Abstract
Ribonucleotides are the most abundant non-canonical component of yeast genomic DNA and their persistence is associated with a distinctive mutation signature characterized by deletion of a single repeat unit from a short tandem repeat. These deletion events are dependent on DNA topoisomerase I (Top1) and are initiated by Top1 incision at the relevant ribonucleotide 3′-phosphodiester. A requirement for the re-ligation activity of Top1 led us to propose a sequential cleavage model for Top1-dependent mutagenesis at ribonucleotides. Here, we test key features of this model via parallel in vitro and in vivo analyses. We find that the distance between two Top1 cleavage sites determines the deletion size and that this distance is inversely related to the deletion frequency. Following the creation of a gap by two Top1 cleavage events, the tandem repeat provides complementarity that promotes realignment to a nick and subsequent Top1-mediated ligation. Complementarity downstream of the gap promotes deletion formation more effectively than does complementarity upstream of the gap, consistent with constraints to realignment of the strand to which Top1 is covalently bound. Our data fortify sequential Top1 cleavage as the mechanism for ribonucleotide-dependent deletions and provide new insight into the component steps of this process.
INTRODUCTION
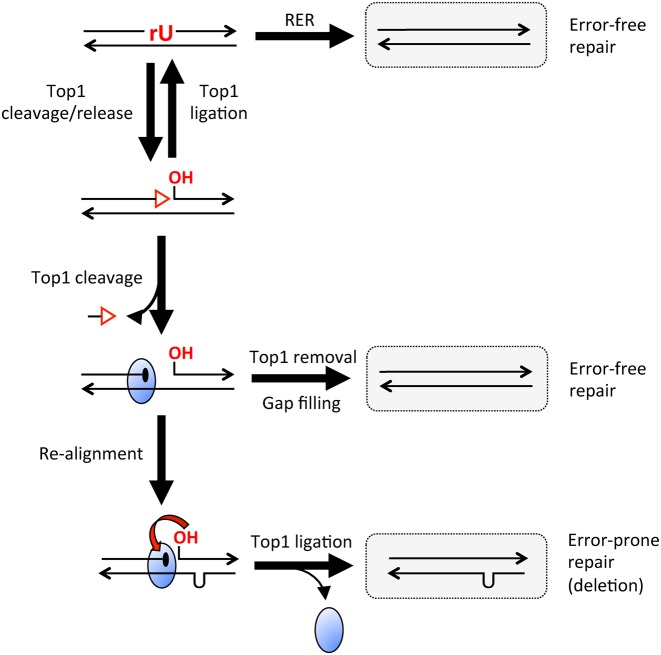
Topoisomerase I (Top1) is a type IB enzyme that removes positive and negative supercoils associated with DNA unwinding during transcription and replication (reviewed by 1). The Top1 reaction comprises two DNA transesterification steps. In the cleavage step, the active site tyrosine of Top1 attacks the phosphodiester backbone of one DNA strand to form a covalent DNA-3′-phosphotyrosyl-enzyme intermediate and a 5′-OH at the resulting DNA nick. The DNA–Top1 adduct structure is referred to as the Top1 cleavage complex (Top1cc). Rotation of the downstream DNA strand about the nick eliminates torsional stress, after which Top1 catalyzes a re-ligation reaction in which the nick-associated 5′-OH attacks the DNA-3′-phosphotyrosyl-enzyme adduct to restore the original DNA phosphodiester. Whereas the substrate for Top1 cleavage is typically a DNA phosphodiester (dN)p(dN), the enzyme also incises at (rN)p(dN) phosphodiesters generated when DNA polymerases occasionally insert ribonucleoside monophosphates (rNMPs) during replicative or repair synthesis (reviewed in 2). When Top1 transesterifies at an (rN)p(dN) site in duplex DNA, the enzyme catalyzes attack of the ribose 2′-OH on the covalent DNA(rN)-3′-phosphotyrosyl-Top1 adduct to release Top1. This leaves a single-strand nick with 2′,3′-cyclic phosphate and 5′-OH termini (Figure 1) (3). The opportunities for Top1-induced breakage at embedded ribonucleotides are normally limited by the error-free ribonucleotide excision repair (RER) surveillance pathway, which is initiated when RNase H2 incises on the 5′-phosphate side of the ribonucleotide (Figure 1) (4).
Figure 1.
Mechanisms for ribonucleotide removal from DNA. A single rU embedded in duplex DNA is indicated in red, as are the ends resulting from Top1 incision. The red triangle corresponds to a 2′,3′-cyclic phosphate and the blue oval to Top1; arrowheads indicate 3′ ends. RER (ribonucleotide excision repair) permanently removes rU from DNA when RNase H2 nicks the backbone on the 5′ side of the ribonucleotide. The resulting 3′ end can be extended by DNA polymerase, displacing rU as part of a 5′ flap that is cleaved by a flap endonuclease. Repair is completed by ligation of the remaining nick. Top1 incision at rU is a reversible reaction, allowing another opportunity for RER to occur. If Top1 cleaves the nicked fragment a second time, the oligo between the two cleavage sites is released, trapping the enzyme as a Top1cc. Enzymatic removal of Top1 from the 3′ end creates a 3′-OH that primes filling of the gap. Alternatively, realignment of complementary strands converts the Top1-generated gap to a nick, thereby facilitating Top1-mediated ligation.
Top1 is generally considered to promote genetic stability by resolving torsional stress, but its activity also can be mutagenic in yeast. This is particularly evident in highly transcribed DNA, where Top1 generates a distinctive mutation signature characterized by the deletion of a repeat unit within a low-copy number tandem repeat (5,6). We previously showed that there are two genetically distinct classes of Top1-dependent deletion hotspots: those that reflect incision at a ribonucleotide and those that likely reflect processing of a stabilized Top1cc (7). The ribonucleotide-dependency of a given deletion hotspot is operationally defined by whether the rate of events is altered in response to varying the amount of ribonucleotides in genomic DNA. This can be done by eliminating RNase H2, which allows misincorporated ribonucleotides to remain in DNA, and/or by altering the level of rNMP incorporation into the genome using steric-gate mutant DNA polymerases (8). As ribonucleotide levels in DNA increase or decrease, Top1-dependent deletion rates increase or decrease, respectively, only at ribonucleotide-dependent hotspots (7,9–11).
We previously proposed a sequential cleavage model for Top1-dependent deletions that initiate at an embedded ribonucleotide (7). As illustrated in Figure 1, Top1 incision/release at a ribonucleotide is followed by a second Top1 cleavage event immediately upstream. Whether or not both cleavages are made by the same enzyme is not known. Spontaneous dissociation of the short, nick-flanked 5′-OH/2′,3′-cyclic phosphate oligonucleotide (oligo) traps the covalent intermediate, leaving a gap between the 5′-OH and the Top1cc formed by the first and second cleavage reactions, respectively. If the resulting gap is part of a tandem repeat, misalignment between complementary DNA strands converts the gap to a nick, thereby facilitating Top1-mediated re-ligation. The reduction in deletion formation that accompanied expression of a catalytically-impaired Top1-T722A protein (12) indicated the importance of ligation activity. A central feature of the sequential cleavage model is that Top1 alone is sufficient to catalyze the ribonucleotide-dependent deletion process, without a need for accessory proteins. Indeed, the 2-base pairs (bp) deletions that occur at dinucleotide repeats were recently reconstituted in vitro with purified yeast or human Top1 (13,14). The initial cleavage/release of Top1 at an (rN)p(dN) site is a reversible process, with Top1 promoting ligation of an artificially generated nick flanked by a 2′,3′-cyclic phosphate and 5′-OH (15). Top1-mediated reconstitution of a ribonucleotide-containing DNA backbone regenerates a substrate for the error-free RER pathway, which permanently removes ribonucleotides from DNA. In contrast to the forward ribonucleotide-cleavage transesterification reaction, the reverse 2′,3′-cyclic phosphate/5′-OH re-ligation reaction does not require the tyrosine nucleophile of Top1, although it does depend on catalysis by other constituents of the active site (14,15). Finally, a 3′-OH created by enzymatic removal of a Top1cc can initiate error-free gap-filling by DNA polymerase (13).
In the current study, ribonucleotide-initiated deletions by Top1 were examined through parallel in vivo and in vitro analyses. By varying the distance between Top1 cleavage sites, we confirm the prediction that inter-nick distance dictates deletion size in vivo and demonstrate an inverse relationship between this distance and deletion rates. The maximum size of Top1-dependent deletions reported previously in vivo was 5 bp (5,11), and our analyses extend this length to 7 bp. Finally, we demonstrate that the position of sequence complementary relative to a Top1cc-generated gap affects the deletion frequency, consistent with constrained realignment of the Top1cc-bound end.
MATERIALS AND METHODS
Yeast strain construction
Strain SJR2261 (MATa ura3-52 ade2-101oc trp1Δ1 lys2Δ::hyg leu2-K:TetR'-Ssn6:LEU2 [pSR857] his4Δ::kan-pTet-LYS2F) is derivative of YPH45 (16), a strain congenic to S288C. This strain contains a pTET-LYS2F ‘forward’ construct near ARS306 on chromosome III, which is oriented so that transcription and replication-fork movement are co-directional (17). SJR2743 is a derivative of SJR2261 that contains the pTET-lys2FΔA746NR allele (10). SJR3464 contains the pTET-lys2FΔA746NR,(CCCTT)2 allele [hereafter referred to as pTET-lys2::(CCCTT)2 allele] and was derived by two-step allele replacement following transformation of SJR2743 with AflII-digested pSR1033. pSR1033 was constructed by ligating BglII-digested pSR963 (18) to annealed oligonucleotides (oligos) 5′-GATCTTTGCAAAGGGAAGGGATGC and 5′-GATCGCATCCCTTCCCTTTGCAAA; the introduced sequence is from URA3 and the (CCCTT)2 hotspot is underlined. The remaining CCCTT-containing alleles were derived from SJR2743 as follows. First, the loxP-hyg-loxP cassette that marks the deleted LYS2 locus was replaced with a natMX4 marker. This allowed subsequent insertion of a CORE-UH cassette containing selectable hyg and counter-selectable URA3-Kl markers (19) into the reversion window of the pTET-lys2FΔA746NR allele. The delitto perfetto method (19) was then used to replace CORE-UH with ∼100 bp duplexes derived by annealing complementary oligos (see Supplementary Table S1 for sequences).
The RNH201 or TOP1 gene was deleted by one-step allele replacement using PCR-generated deletion cassettes amplified from a plasmid containing an appropriate selective marker. The pol2-M644G allele was introduced by two-step allele replacement using AgeI-digested p173-pol2-M644G (11). The mating type of SJR2805 (a top1Δ derivative of SJR2743) was switched to MATα using a pGAL-HO plasmid. This allowed construction of double- and triple-mutant strains by mating, sporulation and tetrad dissection. Names of strains used to generate mutation data are included in Supplementary Table S2.
In vitro reactions
Oligos were synthesized and gel-purified by Integrative DNA Technology (IDT). Oligos were labeled at the 3′-end using [α-32P]cordycepin 5′-triphosphate (PerkinElmer Life Sciences) and terminal deoxynucleotidyl transferase (Invitrogen), or at the 5′-end using [γ-32P]ATP (PerkinElmer Life Sciences) and T4 polynucleotide kinase (New England Biolabs). Radiolabeled oligos were purified using mini Quick Spin Oligo columns (Roche Applied Science) and annealed to the unlabeled, complementary strand at a 1:1 ratio. Nicked substrates were obtained by annealing two short oligos to a longer, complementary oligo at a 1:1:1 ratio.
Cleavage reactions contained 50–200 nM of DNA substrate and 100 nM recombinant yeast Top1 (13) in 10 mM Tris–HCl (pH 7.5), 50 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 15 μg/ml BSA and 10% DMSO. 10 μM camptothecin (CPT) was included as indicated. When ribonucleotide-containing substrates were used, reactions also included 1.6 U/μl RNasin (Promega). Cleavage reactions were carried out at 30°C for 1 h; where indicated, NaCl was added to a final concentration of 0.5 M and incubation was continued at 25°C overnight. Top1 reactions were stopped by addition of SDS to 0.5% final concentration. Human TDP1 (2 μM) was added where indicated and reactions incubated overnight at 25°C. For KOH treatment, samples with or without NaCl addition were further incubated with 0.3 M KOH at 55°C for 1 h or mock treated. Samples and markers were mixed with loading buffer [99.5% (v/v) formamide, 5 mM EDTA, 0.01% (w/v) xylene cyanol, 0.01% (w/v) bromophenol blue] and analyzed by electrophoresis through a denaturing 20%-polyacrylamide gel. Labeled nucleic acids were detected using a Typhoon phosphorimager.
Mutation rates and spectra
Cultures were inoculated either with independent colonies or with 2 × 105 cells from an overnight starter culture. Cells were grown at 30°C in YEPGE (1% yeast extract, 2% Bacto-peptone, 250 μg/ml adenine hemisulfate supplemented with 2% glycerol and 2% ethanol) until saturated. Appropriate dilutions were plated on YEPD (YEP supplemented with 2% dextrose) to determine the total number of viable cells and on synthetic complete dextrose medium lacking lysine (SCD-Lys) to determine the total number of Lys+ revertants in each culture. Mutation rates were calculated using the method of the median (20) and 95% confidence intervals were determined as described previously (21).
To obtain mutation spectra, independent colonies were grown non-selectively as described above and Lys+ revertants were selected on SCD-Lys plates. The portion of LYS2 containing the region of interest was PCR amplified using primers LYSWINF (5′-GCCTCATGATAGTTTTTCTAACAAATACG) and LYSWINR (5′-CCCATCACACATACCATCAAATCCAC). PCR products were sequenced by the Duke University DNA Analysis Facility, Eurofins MWG Operon or Eton Bioscience INC. The rate of the deletion of interest was calculated by multiplying the total Lys+ rate by the proportion of the deletion in the corresponding mutation spectrum (Supplementary Table S2). Two representative spectra are shown in Supplementary Figure S1, illustrating the complexity of mutations detected in the absence of RNase H2.
RESULTS
The ribonucleotide-dependent, 5-bp deletion hotspot used in the current study was initially described in an analysis of forward mutations in a URA3 gene placed in both orientations near ARS306 on chromosome III (11). The 5-bp deletions were observed in a strain background devoid of RNase H2 activity and containing an rNTP-permissive form of DNA polymerase Pol ϵ (pol2-M644G allele). Because Pol ϵ synthesizes DNA primarily during leading-strand synthesis (22,23), orientation specificity of hotspot activity with respect to ARS306 can be used to infer which strand is the target of Top1 cleavage. Importantly, 5-bp deletions were evident when the (CCCTT)2 sequence was generated during the leading-strand synthesis, but not when the complementary sequence (AAGGG)2 was on the nascent leading strand. This asymmetry indicates that it is the (CCCTT)2-containing strand that is nicked by Top1 to generate the corresponding deletion, and we will hereafter refer to this specific hotspot as the (CCCTT)2 hotspot.
(CCCTT)2 is an rNMP-dependent, Top1-dependent deletion hotspot in a frameshift reversion assay
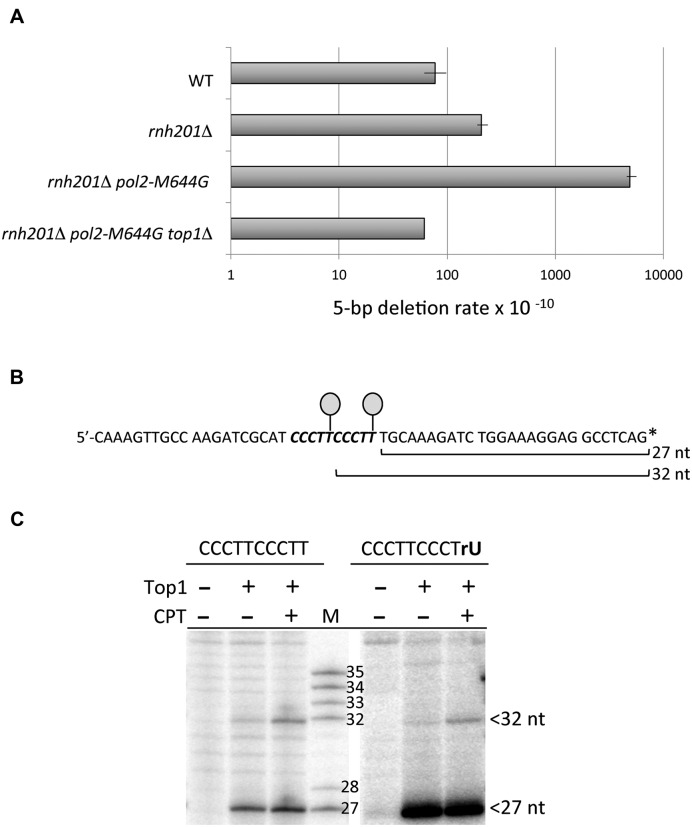
We previously placed lys2 frameshift alleles under control of the highly active TET promoter (pTET) to study the behavior of individual Top1-dependent hotspots (5,7,9,10). A 20-bp fragment of URA3 that encompasses the (CCCTT)2 hotspot was thus introduced into the context of a pTET-lys2 frameshift allele whose reversion selects compensatory, net 2-bp deletions, among which are 5-bp deletions. Our prior studies demonstrated that deletions are much more frequent when the relevant Top1 cleavage sites are on the non-transcribed strand (NTS) of a highly transcribed reporter (10). The (CCCTT)2 hotspot behaved similarly and in all experiments reported here, the (CCCTT)2-containing strand was the NTS. In addition, the pTET-lys2 reporter was inserted near ARS306 so that the transcription machinery and the replication fork were moving in the same direction. This ensured that the (CCCTT)2 sequence was located on the nascent leading strand, where its activity should be affected by the rNTP permissiveness of Pol ϵ. In the resulting pTET-lys2::(CCCTT)2 allele, 5-bp deletions at the hotspot were detected at a low rate (7.7 × 10−9) in a wild-type (WT) background. The deletion rate was elevated ∼3-fold in the absence of RNase H2 (rnh201Δ mutant) and ∼60-fold in an rnh201Δ pol2-M644G double mutant. The 5-bp deletion rate in the double mutant returned to the WT level when TOP1 was deleted (Figure 2A and Supplementary Table S2). These data confirm that 5-bp deletions at the (CCCTT)2 hotspot are both ribonucleotide- and Top1-dependent and that this is a robust system for studying the parameters of hotspot activity.
Figure 2.
(CCCTT)2 is an rNMP-dependent hotspot that contains two Top1 cleavage sites. (A) 5-bp deletions rates in a pTET-lys2 frameshift-reversion assay. No events were observed in the absence of Top1, so the rate was based on one event (see Supplementary Table S2). (B) Sequence of the (CCCTT)2-containing oligo, with the duplication in bold italics. Lollipops indicate the positions of Top1 attachment and the asterisk the radioactive label. (C) Mapping of Top1 incision sites in the 3′-labeled strand shown in (B). Primary cleavage sites in the (CCCTT)2 repeat occur at the terminal thymine of each repeat. The full gel is presented in Supplementary Figure S2. M, size markers; CPT, camptothecin.
Mapping Top1 cleavage sites in the (CCCTT)2 hotspot
The sequential cleavage model predicts that the (CCCTT)2 hotspot should contain two Top1 cleavage sites that are 5 nucleotides (nt) apart. To examine this, we mapped Top1 sites in a 57-bp fragment having the same sequence as the in vivo allele. The sequence of the strand matching the NTS sequence of the pTET-lys2::(CCCTT)2 allele is shown in Figure 2B. This strand was radioactively labeled at either the 3′ or 5′ end, as appropriate, and then annealed to the complementary, unlabeled strand. In the experiment shown in Figure 2C a 3′-labeled DNA duplex was incubated with purified yeast Top1 in the presence or absence of camptothecin (CPT), a drug that stabilizes the Top1cc and enhances the detection of some cleavage sites. Two prominent Top1 cleavage sites within the centrally located (CCCTT)2 repeat were evident, located 27 and 32 nt from the 3′ end. Consistent with our prediction, these two sites are 5 nt apart, with Top1 forming a covalent intermediate with the terminal thymine of each CCCTT repeat. To confirm that the two Top1 cut sites were maintained in the presence of an rNMP at the distal cleavage site, we mapped Top1 incision sites using a 3′-labeled oligo with rU substituted for the terminal T of the 3′ repeat. The positions of the cleavage sites were retained and the amount of the 27-nt fragment was increased, as expected for Top1 release after cyclization to form a rU>p end (Figure 2C and Supplementary Figure S2).
The distance between Top1 cleavage sites affects deletion size and frequency
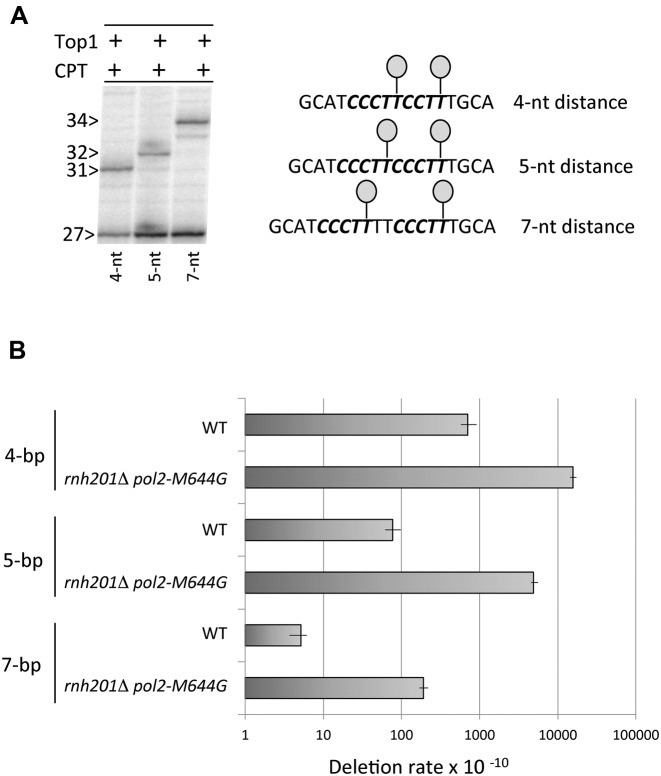
In the sequential cleavage model, a gap is created by spontaneous loss of the oligo between the 5′-OH created by initial Top1 cleavage and the Top1cc created by a second cleavage event. Because retention of the nick-flanked oligo requires persistent hydrogen bonding with the complementary strand, its release should be inversely related to the distance between cleavage sites. In addition, the fewer base pairs that have to be melted to realign the complementary strands and convert the resulting gap to a nick, the more efficient the realignment process should be. These considerations lead to the prediction that the distance between two Top1 cleavage sites should be inversely related to the deletion frequency. To test this, we decreased or increased the distance between the Top1 cleavage sites mapped at the (CCCTT)2 hotspot. The first cytosine of second repeat was deleted to generate a substrate predicted to produce 4-bp deletions: CCCTT*CCTT*, with asterisks indicating predicted Top1 cleavage sites. Alternatively, two thymidines were inserted between the previously mapped cleavage sites to generate a 7-bp deletion substrate of sequence CCCTT*TTCCCTT*. Purified yeast Top1 nicked a 3′-labeled DNA with CCCTTCCTT or CCCTTTTCCCTT substituted for the (CCCTT)2 hotspot at the originally mapped cleavage sites, producing nicks that were 4 or 7 nt apart, respectively (Figure 3A and Supplementary Figure S3).
Figure 3.
Deletion frequency is inversely related to the distance between Top1 cleavage sites. (A) A 3′-labeled strand of the same sequence as in Figure 2B was modified by deleting 1 nt or inserting 2 nt in order to alter the distance between Top1 cleavage sites (lollipops) to 4 or 7 nt, respectively. The distance between the primary cleavage sites was 4, 5 or 7 nt, matching the distance between the terminal thymine of each cleavage site. The full gel is presented in Supplementary Figure S3. (B) Deletion rates in WT and rnh201Δ pol2-M644G strains containing modified (CCCTT)2 hotspots (see Supplementary Table S2).
Following confirmation that the original Top1 cleavage sites were maintained when the distance between them was altered, each modified sequence was transplanted into a pTET-lys2 frameshift allele that can revert via a 4-bp or 7-bp deletion. The 4-bp and 7-bp deletion rates, along with that of 5-bp deletions at the original (CCCTT)2 hotspot, are presented in Figure 3B. Compared to the WT background, 4-bp and 7-bp deletions were highly elevated in an rnh201Δ pol2-M644G background and were absent in an rnh201Δ pol2-M644G top1Δ background (Supplementary Table S2). The salient findings were that the 4-bp deletion rate was 3-fold higher than that of 5-bp deletions, and the 5-bp deletion rate was 26-fold higher than the 7-bp deletion rate (Figure 3B). If the distance between the two Top1 cleavage sites is the primary determinant of deletion size, then we should only observe 5-bp deletions at the (CCCTT)2 hotspot. To test this, we introduced the (CCCTT)2 hotspot into a pTET-lys2 allele that can detect 4-bp deletions. No 4-bp deletions at (CCCTT)2 were observed in this context in an rnh201Δ pol2-M644G background (Supplementary Table S2).
Top1 generates 5-nt deletions at the (CCCTT)2 hotspot in vitro
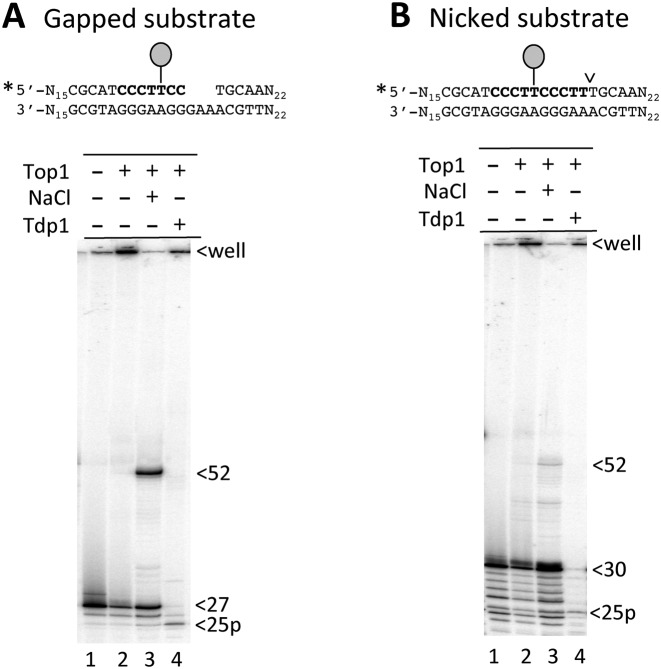
Our model posits that following creation of a nick at an embedded ribonucleotide, Top1 cleaves at an upstream site and becomes trapped when the intervening oligo dissociates from the complementary strand. Top1-mediated ligation then occurs across the gap, a process proposed to be facilitated by misalignment between repeats on complementary strands. These two steps were confirmed at the (CCCTT)2 hotspot using the suicide substrate shown in Figure 4A, which contains a 3-nt gap that eliminates the terminal CTT of the downstream CCCTT repeat. Because of the proximity of the downstream gap, Top1 should become trapped when it cleaves at the upstream CCCTT repeat and the adjacent 2-nt fragment subsequently dissociates. The 27-nt fragment upstream of the gap was labeled at its 5′ end and, as expected by covalent attachment of Top1, less of the labeled strand migrated into the gel (Figure 4A, lane 2). The addition of NaCl, which prevents further cleavage by Top1 but allows an already-formed Top1cc to catalyze ligation, resulted in the disappearance of material in the well and appearance of a 52-nt fragment, which is the size predicted by Top1-mediated ligation across a 5-nt gap (Figure 4A, lane 3). Finally, to define the position of the Top1cc prior to ligation, the cleavage reaction was treated with Tdp1, which hydrolyzes peptides attached to DNA via a 3′-phosphotyrosyl bond. A 25-nt fragment with a 3′ phosphate terminus (25p) was observed, which is the size expected when Top1 cleaves at the terminal T of the upstream CCCTT repeat (Figure 4A, lane 4).
Figure 4.
Purified yeast Top1 generates 5-nt deletions at the (CCCTT)2 repeat in vitro. (A) Reaction of Top1 with a 57-bp, 5′-labeled substrate containing (A) a 3-nt gap in the downstream CCCTT repeat or (B) a nick at the downstream cleavage site. Lollipops indicate the remaining Top1 cleavage site at position 25; ligation in the presence of NaCl generates a 52-nt deletion product. Incubation of the Top1cc intermediate (retained in wells) with Tdp1 releases the enzyme and a 25-nt cleavage product.
A similar reaction was performed using a substrate with a nick at the 3′ end of the downstream CCCTT repeat (Figure 4B). In this case, the starting 5′-labeled fragment upstream of the nick was 30 nt, and enzyme trapping required dissociation of a 5-nt fragment. As with the gapped substrate, the amount of the labeled fragment retained in the well increased upon addition of Top1 to the nicked substrate, and a new 52-nt fragment was generated when NaCl was added to promote ligation. The reduced amount of 52-nt product relative to that obtained with the gapped substrate suggests that Top1 trapping likely diminishes as the distance to a pre-existing nick increases. As noted previously, this could reflect the ease with which a nick-flanked oligo dissociates from the complementary DNA strand. Finally, though we attempted to recapitulate the entire reaction with an appropriately positioned rU (substituted for the terminal dT of the downstream CCCTT repeat), we were unable to convincingly detect the expected 52-nt product (data not shown).
An additional CCCTT repeat facilitates Top1-mediated ligation in vitro and stimulates deletions in vivo
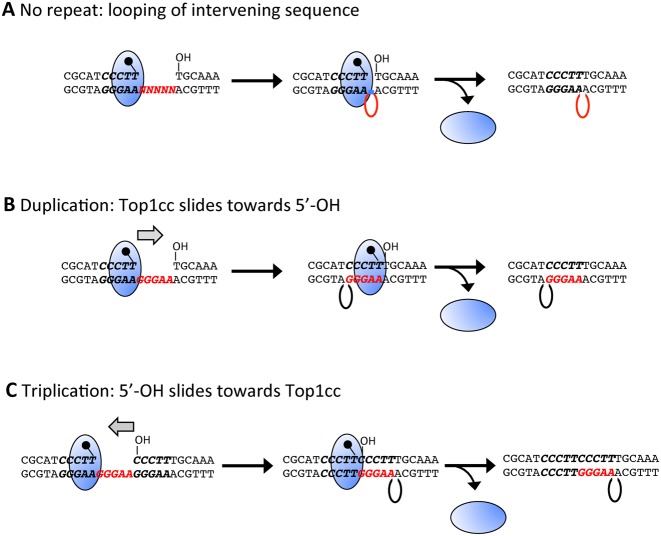
In vitro, Top1 can ligate across single-strand gaps, with the intervening sequence forming a loop (Figure 5A) (24–26). All Top1-dependent deletion hotspots identified in vivo, however, have been perfect or imperfect tandem repeats, suggesting that the repeats have a critical role in the deletion process. In the case of ribonucleotide-dependent hotspots, we proposed that the repeat mediates re-alignment between complementary strands, thereby bringing the downstream 5′-OH and the upstream Top1cc into the correct orientation for efficient ligation. In the case of the (CCCTT)2 hotspot, where Top1 cleaves at the terminal thymine of each repeat, the end to which Top1 is attached would have to ‘slide’ towards the downstream 5′-OH in order to pair with the complementary single strand within the gap (Figure 5B). Because Top1 clamps down on duplex DNA (27), such sliding/realignment likely would be difficult. With a triplication of the CCCTT sequence in which the ribonucleotide resides in the middle repeat, there is an alternative possibility. Namely, the complementary strand of the third repeat can slide towards the Top1cc and pair with the single strand within the gap, bringing the 5′-OH adjacent to the Top1cc (Figure 5C).
Figure 5.
Top1 ligation across a 5-nt gap. (A) No repeat to facilitate strand realignment is present between Top1 cleavage sites. (B) The Top1-linked end realigns to pair with the complementary single strand within the gap created by sequential Top1 cleavage reactions. (C) The 5′-OH end realigns and pairs with the complementary single strand within the gap.
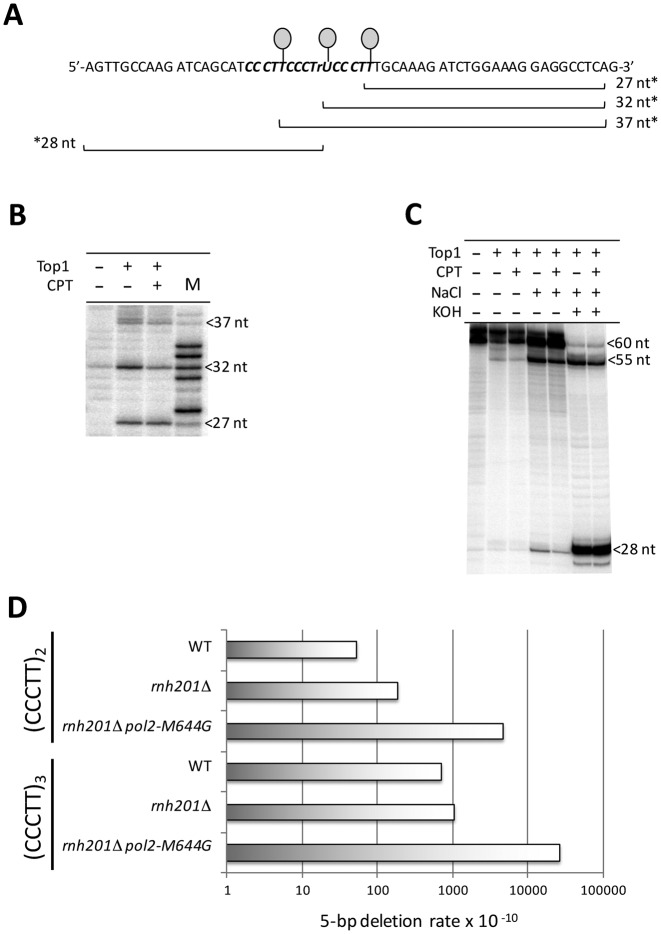
The above considerations suggested that triplication of the CCCTT sequence would be associated with more Top1-dependent deletions than simple duplication of the sequence. This, in turn, could explain why we were unable to recapitulate the entire deletion reaction using a ribonucleotide-containing (CCCTT)2 substrate. We thus introduced a third CCCTT repeat into our in vitro substrate. The length of the resulting duplex was 60 bp and rU replaced the terminal dT of the central repeat (Figure 6A). Using purified yeast Top1, cleavage sites were first mapped using a 3′-labeled (CCCTT)3 substrate. Fragments of 27, 32 and 37 nt were produced, corresponding to Top1 incision at the terminal position of each repeat (Figure 6B and Supplementary Figure S4). The entire deletion process was then examined using a 5′-labeled CCCTTCCCTrUCCCTT strand (Figure 6C). Even in the absence of NaCl to promote Top1-mediated ligation, a weak 55-nt radiolabeled product was detected, which is 5 nt shorter than the starting 60-nt fragment. Following NaCl treatment, the 55-nt species was more abundant and was resistant to KOH treatment, confirming removal of the rU. This result supports a model in which realignment between strands is critical, and suggests that realignment of the 5′-OH end is much more efficient than that of the 3′ end bound by Top1.
Figure 6.
Triplication of CCCTT facilitates 5-bp deletions. (A) Sequence of the (CCCTT)3-containing strand of a 60-bp duplex fragment used in cleavage/ligation reactions. rU is substituted for the terminal thymine of the central repeat. (B) Top1 cleavage of a 3′-labeled oligo generates 27-, 32- and 37-nt fragments that correspond to the primary cleavage sites at the terminal T/rU of each repeat. The full gel is presented in Supplementary Figure S4. (C) Top1 catalyzed deletion of one repeat unit from a 5′-labeled oligo. Addition of NaCl facilitates ligation that produces a 55-nt product consistent with the deletion of a single repeat unit. Treatment of the ligation reaction with KOH nicks the intact, 60-nt fragment at the position of rU and confirms the absence of rU in the 55-nt deletion product. (D) 5-bp deletion rates in a pTET-lys2 reporter containing a duplication or triplication of CCCTT.
To examine whether an additional repeat unit also facilitates ribonucleotide-dependent deletions in vivo, we introduced three copies of CCCTT into an appropriate pTET-lys2 frameshift allele. The corresponding 5-bp deletions were monitored in WT and rnh201Δ pol2-M644G backgrounds. It should be noted that 5-bp deletions within the (CCCTT)3 sequence were not entirely Top1-dependent (see Supplementary Table S2), which likely reflects a contribution of DNA polymerase slippage to deletion formation when there are three tandem repeats. In calculating rates of ribonucleotide-dependent 5-bp deletions, the rate observed in the absence of Top1 was subtracted from the rate in its presence. The rate of ribonucleotide- and Top1-dependent 5-bp deletions was 5.5-fold higher in the (CCCTT)3- than in the (CCCTT)2-containing allele (Figure 6D), providing in vivo evidence that the additional repeat allows realignment of the more mobile 5′-OH end.
DISCUSSION
The sequential cleavage model for ribonucleotide/Top1-dependent deletions posits that a trapped Top1cc mediates ligation to a 5′-OH generated by previous Top1 incision/release at a ribonucleotide. The ability of Top1 alone to mediate the deletion reaction was recently demonstrated in vitro by two independent groups (13,14). Huang et al. used human Top1 protein (14) and a ribonucleotide-independent (AT)2 hotspot previously characterized by us in vivo (7). Human Top1 required a pre-existing nick, which mimics the first Top1 incision at a ribonucleotide, to produce the corresponding 2-nt deletion product in vitro. Sparks and Burgers used purified yeast Top1 (13) and a fragment containing the ribonucleotide-dependent (TC)3 deletion hotspot that also was previously characterized by us in vivo (7). An oligo containing a ribonucleotide at a previously mapped Top1 cleavage site 3′ to the repeat (9) was unable to produce a deletion product, however, and modification of the sequence was required. This underscores the difficulty in correlating Top1 cleavage sites mapped in vitro with their relevance in vivo, and whether Top1 catalyzes ribonucleotide-dependent deletions at a modified (TC)3 hotspot in vivo has not been examined. In the current study, we also demonstrated that yeast Top1 is sufficient to perform the entire sequential cleavage reaction at a CCCTT repeat in vitro and most importantly, that the same sequence is a ribonucleotide- and Top1-dependent hotspot in vivo.
Prior studies with nick-containing suicide substrates suggest that a nick needs to be within 6 nt of a Top1 cleavage site in order to trap the enzyme (24,28,29). We extended the maximum ‘trapping’ distance between cleavage sites to 7 nt, although the corresponding 7-bp deletions occurred very inefficiently. We furthermore demonstrated an inverse relationship between inter-site distance and deletion rate, and suggested this likely reflects the ease of loosing the intervening oligo and/or the stability of the realigned strands. If a single Top1 does both cleavages, an alternative possibility is that the sequential cleavage is more efficient at reduced distances between sites. At present, we do not know whether or not both cleavages are made by the same Top1 enzyme.
Top1 can ligate directly across gaps in vitro (24), but this appears to be too inefficient to detect in vivo, where all of the Top1-dependent deletion hotspots identified to date have been perfect or imperfect tandem repeats (5,6). In the current analysis with CCCTT repeats, Top1 cleavage sites mapped to the terminal nucleotide of the repeat units, but this specific position is not a necessity as long as flanking base complementarity allows realignment that converts a gap to a nick. The relative abilities of the two ends flanking a Top1-generated gap to slide has a particularly profound effect on deletion efficiency, however, with sliding of the Top1cc end being highly disfavored. The number of base pairs that stabilize the realigned end also affects the efficiency of deletion formation in vitro (13), and this would presumably influence deletion efficiency in vivo as well, especially in the case of imperfect repeats. Given the multitude of factors that affect the ribonucleotide/Top1-dependent deletion process in vitro, it is not surprising that only a small subset of tandem repeats accumulate deletions in vivo (5) and that a priori prediction of deletion hotspots is not currently possible.
The parallel in vivo and in vitro analyses presented here provide strong support for a Top1/ribonucleotide-dependent deletion process that follows the steps proposed in the sequential cleavage model (7), and have revealed additional complexities of this process. Given the high conservation of Top1 and DNA metabolic processes, similar reactions at genomic ribonucleotides likely extend to any organism that contains a type IB topoisomerase. Defects in RNase H2 are linked to the human diseases Aicardi-Goutières syndrome (30) and systemic lupus erythematosis (31), and the mutagenic consequences of persistent genomic ribonucleotides may be relevant to the pathogenesis of these diseases.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [GM101690 to S.J.R.]; NIH [GM46330 to S.S.]; NIH [GM32431 to P.M.B.]; NCI Intramural Program, Center for Cancer Research [BC006161 to Y.P.]. Funding for open access charge: NIH [GM101690].
Conflict of interest statement. None declared.
REFERENCES
- 1.Wang J.C. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell. Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 2.Williams J.S., Kunkel T.A. Ribonucleotides in DNA: origins, repair and consequences. DNA Repair. 2014;19:27–37. doi: 10.1016/j.dnarep.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sekiguchi J., Shuman S. Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol. Cell. 1997;1:89–97. doi: 10.1016/s1097-2765(00)80010-6. [DOI] [PubMed] [Google Scholar]
- 4.Sparks J.L., Chon H., Cerritelli S.M., Kunkel T.A., Johansson E., Crouch R.J., Burgers P.M. RNase H2-initiated ribonucleotide excision repair. Mol. Cell. 2012;47:980–986. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lippert M.J., Kim N., Cho J.E., Larson R.P., Schoenly N.E., O'Shea S.H., Jinks-Robertson S. Role for topoisomerase 1 in transcription-associated mutagenesis in yeast. Proc. Natl. Acad. Sci. U.S.A. 2011;108:698–703. doi: 10.1073/pnas.1012363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi T., Burguiere-Slezak G., Van der Kemp P.A., Boiteux S. Topoisomerase 1 provokes the formation of short deletions in repeated sequences upon high transcription in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2011;108:692–697. doi: 10.1073/pnas.1012582108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho J.E., Kim N., Li Y.C., Jinks-Robertson S. Two distinct mechanisms of Topoisomerase 1-dependent mutagenesis in yeast. DNA Repair. 2013;12:205–211. doi: 10.1016/j.dnarep.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nick McElhinny S.A., Watts B.E., Kumar D., Watt D.L., Lundstrom E.B., Burgers P.M., Johansson E., Chabes A., Kunkel T.A. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. U.S.A. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim N., Huang S.N., Williams J.S., Li Y.C., Clark A.B., Cho J.E., Kunkel T.A., Pommier Y., Jinks-Robertson S. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho J.E., Kim N., Jinks-Robertson S. Topoisomerase 1-dependent deletions initiated by incision at ribonucleotides are biased to the non-transcribed strand of a highly activated reporter. Nucleic Acids Res. 2015;43:9306–9313. doi: 10.1093/nar/gkv824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nick McElhinny S.A., Kumar D., Clark A.B., Watt D.L., Watts B.E., Lundstrom E.B., Johansson E., Chabes A., Kunkel T.A. Genome instability due to ribonucleotide incorporation into DNA. Nat. Chem. Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Megonigal M.D., Fertala J., Bjornsti M.A. Alterations in the catalytic activity of yeast DNA topoisomerase I result in cell cycle arrest and cell death. J. Biol. Chem. 1997;272:12801–12808. doi: 10.1074/jbc.272.19.12801. [DOI] [PubMed] [Google Scholar]
- 13.Sparks J.L., Burgers P.M. Error-free and mutagenic processing of topoisomerase 1-provoked damage at genomic ribonucleotides. EMBO J. 2015;34:1259–1269. doi: 10.15252/embj.201490868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang S.Y., Ghosh S., Pommier Y. Topoisomerase I alone is sufficient to produce short DNA deletions and can also reverse nicks at ribonucleotide sites. J. Biol. Chem. 2015;290:14068–14076. doi: 10.1074/jbc.M115.653345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shuman S. Polynucleotide ligase activity of eukaryotic topoisomerase I. Mol. Cell. 1998;1:741–748. doi: 10.1016/s1097-2765(00)80073-8. [DOI] [PubMed] [Google Scholar]
- 16.Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim N., Abdulovic A.L., Gealy R., Lippert M.J., Jinks-Robertson S. Transcription-associated mutagenesis in yeast is directly proportional to the level of gene expression and influenced by the direction of DNA replication. DNA Repair. 2007;6:1285–1296. doi: 10.1016/j.dnarep.2007.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehner K., Jinks-Robertson S. The mismatch repair system promotes Polζ-dependent translesion synthesis in yeast. Proc. Natl. Acad. Sci. U.S.A. 2009;106:5749–5754. doi: 10.1073/pnas.0812715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Storici F., Resnick M.A. The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol. 2006;409:329–345. doi: 10.1016/S0076-6879(05)09019-1. [DOI] [PubMed] [Google Scholar]
- 20.Lea D.E., Coulson C.A. The distribution of the numbers of mutants in bacterial populations. J. Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 21.Spell R.M., Jinks-Robertson S. In: G6enetic Recombination: Reviews and Protocols. Waldman AS, editor. Vol. 262. Totowa: Humana Press; 2004. pp. 3–12. [Google Scholar]
- 22.Johnson R.E., Klassen R., Prakash L., Prakash S. A major role of DNA Polymerase δ in replication of both the leading and lagging DNA strands. Mol. Cell. 2015;59:163–175. doi: 10.1016/j.molcel.2015.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pursell Z.F., Isoz I., Lundstrom E.B., Johansson E., Kunkel T.A. Yeast DNA polymerase ϵ participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henningfeld K.A., Hecht S.M. A model for topoisomerase I-mediated insertions and deletions with duplex DNA substrates containing branches, nicks, and gaps. Biochemistry. 1995;34:6120–6129. doi: 10.1021/bi00018a015. [DOI] [PubMed] [Google Scholar]
- 25.Pourquier P., Jensen A.D., Gong S.S., Pommier Y., Rogler C.E. Human DNA topoisomerase I-mediated cleavage and recombination of duck hepatitis B virus DNA in vitro. Nucleic Acids Res. 1999;27:1919–1925. doi: 10.1093/nar/27.8.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pommier Y., Jenkins J., Kohlhagen G., Leteurtre F. DNA recombinase activity of eukaryotic DNA topoisomerase I; effects of camptothecin and other inhibitors. Mutat. Res. 1995;337:135–145. doi: 10.1016/0921-8777(95)00019-g. [DOI] [PubMed] [Google Scholar]
- 27.Redinbo M.R., Stewart L., Kuhn P., Champoux J.J., Hol W.G. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- 28.Shuman S. Site-specific DNA cleavage by vaccinia virus DNA topoisomerase I. Role of nucleotide sequence and DNA secondary structure. J. Biol. Chem. 1991;266:1796–1803. [PubMed] [Google Scholar]
- 29.Pourquier P., Pilon A.A., Kohlhagen G., Mazumder A., Sharma A., Pommier Y. Trapping of mammalian topoisomerase I and recombinations induced by damaged DNA containing nicks or gaps. Importance of DNA end phosphorylation and camptothecin effects. J. Biol .Chem. 1997;272:26441–26447. doi: 10.1074/jbc.272.42.26441. [DOI] [PubMed] [Google Scholar]
- 30.Crow Y.J., Leitch A., Hayward B.E., Garner A., Parmar R., Griffith E., Ali M., Semple C., Aicardi J., Babul-Hirji R., et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat. Genet. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- 31.Gunther C., Kind B., Reijns M.A., Berndt N., Martinez-Bueno M., Wolf C., Tungler V., Chara O., Lee Y.A., Hubner N., et al. Defective removal of ribonucleotides from DNA promotes systemic autoimmunity. J. Clin. Invest. 2015;125:413–424. doi: 10.1172/JCI78001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.