Abstract
The 5′ m7G cap is an evolutionarily conserved modification of eukaryotic mRNA. Decades of research have established that the m7G cap serves as a unique molecular module that recruits cellular proteins and mediates cap-related biological functions such as pre-mRNA processing, nuclear export and cap-dependent protein synthesis. Only recently has the role of the cap 2′O methylation as an identifier of self RNA in the innate immune system against foreign RNA has become clear. The discovery of the cytoplasmic capping machinery suggests a novel level of control network. These new findings underscore the importance of a proper cap structure in the synthesis of functional messenger RNA. In this review, we will summarize the current knowledge of the biological roles of mRNA caps in eukaryotic cells. We will also discuss different means that viruses and their host cells use to cap their RNA and the application of these capping machineries to synthesize functional mRNA. Novel applications of RNA capping enzymes in the discovery of new RNA species and sequencing the microbiome transcriptome will also be discussed. We will end with a summary of novel findings in RNA capping and the questions these findings pose.
INTRODUCTION
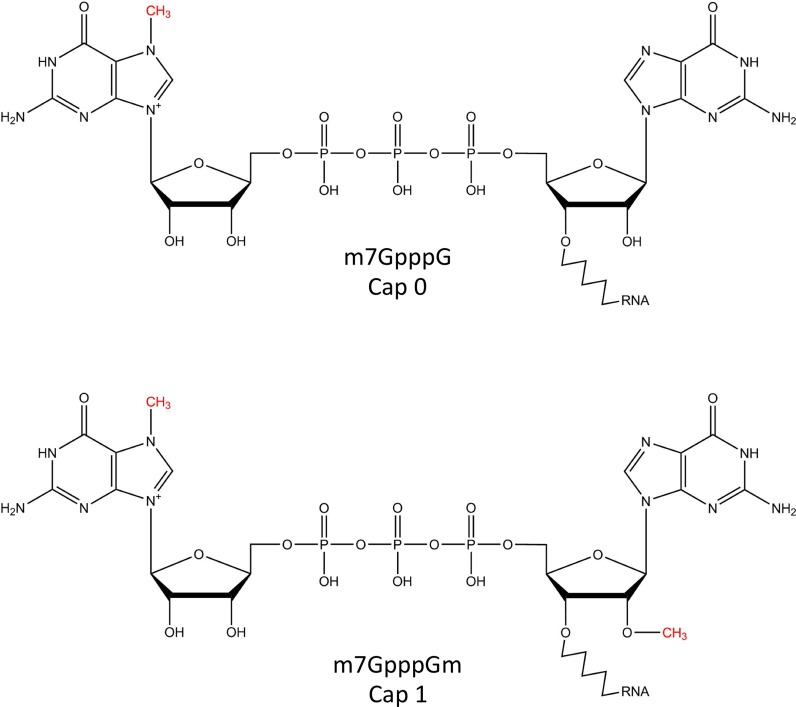
All eukaryotic mRNA contains a cap structure - an N7-methylated guanosine linked to the first nucleotide of the RNA via a reverse 5′ to 5′ triphosphate linkage (Figure 1). In addition to its essential role of cap-dependent initiation of protein synthesis, the mRNA cap also functions as a protective group from 5′ to 3′ exonuclease cleavage and a unique identifier for recruiting protein factors for pre-mRNA splicing, polyadenylation and nuclear export. It also acts as the anchor for the recruitment of initiation factors that initiate protein synthesis and the 5′ to 3′ looping of mRNA during translation. Recent studies have revealed that 2′O methylation of +1 nucleotide (cap 1 structure; Figure 1) is central to the non-self discrimination of innate immune response against foreign RNA (1). Structural studies have shed light on the structural basis of such discrimination (2). The recent characterization of a cytoplasmic (re)-capping complex has added a potentially new layer of control on protein synthesis and RNA-based regulatory network (3).
Figure 1.
mRNA caps in eukaryotes.
Excellent reviews on the biological roles of m7G cap 0 and cap 1 in eukaryotes and the viral RNA capping machineries are available in the literature (4,5). In this review, we will give an update in these areas with a focus on the molecular basis of innate immunity against non-cap 1 5′ structures, cytoplasmic re-capping and the cap quality control machinery. We will also discuss how we can take advantage of the viral capping machineries for the synthesis of functional mRNA, and how RNA capping enzymes can help discover new RNA species and sequence the microbiome transcriptome. We will conclude by overviewing the questions novel findings in RNA capping pose to the larger scientific community.
mRNA capping in eukaryotes
Nuclear RNA capping
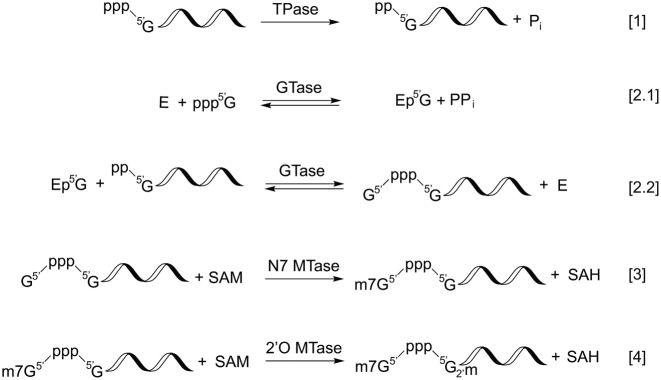
Capping is the first modification made to RNA polymerase II-transcribed RNA and takes place co-transcriptionally in the nucleus as soon as the first 25–30 nts are incorporated into the nascent transcript (6,7). Three enzymatic activities are required to generate the cap 0 structure, namely, RNA triphosphatase (TPase), RNA guanylyltransferase (GTase) and guanine-N7 methyltransferase (guanine-N7 MTase) (Table 1). Each of these enzyme activities carries out an essential step in the conversion of the 5′ triphosphate of nascent RNA to the cap 0 structure. RNA TPase removes the γ-phosphate from the 5′ triphosphate to generate 5′ diphosphate RNA (Figure 2, reaction 1). GTase transfers a GMP group from GTP to the 5′ diphosphate via a lysine-GMP covalent intermediate (Figure 2, reactions 2.1 and 2.2). The guanine-N7 MTase then adds a methyl group to the N7 amine of the guanine cap to form the cap 0 structure (Figure 2, reaction 3). Additionally, the m7G-specific 2′O methyltransferase (2′O MTase) methylates the +1 ribonucleotide at the 2′O position of the ribose to generate the cap 1 structure (Figure 2, reaction 4).
Table 1. Diversity of RNA capping machinery.
|
Figure 2.
Enzymatic steps involved in RNA capping. The RNA triphosphatase activity (TPase) removes the γ-phosphate from 5′ triphosphate, generating a diphosphate 5′ end and inorganic phosphate (reaction [1]). The guanylyltransferase (GTase) activity consumes a GTP molecule and forms a covalent intermediate containing with a lysyl-Nζ-5′-phosphoguanosine (reaction [2.1]). In the presence of a 5′ diphosphate RNA, the GTase activity transfers the 5′-phosphoguanosine (GMP) to the 5′ diphosphate, forming a 5′-5′ triphosphate linkage between the first base of the RNA and the capping base (reaction [2.1]). In the presence of S-adenosylmethionine (SAM), the guanine-N7 methyltransferase (MTase) activity adds a methyl group to N7 amine of the guanosine cap to form the cap 0 structure (reaction [3]). Finally, the m7G cap-specific 2′O MTase modifies the 2′O of +1 ribose and generates the cap 1 structure (reaction [4]).
Although these enzymatic activities are conserved in eukaryotes, the configuration of the capping machineries varies. In Saccharomyces cerevisiae, three separate proteins (Cet1, Ceg1 and Abd1) carry out individual activities (8). In metazoans, the RNA TPase is located at the N-terminus of a bifunctional protein RNA guanylyltransferase and 5′-triphosphatase RNGTT (Mce1 in mice) where the GTase is located at the C-terminus (9,10). A separate protein Hcm1 carries the guanine-N7 MTase activity (11). The human enzymes that methylate the 2′O position of the +1 and +2 ribose to form the cap 1 and cap 2 structures, respectively, have recently been identified (12,13).
First reported in yeast and later shown in mammalian cells, the nuclear RNA capping enzyme interacts with the polymerase subunit of RNA polymerase II complex at phosphorylated Ser5 of the C-terminal heptad repeats (14–16). RNA guanine-N7 methyltransferase also interacts with the RNA polymerase II phosphorylated heptad repeats (reviewed in (17)).
In S. cerevisiae, Cet1 and Ceg1 complexes equilibrate between heterotrimeric (Cet12Ceg1) and heterotetrameric (Cet12Ceg12) forms in vitro (18). A recent report of a cryo-EM-derived model of the active transcribing RNA polymerase II-capping enzyme complex showed that the heterotetrameric form is the major species that interacts with the RNA polymerase (8). Although the interaction between Ceg1 and the phosphorylated RNAP II CTD has been well-studied (16,19), the cryo-EM study showed that Cet1 also forms extensive interactions with the transcribing RNA polymerase II complex outside of CTD. Ceg1 appears to be mobile and adopts multiple conformations in the transcribing complex. Contacts with RNAP subunit Rpb7 have also been observed (8).
Cytoplasmic RNA (re)-capping
Previously thought to take place exclusively in the nucleus, RNA capping has been reported in the cytoplasm of mammalian cells and Trypanosomes. Eukaryotic cells maintain a cytoplasmic pool of uncapped mRNA in the form of messenger ribonucleoprotein (mRNP) in the P-bodies, where storage, deadenylation and decapping of the mRNA can take place (20–22). Uncapped mRNA in P-bodies can re-enter polysome and be translated (21). Interestingly, the length of poly(A) tail of mRNP in P-bodies can become shorter, longer or unchanged for different mRNAs, and the capacity to re-enter polysome depends on the presence of cap but not the length of poly(A) tail (23). From human osteosarcoma U-2 OS cells, a cytoplasmic capping complex composed of RNGTT, a 5′ monophosphate kinase and Nck1 has been found to recap 5′ monophosphate RNA in vitro (3,24,25) (Table 1). While majority of the re-capped RNA are mapped to 5′ CAGE sites, about 25% of them map to internal CAGE sites (26,27) and promoter-associated short RNAs (PASRs) (27,28). This supports the notion that some of the PASRs are processed transcripts capped in the cytoplasm (28). In Trypanosoma brucei, a bifunctional enzyme TbCe1 that phosphorylates 5′ monophosphate and caps the resulting diphosphate RNA in the cytoplasm has recently been identified (29). In vitro, TbCe1 prefers RNA containing the T. brucei splice leader sequence containing 2′O methylation in the first 4 nucleotides, suggesting that previously hypermethylated and decapped T. brucei mRNA is the preferred substrate in vivo (29).
The cytoplasmic re-capping system may represent a novel mRNA inactivation-reactivation mechanism that helps regulate protein synthesis (29,30). Recapping of internal CAGE sites and non-coding RNAs such as PASRs suggests a role of the cap in an unexpected diversity of protein products and yet unknown regulatory networks, respectively (24,26).
The cap quality control system
Pre-mRNA splicing and polyadenylation has been linked to the recently characterized cap quality control mechanism. In mammalian cells, a trifunctional protein DXO/Dom3Z that possesses decapping, pyrophosphohydrolase and 5′ to 3′ exonuclease activities has been reported (31). The enzyme specifically decaps unmethylated cap (GpppN) and degrades the resulting 5′ monophosphate RNA using its 5′ to 3′ exonuclease activity. Knockdown of DXO/Dom3Z results in an increased level of pre-mRNA but not mature methyl-capped mRNA, suggesting that the enzyme actively removes capped but unmethylated pre-mRNA from the cells. The knockdown also leads to defective splicing of the first and subsequent introns, as well as defective cleavage of 3′ polyadenylation site in vivo (31), demonstrating a link between cap N7-methylation and pre-mRNA splicing and polyadenylation.
Saccharomyces cerevisiae has been found to possess two sets of partially redundant machinery for RNA cap quality control. In the Rai1–Rat1 system, Rai1 possesses pyrophosphatase and decapping activity (32). It interacts with Rat1, the yeast homolog of mammalian XRN2, and stimulates the Rat1 exoribonuclease activity (32–34). In addition, S. cerevisiae encodes Dxo1 that possesses both decapping and 5′ to 3′ exonuclease activity (34). While ΔRai1 yeast cells show significant accumulation of unmethylated capped RNA only during glucose or amino acid starvation (32), ΔRai1ΔDox1 yeast cells accumulate unmethylated capped RNA under normal growth conditions (34).
Biological roles of the m7G cap
The m7G cap plays a major role in the coordination of various functional processes that take place throughout the life cycle of mRNA. This is largely attributed to protein factors that bind specifically to the cap structure: the cap-binding complex (CBC) in the nucleus and eIF4E in the cytoplasm.
In the nucleus: mRNA processing and nuclear export
First shown in HeLa cellular extract (35,36) and later in other mammalian systems (37) and S. cerevisiae (38), the m7G cap is required for efficient pre-mRNA splicing. This is mediated through recruiting and binding to the nuclear cap-binding complex (CBC), which orchestrates processes such as spliceosome assembly, 3′ processing, RNA export, miRNA biogenesis and nonsense mediated decay. The composition and functions of CBC have been reviewed elsewhere (39–41). Here we will focus on mRNA cap-mediated processing and nuclear export.
In vivo, mRNA interacts with protein factors throughout its life cycle and should be considered messenger ribonucleoprotein (mRNP). CBC, consisting of CBP80 and CBP20, binds to the m7G cap co-transcriptionally as a CBP80/20 heterodimer. The cap bound-CBC then forms a complex with the U4/U6·U5 snRNPs and initiates splicing (42). CBC also contributes to pre-mRNA 3′ end processing—the cap-bound CBC co-immunoprecipitated with 3′ processing factors bound to the polyadenylation site (43). Depletion of CBC from HeLa cell nuclear extract attenuates the endonucleolytic cleavage step of polyadenylation, which can be restored by the addition of recombinant CBC (43). Through interactions with CBC, the cap is also involved in transcription termination and exosomal degradation (44). These findings highlight the central role of the cap structure in the recruitment of multiple protein complexes that influence the outcome of mRNA splicing and 3′-end formation.
While dispensable for the majority of mRNA export in S. cerevisiae, the interaction of CBC with the m7G cap is essential for nuclear export in higher eukaryotes (45). The directional exit of mRNA, with the 5′ end leading the way (46), is attributed to the interaction of CBC with numerous nuclear export factors such as REF (RNA export factor, also known as Aly) of the transcription export complex TREX (47). The interaction of cap-bound CBC with REF, which also interacts with exon junction complex, is splicing-dependent in vertebrates (48). These export factors directly or indirectly interact with nuclear pore complex (NPC) and direct the mRNP payload to the cytoplasm. For further details about the mechanisms of mRNA processing, nuclear export and its regulation we refer the reader to these recent reviews (41,49,50).
In the cytoplasm: translation initiation, and mRNA pseudo-circularization
The majority of cellular mRNA translation is initiated by the cap-dependent mechanism. Upon exit into the cytoplasm, CBC stays bound to the mRNA cap and recruits eIF4G and RNA helicase eIF4A to the 5′ end of the mRNA. Further recruitment of other initiation factors, such as cap binding protein CBP80/20-dependent translation initiation factor (CTIF), eIF3g and eIF4III, Met-tRNAi and the two ribosomal subunits initiates the CBC-dependent pioneer round of translation, where nonsense-mediated decay takes place (51–56). After the first round of translation, remodeling of the mRNP initiation complex takes place. Importin (IMP)-β binds to IMP-α, a stable binding partner of cap-bound CBC, and initiates the replacement of CBC by eIF4E, which interacts with the eIF4F complex and starts the steady-state rounds of translation (55,57). Recently, advances in cryo-electron topography have allowed imaging of subcellular structures including the polysomes. ER-bound ribosomes are clearly clustered in tandem as polysomes where the mRNA entry and exit channels of adjacent ribosomes are aligned to allow smooth threading of mRNA (58).
When bound to the mRNA cap, eIF4G of the eIF4F complex interacts with poly(A) binding protein PABP1 bound to the poly(A) tail of mRNA and create a pseudo-circular structure of translating mRNA (59,60). The pseudo-circularization of mRNA has been postulated to help ensure that full-length mRNAs are translated and enhance the processivity of the ribosome (61,62). The m7G cap, in essence, is a unique anchor on mRNA where protein factors bind and drive the cap-related functions for the mRNP.
2′O methylated cap (cap 1) as a signature of self RNA
While the m7G cap 0 structure is known to be required for efficient translation of mRNA (63), the biological role of 2′O methylation, outside of c-mos mRNA translation upregulation during oocyte maturation (64), has been obscure until recent work demonstrates that cellular sensors RIG-I and MDA5 and effectors IFIT1 and IFIT5 of the Type I interferon signally pathway act by discriminating cap 1 RNA from others.
Cytoplasmic pattern recognition receptors (PRRs) RIG-I and RIG-I-like receptor MDA5 are sensors that trigger cellular type I interferon (IFN) response to virus infections (65). While MDA5 interacts with long dsRNA (66–70), RIG-I interacts strongly with short dsRNA, and less so with 5′-ppp and 5′-pp ssRNA (71–75). Most significantly, the cap 1 structure abolished the interactions of dsRNA with RIG-I and MDA5 and hence did not activate the IFN signaling pathway (68,76).
Unexpectedly, cap 0 and 5′-ppp dsRNA bind to RIG-I with similar affinity (2). Instead, 2′O methylation of 5′-ppp RNA (5′-ppp(2′OMe)N…) significantly diminishes the binding of the dsRNA to RIG-I and IFN signaling. The interaction with RIG-I and downstream INF induction is further attenuated by the complete cap 1 structure (2,76). In fact, the crystal structure of RIG-I in complex with cap 0 RNA showed that RIG-I made numerous contacts with the 5′ triphosphate but not with the cap structure (2). Mutation of His830, which is in close proximity to the 2′OH of the +1 nucleotide, to alanine rendered RIG-I an ATPase turnover rate similar to that of WT RIG-I on unmethylated dsRNA and triggers the INF response (2). This demonstrated that RIG-I uses His830 as a steric gate to discriminate against cap 1 RNA.
IFN induces the expression and association of IFIT complex, which blocks the translation of mRNA lacking 2′O methylation, leading to the inhibition of viral RNA replication (77–81). West Nile virus ((+)ssRNA genome) lacking 2′O MTase activity was attenuated in primary cells and mice but was virulent in cells with defective IFN signaling pathway (1). IFIT1 and IFIT5, two of the IFIT complex components, interact strongly with 5′-ppp ssRNA (77,81). Similar to RIG-I, crystal structures of IFIT5 in complex with 5′-ppp ssRNAs revealed that the protein interacts directly and indirectly with the 5′ phosphates, consistent with its high affinity interactions with 5′-p, 5′-ppp and cap 0 ssRNA but not with cap 1 ssRNA (78).
The above evidence supports the hypothesis that RIG-I and MDA5 are upstream sensors and that the IFIT complex is a dual sensor-effector of an innate defense system that targets foreign RNA without proper 5′ modifications. An innate immune response built against RNA without 2′O methylation at its cap strongly suggests that this 2′O methylation is a signature of self RNA (1). More detailed reviews of the biological roles of IFIT proteins, RIG-I and MDA5 can be found in the literature (79,82).
Enzymatic activities involved in RNA capping
Conversion of an RNA transcript to cap 0 RNA requires three sequential enzymatic steps: the removal of the 5′ terminal γ-phosphate by RNA triphosphatase activity (TPase), the transfer of a GMP group to the resultant diphosphate 5′ terminus by RNA guanylyltransferase activity (GTase) and the modification of the N7 amine of the guanosine cap by guanine-N7 methyltransferase activity (MTase) (Figure 2).
RNA triphosphatase activity
RNA triphosphatase, categorized as a polynucleotide 5′-phosphatase (EC 3.1.3.33), converts the terminal triphosphate of polyribonucleotides to diphosphate (Reaction 1, Figure 2) and hydrolyzes ribonucleoside triphosphate to diphosphate in vitro. Two structurally and mechanistically distinct groups of enzymes carry out the cap-related RNA TPase activity.
In metazoans, the RNA triphosphatase is independent of divalent metal ions and invariably physically linked to the GTase activity in a bifunctional protein (Mce1 in mice; generally known as RNA guanylyltransferase and 5′-triphosphatase RNGTT in mammals). These metal-independent RNA TPases contain the conserved HCXXXXXR(S/T) motif of the cysteine phosphatase superfamily that includes protein tyrosine phosphatases and phosphoinositide phosphatases. The conserved cysteine is located near the bottom of a deep cleft and forms a covalent cysteinyl phosphoenzyme intermediate with the γ-phosphate during the cleavage of the β–γ phosphoanhydride bond. The deep cleft of the active site is proposed to allow the enzyme to differentiate terminal 5′ triphosphate from 5′ diphosphate of RNA (83).
The RNA TPase of lower eukaryote and most DNA virus capping enzymes are dependent on divalent metal ions for catalysis (4,84,85). Available crystal structures show that these TPases share a β-barrel tunnel structure where the catalysis takes place. The β-barrel structure is the defining structural and catalytic module of the triphosphate tunnel metallozymes (TTMs) whose preferred substrates invariably contain a triphosphate group. In addition to RNA TPases, they include the bacterial class IV adenylate cyclase cyaB, mammalian thiamine triphosphatases (86,87) and inorganic polyphosphatases (88–90). Biochemical and structural evidence suggests different nucleophiles are involved in different enzymes, but these TTMs share the conserved feature of coordinating the metal ions by negatively charged amino acid residues and the positioning and stabilizing the γ-phosphate by positively charged residues (91). Mutation of the charged residues lining the tunnel of the prototypic RNA TPase Cet1 from S. cerevisiae leads to the loss of in vitro TPase activity and a lethal phenotype (84,92).
RNA guanylyltransferase activity
RNA guanylyltransferase, formally known as GTP-RNA guanylyltransferase (EC2.7.7.50) transfers a GMP moiety from GTP to the 5′ diphosphate of TPase-processed RNA, forming a capped 5′ end. RNA GTases belong to the subfamily of nucleotidyl transferases that includes ATP- and NAD+-dependent DNA ligases (93). This class of nucleotidyl transferases are structurally and mechanistically conserved in all domains of life. They catalyze nucleic acid ligation through a two-step mechanism that involves a lysyl-Nζ-linked covalent intermediate and the formation of a 5′-5′ phospho(deoxy)ribose product.
For RNA GTases, the lysine residues within the Kx(D/N)G motif carries out the nucleophilic attack on the α-phosphate of GTP, breaking the α-β phosphoanhydride bond and forming a covalent lysyl-Nζ-GMP intermediate (Figure 2, Reaction 2.1) (94,95). The GMP moiety is then transferred to the 5′ diphosphate to form capped G(5′)ppp(5′)RNA (Figure 2, Reaction 2.2). The GTase reaction is highly reminiscent of the first two steps of the reaction catalyzed by ATP- and NAD+-dependent DNA ligases (DNA 5′ adenylylation). Upon the formation of the covalent lysyl-Nζ-AMP intermediate, these ligases transfer the AMP moiety to the 5′ monophosphate of the DNA, forming the adenylated A(5′)pp(5′)DNA.
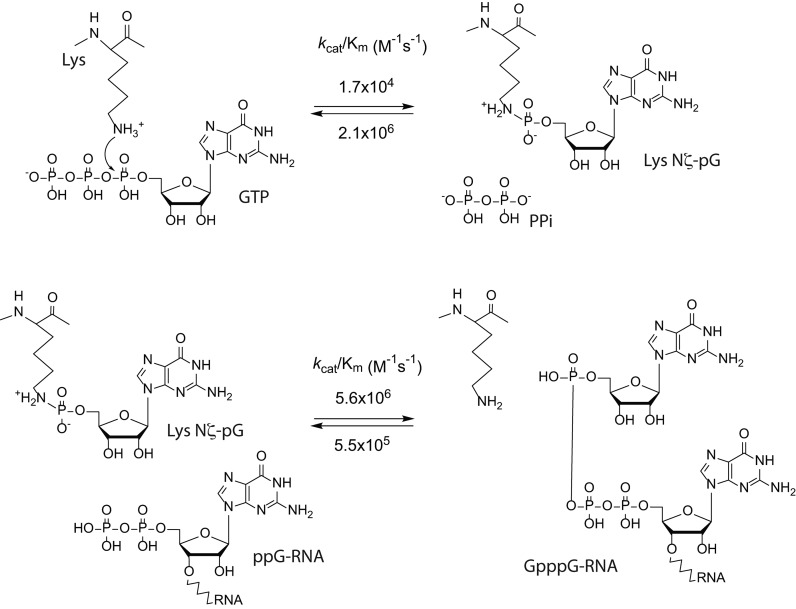
An interesting characteristic of the GTase reaction is its high reversibility. A detailed kinetic and thermodynamic study of Chlorella virus GTase reported that in the absence of a cap-accepting RNA, the reverse reaction of the first half of the reaction (enzyme self-guanylylation; Figure 2, Reaction 2.1) can proceed with a lower substrate concentration and at a much higher rate than the forward reaction (96) (Figure 3). On the other hand, the second order rate constant of the forward reaction of the second half of the reaction (the transfer of GMP to ppRNA; Figure 2, Reaction 2.2) is 10-fold higher than that of the reverse reaction (Figure 3), suggesting that the presence of a cap-accepting RNA drives the GTase reaction forward.
Figure 3.
Guanylyltransferase reaction. The GTase reaction consists of two steps: protein self-guanylylation and the transfer of the GMP group to the 5′ diphosphate RNA. The GTase reaction catalyzed by chlorella virus PBCV-1 capping enzyme is highly reversible. A detailed kinetics study of the forward and reverse reaction of both steps revealed that in the absence of RNA substrate, the kcat/Km value of the reverse reaction of the first step (the pyrophosphorolysis of the lysyl-Nζ-5′-phosphoguanosine intermediate into lysine and GTP) is 120 times higher than that of the forward reaction. The second step (the transfer of the GMP moiety from the lysyl-Nζ-5′-phosphoguanosine to diphosphate 5′ end) is largely forward-tending, with a 10-fold difference in the kcat/Km values (96).
Similar to DNA and RNA ligases, RNA GTases are generally composed of two domains—the N-terminal GTase that contains the conserved KxDG motif and the C-terminal Oligonucleotide/oligosaccharide binding (OB) domain. The OB-folds are made up of <200 residues and are involved in ssDNA-, ssRNA- and protein-protein interactions (97). OB-fold-containing proteins are present in all kingdoms of life and are involved in diverse yet critical activities such as DNA replication, repair, recombination, transcription, cold shock response and telomere maintenance (97–99).
The OB-fold consist of five-stranded beta-barrel capped on one end by an alpha helix located between the third and fourth strands and presents a binding cleft at the other end (99). The loops connecting the β strands can vary in sequence, length, and conformation and may contribute to the binding specificities of the OB folds. The OB folds often present as tandem repeats in proteins where they may confer cooperative binding to the nucleic acids (99).
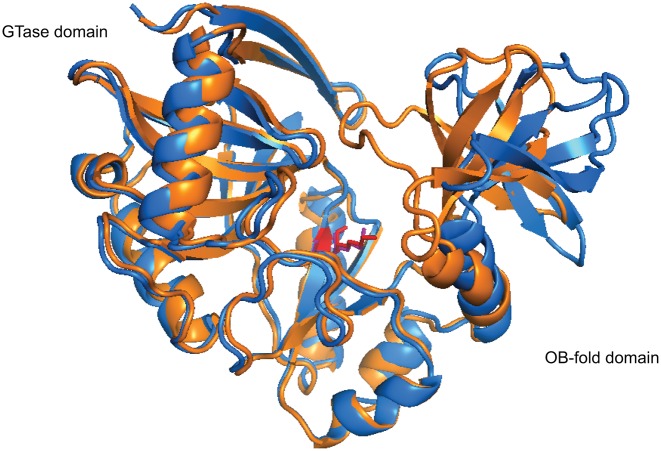
The OB-fold domains are known to remodel the protein structure, the ligand, and in many cases co-fold the interacting parties upon ligand binding (97). Crystal structures of capping enzymes in free, GTP-bound states and the covalent GMP-bound intermediates demonstrates that significant movements of the OB-fold domain relative to the GTase domain is associated with substrate interaction and chemistry steps (93). In the crystal structures of Chlorella virus PBCV-1 GTase, the asymmetric unit consists of two capping enzyme molecules where one exhibits an open conformation with a narrow but deep cleft between the GTase and OB domains. The other molecule adopts a ‘closed’ conformation where the cleft is closed off from the solvent due to rigid movement of the OB domain (Figure 4). Only the closed confirmation is capable of binding the metal ion cofactor and forming the covalent enzyme-GMP intermediate (100). Opened and closed conformers have also been observed in crystal structures of human and Candida albicans GTases (9,100), both of which show an overall structural conservation in the guanine binding pocket. How this OB-fold domain-mediated opening and closing of the active site cleft involves in substrate binding, product release and catalysis of the GTase reaction is still an unanswered question.
Figure 4.
The opened and closed conformations of Chlorella virus PBCV-1 capping enzyme (PDB 1CKM). The PBCV-1 RNA capping enzyme adopts a bilobed structure where the N-terminal guanylyltransferase domain and C-terminal OB fold domain sit on either side of the protein with the active site situated at the interface of the two domains. The asymmetric unit of the crystal structure of Chlorella virus capping enzyme contains two protein units, each exhibiting a different conformation. In the opened conformer (blue), the cleft between the two domains is ∼15 Å at its widest, whereas in the closed conformer (orange), the cleft closes one end off to the solvent completely due to rigid movement of the OB-fold domain. Interestingly, Lys82, the nucleophile that forms the lysyl-Nζ-linkage with incoming GMP (red and violet), shows very limited movement within the active site (100).
RNA guanine-N7 methyltransferase activity
Guanine-N7 MTase (EC 2.1.1.56) catalyzes the transfer of a methyl group from S-adenosyl methionine (SAM) to GpppRNA to form m7GpppRNA and S-adenosyl homocysteine (SAH) (Reaction 3, Figure 2). RNA guanine-N7 MTases are classified as Rossmann fold methyltransferase (RFM) because the SAM-binding domain is structurally similar to the Rossmann fold (101,102).
The transfer of the methyl group is believed to undergo the classic SN2 mechanism exemplified by exocyclic amine DNA methyltransferases. The sulfur-linked methyl carbocation of SAM acts as a strong electrophile whereas the primary amine at the N7 position of the guanosine cap is the nucleophile. In most characterized N6 adenosine DNA MTases, the N6 amine of the deoxyadenosine is invariably in close proximity to a basic amino acid residue that deprotonates the nitrogen as it attacks the electrophilic methyl carbocation (103). Curiously, in the crystal structures of guanine-N7 MTase Ecm1 of the microsporidian parasite Encephalitozoon cuniculi and Bluetongue virus capping enzyme VP4, no direct contacts between the enzyme and the N7 atom or the methyl carbon of SAM can be found. This led to the postulation that RNA guanine-N7 MTases catalyze by coordinating the reacting parties in the correct position instead of stabilizing the transition state or activating the nucleophile (104,105).
As the last step of cap 0 formation, the cap methylation process is regulated by numerous mechanisms. c-Myc protein, known to play a major role in cellular protein synthesis, cell proliferation and transformation, has been shown to upregulate mammalian RNA guanine-N7 MTase (RNMT) activity via upregulating S-adenosyl homocysteine hydrolase (SAHH), which hydrolyzes S-adenosyl homocysteine, a inhibitory byproduct of methyltransferase reaction (106,107). Previously thought to act as a monomer, RNMT has been shown to interact with an uncharacterized protein, RAM/Fam103a1. RAM increases the binding affinity of RNMT to RNA, activates its MTase activity and recruits the RNMT-RAM complex to transcription initiation sites (108,109). RAM is also found to be required for the maintenance mRNA levels, translation and cell viability (108). More recently, the cap methylation process is found to be further regulated by the phosphorylation of RNMT in a cell cycle dependent manner. CDK1-cyclin B1 phosphorylates Thr77 of RNMT during G2/M phase and inhibits RNMT's interaction with KPNA2, a known RNMT inhibitor. RNMT Thr77 phosphorylation leads to an increase of m7G MTase activity at the beginning of G1 phase, presumably to keep up with the surge of mRNA transcription (110). Such control mechanism, along with the cap quality control mechanism mentioned earlier, suggests that the methylation status of the cap is potentially an important regulation point in gene expression.
Viral RNA capping
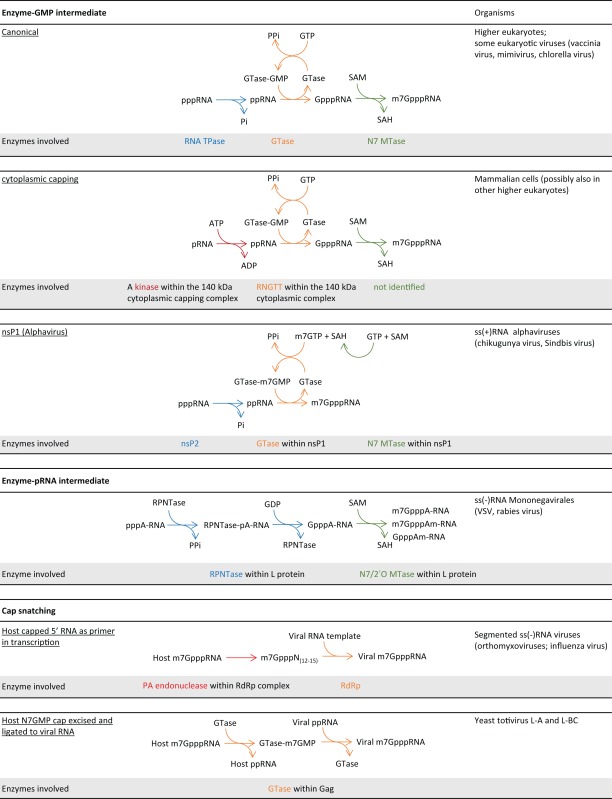
Given the multiple important roles of the cap structure in protein translation and innate immunity, viruses have evolved to produce capped RNA for efficient protein synthesis and evasion of the innate immune response from the host cell. Since the majority of cellular RNA capping takes place in the nucleus, viruses that replicate in the cytoplasm generate their own RNA caps. It is achieved by encoding their own RNA capping machineries or stealing the cap from host mRNA (cap snatching).
Viral RNA capping machineries
As diverse as viruses are, the viral RNA capping machinery is far from monolithic (Table 1). Chlorella virus (dsDNA) has the GTase and TPase activities located in separate proteins. Vaccinia virus (dsDNA) and Bluetongue virus (dsRNA) encode multifunctional proteins that generate cap 0 and cap 1 RNA, respectively. Flavivirus ((+)ssRNA), such as Dengue virus and West Nile virus, and Paramyxoviruses ((-)ssRNA) tether the GTase and MTase activities to their RNA-dependent RNA polymerase (RdRp).
Alphavirus ((+)ssRNA), such as chikungunya virus, has reshuffled the order of the enzyme steps for RNA capping. GTP is first modified and covalently linked to the GTase as m7GMP (111), which is then transferred to the processed ppRNA to form a m7G-cap (112) (Table 1). The alphavirus GTase activity is located on nsP1, which also contains the guanine-N7 MTase activities. The RNA TPase activity is located in nsP2, which is part of the RNA-dependent RNA polymerase complex and the protease that processes the viral polyprotein P1234 into separate non-structural proteins. The capping machinery in Alphavirus is coupled to the viral RdRp activity located in nsP4 through protein-protein interactions (113,114).
Rhabdovirus ((−)ssRNA) adopts a unique RNA capping machinery. Instead of a GTase that forms an enzyme-GMP covalent intermediate, Vesicular Stomatitis Virus encodes a RNA:GDP polyribonucleotidyltransferase (PRNTase) that forms a covalent linkage to the 5′ end of the viral RNA through a monophosphate group. The enzyme then transfers the 5′ monophosphate RNA to GDP, forming the GpppRNA structure, which is further modified to m7G cap 0 and 2′O methylated cap 1 (Table 1). Most interestingly, the 2′O MTase shares the same SAM-binding site with the guanine-N7 MTase and does not require N7 methylation for 2′O methylation. The methylation activities and the PRNTase activity are encoded within the L-protein which also contains the viral RdRp activity (115–118).
Flaviviruses encodes a single MTase that catalyzes the methylation of the N7 amine of the guanosine cap and the 2′O of the +1 nucleoside in a sequential manner (119). Evidence supports a model where the West Nile Virus cap MTase modifies GpppA-RNA to m7GpppA-RNA, which then dissociates and re-associates with another MTase molecule at a GTP-binding pocket for 2′O methylation (120).
Multifunctional capping enzymes
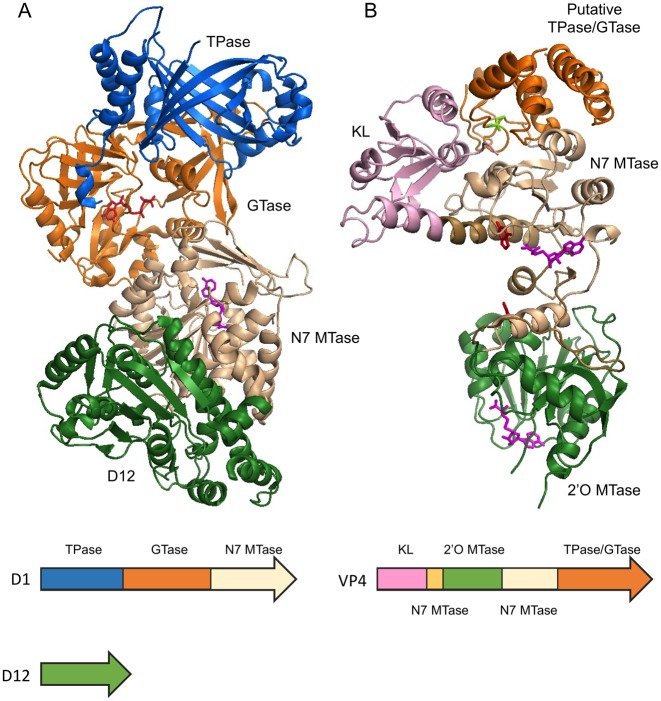
Some viral capping enzymes consolidate all the necessary enzymatic activities of RNA capping to generate cap 0 or cap 1 in a single polypeptide. Vaccinia virus capping enzyme and bluetongue virus capping enzyme are two examples whose full-length protein structures have been solved (Figure 5).
Figure 5.
Multifunctional RNA capping enzymes. (A) Vaccinia capping enzyme structure co-crystallized with GTP and SAH (PDB 4CKB). The enzyme composes of two subunits D1 and D12. The functional domains are ordered as TPase (blue), GTase (orange) and guanine-N7 MTase (beige) from N- to C-terminus in D1. D12 subunit is colored in green. Note the β-barrel characteristic of triphosphate tunnel metaloenzymes (TTM). As indicated by the SAH molecule (magenta), the guanine-N7 MTase active site opens to the back, away from the GTase active site as indicated by the GTP molecule (red). (B) Bluetongue virus capping enzyme VP4 structure. The crystal form that contains two guanine molecules (red) and two SAH molecules (magenta) is shown (PDB 2JHP). The functional domains are arranged from N- to C-terminus in the order of kinase-like (KL) domain (pink), 2′O MTase domain (green), guanine-N7 MTase and the putative combined TPase/GTase domain (orange). The guanine-N7 MTase domain is composed of two discontinuous sequences which are colored in light brown and beige. Two stretches of polypeptide of 10 and 13 amino acid residues within the C-terminal TPase/GTase domain are missing in the structure. The SAH molecules (magenta) clearly identify the active sites of the guanine-N7 MTase and 2′O MTase in the structure. The putative TPase catalytic residue Cys518 is colored in bright green.
Vaccinia virus, the prototypic virus of the poxvirus family, encodes a heterodimeric RNA capping enzyme consisting of large subunit D1 and small subunit D12. Although all three enzyme activities needed to generate cap 0 are located in D1, the guanine-N7 MTase activity requires the association with D12 to function efficiently (121–123). Structural and biochemical data suggests that interaction of D12 with the guanine-N7 MTase domain induces conformational changes necessary for efficient catalysis (124). The vaccinia capping enzyme structure shows that the three enzyme activities are neatly arranged as discrete modules in the order of TPase, GTase and guanine-N7 MTase from N- to C-terminus (Figure 5A). The TPase domain contains an elegant 8-stranded β-barrel structure characteristic of triphosphate tunnel metallozymes (TTMs). The presence of GTP and SAH unambiguously identifies the GTase and MTase active sites. The structure also suggests that the D12 subunit is a dysfunctional 2′O MTase that had become redundant as the virus acquired a separate 2′O MTase VP39 (124,125).
Bluetongue virus capping enzyme VP4 consolidates all 4 enzymatic activities to generate the cap 1 structure (126–128). Crystal structures of VP4 have been solved in the presence of substrate surrogates, namely, the guanine base, GpppG and SAH (105). The protein adopts an elongated shape with the N-terminal and C-terminal domains responsible for homodimerization, which is required for the assembly of VP4 into the viral core-like particle (129). The guanine-N7 MTase and 2′O MTase are well conserved with other known structures but the RNA TPase and GTase domains cannot be unambiguously identified. Biochemical studies have located the lysine responsible for the lysyl-phosphoguanosine intermediate near the C-terminal of the protein (105). However, no conserved GTase fold can be found in the VP4 structures. Most interestingly, the VP4 structure does not contain the β-barrel tunnel structure characteristic of the RNA TPase activity of viral and protozoan capping enzymes. Instead, a putative HCXXXXXR(S/T) motif of the cysteine phosphatase characteristic of metazoan capping apparatus is found co-localized with the putative GTase active site in a cleft near the C-terminus of the protein (105).
Despite the many questions these crystal structures answered, more interesting questions arise. For example, the vaccinia capping enzyme structure shows that the SAH-bound guanine-N7 MTase active site is located on the back side of the cleft where the RNA TPase and GTase active sites are located (125). Does the enzyme undergo a dramatic conformational change or oligomerize to efficiently carry out guanine-N7 MTase activity on the newly capped RNA? Does bluetongue virus capping enzyme indeed have a novel combined GTase and TPase domain containing a cysteine RNA phosphatase? More biochemical evidence and RNA-bound structures will be needed to shed light on these puzzles.
Cap snatching
In lieu of a capping machinery, some RNA viruses steal the cap from the host RNA in a process called cap snatching. In Influenza virus ((−)ssRNA), the RNA-dependent RNA polymerase (RdRp) is a complex of three proteins: polymerase base protein 1 (PB1), polymerase base protein 2 (PB2) and polymerase acidic protein (PA) (130,131). Upon assembly in the nucleus (132), the PB2 subunit binds to the cap structure of host capped RNA. The endonuclease activity of PA then cleaves the first 10–15 nt of the capped RNA, which is then used to prime viral mRNA transcription (133–136) (Table 1). Cap snatching was first demonstrated using human globin mRNA (134,135,137) and had since been presumed to prefer mRNA and pre-mRNA as snatching targets. This presumption has recently been overturned by a study that focuses on capped RNA instead of poly(A) RNA. The study showed that that Influenza A virus prefers non-coding snRNAs U1 and U2 to mRNA or pre-mRNA for snatching (138). Interestingly, a significant portion of the snatched sequences derived from the less understood promoter-associated small RNA (PASRs), which has recently been shown to be capped by the cytoplasmic capping machinery (28) (see Cytoplasmic RNA (re)-capping above). It is still not clear at what stage of snRNA maturation cap snatching takes place, nor if the clipping of the 5′ capped sequences from these regulatory PASRs has any implications in host cells or viral replication (138), but the findings will likely play a major role in understanding virus-host interactions in the regulatory RNA level.
The S. cerevisiae totiviruses L-A and L-BC (dsRNA) take a more direct approach toward cap snatching. The viral Gag protein cleaves the m7GMP group from host RNA and forms a histidyl–m7GMP covalent intermediate. The m7GMP group is then transferred to 5′ diphosphate of the viral transcripts co-transcriptionally (139–141) (Table 1). The similarity of the snatching process to the canonical GTase activity suggests an evolutionary link between totivirus cap snatching and the eukaryotic capping mechanism (142,143).
Applications of RNA capping enzymes
Capping enzymes generate cap 0 structure on RNA carrying a 5′ terminal triphosphate or diphosphate group. As discussed in the previous sections, the cap 0 structure is required for efficient translation of the mRNA in vivo. Vaccinia capping enzyme and 2′O MTase have been used to generate cap 0 and cap 1 structures on in vitro transcripts, which can then be used to transfect eukaryotic cells and drive protein synthesis (more in Exogenous mRNA technologies). The ability to generate cap 0 and cap 1 RNAs in vitro have also been invaluable in researching the role of the G cap and its 2′O methylation in innate immunity (2,68,76,81). The new developments in fluorescently-labeled GTP compatible with the translation machinery has enabled in vitro synthesis of fluorescent-labeled RNA (144). The use of GTP conjugated to desthiobiotin for in vitro RNA capping has allowed enrichment of bacterial transcripts for deep sequencing (145) (more in Cappable-Seq RNA sequencing).
Exogenous mRNA technologies
As the template for protein synthesis, introduction of mRNA into cells is the most intuitive way to drive the expression of target proteins in the cell. Although the transfection of RNA into cells had been reported in the 1990s, RNA-driven gene expression had not been explored in detail largely because of the uncertainty in the stability, efficiency and immunogenicity of exogenous RNA. With better understanding of the biological roles of base and ribose modifications, cellular degradation pathways, combined with technological advances in enzymatic synthesis and modification of RNA and cellular delivery vehicles, the administration of exogenous mRNA has become a viable option to drive protein expression in situ (146). Stem cell reprogramming (147–149), vaccination (150–156) and expression of therapeutic proteins (157–161) are only a few of the ever growing examples of exogenous mRNA technologies (146,157).
As a means of driving target protein expression in situ, administration of RNA offers a number of advantages over DNA. First, there is no concern of unintended integration of the RNA material into the genome as with DNA-based vectors. Second, while DNA vectors need to enter the nucleus to be transcribed into mRNA, which is then exported to the cytoplasm for translation, direct delivery of mRNA into cytoplasm bypasses these hurdles. In addition, it has been shown that the kinetics of protein expression after RNA administration peaks and decays within days, much more rapidly than with DNA-driven protein expression that exhibits a slow decay in weeks (155), making RNA a better option for applications such as vaccination where transient expression is desired. The use of mRNA also allows for simultaneous expression of multiple proteins in situ. As with DNA vectors, mRNA-based therapy or vaccination regimes benefit from the potential for rapid manufacture and dispatch that can be critical in response to disease outbreaks.
Enzymatic synthesis of functional mRNA
To avoid carryover of animal or viral material, it is desirable to manufacture therapeutic RNAs enzymatically using animal-free materials in vitro. In general, a DNA-dependent RNA polymerase transcribes a DNA template containing an appropriate promoter into an RNA transcript. The poly(A) tail can be generated co-transcriptionally by incorporating a poly(T) tract in the template DNA or separately by using a poly(A) polymerase. The transcript may be capped co-transcriptionally by using a cap analog or separately using a capping enzyme. Co-transcriptional capping using a cap analog has the advantage of being a simple workflow. However, one drawback of this approach is the theoretical capping efficiency of <100%, due to competition from the GTP for the starting nucleotide.
Enzymatic capping, usually carried out using Vaccinia capping enzyme, allows for complete capping of the RNA, generating cap 0 RNA. In applications where the innate immune response is to be minimized, the cap 0 structure may be further modified into cap 1 using a cap-specific 2′O methyltransferase. The use of m5CTP and pseudouridine triphosphate instead of CTP and UTP, respectively, in transcription has been shown to reduce the TLR-induced immune response and inhibition of protein synthesis (161–163). On the other hand, induction of a low level of innate response using unmodified RNA or RNA caps without 2′O methylation may be advantageous in applications such as vaccination.
mRNA vaccines
Vaccination via mRNA administration has many positive attributes compared to traditional live attenuated whole organisms. Not only it does not induce anti-vector immunity, mRNA vaccines are a short-lived and often self-limiting source of in situ production of antigen. It has the potential to be regulated and tuned over the RNA function and gene expression. Manufacturing is simple and can response quickly once the genome sequence of the disease causing agent is available (164). Unlike DNA vaccines, the response time of antigen production is faster and more efficient, as mRNA vaccines can be translated directly in the cytosol. There is also no concern of potential genome integration (164).
mRNA Vaccination has shown promising results in influenza virus infection (165,166), allergy (167,168) and tumor regression in animal models (169,170). In situ generation of tumor-specific proteins antigen, rapid induction of T and activation of natural killer cells, infiltration of immune cells into tumor mass have been documented (170). mRNA vaccines targeting prostate cancer and lung cancer have entered phase II, and phase I clinical trial in the US, respectively (171) (clinicaltrials.gov identifiers: NCT01817738, NCT01915524).
mRNA vaccination can be carried out by two means: through the administration of the mRNA vaccine to patients or by transfection of autologous dendritic cells by the mRNA vaccine and re-introducing the activated dendritic cells into the patient (154). The mRNA may be delivered by viral vectors or synthetic carriers. Recent advances in the formulation of synthetic carriers has allowed efficient mRNA vaccine delivery (151,153,156,159), therefore avoiding the use of viral vectors and the risk of contamination with animal materials during manufacturing. The mRNA can be synthesized enzymatically in vitro or produced in situ using self-amplifying mRNA.
Self-amplifying mRNA
Taking advantage of the RNA transcription-capping apparatus of alphavirus, a self-amplifying mRNA technology has been developed as an in situ gene expression vehicle for vaccination. Alphavirus has a (+)ssRNA genome encoding a non-structural pre-protein nsp1–4 for RNA-dependent RNA replication, transcription and capping, and two capsid proteins E2 and E1. By replacing the capsid proteins genes with the gene of interest, capped and polyadenylated self-amplifying RNA constructs were shown to be more potent in eliciting immune response than vaccination through standard mRNA (172), and efficacious in animal models (150,153,166,173). For an in-depth account of the current state of research, pre-clinical trials and manufacturing of mRNA and self-amplifying mRNA, we refer the reader to the following reports (150,174).
Cappable-Seq RNA sequencing
Vaccinia capping enzyme is able to cap RNA using 3′-modified GTP (145). A novel methodology based on this ability was devised to enrich and sequence 5′ triphosphate RNA species from biological samples (145). Termed Cappable-seq, vaccinia capping enzyme is used to cap 5′ triphosphate or diphosphate ends of RNA from a biological sample using 3′- desthiobiotin GTP. The desthiobiotinylated capped RNA species can then be enriched and deep-sequenced. Cappable-seq achieved a 50-fold enrichment of primary transcripts and identified previously unreported transcription start sites (TSS) genome-wide at single base resolution in Escherichia coli (145). The method had also been applied to identify the microbiome transcriptome from mouse cecum samples and for the first time identified TSS in a microbiome. Since ribosomal RNAs are processed and do not have cappable 5′ ends, Cappable-seq also depletes rRNA, which otherwise must be depleted by other means (175). Cappable-seq therefore effectively reduces the complexity of the transcriptome and the cost of sequencing. In addition to producing mRNA molecules in vitro, RNA capping enzymes have the potential to be a versatile RNA 5′ modifying reagent whose utility is just beginning to be appreciated.
CONCLUDING REMARKS
While mRNA cap structures were discovered in the 1970s, their biological roles have only been more deeply understood as the result of recent work. In addition to its role in mRNA export, mRNA maturation and protein synthesis, new understanding of the role of the RNA cap in innate immunity has helped advance the field of synthetic mRNA and their in vivo and therapeutic applications. The innate immune system imposes a strong selective pressure against RNA without a 5′ cap such that viruses have invented and reinvented widely different strategies to cap their RNA. Research on these viral systems have revealed a surprisingly diverse means and molecular machineries to generate capped RNA. It also resulted in applications such as efficient in vitro RNA capping systems and self-amplifying mRNA technology, facilitating large scale production of functional mRNA for in vitro and therapeutic applications. Novel application of RNA capping enzymes has also enabled sequencing and quantification of microbiome transcriptomes.
There are, as always, new questions to be asked as we learn more. RNA capping involves three enzymatic reactions at the 5′-end of a transcript. How is the RNA 5′ terminus transferred or repositioned between the three active sites? Does the enzyme complex go through extensive remodeling or does it form higher order oligomer? On a different front, the discovery of cytoplasmic (re-)capping machinery is exciting and invites further interesting questions. For example, in addition to 5′ CAGE sites, internal CAGE sites and promoter-associated small RNAs (PASRs) makes up a significant portion of cytoplasmic capping target. What processes are these re-capped RNAs involved in? What is the interplay between cytoplasmic (re-)capping and P-body RNA storage and degradation? Is the preferential snatching of cap from snRNA and PASRs by influenza A virus an evolved tactic for the survival and replication of the virus?
Perhaps more surprisingly, RNA caps are more wide-spread and diverse than the m7G cap. In addition to the 2,2,7-trimethylguanosine cap (176,177), the 5′-γ-methyl phosphate cap (178) and the dimethyl monophosphate cap (179), a recent report of novel and uncharacterized caps in mammalian cells begs further investigation of the identities and functions of these novel caps (180). The discovery of NAD- and coenzyme A-capped RNA in bacteria and the identification of NAD-capped small regulatory RNAs in E. coli disrupts the paradigm of the exclusivity of RNA caps in eukaryotes (181–183). What are the roles of NAD and CoA caps? Are they also present in archaea and eukaryotes? These are only some of the exciting questions waiting to be answered.
Acknowledgments
We thank Rich Roberts, Bill Jack and Larry McReynolds for critical reading of the manuscript.
Footnotes
Present address: Anand Ramanathan, Agios Pharmaceuticals, 88 Sidney Street, Cambridge, MA 02139, USA.
FUNDING
New England Biolabs Inc. Funding for open access charge: New England Biolabs Inc.
Conflict of interest statement. G.B. Robb and S.H. Chan are current emloyees of New England Biolabs that markets molecular biology reagents.
REFERENCES
- 1.Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.-Y., Schneller S., Zust R., Dong H., et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Devarkar S.C., Wang C., Miller M.T., Ramanathan A., Jiang F., Khan A.G., Patel S.S., Marcotrigiano J. Structural basis for m7G recognition and 2′-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc. Natl. Acad. Sci. U.S.A. 2016;113:596–601. doi: 10.1073/pnas.1515152113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mukherjee C., Bakthavachalu B., Schoenberg D.R. The cytoplasmic capping complex assembles on adapter protein Nck1 bound to the Proline-rich C-terminus of mammalian capping enzyme. PLoS Biol. 2014;12:e1001933. doi: 10.1371/journal.pbio.1001933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghosh A., Lima C.D. Enzymology of RNA cap synthesis. Wiley Interdiscip. Rev. RNA. 2010;1:152–172. doi: 10.1002/wrna.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Decroly E., Ferron F., Lescar J., Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat. Rev. Microbiol. 2012;10:51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shatkin A.J., Manley J.L. The ends of the affair: capping and polyadenylation. Nat. Struct. Biol. 2000;7:838–842. doi: 10.1038/79583. [DOI] [PubMed] [Google Scholar]
- 7.Moteki S., Price D. Functional coupling of capping and transcription of mRNA. Mol. Cell. 2002;10:599–609. doi: 10.1016/s1097-2765(02)00660-3. [DOI] [PubMed] [Google Scholar]
- 8.Martinez-Rucobo F.W., Kohler R., van de Waterbeemd M., Heck A.J.R., Hemann M., Herzog F., Stark H., Cramer P. Molecular basis of transcription-coupled pre-mRNA capping. Mol. Cell. 2015;58:1079–1089. doi: 10.1016/j.molcel.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Chu C., Das K., Tyminski J.R., Bauman J.D., Guan R., Qiu W., Montelione G.T., Arnold E., Shatkin A.J. Structure of the guanylyltransferase domain of human mRNA capping enzyme. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10104–10108. doi: 10.1073/pnas.1106610108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yue Z., Maldonado E., Pillutla R., Cho H., Reinberg D., Shatkin A.J. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saha N., Schwer B., Shuman S. Characterization of human, Schizosaccharomyces pombe, and Candida albicans mRNA cap methyltransferases and complete replacement of the yeast capping apparatus by mammalian enzymes. J. Biol. Chem. 1999;274:16553–16562. doi: 10.1074/jbc.274.23.16553. [DOI] [PubMed] [Google Scholar]
- 12.Bélanger F., Stepinski J., Darzynkiewicz E., Pelletier J. Characterization of hMTr1, a human Cap1 2′-O-ribose methyltransferase. J. Biol. Chem. 2010;285:33037–33044. doi: 10.1074/jbc.M110.155283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Werner M., Purta E., Kaminska K.H., Cymerman I.A., Campbell D.A., Mittra B., Zamudio J.R., Sturm N.R., Jaworski J., Bujnicki J.M. 2′-O-ribose methylation of cap2 in human: function and evolution in a horizontally mobile family. Nucleic Acids Res. 2011;39:4756–4768. doi: 10.1093/nar/gkr038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCracken S., Fong N., Rosonina E., Yankulov K., Brothers G., Siderovski D., Hessel A., Foster S., Shuman S., Bentley D.L. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho E.-J., Takagi T., Moore C.R., Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh A., Shuman S., Lima C.D. Structural insights to how mammalian capping enzyme reads the CTD code. Mol. Cell. 2011;43:299–310. doi: 10.1016/j.molcel.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cowling V.H. Regulation of mRNA cap methylation. Biochem. J. 2010;425:295–302. doi: 10.1042/BJ20091352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu M., Rajashankar K.R., Lima C.D. Structure of the Saccharomyces cerevisiae Cet1-Ceg1 mRNA capping apparatus. Structure. 2010;18:216–227. doi: 10.1016/j.str.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doamekpor S.K., Sanchez A.M., Schwer B., Shuman S., Lima C.D. How an mRNA capping enzyme reads distinct RNA polymerase II and Spt5 CTD phosphorylation codes. Genes Dev. 2014;28:1323–1336. doi: 10.1101/gad.242768.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Dijk E., Cougot N., Meyer S., Babajko S., Wahle E., Séraphin B. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brengues M. Movement of Eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balagopal V., Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr. Opin. Cell Biol. 2009;21:403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiss D.L., Oman K.M., Dougherty J.A., Mukherjee C., Bundschuh R., Schoenberg D.R. Cap homeostasis is independent of poly(A) tail length. Nucleic Acids Res. 2016;44:304–314. doi: 10.1093/nar/gkv1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schoenberg D.R., Maquat L.E. Re-capping the message. Trends Biochem. Sci. 2009;34:435–442. doi: 10.1016/j.tibs.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otsuka Y., Kedersha N.L., Schoenberg D.R. Identification of a cytoplasmic complex that adds a cap onto 5′-monophosphate RNA. Mol. Cell. Biol. 2009;29:2155–2167. doi: 10.1128/MCB.01325-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiraki T., Kondo S., Katayama S., Waki K., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiss D.L., Oman K., Bundschuh R., Schoenberg D.R. Uncapped 5′ ends of mRNAs targeted by cytoplasmic capping map to the vicinity of downstream CAGE tags. FEBS Lett. 2015;589:279–284. doi: 10.1016/j.febslet.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fejes-Toth K., Sotirova V., Sachidanandam R., Assaf G., Hannon G.J., Kapranov P., Foissac S., Willingham A.T., Duttagupta R., Dumais E., et al. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ignatochkina A. V, Takagi Y., Liu Y., Nagata K., Ho C.K. The messenger RNA decapping and recapping pathway in Trypanosoma. Proc. Natl. Acad. Sci. U.S.A. 2015;112:6967–6972. doi: 10.1073/pnas.1424909112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukherjee C., Patil D.P., Kennedy B.A., Bakthavachalu B., Bundschuh R., Schoenberg D.R. Identification of cytoplasmic capping targets reveals a role for cap homeostasis in translation and mRNA stability. Cell Rep. 2012;2:674–684. doi: 10.1016/j.celrep.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiao X., Chang J.H., Kilic T., Tong L., Kiledjian M. A mammalian pre-mRNA 5′ end capping quality control mechanism and an unexpected link of capping to pre-mRNA processing. Mol. Cell. 2013;50:104–115. doi: 10.1016/j.molcel.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiao X., Xiang S., Oh C., Martin C.E., Tong L., Kiledjian M. Identification of a quality-control mechanism for mRNA 5′-end capping. Nature. 2010;467:608–611. doi: 10.1038/nature09338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiang S., Cooper-Morgan A., Jiao X., Kiledjian M., Manley J.L., Tong L. Structure and function of the 5′–>3′ exoribonuclease Rat1 and its activating partner Rai1. Nature. 2009;458:784–788. doi: 10.1038/nature07731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang J.H., Jiao X., Chiba K., Oh C., Martin C.E., Kiledjian M., Tong L. Dxo1 is a new type of eukaryotic enzyme with both decapping and 5′-3′ exoribonuclease activity. Nat. Struct. Mol. Biol. 2012;19:1011–1017. doi: 10.1038/nsmb.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Konarska M.M., Padgett R.A., Sharp P.A. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984;38:731–736. doi: 10.1016/0092-8674(84)90268-x. [DOI] [PubMed] [Google Scholar]
- 36.Ohno M., Sakamoto H., Shimura Y. Preferential excision of the 5′ proximal intron from mRNA precursors with two introns as mediated by the cap structure. Proc. Natl. Acad. Sci. U.S.A. 1987;84:5187–5191. doi: 10.1073/pnas.84.15.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inoue K., Ohno M., Sakamoto H., Shimura Y. Effect of the cap structure on pre-mRNA splicing in Xenopus oocyte nuclei. Genes Dev. 1989;3:1472–1479. doi: 10.1101/gad.3.9.1472. [DOI] [PubMed] [Google Scholar]
- 38.Fresco L.D., Buratowski S. Conditional mutants of the yeast mRNA capping enzyme show that the cap enhances, but is not required for, mRNA splicing. RNA. 1996;2:584–596. [PMC free article] [PubMed] [Google Scholar]
- 39.Topisirovic I., Svitkin Y. V, Sonenberg N., Shatkin A.J. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip. Rev. RNA. 2011;2:277–298. doi: 10.1002/wrna.52. [DOI] [PubMed] [Google Scholar]
- 40.Gonatopoulos-Pournatzis T., Cowling V.H. Cap-binding complex (CBC) Biochem. J. 2014;457:231–242. doi: 10.1042/BJ20131214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hocine S., Singer R.H., Grunwald D. RNA processing and export. Cold Spring Harb. Perspect. Biol. 2010;2:a000752. doi: 10.1101/cshperspect.a000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pabis M., Neufeld N., Steiner M.C., Bojic T., Shav-Tal Y., Neugebauer K.M. The nuclear cap-binding complex interacts with the U4/U6·U5 tri-snRNP and promotes spliceosome assembly in mammalian cells. RNA. 2013;19:1054–1063. doi: 10.1261/rna.037069.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flaherty S.M., Fortes P., Izaurralde E., Mattaj I.W., Gilmartin G.M. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc. Natl. Acad. Sci. U.S.A. 1997;94:11893–11898. doi: 10.1073/pnas.94.22.11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andersen P.R., Domanski M., Kristiansen M.S., Storvall H., Ntini E., Verheggen C., Schein A., Bunkenborg J., Poser I., Hallais M., et al. The human cap-binding complex is functionally connected to the nuclear RNA exosome. Nat. Struct. Mol. Biol. 2013;20:1367–1376. doi: 10.1038/nsmb.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Izaurralde E., Adam S. Transport of macromolecules between the nucleus and the cytoplasm. RNA. 1998;4:351–364. [PMC free article] [PubMed] [Google Scholar]
- 46.Daneholt B. Assembly and transport of a premessenger RNP particle. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7012–7017. doi: 10.1073/pnas.111145498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nojima T., Hirose T., Kimura H., Hagiwara M. The interaction between cap-binding complex and RNA export factor is required for intronless mRNA export. J. Biol. Chem. 2007;282:15645–15651. doi: 10.1074/jbc.M700629200. [DOI] [PubMed] [Google Scholar]
- 48.Cheng H., Dufu K., Lee C.-S., Hsu J.L., Dias A., Reed R. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 49.Carmody S.R., Wente S.R. mRNA nuclear export at a glance. J. Cell Sci. 2009;122:1933–1937. doi: 10.1242/jcs.041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erkmann J.A., Kutay U. Nuclear export of mRNA: from the site of transcription to the cytoplasm. Exp. Cell Res. 2004;296:12–20. doi: 10.1016/j.yexcr.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 51.Fortes P., Inada T., Preiss T., Hentze M.W., Mattaj I.W., Sachs A.B. The yeast nuclear cap binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol. Cell. 2000;6:191–196. [PubMed] [Google Scholar]
- 52.Chiu S.-Y., Lejeune F., Ranganathan A.C., Maquat L.E. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev. 2004;18:745–754. doi: 10.1101/gad.1170204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Halstead J.M., Lionnet T., Wilbertz J.H., Wippich F., Ephrussi A., Singer R.H., Chao J.A. An RNA biosensor for imaging the first round of translation from single cells to living animals. Science. 2015;347:1367–1671. doi: 10.1126/science.aaa3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choe J., Ryu I., Park O.H., Park J., Cho H., Yoo J.S., Chi S.W., Kim M.K., Song H.K., Kim Y.K. eIF4AIII enhances translation of nuclear cap-binding complex-bound mRNAs by promoting disruption of secondary structures in 5′UTR. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E4577–E4586. doi: 10.1073/pnas.1409695111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maquat L.E., Hwang J., Sato H., Tang Y. CBP80-promoted mRNP rearrangements during the pioneer round of translation, nonsense-mediated mRNA decay, and thereafter. Cold Spring Harb. Symp. Quant. Biol. 2010;75:127–134. doi: 10.1101/sqb.2010.75.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choe J., Oh N., Park S., Lee Y.K., Song O.-K., Locker N., Chi S.-G., Kim Y.K. Translation initiation on mRNAs bound by nuclear cap-binding protein complex CBP80/20 requires interaction between CBP80/20-dependent translation initiation factor and eukaryotic translation initiation factor 3g. J. Biol. Chem. 2012;287:18500–18509. doi: 10.1074/jbc.M111.327528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maquat L.E., Tarn W.-Y., Isken O. The pioneer round of translation: features and functions. Cell. 2010;142:368–374. doi: 10.1016/j.cell.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mahamid J., Pfeffer S., Schaffer M., Villa E., Danev R., Kuhn Cuellar L., Forster F., Hyman A.A., Plitzko J.M., Baumeister W. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science. 2016;351:969–972. doi: 10.1126/science.aad8857. [DOI] [PubMed] [Google Scholar]
- 59.Preiss T., Hentze M.W. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature. 1998;392:516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 60.Preiss T., Muckenthaler M., Hentze M.W. Poly(A)-tail-promoted translation in yeast: implications for translational control. RNA. 1998;4:1321–1331. doi: 10.1017/s1355838298980669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Preiss T., Hentze M.W. From factors to mechanisms: translation and translational control in eukaryotes. Curr. Opin. Genet. Dev. 1999;9:515–521. doi: 10.1016/s0959-437x(99)00005-2. [DOI] [PubMed] [Google Scholar]
- 62.Smith R.W.P., Blee T.K.P., Gray N.K. Poly(A)-binding proteins are required for diverse biological processes in metazoans. Biochem. Soc. Trans. 2014;42:1229–1237. doi: 10.1042/BST20140111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muthukrishnan S., Moss B., Cooper J.A., Maxwell E.S. Influence of 5′-terminal cap structure on the initiation of translation of vaccinia virus mRNA. J. Biol. Chem. 1978;253:1710–1715. [PubMed] [Google Scholar]
- 64.Kuge H. Cap ribose methylation of c-mos mRNA stimulates translation and oocyte maturation in Xenopus laevis. Nucleic Acids Res. 1998;26:3208–3214. doi: 10.1093/nar/26.13.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Loo Y.-M., Fornek J., Crochet N., Bajwa G., Perwitasari O., Martinez-Sobrido L., Akira S., Gill M.A., García-Sastre A., Katze M.G., et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peisley A., Wu B., Yao H., Walz T., Hur S. RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Mol. Cell. 2013;51:573–583. doi: 10.1016/j.molcel.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 67.Binder M., Eberle F., Seitz S., Mücke N., Hüber C.M., Kiani N., Kaderali L., Lohmann V., Dalpke A., Bartenschlager R. Molecular mechanism of signal perception and integration by the innate immune sensor retinoic acid-inducible gene-I (RIG-I) J. Biol. Chem. 2011;286:27278–27287. doi: 10.1074/jbc.M111.256974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Züst R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J., Szretter K.J., Baker S.C., Barchet W., Diamond M.S., et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bruns A.M., Leser G.P., Lamb R.A., Horvath C.M. The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5-RNA interaction and filament assembly. Mol. Cell. 2014;55:771–781. doi: 10.1016/j.molcel.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peisley A., Jo M.H., Lin C., Wu B., Orme-Johnson M., Walz T., Hohng S., Hur S. Kinetic mechanism for viral dsRNA length discrimination by MDA5 filaments. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E3340–E3349. doi: 10.1073/pnas.1208618109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kolakofsky D., Kowalinski E., Cusack S. A structure-based model of RIG-I activation. RNA. 2012;18:2118–2127. doi: 10.1261/rna.035949.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Y., Ludwig J., Schuberth C., Goldeck M., Schlee M., Li H., Juranek S., Sheng G., Micura R., Tuschl T., et al. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat. Struct. Mol. Biol. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiang F., Ramanathan A., Miller M.T., Tang G.-Q., Gale M., Patel S.S., Marcotrigiano J. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ramanathan A., Devarkar S.C., Jiang F., Miller M.T., Khan A.G., Marcotrigiano J., Patel S.S. The autoinhibitory CARD2-Hel2i Interface of RIG-I governs RNA selection. Nucleic Acids Res. 2016;44:896–909. doi: 10.1093/nar/gkv1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goubau D., Schlee M., Deddouche S., Pruijssers A.J., Zillinger T., Goldeck M., Schuberth C., Van der Veen A.G., Fujimura T., Rehwinkel J., et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature. 2014;514:372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schuberth-Wagner C., Ludwig J., Bruder A.K., Herzner A.-M., Zillinger T., Goldeck M., Schmidt T., Schmid-Burgk J.L., Kerber R., Wolter S., et al. A conserved histidine in the RNA sensor RIG-I controls immune tolerance to N1-2′O-methylated self RNA. Immunity. 2015;43:41–51. doi: 10.1016/j.immuni.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pichlmair A., Lassnig C., Eberle C.-A., Górna M.W., Baumann C.L., Burkard T.R., Bürckstümmer T., Stefanovic A., Krieger S., Bennett K.L., et al. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat. Immunol. 2011;12:624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- 78.Abbas Y.M., Pichlmair A., Górna M.W., Superti-Furga G., Nagar B. Structural basis for viral 5′-PPP-RNA recognition by human IFIT proteins. Nature. 2013;494:60–64. doi: 10.1038/nature11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hyde J.L., Diamond M.S. Innate immune restriction and antagonism of viral RNA lacking 2′-O methylation. Virology. 2015;479-480:66–74. doi: 10.1016/j.virol.2015.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Diamond M.S. IFIT1: A dual sensor and effector molecule that detects non-2′-O methylated viral RNA and inhibits its translation. Cytokine Growth Factor Rev. 2014;25:543–550. doi: 10.1016/j.cytogfr.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kumar P., Sweeney T.R., Skabkin M.A., Skabkina O. V, Hellen C.U.T., Pestova T. V. Inhibition of translation by IFIT family members is determined by their ability to interact selectively with the 5′-terminal regions of cap0-, cap1- and 5′ppp- mRNAs. Nucleic Acids Res. 2014;42:3228–3245. doi: 10.1093/nar/gkt1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schlee M. Master sensors of pathogenic RNA - RIG-I like receptors. Immunobiology. 2013;218:1322–1335. doi: 10.1016/j.imbio.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Changela A., Ho C.K., Martins A., Shuman S., Mondragón A. Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme. EMBO J. 2001;20:2575–2586. doi: 10.1093/emboj/20.10.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bisaillon M., Shuman S. Structure-function analysis of the active site tunnel of yeast RNA triphosphatase. J. Biol. Chem. 2001;276:17261–17266. doi: 10.1074/jbc.M100980200. [DOI] [PubMed] [Google Scholar]
- 85.Gong C., Shuman S. Chlorella virus RNA triphosphatase. Mutational analysis and mechanism of inhibition by tripolyphosphate. J. Biol. Chem. 2002;277:15317–15324. doi: 10.1074/jbc.M200532200. [DOI] [PubMed] [Google Scholar]
- 86.Song J., Bettendorff L., Tonelli M., Markley J.L. Structural basis for the catalytic mechanism of mammalian 25-kDa thiamine triphosphatase. J. Biol. Chem. 2008;283:10939–10948. doi: 10.1074/jbc.M709675200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Delvaux D., Kerff F., Murty M.R.V.S., Lakaye B., Czerniecki J., Kohn G., Wins P., Herman R., Gabelica V., Heuze F., et al. Structural determinants of specificity and catalytic mechanism in mammalian 25-kDa thiamine triphosphatase. Biochim. Biophys. Acta. 2013;1830:4513–4523. doi: 10.1016/j.bbagen.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 88.Jain R., Shuman S. Polyphosphatase activity of CthTTM, a bacterial triphosphate tunnel metalloenzyme. J. Biol. Chem. 2008;283:31047–31057. doi: 10.1074/jbc.M805392200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moeder W., Garcia-Petit C., Ung H., Fucile G., Samuel M.A., Christendat D., Yoshioka K. Crystal structure and biochemical analyses reveal that the Arabidopsis triphosphate tunnel metalloenzyme AtTTM3 is a tripolyphosphatase involved in root development. Plant J. 2013;76:615–626. doi: 10.1111/tpj.12325. [DOI] [PubMed] [Google Scholar]
- 90.Delvaux D., Murty M.R.V.S., Gabelica V., Lakaye B., Lunin V. V, Skarina T., Onopriyenko O., Kohn G., Wins P., De Pauw E., et al. A specific inorganic triphosphatase from Nitrosomonas europaea: structure and catalytic mechanism. J. Biol. Chem. 2011;286:34023–34035. doi: 10.1074/jbc.M111.233585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martinez J., Truffault V., Hothorn M. Structural determinants for substrate binding and catalysis in triphosphate tunnel metalloenzymes. J. Biol. Chem. 2015;290:23348–23360. doi: 10.1074/jbc.M115.674473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lima C.D., Wang L.K., Shuman S. Structure and mechanism of yeast RNA triphosphatase: an essential component of the mRNA capping apparatus. Cell. 1999;99:533–543. doi: 10.1016/s0092-8674(00)81541-x. [DOI] [PubMed] [Google Scholar]
- 93.Shuman S., Lima C.D. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr. Opin. Struct. Biol. 2004;14:757–764. doi: 10.1016/j.sbi.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 94.Shuman S., Liu Y., Schwer B. Covalent catalysis in nucleotidyl transfer reactions: essential motifs in Saccharomyces cerevisiae RNA capping enzyme are conserved in Schizosaccharomyces pombe and viral capping enzymes and among polynucleotide ligases. Proc. Natl. Acad. Sci. U.S.A. 1994;91:12046–12050. doi: 10.1073/pnas.91.25.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shuman S., Hurwitz J. Mechanism of mRNA capping by vaccinia virus guanylyltransferase: characterization of an enzyme–guanylate intermediate. Proc. Natl. Acad. Sci. U.S.A. 1981;78:187–191. doi: 10.1073/pnas.78.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Soulière M.F., Perreault J.-P., Bisaillon M. Kinetic and thermodynamic characterization of the RNA guanylyltransferase reaction. Biochemistry. 2008;47:3863–3874. doi: 10.1021/bi702054a. [DOI] [PubMed] [Google Scholar]
- 97.Theobald D.L., Mitton-Fry R.M., Wuttke D.S. Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Arcus V. OB-fold domains: a snapshot of the evolution of sequence, structure and function. Curr. Opin. Struct. Biol. 2002;12:794–801. doi: 10.1016/s0959-440x(02)00392-5. [DOI] [PubMed] [Google Scholar]
- 99.Flynn R.L., Zou L. Oligonucleotide/oligosaccharide-binding fold proteins: a growing family of genome guardians. Crit. Rev. Biochem. Mol. Biol. 2010;45:266–275. doi: 10.3109/10409238.2010.488216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Håkansson K., Doherty A.J., Shuman S., Wigley D.B. X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell. 1997;89:545–553. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- 101.Schubert H.L., Blumenthal R.M., Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Byszewska M., Śmietański M., Purta E., Bujnicki J.M. RNA methyltransferases involved in 5′ cap biosynthesis. RNA Biol. 2014;11:1597–1607. doi: 10.1080/15476286.2015.1004955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bheemanaik S., Reddy Y.V.R., Rao D.N. Structure, function and mechanism of exocyclic DNA methyltransferases. Biochem. J. 2006;399:177–190. doi: 10.1042/BJ20060854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fabrega C., Hausmann S., Shen V., Shuman S., Lima C.D. Structure and mechanism of mRNA cap (guanine-N7) methyltransferase. Mol. Cell. 2004;13:77–89. doi: 10.1016/s1097-2765(03)00522-7. [DOI] [PubMed] [Google Scholar]
- 105.Sutton G., Grimes J.M., Stuart D.I., Roy P. Bluetongue virus VP4 is an RNA-capping assembly line. Nat. Struct. Mol. Biol. 2007;14:449–451. doi: 10.1038/nsmb1225. [DOI] [PubMed] [Google Scholar]
- 106.Fernandez-Sanchez M.E., Gonatopoulos-Pournatzis T., Preston G., Lawlor M.A., Cowling V.H. S-adenosyl homocysteine hydrolase is required for Myc-induced mRNA cap methylation, protein synthesis, and cell proliferation. Mol. Cell. Biol. 2009;29:6182–6191. doi: 10.1128/MCB.00973-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cole M.D., Cowling V.H. Specific regulation of mRNA cap methylation by the c-Myc and E2F1 transcription factors. Oncogene. 2009;28:1169–1175. doi: 10.1038/onc.2008.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gonatopoulos-Pournatzis T., Dunn S., Bounds R., Cowling V.H. RAM/Fam103a1 is required for mRNA cap methylation. Mol. Cell. 2011;44:585–596. doi: 10.1016/j.molcel.2011.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Aregger M., Cowling V.H. Human cap methyltransferase (RNMT) N-terminal non-catalytic domain mediates recruitment to transcription initiation sites. Biochem. J. 2013;455:67–73. doi: 10.1042/BJ20130378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Aregger M., Kaskar A., Varshney D., Fernandez-Sanchez M.E., Inesta-Vaquera F.A., Weidlich S., Cowling V.H. CDK1-Cyclin B1 activates RNMT, coordinating mRNA cap methylation with G1 phase transcription. Mol. Cell. 2016;61:734–746. doi: 10.1016/j.molcel.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ahola T., Kääriäinen L. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc. Natl. Acad. Sci. U.S.A. 1995;92:507–511. doi: 10.1073/pnas.92.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vasiljeva L., Merits A., Auvinen P., Kääriäinen L. Identification of a novel function of the alphavirus capping apparatus. RNA 5′-triphosphatase activity of Nsp2. J. Biol. Chem. 2000;275:17281–17287. doi: 10.1074/jbc.M910340199. [DOI] [PubMed] [Google Scholar]
- 113.Shirako Y., Strauss E.G., Strauss J.H. Suppressor mutations that allow sindbis virus RNA polymerase to function with nonaromatic amino acids at the N-terminus: evidence for interaction between nsP1 and nsP4 in minus-strand RNA synthesis. Virology. 2000;276:148–160. doi: 10.1006/viro.2000.0544. [DOI] [PubMed] [Google Scholar]
- 114.Fata C.L., Sawicki S.G., Sawicki D.L. Modification of Asn374 of nsP1 suppresses a Sindbis virus nsP4 minus-strand polymerase mutant. J. Virol. 2002;76:8641–8649. doi: 10.1128/JVI.76.17.8641-8649.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li J., Wang J.T., Whelan S.P.J. A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc. Natl. Acad. Sci. U.S.A. 2006;103:8493–8498. doi: 10.1073/pnas.0509821103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ogino T., Banerjee A.K. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol. Cell. 2007;25:85–97. doi: 10.1016/j.molcel.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 117.Ogino T., Yadav S.P., Banerjee A.K. Histidine-mediated RNA transfer to GDP for unique mRNA capping by vesicular stomatitis virus RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 2010;107:3463–3468. doi: 10.1073/pnas.0913083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ogino T. In vitro capping and transcription of rhabdoviruses. Methods. 2013;59:188–198. doi: 10.1016/j.ymeth.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dong H., Ray D., Ren S., Zhang B., Puig-Basagoiti F., Takagi Y., Ho C.K., Li H., Shi P.Y. Distinct RNA elements confer specificity to flavivirus RNA cap methylation events. J. Virol. 2007;81:4412–4421. doi: 10.1128/JVI.02455-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dong H., Ren S., Zhang B., Zhou Y., Puig-Basagoiti F., Li H., Shi P.-Y. West Nile virus methyltransferase catalyzes two methylations of the viral RNA cap through a substrate-repositioning mechanism. J. Virol. 2008;82:4295–4307. doi: 10.1128/JVI.02202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Higman M.A., Christen L.A., Niles E.G. The mRNA (guanine-7-)methyltransferase domain of the vaccinia virus mRNA capping enzyme. Expression in Escherichia coli and structural and kinetic comparison to the intact capping enzyme. J. Biol. Chem. 1994;269:14974–14981. [PubMed] [Google Scholar]
- 122.Mao X., Shuman S. Intrinsic RNA (guanine-7) methyltransferase activity of the vaccinia virus capping enzyme D1 subunit is stimulated by the D12 subunit. Identification of amino acid residues in the D1 protein required for subunit association and methyl group transfer. J. Biol. Chem. 1994;269:24472–24479. [PubMed] [Google Scholar]
- 123.Schwer B., Shuman S. Genetic analysis of poxvirus mRNA cap methyltransferase: Suppression of conditional mutations in the stimulatory D12 subunit by second-site mutations in the catalytic D1 subunit. Virology. 2006;352:145–156. doi: 10.1016/j.virol.2006.03.050. [DOI] [PubMed] [Google Scholar]
- 124.De La Pea M., Kyrieleis O.J.P., Cusack S. Structural insights into the mechanism and evolution of the vaccinia virus mRNA cap N7 methyl-transferase. EMBO J. 2007;26:4913–4925. doi: 10.1038/sj.emboj.7601912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kyrieleis O.J., Chang J., de la Pena M., Shuman S., Cusack S. Crystal structure of vaccinia virus mRNA capping enzyme provides insights into the mechanism and evolution of the capping apparatus. Structure. 2014;22:452–465. doi: 10.1016/j.str.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Le Blois H., French T., Mertens P.P., Burroughs J.N., Roy P. The expressed VP4 protein of bluetongue virus binds GTP and is the candidate guanylyl transferase of the virus. Virology. 1992;189:757–761. doi: 10.1016/0042-6822(92)90600-t. [DOI] [PubMed] [Google Scholar]
- 127.Martinez-Costas J., Sutton G., Ramadevi N., Roy P. Guanylyltransferase and RNA 5′-triphosphatase activities of the purified expressed VP4 protein of bluetongue virus. J. Mol. Biol. 1998;280:859–866. doi: 10.1006/jmbi.1998.1926. [DOI] [PubMed] [Google Scholar]
- 128.Ramadevi N., Burroughs N.J., Mertens P.P., Jones I.M., Roy P. Capping and methylation of mRNA by purified recombinant VP4 protein of bluetongue virus. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13537–13542. doi: 10.1073/pnas.95.23.13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ramadevi N., Rodriguez J., Roy P. A leucine zipper-like domain is essential for dimerization and encapsidation of bluetongue virus nucleocapsid protein VP4. J. Virol. 1998;72:2983–2990. doi: 10.1128/jvi.72.4.2983-2990.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sugiyama K., Obayashi E., Kawaguchi A., Suzuki Y., Tame J.R.H., Nagata K., Park S.-Y. Structural insight into the essential PB1-PB2 subunit contact of the influenza virus RNA polymerase. EMBO J. 2009;28:1803–1811. doi: 10.1038/emboj.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Reich S., Guilligay D., Pflug A., Malet H., Berger I., Crépin T., Hart D., Lunardi T., Nanao M., Ruigrok R.W.H., et al. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature. 2014;516:361–366. doi: 10.1038/nature14009. [DOI] [PubMed] [Google Scholar]
- 132.Huet S., Avilov S. V, Ferbitz L., Daigle N., Cusack S., Ellenberg J. Nuclear import and assembly of influenza A virus RNA polymerase studied in live cells by fluorescence cross-correlation spectroscopy. J. Virol. 2010;84:1254–1264. doi: 10.1128/JVI.01533-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dias A., Bouvier D., Crepin T., McCarthy A.A., Hart D.J., Baudin F., Cusack S., Ruigrok R.W. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature. 2009;458:914–918. doi: 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- 134.Plotch S.J., Bouloy M., Krug R.M. Transfer of 5′-terminal cap of globin mRNA to influenza viral complementary RNA during transcription in vitro. Proc. Natl. Acad. Sci. U.S.A. 1979;76:1618–1622. doi: 10.1073/pnas.76.4.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Plotch S.J., Bouloy M., Ulmanen I., Krug R.M. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981;23:847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- 136.Ruigrok R.W., Crepin T., Hart D.J., Cusack S. Towards an atomic resolution understanding of the influenza virus replication machinery. Curr. Opin. Struct. Biol. 2010;20:104–113. doi: 10.1016/j.sbi.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 137.Krug R.M., Broni B.A., Bouloy M. Are the 5′ ends of influenza viral mRNAs synthesized in vivo donated by host mRNAs? Cell. 1979;18:329–334. doi: 10.1016/0092-8674(79)90052-7. [DOI] [PubMed] [Google Scholar]
- 138.Gu W., Gallagher G.R., Dai W., Liu P., Li R., Trombly M.I., Gammon D.B., Mello C.C., Wang J.P., Finberg R.W. Influenza A virus preferentially snatches noncoding RNA caps. RNA. 2015;21:2067–2075. doi: 10.1261/rna.054221.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fujimura T., Esteban R. Cap snatching of yeast L-A double-stranded RNA virus can operate in trans and requires viral polymerase actively engaging in transcription. J. Biol. Chem. 2012;287:12797–12804. doi: 10.1074/jbc.M111.327676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fujimura T., Esteban R. Cap-snatching mechanism in yeast L-A double-stranded RNA virus. Proc. Natl. Acad. Sci. U.S.A. 2011;108:17667–17671. doi: 10.1073/pnas.1111900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fujimura T., Esteban R. Cap snatching in yeast L-BC double-stranded RNA totivirus. J. Biol. Chem. 2013;288:23716–23724. doi: 10.1074/jbc.M113.490953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Blanc A., Goyer C., Sonenberg N. The coat protein of the yeast double-stranded RNA virus L-A attaches covalently to the cap structure of eukaryotic mRNA. Mol. Cell. Biol. 1992;12:3390–3398. doi: 10.1128/mcb.12.8.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Blanc A., Ribas J.C., Wickner R.B., Sonenberg N. His-154 is involved in the linkage of the Saccharomyces cerevisiae L-A double-stranded RNA virus Gag protein to the cap structure of mRNAs and is essential for M1 satellite virus expression. Mol. Cell. Biol. 1994;14:2664–2674. doi: 10.1128/mcb.14.4.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gunawardana D., Domashevskiy A. V, Gayler K.R., Goss D.J. Efficient preparation and properties of mRNAs containing a fluorescent cap analog: Anthraniloyl-m(7)GpppG. Transl. (Austin, Tex.) 2015;3:e988538. doi: 10.4161/21690731.2014.988538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ettwiller L., Buswell J., Yigit E., Schildkraut I. A novel enrichment strategy reveals unprecedented number of novel transcription start sites at single base resolution in a model prokaryote and the gut microbiome. BMC Genomics. 2016;17:199. doi: 10.1186/s12864-016-2539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Quabius E.S., Krupp G. Synthetic mRNAs for manipulating cellular phenotypes: an overview. N. Biotechnol. 2015;32:229–235. doi: 10.1016/j.nbt.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 147.Hansson M.L., Albert S., González Somermeyer L., Peco R., Mejía-Ramírez E., Montserrat N., Izpisua Belmonte J.C. Efficient delivery and functional expression of transfected modified mRNA in human embryonic stem cell-derived retinal pigmented epithelial cells. J. Biol. Chem. 2015;290:5661–5672. doi: 10.1074/jbc.M114.618835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Warren L., Manos P.D., Ahfeldt T., Loh Y.-H., Li H., Lau F., Ebina W., Mandal P.K., Smith Z.D., Meissner A., et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Avci-Adali M., Behring A., Keller T., Krajewski S., Schlensak C., Wendel H.P. Optimized conditions for successful transfection of human endothelial cells with in vitro synthesized and modified mRNA for induction of protein expression. J. Biol. Eng. 2014;8:8. doi: 10.1186/1754-1611-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Deering R.P., Kommareddy S., Ulmer J.B., Brito L.A., Geall A.J. Nucleic acid vaccines: prospects for non-viral delivery of mRNA vaccines. Expert Opin. Drug Deliv. 2014;11:885–899. doi: 10.1517/17425247.2014.901308. [DOI] [PubMed] [Google Scholar]
- 151.Pardi N., Tuyishime S., Muramatsu H., Kariko K., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Weissman D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Weiner D.B. RNA-based vaccination: sending a strong message. Mol. Ther. 2013;21:506–508. doi: 10.1038/mt.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Brito L.A., Chan M., Shaw C.A., Hekele A., Carsillo T., Schaefer M., Archer J., Seubert A., Otten G.R., Beard C.W., et al. A cationic nanoemulsion for the delivery of next-generation RNA vaccines. Mol. Ther. 2014;22:2118–2129. doi: 10.1038/mt.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Van Lint S., Heirman C., Thielemans K., Breckpot K. mRNA: From a chemical blueprint for protein production to an off-the-shelf therapeutic. Hum. Vaccin. Immunother. 2013;9:265–274. doi: 10.4161/hv.22661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Probst J., Weide B., Scheel B., Pichler B.J., Hoerr I., Rammensee H.-G., Pascolo S. Spontaneous cellular uptake of exogenous messenger RNA in vivo is nucleic acid-specific, saturable and ion dependent. Gene Ther. 2007;14:1175–1180. doi: 10.1038/sj.gt.3302964. [DOI] [PubMed] [Google Scholar]
- 156.Phua K.K.L., Staats H.F., Leong K.W., Nair S.K. Intranasal mRNA nanoparticle vaccination induces prophylactic and therapeutic anti-tumor immunity. Sci. Rep. 2014;4:5128. doi: 10.1038/srep05128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tavernier G., Andries O., Demeester J., Sanders N.N., De Smedt S.C., Rejman J. mRNA as gene therapeutic: how to control protein expression. J. Control. Release. 2011;150:238–247. doi: 10.1016/j.jconrel.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 158.Hecker J.G., Hall L.L., Irion V.R. Nonviral gene delivery to the lateral ventricles in rat brain: initial evidence for widespread distribution and expression in the central nervous system. Mol. Ther. 2001;3:375–384. doi: 10.1006/mthe.2001.0272. [DOI] [PubMed] [Google Scholar]
- 159.Anderson D.M., Hall L.L., Ayyalapu A.R., Irion V.R., Nantz M.H., Hecker J.G. Stability of mRNA/cationic lipid lipoplexes in human and rat cerebrospinal fluid: methods and evidence for nonviral mRNA gene delivery to the central nervous system. Hum. Gene Ther. 2003;14:191–202. doi: 10.1089/10430340360535751. [DOI] [PubMed] [Google Scholar]
- 160.Kormann M.S.D., Hasenpusch G., Aneja M.K., Nica G., Flemmer A.W., Herber-Jonat S., Huppmann M., Mays L.E., Illenyi M., Schams A., et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- 161.Karikó K., Muramatsu H., Keller J.M., Weissman D. Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol. Ther. 2012;20:948–953. doi: 10.1038/mt.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 163.Anderson B.R., Muramatsu H., Nallagatla S.R., Bevilacqua P.C., Sansing L.H., Weissman D., Karikó K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Geall A.J., Ulmer J.B. Introduction to RNA-based vaccines and therapeutics. Expert Rev. Vaccines. 2015;14:151–152. doi: 10.1586/14760584.2015.1001244. [DOI] [PubMed] [Google Scholar]
- 165.Petsch B., Schnee M., Vogel A.B., Lange E., Hoffmann B., Voss D., Schlake T., Thess A., Kallen K.-J., Stitz L., et al. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat. Biotechnol. 2012;30:1210–1216. doi: 10.1038/nbt.2436. [DOI] [PubMed] [Google Scholar]
- 166.Hekele A., Bertholet S., Archer J., Gibson D.G., Palladino G., Brito L.A., Otten G.R., Brazzoli M., Buccato S., Bonci A., et al. Rapidly produced SAM® vaccine against H7N9 influenza is immunogenic in mice. Emerg. Microbes Infect. 2013;2:e52. doi: 10.1038/emi.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Weiss R., Scheiblhofer S., Roesler E., Ferreira F., Thalhamer J. Prophylactic mRNA vaccination against allergy. Curr. Opin. Allergy Clin. Immunol. 2010;10:567–574. doi: 10.1097/ACI.0b013e32833fd5b6. [DOI] [PubMed] [Google Scholar]
- 168.Roesler E., Weiss R., Weinberger E.E., Fruehwirth A., Stoecklinger A., Mostböck S., Ferreira F., Thalhamer J., Scheiblhofer S. Immunize and disappear-safety-optimized mRNA vaccination with a panel of 29 allergens. J. Allergy Clin. Immunol. 2009;124:e1–e11. doi: 10.1016/j.jaci.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 169.Fotin-Mleczek M., Zanzinger K., Heidenreich R., Lorenz C., Kowalczyk A., Kallen K.-J., Huber S.M. mRNA-based vaccines synergize with radiation therapy to eradicate established tumors. Radiat. Oncol. 2014;9:180. doi: 10.1186/1748-717X-9-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Fotin-Mleczek M., Zanzinger K., Heidenreich R., Lorenz C., Thess A., Duchardt K.M., Kallen K.-J. Highly potent mRNA based cancer vaccines represent an attractive platform for combination therapies supporting an improved therapeutic effect. J. Gene Med. 2012;14:428–439. doi: 10.1002/jgm.2605. [DOI] [PubMed] [Google Scholar]
- 171.Kübler H., Scheel B., Gnad-Vogt U., Miller K., Schultze-Seemann W., Vom Dorp F., Parmiani G., Hampel C., Wedel S., Trojan L., et al. Self-adjuvanted mRNA vaccination in advanced prostate cancer patients: a first-in-man phase I/IIa study. J. Immunother. cancer. 2015;3:26. doi: 10.1186/s40425-015-0068-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Johanning F.W., Conry R.M., LoBuglio A.F., Wright M., Sumerel L.A., Pike M.J., Curiel D.T. A Sindbis virus mRNA polynucleotide vector achieves prolonged and high level heterologous gene expression in vivo. Nucleic Acids Res. 1995;23:1495–1501. doi: 10.1093/nar/23.9.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Geall A.J., Verma A., Otten G.R., Shaw C.A., Hekele A., Banerjee K., Cu Y., Beard C.W., Brito L.A., Krucker T., et al. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. U.S.A. 2012;109:14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Brito L.A., Kommareddy S., Maione D., Uematsu Y., Giovani C., Berlanda Scorza F., Otten G.R., Yu D., Mandl C.W., Mason P.W., et al. Self-amplifying mRNA vaccines. Adv. Genet. 2015;89:179–233. doi: 10.1016/bs.adgen.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 175.Adiconis X., Borges-Rivera D., Satija R., DeLuca D.S., Busby M.A., Berlin A.M., Sivachenko A., Thompson D.A., Wysoker A., Fennell T., et al. Comparative analysis of RNA sequencing methods for degraded or low-input samples. Nat. Methods. 2013;10:623–629. doi: 10.1038/nmeth.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Qiu Z.R., Shuman S., Schwer B. An essential role for trimethylguanosine RNA caps in Saccharomyces cerevisiae meiosis and their requirement for splicing of SAE3 and PCH2 meiotic pre-mRNAs. Nucleic Acids Res. 2011;39:5633–5646. doi: 10.1093/nar/gkr083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Chang J., Schwer B., Shuman S. Mutational analyses of trimethylguanosine synthase (Tgs1) and Mud2: proteins implicated in pre-mRNA splicing. RNA. 2010;16:1018–1031. doi: 10.1261/rna.2082610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Cosgrove M.S., Ding Y., Rennie W.A., Lane M.J., Hanes S.D. The Bin3 RNA methyltransferase targets 7SK RNA to control transcription and translation. Wiley Interdiscip. Rev. RNA. 2012;3:633–647. doi: 10.1002/wrna.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Xhemalce B., Robson S.C., Kouzarides T. Human RNA methyltransferase BCDIN3D regulates microRNA processing. Cell. 2012;151:278–288. doi: 10.1016/j.cell.2012.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Abdelhamid R.F., Plessy C., Yamauchi Y., Taoka M., de Hoon M., Gingeras T.R., Isobe T., Carninci P. Multiplicity of 5′ cap structures present on short RNAs. PLoS One. 2014;9:e102895. doi: 10.1371/journal.pone.0102895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Chen Y.G., Kowtoniuk W.E., Agarwal I., Shen Y., Liu D.R. LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat. Chem. Biol. 2009;5:879–881. doi: 10.1038/nchembio.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kowtoniuk W.E., Shen Y., Heemstra J.M., Agarwal I., Liu D.R. A chemical screen for biological small molecule–RNA conjugates reveals CoA-linked RNA. Proc. Natl. Acad. Sci. U.S.A. 2009;106:7768–7773. doi: 10.1073/pnas.0900528106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Cahová H., Winz M.-L., Höfer K., Nübel G., Jäschke A. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature. 2015;519:374–377. doi: 10.1038/nature14020. [DOI] [PubMed] [Google Scholar]