Abstract
DNA damage tolerance pathways allow cells to duplicate their genomes despite the presence of replication blocking lesions. Cells possess two major tolerance strategies, namely translesion synthesis (TLS) and homology directed gap repair (HDGR). TLS pathways involve specialized DNA polymerases that are able to synthesize past DNA lesions with an intrinsic risk of causing point mutations. In contrast, HDGR pathways are essentially error-free as they rely on the recovery of missing information from the sister chromatid by RecA-mediated homologous recombination. We have investigated the genetic control of pathway choice between TLS and HDGR in vivo in Escherichia coli. In a strain with wild type RecA activity, the extent of TLS across replication blocking lesions is generally low while HDGR is used extensively. Interestingly, recA alleles that are partially impaired in D-loop formation confer a decrease in HDGR and a concomitant increase in TLS. Thus, partial defect of RecA's capacity to invade the homologous sister chromatid increases the lifetime of the ssDNA.RecA filament, i.e. the ‘SOS signal’. This increase favors TLS by increasing both the TLS polymerase concentration and the lifetime of the TLS substrate, before it becomes sequestered by homologous recombination. In conclusion, the pathway choice between error-prone TLS and error-free HDGR is controlled by the efficiency of homologous recombination.
INTRODUCTION
Despite efficient DNA repair mechanisms that excise most DNA lesions prior to replication, some lesions persist and interfere with the process of replication. Cells possess pathways to deal with DNA lesions during replication; these pathways do not remove the lesions but merely allow replication to cope with them and are thus referred to as DNA damage tolerance (DDT) pathways. Two major DDT strategies have been identified: (i) translesion synthesis (TLS) a process during which a specialized DNA polymerase is recruited to copy past the DNA Damage with an inherent risk of introducing a mutation in the newly synthesised strand and (ii) damage avoidance (DA) strategies, that encompass all non-TLS events (1). The most frequent DA pathway is Homology Directed Gap Repair (HDGR) that involves repair of the gaps formed upon replication of lesion containing DNA via RecA-mediated homologous recombination with the sister chromatid (1–5).
We have recently established an assay allowing to quantify the relative extent that Escherichia coli cells use TLS and DA to tolerate replication-blocking lesions in vivo (6). Under normal, non-SOS-induced, growth conditions, TLS is used with parsimony, representing ≈1–2% of tolerance events while increasing up to 10–40% under genotoxic stress conditions (7). Consequently, from 60 to 99% of tolerance events involve DA events. We have shown that a rate-limiting step for TLS usage is the concentration of the specialized DNA polymerases (7).
When non-coding lesions impede DNA synthesis in one strand, the replicative helicase keeps unwinding the parental duplex and complementary strand synthesis continues essentially unperturbed (8,9). Repriming the lesion-containing strand downstream from the offending lesions (10) restores the fork and generates single-stranded gaps that become covered with RecA protein (11). The single-stranded RecA filaments thus formed constitute the signal for SOS activation, the substrate for TLS and the key intermediate to start gap repair via homologous recombination (12). Indeed, formation of a D-loop between the single-stranded DNA RecA-filament and its homologous sister duplex is thought to initiate the HDGR pathway (13–15).
In the present paper, our goal is to investigate the way DDT pathways are interconnected in E. coli. For this purpose we determined lesion tolerance usage, in E. coli strains carrying recA alleles that are partially defective in recombination but not in SOS induction (16). Compared to a strain wild type for the recA gene, strains carrying recA alleles with reduced recombination activity exhibit a robust increase in TLS at the expense of DA events. We attribute this increase in TLS to the increase in lifetime of the ssDNA.RecA filament. This increase in the so-called ‘SOS signal’, favors TLS by increasing both the TLS polymerase concentration and the lifetime of the TLS substrate, before it becomes sequestered by homologous recombination. The role, on the extent of TLS, of increased persistence of the TLS substrate was further evidenced in recF (or recO) strains. In these stains, delayed kinetics ssDNA.RecA filament formation (17) increases the lifetime of the TLS substrate and results in an increase in TLS in the absence of any confounding effect due to TLS polymerase concentration.
MATERIALS AND METHODS
Strains
All strains used in the present study for site-specific recombination are derivative of strains FBG151/2 that allows site-specific integration of the lesion-containing plasmid into lagging (FBG151) or leading (FBG152) strands (6,18). Gene disruptions in recA, mutS, uvrA, polB, recF, umuDC, sulA and lexA were achieved by the one-step PCR method (19). The FBG151/2 derived strains were constructed by P1 transduction. Strains used in this study are listed in the Table 1. Mutated alleles (RecA-G288Y, RecA-Q300A and RecA-N304D) were all expressed from a single copy F plasmid pGE523 in the strains EVP122/3 and EVP363/4. The recA gene is under control of its natural promoter (16). These RecA plasmids were a kind gift of Olivier Lichtarge (Baylor, Houston, USA).
Table 1. Strains used in this study.
| Name | Short name | Genotype |
|---|---|---|
| EVP22/3 | parental strain | FBG151/2 uvrA::frt mutS::frt |
| EVP49/50 | recF | FBG151/2 uvrA::frt mutS::frt recF::Cm |
| EVP100 | polB | FBG152 uvrA::frt mutS::frt polB::frt |
| EVP112/3 | lexA(def) | FBG151/2 uvrA::frt mutS::frt sulA::frt lexA::frt |
| EVP119 | recA lexA(def) | FBG152 uvrA::frt mutS::frt sulA::frt lexA::frt recA::frt |
| EVP122/3 | recA | FBG151/2 uvrA::frt mutS::frt recA::frt |
| EVP146/7 | recO | FBG151/2 uvrA::frt mutS::frt recO::frt |
| EVP268/9 | dinB | FBG151/2 uvrA::frt mutS::frt dinB::frt |
| EVP290 | Oc-umuD'C | FBG152 uvrA::frt mutS::frt Oc-umuD‘C |
| EVP363/4 | polB umuDC | FBG151/2 uvrA::frt mutS::frt polB::frt recA::frt, umuDC::frt |
All strains carry plasmid either pVP135 (kanamycin resistance) or pKN13 (chloramphenicol resistance) that allows the expression of the phage lambda integrase-excisionase under the control of IPTG. Following the site-specific recombination reaction, the lesions [G-BaP, G-AAF, TT-CPD or TT(6-4)] are located either in the lagging strand of strain FBG151 or in the leading strand of strain FBG152. As we have not observed any difference in DDT pathways during bypass of G-BaP and AAF in lagging or leading strands, we plotted in the graphs average value of pooled data for leading and lagging strands. The DDT profile in the graph for TT-CPD and TT(6-4) contains data with the lesion located in the leading strand.
Strain expressing umuD'C from a constitutive promoter was constructed as follows: a single mutation in the SOS box (CTGT → CTAT) was introduced by site directed mutagenesis on plasmid pRW134 (20). This mutation was reported to inactivate the SOS box (21). We inserted the Kan/frt cassette from pKD13 (19) upstream of the promoter region into the PmlI restriction site. The ScaI-EcoRI fragment containing the Kan-Oc-umuD'C region was introduced into the chromosome leading to strain EVP272 (MG1655 kan::Oc-umuD'C).
Preparation of competent cells
One hundred millilitres of LB containing kanamycin (or chloramphenicol) to maintain plasmid pVP135 (or pKN13) that expresses the int-xis operon and 200 μM of IPTG are inoculated with 500 μl of an overnight starter. When the culture reaches OD600 ≈ 0.5, cells are resuspended twice in water, once in 10% glycerol and finally re-suspended in 200 μl of 10% glycerol and frozen in 40 μl aliquots.
Plasmids for integration
Vectors carrying a single lesion for integration were constructed as described previously (6) following the gap-duplex method (22). A 13-mer oligonucleotide, 5′-GCAAGTTAACACG, containing no lesion, a TT-CPD or a TT(6-4) lesion (underlined) was inserted into the gapped-duplex pGP1/2 leading to an in frame lacZ gene. A 13-mer oligonucleotide, 5′-GAAGACCTGCAGG, containing no lesion or a G-BaP lesion (underlined) was inserted into the gapped-duplex pVP143/pGP9 leading to an in frame lacZ gene. Since the G-AAF lesion can be bypassed by two distinct pathways (23), two vectors were constructed to monitor all TLS events. A 15-mer oligonucleotide containing or not a single G-AAF adduct (underlined) in the NarI site (ATCACCGGCGCCACA) was inserted into a gapped-duplex pVP141/142 or pVP143/144, leading respectively to an in frame lacZ gene, and a +2 frameshift lacZ. Therefore, the construct pVP141/142 Nar3AAF/Nar+3 monitors TLS0 events, while pVP143/144 Nar3AAF/Nar+3 monitors TLS-2 events.
Integration protocol – measurement of lesion tolerance pathways: TLS and damage avoidance (DA)
To the 40 μl aliquot of competent cells, 1ng of the lesion carrying vector mixed with 1 ng of the internal standard (pVP146) is added, and electroporated (in a GenePulser Xcell™ from BioRad, 2.5 kV – 25 μF – 200 Ω). One millilitre of SOC containing 200 μM IPTG is then added and cells are incubated 1h at 37°C. Part of the cells are plated on LB + 10 μg/ml tetracycline to measure the transformation efficiency of plasmid pVP146, and the rest is plated on LB + 50 μg/ml ampicillin and 80 μg/ml X-gal to select for integrants (Amp®) and TLS events (blue colonies). Cells were diluted and plated using the automatic serial diluter and plater easySpiral Dilute (Interscience). The integration rate is about 2000 clones per picogram of vector for our parental strain.
In order to measure TLS0, TLS-2 and DA at the G-AAF lesion in a given strain, four integration experiments are performed with the following heteroduplex vectors: pVP141/142Nar0/Nar+3, pVP141/142Nar3AAF/Nar+3, pVP143/144Nar0/Nar+3 and pVP143/144Nar3AAF/Nar+3. Following the integration of pVP141/142Nar3AAF/Nar+3, blue colonies represent TLS0 events, whereas white colonies represent the sum of TLS-2 and DA events. Similarly, the integration of pVP143/144Nar0/Nar+3 allows score TLS-2 events as blue colonies, whereas white colonies reflect the sum of TLS0 and DA events. The integration efficiencies of damaged vectors compared to their non-damaged homologs, and normalized by the transformation efficiency of pVP146 plasmid in the same electroporation experiment, give the overall rate of tolerance of the lesion. DA is scored as the overall lesion tolerance rate from which is subtracted TLS0 and TLS-2. Each experiment is repeated three to five times with at least two different batches of competent cells.
TLS at the G-BaP, TT-CPD and TT(6-4) lesions is scored by similar manner. Integrations experiments are performed with the corresponding heteroduplex vector (see above). Following the integration of the heteroduplex vector, we have scored blue colonies representing TLS0. DA is scored as the overall lesion tolerance rate from which is subtracted TLS0. Each experiment is repeated three to five times with at least two different batches of competent cells.
Expression of RecA(wt), RecA-G288Y, RecA-Q300A and RecA-N304D
Plasmids for expression of RecA-G288Y, RecA-Q300A and RecA-N304D were obtained by the cloning of a BlpI-EcoRI fragment from pGE591 recA(wt), recA-G288Y, recA-Q300A or recA-N304D into the vector pROEX carrying polyhistidine tagged recA(wt).
The plasmids were transformed into DH5α cells and proteins were expressed by growth in Luria broth medium at 37°C to OD600nm = 0.5 and induced by addition of 1 mM IPTG and further incubated at 15°C for 16 h. Cells were harvested, lysed in Ni-A buffer (20 mM Tris, pH8.0, 1 M NaCl, 10% glycerol, 20 mM imidazole and 2 mM β-mercaptoethanol), clarified by high speed centrifugation, and loaded on a column (HisTrap HP; GE Healthcare). The column was washed extensively with Ni-A buffer, and the proteins were eluted with Ni-B buffer (20 mM Tris, pH 8.0, 1 M NaCl, 10% glycerol, 250 mM imidazole and 2 mM β-mercaptoethanol). The purified protein was dialyzed against buffer D (20 mM Tris, pH 7.5, 0,3 M KCl, 10% glycerol, 1 mM EDTA and 1 mM DTT) and aliquoted and stored at −80°C.
D-loop assay
RecA(wt), RecA-G288Y, RecA-Q300A or RecA-N304D (1µM) were incubated with ssDNA (60-mer with Cy5 tag at 5′ end, modified oligonucleotide SK3 from (24): 5′-GCAGTGGCAGATTGTACTGAGAGTGCACCATATGCGGTGTGAAATACCGCACAGATGCGT-3′) in 30 mM Tris–HCl (pH7,5), 1 mM ATPγS, 100 μg/ml BSA, 1 mM DTT, 15 mM KCl (added with the recA stock) and 10 mM MgCl2 for 5 min to allow filament formation. The addition of equimolar quantity of pUC19 to incubation mixture initiated D-loop formation. The samples were incubated from 0 to 40 min at 37°C. The reaction was stopped by adding 1 mg/ml final of proteinase K and 1% (w/v) SDS and incubating by 10 min at 37°C. Reaction products were fractionated by 0.6% agarose gel electrophoresis in Tris–borate buffer.
Gel retardation assay
Different concentration of RecA(wt), RecA-G288Y, RecA-Q300A or RecA-N304D (0–2 μM) was incubated with ssDNA oligonucleotide-modified SK3 mentioned above (0.05 μM) in 30 mM Tris–HCl (pH 7.5), 1 mM ATPγS, 100 μg/ml BSA, 1 mM DTT, 0–30 mM KCl (added with the recA stock) and 10 mM MgCl2 for 15 min to allow filament formation. Reaction products were fractionated by 10% acrylamide gel electrophoresis in Tris–borate buffer.
Immunoblots
Strains EVP23, EVP113, EVP123+recA-G288Y, EVP123+recA-Q300A, EVP123+recA-N304D, EVP123+recA, EVP118+recA-G288Y, EVP118+recA-Q300A, EVP118+recA-N304D and EVP118-recA were grown to exponential phase. Cells were lysed for 30 min at 37°C in lysis buffer (50 mM Tris–HCl pH 7.5; 100 mM KCl; 1 mM EDTA, 1 mM DTT; 10% glycerol; 0.1% NP40 substitute, 0.1 mM PMSF; 20 μg/ml lysozyme; 10 μg/ml RNAse; protease inhibitors from Roche). The samples were sonicated and the supernatant was stored at −80°C.
Samples were diluted in ratio 1:1 with Laemmli sample buffer (62.5 mM Tris–HCl pH 6.8; 25% glycerol; 2% SDS; 5% β-mercapto ethanol; bromophenol blue), boiled 4 min.
RecA immunoblot: The equivalent of 0.002 OD600 were loaded on a MiniProtean TGX strain-free any Kd gel (BioRad). Anti-RecA antibody from Abcam® (ab63797) was used.
UmuD immunoblot: The equivalent of 0.2 OD600 were loaded on a MiniProtean TGX strain-free any Kd gel (BioRad). Purified anti-UmuD antibody provided by R. Woodgate was used.
LexA immunoblot: The equivalent of 0.2 OD600 were loaded on a MiniProtean TGX strain-free any Kd gel (BioRad). Anti-LexA antibody LexA(D-19) from Santa Cruz Biotechnology was used.
Irradiation by UV light
Cells were grown to OD600 = 0.4, then resuspended and diluted 20 times in 10 mM MgSO4. Dilluted cells were irradiated with 254-nm UV light by different doses: 0.25–120 J/m2. Cells were plated on LB-agar plates and their survival compared to non-irradiated control.
RESULTS
Characterization of putative recA alleles, impaired in homologous recombination and proficient in SOS induction
The RecA protein plays multiple roles in cells: it is the main player in homologous recombination, it regulates the DNA Damage Response (through SOS induction) and is essential for activation of the major, Pol V-mediated TLS pathway (25–27). In a recent study, Lichtarge et al. (16) applied a phylogenomic method, called Evolutionary Trace, to ≈200 bacterial RecA homologs to map the conserved residues at the surface of the protein. Based on this approach the authors identified a class of three recA point mutants with a specific defect in HR but not in SOS induction (ET-site2 separation of function mutants). These authors used P1 transduction efficiency to monitor HR. The authors found that three recA alleles (G288Y, Q300A and N304D) were strongly deficient in HR (less than 20% of recA+ activity). Analysis of whole cell extracts of all three recA strains by western blot (WB), shows normal LexA cleavage and RecA up-regulation following SOS induction by nalidixic acid as in a wild-type recA (16). We decided to express, purify and characterize these three alleles biochemically. In addition, we also provide further in vivo characterization of these recA allele strains.
In vitro characterisation
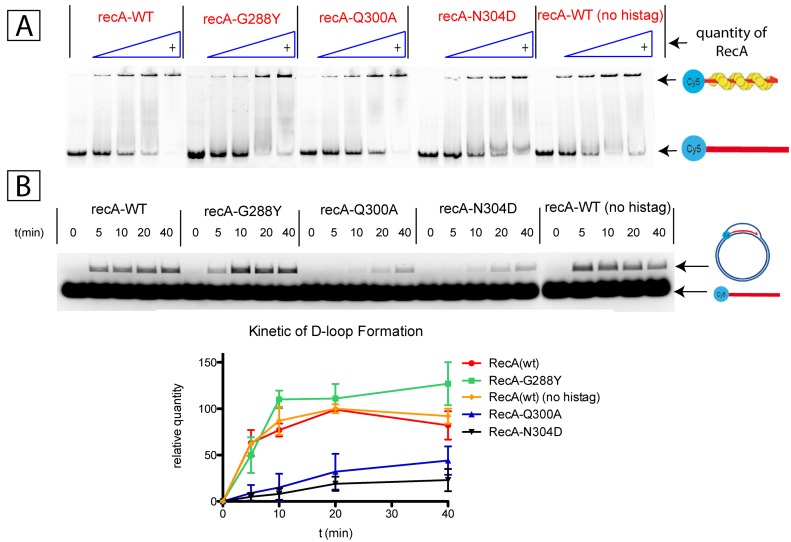
In order to further characterize these alleles, we expressed, purified N-terminally histidine-tagged RecA mutant proteins and tested their ability to form single-stranded DNA–RecA filament by incubating a single-stranded 60-mer oligonucleotide with increasing concentrations of RecA protein (Figure 1A). ssDNA–RecA filaments were visualised by retardation on acrylamide gel electrophoresis. All three RecA alleles (G288Y, Q300A and N304D) exhibit a RecA concentration dependent ssDNA filament formation similar to tagged or non-tagged wild type RecA+ (Figure 1A).
Figure 1.
(A) Gel retardation assay demonstrating the interaction between ssDNA oligonucleotide and increasing quantity of his-tagged forms of RecA(wt), RecA-G288Y, RecA-Q300A, RecA-N304D and non-tagged RecA(wt). 0.05μM ssDNA (60mer) was incubated with 0; 0.25; 0.5; 1 and 2 μM RecA-WT or RecA allele. RecA interaction with oligonucleotide is affected neither by the specific recA mutations nor by the presence of polyhistidine tag. (B) Kinetic of strand exchange (D-loop formation) promoted by his-tagged forms of RecA(wt), RecA-G288Y, RecA-Q300A, RecA-N304D and non-tagged RecA(wt). The D-loop formation was analyzed 0, 5, 10, 20 and 40 min after initiation of dsDNA uptake into recA filament. The graph represents the average and standard deviation of three independent experiments.
The different RecA alleles were then tested for their capacity to form D-loops using a 5′-Cy5-end-labelled 60-mer oligonucleotide pre-incubated with RecA and a supercoiled plasmid that contains a region of sequence homology with the oligonucleotide (Figure 1B) (24,28). The kinetics of D-loop formation was monitored by the appearance of fluorescently labelled plasmid DNA. Both Q300A and N304D RecA alleles exhibit a strongly reduced capacity of D-loop formation compared to wild type RecA. In contrast, allele G288Y appears to be slightly more efficient than wt RecA for D-loop formation in good agreement with its UV survival phenotype (see below).
In vivo characterisation
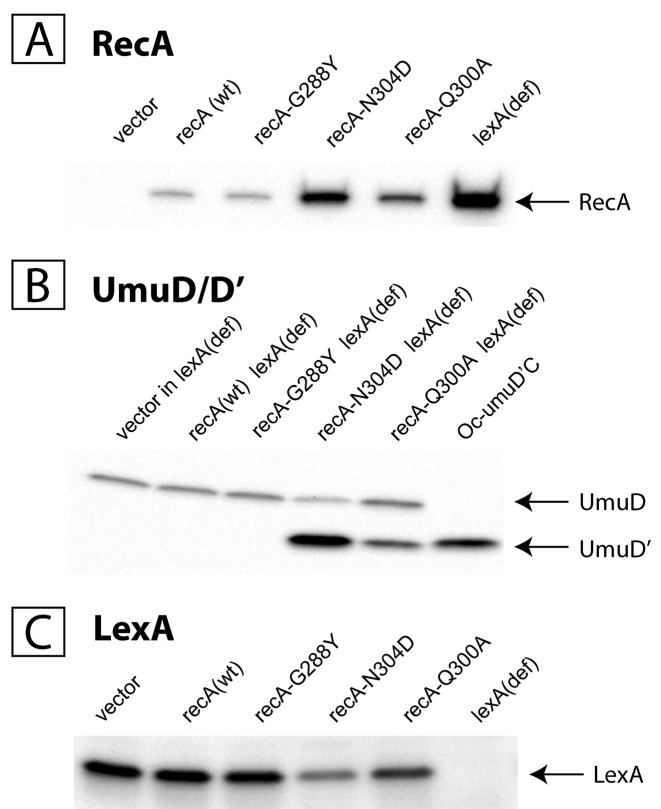
The different recA strains were characterized with respect to their UV sensitivity by measuring the fraction of colonies surviving following exposure at a given UV dose compared to the non-irradiated strain (Table 2). Strains expressing Q300A and N304D recA alleles exhibit severe UV sensitivity while the strain expressing recA G288Y was found to be at least as UV-resistant as a wild type recA strain, in good agreement with Adikesavan et al. (16). Strains expressing Q300A and N304D RecA alleles exhibit a partially induced SOS phenotype in the absence of genotoxic stress as evidenced by RecA protein expression level, as well as LexA and UmuD cleavage (Figure 2A–C). Indeed, the level of RecA protein is significantly increased in recA N304D and to a lesser extent in Q300A strains, while it remains expressed at wild-type levels in G288Y. Similarly, when analysing the status of UmuD/D’ by WB in whole cell extracts, the extent of UmuD cleavage is significantly increased in recA N304D and to a lesser extent in Q300A strains. Conversely, the amount of uncleaved LexA protein, as detected by WB, is meaningfully lower in strains N304D and less so in strain Q300A compared to wild type. Taken together these data suggest that recA strain N304D and to a lesser extent strain Q300A are partially SOS-induced. These results are in contrast with the observations of Lichtarge et al. (16) who did not observe an increase in RecA expression nor in LexA cleavage in strains N304D and Q300A. This difference may be ascribed to the fact that our strains are all nucleotide excision repair defective (uvrA), a situation known to trigger spontaneous SOS induction (29).
Table 2. UV survival of recA allele strains.
| ΔrecA | recA+ | G288Y | Q300A | N304D | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Dose (J/m2) | 0.25 | 0.75 | 60 | 90 | 90 | 120 | 3 | 5 | 1 | 2 |
| Survival (%) | 20% | 1.6% | 40% | 18% | 43% | 35% | 20% | 8.5% | 15% | 4% |
Figure 2.
Expression of RecA (A) cleavage of UmuD (B) and of LexA (C) in vivo. All cell extracts are made from cultures grown under standard conditions without genotoxic stress. (A) The level of expression of RecA in recA-Q300A and N304D extracts is intermediate between wt recA+ and lexA(Def) strains, illustrating partial SOS induction in Q300A and N304D strains. No induction is seen in allele G288Y. A similar conclusion can be reached from experiments monitoring UmuD cleavage into UmuD’ (B) or the disappearance of the intact form of LexA (C). All three assays suggest a relatively modest SOS induction in strain Q300A and a more robust one in strain N304D. oc-umuD'C is a strain that expresses chromosomal umuD'C under the control of a constitutive promoter.
In conclusion, the D-loop formation data suggest that recA alleles Q300A and N304D are likely to be affected in gap repair while allele G288Y appears to be wild type. The D-loop formation ability parallels the UV sensitivity data as both Q300A and N304D are UV sensitive while G288Y is not. In agreement with published data (16), the present biochemical and in vivo characterization of the recA alleles suggest that both Q300A and N304D recA alleles are partially deficient in homologous recombination. In addition, most likely due to increased persistence of the single-stranded DNA RecA-filament, these strains exhibit a partially-induced SOS phenotype.
We would like to stress that recA allele G288Y represents an interesting separation-of-function allele proficient in gap-filling repair but impaired in double-strand break repair. Indeed all three recA alleles were initially found to be impaired in P1 transduction (16). While P1 transduction involves the capacity of RecA to promote double-strand break repair via the recBCD pathway, resistance to UV relates to post-replicative gap-filling repair via the recF pathway.
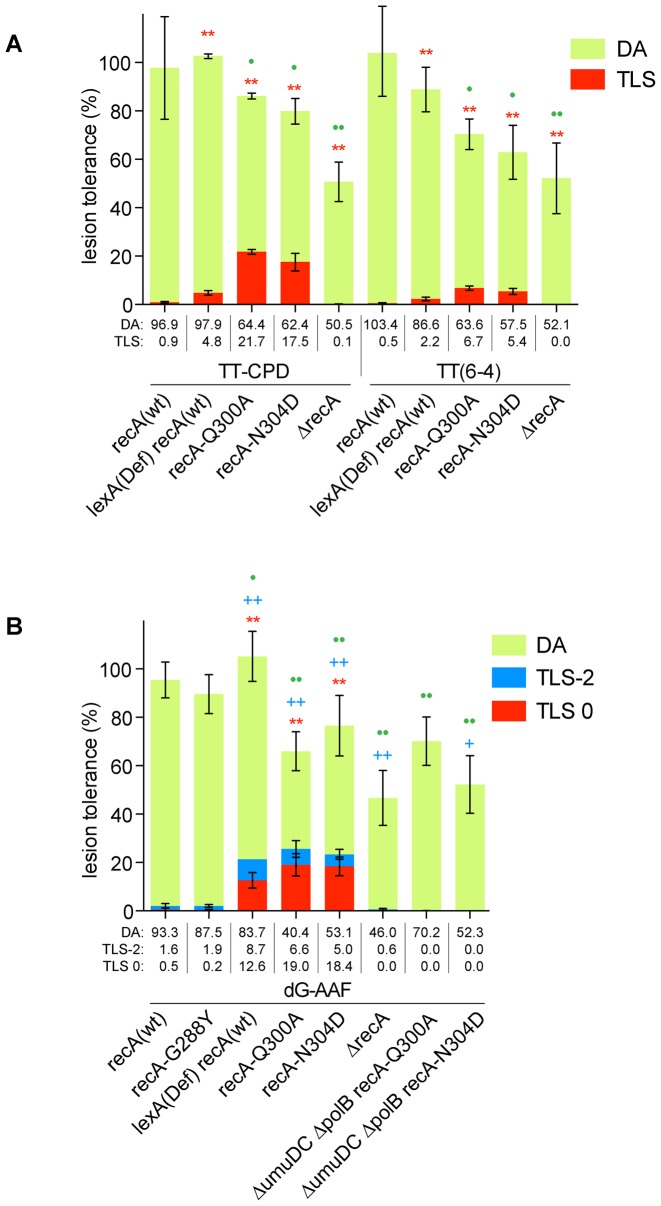
RecA alleles (Q300A and N304D) exhibit a robust increase in TLS linked to a decrease in Damage Avoidance (Figure 3A and B)
Figure 3.
DNA Damage tolerance of typical replication blocking lesions in partially recombination defective strains: partition between TLS and damage avoidance (DA): (A) major UV lesions, TT-CPD and TT(6-4) and (B) G-AAF. Tolerance events (%) are determined as the efficiency of site-specific integration of a single lesion-containing construct relative to the same amount of damage-free control construct. DA and TLS events are determined as specified in Materials and Methods. DDT profiles in ΔrecA strain were published previously in (6) and found to be mainly due to bacteria that could divide and proliferate following replication of the non-damaged chromatid only (1). Tolerance events for G-AAF in wt was previously published in (7). The data represent the average value and standard deviation of at least 3 independent experiments. T-test was performed to compare values from the different mutants to the recA(wt) strain. For TLS0: *P < 0.05; **P < 0.005. For TLS-2: +P < 0.05; ++P < 0.005; for DA: •P < 0.05; ••P < 0.005.
In order to analyse the pathway choice between TLS and DA, we introduced a single replication-blocking lesion at a specific locus in the E. coli genome via the phage lambda site-specific recombination reaction (6). Throughout the paper we use a strain deficient in both nucleotide excision repair (uvrA mutation) and in mismatch repair (mutS) to prevent the repair of the single lesion and of the strand marker located opposite the lesion. This strain is thereafter referred to as the parental strain. We determined the proportion of DDT events for three strong replication blocks, namely N-2-acetylaminofluorene (AAF) covalently linked to the C-8 position of guanine (G-AAF), and two major UV-induced lesions cyclobutane pyrimidine dimer (TT-CPD) and thymine-thymine pyrimidine(6-4)pyrimidone photoproduct TT(6-4). Compared to wild-type recA, alleles Q300A and N304D show a strong increase in TLS events for all three lesions. For the two UV-induced lesions, the increase in TLS between the parental and the two recA alleles (Q300A and N304D) strains is ≈ 20 and 10-fold for TT-CPD and TT(6-4), respectively (Figure 3A). The stimulation of TLS by the two recA alleles (Q300A and N304D) is actually much stronger than the stimulation observed in a lexA(Def) strain (4–5-fold). It should also be noted that there is a modest but clear reduction in the extent of survival in the two recA alleles (70–80%). This reduction is less severe than in a delta recA strain (40–50% reduction) (6). Not surprisingly, TLS is fully defective in a delta recA strain due to the absolute requirement of RecA for Pol V activation (25–27).
We also measured the tolerance of a G-AAF adduct located within the NarI sequence context in the recombination defective recA alleles (Figure 3B). In the NarI sequence context, the G-AAF adduct is bypassed either via an error-free pathway (TLS0) by Pol V or through a −2 frameshift pathway (TLS-2) when the adduct is bypassed by Pol II (23). Compared to the parental strain, in recA allele strains Q300A and N304D, TLS events are stimulated, >20-fold and ≈5 fold for TLS0 and TLS-2, respectively. In agreement with the properties described above, allele G288Y behaves like wt recA (Figure 3B). Both TLS pathways are fully suppressed upon inactivation of the corresponding TLS polymerase genes umuDC (Pol V) and polB (Pol II) (Figure 3B).
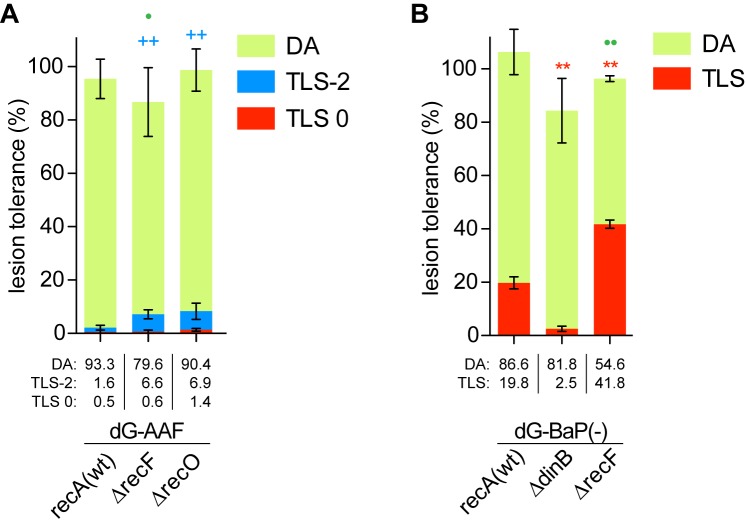
DDT profile in a recF background
In E. coli, recA-mediated homologous recombination entails two major sub-pathways, namely the recF and recBCD pathways. While the recBCD pathway mostly mediates double strand break repair (30–32), the recF pathway is mainly involved in post replicative daughter strand gap filling repair (17,33). On a biochemical level, the RecFOR proteins are known to accelerate single-stranded DNA-RecA filament formation by mediating the displacement of SSB (17). Therefore, recF strains exhibit a delay in the SOS response (34–36). We have previously shown that UV-induced mutagenesis is strongly reduced in recFOR strains using both a chromosomal rifR assay and a plasmid-borne single lesion bypass assay; this effect reflects a delay in Pol V activation by RecA (37,38). In contrast, as the TLS activity of Pol II does not depend on RecA, Pol II mediated TLS events should not be affected by delayed single-stranded RecA filament formation. In fact, rather than being decreased, we observe that TLS-2 events are increased about 4-fold in a recF strain compared to wild type (Figure 4A). A similar increase in TLS-2 is also observed in a recO strain (Figure 4A). In an attempt to generalize the notion that TLS may increase in a recF strain as a result of increased persistence of the TLS substrate, we investigated the DDT profile of another lesion that solely relies on Pol IV. Metabolically activated benzo(a)pyrene (BaP) reacts with the N2 position of guanine forming two stereoisomeric adducts, (−) trans-anti-BaP-N2dG and (+) trans-anti-BaP-N2dG, respectively (39). The (−) trans-anti-BaP-N2dG adduct (abbreviated dG-BaP(−)) is specifically bypassed by Pol IV (39–41). In a recF strain, TLS across the dG-BaP(−) adduct increases from 20% in the wild type strain to 40% in the corresponding recF strain (Figure 4B).
Figure 4.
DNA damage tolerance of (A) G-AAF and (B) BaP(−) in the strains delayed for recombination: partition between TLS and DA. Tolerance events (%) are determined as the efficiency of site-specific integration of a single lesion-containing construct relative to the same amount of damage-free control construct. DA and TLS events are determined as specified in Materials and Methods section. The data represent the average value and standard deviation of at least three independent experiments. t-test was performed to compare values from the different mutants to the recA(wt) strain. For TLS0: *P < 0.05; **P < 0.005. For TLS-2: +P < 0.05; ++P < 0.005; for DA: •P < 0.05; ••P < 0.005.
DISCUSSION AND CONCLUSIONS
The aim of the present work is to investigate the interplay between the two major DNA Damage Tolerance pathways, namely between TLS and HDGR. When a lesion blocks the advancing replicative DNA polymerase, on-going unwinding of the parental DNA strands by the replicative helicase and downstream re-priming (10,42) generate ssDNA gaps. The size of these gaps has been estimated to be in the range of 1–2 kb on the leading strand and of smaller size on the lagging strand (9). These gaps, initially covered by SSB, are converted into ssDNA.RecA filament mainly with the help of the RecFOR mediator proteins. Subsequently, the gaps are either filled-in by translesion synthesis (TLS) or repaired via homology directed gap repair (HDGR).
Filling of these gaps by TLS requires a specialized DNA polymerase to bind and extend the primer terminus at the lesion site. Binding of the TLS polymerase to the β-clamp confers them limited processivity that is essential to produce a short patch of DNA synthesis across and beyond the lesion (‘TLS patch’). The patch made by the TLS polymerase needs to be long enough to escape degradation by the proofreading function associated with the replicative DNA polymerase (43). Following that step, a replicative DNA polymerase, possibly Pol III core, will take over to complete synthesis of the bulk of the gap.
On the other hand, the repair of the gaps by recombination involves a strand exchange reaction with the sister chromatid that is clearly dependent upon RecA (1,15). While the different steps involved in the HDGR pathway are not yet known precisely, it is likely that the first step involves a strand invasion of the ssDNA.RecA filament into the sister chromatid thus forming a D-loop. In addition, the RecA protein provides an overall stabilisation of the transiently stalled replication fork (44,45). Thus to investigate the effect of RecA on HDGR we used recA allele strains that are specifically defective in strand invasion.
In E. coli, TLS events always represent a minor DNA damage tolerance pathway compared to DA. Under ‘normal’, non-stressed, conditions, the basal level of TLS polymerase only allows 1–2% of the gaps to be filled by TLS. Induction of the SOS system triggers TLS polymerase expression and leads to a ≈10-fold increase in TLS events. In the absence of TLS polymerase expression, i.e. in TLS deletion mutants, all tolerance events result from DA ((7) and Figure 3B). In contrast, when the TLS polymerases are overexpressed TLS events can reach 100% (7).
Interplay between these two pathways entails a strong chronological component, TLS acting first followed by DA (7). In the present paper, we implemented two sets of experiments aimed at impairment of the onset of HR, first we used recA point mutation alleles that are deficient in D-loop formation (Figure 3) and second we used recFOR mutants known to exhibit a slower kinetics in ssDNA.RecA filament formation (Figure 4).
recA alleles Q300A and N304D are deficient in D-loop formation in comparison to wild-type recA. For both UV-induced lesions TT-CPD and TT(6-4), as well as for G-AAF we observe a robust (10–20-fold) increase in TLS. It should be noted that the overall level of survival is slightly affected in the recA Q300A and N304D strains (70-80%, Figure 3A and B), implying that the partial defect in recombination is largely but not fully compensated by the increase in TLS. The increase in TLS can be attributed to the increase in lifetime of the ssDNA.RecA filament, the so-called ‘SOS signal’. In fact, the increased persistence of the SOS signal only partially triggers SOS induction as judged by the level of RecA expression, and the level of UmuD and LexA cleavage (Figure 2A–C). We propose that the increased persistence of the SOS signal favors TLS by increasing both the TLS polymerase concentration and by extending the lifetime of the TLS substrate, as confirmed by data in the recF strain (see below). Subsequently, when the substrate for TLS becomes sequestered by D-loop formation, the TLS pathway is shut off.
The role of increased persistence of the TLS substrate on the extent of TLS was directly evidenced in recF (or recO) strains. Upon formation of ssDNA gaps downstream from replication-blocking lesions, the so-called RecFOR mediator proteins facilitate the formation of the ssDNA.RecA filament by helping SSB protein displacement (17). In a recF (or recO) strain, ssDNA.RecA filament formation is thus retarded leading to a delay in HR-mediated gap repair. We tested the time window hypothesis for lesions that rely on either DNA Pol II or Pol IV for TLS as these two polymerases, in contrast to Pol V, exhibit a significant basal expression under non-SOS induced conditions (35 molecules of Pol II/cell; 250 molecules of Pol IV/cell) and do not require RecA for activation (45). Both Pol II-dependent bypass of G-AAF and Pol IV-dependent bypass of dG-BaP(−) are strongly increased in a recF strain compared to the parental strain.
In a strain totally devoid of RecA protein, i.e. in a recA deletion strain [Figure 3, (6)], one might have expected that, in the absence of HR, the time window for TLS would remain open much longer thus leading to even higher levels of TLS. However, this is not the case since in a recA strain, we observe a strongly decreased overall survival level together with a relatively low fraction of TLS for both Pol II-dependent bypass of G-AAF and PolIV-dependent bypass of dG-BaP(-). We suggest that since the RecA protein is structurally required to prevent fork collapse (44,45), it confers stability to the post-replicative gaps that is essential for both TLS and HDGR pathways.
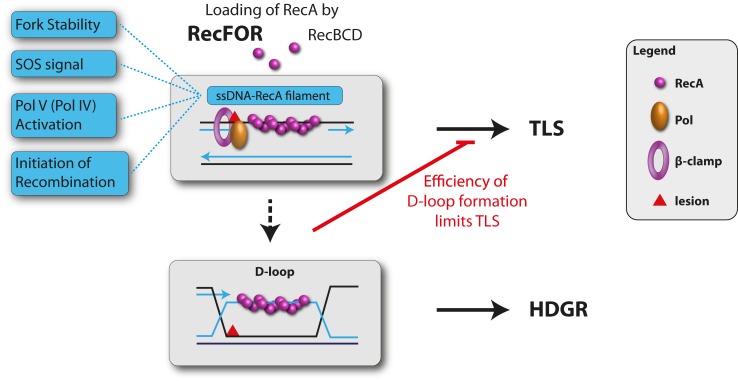
Taken together, our data suggest the following scenario (Figure 5): when the genome is damaged, the replication skips over replication-blocking lesions via downstream re-priming leaving single-stranded gaps behind the fork. These single-stranded DNA gaps become converted into ssDNA.RecA filaments, the so-called SOS signal; loading of RecA on single-stranded DNA gaps is catalyzed by RecFOR recombination mediator proteins that help displace SSB from ssDNA. Recent evidence suggests that RecBCD partially contributes to ssDNA.RecA filament formation as well (47). Once formed, the ssDNA.RecA filament plays several important roles: (i) SOS induction leading to increased TLS polymerase expression, (ii) activation of Pol V, (iii) stability of the fork and (iv) initiation of homologous recombination. In the present paper we show that TLS is increased in strains carrying recA alleles that are partially defective in D-loop formation. This increase in TLS is due to increased persistence of the SOS signal that in turn induces both TLS polymerase expression and the time during which the TLS substrate persists. The role of increased persistence of the TLS substrate on the extent of TLS, was directly evidenced in recF (or recO) strains. As soon as the ssDNA.RecA filament invades the homologous sister chromatid and forms a D-loop, i.e. an early HDGR intermediate, the TLS reaction is turned off by mere substrate sequestration. This model defines a chronological competition between TLS and HDGR, the time window for TLS closes upon initiation of recombination.
Figure 5.
Crosstalk between TLS and HDGR: When the fork encounters a replication-blocking lesion in one of the template strands, it skips over the lesion via downstream re-priming leaving a single-stranded gap. These single-stranded DNA gaps become converted into ssDNA.RecA filaments, the so-called SOS signal; loading of RecA on single-stranded DNA gaps is catalyzed by RecFOR recombination mediator proteins that help displace SSB from ssDNA. Recent evidence suggests that RecBCD partially contributes to ssDNA.RecA filament formation as well (47). Once formed, the ssDNA.RecA filament plays several important roles: (i) SOS induction leading to increased TLS polymerase expression, (ii) activation of Pol V, i(ii) stability of the fork and (iv) initiation of homologous recombination. In the present paper we show that TLS is increased in strains carrying recA alleles that are partially defective in D-loop formation. This increase in TLS is due to increased persistence of the ssDNA.RecA filament that in turn induces both TLS polymerase expression and the time during which the TLS substrate persists. The role of increased persistence of the TLS substrate on the extent of TLS, was also directly evidenced in recF (or recO) strains (see text). As soon as the ssDNA.RecA filament invades the homologous sister chromatid and forms a D-loop, i.e. an early HDGR intermediate, the TLS reaction is turned off by mere substrate sequestration. This model defines a chronological competition between TLS and HDGR, the time window for TLS closes upon initiation of recombination.
Acknowledgments
We thank our team members Gaëlle Philippin for the construction of plasmids and Luisa Laureti for critical reading of the manuscript. Edwige Garcin, Stéphanie Bernard and Mauro Modesti (CRCM) are gratefully acknowledged for their help with RecA modelling and purification. O. Lichtarge (Baylor College of Medecine, Houston) is acknowledged for the RecA allele plasmids. R. Woodgate is acknowledged for antibody against UmuD.
Footnotes
Present address: Robert P. Fuchs, Harvard Medical School, Boston, MA, USA.
FUNDING
Équipe labellisée Ligue Contre le Cancer; Agence Nationale de la Recherche (ANR) grant ForkRepair [ANR 11 BSV8 017 01]; GenoBlock [ANR-14-CE09-0010-01]; Fondation ARC pour la recherche sur le cancer (to K.N.). Funding for open access charge: ANR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Laureti L., Demol J., Fuchs R.P., Pagès V. Bacterial proliferation: keep dividing and don't mind the gap. PLoS Genet. 2015;11:e1005757. doi: 10.1371/journal.pgen.1005757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedberg E.C. Suffering in silence: the tolerance of DNA damage. Nat. Rev. Mol. Cell Biol. 2005;6:943–953. doi: 10.1038/nrm1781. [DOI] [PubMed] [Google Scholar]
- 3.Sale J.E. Competition, collaboration and coordination – determining how cells bypass DNA damage. J. Cell Sci. 2012;125:1633–1643. doi: 10.1242/jcs.094748. [DOI] [PubMed] [Google Scholar]
- 4.Rupp W.D., Wilde C.E., Reno D.L., Howard-Flanders P. Exchanges between DNA strands in ultraviolet-irradiated Escherichia coli. J. Mol. Biol. 1971;61:25–44. doi: 10.1016/0022-2836(71)90204-x. [DOI] [PubMed] [Google Scholar]
- 5.Lusetti S.L., Cox M.M. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu. Rev. Biochem. 2002;71:71–100. doi: 10.1146/annurev.biochem.71.083101.133940. [DOI] [PubMed] [Google Scholar]
- 6.Pagès V., Mazón G., Naiman K., Philippin G., Fuchs R.P. Monitoring bypass of single replication-blocking lesions by damage avoidance in the Escherichia coli chromosome. Nucleic Acids Res. 2012;40:9036–9043. doi: 10.1093/nar/gks675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naiman K., Philippin G., Fuchs R.P., Pagès V. Chronology in lesion tolerance gives priority to genetic variability. Proc. Natl. Acad. Sci. U.S.A. 2014;111:5526–5531. doi: 10.1073/pnas.1321008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pagès V., Fuchs R.P. Uncoupling of leading- and lagging-strand DNA replication during lesion bypass in vivo. Science. 2003;300:1300–1303. doi: 10.1126/science.1083964. [DOI] [PubMed] [Google Scholar]
- 9.Higuchi K., Katayama T., Iwai S., Hidaka M., Horiuchi T., Maki H. Fate of DNA replication fork encountering a single DNA lesion during oriC plasmid DNA replication in vitro. Genes Cells. 2003;8:437–449. doi: 10.1046/j.1365-2443.2003.00646.x. [DOI] [PubMed] [Google Scholar]
- 10.Yeeles J.T.P., Marians K.J. Dynamics of leading-strand lesion skipping by the replisome. Mol. Cell. 2013;52:855–865. doi: 10.1016/j.molcel.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rupp W.D. Early days of DNA repair: discovery of nucleotide excision repair and homology-dependent recombinational repair. Yale J. Biol. Med. 2013;86:499–505. [PMC free article] [PubMed] [Google Scholar]
- 12.Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kowalczykowski S.C., Dixon D.A, Eggleston A.K., Lauder S.D., Rehrauer W.M. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McEntee K., Weinstock G.M., Lehman I.R. Initiation of general recombination catalyzed in vitro by the recA protein of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1979;76:2615–2619. doi: 10.1073/pnas.76.6.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith GR. Mechanism and control of homologous recombination in Escherichia coli. Annu. Rev. Genet. 1987;21:179–201. doi: 10.1146/annurev.ge.21.120187.001143. [DOI] [PubMed] [Google Scholar]
- 16.Adikesavan A.K., Katsonis P., Marciano D.C., Lua R., Herman C., Lichtarge O. Separation of recombination and SOS response in Escherichia coli RecA suggests LexA interaction sites. PLoS Genet. 2011;7:e1002244. doi: 10.1371/journal.pgen.1002244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morimatsu K., Kowalczykowski S.C. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: A universal step of recombinational repair. Mol. Cell. 2003;11:1337–1347. doi: 10.1016/s1097-2765(03)00188-6. [DOI] [PubMed] [Google Scholar]
- 18.Esnault E., Valens M., Espéli O., B F. Chromosome structuring limits genome plasticity in Escherichia coli. PLoS Genet. 2007;3:e226. doi: 10.1371/journal.pgen.0030226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ennis D.G., Levine A.S., Koch W.H., Woodgate R. Analysis of recA mutants with altered SOS functions. Mutat. Res. DNA Repair. 1995;336:39–48. doi: 10.1016/0921-8777(94)00045-8. [DOI] [PubMed] [Google Scholar]
- 21.Sommer S., Knezevic J., Bailone A., Devoret R. Induction of only one SOS operon, umuDC, is required for SOS mutagenesis in Escherichia coli. Mol. Gen. Genet. 1993;239:137–144. doi: 10.1007/BF00281612. [DOI] [PubMed] [Google Scholar]
- 22.Koehl P., Burnouf D., Fuchs R.P. Construction of plasmids containing a unique acetylaminofluorene adduct located within a mutation hot spot. A new probe for frameshift mutagenesis. J. Mol. Biol. 1989;207:355–364. doi: 10.1016/0022-2836(89)90259-3. [DOI] [PubMed] [Google Scholar]
- 23.Napolitano R., Janel-Bintz R., Wagner J., Fuchs R.P. All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J. 2000;19:6259–6265. doi: 10.1093/emboj/19.22.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazin A.V., Zaitseva E., Sung P., Kowalczykowski S.C. Tailed duplex DNA is the preferred substrate for Rad51 protein-mediated homologous pairing. EMBO J. 2000;19:1148–1156. doi: 10.1093/emboj/19.5.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujii S., Gasser V., Fuchs R.P. The biochemical requirements of DNA polymerase V-mediated translesion synthesis revisited. J. Mol. Biol. 2004;341:405–417. doi: 10.1016/j.jmb.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 26.Nohmi T., Battista J.R., Dodson L.A., Walker G.C. RecA-mediated cleavage activates UmuD for mutagenesis: mechanistic relationship between transcriptional derepression and posttranslational activation. Proc. Natl. Acad. Sci. U.S.A. 1988;85:1816–1820. doi: 10.1073/pnas.85.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang Q., Karata K., Woodgate R., Cox M.M., Goodman M.F. The active form of DNA polymerase V is UmuD’(2)C-RecA-ATP. Nature. 2009;460:359–363. doi: 10.1038/nature08178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konforti B.B., Davis R.W. DNA substrate requirements for stable joint molecule formation by the RecA and single-stranded DNA-binding proteins of Escherichia coli. J. Biol. Chem. 1991;266:10112–10121. [PubMed] [Google Scholar]
- 29.Foster P.L. Escherichia coli strains with multiple DNA repair defects are hyperinduced for the SOS response. J. Bacteriol. 1990;172:4719–4720. doi: 10.1128/jb.172.8.4719-4720.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuzminov A., Stahl F.W. Double-strand end repair via the RecBC pathway in Escherichia coli primes DNA replication Double-strand end repair via the RecBC pathway in Escherichia coli primes DNA replication. Genes Dev. 1999;13:345–356. doi: 10.1101/gad.13.3.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Churchill J.J., Anderson D.G., Kowalczykowski S.C. The RecBC enzyme loads recA protein onto ssDNA asymmetrically and independently of chi, resulting in constitutive recombination activation. Genes Dev. 1999;13:901–911. doi: 10.1101/gad.13.7.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson D.G., Kowalczykowski S.C. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a chi-regulated manner. Cell. 1997;90:77–86. doi: 10.1016/s0092-8674(00)80315-3. [DOI] [PubMed] [Google Scholar]
- 33.Courcelle J., Carswell-Crumpton C., Hanawalt P.C. recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1997;94:3714–3719. doi: 10.1073/pnas.94.8.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thoms B., Wackernagel W. Regulatory role of recF in the SOS response of Escherichia coli: Impaired induction of SOS genes by UV irradiation and nalidixic acid in a recF mutant. J. Bacteriol. 1987;169:1731–1736. doi: 10.1128/jb.169.4.1731-1736.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whitby M.C., Lloyd R.G. Altered SOS induction associated with mutations in recF, recO and recR. Mol. Gen. Genet. 1995;246:174–179. doi: 10.1007/BF00294680. [DOI] [PubMed] [Google Scholar]
- 36.Hegde S., Sandler S.J., Clark A.J., Madiraju M.V.V.S. recO and recR mutations delay induction of the SOS response in Escherichia coli. MGG Mol. Gen. Genet. 1995;246:254–258. doi: 10.1007/BF00294689. [DOI] [PubMed] [Google Scholar]
- 37.Fujii S., Isogawa A., Fuchs R.P. RecFOR proteins are essential for Pol V-mediated translesion synthesis and mutagenesis. EMBO J. 2006;25:5754–5763. doi: 10.1038/sj.emboj.7601474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujii S., Fuchs R.P. Biochemical basis for the essential genetic requirements of RecA and the beta-clamp in Pol V activation. Proc. Natl. Acad. Sci. U.S.A. 2009;106:14825–14830. doi: 10.1073/pnas.0905855106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seo K.Y., Nagalingam A., Miri S., Yin J., Chandani S., Kolbanovskiy A., Shastry A., Loechler E.L. Mirror image stereoisomers of the major benzo[a]pyrene N2-dG adduct are bypassed by different lesion-bypass DNA polymerases in E. coli. DNA Repair (Amst.) 2006;5:515–522. doi: 10.1016/j.dnarep.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 40.Shen X., Sayer J.M., Kroth H., Ponten I., O'Donnell M., Woodgate R., Jerina D.M., Goodman M.F. Efficiency and accuracy of SOS-induced DNA polymerases replicating benzo[a]pyrene-7, 8-diol 9, 10-epoxide A and G adducts. J. Biol. Chem. 2002;277:5265–5274. doi: 10.1074/jbc.M109575200. [DOI] [PubMed] [Google Scholar]
- 41.Ikeda M., Furukohri A., Philippin G., Loechler E., Akiyama M.T., Katayama T., Fuchs R.P., Maki H. DNA polymerase IV mediates efficient and quick recovery of replication forks stalled at N2-dG adducts. Nucleic Acids Res. 2014;42:8461–8472. doi: 10.1093/nar/gku547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heller R.C., Marians K.J. Replication fork reactivation downstream of a blocked nascent leading strand. Nature. 2006;439:557–562. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]
- 43.Fujii S., Fuchs R.P. Defining the position of the switches between replicative and bypass DNA polymerases. EMBO J. 2004;23:4342–4352. doi: 10.1038/sj.emboj.7600438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Courcelle J., Hanawalt P.C. RecA-dependent recovery of arrested DNA replication forks. Annu. Rev. Genet. 2003;37:611–646. doi: 10.1146/annurev.genet.37.110801.142616. [DOI] [PubMed] [Google Scholar]
- 45.Fuchs R.P., Fujii S. Translesion DNA synthesis and mutagenesis in prokaryotes. Cold Spring Harb. Perspect. Biol. 2013;5:1–22. doi: 10.1101/cshperspect.a012682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robu M.E., Inman R.B., Cox M.M. RecA protein promotes the regression of stalled replication forks in vitro. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8211–8218. doi: 10.1073/pnas.131022698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pagès V. Single-strand gap repair involves both RecF and RecBCD pathways. Curr. Genet. 2016 doi: 10.1007/s00294-016-0575-5. doi:10.1007/s00294-016-0575-5. [DOI] [PubMed] [Google Scholar]