Abstract
RNA 2′-O-methylation is one of the ubiquitous nucleotide modifications found in many RNA types from Bacteria, Archaea and Eukarya. RNAs bearing 2′-O-methylations show increased resistance to degradation and enhanced stability in helices. While the exact role of each 2′-O-Me residue remained elusive, the catalytic protein Fibrillarin (Nop1 in yeast) responsible for 2′-O-methylation in eukaryotes, is associated with human pathologies. Therefore, there is an urgent need to precisely map and quantify hundreds of 2′-O-Me residues in RNA using high-throughput technologies. Here, we develop a reliable protocol using alkaline fragmentation of total RNA coupled to a commonly used ligation approach, and Illumina sequencing. We describe a methodology to detect 2′-O-methylations with high sensitivity and reproducibility even with limited amount of starting material (1 ng of total RNA). The method provides a quantification of the 2′-O-methylation occupancy of a given site, allowing to detect relatively small changes (>10%) in 2′-O-methylation profiles. Altogether this technique unlocks a technological barrier since it will be applicable for routine parallel treatment of biological and clinical samples to decipher the functions of 2′-O-methylations in pathologies.
INTRODUCTION
Post-transcriptional RNA modifications are omnipresent in cellular RNAs and are extremely widespread in all domains of life. A number of specific and rather sophisticated modifications are present only in a selected few RNA species (like hypermodified nucleotides in tRNA for example), while many chemically more simple modifications are typically widely distributed among all RNA species and in all living organisms. One such very common and widespread modification is the methylation of the ribose 2′-OH moiety, which was found in tRNA, rRNA, snRNAs, mRNAs, as well as, in miRNAs (1–4).
Initially, RNA 2′-O-methylation was shown to play a role in RNA stabilization under extreme temperatures and pH, since the replacement of the 2′-OH by 2′-O-Me considerably reduces the reactivity of the oxygen atom and thus prevents a nucleophilic attack and subsequent cleavage of the adjacent phosphodiester bond (5). More recently, it became clear that the functions of 2′-O-methylation spread well beyond a simple RNA stabilization and that these nucleotides are involved in regulation of gene expression as well as various other cellular processes (6–8). For example, bacterial resistance to aminoglycoside antibiotics can be mediated by additional 2′-O-Me residues appearing in 16S or 23S rRNA under environmental pressure. 2′-O-Methylation of m7G-cap adjacent residues in eukaryotic mRNA and some Gm residues in rRNA and tRNA serve as innate immunity markers (9,10). Some eukaryotic and viral mRNAs are 2′-O-methylated at 5′-cap adjacent and internal positions, thereby regulating their translation (11–14).
Formation of 2′-O-Me residues in RNA is ensured either by stand-alone modification enzymes which recognize directly their target, or via a complex snoRNP machinery involving the catalytic protein (Nop1/Fibrillarin) (15), several auxiliary proteins and box C/D snoRNA guides, each specific for a given RNA position (4,16,17). Stand-alone protein enzymes for 2′-O-methylation are widely used in Bacteria (18,19), while almost all 2′-O-methylations in Eukarya and Archaea are incorporated using the C/D snoRNP machinery (20,21).
Three major approaches have been proposed to detect 2′-O-methylated residues in mature cellular RNAs. Some of them, including in particular reverse transcription (RT) at low dNTP concentrations ([dNTP]), have been extensively used in the past, and allowed the establishment of 2′-O-methylation profiles for rRNAs from various species and also for some snRNAs (22,23). More recently, the combination of RT at low [dNTP] with quantitative PCR was also used for relative quantification of 2′-O-Me residues (24). However, all these methods are extremely laborious and time-consuming and only allow site-by-site detection and measurement of 2′-O-methylation. Bioinformatic approaches for the prediction of 2′-O-Me residues have been developed based on careful identification of C/D-box sRNA guides, e.g. for the deduction of ribose methylation patterns of rRNAs in various Archaea (25).
Last year, a conceptually new method was published that combined alkaline RNA fragmentation and high-throughput sequencing. Since the phosphodiester bond located at the 3′ of a 2′-O-Me nucleotide is relatively resistant to a nucleophilic cleavage under alkaline conditions, such RNA fragments starting or ending at the +1 nucleotide relative to the modified residue, were shown to be excluded from a sequencing library constructed by adapter ligation to samples of randomly fragmented RNA. This concept was successfully applied to map all known 2′-O-methylations in yeast rRNA and furthermore discovered one additional previously un-annotated site in 18S rRNA. However, the approach was based on a rather particular ligation procedure, conceived for coupling to high-throughput sequencing on the Ion Torrent system, and required additional steps for preparation of home-made 3′- and 5′-adapters as well as a mutant RNA ligase (26). Since fragmentation of RNA is random and irregular, a rather high read coverage was required for robust analysis of 2′-O-methylations using this approach. Thus, the method is generally limited to highly abundant cellular RNAs, like for instance, rRNA, since several μg of starting material are required for analysis.
This method added 2′-O-Me modifications to the limited subset of RNA modifications that can be detected and mapped by high-throughput techniques. Pioneering studies in this area already described the mapping of m5C (27–29), m6A (30–32), pseudouridine (Ψ) (33–35) and m1A (36–39) in RNAs, and, depending on the nature of the technique used, those methods are suitable for transcriptome-wide detection, or limited to highly abundant RNAs.
The goal of the present work was the development of a high-throughput technique coupled to a more standard sequencing procedure than Ion Torrent, in order to facilitate and speed-up the mapping of 2′-O-methylations in RNAs. Despite the existence of several possible approaches for 2′-O-methylation detection by conventional methods, the added value of high-throughput approaches is the simultaneous detection of hundreds of modification sites using a limited amount of biological material.
In this method article we provide a detailed description and optimization of the RiboMethSeq method based on alkaline hydrolysis, now coupled to a standard library preparation protocol from Illumina. Yeast rRNA was used as a model to demonstrate that RiboMethSeq can be robustly applied to detection and even relative quantification of 2′-O-Me sites and has an acceptable performance with a very small amount of starting RNA material (∼1 ng).
MATERIALS AND METHODS
Yeast strains and cultures
Yeast strains used in this study were obtained from the EUROSCARF collection (Germany, see Supplementary Table S1 in Supporting information). Cells were grown in standard Yeast Extract/Peptone/Dextrose (YPD) or in Synthetic/Dextrose media (SDM) to 0.6–0.7 OD600 for exponential phase and to 5.5–6.0 OD600 for stationary phase.
RNA extraction
Total RNA from yeast cells from stationary or exponential phases were isolated using hot acid phenol (40). RNA concentration was measured on Nanodrop 2000 and RNA quality was checked by capillary electrophoresis using a PicoRNA chip on Bioanalyzer 2100 (Agilent, USA).
Synthetic 2′-O-methylated RNA oligonucleotide
Modified and unmodified synthetic RNA oligonucleotides of 39 nt derived from 5′-sequence of tetracycline binding riboswitch were ordered from Dharmacon, USA. The site of 2′-O-methylation (or its equivalent in non-modified molecule) and its 3′-adjacent neighbor were made random. The sequence of modified oligonucleotide is: 5′-CCUUAAGGCNmNAUAACAUACCAGAUCGCCACCCGCGCUC-3′.
RNA fragmentation conditions
RNA (1–250 ng) was subjected to either alkaline hydrolysis or metal ion-based RNA cleavage (magnesium or zinc ions). RNA hydrolysis was performed in 50 mM bicarbonate buffer pH 9.2 for 4–10 min at 95°C or in 100 mM Tris–HCl buffer, pH 8.0, containing either 0.1 mM ZnCl2 or 2 mM MgCl2 for 2 or 3 min, respectively. The reaction was stopped by ethanol precipitation using 3 M Na-OAc, pH 5.2 and glycoblue as a carrier in liquid nitrogen. In addition, the zinc-based RNA cleavage was stopped by addition of 2 μl of 0.5 M EDTA pH 8.0 before ethanol precipitation. After centrifugation, the pellet was washed with 80% ethanol and resuspended in nuclease-free water. The sizes of generated RNA fragments were assessed by capillary electrophoresis using a PicoRNA chip on Bioanalyzer 2100 (Agilent, USA) and were ranging from 30 to 200 nt.
Synthetic RNA oligonucleotides were fragmented in 50 mM bicarbonate buffer pH 9.2 for 20 min at 95°C and processed as described above.
End repair
RNA fragments without any gel-purification step were directly 3′-end dephosphorylated using 5 U of Antarctic Phosphatase (NEB, UK) for 30 min at 37°C. After inactivation of the phosphatase for 5 min at 70°C, RNA fragments were phosphorylated at the 5′-end using T4 PNK and 1 mM ATP for 1h at 37°C. End-repaired RNA fragments were then purified using RNeasy MinElute Cleanup kit (QIAGEN, Germany) according to the manufacturer's recommendations except that 675 μl of 96% ethanol were used for RNA binding. Elution was performed in 10 μl of nuclease-free water.
Library preparation
RNA fragments were converted to library using NEBNext® Small RNA Library kit (NEB ref E7330S, UK or equivalent from Illumina, USA) following the manufacturer's instructions. DNA library quality was assessed using a High Sensitivity DNA chip on a Bioanalyzer 2100. Library quantification was done using a fluorometer (Qubit 2.0 fluorometer, Invitrogen, USA).
Sequencing
Libraries were multiplexed and subjected for high-throughput sequencing using an Illumina HiSeq 1000 instrument with a 50 bp single-end read mode (or using an Illumina MiSeq for paired-end read runs). Since clustering of short fragments was very efficient, libraries were loaded at 8 pM concentration per lane.
Bioinformatics pipeline
Initial trimming of adapter sequence was done using Trimmomatic-0.32 (41) with the following parameters: LEADING:30 TRAILING:30 SLIDINGWINDOW:4:15 MINLEN:17 AVGQUAL:30. Alignment to the reference rRNA sequence was done by Bowtie2 (ver 2.2.4) in End-to-End mode and k = 1. 5′-end counting was done directly on *.sam file using a dedicated Unix script. Simultaneous 5′- and 3′-ends counting was done by bedtools v2.25.0 after conversion to *.bed file. Final analysis was performed by calculation of score MAX for detection of 2′-O-Me residues, and MethScore for their quantification.
Calculation of score MAX and MethScore
To calculate score MAX, the relative change of end coverage position by position was calculated in 5′→3′ and reverse direction. The relative change was normalized to average values for -6 and +6 nucleotides. The normalized relative change for 5′→3′ and reverse direction were averaged and the maximal value between the average and normalized relative change was retained (score MAX). MethScore was calculated essentially as described in (26) for ScoreC using the same relative impact of neighboring nucleotides.
Mung Bean Assay and mass spectrometry analysis
A mung bean nuclease protection assay was performed as previously described (26,42). A complementary DNA oligonucleotide (50 nt) was used for hybridization and protection of the region 2181–2230 of yeast 25S rRNA (see Supporting information). The protected fragment was gel-purified, digested to nucleosides and subjected to LC–MS/MS analysis as described previously (43,44) (for further details see Supporting information).
RESULTS AND DISCUSSION
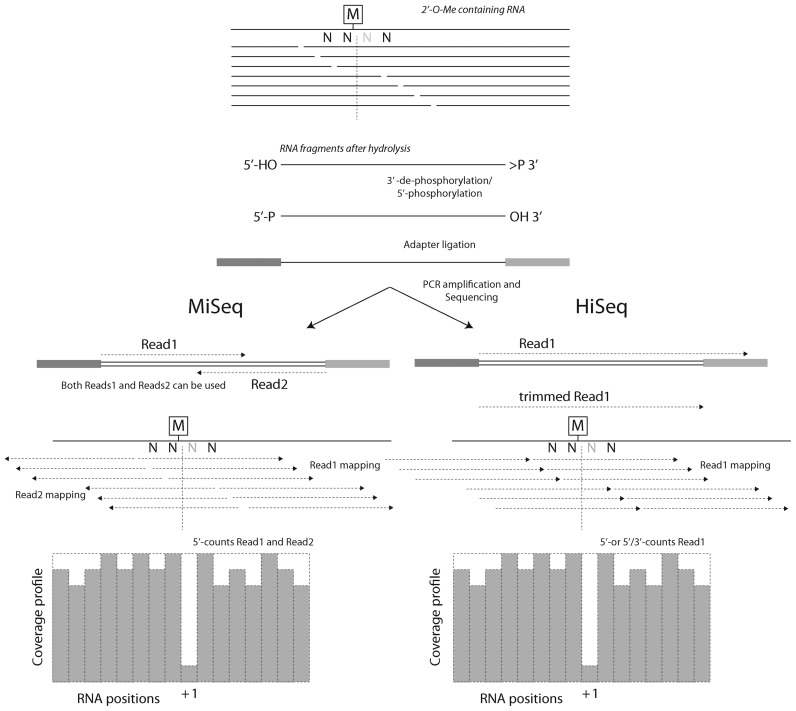
The method for detection of 2′-O-methylated residues described in this paper is based on differential cleavage of RNA phosphodiester bond 3′-adjacent to methylated or unmethylated ribose, as proposed by Birkedal et al. (26). Basically, after fragmentation under alkaline conditions, the RNA fragments starting or ending 3′ to a 2′-O-Me residue are strongly underrepresented or entirely excluded from the resulting pool. A corresponding RNA fragmentation profile can be deduced by conversion of these fragments to sequencing amplicons, followed by sequencing, mapping to the reference and quantitative analysis of the number of fragments starting upstream or downstream, respectively (5′- and 3′-ends) of each RNA position (Figure 1).
Figure 1.
General overview of the RiboMethSeq protocol. RNA containing 2′-O-Me residues is randomly fragmented, 5′- and 3′-ends are repaired and adapters are ligated to both extremities. After amplification and barcoding, amplicons are subjected to Illumina sequencing. MiSeq sequencing (left) generally provides paired-end reads, while HiSeq sequencing is generally performed in a single-read mode (right). Orientations and mapping of reads are indicated. The 2′-O-Me residues protect the 3′-adjacent phosphodiester bond from cleavage, generating a typical gap in 5′-/3′-ends coverage profile.
We first explored different fragmentation approaches, which could potentially be used for detection of 2′-O-Me residues in RNA using high-throughput sequencing. As a model RNA, yeast Saccharomyces cerevisiae rRNAs were used, which contain 18 known 2′-O-Me sites in 18S rRNA and 37 in 25S rRNA. No 2′-O-Me residues were reported in 5S and 5.8S rRNAs in S. cerevisiae. A synthetic RNA oligonucleotide bearing a unique 2′-O-methylated residue was used to evaluate sequence context effects for quantification of modification.
Appropriate library preparation protocols
To convert RNA fragments to a sequencing library, several alternative approaches are available. A direct ligation protocol using 2′,3′-cyclophosphates and 5′-OH extremities in RNA fragments renders de-phosphorylation/phosphorylation steps unnecessary and may reduce ligation biases (26). However, this direct ligation protocol, as used in the original RiboMethSeq approach, required a particular mutant RNA ligase and a specially prepared 5′-adaptor with a 2′,3′-cyclophosphate, making the method difficult to implement in a non RNA-expert laboratory. In our experimental approach we used standard commercially available protocols for library generation. For fragmentation-based protocols, the criterion of choice is the exact end mapping of the fragments, since the quality of the results directly depends on the precise quantification of cleaved versus uncleaved phosphodiester bonds (Figure 1). For this reason, the NEBNext® Small RNA Library Kit was selected, since here the 3′- and 5′-adapters are directly ligated to the RNA fragments, after a mandatory de-phosphorylation/phosphorylation step. Other commonly used methods including cDNA 3′-end tailing (45) or template-directed extension with a terminal-tagging oligonucleotide (TTO) (46) may not be optimal, since such protocols do not provide single nucleotide resolution at RNA 5′-ends.
In addition to a precise resolution mapping of 5′- and/or 3′-ends, all fragmentation protocols require a rather important depth of sequencing reads to calculate the characteristic drop in coverage profile. Thus fragmentation protocols are particularly suitable for the analysis of highly abundant or purified or, at least, enriched RNAs.
Comparison of RNA fragmentation techniques
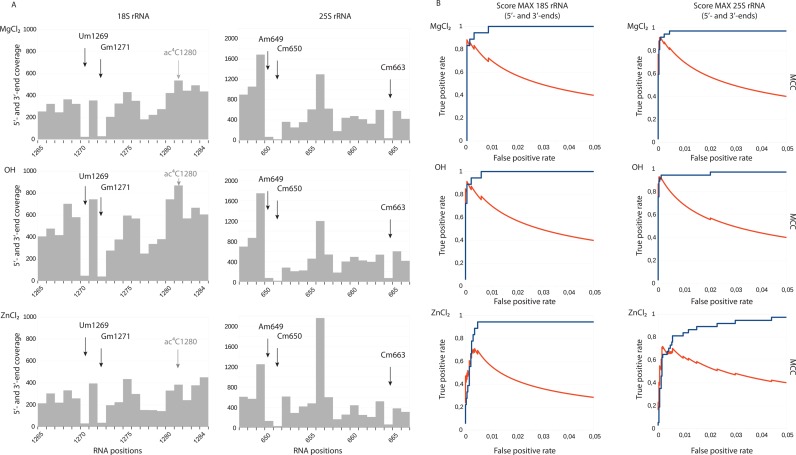
RNA fragmentation can be achieved using different reagents. In this study we tested three of them: sodium carbonate at pH 9.2, MgCl2 and ZnCl2 at neutral conditions (see Materials and Methods for further details) using yeast S. cerevisiae 18S and 25S rRNAs. The RNA fragments generated were 3′-dephosphorylated/5′-phosphorylated and converted to Illumina sequencing amplicons. Sequencing was performed using the paired-end (PE) mode on MiSeq in order to precisely map 5′- and 3′-ends of each RNA fragment. Details on sequencing libraries and on bioinformatic treatment are described in Materials and Methods and in Supplementary Table S2. Representative profiles obtained for cumulative 5′- and 3′-end coverage for all three fragmentation protocols are shown in Figure 2A. Each type of fragmentation allowed the detection of a characteristic drop in end's coverage corresponding to 2′-O-methylated residues, even though the coverage profile was slightly different between the treatments. In order to evaluate the performance of fragmentation protocols in detection of known 2′-O-Me residues in yeast rRNA, the so-called score MAX was calculated, which represents the variation of end-coverage at a given position compared to its environment (for more details see Materials and Methods). RNA positions were sorted using a decreasing score MAX and for comparative assessment, the corresponding Receiver Operating Characteristic (ROC) curves and Matthews Correlation Coefficient (MCC) for the score MAX for different parameters (see Material and Methods and Figure 2B) were plotted. Significant parameters commonly used to assess performance, including accuracy (ACC), positive predictive value (PPV), false discovery rate (FDR), sensitivity, specificity and area under curve (AUC) for the maximal value of MCC coefficient are given in Supplementary Table S3. The general shape of ROC curves and calculated ACC and AUC showed that the three fragmentation protocols allowed detection of 2′-O-Me residues with an excellent accuracy (AUC > 0.97). However, the detailed inspection of associated parameters revealed that the best results were obtained for both MgCl2 and sodium bicarbonate buffer (OH−) fragmentation methods.
Figure 2.
2′-O-Methylation analysis using RNA fragmentation with MgCl2, alkaline bicarbonate buffer (OH−) and ZnCl2. Panel (A) Typical fragmentation profiles for regions of yeast 18S and 25S rRNA are shown. Arrows indicate the +1 positions for 2′-O-Me residues. 18S rRNA contains another RNA modification, ac4C, which is present nearby, but does not generate a signal (shown in gray). Panel (B) Performance of each method was compared by calculation of score MAX to detect known 2′-O-methylations followed by construction of Receiver Operating Characteristic (ROC) curves (blue). Matthews Correlation Coefficient (MCC) is traced in red on the same graph.
The lowest FDR < 0.06 and the best precision (0.944) was observed for OH− hydrolysis, with 34 true positives (TP) and only two false positive (FP) signals in 25S rRNA, and 16 TP with one FP in 18S rRNA. MgCl2 fragmentation gave 34TP/3FP in 25S rRNA and 15TP/1FP in 18S rRNA. Of note, all calculations of the performance were done under the assumption that all 2′-O-Me sites were fully modified, which later turned out not be true for certain positions (see below). De novo detection of such hypomodified positions is indeed problematic, since their score MAX is similar to many false-positives signals.
Unexpectedly, the very popular ZnCl2 RNA fragmentation protocol turned out to be far less appropriate for 2′-O-Me residues detection since it exhibited rather poor sensitivity and precision (24TP/6FP in 25S rRNA and 16TP/12FP in 18S rRNA), compared to MgCl2 and OH− hydrolysis (see Supplementary Table S3). This comparison demonstrated that the technique used for RNA fragmentation has a great influence on the results obtained. We also explored the possibility to cumulate the data obtained with different types of hydrolysis for the same RNA, but this approach did not improve the final results (data not shown).
False positive signals and other modified nucleotides in RiboMethSeq
A part of the false positive signals observed in RiboMethSeq corresponds to nucleotides 5′-adjacent to existing 2′-O-Me residues (e.g. positions 1126 and 1428 in 18S rRNA), or to other modified nucleotides in yeast rRNA. As observed previously (26), some pseudouridine residues gave positive signals (e.g. Ψ1187 in 18S rRNA, Ψ2260 and Ψ2416 in 25S rRNA). This was most probably due to a bias in hydrolysis, 5′-phosphorylation and/or 5′-adapter ligation (see Supplementary Figure S1). Non-stoichiometric phosphorylation of 5′-Ψ residues in RNAs was already observed in the past, notably in RNA post-labeling techniques (47). Other RT-silent base methylations and further modifications including m7G, m5C, m5U or ac4C (see Figure 2A) do not show any particular signature, while RT-arresting residues might be anticipated to generate false positive signals, like m1A2142 in 25S rRNA (not shown). However, this was not the case for m1acp3Ψ (see Supplementary Figure S1).
Finally, other false positives observed stem from gradual (and not sudden) decrease of 5′-end coverage over several nucleotides in certain regions (e.g. positions 583, 2669, 2689 in 25S rRNA) and this drop may be interpreted as a false positive signal when using score MAX.
Use of 5′- and 3′-end coverage
In principle, a calculation of the score MAX may be based independently on either 5′-end coverage, or on 3′-end coverage, or on both. The separate analysis of 5′- and 3′-end coverage revealed that the data obtained for 3′-ends gave a higher FDR, as well as a lower PPV compared to 5′-ends, most probably due to a bias in hydrolysis and 3′-ligation. OH- fragmentation gave better results, compared to the other two methods (see Supplementary Figure S2). This is likely due to a more evenly distributed coverage pattern, resulting from largely unspecific cleavage. In contrast, divalent cations like Mg2+ or Zn2+ are known to bind to certain RNA structures with differential affinity, a likely cause for specific cleavages (48), which are, in the current situation, a detrimental feature. Another interesting observation for MgCl2 and OH- fragmentation protocols is, that 5′-ends by themselves provide high quality results, which compare favorably with those obtained from cumulative 5′- and 3′-ends coverage.
Taking this information into account, another OH−fragmented RNA library preparation was conducted with extended time of hydrolysis, leading to an increased fraction of shorter RNA fragments (<50 nts). This preparation was converted into the corresponding library and sequenced on HiSeq1000 using a single-read 50 nts (SR50) sequencing mode. In these conditions, only 50 nts reads were obtained, but for the fraction of RNA fragments used for library preparation which was shorter than 50 nts, these reads contained both, the 5′- and the 3′-end information and it was thus possible to use both for score calculation. However, the precision of 3′-end mapping depended on the recognition and exact trimming of the adapter sequence during the bioinformatic treatment (see Figure 1). To evaluate if a single-read HiSeq sequencing mode could be used to improve quality parameters over those obtained from the paired-end MiSeq sequencing mode, we analyzed the discrimination of 2′-O-Me signals in rRNA using either the complete set of reads obtained for the sample (only 5′-end information was used) or only a subset of shorter reads <50 nts (in this case 3′-ends of RNA fragments generated by hydrolysis were used).
The results for the shorter reads shown in Supplementary Figure S3 again revealed, as already observed, a strong bias in the 3′-ends coverage, while 5′-ends only or combination of 5′- and 3′-ends performed much better. Again, the global results obtained with a complete subset of 5′-ends and with the combination of 5′- and 3′-ends were rather similar, as reflected by similar values for MCC and ratio FP/TP.
This observation is important since it validates the use of SR50 sequencing runs on HiSeq or similar machines and allows to restrict the analysis to 5′-end coverage for equally efficient 2′-O-Me residue mapping. Further RiboMethSeq experiments where therefore conducted using only 5′-end coverage obtained from SR50 sequencing.
Number of reads and required minimal coverage
The initial tests described above had been performed using MiSeq-sequenced libraries with an average coverage of about 750x, where the coverage was calculated as number of reads (or the respective 5′-ends)/RNA position. Thus, ∼4 mln reads were used for analysis. In order to determine the importance of sequencing coverage, one of the libraries was further sequenced on HiSeq 1000 in SR50 mode, where around 80 mln reads (coverage ∼ 15 000×) were obtained. Several subsets (4 mln and 12 mln of reads) from the same dataset were also used to compare results based on variegated coverage levels. The results obtained from 12 min versus 80 mln reads were very similar in quality (Supplementary Figure S4), an analysis based on the 4 mln read subset was clearly less accurate with respect to representative information on all known modified rRNA positions. This identified an average coverage as sufficient for effective analysis, thereby reducing the sequencing cost. A decrease of the coverage below 750× should be avoided, since statistical distribution inevitably produces sites of low coverage in the single digit range, such that the methylation status cannot efficiently be deduced via score MAX for certain positions in rRNA. Optimal coverage of 5′-ends for 2′-O-Me residues detection was found to be around 1000–1500× compared to the target RNA, this value generally provided a minimal coverage of ∼10 reads at all modified positions. However, the coverage required depends on the RNA sequence and may need adjustment depending on the regularity of fragmentation.
Taken together, our data show that the great majority of 2′-O-Me residues in yeast rRNA can be detected using this high-throughput approach combining alkaline fragmentation followed by library preparation and Illumina sequencing. Analysis of the 5′-end coverage is appropriate to detect known 2′-O-Me residues and to confirm their exact positions.
Analysis of technical and biological variability
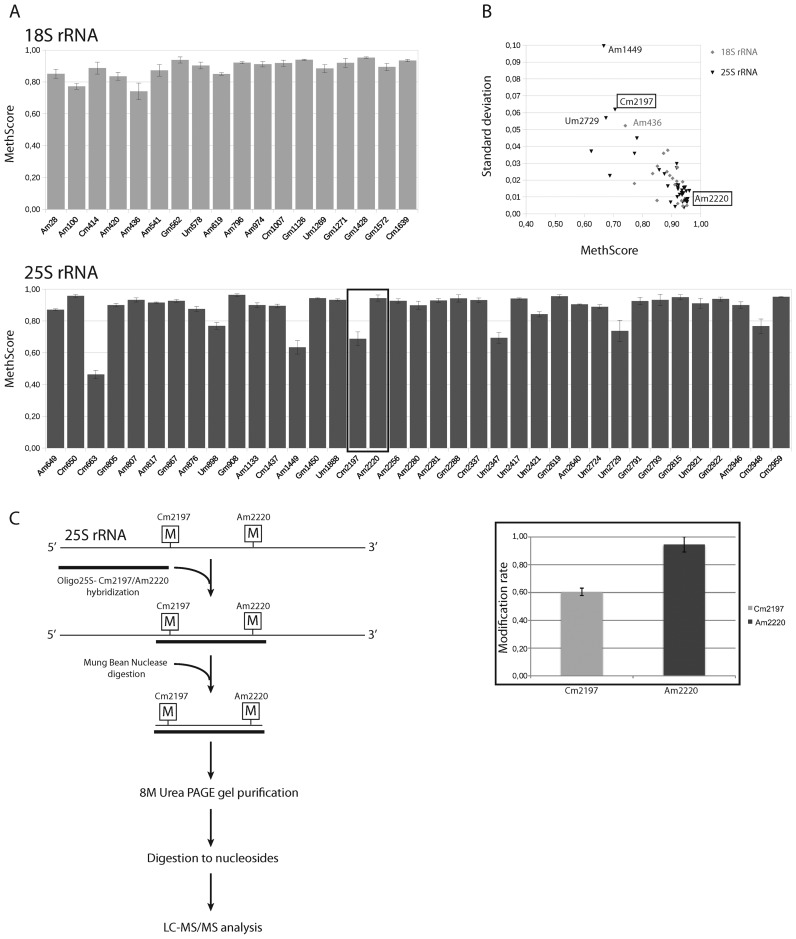
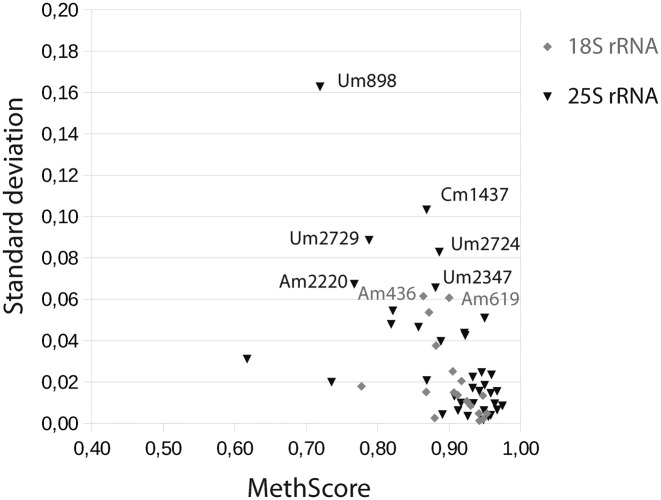
In order to measure the technical reproducibility of the RiboMethSeq approach, the same sample of yeast total RNA was processed in triplicate for fragmentation, library preparation and sequencing. The shape of ROC curves plotted for score MAX and the associated parameters are very similar for the three technical triplicates (not shown), demonstrating that detection of 2′-O-Me residues is robust and reproducible. In order to provide a quantitative measurement of 2′-O-methylation at a given position, we calculated another score (MethScore, see Materials and Methods). This score shows a theoretical linear dependence from 0 to 1.0 between the depth of the gap in the end-coverage profile and the expected 2′-O-methylation rate of the nucleotide. The MethScores calculated for three technical replicates show a rather low dispersion, the mean standard deviation (SD) values for 18S rRNA were only 1.35%, and 1.8% for 25S rRNA, respectively, and the maximum values were 3.9% and 6.7%, respectively. Positions with a low MethScore, like Am100 and Am436 in 18S rRNA as well as Cm663, Am1449, Cm2197, Um2347, Um2729 and Cm2948 in 25S rRNA, showed the highest variability, while the dispersion was lower (Supplementary Figure S5) for positions with a MethScore close to 1.0. The most variable position was Um2729 in 25S rRNA with a SD >5%, for all other positions the SD was <5%.
In order to evaluate the biological variability, RiboMethSeq profiles were compared of RNA preparations from the WT BY4741 strain and from 4 different yeast strains bearing deletions in non-essential genes encoding known tRNA modification enzymes, none of which showed a growth phenotype in YPD media. We used strains with deletions of TRM2 (m5U54), TRM3 (Gm18), TRM8 (m7G47) and TRM44 (Um44) (see Supplementary Table S1). As shown in Figure 3, all strains show almost identical MethScore profiles for 18S and 25S rRNAs, with mean values close to the WT strain (see Supplementary Figure S5). For these samples, the MethScore values showed somewhat more variability than for technical replicates, but the mean SD is only 2.1% for 18S rRNA, and 2.0% for 25S rRNA (Figure 3). As already observed for technical replicates, the most variable positions were: Am1449, Cm2197 and Um2729 in 25S rRNA and Am436 in 18S rRNA.
Figure 3.
Biological variability of MethScore observed between wild type (WT) and four yeast strains with chromosomal deletions of genes encoding different tRNA-specific methyltransferases. Panel (A) The MethScore values for each modified position in 18S and 25S rRNA. For each strain MethScore values were calculated position by position and used for calculation of the mean values as well as standard deviation (SD). Graph shows the values of MethScore, error bars represent SD for each position. Methylation level for positions Cm2197 and Am2220 (boxed) was verified by LC–MS/MS (see panel C). Panel (B) Graph showing the dispersion of the observed MethScore values for 18S rRNA (gray) and 25S rRNA (black) and their corresponding SD values. Positions with a lower MethScore show a higher variability, as attested by higher SD values. Panel (C) Experimental workflow for isolation of 25S rRNA fragment followed by LC–MS/MS analysis (left). Methylation levels for positions Cm2197 and Am2220 measured by LC–MS/MS (right). Error bars are calculated for technical triplicate.
The majority of 2′-O-Me sites in rRNA showed a MethScore > 0.8, indicating a close-to-complete methylation status, while two sites in 18S and seven sites in 25S rRNA were partially methylated according to the MethScore. These included Am100 in 18S rRNA (MethScore 0.77), which was previously shown to be incompletely methylated using HPLC-MS/MS quantification (68% of methylation) (42).
To further underscore the suitability of this approach for a quantitative assessment of 2′-O-methylation occupancy, we isolated a fragment containing Cm2197 and Am2220 from the WT 25S rRNA by Mung Bean Nuclease assay (42) (Figure 3C). The isolated fragment was subjected to LC–MS/MS analysis (43,44). Quantification of Cm2197 and Am2220 was done as described in Supporting information. The results presented in Figure 3C and Supplementary Figure S6 show 61% ribose methylation at Cm2197 and 95% at Am2220. These data is in good agreement with our RiboMethSeq results (MethScore 0.71 and 0.96, respectively).
Altogether, these data demonstrate that RiboMethSeq provides a robust and reproducible detection as well as a quantitative assessment of the 2′-O-Me status of a given residue in rRNA.
RNA modification is now considered as a highly dynamic process actively contributing to regulation of gene expression. Recent publications clearly demonstrated that the modification profiles of eukaryotic transcriptomes are subject to rapid changes under different growth conditions (33,35,36,38,49,50). In addition, it was observed that nutrient starvation in yeast affected localization of C/D-box snoRNP proteins. This type of stress could therefore reasonably be suspected to affect the 2′-O-Me profile of rRNA (51).
Using the developed RiboMethSeq approach we analyzed the yeast rRNA 2′-O-methylations under various growth conditions (in rich and poor growth media during exponential and stationary growth phases). The results presented in Supplementary Figure S7 showed, that the 2′-O-Me profile of yeast rRNA did not vary substantially, but rather appeared to be of constitutive character under different growth conditions. Very similar MethScore profiles were obtained for all four datasets, which once more, albeit indirectly underscored the robustness of the applied 2′-O-Me residue detection protocol.
RNA quantity required for analysis
To determine the minimum amount of input RNA required for efficient RiboMethSeq analysis, we compared data derived from analysis of the standard amount of starting material (250 ng) employed so far to and such obtained from as little as 1 ng of yeast total RNA. Samples were treated independently for fragmentation, library preparation and sequencing. The results of RiboMethSeq analysis are presented on Figure 4. Briefly summarized, even 1 ng of total RNA was found to be still sufficient for precise mapping of 2′-O-Me residues, and the overall variability of the MethScore data was well within the range determined for variability of technical and biological replicates. However, some positions started to show somewhat stronger variations, like Um898 and Cm1437 in 25S rRNA with a SD value >10%. Other positions in both 18S and 25S rRNA showed consistent values of MethScore. In our opinion, 1 ng represents the lower limit for a representative amplification during library preparation. In routine analysis we recommend to use 10–100 ng of total RNA as input material for rRNA analysis. Altogether this data showed that as little as 1 ng of total RNA sufficed for reliable detection of 2′-O-Me residues in yeast rRNA. This is an important advantage in the analysis of rRNA 2′-O-methylation profiles for precious biological or clinical samples.
Figure 4.
RiboMethSeq analysis performed with a reduced amount of input RNA. The results of MethScore values were calculated for all modified positions in yeast 18S and 25S rRNA for 1 ng and for 250 ng of input total RNA. Mean values obtained with their SD values are traced on the graph, 18S data are in gray, 25S data are in black, respectively.
2′-O-Me residues are also present in few yeast tRNAs, notably at positions 18 (Gm18, formed by Trm3) and 44 (Um44, formed by Trm44). In order to extend the application of RiboMethSeq to these less abundant RNAs, we extracted the reads corresponding to tRNAHis containing Gm18 (estimated amount 0.5–1 ng in total RNA fraction used for analysis). About 20 000 reads for WT and ΔTRM3 were used for analysis and MethScores for nucleotides in the region containing Gm18 were calculated and compared. Supplementary Figure S8 shows the 5′-ends profiles for Gm18 in yeast tRNAHis in WT and ΔTRM3 strains. These data demonstrated that, even at limiting coverage and irregular cleavage, Gm18 could be detected in tRNA that is significantly less abundant than rRNA. This detection was efficient in the WT strain but not in the mutant, confirming the absence of Gm18.
We conclude that the RiboMethSeq approach can also be successfully applied for detection and quantification of 2′-O-methylations in RNA species that are significantly less abundant than rRNA.
Analysis of 2′-O-methylation profiles in C/D-box snoRNA-deleted strains
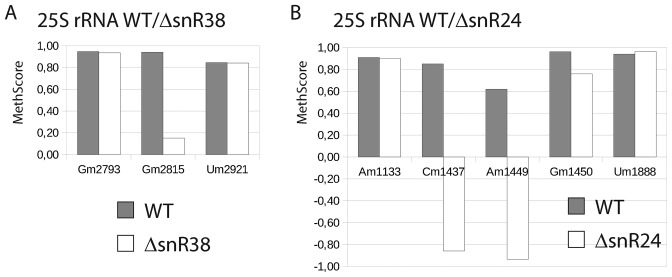
Ribose methylation of eukaryotic rRNA is ensured by C/D-box snoRNP complexes. Fibrillarin (Nop1 in yeast) is the catalytic subunit in these complexes and variable snoRNA guides serve to define the to-be-methylated RNA positions. In order to validate the capacity of RiboMethSeq to detect changes in rRNA modification profile, we used two yeast strains bearing deletions of non-essential genes with intronic C/D-box snoRNAs. These two snoRNAs were known to target individual positions in 25S rRNA: Gm2815 for snR38 and Cm1437/Am1449/Gm1450 for snR24. As shown in Figure 5, deletion of snR38 led to a reduced MethScore for G2815 (from 0.94 for WT to 0.15 for mutant), while the signals for all other 2′-O-methylated residues remained constant. Deletion of C/D-box snoRNA snR24 led to a negative MethScore at positions 1437 and 1449, and a visible decrease for position 1450. The somewhat counter-intuitive negative MethScore, which is otherwise expected to vary from 0 to 1.0, was identified as an artifact originating from locally irregular fragmentation profiles that are common to highly structured RNAs such as the one under investigation. For a cleavage efficiency that is significantly higher at a given position than at the surrounding nucleotides, the algorithm calculates negative MethScore. This is also the case when the coverage changes considerably within the 12 neighboring nucleotides used for MethScore calculation. Hence, the MethScores for positions 1437 and 1449 vary from negative values to 1.0 (see also below for quantification of 2′-O-methylation). The calculated value of MethScore for the position 1451 (corresponding to Gm1450) in WT and in ΔsnR24 strains was affected both by the presence of a neighboring 2′-O-methylated nucleotide (Am1449) as well as by very efficient cleavage at the following position (A1452). A careful inspection of the profiles revealed that the protection efficiency drops by a factor of ∼3 for both positions 1450/1451 (Am1449/Gm1450) in the ΔsnR24 strain compared to WT, while the cleavages at the surrounding nucleotides remained constant (data not shown). However, even in the absence of methylation Gm1450, the cleavage at position 1451 remained relatively low, generating an artificially high MethScore. This leads to conclude, that in the case of very irregular cleavage profiles in highly structured RNA regions, the MethScore can behave abnormally and does not necessarily approach a zero value even in the absence of 2′-O-methylation.
Figure 5.
Validation of the RiboMethSeq approach using yeast deleted strains deficient in 2′-O-methylation residue at certain positions in 25S rRNA. Two yeast strains with deletions of snR24 and snR38, together with the corresponding WT strain, were subjected to RiboMethSeq. Variations of MethScore values are observed upon deletion of snR38, responsible for modification of position Gm2815 (left) and snR24, implicated for modification of positions Cm1437, Am1449 and Cm1450 (right). Two neighboring positions around of the affected modification site(s) are also shown for comparison. In gray, values obtained for WT yeast strain, in white, values for corresponding snoRNA-deleted mutant.
Altogether these data confirm that the RiboMethSeq approach reliably detects the changes in the nucleotide methylation status.
Quantification of 2′-O-methylated residues
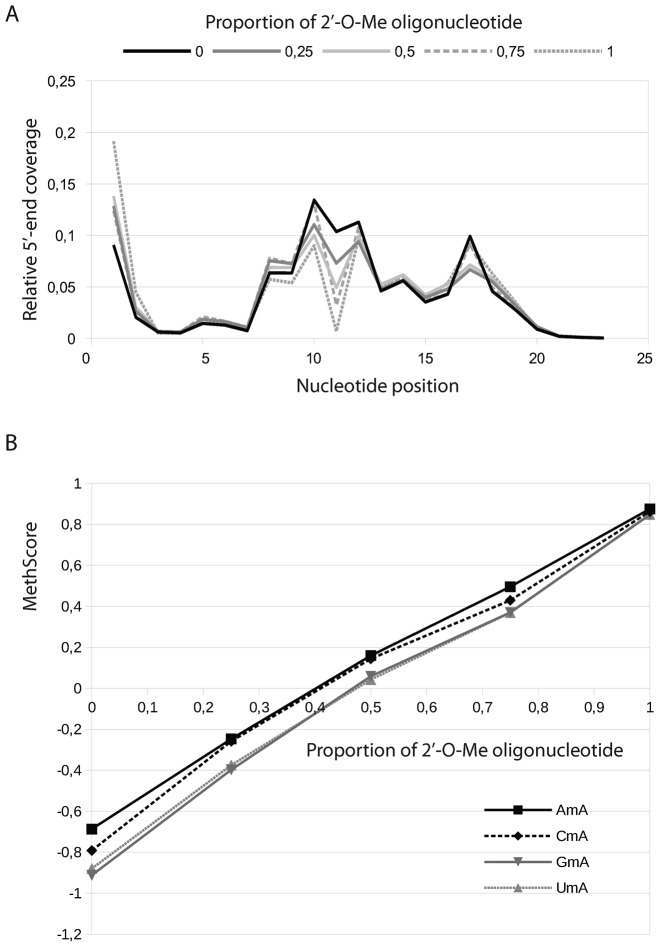
For more precise insight into quantification by RiboMethSeq with respect to sequence context, we analyzed synthetic RNA oligonucleotides containing a single 2′-O-Me residue. An unmodified RNA version with an identical sequence was used as a control. To evaluate the importance of the nucleotide context at the methylated and 3′-neighboring position, synthetic RNA oligonucleotides containing a randomized sequence NmN were analyzed. The RiboMethSeq protocol was applied to pure 2′-O-methylated and unmethylated RNA oligonucleotides as well as to different ratios of both. After sequencing, the reads were mapped to all 16 possible reference sequences, mismatched alignments were removed and 5′-ends counting followed by MethScore calculation was performed.
The 5′-end coverage of nucleotides in the short synthetic oligonucleotide was found to be quite irregular, yet representative compared to profiles obtained for yeast rRNAs. The results presented in Figure 6 showed that the MethScore was indeed linearly depending on the proportion of the methylated oligonucleotide present in the mixture for almost all nucleotide contexts, and its value ranged from -1.5 for unmethylated oligo to ∼0.9 for pure methylated RNA. Surprisingly, only NmG contexts behave slightly differently. The MethScore still showed a linear dependence on the methylation rate, but the maximal value even for completely methylated RNA was only ∼0.8. However, this behavior seemed to be restricted to synthetic oligonucleotides and was not observed for any NmG context found in yeast rRNA.
Figure 6.
Analysis of 2′-O-methylation rate using synthetic modified and unmodified RNA oligonucleotides. Panel (A) Superposition of 5′-end coverage profiles obtained for modified oligonucleotide, its unmodified counterpart, as well as for mixtures 25:75, 50:50 and 75:25 of both. The 2′-O-Me signal appears at position 11 (modified Nm10 in RNA oligonucleotide). Panel (B) representative traces of MethScore values for different modified nucleotides in the same environment (contexts AmA, CmA, GmA and UmA are shown) in relation to the proportion of modified RNA oligonucleotide in the mixture.
This data demonstrated once more, that the RiboMethSeq approach can be used for a relative quantification of 2′-O-Me sites in RNA and is relatively sensitive to the methylation rate. Thus, even small changes in 2′-O-methylation rate (>10%) could be detected and quantified. Nevertheless, other independent approaches (e.g. HPLC or HPLC-MS/MS) may be necessary to confirm the absolute rate of modification at a given position identified by RiboMethSeq. Taking into account the precision of other analytical methods used for quantification of modified nucleotides (∼10%), RiboMethSeq gave very comparable results.
Comparison with the previously published RiboMethSeq protocol
The original protocol for RiboMethSeq was published in 2015 (26). This protocol and our modified approach can now be compared under different aspects, including library preparation protocol, amount of required input material as well as precision and accuracy (see Table S4 in Supporting information). The main differences were identified as follows: the previously published Ion Torrent-based protocol used a proprietary direct ligation method, multiple gel purification steps and thus required at least 1–10 μg of RNA as starting material. In our Illumina-based approach we used a standard library preparation protocol widely applied for small RNA analysis, no gel purification was required and thus the minimal amount of starting RNA was found to be 1000× lower (∼1–10 ng). The precision (FDR/accuracy) and number of True/False positives are quite comparable for both protocols.
CONCLUSION
In this work, we describe a method for high-throughput mapping and relative quantification of 2′-O-Me residues in highly abundant or purified/enriched RNAs. Significant improvements over its predecessor (26) include a very low amount of input RNA (minimum 1 ng) required, which is well suitable for precious biological samples or rare purified RNA species. Our study carefully characterizes various aspects ranging from data analysis to sample preparation, and based on this demonstrates that 5′-end coverage alone is generally sufficient for analysis, allowing researchers to use the most common SR50 Illumina sequencing runs for analysis. This type of sequencing can be performed without further adjustments using the whole range of Illumina sequencers, starting from MiSeq and NextSeq to the most recent HiSeq 3000 or 4000 machines. Our method uses common and commercially available procedures for library preparation, does not require gel-purification steps of RNA and can be used for routine analysis of biological and clinical samples to study 2′-O-Me residue profiles.
Supplementary Material
Acknowledgments
We thank P. Keller (IPB, Mainz, Germany) for help in LC–MS/MS measurements of rRNA 2′-O-methylations, J.-J. Diaz lab members (CRCL, Lyon, France), S. Sharma and D.L.J. Lafontaine (ULB, Brussels, Belgium) for fruitful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
ANR-DFG grant HTRNAMod [ANR-13-ISV8-0001/HE 3397/8-1]. Funding for open access charge: ANR-DFG grant HTRNAMod [ANR-13-ISV8-0001/HE 3397/8-1].
Conflict of interest statement. None declared.
REFERENCES
- 1.Machnicka M.A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K.M., et al. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res. 2013;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Motorin Y., Helm M. RNA nucleotide methylation. Wiley Interdiscip. Rev. RNA. 2011;2:611–631. doi: 10.1002/wrna.79. [DOI] [PubMed] [Google Scholar]
- 3.Tycowski K.T., Smith C.M., Shu M.D., Steitz J.A. A small nucleolar RNA requirement for site-specific ribose methylation of rRNA in Xenopus. Proc. Natl. Acad. Sci. U.S.A. 1996;93:14480–14485. doi: 10.1073/pnas.93.25.14480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tycowski K.T., You Z.H., Graham P.J., Steitz J.A. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol. Cell. 1998;2:629–638. doi: 10.1016/s1097-2765(00)80161-6. [DOI] [PubMed] [Google Scholar]
- 5.Motorin Y., Helm M. tRNA stabilization by modified nucleotides. Biochemistry. 2010;49:4934–4944. doi: 10.1021/bi100408z. [DOI] [PubMed] [Google Scholar]
- 6.Ge J., Liu H., Yu Y.-T. Regulation of pre-mRNA splicing in Xenopus oocytes by targeted 2′-O-methylation. RNA. 2010;16:1078–1085. doi: 10.1261/rna.2060210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ji L., Chen X. Regulation of small RNA stability: methylation and beyond. Cell Res. 2012;22:624–636. doi: 10.1038/cr.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao X., Yu Y.-T. Targeted pre-mRNA modification for gene silencing and regulation. Nat. Methods. 2008;5:95–100. doi: 10.1038/nmeth1142. [DOI] [PubMed] [Google Scholar]
- 9.Gehrig S., Eberle M.-E., Botschen F., Rimbach K., Eberle F., Eigenbrod T., Kaiser S., Holmes W.M., Erdmann V.A., Sprinzl M., et al. Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity. J. Exp. Med. 2012;209:225–233. doi: 10.1084/jem.20111044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rimbach K., Kaiser S., Helm M., Dalpke A.H., Eigenbrod T. 2′-O-Methylation within Bacterial RNA Acts as Suppressor of TLR7/TLR8 Activation in Human Innate Immune Cells. J. Innate Immun. 2015;7:482–493. doi: 10.1159/000375460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.-Y., Schneller S., Zust R., Dong H., et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuge H., Brownlee G.G., Gershon P.D., Richter J.D. Cap ribose methylation of c-mos mRNA stimulates translation and oocyte maturation in Xenopus laevis. Nucleic Acids Res. 1998;26:3208–3214. doi: 10.1093/nar/26.13.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahmeh A.A., Li J., Kranzusch P.J., Whelan S.P.J. Ribose 2′-O methylation of the vesicular stomatitis virus mRNA cap precedes and facilitates subsequent guanine-N-7 methylation by the large polymerase protein. J. Virol. 2009;83:11043–11050. doi: 10.1128/JVI.01426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szretter K.J., Daniels B.P., Cho H., Gainey M.D., Yokoyama W.M., Gale M., Virgin H.W., Klein R.S., Sen G.C., Diamond M.S. 2′-O methylation of the viral mRNA cap by West Nile virus evades ifit1-dependent and -independent mechanisms of host restriction in vivo. PLoS Pathog. 2012;8:e1002698. doi: 10.1371/journal.ppat.1002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tollervey D., Lehtonen H., Jansen R., Kern H., Hurt E.C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- 16.Lafontaine D.L., Tollervey D. Birth of the snoRNPs: the evolution of the modification-guide snoRNAs. Trends Biochem. Sci. 1998;23:383–388. doi: 10.1016/s0968-0004(98)01260-2. [DOI] [PubMed] [Google Scholar]
- 17.Tollervey D., Kiss T. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 18.Hori H., Suzuki T., Sugawara K., Inoue Y., Shibata T., Kuramitsu S., Yokoyama S., Oshima T., Watanabe K. Identification and characterization of tRNA (Gm18) methyltransferase from Thermus thermophilus HB8: domain structure and conserved amino acid sequence motifs. Genes Cells Devoted Mol. Cell. Mech. 2002;7:259–272. doi: 10.1046/j.1365-2443.2002.00520.x. [DOI] [PubMed] [Google Scholar]
- 19.Persson B.C., Jäger G., Gustafsson C. The spoU gene of Escherichia coli, the fourth gene of the spoT operon, is essential for tRNA (Gm18) 2′-O-methyltransferase activity. Nucleic Acids Res. 1997;25:4093–4097. doi: 10.1093/nar/25.20.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rashid R., Aittaleb M., Chen Q., Spiegel K., Demeler B., Li H. Functional requirement for symmetric assembly of archaeal box C/D small ribonucleoprotein particles. J. Mol. Biol. 2003;333:295–306. doi: 10.1016/j.jmb.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 21.Tran E.J., Zhang X., Maxwell E.S. Efficient RNA 2′-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C’/D’ RNPs. EMBO J. 2003;22:3930–3940. doi: 10.1093/emboj/cdg368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maden B.E. Mapping 2′-O-methyl groups in ribosomal RNA. Methods (San Diego, Calif) 2001;25:374–382. doi: 10.1006/meth.2001.1250. [DOI] [PubMed] [Google Scholar]
- 23.Maden B.E., Corbett M.E., Heeney P.A., Pugh K., Ajuh P.M. Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie. 1995;77:22–29. doi: 10.1016/0300-9084(96)88100-4. [DOI] [PubMed] [Google Scholar]
- 24.Dong Z.-W., Shao P., Diao L.-T., Zhou H., Yu C.-H., Qu L.-H. RTL-P: a sensitive approach for detecting sites of 2′-O-methylation in RNA molecules. Nucleic Acids Res. 2012;40:e157. doi: 10.1093/nar/gks698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dennis P.P., Tripp V., Lui L., Lowe T., Randau L. C/D box sRNA-guided 2′-O-methylation patterns of archaeal rRNA molecules. BMC Genomics. 2015;16:632. doi: 10.1186/s12864-015-1839-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Birkedal U., Christensen-Dalsgaard M., Krogh N., Sabarinathan R., Gorodkin J., Nielsen H. Profiling of ribose methylations in RNA by high-throughput sequencing. Angew. Chem. Int. Ed. Engl. 2015;54:451–455. doi: 10.1002/anie.201408362. [DOI] [PubMed] [Google Scholar]
- 27.Edelheit S., Schwartz S., Mumbach M.R., Wurtzel O., Sorek R. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet. 2013;9:e1003602. doi: 10.1371/journal.pgen.1003602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hussain S., Aleksic J., Blanco S., Dietmann S., Frye M. Characterizing 5-methylcytosine in the mammalian epitranscriptome. Genome Biol. 2013;14:215. doi: 10.1186/gb4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Squires J.E., Patel H.R., Nousch M., Sibbritt T., Humphreys D.T., Parker B.J., Suter C.M., Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen K., Lu Z., Wang X., Fu Y., Luo G.-Z., Liu N., Han D., Dominissini D., Dai Q., Pan T., et al. High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. Angew. Chem. Int. Ed Engl. 2015;54:1587–1590. doi: 10.1002/anie.201410647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M., et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 32.Dominissini D., Moshitch-Moshkovitz S., Salmon-Divon M., Amariglio N., Rechavi G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat. Protoc. 2013;8:176–189. doi: 10.1038/nprot.2012.148. [DOI] [PubMed] [Google Scholar]
- 33.Carlile T.M., Rojas-Duran M.F., Zinshteyn B., Shin H., Bartoli K.M., Gilbert W.V. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lovejoy A.F., Riordan D.P., Brown P.O. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One. 2014;9:e110799. doi: 10.1371/journal.pone.0110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwartz S., Bernstein D.A., Mumbach M.R., Jovanovic M., Herbst R.H., León-Ricardo B.X., Engreitz J.M., Guttman M., Satija R., Lander E.S., et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dominissini D., Nachtergaele S., Moshitch-Moshkovitz S., Peer E., Kol N., Ben-Haim M.S., Dai Q., Di Segni A., Salmon-Divon M., Clark W.C., et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530:441–446. doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hauenschild R., Tserovski L., Schmid K., Thüring K., Winz M.-L., Sharma S., Entian K.-D., Wacheul L., Lafontaine D.L.J., Anderson J., et al. The reverse transcription signature of N-1-methyladenosine in RNA-Seq is sequence dependent. Nucleic Acids Res. 2015;43:9950–9964. doi: 10.1093/nar/gkv895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X., Xiong X., Wang K., Wang L., Shu X., Ma S., Yi C. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat. Chem. Biol. 2016;12:311–316. doi: 10.1038/nchembio.2040. [DOI] [PubMed] [Google Scholar]
- 39.Tserovski L., Marchand V., Hauenschild R., Blanloeil-Oillo F., Helm M., Motorin Y. High-throughput sequencing for 1-methyladenosine (m(1)A) mapping in RNA. Methods. 2016 doi: 10.1016/j.ymeth.2016.02.012. doi:10.1016/j.ymeth.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 40.Collart M.A., Oliviero S. Preparation of yeast RNA. Curr. Protoc. Mol. Biol. 2001 doi: 10.1002/0471142727.mb1312s23. doi:10.1002/0471142727.mb1312s23. [DOI] [PubMed] [Google Scholar]
- 41.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buchhaupt M., Sharma S., Kellner S., Oswald S., Paetzold M., Peifer C., Watzinger P., Schrader J., Helm M., Entian K.-D. Partial methylation at Am100 in 18S rRNA of baker's yeast reveals ribosome heterogeneity on the level of eukaryotic rRNA modification. PLoS One. 2014;9:e89640. doi: 10.1371/journal.pone.0089640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crain P.F. Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry. Methods Enzymol. 1990;193:782–790. doi: 10.1016/0076-6879(90)93450-y. [DOI] [PubMed] [Google Scholar]
- 44.Thüring K., Schmid K., Keller P., Helm M. Analysis of RNA modifications by liquid chromatography-tandem mass spectrometry. Methods. 2016 doi: 10.1016/j.ymeth.2016.03.019. doi:10.1016/j.ymeth.2016.03.019. [DOI] [PubMed] [Google Scholar]
- 45.Land H., Grez M., Hauser H., Lindenmaier W., Schütz G. 5′-Terminal sequences of eucaryotic mRNA can be cloned with high efficiency. Nucleic Acids Res. 1981;9:2251–2266. doi: 10.1093/nar/9.10.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sooknanan R., Pease J., Doyle K. Novel methods for rRNA removal and directional, ligation-free RNA-seq library preparation. Nat. Methods. 2010;7 http://www.nature.com/nmeth/journal/v7/n10/pdf/nmeth.f.313.pdf [Google Scholar]
- 47.Kuchino Y., Borek E., Grunberger D., Mushinski J.F., Nishimura S. Changes of post-transcriptional modification of wye base in tumor-specific tRNAPhe. Nucleic Acids Res. 1982;10:6421–6432. doi: 10.1093/nar/10.20.6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsuo M., Yokogawa T., Nishikawa K., Watanabe K., Okada N. Highly specific and efficient cleavage of squid tRNA(Lys) catalyzed by magnesium ions. J. Biol. Chem. 1995;270:10097–10104. doi: 10.1074/jbc.270.17.10097. [DOI] [PubMed] [Google Scholar]
- 49.Meyer K.D., Jaffrey S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014;15:313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yi C., Pan T. Cellular dynamics of RNA modification. Acc. Chem. Res. 2011;44:1380–1388. doi: 10.1021/ar200057m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kakihara Y., Makhnevych T., Zhao L., Tang W., Houry W.A. Nutritional status modulates box C/D snoRNP biogenesis by regulated subcellular relocalization of the R2TP complex. Genome Biol. 2014;15:404. doi: 10.1186/s13059-014-0404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.