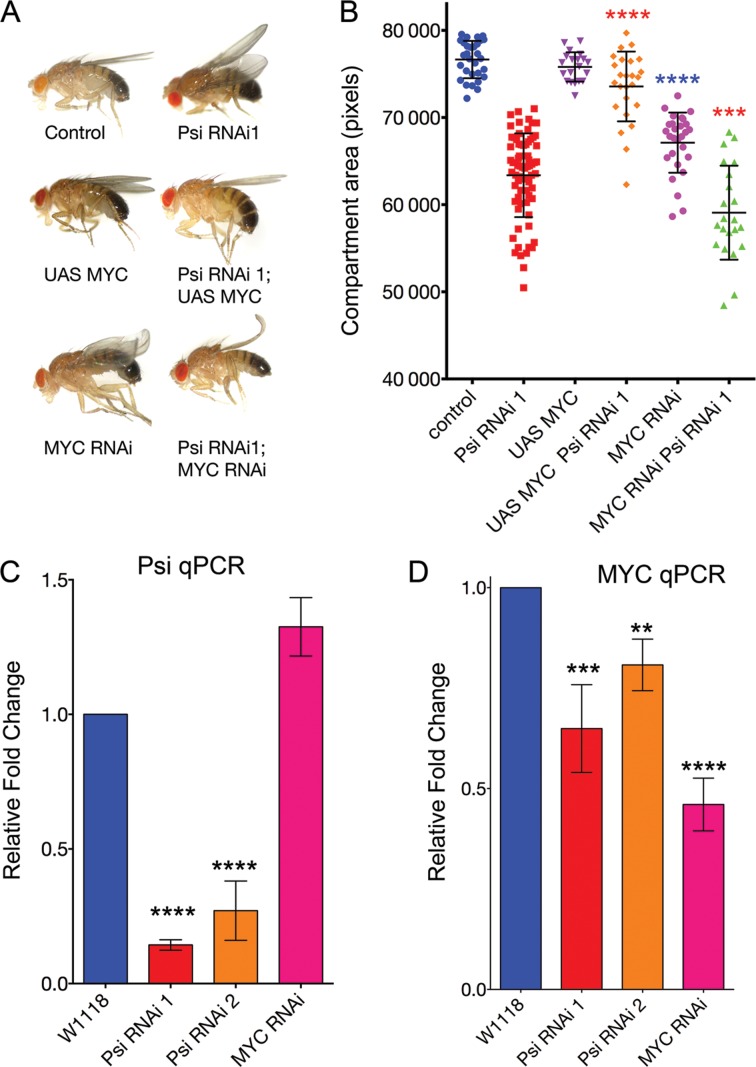
Knockdown of Psi in the dorsal compartment of the larval wing imaginal disc with the serrate-GAL4 (ser-GAL4) driver results in a curled adult wing phenotype (Figure 1A). Similar ‘wings up’ phenotypes have been reported previously following manipulation of growth regulators in the dorsal wing compartment of the larval wing disc, which develops into the top layer of the adult wing (37). Impaired growth of the top sheet of the wing bilayer causes wing cupping and bending due to torsional strain associated with a comparatively smaller top sheet juxtaposing a larger bottom sheet. We observed this phenotype using 2 alternate and non-overlapping UAS-Psi RNAi lines (Figure 1A), and confirmed depletion of Psi protein in the dorsal compartment using our Psi antibody (Figure 1B). Quantification of a defined area of the adult wing revealed a significant reduction in wing size (Figure 1C, P < 0.0001 for Psi RNAi 1 and Psi RNAi 2 compared with control, and see Supplementary Table S2). As each wing-blade cell protrudes a single hair, counting hair numbers within a fixed area provides a measure of cell size, i.e. increased wing hairs indicate reduced cell size. In line with Psi being essential for cell growth we observed a significant increase in wing hairs (Figure 1D, P < 0.0001 for Psi RNAi 1 compared with control within the central region posterior of wing vein L5, and see Supplementary Table S3). Psi depletion therefore resulted in impaired wing size, primarily as a consequence of reduced cell growth.
Figure 1.
Psi is essential for cell and tissue growth. Ser-GAL4 driven Psi RNAi in the larval wing using two alternate and non-overlapping RNAi lines reduces dorsal compartment size due to impaired cell growth. (A) Psi RNAi knockdown results in a ‘wings up’ phenotype in the adult. (B) Psi antibody staining in the larval wing imaginal discs following Psi knockdown. (C) Quantification of the posterior compartment defined by L5 vein in the adult wing compartment. (D) Cell size via wing hair number within a fixed area in the central region posterior of wing vein LV.
Psi interacts with RNA Pol II machinery including the transcriptional Mediator complex
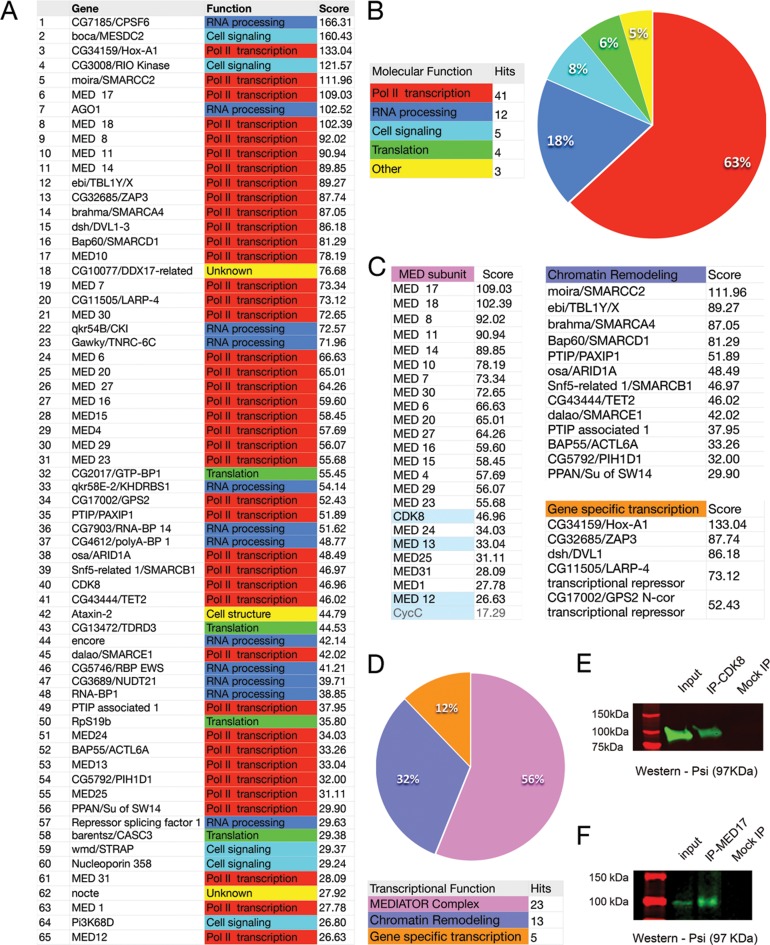
The large scale DPiM Co-IP mass spectrometry study generated a significant metazoan protein map; and included rigorous statistical analysis and follow up validation of interaction networks (11). The top 65 Psi interactors identified following in silico mining of these data are listed in Figure 2A (see also Supplementary Table S1) and the percentage of hits comprising major functional classes summarized in Figure 2B. Strikingly, 41 (or 63%) of these Psi-interactors fell into ontology classes associated with RNA Pol II-dependent transcription. In accordance with previously reported RNA-binding roles for Psi (1), the majority of the remaining Psi interactors were implicated in RNA processing (18%) and/or mRNA translation (6%) (Figure 2A and B). These data suggest that in addition to previously reported roles in mRNA processing, Psi might interact with the RNA Pol II machinery to regulate transcription. To further interrogate potential transcriptional roles, the 41 interactors with designated functions in RNA Pol II transcription were divided into sub-classes. Strikingly, 23 of the 41 transcriptional class interactors (56%) comprised subunits of the Mediator (MED) complex (Figure 2C and D). The remaining 18 interactors were either part of the chromatin-remodeling machinery (32%) or gene specific transcriptional regulators (12%) (Figure 2C and D).
Figure 2.
Psi interacts with RNA Pol II transcriptional machinery, including most Mediator subunits. (A) List of top 65 Psi interactors from the Drosophila Protein Interaction Map (DPiM) Co-IP mass spectrometry data set. (B) Summary of functional classes for the top 65 Psi-interactors showing 63% are implicated in RNA Pol II-dependent transcription. (C and D) Subdivision of the RNA Pol II associated interactors showing 56% comprise the transcriptional mediator complex (MED), 32% are chromatin remodelling factors and 12% other transcription factors. See also Supplementary Table S1. Psi physically interacts with Mediator in vivo. (E and F) Co-IP of endogenous MED subunits and Psi from wild type third instar larval tissue. (E) IP with anti-CDK8 and blot with Psi; (F) IP with MED17 and blot with Psi.
As the DPiM studies (11) that detected Psi in complex with most MED subunits were performed in vitro using overexpressed tagged protein in Drosophila S2 culture cells, we first confirmed that key MED subunits could be detected in complex with Psi in vivo, by conducting Co-immunoprecipitation (Co-IP) from wild type 3rd instar larval imaginal disc lysates (Figure 2E and F). In general, the MED complex can behave as either an activator or inhibitor of RNA Pol II-dependent transcription. The ‘small’ or core MED complex is required for activation of RNA Pol II transcription. The ‘large’ complex has been predominantly characterised as a transcriptional repressor and is comprised of an additional 4 proteins; the kinase module comprising the Cyclin dependent kinase complex (CDK8/CycC) and 2 additional MED subunits (MED12 and MED13) (20). As the DPiM detected Psi in complex with both core subunits of the MED complex and the CDK8/CycC module, we conducted Co-IP with either anti-CDK8 or anti-MED17 (a gift from Michael Marr) and Western blot with an anti-Psi antibody. The 97 kDa band for Psi was detected in association with the kinase module subunit of the MED complex, i.e. the CDK8 subunit (Figure 2E) and using the MED17 antibody we were able to IP endogenous Psi from wild type larvae lysates (Figure 2F, representative blots from at least 3 biological replicates). Thus Psi could be detected in association with endogenous MED subunits from both the kinase module and the core MED complex in vivo, however, further studies are required to determine whether Psi directly interacts with these mediator subunits.
Psi-dependent growth is sensitive to MED abundance
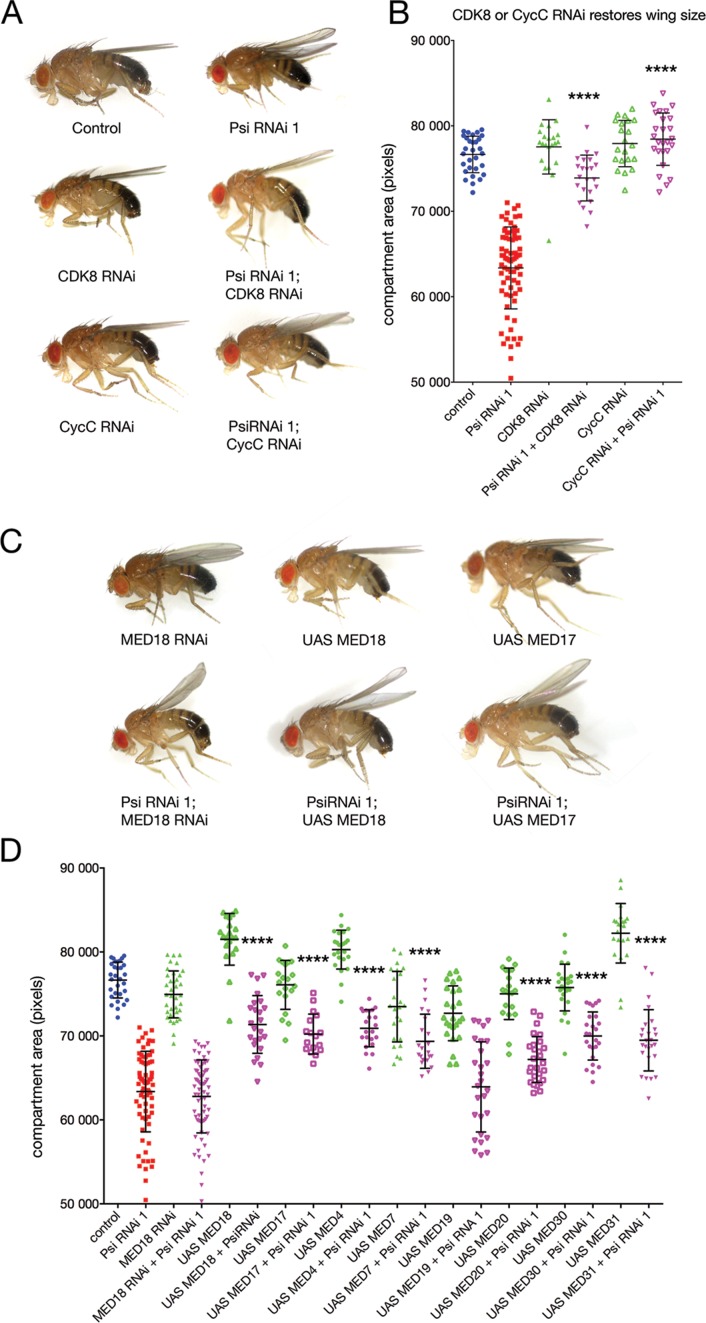
To determine the importance of the Psi-MED interaction to development, we tested whether either transcriptional core or kinase MED subunits were capable of modifying the Psi ‘wings up’ phenotype. We predicted suppressors of Psi-dependent growth would increase the size of the dorsal sheet to bend wings down, i.e. to become more like wild type (i.e. through loss of an inhibitor or increased abundance of an activator), while enhancers (i.e. loss of an activator or increased abundance of an inhibitor) of Psi-driven growth would result in further wing bending. Interestingly, genetic manipulation of core and kinase module subunits of MED differentially modified the Psi RNAi phenotype (Figure 3). Co-knockdown of CDK8 or CycC suppressed the bending of the Psi adult wings and significantly increased compartment size (Figure 3A and B P < 0.0001 for Psi RNAi + CDK8 RNAi and Psi RNAi + CycC RNAi compared with Psi RNAi alone, see Supplementary Tables S4 and S5), consistent with the large MED module normally inhibiting Psi-dependent growth.
Figure 3.
Impaired growth in the Psi knockdown wing is sensitive to MED abundance. (A and B) Co-knockdown of the kinase module subunits (CDK8 or CycC) suppressed the Psi RNAi wings up phenotype. (C and D) MED18 RNAi alone reduced wing area (P = 0.0074) but co-knockdown did not significantly alter the Psi phenotype. Psi knockdown in the wing was suppressed by overexpression of MED4, MED7, MED17, MED19, MED20, MED30 and MED31. Overexpression of certain core MED subunits alone (MED4, MED18, MED31) also increased wing size.
Although there was no obvious wing bending following MED18 knockdown alone we observed a significant reduction in compartment size (Figure 3C and D, P < 0.0074). As the MED complex is required for modulating all RNA Pol II-dependent transcription, the reduced wing phenotype associated with depletion of the MED18 subunit alone may reflect this more general role. Co-knockdown of MED18 and Psi did not significantly further reduce compartment size, however, MED18 overexpression suppressed the Psi wings up phenotype (Figure 3C and D for Psi RNAi + MED18 RNAi compared with Psi RNAi alone, and see Supplementary Table S6). Further analyses for additional components of the core MED complex revealed that the impaired growth associated with Psi knockdown in the wing was suppressed by overexpression of MED4, MED7, MED17, MED19, MED20, MED30 and MED31 (Figure 3D and see Supplementary Table S6). Together with the ability to Co-IP Psi with antibodies to the MED complex these data demonstrate both core and kinase module subunits of MED physically interact with Psi, and that Psi-dependent growth is sensitive to MED abundance.
Psi is required for maintaining endogenous levels of dMYC mRNA and impaired growth in Psi knockdown depends on dMYC reduction
Given mammalian FBP1 has been implicated in controlling activated MYC transcription in vitro (4–9) and the observation that MYC is a potent driver of cell and tissue growth (29,30,38), we tested whether the decreased cell and tissue growth associated with the Psi ‘wings up’ phenotype might be sensitive to abundance of Drosophila MYC, dMYC. In accordance with the reduced compartment size following Psi knockdown in the wing being dMYC-dependent, dMYC overexpression suppressed the ‘wings up’ phenotype. Furthermore co-knockdown of dMYC using RNAi enhanced the Psi RNAi phenotype (Figure 4A and B, and see Supplementary Table S7). To test whether impaired wing growth was associated with decreased dMYC abundance, we ubiquitously depleted Psi and measured dMYC mRNA in wing imaginal discs by qPCR. Psi mRNA abundance was not decreased following dMYC RNAi, however, following a significant reduction in Psi mRNA (Figure 4C, P < 0.0001 for Psi RNAi 1 and 2 compared with control, Supplementary Table S8), dMYC mRNA was significantly decreased (Figure 4D, P = 0.0006 for Psi RNAi 1 and P = 0.0066 for Psi RNAi 2 compared with control, Supplementary Table S9). Taken together these data suggest that Psi is required for maintaining dMYC mRNA at endogenous levels and, thus, for cell growth during Drosophila wing development.
Figure 4.
Psi is required for maintaining endogenous dMYC mRNA in larval wings and the impaired growth in Psi knockdown wings depends on down regulation of dMYC. (A and B) ser-GAL4-driven dMYC overexpression alone did not increase wing size, but suppressed the Psi RNAi wings up phenotype. dMYC RNAi driven by ser-GAL4 alone reduced compartment size to result in a wings up phenotype, and also enhanced the Psi RNAi phenotype. (C and D) qPCR for Psi (C) and dMYC (D) following tubulin-GAL4 driven knockdown of UAS-RNAi lines (as marked) in larval wing discs.
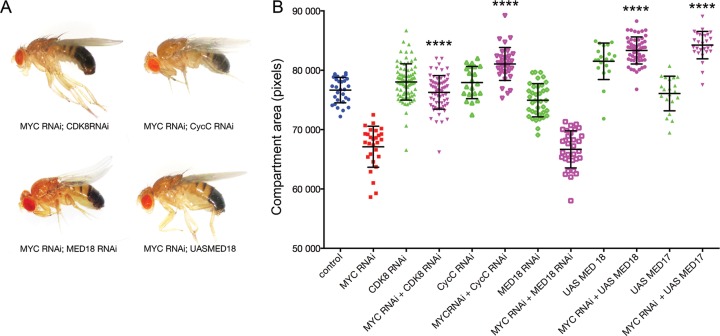
dMYC-dependent wing growth is sensitive to MED abundance/activity
As observed for Psi RNAi (Figure 1A), depletion of dMYC in the dorsal wing compartment (i.e. Ser-GAL4 driven UAS-dMYC RNAi) resulted in a ‘wings up’ phenotype (Figure 4A). Given the observation that Psi-dependent growth was modulated by MED and dMYC (Figures 3 and 4, respectively), we hypothesised that dMYC-dependent growth might be sensitive to MED activity/abundance. We therefore tested whether impaired wing growth associated with dMYC RNAi was modified by the core or kinase module MED subunits. As observed for Psi knockdown (Figure 3), manipulation of core and kinase module subunits of MED differentially modified the dMYC RNAi wings up phenotype (Figure 5). Co-knockdown of CDK8 or CycC suppressed adult wing bending associated with dMYC RNAi and significantly increased compartment size (Figure 5A and B, P < 0.0001 for dMYC RNAi compared with dMYC RNAi + CDK8 RNAi and dMYC RNAi + CycC RNAi, Supplementary Table S10), consistent with the CDK8/CycC module normally acting as a negative regulator of dMYC-dependent growth. Although MED18 co-knockdown did not significantly further reduce compartment size (as also observed for MED18 co-knockdown with Psi, Figure 3C and D), MED18 or MED 17 overexpression suppressed the dMYC RNAi small wing phenotype (Figure 5A and B). Thus dMYC-dependent tissue growth was sensitive to MED abundance; wing growth was restored following depletion of kinase module CDK8/CycC subunits or via overexpression of core MED subunits.
Figure 5.
Impaired growth in the dMYC knockdown wing is sensitive to MED. (A and B) The dMYC RNAi wings up phenotype (see Figure 4A) was suppressed by depletion of the kinase module MED subunits, CDK8 and CycC, and by overexpression of the core MED subunits MED 17 and 18, but was not modified by MED18 knockdown.
Psi regulates transcription in vitro
To determine whether Psi might regulate dMYC by behaving as a transcriptional activator in vitro we transfected HeLa cells with GAL4 DNA binding domain-Psi fusion constructs and a reporter plasmid (pLGB2) containing a single DNA binding site for the Gal4 transcriptional activator 5′ of the minimal TATAA box sequence (13 bp from E1B, for loading of RNA Pol II and the basic transcriptional machinery) and upstream of a luciferase reporter (Figure 6A). Strikingly, Psi activates more strongly than control transactivators (E1A and VP16) for this minimal promoter (Figure 6C, see Supplementary Table S11). Alignment of the mammalian FBP-KH domain protein FBP1 with Psi revealed regions of homology in addition to the KH domains, i.e. the N-terminus (N-box) and C-terminus (tyrosine diad YM-motifs) (Figure 6B). To determine whether particular regions of Psi might be important for transactivation we therefore conducted a deletion series spanning both C-terminal and N-terminal domains (Figure 6B). Deletion of the C terminal YM2 domain, alone (αα1-610, orange bar in 6B and D) or together with the YM1 domain (αα1-591, green bar in 6B and D), abolished the transactivation capacity of Psi (Figure 6D). Interestingly, N-terminal constructs containing even a single C-terminal Y motif were capable of activating transcription (αα659-610, pink bar in 6B and D). The strongest luciferase activity was detected for N-terminal constructs containing both the YM1 or YM2 repeat domains (αα659–676, indigo bar in Figure 6B and D). Together these data suggest that full-length Psi has potent capacity as a transcriptional activator, which requires the C terminal YM1 and YM2 repeat domains. However, the presence of the N-terminal α-helix and KH-containing ssDNA/RNA binding domains diminished transactivation associated with the C-terminal domain (Figure 6B and D, e.g. compare Psi full length with αα659–676).
The interaction between FBP and the active MYC promoter will be dynamic and encompass both ssDNA and protein interfaces of the transcriptional machinery, including the multi-subunit mediator and general transcription factor complex (TFIIH). The observation that C-terminal constructs transactivate more strongly than full length Psi might suggest the N-terminus normally represses Psi-dependent transcription, either directly by masking the activation module and/or via interaction with a second transcriptional repressor protein. In light of this possibility, FBP1 has been shown to interact directly with the RNA recognition motif (RRM) containing protein FUSE Interacting Repressor (FIR), which also binds ssDNA to repress transcription (3,8,25,39). In humans the N-terminal domain in FBP1 is required for the interaction with FIR in vitro (8), which is supported by NMR studies demonstrating the second RNA recognition motif (RRM2) of FIR interacts with the N-terminal α-helix (or N-box) of FBP (3). Cellular studies have demonstrated that this interaction between FIR and the FBP1 N-box is essential for FIR recruitment to FUSE DNA and thus transcriptional repression (8). However, further studies are required to determine the likely cause(s) of the variation in transcriptional capacity associated with the different Psi deletion constructs.
Psi depletion reduced Ser 5 and Ser2 RNA Pol II abundance on the dMYC promoter
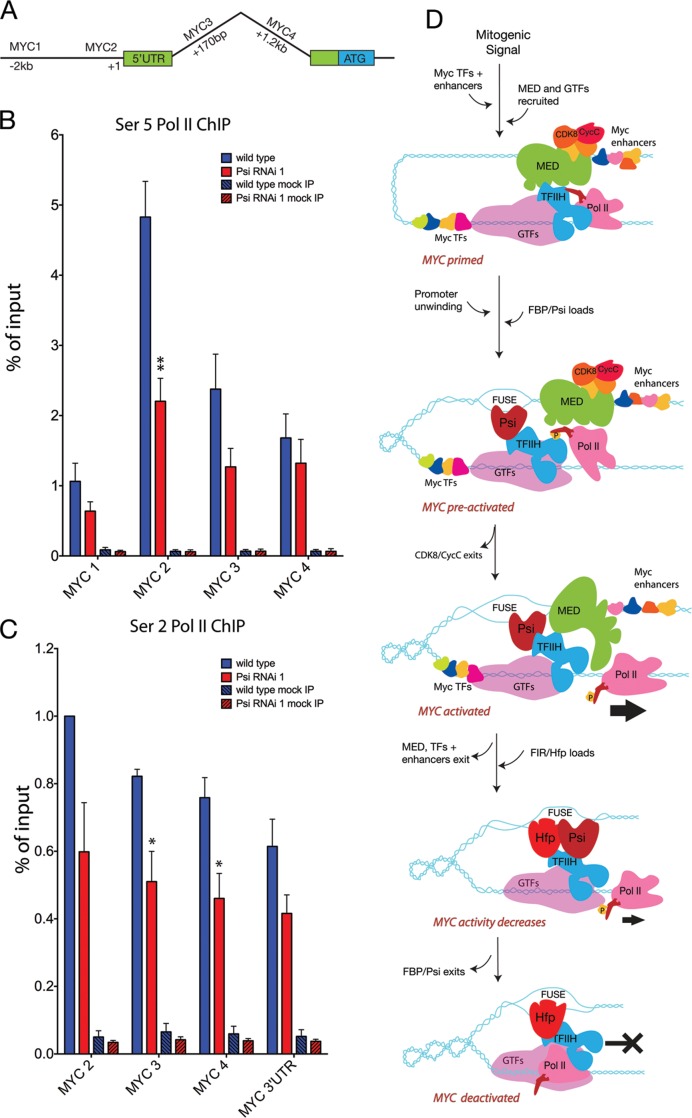
To investigate the capacity of Psi to alter dMYC transcription in vivo we examined RNA Pol II activity on the dMYC promoter. The transcription cycle in all eukaryotes generally occurs as follows: (i) chromatin remodeling proteins and gene specific transcription factors (TFs) engage, (ii) the Mediator complex, the general transcription initiation factors (GTFs, e.g. TFIIH), and the hypophosphorylated RNA Pol II holoenzyme are recruited to form the pre-initiation complex (PIC) (40,41) and (iii) initiation of transcription and release of Pol II allows elongation. Activation of Pol II requires phosphorylation of the carboxyl-terminal domain (CTD) of the largest RNA Pol II subunit at (i) Ser-5 for initiation and promoter escape and (ii) Ser-2 for transcript elongation (42,43).
Ex vivo studies have shown that in response to serum stimulation of previously starved human fibroblasts, FBP1 physically interacts with single-stranded DNA upstream of the MYC transcriptional start site, which precedes release of paused RNA Pol II and activation of MYC expression (5). We therefore examined enrichment for Ser 5 RNA Pol II on the dMYC promoter in larval imaginal tissues following depletion of Psi. ChIP for control tissue revealed a peak for Ser 5 Pol II proximal to the dMYC TSS (Figure 7A and B, see Supplementary Table S12). In comparison, Psi knockdown resulted in a significant decrease in Ser 5 RNA Pol II enrichment proximal to the dMYC TSS (MYC2 amplicon, Figure 7B, see Supplementary Table S12). Thus, Psi was required to maintain the pool of Ser 5 phosphorylated RNA Pol II on the dMYC promoter.
Figure 7.
Psi is required for Ser5 RNA Pol II enrichment across the dMYC promoter and Ser 2 across the body of the dMYC gene. (A) Schematic of dMYC showing the position of the amplicons used for qPCR. (B) Ser 5 RNA Pol II ChIP (control and Psi RNAi); (C) Ser 2 RNA Pol II ChIP (control and Psi RNAi). (D) Model. During the initial stages of the transcription cycle the MED complex and hypophosphorylated RNA Pol II holoenzyme are recruited to form the pre-initiation complex (PIC). Following receipt of the appropriate growth stimulating signals, dMYC enhancers are activated and GTFs (particularly TFIIH) load to drive Ser-5 phosphorylation, activation of RNA Pol II and promoter escape. Increased Pol II activity results in conformational changes in the dMYC promoter, including supercoiling and generation of the single-stranded FUSE. Structural changes following Psi binding modulate promoter architecture to facilitate exit of the CDK8 module and maximise RNA Pol II escape and dMYC transcription. Recruitment of FIR/Hfp inactivates RNA Pol II and returns MYC to basal levels.
The significant decrease in dMYC mRNA in wing discs from these animals is consistent with the reduced Ser 5 RNA Pol II, and would suggest the decrease in RNA Pol II is not due to precocious Pol II escape and increased elongation. Indeed, we also observed a significant decrease in elongating (Ser 2 phosphorylated RNA Pol II) in Psi depleted larval tissues (Figure 7C, see Supplementary Table S12). Together the reduction in Ser 5 and Ser 2 RNA Pol II, and decrease in dMYC mRNA abundance are consistent with either diminished loading and/or phosphorylation of RNA Pol II, either at the initiation/promoter escape step alone (and thus leading to a follow-on reduction in elongation), or also at the elongation step. However, the observation that Psi activates from a single DNA binding site within the minimal promoter (Figure 6, as observed for FBP1 (44)), compared with most G4-activators that require tandem sites, suggests that Psi is unlikely to act at the level of Pol II recruitment, but downstream of PIC assembly.
DISCUSSION
The capacity to integrate extracellular growth and developmental signals is fundamental to coordinated growth of organs and tissues in multicellular animals. Although the MED complex is required for most (if not all) RNA Pol II dependent transcription, the CDK8 module in particular has been noted for its capacity to sense developmental and environmental cues to activate specific transcriptional networks (14–17). In Drosophila, MED responds to specific developmental networks to control patterning of the wing imaginal disc (16,21–24). Consistent with specific roles in regulating growth networks, CDK8 has also been implicated as a negative regulator of tissue growth in mice (45). Our observation that Psi contains potent transcriptional activation capacity suggest Psi helps activate endogenous RNA Pol II activity on dMYC to modulate dMYC transcription and control tissue growth during development. Together with the impaired growth phenotype associated with Psi depletion being suppressed following either co-depletion of subunits from the transcriptionally repressive CDK8/CycC kinase module or overexpression of core subunits, our findings suggests Psi/dMYC-dependent tissue growth depends on MED abundance and activity.
Thus, in Drosophila Psi and MED integrate growth signals to maintain developmentally regulated MYC transcription, cell and tissue growth. Mammalian signaling networks are well known to co-ordinate transcriptional up- or down-regulation of many growth and cell cycle genes, particularly MYC. Ex vivo studies from the early 80s demonstrated that serum stimulation of mammalian tissue culture cells results in rapid activation of MYC transcription (46). Subsequently, extensive studies revealed that torsional stress and strain on dsDNA in the MYC promoter following initiation of MYC transcription results in melting of the double stranded FUSE (the Far Upstream Sequence Element/FUSE −1.7 kb upstream of the major MYC transcription start site) into ssDNA (6,8,27). Moreover, maximal activation of MYC transcription correlates with dissociation of RNA Pol II from the MYC TSS and recruitment of FBP1 (5), which has been extensively characterized for specificity in binding the single stranded FUSE structure (25–27,39,47). The rapid release of RNA Pol II prior to the peak in MYC mRNA levels was associated with maximal enrichment for FBP1, consistent with FBP1 promoting RNA Pol II release to hyperactivate MYC transcription (5).
Based on these observations, and our current study in Drosophila, we propose the following model for the action of Psi/FBP1 in activated MYC transcription (Figure 7D). MED will first integrate the activity of MYC enhancers, stimulated by growth or developmental signals, with the general transcription factors (GTFs) and RNA Pol II to form the PIC. In a signaling environment conducive to MYC transcription, MED will recruit TFIIH and stabilise the PIC (48,49). MED will also bring TFIIH kinase activity into close proximity with the CTD of RNA Pol II (49), consistent with MED stimulating TFIIH-dependent CTD phosphorylation and RNA Pol II promoter clearance (40,41,48–50).
The increased RNA Pol II activity will result in conformational changes in the MYC promoter, including supercoiling to cause torsional stress and generation of the single-stranded FUSE, which binds FBP1/Psi. FBP1 can also directly interact with the XPB helicase subunit of TFIIH, and this interaction is required for formation of the promoter loop between RNA Pol II and FUSE (5). Psi is found in complex with the kinase module subunits of MED, thus we predict Psi/FBP1 will first interact with the preactivated MYC promoter, i.e. with the large MED complex still in residence. Structural changes in ssDNA following Psi/FBP1 loading will modulate promoter architecture further to facilitate exit of the CDK8 module, thus maximising MED-driven RNA Pol II activity and MYC transcription. At this stage we cannot make conclusions on whether Psi/FBP1 interacts directly with a given MED subunit, or indirectly via the TFIIH complex, however, we predict the transition to a maximally activated MYC promoter will be dependent on Psi/FBP1.
Subsequently, FIR/Hfp will be recruited to the MYC promoter via binding to ssDNA, FBP1 and TFIIH to facilitate FBP1 exit, inactivation of RNA Pol II and return of MYC transcription to basal levels. Moreover, RNA interference studies suggest FIR is required for repression of MYC transcription (5) and is dysregulated in cancers with associated elevation of MYC (51). Our previous Drosophila studies suggested the RRM protein with most similarity to FIR, Half Pint (Hfp), is essential for developmentally driven downregulation of dMYC transcription in vivo (36,52,53). Moreover, as observed for FIR (5,34), Hfp interacts with the XPB helicase component of the general transcription factor TFIIH to maintain a pool of engaged RNA Pol II on the MYC promoter, consistent with poised RNA Pol II being required to attenuate MYC transcription (36).
Our analysis of the Drosophila protein interaction map revealed the XPB/Haywire subunit of TFIIH was not one of the 3488 bait proteins in the DPiM screen; nor was XPB/Haywire detected as one of the 4927 prey proteins with 0.8% (or less) false discovery. This was despite known interactors (including other TFIIH complex subunits such as CycH, Cdk7 and MAT1) being included as bait in the DPiM screen. Moreover, CycH, and Cdk7 only detected other CAK subunits (MAT1 and Cdk7 or CycH), but did not detect any of the core subunits (e.g. dXPB/Haywire). MAT1 as bait detected Cdk7, CycH and XPD from the core, but no other subunits. Thus, the DPiM is not saturating, in particular the core TFIIH subunits appear to be under represented, and further studies are required to establish whether Psi interacts with XBP/TFIIH.
Our studies demonstrate nuanced mechanisms of MYC transcriptional regulation, requiring interaction between the MED complex and ssDNA binding proteins are essential for normal tissue growth. Further studies are required to determine whether human FBP1 also interacts with MED as we predict this interaction will also modulate expression of the MYC oncogene. As even subtle increases in MYC expression (>2-fold) can promote the cell and tissue overgrowth fundamental to cancer initiation and progression, these observations will have implications for human disease (28,54).
Supplementary Material
Acknowledgments
The authors thank Michael Marr for the MED17 and MED26 antibodies. We thank Bloomington and VDRC stock centres for Drosophila strains and the DSHB for antibodies.
Author contributions: L.G, J.E.A.L., O.Z., N.C.M., Z.N., G.P. T.W., R.L. and L.M.P. conceived experiments, designed the experiments, performed the experiments, analysed the data and assisted with drafting the manuscript. R.D.H. and D.L.L. conceived experiments, contributed reagents and assisted with drafting the manuscript. L.M.Q. conceived experiments, designed experiments, analysed data and drafted the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Project Grants and a Senior Research Fellowship from the National Health and Medical Research Council of Australia (to L.Q and R.H.); NIH (to D.L.); Cancer Council of Victoria (to L.Q.). Funding for open access charge: Cancer Council Victoria.
Conflict of interest statement. None declared.
REFERENCES
- 1.Siebel C.W., Kanaar R., Rio D.C. Regulation of tissue-specific P-element pre-mRNA splicing requires the RNA-binding protein PSI. Genes Dev. 1994;8:1713–1725. doi: 10.1101/gad.8.14.1713. [DOI] [PubMed] [Google Scholar]
- 2.Min H., Turck C.W., Nikolic J.M., Black D.L. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- 3.Cukier C.D., Hollingworth D., Martin S.R., Kelly G., Díaz-Moreno I., Ramos A. Molecular basis of FIR-mediated c-myc transcriptional control. Nat. Struct. Mol. Biol. 2010;17:1058–1064. doi: 10.1038/nsmb.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J., Chen Q.M. Far upstream element binding protein 1: a commander of transcription, translation and beyond. Oncogene. 2013;32:2907–2916. doi: 10.1038/onc.2012.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J., Kouzine F., Chung H.J., Elisha-Feil Z., Weber A., Levens D. The FUSE/FBP/FIR/TFIIH system is a molecular machine programming a pulse of c-myc expression. EMBO J. 2006;25:2119–2130. doi: 10.1038/sj.emboj.7601101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung H.J., Levens D. c-myc expression: keep the noise down! Mol. Cells. 2005;20:157–166. [PubMed] [Google Scholar]
- 7.Liu J., Akoulitchev S., Weber A., Ge H., Chuikov S., Libutti D., Wang X.W., Conaway J.W., Harris C.C., Conaway R.C., et al. Defective interplay of activators and repressors with TFIH in xeroderma pigmentosum. Cell. 2001;104:353–363. doi: 10.1016/s0092-8674(01)00223-9. [DOI] [PubMed] [Google Scholar]
- 8.Chung H.-J., Liu J., Dundr M., Nie Z., Sanford S., Levens D. FBPs are calibrated molecular tools to adjust gene expression. Mol. Cell Biol. 2006;26:6584–6597. doi: 10.1128/MCB.00754-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He L., Liu J., Collins I., Sanford S., Levens D. Loss of FBP function arrests cellular proliferation and extinguishes c-myc expression. EMBO J. 2000;19:1034–1044. doi: 10.1093/emboj/19.5.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brookfield J.F., Montgomery E., Langley C.H. Apparent absence of transposable elements related to the P elements of D. melanogaster in other species of Drosophila. Nature. 1984;310:330–332. doi: 10.1038/310330a0. [DOI] [PubMed] [Google Scholar]
- 11.Guruharsha K.G., Rual J.-F., Zhai B., Mintseris J., Vaidya P., Vaidya N., Beekman C., Wong C., Rhee D.Y., Cenaj O., et al. A protein complex network of Drosophila melanogaster. Cell. 2011;147:690–703. doi: 10.1016/j.cell.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boube M., Joulia L., Cribbs D.L., Bourbon H.-M. Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell. 2002;110:143–151. doi: 10.1016/s0092-8674(02)00830-9. [DOI] [PubMed] [Google Scholar]
- 13.Bourbon H.-M. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 2008;36:3993–4008. doi: 10.1093/nar/gkn349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van de Peppel J., Kettelarij N., van Bakel H., Kockelkorn T.T.J.P., van Leenen D., Holstege F.C.P. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol. Cell. 2005;19:511–522. doi: 10.1016/j.molcel.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 15.Kim S., Xu X., Hecht A., Boyer T.G. Mediator is a transducer of Wnt/beta-catenin signaling. J. Biol. Chem. 2006;281:14066–14075. doi: 10.1074/jbc.M602696200. [DOI] [PubMed] [Google Scholar]
- 16.Carrera I., Janody F., Leeds N., Duveau F., Treisman J.E. Pygopus activates Wingless target gene transcription through the mediator complex subunits Med12 and Med13. Proc. Natl. Acad. Sci. U.S.A. 2008;105:6644–6649. doi: 10.1073/pnas.0709749105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Firestein R., Bass A.J., Kim S.Y., Dunn I.F., Silver S.J., Guney I., Freed E., Ligon A.H., Vena N., Ogino S., et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding N., Zhou H., Esteve P.-O., Chin H.G., Kim S., Xu X., Joseph S.M., Friez M.J., Schwartz C.E., Pradhan S., et al. Mediator links epigenetic silencing of neuronal gene expression with x-linked mental retardation. Mol. Cell. 2008;31:347–359. doi: 10.1016/j.molcel.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gwack Y., Baek H.J., Nakamura H., Lee S.H., Meisterernst M., Roeder R.G., Jung J.U. Principal role of TRAP/mediator and SWI/SNF complexes in Kaposi's sarcoma-associated herpesvirus RTA-mediated lytic reactivation. Mol. Cell Biol. 2003;23:2055–2067. doi: 10.1128/MCB.23.6.2055-2067.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allen B.L., Taatjes D.J. The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol. 2015;16:155–166. doi: 10.1038/nrm3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janody F., Martirosyan Z., Benlali A., Treisman J.E. Two subunits of the Drosophila mediator complex act together to control cell affinity. Development. 2003;130:3691–3701. doi: 10.1242/dev.00607. [DOI] [PubMed] [Google Scholar]
- 22.Janody F., Treisman J.E. Requirements for mediator complex subunits distinguish three classes of notch target genes at the Drosophila wing margin. Dev. Dyn. 2011;240:2051–2059. doi: 10.1002/dvdy.22705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marr S.K., Lis J.T., Treisman J.E., Marr M.T. The metazoan-specific mediator subunit 26 (Med26) is essential for viability and is found at both active genes and pericentric heterochromatin in Drosophila melanogaster. Mol. Cell Biol. 2014;34:2710–2720. doi: 10.1128/MCB.01365-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boube M., Hudry B., Immarigeon C., Carrier Y., Bernat-Fabre S., Merabet S., Graba Y., Bourbon H.-M., Cribbs D.L. Drosophila melanogaster Hox transcription factors access the RNA polymerase II machinery through direct homeodomain binding to a conserved motif of mediator subunit Med19. PLoS Genet. 2014;10:e1004303. doi: 10.1371/journal.pgen.1004303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braddock D.T., Louis J.M., Baber J.L., Levens D., Clore G.M. Structure and dynamics of KH domains from FBP bound to single-stranded DNA. Nature. 2002;415:1051–1056. doi: 10.1038/4151051a. [DOI] [PubMed] [Google Scholar]
- 26.Braddock D.T., Baber J.L., Levens D., Clore G.M. Molecular basis of sequence-specific single-stranded DNA recognition by KH domains: solution structure of a complex between hnRNP K KH3 and single-stranded DNA. EMBO J. 2002;21:3476–3485. doi: 10.1093/emboj/cdf352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duncan R., Bazar L., Michelotti G., Avigan M., Levens D. A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev. 1994;8:465–480. doi: 10.1101/gad.8.4.465. [DOI] [PubMed] [Google Scholar]
- 28.Levens D. You don't muck with MYC. Genes Cancer. 2010;1:547–554. doi: 10.1177/1947601910377492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnston L.A., Prober D.A., Edgar B.A., Eisenman R.N., Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98:779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grewal S.S., Li L., Orian A., Eisenman R.N., Edgar B.A. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat. Cell Biol. 2005;7:295–302. doi: 10.1038/ncb1223. [DOI] [PubMed] [Google Scholar]
- 31.Nie Z., Hu G., Wei G., Cui K., Yamane A., Resch W., Wang R., Green D.R., Tessarollo L., Casellas R., et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin C.Y., Lovén J., Rahl P.B., Paranal R.M., Burge C.B., Bradner J.E., Lee T.I., Young R.A. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dietzl G., Chen D., Schnorrer F., Su K.C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 34.Liu J., He L., Collins I., Ge H., Libutti D., Li J., Egly J.M., Levens D. The FBP interacting repressor targets TFIIH to inhibit activated transcription. Mol. Cell. 2000;5:331–341. doi: 10.1016/s1097-2765(00)80428-1. [DOI] [PubMed] [Google Scholar]
- 35.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. doi:10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J.E.A., Mitchell N.C., Zaytseva O., Chahal A., Mendis P., Cartier-Michaud A., Parsons L.M., Poortinga G., Levens D.L., Hannan R.D., et al. Defective Hfp-dependent transcriptional repression of dMYC is fundamental to tissue overgrowthin Drosophila XPB models. Nat. Commun. 2015;6:1–12. doi: 10.1038/ncomms8404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barcelo H., Stewart M.J. Altering Drosophila S6 kinase activity is consistent with a role for S6 kinase in growth. Genesis. 2002;34:83–85. doi: 10.1002/gene.10132. [DOI] [PubMed] [Google Scholar]
- 38.Poortinga G., Quinn L.M., Hannan R.D. Targeting RNA polymerase I to treat MYC-driven cancer. Oncogene. 2014;34:403–412. doi: 10.1038/onc.2014.13. [DOI] [PubMed] [Google Scholar]
- 39.Benjamin L.R., Chung H.J., Sanford S., Kouzine F., Liu J., Levens D. Hierarchical mechanisms build the DNA-binding specificity of FUSE binding protein. Proc. Natl. Acad. Sci. U.S.A. 2008;105:18296–18301. doi: 10.1073/pnas.0803279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ebmeier C.C., Taatjes D.J. Activator-Mediator binding regulates Mediator-cofactor interactions. Proc. Natl. Acad. Sci. U.S.A. 2010;107:11283–11288. doi: 10.1073/pnas.0914215107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taatjes D.J. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem. Sci. 2010;35:315–322. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirose Y., Ohkuma Y. Phosphorylation of the C-terminal domain of RNA polymerase II plays central roles in the integrated events of eucaryotic gene expression. J. Biochem. 2007;141:601–608. doi: 10.1093/jb/mvm090. [DOI] [PubMed] [Google Scholar]
- 43.Larivière L., Seizl M., Cramer P. A structural perspective on Mediator function. Curr. Opin. Cell Biol. 2012;24:305–313. doi: 10.1016/j.ceb.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 44.Duncan R., Collins I., Tomonaga T., Zhang T., Levens D. A unique transactivation sequence motif is found in the carboxyl-terminal domain of the single-strand-binding protein FBP. Mol. Cell Biol. 1996;16:2274–2282. doi: 10.1128/mcb.16.5.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akoulitchev S., Chuikov S., Reinberg D. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature. 2000;407:102–106. doi: 10.1038/35024111. [DOI] [PubMed] [Google Scholar]
- 46.Kelly K., Cochran B.H., Stiles C.D., Leder P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell. 1983;35:603–610. doi: 10.1016/0092-8674(83)90092-2. [DOI] [PubMed] [Google Scholar]
- 47.Kouzine F., Sanford S., Elisha-Feil Z., Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat. Struct. Mol. Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 48.Esnault C., Ghavi-Helm Y., Brun S., Soutourina J., Van Berkum N., Boschiero C., Holstege F., Werner M. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol. Cell. 2008;31:337–346. doi: 10.1016/j.molcel.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 49.Plaschka C., Larivière L., Wenzeck L., Seizl M., Hemann M., Tegunov D., Petrotchenko E.V., Borchers C.H., Baumeister W., Herzog F., et al. Architecture of the RNA polymerase II-Mediator core initiation complex. Nature. 2015;518:376–380. doi: 10.1038/nature14229. [DOI] [PubMed] [Google Scholar]
- 50.Max T., Søgaard M., Svejstrup J.Q. Hyperphosphorylation of the C-terminal repeat domain of RNA polymerase II facilitates dissociation of its complex with mediator. J. Biol. Chem. 2007;282:14113–14120. doi: 10.1074/jbc.M701345200. [DOI] [PubMed] [Google Scholar]
- 51.Matsushita K., Tomonaga T., Shimada H., Shioya A., Higashi M., Matsubara H., Harigaya K., Nomura F., Libutti D., Levens D., et al. An essential role of alternative splicing of c-myc suppressor FUSE-binding protein-interacting repressor in carcinogenesis. Cancer Res. 2006;66:1409–1417. doi: 10.1158/0008-5472.CAN-04-4459. [DOI] [PubMed] [Google Scholar]
- 52.Quinn L.M., Dickins R.A., Coombe M., Hime G.R., Bowtell D.D.L., Richardson H. Drosophila Hfp negatively regulates dmyc and stg to inhibit cell proliferation. Development. 2004;131:1411–1423. doi: 10.1242/dev.01019. [DOI] [PubMed] [Google Scholar]
- 53.Mitchell N.C., Johanson T.M., Cranna N.J., Er A.L.J., Richardson H.E., Hannan R.D., Quinn L.M. Hfp inhibits Drosophila myc transcription and cell growth in a TFIIH/Hay-dependent manner. Development. 2010;137:2875–2884. doi: 10.1242/dev.049585. [DOI] [PubMed] [Google Scholar]
- 54.Eilers M., Eisenman R.N. Myc's broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.