Abstract
Release of RNA polymerase II (Pol II) from promoter-proximal pausing has emerged as a critical step regulating gene expression in multicellular organisms. The transition of Pol II into productive elongation requires the kinase activity of positive transcription elongation factor b (P-TEFb), which is itself under a stringent control by the inhibitory 7SK small nuclear ribonucleoprotein (7SK snRNP) complex. Here, we provide an overview on stimulating Pol II pause release by P-TEFb and on sequestering P-TEFb into 7SK snRNP. Furthermore, we highlight mechanisms that govern anchoring of 7SK snRNP to chromatin as well as means that release P-TEFb from the inhibitory complex, and propose a unifying model of P-TEFb activation on chromatin. Collectively, these studies shine a spotlight on the central role of RNA binding proteins (RBPs) in directing the inhibition and activation of P-TEFb, providing a compelling paradigm for controlling Pol II transcription with a non-coding RNA.
INTRODUCTION
‘If you're going to try, go all the way. Otherwise, don't even start.’ With these words, a late American poet Charles Bukowski has unwittingly commented on an important regulatory step in gene transcription by Pol II in multicellular organisms. Namely, to respond to intra- and extra-cellular signals, cells possess a highly sophisticated transcription apparatus, wherein Pol II is the principle player. With the help of an extensive entourage of general and gene-specific transcription factors, Pol II goes through a series of specific steps while transcribing protein-coding and non-protein coding genes. These steps include loading of Pol II to gene promoter, initiation of RNA synthesis from transcription start site (TSS), promoter-proximal pausing (herein referred to as pausing), elongation along the length of a gene, termination soon after transcription end site (TES) and eventual re-initiation.
Up to the turn of the new millennium, a predominantly held view based mainly on yeast models of gene transcription claimed that Pol II loading to promoters was the main rate-limiting step in the Pol II transcription cycle. However, pioneering work in late 1980's and early 90's on human MYC and FOS (1,2), fruit fly HSP70 (3), as well as the human immunodeficiency virus type 1 (HIV-1) transcription (4) began to challenge this prevailing paradigm. These studies showed that Pol II could start with transcription but fail to generate full-length transcripts in the absence of additional stimuli due to a block in early elongation. Finally, with the help of various genome-wide approaches which track Pol II along DNA as well as nascent transcripts, investigators have revealed that once it engages transcription, the bulk of Pol II is found at promoter-proximal pause sites, 30–60 nucleotides downstream of TSS (5,6). Thus, the great deal of regulation of gene expression, particularly at developmental genes and those involved in stimulus-controlled pathways (7), takes place at the level of Pol II pausing and Pol II release into early transcription elongation. In this survey, we first review the fundamentals of gene transcription control by P-TEFb and its sequestration within the inhibitory 7SK snRNP complex. We next focus on recent developments in the field which are beginning to reveal mechanisms that anchor 7SK snRNP to chromatin and liberate P-TEFb from this inhibition, illustrating a highly controlled step in the transition from paused to elongating Pol II.
Stimulating Pol II pause release with P-TEFb
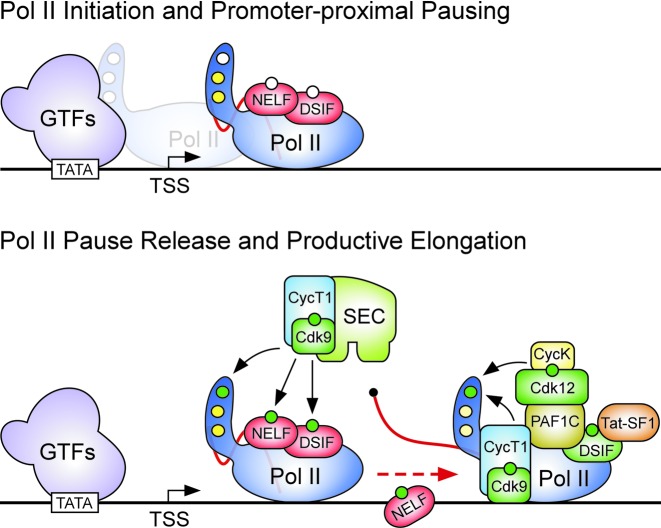
The critical factor that triggers the release of paused Pol II into productive elongation at almost all metazoan genes is P-TEFb (8,9). It is an evolutionarily conserved heterodimeric cyclin-dependent kinase (Cdk9/CycT), which is in humans composed of the catalytic Cdk9 and a regulatory CycT1, T2a or T2b subunits. After its initial identification from Drosophila nuclear extracts (10), P-TEFb rose to prominence because it was discovered to be an essential cellular co-factor for stimulating HIV-1 gene expression by the viral transactivator of transcription (Tat) (11–13). Without Tat, Pol II gets loaded to and initiates transcription from the viral promoter but pauses soon afterward due to the actions of multi-subunit negative transcription elongation factors (N-TEFs) NELF and DSIF (14,15). Thus only short viral transcripts are produced, measuring in length barely enough to contain transactivation response (TAR) element (16), a stable stem-loop structure that constitutes another key component for HIV-1 transcription. To overcome the early transcriptional blockade, Tat employs its activation domain (AD) to recruit P-TEFb to Pol II via TAR (12), whereby Cdk9 meets its three principal substrate targets: Y1S2P3T4S5P6S7 heptapeptide repeats of the C-terminal domain (CTD) of Pol II's biggest subunit Rbp1, as well as NELF-E and Spt5 subunits of NELF and DSIF, respectively.
Although P-TEFb may target additional components influencing Pol II pausing (17), phosphorylation events exemplified at HIV-1 define Pol II transition from pausing into productive elongation at most genes in multicellular organisms (Figure 1). While P-TEFb catalyzes the CTD Serine 2 phosphorylation (P-Ser2) to tether additional elongation, RNA maturation and chromatin modifying factors to Pol II (18–20), NELF-E phosphorylation leads to NELF dissociation from it (21). Furthermore, Spt5 phosphorylation transforms DSIF into a positive elongation factor, attracting Tat-SF1 and PAF1 complex (PAF1C) to elongating Pol II (22,23). Subsequently, PAF1C can in turn recruit the Cdk12/CycK complex (24), which could in concert with P-TEFb contribute to the progressive levels of Pol II P-Ser2 along gene's body and proper Pol II termination (17,25,26). In contrast to the wealth of in vivo data showing that P-TEFb catalyzes the P-Ser2 mark (20,26–33), purified P-TEFb exhibited a robust P-Ser5 activity in vitro (34,35), possibly reflecting the lack of cellular complexity in the in vitro systems.
Figure 1.
P-TEFb kinase stimulates the transition of Pol II from promoter-proximal pausing into productive elongation. Top: recruitment of Pol II by general transcription factors (GTFs) through TATA-binding protein of the core promoter recognition complex TFIID results in the formation of a PIC. The CTD of Pol II is represented as a tail, wherein white circles depict unphosphorylated Serine residues at position 2, 5 and 7 within the consensus heptapeptide repeat. Black arrow depicts transcription start site (TSS). While rapid Pol II initiation ensues (depicted in light blue), Pol II pauses (depicted in blue) at promoter-proximal pause sites due to the actions of NELF and DSIF which are both in non-phosphorylated forms (white circles). At this stage, the CTD has been phosphorylated already by Cdk7 of TFIIH at Ser5 and Ser7 (yellow circles). A short RNA transcript is depicted in red. Bottom: catalytically active P-TEFb within SEC stimulates the release of paused Pol II into elongation (dashed red arrow) by phosphorylating the CTD at Ser2, NELF and DSIF (green circles). While phosphorylated NELF dissociates from Pol II, phosphorylated DSIF becomes a positive elongation factor and recruits Tat-SF1 and PAF1C to elongating Pol II. In turn, PAF1C can promote the recruitment of Cdk12/CycK, which in concert with P-TEFb catalyzes P-Ser2 during Pol II elongation, yielding an increasingly longer RNA transcript (red).
The demonstration that HIV-1 Tat stimulated Pol II elongation through P-TEFb ushered in an era of efforts aiming to reveal P-TEFb tethering mechanisms to endogenous genes. As has the number of genes requiring P-TEFb for paused Pol II release grown to encompass almost an entire gene collection (32,36), so has the variety of factors promoting recruitment of P-TEFb to genes. Among others, these include classical transcription factors and co-activators such as NF-kB, CIITA, p53 and c-Myc (32,37–39), the BET family acetyl-lysine recognizing chromatin adaptor protein Brd4 (40–42), and the nuclear cap-binding protein complex which links pre-mRNA capping to transcript elongation (30,43).
Importantly, P-TEFb has been subsequently identified as an integral component of many super elongation complexes (SECs), which can be formed by different combinations of the ELL family members (ELL1, ELL2, ELL3), the AF4/FMR2 SEC-scaffolding family members AFF1 and AFF4, as well as ENL and AF9 proteins (9,44–47). Of note, ELL1, AFF1, AFF4, ENL and AF9 have been described previously as fusion partners of the DNA-binding domain of the mixed lineage leukemia (MLL) gene as a result of in-frame gene translocations (48). In turn, the potent Pol II elongation stimulatory activity of SECs is mis-targeted to MLL-dependent genes, causing childhood hematological malignancies. In addition, Tat can deliver an entire SEC containing AFF1 as a scaffolding component via TAR for productive HIV-1 transcription (49). Under normal circumstances, recruitment of P-TEFb as a part of many SECs can be accomplished by the Pol II CTD interacting Mediator and Integrator complexes (50,51), the BET family protein member Brd4 (52) and PAF1C (53).
KEEPING P-TEFb IN CHECK BY 7SK snRNP
Befitting the vital role of P-TEFb in Pol II transcription, its kinase activity is kept under a tight control in metazoan cells (5,9). The first glimpse into the negative regulation came by groundbreaking studies showing that an abundant, Pol III-transcribed non-coding 7SK small nuclear RNA (7SK) of 330–332 nucleotides in length (54,55) and of hitherto mysterious function binds to P-TEFb and inhibits its kinase activity (56,57). Moreover, this work documented that chemical inhibition of Pol II transcription by 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB) and actinomycin D (ActD) or ultraviolet (UV) irradiation activates P-TEFb by dissociating it from 7SK in a reversible manner, underscoring that regulated de-repression of P-TEFb could be an important way to fine-tune Pol II pause release in response to diverse sets of circumstances. Of note, the portion of P-TEFb bound to 7SK varies between different cell types: while this fraction is about half in growing HeLa cells (56,57), it reaches close to 90% in Jurkat T cells or human primary blood lymphocytes (58,59).
Subsequently, it has become apparent that 7SK does not impact P-TEFb by itself but rather serves as a scaffold for proteins which enable its own stability and inhibition of P-TEFb (Figure 2 and Table 1). In addition to 7SK and P-TEFb, the canonical 7SK snRNP complex is composed of three RBPs: P-TEFb inhibitor HEXIM1 (and/or its paralog HEXIM2), LA-related protein family member LARP7 and the 7SK γ-methylphosphate capping enzyme MePCE (5,9,60–63). Finally, recent evidence suggests that AFF1 and AFF4 accompany P-TEFb in 7SK snRNP as well (49). Structure of the inhibitory complex has not been solved, and conclusions from previous studies differ in what the exact structure of 7SK within the snRNP might be (64–66). Nevertheless, 7SK was proposed to form complex and dynamic secondary structures that may consist of either eight highly conserved structural motifs (M1-8) or four stem loops (SL1-4), creating binding surfaces for RBPs. Of note, outside of the canonical 7SK snRNP, 7SK can be also assembled within alternative snRNPs (Table 1; for details, see text below).
Figure 2.
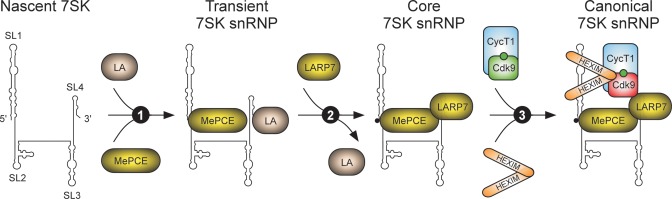
Biogenesis of core and canonical 7SK snRNP. Following the transcription of RN7SK by Pol III, nascent 7SK folds into a structure that may consist of four stem loops (SL1-4) as displayed on the left. LA and MePCE prevent 7SK nucleolytic degradation by binding its 3′-terminal U-rich sequence and by binding the basal part of the 7SK SL1 and capping the 5′ end of 7SK, respectively (Step 1). The capping (depicted as black circle) might provoke the replacement of LA in the transient 7SK snRNP by LARP7, which in addition to the 3′-terminal U-rich sequence binds 7SK SL4, ensuring 7SK stability. Moreover, LARP7 binds MePCE, provoking tighter LARP7-7SK interaction and inhibition of the MePCE capping activity, yielding a stable core 7SK snRNP (Step 2). Finally, HEXIM1/2 dimers bind the proximal and distal parts of 7SK SL1 through their ARMs, leading to the exposure of the dimeric coiled-coil binding surface for active P-TEFb (Cdk9 is in green) phosphorylated at Threonine 186 (depicted as green circle on Cdk9). Inhibition of P-TEFb (Cdk9 is in red) by HEXIM1/2 ensues, concluding formation of the canonical 7SK snRNP (Step 3).
Table 1. 7SK forms canonical and alternative snRNPs in cells.
| Canonical 7SK snRNP | Factor | Mode of action |
| Core 7SK snRNP | 7SK | RNA scaffold of the complex. |
| LARP7 | Binds 3′ end and SL4 of 7SK to enable 7SK stability and binds P-TEFb to promote its incorporation into 7SK snRNP. | |
| MePCE | Binds the basal part in SL1 of 7SK and caps 7SK at 5′ end to enable 7SK and LARP7 stability. | |
| P-TEFb inhibitors | HEXIM1/2 | Binds proximal and distal parts in SL1 of 7SK to bind and inhibit P-TEFb. |
| P-TEFb | Cdk9 | Catalytic subunit of P-TEFb that is inhibited by HEXIM1/2. |
| CycT1/T2a/T2b | Regulatory subunit of P-TEFb that provides binding surface for HEXIM1/2. | |
| P-TEFb auxiliary factor | AFF1, AFF4 | Binds P-TEFb and serves as a scaffold for SEC assembly. |
| 7SK-hnRNP snRNP | Factor | Mode of action |
| Core 7SK snRNP | 7SK, LARP7, MePCE | Platform for hnRNPs and RNA helicase A upon inhibition of Pol II transcription. |
| hnRNPs and RNA helicase A | hnRNP A1, A2/B1, Q, R and RNA helicase A | Bind SL3 of 7SK to enable the release of P-TEFb and HEXIM1 from the canonical 7SK snRNP. |
| 7SK–BAF snRNP | Factor | Mode of action |
| 7SK | 7SK | Scaffolds BAF complex with Pol II and promotes occupancy and function of BAF at enhancers. |
| BAF | subunits of BAF complex | Inhibits Pol II transcription at enhancers. |
Table summarizes three known 7SK-containing snRNPs, along with their composition and mode of action for each factor.
Biogenesis of the canonical 7SK snRNP particle is preceded by the assembly of core 7SK snRNP, consisting of 7SK, LARP7 and MePCE (27,67) (Figure 2). Namely, all 7SK molecules within the core are bound stably to LARP7 and MePCE as depletion of either one of them via RNAi triggers 7SK degradation in cells (60,61,68). Moreover, cell stress-inducing agents that release P-TEFb and HEXIM1/2 from 7SK snRNP leave core 7SK snRNP intact, allowing for prompt re-association of these factors once cell stress subsides (27,67). Current evidence suggest that upon 7SK synthesis, LA protein stabilizes it by binding its 3′-terminal U-rich sequence (69,70), while MePCE binds to the basal part of the 7SK SL1 and caps 7SK at its 5′ end to enable its stability (71). Perhaps due to the capping (72), LA leaves this transient 7SK snRNP complex and is replaced by LARP7, which employs its La and RRM1 motifs to bind the 7SK 3′-terminal U-rich sequence and the preceding SL4, protecting 7SK from nucleolytic degradation (27,60,71,73). Upon this binding event, affinity between LARP7 and MePCE increases, which in return provokes the tighter LARP7-7SK interaction and inhibits the capping activity of MePCE (27,67). This inhibition effectively prevents 7SK de-capping reaction and concludes formation of core 7SK snRNP.
Upon the assembly, core 7SK snRNP sets the stage for incorporation and subsequent inhibition of P-TEFb. Critical factors for both steps turn out to be HEXIM proteins, which are sequestered into the snRNP as homo- or hetero-dimers upon binding 7SK (62,63,74–76). Specifically, HEXIM1 dimers bind cooperatively to a proximal and distal part of 7SK SL1 through arginine-rich RNA-binding motif (ARM) which is positioned within centrally located HEXIM1's positively charged region (62,77–79). As a result, HEXIM1 auto-inhibitory conformation mediated by the N-terminal domain and negatively charged region is relieved (62,75,77), exposing the C-terminal dimeric coiled-coil region that provides the binding surface for the CycT1 subunit of P-TEFb (74,80,81). Inhibition of P-TEFb by HEXIM1 ensues, most likely via blocking ATP binding to Cdk9 (75). In addition, through its C-terminal RRM3 motif, LARP7 interacts with Cdk9 to further stabilize P-TEFb incorporation (27,71,82). Finally, Cdk9 T loop phosphorylation at Threonine 186 (P-Thr186) is required for the assembly of P-TEFb into 7SK snRNP (75,83). Because this phosphorylation is the hallmark of activation of any cyclin-dependent kinase, P-TEFb within 7SK snRNP is in a pre-activated form, fully ready to promote Pol II pause release upon its liberation from the inhibitory complex.
ANCHORING 7SK snRNP TO CHROMATIN
That 7SK snRNP represents the major cellular reservoir of transcriptionally inactive but poised P-TEFb has raised an important question of how does the inhibitory complex engage with transcriptional machinery to enable the transition into productive Pol II elongation. A previous work reported that in contrast to all other abundant snRNPs, 7SK snRNP is not tightly associated with any nuclear compartment including chromatin (84). The study gave rise to a view that simple diffusion of this abundant snRNP may provide a basis for delivering P-TEFb, once released from 7SK snRNP, to any genomic location in a timely manner. However, a mass-spectrometry analysis of factors bound to transcriptionally engaged Pol II detected the presence of Cdk9 as well as HEXIM1 and 7SK (85). In a separate report, P-TEFb was found to assemble into pre-initiation complex (PIC) with non-phosphorylated Pol II and many other factors including the 7SK snRNP components HEXIM1, LARP7 and 7SK (86). Correspondingly, P-TEFb isolated from the PIC failed to phosphorylate the CTD of Pol II in vitro. Together, these studies illustrate that a fraction of P-TEFb held within the inhibitory 7SK snRNP interacts with Pol II, suggesting that the inactive pool of P-TEFb could be embedded with Pol II on chromatin.
The first example of 7SK snRNP anchoring to chromatin came from studying the usual suspect, HIV-1 (86,87). This work revealed HEXIM1 and LARP7 co-occupying the viral core promoter with P-TEFb, and this same scenario was soon to be recapitulated at promoters of stimulus-dependent primary response genes (PRGs) (88), providing evidence that 7SK snRNP tethered to chromatin could serve as a source of P-TEFb stimulating Pol II escape from pausing. Importantly, a recent study has put this initial discoveries into a genome-wide context (89,90). Through chromatin isolation by RNA purification and subsequent sequencing (ChIRP-seq), the authors observed extensive 7SK occupancy along the entire transcribed loci of about 3500 active protein-coding genes in murine embryonic stem (mES) cells and human cells, with prominent and minor peaks encompassing TSSs and TESs, respectively, largely mirroring Pol II occupancy. In addition, higher levels of 7SK were detected at active enhancers and in particular at densely clustered enhancer elements also known as super enhancers (SEs), collectively invoking a role for 7SK in controlling Pol II transcription across genome. Of note, compared to promoters, the 7SK at SEs was not associated with proportional levels of N-TEFs and the canonical 7SK snRNP component HEXIM1, implicating 7SK in impacting transcription beyond Pol II pause release.
Considering the Pol II-7SK snRNP interaction studies and co-occupancy of 7SK with Pol II at transcribed regions, Pol II could be envisioned as a platform for 7SK snRNP recruitment. However, recent findings describing three recruitment mechanisms through factors defined herein as 7SK snRNP chromatin adaptor factors (Ch-AFs) paint an increasingly exciting and refined picture (Table 2).
Table 2. Mechanisms directing the anchoring and regulation of canonical 7SK snRNP on chromatin.
| Anchoring 7SK snRNP to chromatin | ||
| Chromatin adaptor factors (Ch-AFs) | Factor | Mode of action |
| Repressive chromatin mark | H4R3me2(s) | Binds first half of 7SK to recruit 7SK snRNP to A-PEs. |
| Transcription regulator | KAP1 (TRIM28 or TIF1β) | Binds LARP7 to recruit 7SK snRNP to promoters. |
| Non-histone chromatin protein | HMGA1 | Binds CTIP2 and SL2 of 7SK to recruit 7SK snRNP to promoters. |
| Releasing P-TEFb from 7SK snRNP | ||
| Signal transduction pathways | Factor | Mode of action |
| T-cell receptor signaling | PKCθ | Phosphorylates HEXIM1 Ser158 to prevent 7SK binding. |
| ERK | ERK-dependent kinase phosphorylates HEXIM1 Tyr271 and Tyr274 to prevent CycT1/P-TEFb binding. | |
| PI3K/Akt signaling | Akt | Phosphorylates HEXIM1 Ser270 and Thr278 to prevent CycT1/P-TEFb binding. |
| Ca2+-calmodulin-PP2B signaling | PP2B + PP1α | PP1α dephosphorylates P-Thr186 in Cdk9 after PP2B remodels 7SK snRNP. |
| Ca2+-dependent calpain 2 activation | Calpain 2 | Cleaves MePCE to destabilize core 7SK snRNP. |
| P-TEFb release factors (P-REFs) | Factor | Mode of action |
| HIV-1 transactivator of transcription | Tat | Binds the distal part in SL1 of 7SK and CycT1 to dislodge HEXIM1; recruits PPM1G for P-Thr186 Cdk9 dephosphorylation. |
| HTLV-1 transactivator of transcription | Tax | Binds CycT1 to dislodge HEXIM1. |
| Inducible transcription factor | NF-κB | Recruits PPM1G for P-Thr186 Cdk9 dephosphorylation. |
| Splicing and transcription regulator | SRSF2 | Binds SL3 of 7SK and nascent RNA to trigger the release. |
| DEAD-box RNA helicase | DDX21 | Binds SL3 of 7SK and changes conformation of 7SK. |
| hnRNPs and RNA helicase A | hnRNP A1, A2/B1, Q, R and RNA helicase A | Bind SL3 of 7SK to enable the subsequent release. |
| Histone demethylase | JMJD6 | Demethylates H4R3me2(s) and 7SK cap at A-PEs. |
| Histone acetyltransferase | p300 | Acetylates CycT1 to dislodge HEXIM1. |
Table summarizes principal ways by which 7SK snRNP is anchored to chromatin and by which P-TEFb is released from 7SK snRNP for promoting Pol II escape from pausing. For each factor, the mode of action is provided. The acronym SL stands for stem loop in 7SK.
First, 5′-terminal half of 7SK binds directly and independently of HEXIM1 to the repressive H4R3me2(s) chromatin mark at a sub-class of enhancers co-occupied by Brd4 and jumonji C domain-containing protein JMJD6 (90). This cohort of enhancers, also referred to as anti-pause enhancers (A-PEs) loop through Mediator to promoters to facilitate P-TEFb-dependent Pol II pause release at about thousand human genes. Second, 7SK snRNP could be loaded to promoter-proximal regions containing paused Pol II by the Kruppel-associated box (KRAB)-interacting protein KAP1 (91), also referred to as TRIM28 or TIF1β, which has been known mostly for promoting chromatin-based epigenetic silencing (92). In response to stimulation, KAP1 functioned as a Ch-AF that continuously tethered 7SK snRNP to promoters of PRGs via binding the ubiquitous 7SK snRNP component LARP7, enabling P-TEFb recruitment, Pol II elongation and PRGs expression. Judging from the genome-wide presence of KAP1 with 7SK snRNP components Cdk9, LARP7 and HEXIM1 in the vicinity of paused Pol II, the authors estimated that up to 70% of all genes experiencing pausing could be controlled via the KAP1-7SK snRNP axis. Further work is required to determine whether KAP1 regulates transcription of so many genes through 7SK snRNP. Importantly, a preceding study also identified KAP1 as a regulator of Pol II pausing (93). Here, phosphorylation of KAP1 at Serine 824 determined its influence on target gene transcription by Pol II. While the non-phosphorylated form of KAP1 augmented Pol II pausing, the phosphorylated KAP1 stimulated Pol II pause release for productive transcription, suggesting that this post-translational modification (PTM) might promote the promoter-proximal phosphorylation actions by P-TEFb. It remains to be tested, however, whether there is any connection between KAP1 phosphorylation, 7SK snRNP tethering and P-TEFb activation. Finally, a specific subset of 7SK snRNP contains a transcriptional repressor, C2H2-type zing-finger protein CTIP2, which interacts with HEXIM1 and the 7SK SL2 to augment inhibition of P-TEFb (94). Recruitment of this particular 7SK snRNP to HIV-1 and specific human gene promoters could be achieved via the non-histone chromatin protein HMGA1 that attracts the snRNP by binding CTIP2 and the 7SK SL2 (95).
Taking together, anchoring 7SK snRNP to promoters and enhancers by Ch-AFs represents an important step in controlling Pol II pause release by P-TEFb, providing an opportunity for additional regulatory layers impacting this critical checkpoint in metazoan gene transcription. In addition, as global disentanglement of P-TEFb from 7SK snRNP can be associated with hypertrophic and hyperproliferative diseases such as cardiac hypertrophy and cancer (96,97), supplying the pre-activated pool of P-TEFb to paused Pol II at specific genes sets the stage for a controlled use of P-TEFb by the subsequent transcriptional regulators.
LIBERATING P-TEFb FROM 7SK snRNP
Having realized the critical importance of P-TEFb in Pol II elongation and that its major pool resides within 7SK snRNP, many investigators have turned their attention to elucidate mechanisms governing P-TEFb activation through its release from the inhibitory snRNP (Table 2).
Release of P-TEFb by cellular signal transduction pathways
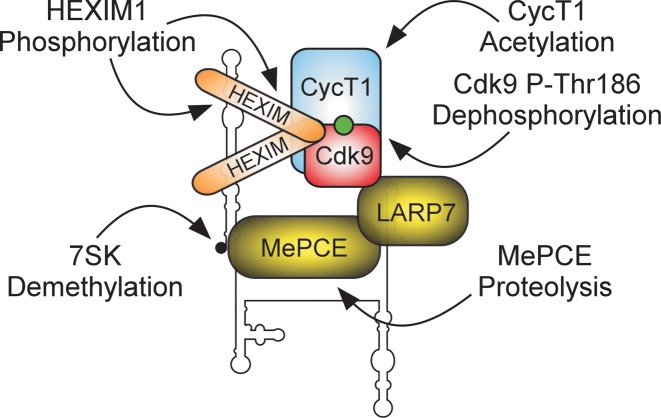
First, diverse cellular signal transduction pathways converge on almost all 7SK snRNP components to disable key mechanisms governing the assembly of P-TEFb into 7SK snRNP. As a result, specific PTMs occurring on Hexim1, Cdk9 and CycT1 as well as MePCE degradation provoke P-TEFb liberation (Figure 3). In the case of HEXIM1, its ability to bind 7SK and CycT1 is compromised. Specifically, in response to T-cell receptor (TCR) signaling, protein kinase C θ (PKCθ) phosphorylated Serine 158 within the positively-charged 7SK binding region of HEXIM1, antagonizing HEXIM1-7SK interaction and thus the subsequent P-TEFb binding and inhibition by HEXIM1 (98). In addition, phosphorylation of multiple residues within the unstructured HEXIM1 region just before the C-terminal coiled-coil CycT1 binding domain interferes with HEXIM1-P-TEFb interaction. Here, hexamethylene bisacetamide (HMBA) treatment triggered phosphorylation of Serine 270 and Threonine 278 via PI3K/Akt pathway (99). Moreover, activating TCR signaling stimulated extracellular-signal-regulated kinase (ERK), leading to Tyrosine 271 and 274 phosphorylation (100,101). Importantly, expression of mutant HEXIM1 proteins that could no longer become phosphorylated at the above residues inhibited the global release of P-TEFb from 7SK snRNP and P-TEFb occupancy at HIV-1 promoter, limiting viral transcription (99,101).
Figure 3.
Summary of post-translational modifications and proteolysis of 7SK snRNP components that provoke the release of P-TEFb. The model represents the canonical 7SK snRNP complex, in which 7SK, LARP7 and MePCE form core 7SK snRNP that enables binding and inhibition of P-TEFb (Cdk9 is in red) by HEXIM1/2. The arrows depict stimulus-dependent PTMs of HEXIM1, CycT1 and Cdk9, which lead to the release of P-TEFb by disabling key steps that promote the assembly of P-TEFb into 7SK snRNP. Whereas the PTMs in HEXIM1, CycT1 and Cdk9 leave core 7SK snRNP intact, demethylation of 7SK and proteolysis of MePCE destabilize the core, yielding irreversible P-TEFb activation.
Concerning P-TEFb, its presence and thus inhibition within 7SK snRNP is controlled directly through dephosphorylation and acetylation of Cdk9 and CycT1, respectively. Following UV irradiation or HMBA exposure, a key role in P-TEFb activation for the calcium (Ca2+)-calmodulin-protein phosphatase 2B (PP2B) signaling pathway and protein phosphatase 1α (PP1α) was uncovered (102). Therein, activated by UV or HMBA and facilitated by a PP2B-induced conformational change in 7SK snRNP, PP1α dephosphorylated P-Thr186 in Cdk9, inducing P-TEFb release. Of note, Threonine 186 has to be re-phosphorylated before P-TEFb could promote Pol II elongation. Finally, histone acetyltransferase p300 acetylated four Lysine residues within the predicted CycT1 coiled-coil region, triggering P-TEFb dissociation from HEXIM1 and thus 7SK snRNP by as of yet undefined mechanism (103).
In the cases of global activation of P-TEFb described thus far, P-TEFb release from the inhibitory grip by 7SK snRNP is reversible. This is because the core 7SK snRNP remains intact, serving as a platform that enables subsequent reassembly of HEXIM1 and P-TEFb. However, P-TEFb activation could also be irreversible due to destabilization of core 7SK snRNP. This was demonstrated in the context of megakaryocyte development, a process characterized by cellular enlargement and nuclear polyploidization (104). Early in megakaryopoiesis, high levels of Ca2+ provoked upregulation and activation of a Ca2+-dependent protease, calpain 2, which cleaved MePCE directly. Concomitantly, LARP7 was downregulated in a calpain 2-independent manner, fully destabilizing 7SK, leading to a global P-TEFb release and transcriptional upregulation of cytoskeletal regulatory factors that are essential for megakaryocyte morphogenesis.
Release of P-TEFb by HIV-1 Tat
In addition to cell signaling cascades that result in the PTMs or proteolysis of 7SK snRNP components, transcriptional regulators defined herein as P-TEFb release factors (P-REFs) can directly liberate P-TEFb from 7SK snRNP to stimulate Pol II pause release. This paradigm has been best illustrated by the way HIV-1 Tat hijacks P-TEFb for optimal viral transcription. Following initial observations that HIV-1 infection stimulated global release of P-TEFb from 7SK snRNP, Tat by itself also released P-TEFb from 7SK snRNP or prevented its sequestration into it in vivo and in vitro (58,105). Further mechanistic studies have unveiled key steps by which HIV-1 Tat and TAR transferred the repressed P-TEFb to nascent viral transcripts for their completion. The mechanisms involved are grounded by the obvious similarity between the HIV-1 TAR-Tat-P-TEFb and host cell 7SK-HEXIM1-P-TEFb systems. Namely, the TAR-binding ARM of Tat is very similar to the first half of 7SK-binding ARM of HEXIM1 (79). Furthermore, the 7SK SL1 carries proximal and distal HEXIM1-binding segments that are highly similar to the consensus minimal structure of the HIV-1 TAR (78), and both Tat and HEXIM1 bind CycT1 in a mutually-exclusive manner due to the shared CycT1 binding site (106,107). While through its ARM, Tat displaced HEXIM1 from 7SK by interacting with the evolutionarily conserved distal part of the 7SK SL1 (78), integrity of Tat's AD was critical for outcompeting HEXIM1 from the CycT1 subunit of P-TEFb, which stems from about 10-fold higher affinity of Tat for CycT1 (58). Moreover, the Tat-mediated release of P-TEFb from 7SK snRNP was assisted by AFF1 which further increased the affinity between Tat and P-TEFb (49). Thus, Tat releases P-TEFb through targeting two principal steps that govern its presence within 7SK snRNP. Of note, the human T-lymphotropic virus type 1 transcriptional activator Tax also utilizes P-TEFb for viral transcription and displaces P-TEFb from 7SK snRNP through binding CycT1 (108,109), suggesting that P-TEFb liberation from 7SK snRNP could be a common theme supporting replication of different viruses in host cells.
Furthermore, at least two additional mechanisms could play a role in promoting the transfer of P-TEFb by Tat from 7SK snRNP to TAR at proviral promoter within chromatin. As already pointed out, Tat could assemble into 7SK snRNP within the PIC at viral promoter. The local release of P-TEFb from the snRNP by Tat, however, occurred only as TAR emerged on the nascent viral transcript from paused Pol II (86). Thus, the inhibitory 7SK snRNP is released by competitive binding of the Tat:P–TEFb complex to TAR as it is transcribed, activating P-TEFb and effectively timing the switch between Pol II pausing and elongation. Finally, Tat facilitated extraction of P-TEFb from 7SK snRNP by recruiting the metal-dependent PPM1G/PP2Cγ phosphatase to HIV-1 promoter, which dephosphorylated the P-Thr186 in Cdk9 (88). Likewise, Tat might recruit PP1γ to 7SK snRNP to promote P-TEFb release and HIV-1 transcription. Namely, Tat was reported to interact with PP1γ directly and this PP1 isoform could also dephosphorylate the P-Thr186 in Cdk9 (110–112).
Release of P-TEFb by cellular RNA-binding proteins and transcription regulators
Historically, studying pathogen:host cell interactions has disclosed vital regulatory mechanisms of host cells, for the invading microbes usually co-opt them to their own advantage. Following the suit, many cellular regulatory proteins have been discovered that directly impact 7SK snRNP to promote P-TEFb activation and subsequent Pol II elongation.
By analyzing how abrupt Pol II inhibition by DRB and ActD affected the global pool of 7SK snRNP, investigators realized that the released P-TEFb and HEXIM1 proteins were replaced by a series of RBPs that included heterogeneous ribonuclear protein (hnRNP) A1, A2/B1, Q, R and RNA helicase A (113,114). The RBPs within this alternative 7SK complex termed herein 7SK-hnRNP snRNP assembled onto core 7SK snRNP by binding SL3 of 7SK (114). Critically, a combined knockdown of HNRNPA1 and HNRNPA2 or deletion of the 7SK SL3 precluded the release of P-TEFb and HEXIM1 in cells with inhibited transcription, indicating that the binding of the newly identified RBPs with 7SK is a pre-requisite for P-TEFb activation (113,114). Because the hnRNPs assemble with the freshly made transcripts and their free pool increases while Pol II is inhibited, these studies are consistent with a model in which the global steady-state pool of P-TEFb released from 7SK snRNP corresponds to the overall transcriptional demand of the cell.
Importantly, a series of fresh studies have shined a spotlight on how P-REFs, including individual RBPs and transcription regulators, could impact 7SK snRNP anchored to promoters and enhancers to provoke P-TEFb activation and Pol II elongation at specific sets of genes.
First, SRSF2 (formerly known as SC35), a member of the SR family of proteins which are normally involved in multiple RNA metabolism steps including constitutive and regulated splicing, was discovered to promote Pol II elongation at about thousand murine genes in a way that is reminiscent to the HIV-1 Tat/TAR system (29). SRSF2 was found to preferentially occupy active gene promoters, and its depletion decreased global levels of Pol II P-Ser2, causing a defect in Pol II pause release. Intriguingly, SRSF2 interacted directly with 7SK in living cells, which in turn promoted the binding between SRSF2 and Pol II as well as promoter occupancy of SRSF2 at its several target genes. Moreover, SRSF2 bound the 7SK SL3 and was assembled into the canonical 7SK snRNP, in effect mirroring the Tat-7SK snRNP-Pol II assembly at promoters. In contrast to the earlier observation in human cells, about 50% of 7SK snRNP was found to be associated with the chromatin fraction. Critically, the addition of RNA with high-affinity binding site for SRSF2 triggered the release of SRSF2 together with P-TEFb from 7SK snRNP in vitro, and further in vivo evidence suggested that single-stranded SRSF2 binding sites on nascent transcripts near TSSs promoted Pol II pause release through SRSF2. Thus, similar to HIV-1 transcription, it appears that emergence of RNA binding site from Pol II transcript serves as a switch that liberates SRSF2 and P-TEFb from 7SK snRNP, triggering productive Pol II elongation.
In addition to the SR protein SRSF2, the promoter-bound DEAD-box RNA helicase DDX21 is another member of P-REFs that could stimulate Pol II transcription through 7SK snRNP (115). By occupying the Pol I-transcribed rDNA locus in the nucleolus and more than three thousand active gene promoters with enriched levels of Pol II, DDX21 coordinates multiple steps of ribosome biogenesis in human cells. To control specifically Pol II genes, DDX21 bound 7SK mostly just before and within SL3 and, as a regulatory component of 7SK snRNP, was recruited to the promoters of genes encoding ribosomal proteins and snoRNAs through 7SK. Importantly, DDX21 promoted gene expression in a helicase-dependent manner, and through its active helicase domain, it induced the release of P-TEFb from 7SK snRNP in vitro. Hence, this work suggested that by directly impacting the conformation of 7SK within the snRNP, DDX21 provoked P-TEFb activation and Pol II pause release at target genes.
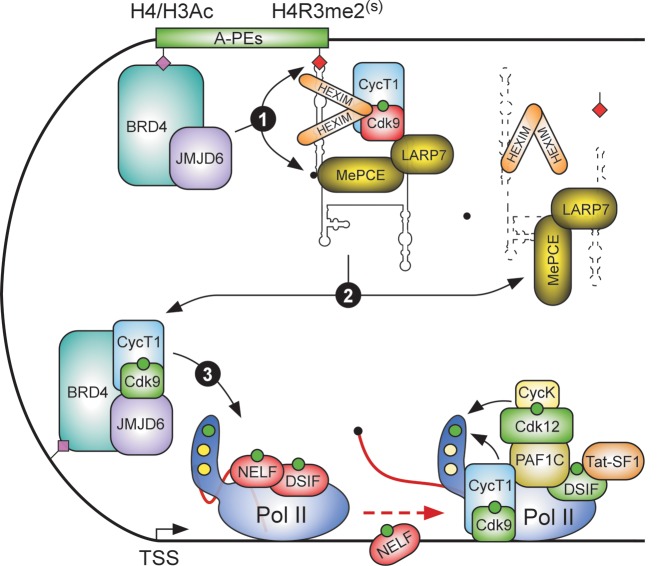
While SRSF2 and DDX21 stimulated P-TEFb activation on chromatin without impacting the stability of 7SK snRNP, P-TEFb release from the chromatin-anchored 7SK snRNP at Brd4-JMJD6 co-bound A-PEs is dramatically different (Figure 4). There, the demethylase activity of JMJD6 targeted two key steps that otherwise promoted the anchoring of 7SK snRNP and inhibition of P-TEFb at A-PEs (90). First, JMJD6 demethylated H4R3me2(s), erasing the repressive chromatin mark bound directly by 7SK. Second, via its demethylase activity, JMJD6 de-capped 7SK by removing the methyl group from the γ-phosphate of the first 5′ nucleotide of 7SK. Thus, the dual demethylase activity of JMJD6 prohibited the anchoring of 7SK snRNP at A-PEs and destabilized 7SK, releasing P-TEFb that led to increased Pol II P-Ser2 levels. Correspondingly, knockdown of JMJD6 as well as BRD4, of which product recruits JMJD6 to A-PEs, led to increased 7SK and HEXIM1 occupancy at A-PEs, preventing the release of Pol II from pausing at target genes. Illustrating further the role of the repressive chromatin mark in the dynamics of Pol II pause release at the enhancers, knockdown of PRMT5, which encodes the H4R3me2(s) methyltransferase, stimulated Pol II elongation. Importantly, once released from A-PEs, Brd4 and JMJD6 used their respective P-TEFb interacting domains (PIDs) (90,116) to capture active P-TEFb at target promoters for Pol II pause release. Besides promoting the capturing, Brd4 might employ its PID to further enhance the kinase activity of P-TEFb (117).
Figure 4.
Brd4 and the dual enzymatic activities of JMJD6 target 7SK snRNP at anti-pause enhancers to trigger Pol II pause release. A-PEs (green) contain either tetra-acetylated histone H4 (Lys5, 8, 12 and 16) or di-acetylated histone H3 (Lys9 and Lys14) (H4/H3Ac), as well as symmetric di-methylated histone H4 (Arg3) (H4R3me2(s)). While the H4/H3Ac mark recruits JMJD6 via Brd4, the H4R3me2(s) mark is bound directly by the 7SK SL1 to tether 7SK snRNP harboring inactive P-TEFb (Cdk9 is in red). The A-PEs loop to target gene promoters through Mediator (not shown) and stimulate Pol II pause release via a series of events. Through its histone demethylase activity targeting H4R3me2(s) and RNA demethylase activity targeting the 5′ 7SK methyl-group in its cap structure (Step 1), JMJD6 ablates the anchoring of 7SK snRNP at A-PEs and destabilizes 7SK, respectively. These actions disintegrate 7SK snRNP and release active P-TEFb (Cdk9 is in green), which is captured cooperatively by Brd4 and JMJD6 at target gene promoters containing the active H4/H3Ac chromatin mark (Step 2). Finally, P-TEFb phosphorylates its three main targets in paused Pol II complex (Step 3), stimulating Pol II elongation.
Finally, like Tat at HIV-1 promoter, the activated NF-κB could release P-TEFb from KAP1-tethered 7SK snRNP at PRGs through recruiting the metal-dependent PPM1G/PP2Cγ phosphatase, which dephosphorylated the P-Thr186 in Cdk9 (88,118). Furthermore, upon P-TEFb release at NF-κB-dependent DNA damage-induced genes, PPM1G bound 7SK and HEXIM1, effectively blocking reassembly of P-TEFb into 7SK snRNP to sustain productive Pol II elongation for duration of the stimulus (118).
ROLES OF 7SK snRNP BEYOND POL II PAUSE RELEASE
Although the role of 7SK snRNP in controlling the escape of Pol II from pausing through P-TEFb has received tremendous attention and is arguably the dominant function of the snRNP, it is becoming increasingly clear that 7SK or other components of the canonical 7SK snRNP impact additional steps in Pol II transcription and gene expression. As already pointed out, 7SK occupancy resembles that of Pol II along protein-coding genes (89), raising the possibility that 7SK could affect processes occurring after Pol II pause release. Indeed, by destabilizing core 7SK snRNP, the active P-TEFb could stimulate pre-mRNA alternative splicing by augmenting recruitment of SRSF1 (formerly known as SF2/ASF) and general splicing machinery via increased Pol II P-Ser2 levels (27). In addition, 7SK depletion led to failed Pol II termination at thousands of genes in mES cells (119). Although recent evidence described contributions of P-TEFb in controlling Pol II termination around TESs (17,26), more work needs to be done to reveal how exactly does 7SK prevent read-through transcription.
The recent 7SK ChIRP-seq study revealed a novel role for 7SK in guarding genomic stability and Pol II initiation (89). Namely, the loss of 7SK provoked an increase of convergent transcription at promoters, enhancers and particularly at SEs, which could lead to previously described AID-initiated genomic instability (120). Indeed, augmented DNA-damage signaling was observed at the sites. Importantly, 7SK at enhancers and SEs restricted not only elongation but also initiation by Pol II, clearly distinguishing these transcribed loci from promoters where 7SK controlled Pol II pause release. The authors ascribed the new function of 7SK at enhancers to its interaction with the mammalian ATP-dependent nucleosome-remodeling BAF complex. The canonical 7SK snRNP and the 7SK–BAF snRNP were found to be mutually exclusive, and by and large they resided at promoters and enhancers, respectively. Although only a minor fraction of cellular 7SK was found to be associated with BAF components, the high cellular abundance of 7SK could still translate into about 10 000 7SK–BAF snRNPs per cell, potentially impacting thousands of enhancers across the genome. Curiously, LARP7 and MePCE failed to co-purify with BAF, suggesting that 7SK bound to BAF may be stabilized through alternative means. Surprisingly, JQ1, a small molecule bromodomain inhibitor which is highly specific for dislodging Brd4 from chromatin (121), interfered with targeting 7SK to enhancers genome wide. Because previous body of work has never linked Brd4 to any 7SK complexes, future experiments are needed to examine if Brd4 plays a direct role in promoting the 7SK enhancer occupancy. Functionally, depletion of 7SK caused stronger central nucleosome positioning at enhancers and 7SK was found to be important for scaffolding of BAF with Pol II as well as for global BAF recruitment to enhancers, suggesting that 7SK facilitates action of BAF, a known inhibitor of enhancer RNA transcription (122).
Finally, mounting evidence indicates that genes encoding the canonical 7SK snRNP components LARP7 and HEXIM1 are tumor suppressors. Whereas LARP7 downregulation in breast cancer has been proposed to promote tumorigenesis by transcriptional activation of key regulators stimulating epithelial–mesenchymal transition (96), a recent study on ubiquitous HEXIM1 silencing in melanoma has revealed a dual role of HEXIM1 in suppressing genesis of this cancer (123). While increased HEXIM1 levels did lead to a defect in paused Pol II release at tumorigenic genes through decreased 7SK snRNP promoter anchoring, HEXIM1 outside of the 7SK snRNP bound to many mRNAs including anti-tumorigenic transcripts. The binding increased stability of examined transcripts, highlighting how under certain circumstances HEXIM1 not only inhibits transcription but also promotes gene expression in a post-transcriptional manner.
CONCLUSION AND PERSPECTIVES
In keeping with the vital role of P-TEFb in transcription cycle by Pol II, cells have devised the robust 7SK snRNP control system to inhibit P-TEFb and enable its delivery in a pre-activated form to gene promoters for stimulating Pol II pause release. A view that has emerged from many recent reports is that the canonical 7SK snRNP, once assembled, should not be perceived as an isolated RNP particle roaming the nucleoplasm, waiting for a signal to eject P-TEFb. True, a strategy to release P-TEFb globally, including from the non-chromatin associated pool of 7SK snRNP, could be a part of certain gene expression program such as the one enabling cell proliferation, megakaryopoiesis or recovery from UV irradiation-induced DNA damage. Rather than being inert, 7SK snRNP is contacted by a growing number of auxiliary factors, which deliver 7SK snRNP to gene promoters or enhancers and stimulate P-TEFb activation at appropriate circumstances, respectively, for a controlled gene-specific transition of Pol II from pausing into productive elongation.
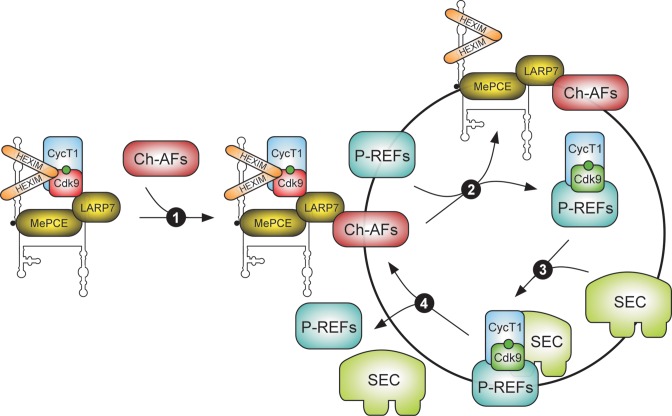
Taking together, we propose a unifying model of P-TEFb activation on chromatin (Figure 5). In principle, each and every major step in the model, including the anchoring of 7SK snRNP to chromatin by Ch-AFs, release of P-TEFb from 7SK snRNP by P-REFs, assembly of P-TEFb with transcription factor and SECs, and sequestration of P-TEFb back into 7SK snRNP upon transcription shutdown, could be regulated. Hence, many questions remain unanswered and interrogating this model warrants intensive future experimentation.
Figure 5.
A model of P-TEFb activation on chromatin by 7SK snRNP chromatin adaptor and P-TEFb release factors. The model depicts the P-TEFb activation and de-activation cycle at gene promoter. Specific 7SK snRNP chromatin adaptor factors (Ch-AFs), i.e. a regulatory protein or a chromatin mark, direct the anchoring of a fraction of nucleoplasmic 7SK snRNP to chromatin, harboring inactive P-TEFb (Cdk9 is in red) (Step 1). Upon diverse circumstances, such as cell signaling or emergent nascent transcript, P-TEFb release factors (P-REFs) converge on 7SK snRNP to trigger the liberation of active P-TEFb (Cdk9 is in green) (Step 2). Subsequently, P-TEFb is assembled into SEC and could in concert with the dedicated transcription factor or P-REF promote the release of Pol II from pausing (Step 3). Finally, upon the cessation of the signal stimulating gene transcription, P-TEFb is re-sequestered into 7SK snRNP anchored at promoter (Step 4). Chromatin is represented as a black circle. For clarity, Pol II is omitted from the model. Each of the steps could be subject to control, and for simplicity, possible regulatory mechanisms are not depicted.
For example, are there mechanisms in place that could stimulate or ablate an interaction between 7SK snRNP and its chromatin adaptor protein such as KAP1, or an interaction between the already assembled 7SK snRNP–KAP1 complex and chromatin? Staying with the theme, how exactly is KAP1 recruited to paused Pol II, and are there additional Ch-AFs that tether 7SK snRNP to promoters or enhancers? Intriguingly, TIF1γ, a protein of the four member TIF1 family that includes KAP1, promotes the recruitment of stimulatory Pol II elongation factors including P-TEFb to erythroid genes (124), suggesting that TIF1γ could indeed be another Ch-AF mediating recruitment of 7SK snRNP to chromatin. Likewise, given that P-TEFb release factors SRSF2 and DDX21 require 7SK for their promoter occupancy, is KAP1 or another Ch-AF involved in their recruitment?
Another important and little understood issue is how does P-TEFb, subsequently to its release from 7SK snRNP on chromatin, become a part of SEC? One attractive possibility is a scenario that might take place in the HIV-1 system, whereby Tat releases P-TEFb as a hetero-trimer containing AFF1 (49), which could then serve as a link for the SEC assembly. Whether AFF1 is globally present at chromatin-anchored 7SK snRNP and if cellular RBPs and transcription factors also capture P-TEFb as a trimer remains to be determined. More generally, is chromatin anchoring of P-TEFb within 7SK snRNP a vital pre-requisite for the ability of numerous transcription factors to employ P-TEFb for stimulating Pol II pause release? Of note, as the other major pool of P-TEFb (41,42), the Brd4-containing P-TEFb complex might also serve as a source of P-TEFb on chromatin. Curiously, the scaffolding SEC components AFF1 and AFF4 were also detected within the Brd4-P-TEFb complex (49), indicating that they might direct SEC assembly upon the possible release of the P-TEFb-AFF1/4 module from Brd4 by transcription factors. Indeed, HIV-1 Tat released P-TEFb from Brd4 (41), suggesting a model in which SEC could be employed at target cellular genes independently of the 7SK snRNP route. Finally, given the sizable nucleoplasmic pools of 7SK snRNP and Brd4–P-TEFb complexes, it remains possible that viral and cellular activators could also get a handle on P-TEFb off chromatin.
Concerning the 7SK–BAF snRNP, it would be informative to determine the molecular basis for 7SK-mediated scaffolding of this novel snRNP with Pol II, and why is only a fraction of about 150 000 chromatin-bound BAF complexes associated with 7SK? Finally, is 7SK–BAF snRNP also subject to such an elaborate regulation as the canonical 7SK snRNP? Given that 7SK–BAF snRNP restricted Pol II initiation and that 7SK assumes different conformations within the two 7SK-containing snRNPs (89), it seems likely that mechanisms other than those impacting the canonical 7SK snRNP regulate 7SK:BAF interaction. Addressing these and many other outstanding challenges, including obtaining structures of various 7SK-containing snRNPs, will undoubtedly move the field to a greater understanding of a mystery controlled by the string of nucleotides that goes by the name of 7SK.
Acknowledgments
We thank Tina Lenasi for critical reading of the manuscript and apologize to many colleagues for not being able to cite all relevant studies due to space limitations.
FUNDING
Academy of Finland [1263825 to M.B.]; Sigrid Juselius Foundation [4704610 to M.B.]; University of Helsinki Three-year Research Grant [490123 to M.B.]. Funding for open access charge: Sigrid Juselius Foundation [4704610 to M.B].
Conflict of interest statement. None declared.
REFERENCES
- 1.Strobl L.J., Eick D. Hold back of RNA polymerase II at the transcription start site mediates down-regulation of c-myc in vivo. EMBO J. 1992;11:3307–3314. doi: 10.1002/j.1460-2075.1992.tb05409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krumm A., Meulia T., Brunvand M., Groudine M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 1992;6:2201–2213. doi: 10.1101/gad.6.11.2201. [DOI] [PubMed] [Google Scholar]
- 3.Rougvie A.E., Lis J.T. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- 4.Kao S.-Y., Calman A.F., Luciw P.A., Peterlin B.M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- 5.Guo J., Price D.H. RNA Polymerase II Transcription Elongation Control. Chem. Rev. 2013;113:8583–8603. doi: 10.1021/cr400105n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jonkers I., Lis J.T. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015;16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adelman K., Lis J.T. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price D.H. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 2000;20:2629–2634. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Q., Li T., Price D.H. RNA polymerase II elongation control. Annu. Rev. Biochem. 2012;81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marshall N.F., Price D.H. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 11.Mancebo H.S., Lee G., Flygare J., Tomassini J., Luu P., Zhu Y., Peng J., Blau C., Hazuda D., Price D., et al. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei P., Garber M.E., Fang S.M., Fischer W.H., Jones K.A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 13.Lu H., Li Z., Xue Y., Zhou Q. Viral-host interactions that control HIV-1 transcriptional elongation. Chem. Rev. 2013;113:8567–8582. doi: 10.1021/cr400120z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wada T., Takagi T., Yamaguchi Y., Ferdous A., Imai T., Hirose S., Sugimoto S., Yano K., Hartzog G.A., Winston F., et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamaguchi Y., Takagi T., Wada T., Yano K., Furuya A., Sugimoto S., Hasegawa J., Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 16.Palangat M., Meier T.I., Keene R.G., Landick R. Transcriptional pausing at +62 of the HIV-1 nascent RNA modulates formation of the TAR RNA structure. Mol. Cell. 1998;1:1033–1042. doi: 10.1016/s1097-2765(00)80103-3. [DOI] [PubMed] [Google Scholar]
- 17.Sanso M., Levin R.S., Lipp J.J., Wang V.Y., Greifenberg A.K., Quezada E.M., Ali A., Ghosh A., Larochelle S., Rana T.M., et al. P-TEFb regulation of transcription termination factor Xrn2 revealed by a chemical genetic screen for Cdk9 substrates. Genes Dev. 2016;30:117–131. doi: 10.1101/gad.269589.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bentley D.L. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 2014;15:163–175. doi: 10.1038/nrg3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lenasi T., Barboric M. P-TEFb stimulates transcription elongation and pre-mRNA splicing through multilateral mechanisms. RNA Biol. 2010;7:145–150. doi: 10.4161/rna.7.2.11057. [DOI] [PubMed] [Google Scholar]
- 20.Schuller R., Forne I., Straub T., Schreieck A., Texier Y., Shah N., Decker T.M., Cramer P., Imhof A., Eick D. Heptad-specific phosphorylation of RNA polymerase II CTD. Mol. Cell. 2016;61:305–314. doi: 10.1016/j.molcel.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 21.Fujinaga K., Irwin D., Huang Y., Taube R., Kurosu T., Peterlin B.M. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol. Cell. Biol. 2004;24:787–795. doi: 10.1128/MCB.24.2.787-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y., Yamaguchi Y., Tsugeno Y., Yamamoto J., Yamada T., Nakamura M., Hisatake K., Handa H. DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Genes Dev. 2009;23:2765–2777. doi: 10.1101/gad.1834709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wada T., Takagi T., Yamaguchi Y., Watanabe D., Handa H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 1998;17:7395–7403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu M., Yang W., Ni T., Tang Z., Nakadai T., Zhu J., Roeder R.G. RNA polymerase II-associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II. Science. 2015;350:1383–1386. doi: 10.1126/science.aad2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davidson L., Muniz L., West S. 3′ end formation of pre-mRNA and phosphorylation of Ser2 on the RNA polymerase II CTD are reciprocally coupled in human cells. Genes Dev. 2014;28:342–356. doi: 10.1101/gad.231274.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laitem C., Zaborowska J., Isa N.F., Kufs J., Dienstbier M., Murphy S. CDK9 inhibitors define elongation checkpoints at both ends of RNA polymerase II-transcribed genes. Nat. Struct. Mol. Biol. 2015;22:396–403. doi: 10.1038/nsmb.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barboric M., Lenasi T., Chen H., Johansen E.B., Guo S., Peterlin B.M. 7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development. Proc. Natl. Acad. Sci. U.S.A. 2009;106:7798–7803. doi: 10.1073/pnas.0903188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guiguen A., Soutourina J., Dewez M., Tafforeau L., Dieu M., Raes M., Vandenhaute J., Werner M., Hermand D. Recruitment of P-TEFb (Cdk9-Pch1) to chromatin by the cap-methyl transferase Pcm1 in fission yeast. EMBO J. 2007;26:1552–1559. doi: 10.1038/sj.emboj.7601627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji X., Zhou Y., Pandit S., Huang J., Li H., Lin C.Y., Xiao R., Burge C.B., Fu X.-D. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell. 2013;153:855–868. doi: 10.1016/j.cell.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenasi T., Peterlin B.M., Barboric M. Cap-binding protein complex links pre-mRNA capping to transcription elongation and alternative splicing through positive transcription elongation factor b (P-TEFb) J. Biol. Chem. 2011;286:22758–22768. doi: 10.1074/jbc.M111.235077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ni Z., Schwartz B.E., Werner J., Suarez J.R., Lis J.T. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell. 2004;13:55–65. doi: 10.1016/s1097-2765(03)00526-4. [DOI] [PubMed] [Google Scholar]
- 32.Rahl P.B., Lin C.Y., Seila A.C., Flynn R.A., McCuine S., Burge C.B., Sharp P.A., Young R.A. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shim E.Y., Walker A.K., Shi Y., Blackwell T.K. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 2002;16:2135–2146. doi: 10.1101/gad.999002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Czudnochowski N., Bosken C.A., Geyer M. Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nat. Commun. 2012;3:842. doi: 10.1038/ncomms1846. [DOI] [PubMed] [Google Scholar]
- 35.Liang K., Gao X., Gilmore J.M., Florens L., Washburn M.P., Smith E., Shilatifard A. Characterization of human cyclin-dependent kinase 12 (CDK12) and CDK13 complexes in C-terminal domain phosphorylation, gene transcription, and RNA processing. Mol. Cell. Biol. 2015;35:928–938. doi: 10.1128/MCB.01426-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayer A., di Iulio J., Maleri S., Eser U., Vierstra J., Reynolds A., Sandstrom R., Stamatoyannopoulos J.A., Churchman L.S. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell. 2015;161:541–554. doi: 10.1016/j.cell.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barboric M., Nissen R.M., Kanazawa S., Jabrane-Ferrat N., Peterlin B.M. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- 38.Gomes N.P., Bjerke G., Llorente B., Szostek S.A., Emerson B.M., Espinosa J.M. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 2006;20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanazawa S., Okamoto T., Peterlin B.M. Tat competes with CIITA for the binding to P-TEFb and blocks the expression of MHC class II genes in HIV infection. Immunity. 2000;12:61–70. doi: 10.1016/s1074-7613(00)80159-4. [DOI] [PubMed] [Google Scholar]
- 40.Hargreaves D.C., Horng T., Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Z., Yik J.H., Chen R., He N., Jang M.K., Ozato K., Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 42.Jang M.K., Mochizuki K., Zhou M., Jeong H.S., Brady J.N., Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 43.Hossain M.A., Chung C., Pradhan S.K., Johnson T.L. The yeast cap binding complex modulates transcription factor recruitment and establishes proper histone H3K36 trimethylation during active transcription. Mol. Cell. Biol. 2013;33:785–799. doi: 10.1128/MCB.00947-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He N., Liu M., Hsu J., Xue Y., Chou S., Burlingame A., Krogan N.J., Alber T., Zhou Q. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol. Cell. 2010;38:428–438. doi: 10.1016/j.molcel.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin C., Smith E.R., Takahashi H., Lai K.C., Martin-Brown S., Florens L., Washburn M.P., Conaway J.W., Conaway R.C., Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mueller D., Garcia-Cuellar M.P., Bach C., Buhl S., Maethner E., Slany R.K. Misguided transcriptional elongation causes mixed lineage leukemia. PLoS Biol. 2009;7:e1000249. doi: 10.1371/journal.pbio.1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sobhian B., Laguette N., Yatim A., Nakamura M., Levy Y., Kiernan R., Benkirane M. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol. Cell. 2010;38:439–451. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith E., Lin C., Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 2011;25:661–672. doi: 10.1101/gad.2015411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu H., Li Z., Xue Y., Schulze-Gahmen U., Johnson J.R., Krogan N.J., Alber T., Zhou Q. AFF1 is a ubiquitous P-TEFb partner to enable Tat extraction of P-TEFb from 7SK snRNP and formation of SECs for HIV transactivation. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E15–E24. doi: 10.1073/pnas.1318503111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gardini A., Baillat D., Cesaroni M., Hu D., Marinis J.M., Wagner E.J., Lazar M.A., Shilatifard A., Shiekhattar R. Integrator regulates transcriptional initiation and pause release following activation. Mol. Cell. 2014;56:128–139. doi: 10.1016/j.molcel.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takahashi H., Parmely T.J., Sato S., Tomomori-Sato C., Banks C.A., Kong S.E., Szutorisz H., Swanson S.K., Martin-Brown S., Washburn M.P., et al. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell. 2011;146:92–104. doi: 10.1016/j.cell.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dawson M.A., Prinjha R.K., Dittmann A., Giotopoulos G., Bantscheff M., Chan W.I., Robson S.C., Chung C.W., Hopf C., Savitski M.M., et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He N., Chan C.K., Sobhian B., Chou S., Xue Y., Liu M., Alber T., Benkirane M., Zhou Q. Human Polymerase-Associated Factor Complex (PAFc) connects the Super Elongation Complex (SEC) to RNA polymerase II on chromatin. Proc. Natl. Acad. Sci. U.S.A. 2011;108:E636–E645. doi: 10.1073/pnas.1107107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perumal K., Sinha K., Henning D., Reddy R. Purification, characterization, and cloning of the cDNA of human signal recognition particle RNA 3′-adenylating enzyme. J. Biol. Chem. 2001;276:21791–21796. doi: 10.1074/jbc.M101905200. [DOI] [PubMed] [Google Scholar]
- 55.Sinha K.M., Gu J., Chen Y., Reddy R. Adenylation of small RNAs in human cells. Development of a cell-free system for accurate adenylation on the 3′-end of human signal recognition particle RNA. J. Biol. Chem. 1998;273:6853–6859. doi: 10.1074/jbc.273.12.6853. [DOI] [PubMed] [Google Scholar]
- 56.Nguyen V.T., Kiss T., Michels A.A., Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 57.Yang Z., Zhu Q., Luo K., Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 58.Barboric M., Yik J.H., Czudnochowski N., Yang Z., Chen R., Contreras X., Geyer M., Matija Peterlin B., Zhou Q. Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res. 2007;35:2003–2012. doi: 10.1093/nar/gkm063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim Y.K., Mbonye U., Hokello J., Karn J. T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway. J. Mol. Biol. 2011;410:896–916. doi: 10.1016/j.jmb.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He N., Jahchan N.S., Hong E., Li Q., Bayfield M.A., Maraia R.J., Luo K., Zhou Q. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol. Cell. 2008;29:588–599. doi: 10.1016/j.molcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jeronimo C., Forget D., Bouchard A., Li Q., Chua G., Poitras C., Therien C., Bergeron D., Bourassa S., Greenblatt J., et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Michels A.A., Fraldi A., Li Q., Adamson T.E., Bonnet F., Nguyen V.T., Sedore S.C., Price J.P., Price D.H., Lania L., et al. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 2004;23:2608–2619. doi: 10.1038/sj.emboj.7600275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yik J.H., Chen R., Nishimura R., Jennings J.L., Link A.J., Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 64.Krueger B.J., Varzavand K., Cooper J.J., Price D.H. The mechanism of release of P-TEFb and HEXIM1 from the 7SK snRNP by viral and cellular activators includes a conformational change in 7SK. PLoS One. 2010;5:e12335. doi: 10.1371/journal.pone.0012335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marz M., Donath A., Verstraete N., Nguyen V.T., Stadler P.F., Bensaude O. Evolution of 7SK RNA and its protein partners in metazoa. Mol. Biol. Evol. 2009;26:2821–2830. doi: 10.1093/molbev/msp198. [DOI] [PubMed] [Google Scholar]
- 66.Wassarman D.A., Steitz J.A. Structural analyses of the 7SK ribonucleoprotein (RNP), the most abundant human small RNP of unknown function. Mol. Cell. Biol. 1991;11:3432–3445. doi: 10.1128/mcb.11.7.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xue Y., Yang Z., Chen R., Zhou Q. A capping-independent function of MePCE in stabilizing 7SK snRNA and facilitating the assembly of 7SK snRNP. Nucleic Acids Res. 2010;38:360–369. doi: 10.1093/nar/gkp977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krueger B.J., Jeronimo C., Roy B.B., Bouchard A., Barrandon C., Byers S.A., Searcey C.E., Cooper J.J., Bensaude O., Cohen E.A., et al. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 2008;36:2219–2229. doi: 10.1093/nar/gkn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bayfield M.A., Yang R., Maraia R.J. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs) Biochim. Biophys. Acta. 2010;1799:365–378. doi: 10.1016/j.bbagrm.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chambers J.C., Kurilla M.G., Keene J.D. Association between the 7 S RNA and the lupus La protein varies among cell types. J. Biol. Chem. 1983;258:11438–11441. [PubMed] [Google Scholar]
- 71.Muniz L., Egloff S., Kiss T. RNA elements directing in vivo assembly of the 7SK/MePCE/Larp7 transcriptional regulatory snRNP. Nucleic Acids Res. 2013;41:4686–4698. doi: 10.1093/nar/gkt159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bhattacharya R., Perumal K., Sinha K., Maraia R., Reddy R. Methylphosphate cap structure in small RNAs reduces the affinity of RNAs to La protein. Gene Expr. 2002;10:243–253. doi: 10.3727/000000002783992398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Uchikawa E., Natchiar K.S., Han X., Proux F., Roblin P., Zhang E., Durand A., Klaholz B.P., Dock-Bregeon A.C. Structural insight into the mechanism of stabilization of the 7SK small nuclear RNA by LARP7. Nucleic Acids Res. 2015;43:3373–3388. doi: 10.1093/nar/gkv173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blazek D., Barboric M., Kohoutek J., Oven I., Peterlin B.M. Oligomerization of HEXIM1 via 7SK snRNA and coiled-coil region directs the inhibition of P-TEFb. Nucleic Acids Res. 2005;33:7000–7010. doi: 10.1093/nar/gki997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li Q., Price J.P., Byers S.A., Cheng D., Peng J., Price D.H. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J. Biol. Chem. 2005;280:28819–28826. doi: 10.1074/jbc.M502712200. [DOI] [PubMed] [Google Scholar]
- 76.Yik J.H., Chen R., Pezda A.C., Zhou Q. Compensatory contributions of HEXIM1 and HEXIM2 in maintaining the balance of active and inactive positive transcription elongation factor b complexes for control of transcription. J. Biol. Chem. 2005;280:16368–16376. doi: 10.1074/jbc.M500912200. [DOI] [PubMed] [Google Scholar]
- 77.Barboric M., Kohoutek J., Price J.P., Blazek D., Price D.H., Peterlin B.M. Interplay between 7SK snRNA and oppositely charged regions in HEXIM1 direct the inhibition of P-TEFb. EMBO J. 2005;24:4291–4303. doi: 10.1038/sj.emboj.7600883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Muniz L., Egloff S., Ughy B., Jády B.E., Kiss T. Controlling cellular P-TEFb Activity by the HIV-1 transcriptional transactivator Tat. PLoS Pathog. 2010;6:e1001152. doi: 10.1371/journal.ppat.1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yik J.H., Chen R., Pezda A.C., Samford C.S., Zhou Q. A human immunodeficiency virus type 1 Tat-like arginine-rich RNA-binding domain is essential for HEXIM1 to inhibit RNA polymerase II transcription through 7SK snRNA-mediated inactivation of P-TEFb. Mol. Cell. Biol. 2004;24:5094–5105. doi: 10.1128/MCB.24.12.5094-5105.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bigalke J.M., Dames S.A., Blankenfeldt W., Grzesiek S., Geyer M. Structure and Dynamics of a Stabilized Coiled-Coil Domain in the P-TEFb Regulator Hexim1. J. Mol. Biol. 2011;414:639–653. doi: 10.1016/j.jmb.2011.10.022. [DOI] [PubMed] [Google Scholar]
- 81.Dames S.A., Schonichen A., Schulte A., Barboric M., Peterlin B.M., Grzesiek S., Geyer M. Structure of the Cyclin T binding domain of Hexim1 and molecular basis for its recognition of P-TEFb. Proc. Natl. Acad. Sci. U.S.A. 2007;104:14312–14317. doi: 10.1073/pnas.0701848104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Markert A., Grimm M., Martinez J., Wiesner J., Meyerhans A., Meyuhas O., Sickmann A., Fischer U. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep. 2008;9:569–575. doi: 10.1038/embor.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen R., Yang Z., Zhou Q. Phosphorylated positive transcription elongation factor b (P-TEFb) is tagged for inhibition through association with 7SK snRNA. J. Biol. Chem. 2004;279:4153–4160. doi: 10.1074/jbc.M310044200. [DOI] [PubMed] [Google Scholar]
- 84.Biglione S., Byers S.A., Price J.P., Nguyen V.T., Bensaude O., Price D.H., Maury W. Inhibition of HIV-1 replication by P-TEFb inhibitors DRB, seliciclib and flavopiridol correlates with release of free P-TEFb from the large, inactive form of the complex. Retrovirology. 2007;4:47. doi: 10.1186/1742-4690-4-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Das R., Yu J., Zhang Z., Gygi M.P., Krainer A.R., Gygi S.P., Reed R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol. Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 86.D'Orso I., Frankel A.D. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat. Struct. Mol. Biol. 2010;17:815–821. doi: 10.1038/nsmb.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barboric M., Lenasi T. Kick-sTARting HIV-1 transcription elongation by 7SK snRNP deporTATion. Nat. Struct. Mol. Biol. 2010;17:928–930. doi: 10.1038/nsmb0810-928. [DOI] [PubMed] [Google Scholar]
- 88.McNamara R.P., McCann J.L., Gudipaty S.A., D'Orso I. Transcription factors mediate the enzymatic disassembly of promoter-bound 7SK snRNP to locally recruit P-TEFb for transcription elongation. Cell Rep. 2013;5:1256–1268. doi: 10.1016/j.celrep.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Flynn R.A., Do B.T., Rubin A.J., Calo E., Lee B., Kuchelmeister H., Rale M., Chu C., Kool E.T., Wysocka J., et al. 7SK-BAF axis controls pervasive transcription at enhancers. Nat. Struct. Mol. Biol. 2016;23:231–238. doi: 10.1038/nsmb.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu W., Ma Q., Wong K., Li W., Ohgi K., Zhang J., Aggarwal A., Rosenfeld M.G. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013;155:1581–1595. doi: 10.1016/j.cell.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McNamara R.P., Reeder J.E., McMillan E.A., Bacon C.W., McCann J.L., D'Orso I. KAP1 recruitment of the 7SK snRNP complex to promoters enables transcription elongation by RNA polymerase II. Mol. Cell. 2016;61:39–53. doi: 10.1016/j.molcel.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Iyengar S., Farnham P.J. KAP1 protein: an enigmatic master regulator of the genome. J. Biol. Chem. 2011;286:26267–26276. doi: 10.1074/jbc.R111.252569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bunch H., Zheng X., Burkholder A., Dillon S.T., Motola S., Birrane G., Ebmeier C.C., Levine S., Fargo D., Hu G., et al. TRIM28 regulates RNA polymerase II promoter proximal pausing and pause release. Nat. Struct. Mol. Biol. 2014;21:876–883. doi: 10.1038/nsmb.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cherrier T., Le Douce V., Eilebrecht S., Riclet R., Marban C., Dequiedt F., Goumon Y., Paillart J.-C., Mericskay M., Parlakian A., et al. CTIP2 is a negative regulator of P-TEFb. Proc. Natl. Acad. Sci. U.S.A. 2013;110:12655–12660. doi: 10.1073/pnas.1220136110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Eilebrecht S., Le Douce V., Riclet R., Targat B., Hallay H., Van Driessche B., Schwartz C., Robette G., Van Lint C., Rohr O., et al. HMGA1 recruits CTIP2-repressed P-TEFb to the HIV-1 and cellular target promoters. Nucleic Acids Res. 2014;42:4962–4971. doi: 10.1093/nar/gku168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ji X., Lu H., Zhou Q., Luo K. LARP7 suppresses P-TEFb activity to inhibit breast cancer progression and metastasis. Elife. 2014;3:e02907. doi: 10.7554/eLife.02907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sano M., Abdellatif M., Oh H., Xie M., Bagella L., Giordano A., Michael L.H., DeMayo F.J., Schneider M.D. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat. Med. 2002;8:1310–1317. doi: 10.1038/nm778. [DOI] [PubMed] [Google Scholar]
- 98.Fujinaga K., Barboric M., Li Q., Luo Z., Price D.H., Peterlin B.M. PKC phosphorylates HEXIM1 and regulates P-TEFb activity. Nucleic Acids Res. 2012;40:9160–9170. doi: 10.1093/nar/gks682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Contreras X., Barboric M., Lenasi T., Peterlin B.M. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog. 2007;3:e146. doi: 10.1371/journal.ppat.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim Y.K., Mbonye U., Hokello J., Karn J. T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway. J. Mol. Biol. 2011;410:896–916. doi: 10.1016/j.jmb.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mbonye U.R., Wang B., Gokulrangan G., Chance M.R., Karn J. Phosphorylation of HEXIM1 at Tyr271 and Tyr274 promotes release of P-TEFb from the 7SK snRNP complex and enhances proviral HIV gene expression. Proteomics. 2015;15:2078–2086. doi: 10.1002/pmic.201500038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen R., Liu M., Li H., Xue Y., Ramey W.N., He N., Ai N., Luo H., Zhu Y., Zhou N., et al. PP2B and PP1α cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca(2+) signaling. Genes Dev. 2008;22:1356–1368. doi: 10.1101/gad.1636008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cho S., Schroeder S., Kaehlcke K., Kwon H.S., Pedal A., Herker E., Schnoelzer M., Ott M. Acetylation of cyclin T1 regulates the equilibrium between active and inactive P-TEFb in cells. EMBO J. 2009;28:1407–1417. doi: 10.1038/emboj.2009.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Elagib K.E., Rubinstein J.D., Delehanty L.L., Ngoh V.S., Greer P.A., Li S., Lee J.K., Li Z., Orkin S.H., Mihaylov I.S., et al. Calpain 2 activation of P-TEFb drives megakaryocyte morphogenesis and is disrupted by leukemogenic GATA1 mutation. Dev. Cell. 2013;27:607–620. doi: 10.1016/j.devcel.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sedore S.C., Byers S.A., Biglione S., Price J.P., Maury W.J., Price D.H. Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR. Nucleic Acids Res. 2007;35:4347–4358. doi: 10.1093/nar/gkm443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Michels A.A., Nguyen V.T., Fraldi A., Labas V., Edwards M., Bonnet F., Lania L., Bensaude O. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol. Cell. Biol. 2003;23:4859–4869. doi: 10.1128/MCB.23.14.4859-4869.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schulte A., Czudnochowski N., Barboric M., Schonichen A., Blazek D., Peterlin B.M., Geyer M. Identification of a cyclin T-binding domain in Hexim1 and biochemical analysis of its binding competition with HIV-1 Tat. J. Biol. Chem. 2005;280:24968–24977. doi: 10.1074/jbc.M501431200. [DOI] [PubMed] [Google Scholar]
- 108.Cho W.K., Jang M.K., Huang K., Pise-Masison C.A., Brady J.N. Human T-lymphotropic virus type 1 Tax protein complexes with P-TEFb and competes for Brd4 and 7SK snRNP/HEXIM1 binding. J. Virol. 2010;84:12801–12809. doi: 10.1128/JVI.00943-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhou M., Lu H., Park H., Wilson-Chiru J., Linton R., Brady J.N. Tax interacts with P-TEFb in a novel manner to stimulate human T-lymphotropic virus type 1 transcription. J. Virol. 2006;80:4781–4791. doi: 10.1128/JVI.80.10.4781-4791.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ammosova T., Jerebtsova M., Beullens M., Lesage B., Jackson A., Kashanchi F., Southerland W., Gordeuk V.R., Bollen M., Nekhai S. Nuclear targeting of protein phosphatase-1 by HIV-1 Tat protein. J. Biol. Chem. 2005;280:36364–36371. doi: 10.1074/jbc.M503673200. [DOI] [PubMed] [Google Scholar]
- 111.Ammosova T., Obukhov Y., Kotelkin A., Breuer D., Beullens M., Gordeuk V.R., Bollen M., Nekhai S. Protein phosphatase-1 activates CDK9 by dephosphorylating Ser175. PLoS One. 2011;6:e18985. doi: 10.1371/journal.pone.0018985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ammosova T., Yedavalli V.R., Niu X., Jerebtsova M., Van Eynde A., Beullens M., Bollen M., Jeang K.T., Nekhai S. Expression of a protein phosphatase 1 inhibitor, cdNIPP1, increases CDK9 threonine 186 phosphorylation and inhibits HIV-1 transcription. J. Biol. Chem. 2011;286:3798–3804. doi: 10.1074/jbc.M110.196493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Barrandon C., Bonnet F., Nguyen V.T., Labas V., Bensaude O. The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK RNA relies upon formation of hnRNP-7SK RNA complexes. Mol. Cell. Biol. 2007;27:6996–7006. doi: 10.1128/MCB.00975-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Van Herreweghe E., Egloff S., Goiffon I., Jády B.E., Froment C., Monsarrat B., Kiss T. Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J. 2007;26:3570–3580. doi: 10.1038/sj.emboj.7601783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Calo E., Flynn R.A., Martin L., Spitale R.C., Chang H.Y., Wysocka J. RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature. 2015;518:249–253. doi: 10.1038/nature13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bisgrove D.A., Mahmoudi T., Henklein P., Verdin E. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc. Natl. Acad. Sci. U.S.A. 2007;104:13690–13695. doi: 10.1073/pnas.0705053104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Itzen F., Greifenberg A.K., Bosken C.A., Geyer M. Brd4 activates P-TEFb for RNA polymerase II CTD phosphorylation. Nucleic Acids Res. 2014;42:7577–7590. doi: 10.1093/nar/gku449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gudipaty S.A., McNamara R.P., Morton E.L., D'Orso I. PPM1G binds 7SK RNA and hexim1 To Block P-TEFb assembly into the 7SK snRNP and sustain transcription elongation. Mol. Cell. Biol. 2015;35:3810–3828. doi: 10.1128/MCB.00226-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Castelo-Branco G., Amaral P.P., Engström P.G., Robson S.C., Marques S.C., Bertone P., Kouzarides T. The non-coding snRNA 7SK controls transcriptional termination, poising, and bidirectionality in embryonic stem cells. Genome Biol. 2013;14:1–18. doi: 10.1186/gb-2013-14-9-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Meng F.L., Du Z., Federation A., Hu J., Wang Q., Kieffer-Kwon K.R., Meyers R.M., Amor C., Wasserman C.R., Neuberg D., et al. Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell. 2014;159:1538–1548. doi: 10.1016/j.cell.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Filippakopoulos P., Qi J., Picaud S., Shen Y., Smith W.B., Fedorov O., Morse E.M., Keates T., Hickman T.T., Felletar I., et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hainer S.J., Gu W., Carone B.R., Landry B.D., Rando O.J., Mello C.C., Fazzio T.G. Suppression of pervasive noncoding transcription in embryonic stem cells by esBAF. Genes Dev. 2015;29:362–378. doi: 10.1101/gad.253534.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tan J.L., Fogley R.D., Flynn R.A., Ablain J., Yang S., Saint-André V., Fan Z.P., Do B.T., Laga A.C., Fujinaga K., et al. Stress from nucleotide depletion activates the transcriptional regulator HEXIM1 to suppress melanoma. Mol. Cell. 62:34–46. doi: 10.1016/j.molcel.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bai X., Kim J., Yang Z., Jurynec M.J., Akie T.E., Lee J., LeBlanc J., Sessa A., Jiang H., DiBiase A., et al. TIF1γ controls erythroid cell fate by regulating transcription elongation. Cell. 2010;142:133–143. doi: 10.1016/j.cell.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]